- Institute of Molecular Biology and Pathology, National Research Council (CNR), c/o Department of Biology and Biotechnology, Sapienza University of Rome, Rome, Italy
The Aurora kinase A (AURKA) is involved in different aspects of mitotic control, from mitotic entry to cytokinesis. Consistent with its pleiotropic roles, several AURKA interactors are able to modulate its activity, the best characterized being the microtubule-binding protein TPX2, the centrosomal protein Cep192, and Bora. Bora has been described as an essential cofactor of AURKA for phosphorylation-mediated activation of the mitotic kinase polo-like kinase 1 (Plk1) at the G2/M transition. A complex AURKA/Plk1 signaling axis is emerging, with multiple involved actors; recent data suggest that this control network is not restricted to mitotic entry only, but operates throughout mitosis. Here, we integrate available data from the literature to depict the complex interplay between AURKA and Plk1 in G2 and mitosis and how it contributes to their mitotic functions. We will particularly focus on how the activity of specifically localized AURKA/Plk1 pools is modulated in time and space by their reciprocal regulation to ensure the timely and coordinated unfolding of downstream mitotic events.
Introduction
About 20 years ago, two loci encoding for serine–threonine kinases required for correct spindle pole assembly were described in Drosophila and named “polo” and “aurora” (1–3); these were the forefathers of the corresponding kinase families, now well characterized as key regulators of the cell cycle and mitotic division. Aurora and polo kinases are evolutionary highly conserved, from yeast to mammals (4, 5), and homologs of the originally identified Drosophila genes were described in humans as Aurora2 (now AURKA) and polo-like kinase 1 (Plk1), respectively (6–9). Besides the spindle pole phenotypes, several common features led to association of the two kinases, since their discovery. Both display cell cycle-regulated expression (6, 9), with upregulation of mRNAs in the late S and G2 phases ensured by shared transcriptional mechanisms, such as activation by E2F factors (10, 11) and G1-specific repression through CDE/CHR elements (12, 13). Protein levels peak at G2 and mitosis, paralleled by the activation of kinase enzymatic function (9, 14), and drop in a highly coordinated manner at mitotic exit by proteasome-dependent degradation (15). Both kinases localize at centrosomes and spindle poles, although they also display nonoverlapping localization sites, with AURKA associated to spindle pole microtubules, and Plk1 residing at kinetochores; both are also found at the spindle midzone and midbody at ana–telophase (16, 17). Functionally, both AURKA and Plk1 are involved in control of mitotic entry, with an essential role during recovery from DNA damage checkpoint-mediated G2 arrest, and in several aspects of mitotic progression (18–21). Finally, ever since their discovery it has been evident that cancer cells frequently display altered levels of AURKA and Plk1 (7–9, 22) and that downregulating their expression yields antiproliferative effects (23–25); indeed, both kinases are actively studied as potential anticancer targets (26, 27). All these similarities suggested direct links between AURKA and Plk1, which started to come out only in the last 10 years. Here, we review data about the interplay of AURKA and Plk1, focusing on the emerging view of how this can contribute to AURKA activation at distinct subcellular sites and in different cell cycle windows, thus finely coordinating downstream mitotic events.
Activation Mechanisms for AURKA and Plk1
Phosphorylation of a threonine residue within the activation loop of AURKA and Plk1 kinases, Thr-288 and Thr-210, respectively, is crucial for their enzymatic activity (28, 29). Phosphorylation of Plk1Thr-210 occurs upon release of an inhibitory intramolecular interaction between the N-terminal catalytic domain and the C-terminal “polo-box” domain (PBD). The latter is a phosphoserine/threonine recognition domain; its binding to target phosphopeptides, mainly generated by the cdk1 kinase, impairs the interaction with the catalytic domain, thus triggering Plk1 activation (30, 31). Plk1 activation mechanism, thus, relies on making the region where Thr-210 lies accessible; Thr-210 can then be phosphorylated by an upstream kinase (see the following sections).
Data collected so far indicate a more complex mechanism for AURKA activation. AURKAThr-288 lies within an AURKA consensus motif and is therefore regarded as an autophosphorylation site. It is still debated whether autophosphorylation is achieved by an intra- or intermolecular reaction, and conformational shifts as well as dimerization appear to underlie different activation states (32–34). Indeed, data in the literature indicate multiple binding partners (see the following sections) that are able to stimulate AURKA activity without a direct enzymatic action but rather by inducing specific conformational transitions. These observations suggest that cells need to manage distinct pools of AURKA, acting at distinct subcellular sites and displaying different extents of activity.
Interestingly, although activation mechanisms for AURKA and Plk1 are distinct, coupling intracellular localization with function appears to be a conserved feature: for Plk1, the PBD is also required for correct targeting of the kinase to centrosomes, kinetochores, and spindle midzone (35, 36), and the major AURKA activators, namely Cep192 and TPX2, mediate AURKA binding to centrosomes and microtubules, respectively (37–39).
The AURKA/Plk1/Bora Axis and Mitotic Entry
The direct link between AURKA and Plk1 came with the identification of AURKA as the upstream kinase responsible of phosphorylation of Thr-210 in the Plk1 activation loop, an event requiring the presence of the coactivating protein Bora (19, 40) (Figure 1, upper box). Distinctly from other AURKA activators, Bora does not modify AURKA activity per se but rather interferes with the intramolecular interaction between the catalytic domain of Plk1 and the PBD, so to render Thr-210 accessible (40). Consistently, Bora does not significantly increase AURKA activity toward substrates other than Plk1 (19, 40), and the extent of activation of AURKA coimmunoprecipitated with Bora, as assessed by p-Thr-288, is by far lower than that associated with the fractions immunoprecipitated with TPX2 or Cep192 (41, 42). Although low, this activity may suffice to trigger what was defined as the “outer feedback loop” through which AURKA, Plk1, and cdk1 activate each other (43). Phosphorylation of Bora at Ser-252 (human) by cdk1 creates a PBD-docking site and promotes Bora/Plk1 interaction (Figure 1); consistently, phosphorylation of Bora by cdk1 enhances its ability to stimulate AURKA-mediated Plk1 activation (41, 44). A second residue on human Bora, i.e., Thr-52, is responsive to cdk1: GST-tagged human Bora carrying Thr-52 substitution to alanine is destabilized in CSF-arrested Xenopus oocytes extracts (45), thus suggesting that cdk1 phosphorylation plays also a role in protecting Bora from degradation. An opposite effect is mediated by Plk1 in that Plk1 phosphorylation of Bora in the 496-DSGYNT-501 degron triggers Bora degradation through the SCF-β-TrCP pathway (41, 46) about 2 h before mitotic entry (45, 47). Consistently, a decreased interaction between Plk1 and Bora, by mutating the previously mentioned BoraSer-252 to Alanine, influences Bora stability: (i) it prevents GST-Bora degradation in CSF extracts (45) and (ii) in human cells, it impairs the interaction between Bora and β-TrCP (41). In addition, it prevents Bora accumulation induced – as a result of a dominant-negative effect – by kinase-dead Plk1 (41). The opposite effects of cdk1-mediated phosphorylation of Bora on Thr-52 and Ser-252 suggest that timely degradation of Bora constitutes a strictly controlled event; the balance between phosphorylation of Thr-52 by cdk1 and on the degron sequence by Plk1 may determine when the switch toward SCF-β-TrCP-mediated degradation of Bora occurs (Figure 1).
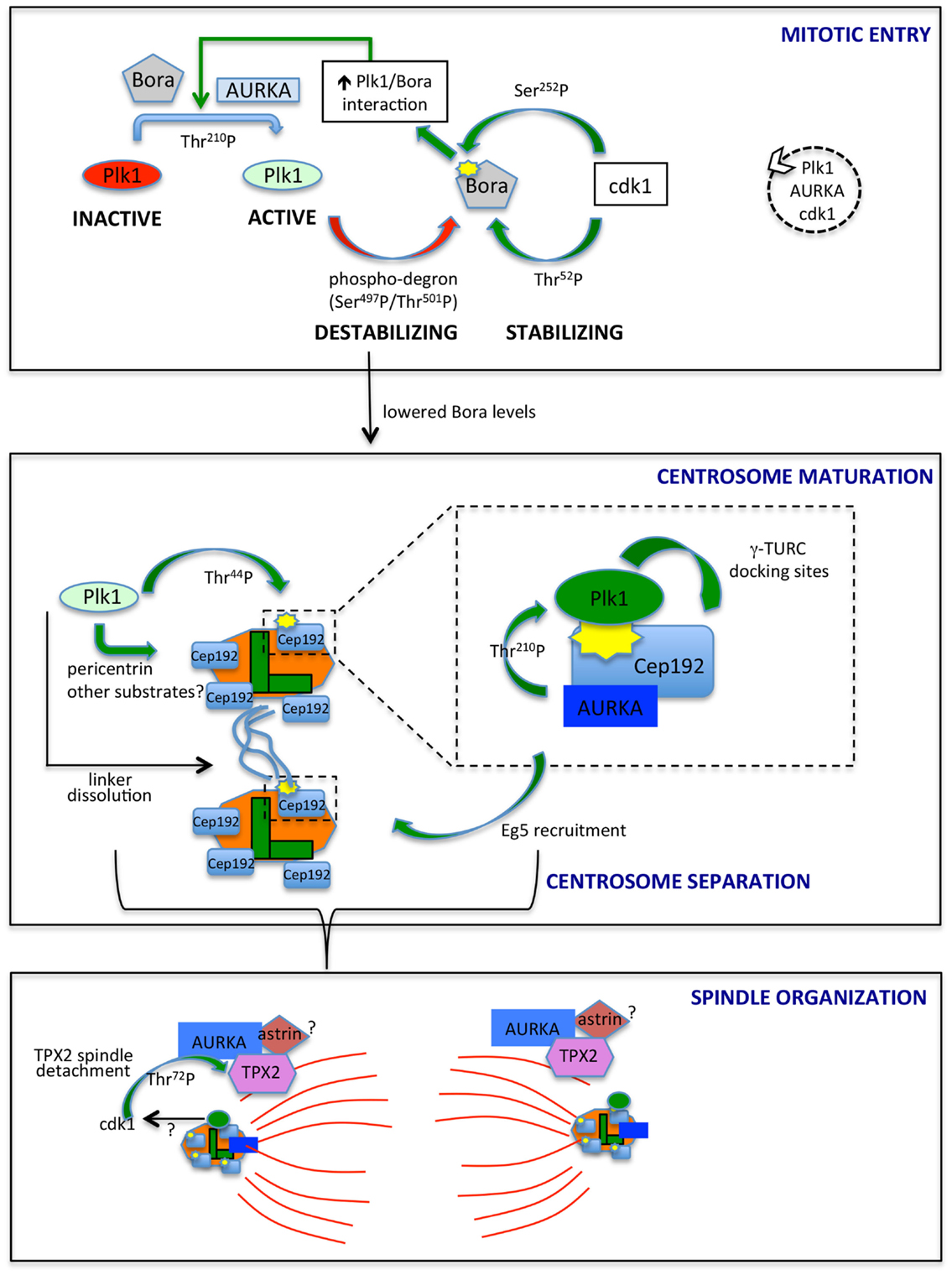
Figure 1. AURKA and Plk1 in mitotic entry and spindle formation. The best characterized links between AURKA and Plk1 are schematized. In mitotic entry (upper box), the combined action of AURKA and Bora activates Plk1, while antagonistic phosphorylation events by Plk1 and cdk1 control Bora stability. The dashed circle on the right indicates the ongoing feedback loop leading to the activation of Plk1, AURKA, and cdk1. Lowered Bora levels enable the interaction of AURKA with Cep192 (central box) and TPX2 (lower box), at centrosomes (centrioles, green; PCM, orange) and microtubules (red), respectively. The enlargement in the central box depicts the scaffolding function of Cep192, leading to recruitment of AURKA and Plk1, activation of the latter and generation by activated Plk1 of γ-TURC-docking sites, with consequent centrosome maturation. Note that Cep192-bound AURKA is activated in a dimeric form, although not represented here to simplify the scheme. Cep192/Plk1/AURKA also contributes to centrosome separation via Eg5 recruitment, and Plk1 independently participates to this process by triggering centrosome linker (light blue lines) dissolution. Separated centrosomes nucleate spindle microtubules that are organized, among others, by AURKA/TPX2 complexes, possibly bound to astrin (lower box). cdk1 phosphorylation of TPX2, possibly influenced by Plk1 activity, yields decreased binding to microtubules. Centrosomal proteins in the lower panel are schematized as in the upper ones, although for space reasons their names are not indicated. The yellow symbols identify PBD-docking sites. Green arrows indicate positive regulatory events, while red arrows represent negative ones. Phosphorylated residues or domains are indicated on the arrows. The different intensities of colors for Plk1 and AURKA denote a different extent of activity.
The Spatiotemporal Level of Bora/AURKA/Plk1 Regulation
The bulk of cycB1/cdk1 complexes is cytoplasmic until prophase, when it promotes its own translocation to the nucleus (48, 49). On the other hand, although Thr-210-phosphorylated Plk1 is first detected at centrosomes, results obtained using a FRET biosensor suggest that Plk1 kinase activity first increases in the nucleus and raises in the cytoplasm only 2 h before mitotic entry (19, 45, 47), at a time that coincides with the onset of Bora degradation (45, 47). Together with the recent observation that Bora is prevalently cytoplasmic in mammalian cells (47), these data suggest that cdk1 and Plk1 activities antagonistically modulate Bora levels, with cdk1-mediated Thr-52 phosphorylation protecting Bora from degradation until cytoplasmic Plk1 activity raises. A potential player in this regulatory mechanism is the peptydylprolyl isomerase Pin1, a modulator of the G2/M transition, which promotes Bora degradation (50) and whose activity and stability are controlled by AURKA and Plk1, respectively (50, 51); further studies are needed to understand how these molecular events interplay in regulating mitotic entry. Phosphatases acting both on kinases themselves and on their substrates, with time- and space-dependent selectivities (52), are also expected to play a role in this fine-tuned regulation. The key serine–threonine phosphatases that counteract mitotic kinase activity are PP1 subunits and PP2A complexes (53, 54). Potentially relevant to Bora degradation, PP2A activity, which is able to counteract Plk1 and cdk1 substrate phosphorylation, is inhibited in the cytoplasm by the Mastl/Greatwall kinase before mitotic entry (52). Translocation of nuclear Greatwall to the cytoplasm is promoted by both cdk1 and Plk1 (55, 56): this mechanism may ensure that phosphorylation of Plk1 cytoplasmic substrates, such as Bora, only accumulates subsequent to Plk1 activation in the nucleus and to cdk1 nuclear import. Whether a differential specificity of action of phosphatases on the different Bora residues phosphorylated by cdk1 and Plk1 exists is an open question that may provide further hints on the time-dependent regulation of Bora stability. As also recently proposed by Bruinsma and colleagues (47), differentially localized phosphatase activity may generally contribute to time-dependent compartmentalization of Plk1 activity, thus explaining why the latter is first observed in the nucleus, although Bora is reported to be strictly cytoplasmic and the extent and timing of AURKA nuclear entry is poorly characterized. We also noticed that the NLS sequences described for Plk1 fall within the catalytic and polo-box domains (57, 58) (Figure 2), raising the possibility that formation of import complexes in the cytoplasm impairs Plk1 kinase function, which would be only released in the nucleus. Modulated interaction between Plk1 and importins may therefore contribute to the switch to cytoplasmic Plk1 activity 2 h before mitotic entry: indeed Ser-137 within one of the NLS sequences (Figure 2) is phosphorylated in vivo and this is described as an activating event for Plk1, although so far described only in late mitosis (29, 59). Alternatively, over time, increased cdk1-generated PBD-docking sites on Plk1 cytoplasmic substrates could retain Plk1 in the cytoplasm by competing with importins for Plk1 binding.
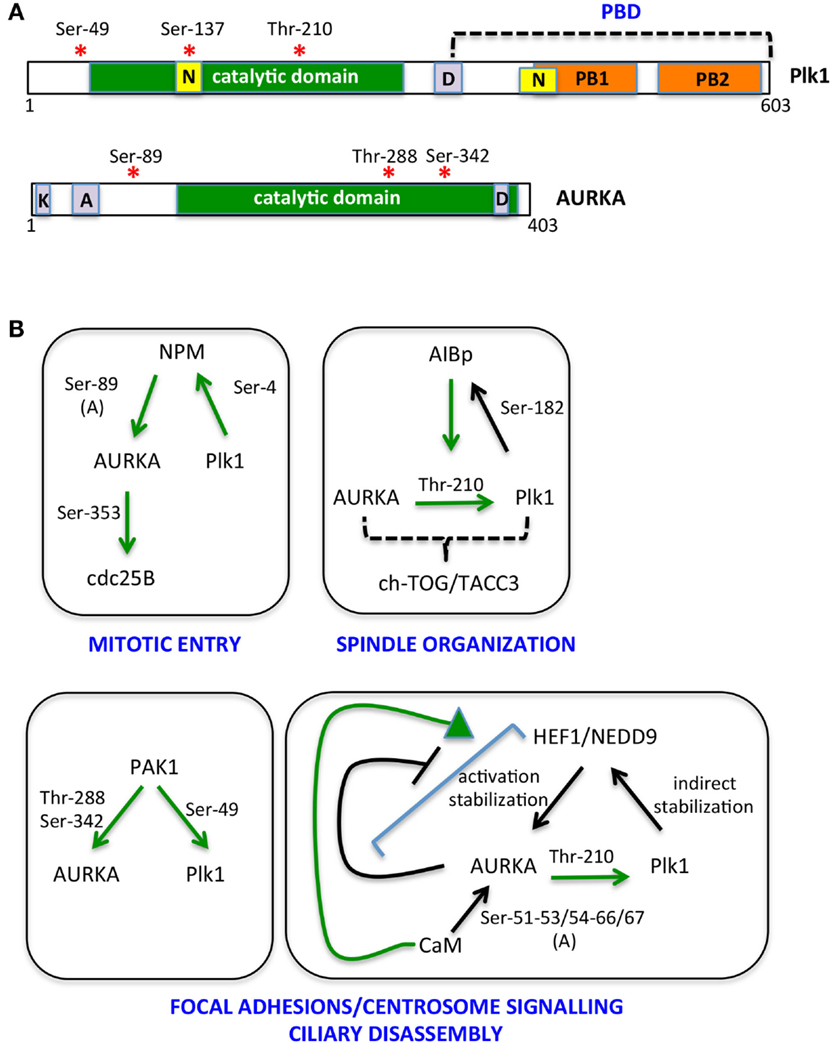
Figure 2. The multiple AURKA and Plk1 activating networks. (A) A schematic representation of Plk1 and AURKA kinases is shown with activating phosphorylation sites indicated by red asterisks. Yellow boxes represent nuclear localization sequences (N); orange ones are polo boxes (PB1 and 2); and violet ones are degradation motifs (D, destruction box; K, KEN box; A, A box). Catalytic domains are in green. (B) Networks involving NPM, AIBp, PAK1, HEF1/NEDD9, and calmodulin (CaM) as activators and/or substrates of AURKA and Plk1 are represented. Green arrows indicate direct and clarified activating events; phosphorylated residues are indicated; and (A) denotes induction of an autophosphorylation event. Downstream substrates and proposed regulated processes are included.
Changing Interactors for Progressing Through Mitosis
What is the functional significance of Bora degradation before mitotic entry by the same protein (Plk1) that it activates? A possible explanation is that the cdk1/AURKA/Plk1 signaling cascade generating the mitotic entry signal (43) must timely switch toward other pathways to sustain spindle assembly and mitotic progression. Evidence summarized below supports the notion that lowering Bora levels is necessary to make AURKA available to other partners. Immunoprecipitation experiments indicate that AURKA complexes containing Bora or TPX2 are distinct and that artificially increasing Bora levels – through Plk1 inactivation – changes the stoichiometry and decreases the amount of TPX2 bound to AURKA (41). In addition, AURKA localization to spindle poles, mediated by Cep192 and TPX2 (see below), is altered when Bora levels are increased by overexpression or by Plk1 inactivation (41). This is likely accounting for the proposed role of Plk1 in AURKA centrosomal localization (38, 60) and further indicates that the Bora/Plk1 complex is able to compete with other AURKA activating/localizing partners.
Together, these observations suggest that AURKA activity initially needs to be focused toward the Plk1 kinase; this activates the AURKA–Plk1–cdk1 loop, until a threshold is reached and the cell is committed toward mitosis (43, 61). Now AURKA and Plk1 kinases must be properly redirected toward their mitotic activators and substrates to coordinate mitotic entry with centrosomal and spindle processes (Figure 1). How does Plk1 remain active in mitosis when Bora is degraded? On the one hand, the accessibility of Thr-210 may not represent a limiting factor in mitosis, when high cdk1 activity creates abundant PBD-docking sites. On the other hand, recent data indicate that although Bora levels are strongly reduced in mitosis, a residual fraction exists (45), and it is responsible of Plk1 activation throughout the division process (62). An independent protein, Furry, has been described to activate Plk1 through AURKA, with a mechanism comparable to Bora (63). It will be interesting to investigate whether this redundancy underlies subcellular, temporal, or cell-type specificity. Most importantly, Cep192 emerging scaffolding functions may bypass the requirement for Bora in the AURKA/Plk1 axis at centrosomes.
The AURKA/Plk1/Cep192 Axis Controls Centrosome Maturation and Separation
The drop in Bora levels following Plk1 activation may ensure that centrosomal processes leading to spindle assembly, depending on other AURKA containing complexes, start only when the mitotic entry signaling cascade is fully active. The centrosomal protein Cep192, involved in both centrosome maturation and separation (39, 64), appears as a key coordinator of AURKA and Plk1 activity at this stage. Cep192 was first shown to trigger dimerization-driven AURKA activation at centrosomes in Xenopus egg extracts (65) and was later confirmed as a key AURKA centrosomal activator in mammalian cells (42, 66). Cep192-bound AURKA is highly active compared to the Bora- or TPX2-bound pools (42, 65). In human cells, the interaction between AURKA and Cep192 is reported from S phase (42); the strong increase in centrosomal Cep192 at mitotic entry, just before centrosome separation (39, 64), suggests that more Cep192–AURKA centrosomal complexes exist at this stage, in agreement with the proposed requirement of freeing AURKA from Bora-containing complexes. Importantly, Plk1 has recently been shown to be a part of the AURKA/Cep192 axis driving centrosome maturation (Figure 1, central box): Cep192 acts as a scaffold for both Plk1 and AURKA and is the key recruiting factor for the kinases at centrosomes, with Plk1 binding following that of AURKA (39, 42, 66). Cep192 brings AURKA and Plk1 in close proximity thereby enabling Plk1Thr-210 phosphorylation (42, 66). AURKA-activated Plk1 creates its own PBD-docking site on Cep192 by phosphorylating Cep192Thr-44 (42, 66); a subsequent AURKA-independent PBD-docking site centered on Cep192Ser-995 has been reported (42), although the separation of the functional roles of Thr-44 and Ser-995 needs further investigation.
It could be speculated that preceding activation by Bora/AURKA generates the low Plk1 activity required for initial phosphorylation of Thr-44, while ensuing stabilization of Cep192/Plk1/AURKA complexes (42), where AURKA activity is higher, boosts the signaling cascade leading to centrosome maturation. Plk1 is required for Cep192 centrosomal localization, partly through phosphorylation of pericentrin (67, 68), supporting the hypothesis that an initial Cep192-independent Plk1 activation triggers a subsequent and more sustained Cep192-mediated one. Cep192/AURKA-activated Plk1 in turn phosphorylates Cep192 to generate γ-TURC-docking sites and induce the sudden increase in pericentriolar material (PCM) characterizing centrosome maturation (42, 66) (Figure 1, central box).
Centrosome separation requires linker dissolution and Eg5-mediated centrosome movement, both involving Plk1 (69–71). While linker dissolution does not require Cep192, the observation that loss of Cep192 impairs Eg5 centrosomal localization and centrosome separation (39, 66) suggests that the role of Plk1 in centrosome movement passes through the Cep192/AURKA axis, with a key upstream involvement of centrosomal cyclin B2/cdk1 (72) (Figure 1, central box).
Cep192 complexes identify an AURKA pool clearly distinct from the microtubule- and TPX2-bound one: (i) the AURKA/Cep192 interaction occurs also in the absence of microtubules (42); (ii) Cep192 and TPX2 bind to the same region of AURKA and are detected in independent AURKA complexes (65); (iii) Cep192-loaded beads recapitulate in CSF-arrested Xenopus oocytes extracts the functions as microtubule-organizing center (MTOC) of AURKA-loaded beads but not their ability of RanGTP-induced spindle organization (66, 73). TPX2 is a RanGTP-regulated factor (74); these observations together suggest that the pools of AURKA bound to Cep192 and TPX2 are functionally separated and involved in centrosome maturation and spindle assembly, respectively. The observation that both Cep192/AURKA and TPX2 regulate Eg5 activity (66, 75) may reflect independent functions in centrosome separation or an interplay of the two pools of AURKA in this process yet to be unveiled.
Microtubule-Associated AURKA Pools and Spindle Organization
Microtubule-organizing functions of AURKA are less obviously linked to Plk1 activity. AURKA localization to microtubules is mediated by the microtubule-binding protein TPX2 (37, 38), which also activates AURKA by stabilizing the active conformation and making AURKAThr-288 inaccessible to the PP1 phosphatase (76). In addition, TPX2 protects AURKA from APC/CCdh1 proteasome-dependent degradation in G2 and early mitosis, with TPX2 depletion impairing accumulation of high levels of AURKA in prometaphase (77). Xenopus Plx1 has been shown to phosphorylate TPX2 on Ser-204, with a positive effect on TPX2-mediated AURKA activation (78). A corresponding mechanism has not been explored in mammalian cells given the poor conservation of the phosphorylated site. Yet, phosphoproteomic screenings identified TPX2 in vivo phosphosites that are likely to be phosphorylated by Plk1 (79, 80). Furthermore, TPX2 abnormally accumulates at spindle poles in Plk1-interfered mitoses (38), and recent data show that cdk1-mediated phosphorylation of TPX2Thr-72 negatively modulates TPX2 association to the mitotic spindle (81). It is, therefore, conceivable that Plk1 activity at mitotic centrosomes, through its effects on cdk1, influences TPX2 mobility at spindle poles (Figure 1, lower box).
Astrin is an independent regulator of AURKA localization at microtubules, with no effect on the kinase activity (82); RNA interference-mediated depletion of astrin induces spindle defects reminiscent of those observed following AURKA inactivation (82, 83). Astrin localization to the spindle is in turn mediated by TPX2 (82). Interestingly, Plk1 has also been detected in astrin–kinastrin complexes in mitotic cell extracts (84).
Together these observations suggest that exploring the interplay between AURKA, TPX2, Plk1, and astrin deserves further investigation and may improve our understanding of AURKA spindle-organizing functions.
The Growing Network of AURKA Activators
Additional activators of AURKA at centrosomes and spindle poles have been described, many being also functionally linked to Plk1 (Figure 2).
Nucleophosmin (NPM) activates AURKA by stimulating a newly identified autophosphorylation event, on Ser-89 (85). Phosphorylation of NPM by Plk1 is required for its mitotic functions (86), while NPM depletion does not affect Plk1Thr-210 phosphorylation (85). These observations suggest that NPM-activated AURKA is generated when Plk1 activation has become prevalently AURKA independent; alternatively, since the only AURKA substrate affected by NPM depletion is so far CDC25B, NPM may provide AURKA specificity of action toward a limited set of substrates.
The AIBp protein, colocalizing with AURKA at centrosomes and spindle poles, has recently been reported as an AURKA regulator, relevant for Plk1 activation and in turn a substrate of it; the observation that localization of the downstream AURKA targets TACC3 and ch-TOG is affected by AIBp depletion, while PCM recruitment is not, together with the associated spindle pole phenotypes, suggest an involvement of AIBp in the spindle-organizing functions of AURKA (87).
AURKA activators also include proteins that localize both at focal adhesions and centrosomes, in particular the PAK1 kinase and the HEF1/NEDD9 scaffolding protein (88, 89). PAK1 promotes AURKA activation by directly phosphorylating Thr-288 and Ser-342 (89) and also phosphorylates Plk1Ser-49, an event that contributes to its activation (90). HEF1/NEDD9 promotes the catalytic activity of AURKA (88) and also stabilizes it (91); the interaction between AURKA and HEF1/NEDD9 is favored by CaM (92), while it is inhibited by AURKA phosphorylation of HEF1/NEDD9 (88), indicating the presence of a negative feedback loop. Plk1 in turn indirectly regulates HEF1/NEDD9 stability, with deriving increased AURKA activity signaling back on Plk1 activation (93). The focal adhesion localization of PAK1 and HEF1/NEDD9 suggests that they define a pool of AURKA responsible of a signaling path that links loss of cell adhesion – typical of the cell division process – with mitotic centrosomal events and mitotic entry (94, 95). This pool appears also to be involved in the non-mitotic role of AURKA in cilia disassembly at cell cycle reentry from G0 (92, 96), a process that also requires Plk1 activity (93). An additional interactor of AURKA involved in both cell–cell adhesion and cell proliferation and survival (97) is Ajuba. The interaction between AURKA and Ajuba was first described in human cells (98), where it was shown as a key AURKA-activating step at G2 centrosomes (98). Recent data suggest that the activation mechanism relies on the ability of Ajuba, upon binding to AURKA N-terminus, to prevent an inhibitory intramolecular interaction between the N- and C-termini of the kinase (99); in addition, the subsequent binding of a distinct Ajuba domain to the C-terminus of AURKA directly stimulates kinase activity (99). A role of Ajuba in AURKA regulation has been confirmed in Drosophila neuroblasts, although data indicate an effect on localization, rather than activation, of the kinase (100). Organism and/or cell-type specificity may account for the observed differences, although cell cycle- (G2 vs. mitosis) or reporter- (phospho-AURKA/phospho-H3 vs. phospho-TACC3) dependent effects may also be envisaged.
Conclusion
Several AURKA activators have been described at centrosomes and microtubules, and evidence exist that they create independent complexes with the kinase. The scaffolding functions of some of them and the finding of specific phospho-AURKA fractions depending on the bound activator suggest that distinct interactors define specific AURKA pools with differential kinase activity and/or substrate specificity. More interconnected analyses of the different AURKA pools and a better spatiotemporal resolution of their formation during the cell cycle are expected to uncover in the next years how they ensure tight coordination of downstream events. Plk1 is a key substrate of AURKA and at the same time a major regulator of the multiple AURKA activators: besides contributing to generate an activation feedback loop that reinforces AURKA and Plk1 activities at mitotic entry, this is also emerging as a mechanism to impart time-dependent regulation to the unfolding of AURKA-regulated events. Exploring the contribution of the AURKA/Plk1 axis in mitotic control, including in newly identified mitotic functions of AURKA (101–104), is a promising field of investigation for the future.
Author Contributions
IAA critically analyzed the literature, contributed to article writing, and prepared figures. FDM critically analyzed the literature and discussed extensively review structure, contents, and models. GG conceived the structure of the review, critically analyzed the literature, and wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Work in the laboratory is supported by the Italian Association for Cancer Research (MFAG-13350) and CNR-InterOmics Flagship Project “Regulatory nodes in mitosis; a systems analysis of importin beta interactors in mitotic cells.” IAA is supported by a Fondazione Umberto Veronesi fellowship.
References
1. Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, et al. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev (1991) 5:2153–65. doi:10.1101/gad.5.12a.2153
2. Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell (1995) 81:95–105. doi:10.1016/0092-8674(95)90374-7
3. Sunkel CE, Glover DM. Polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci (1998) 89:25–38.
4. Brown JR, Koretke KK, Birkeland ML, Sanseau P, Patrick DR. Evolutionary relationships of aurora kinases: implications for model organism studies and the development of anti-cancer drugs. BMC Evol Biol (2004) 4:39. doi:10.1186/1471-2148-4-39
5. de Cárcer G, Manning G, Malumbres M. From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle (2011) 10:2255–62. doi:10.4161/cc.10.14.16494
6. Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J Cell Sci (1994) 107:1509–17.
7. Holtrich U, Wolf G, Bräuninger A, Karn T, Böhme B, Rübsamen-Waigmann H, et al. Induction and down-regulation of PLK, a human serine/threonine kinase expressed in proliferating cells and tumors. Proc Natl Acad Sci U S A (1994) 91:1736–40. doi:10.1073/pnas.91.5.1736
8. Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene (1997) 14:2195–200. doi:10.1038/sj.onc.1201065
9. Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J (1998) 17:3052–65. doi:10.1093/emboj/17.11.3052
10. He L, Yang H, Ma Y, Pledger WJ, Cress WD, Cheng JQ. Identification of Aurora-A as a direct target of E2F3 during G2/M cell cycle progression. J Biol Chem (2008) 283:31012–20. doi:10.1074/jbc.M803547200
11. Tategu M, Nakagawa H, Sasaki K, Yamauchi R, Sekimachi S, Suita Y, et al. Transcriptional regulation of human polo-like kinases and early mitotic inhibitor. J Genet Genomics (2008) 35:215–24. doi:10.1016/S1673-8527(08)60030-2
12. Uchiumi T, Longo DL, Ferris DK. Cell cycle regulation of the human polo-like kinase (PLK) promoter. J Biol Chem (1997) 272:9166–74. doi:10.1074/jbc.272.14.9166
13. Tanaka M, Ueda A, Kanamori H, Ideguchi H, Yang J, Kitajima S, et al. Cell-cycle-dependent regulation of human Aurora A transcription is mediated by periodic repression of E4TF1. J Biol Chem (2002) 277:10719–26. doi:10.1074/jbc.M108252200
14. Golsteyn RM, Mundt KE, Fry AM, Nigg EA. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J Cell Biol (1995) 129:1617–28. doi:10.1083/jcb.129.6.1617
15. Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. J Cell Biol (2004) 164:233–41. doi:10.1083/jcb.200309035
16. Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol (2001) 2:21–32. doi:10.1038/35048096
17. Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol (2003) 4:842–54. doi:10.1038/nrm1245
18. van Vugt MA, Brás A, Medema RH. Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell (2004) 15:799–811. doi:10.1016/j.molcel.2004.07.015
19. Macůrek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, et al. Polo-like kinase-1 is activated by Aurora A to promote checkpoint recovery. Nature (2008) 455:119–23. doi:10.1038/nature07185
20. Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem Sci (2012) 37:534–42. doi:10.1016/j.tibs.2012.09.005
21. Nikonova AS, Astsaturov I, Serebriiskii IG, Dunbrack RL Jr, Golemis EA. Aurora A kinase (AURKA) in normal and pathological cell division. Cell Mol Life Sci (2013) 70:661–87. doi:10.1007/s00018-012-1073-7
22. Lens SM, Voest EE, Medema RH. Shared and separate functions of polo-like kinases and aurora kinases in cancer. Nat Rev Cancer (2010) 10:825–41. doi:10.1038/nrc2964
23. Spänkuch-Schmitt B, Bereiter-Hahn J, Kaufmann M, Strebhardt K. Effect of RNA silencing of polo-like kinase-1 (PLK1) on apoptosis and spindle formation in human cancer cells. J Natl Cancer Inst (2002) 94:1863–77. doi:10.1093/jnci/94.24.1863
24. Liu X, Erikson RL. Polo-like kinase (Plk)1 depletion induces apoptosis in cancer cells. Proc Natl Acad Sci U S A (2003) 100:5789–94. doi:10.1073/pnas.1031523100
25. Hata T, Furukawa T, Sunamura M, Egawa S, Motoi F, Ohmura N, et al. RNA interference targeting aurora kinase a suppresses tumor growth and enhances the taxane chemosensitivity in human pancreatic cancer cells. Cancer Res (2005) 65:2899–905. doi:10.1158/0008-5472.CAN-04-3981
26. Doménech E, Malumbres M. Mitosis-targeting therapies: a troubleshooting guide. Curr Opin Pharmacol (2013) 13:519–28. doi:10.1016/j.coph.2013.03.011
27. Marzo I, Naval J. Antimitotic drugs in cancer chemotherapy: promises and pitfalls. Biochem Pharmacol (2013) 86:703–10. doi:10.1016/j.bcp.2013.07.010
28. Walter AO, Seghezzi W, Korver W, Sheung J, Lees E. The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene (2000) 19:4906–16. doi:10.1038/sj.onc.1203847
29. Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem (2002) 277:44115–20. doi:10.1074/jbc.M202172200
30. Elia AE, Cantley LC, Yaffe MB. Proteomicscreen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science (2003) 299:1228–31. doi:10.1126/science.1079079
31. Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. The molecular basis forphosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell (2003) 115:83–95. doi:10.1016/S0092-8674(03)00725-6
32. Dodson CA, Bayliss R. Activation of Aurora-A kinase by protein partner binding and phosphorylation are independent and synergistic. J Biol Chem (2012) 287:1150–7. doi:10.1074/jbc.M111.312090
33. Dodson CA, Yeoh S, Haq T, Bayliss R. A kinetic test characterizes kinase intramolecular and intermolecular autophosphorylation mechanisms. Sci Signal (2013) 6:ra54. doi:10.1126/scisignal.2003910
34. Zorba A, Buosi V, Kutter S, Kern N, Pontiggia F, Cho YJ, et al. Molecular mechanism of Aurora A kinase autophosphorylation and its allosteric activation by TPX2. Elife (2014) 27:e02667. doi:10.7554/eLife.02667
35. Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci U S A (1998) 95:9301–6. doi:10.1073/pnas.95.16.9301
36. Seong YS, Kamijo K, Lee JS, Fernandez E, Kuriyama R, Miki T, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem (2002) 277:32282–93. doi:10.1074/jbc.M202602200
37. Kufer TA, Silljé HH, Körner R, Gruss OJ, Meraldi P, Nigg EA. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol (2002) 158:617–23. doi:10.1083/jcb.200204155
38. De Luca M, Lavia P, Guarguaglini G. A functional interplay between Aurora-A, Plk1 and TPX2 at spindle poles: Plk1 controls centrosomal localization of Aurora-A and TPX2 spindle association. Cell Cycle (2006) 5:296–303. doi:10.4161/cc.5.3.2392
39. Gomez-Ferreria MA, Rath U, Buster DW, Chanda SK, Caldwell JS, Rines DR, et al. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr Biol (2007) 17:1960–6. doi:10.1016/j.cub.2007.10.019
40. Seki A, Coppinger JA, Jang CY, Yates JR, Fang G. Bora and the kinase Aurora A cooperatively activate the kinase Plk1 and control mitotic entry. Science (2008) 320:1655–8. doi:10.1126/science.1157425
41. Chan EHY, Santamaria A, Silljé HHW, Nigg EA. Plk1 regulates mitotic Aurora A function through βTrCP-dependent degradation of hBora. Chromosoma (2008) 117:457–69. doi:10.1007/s00412-008-0165-5
42. Meng L, Park JE, Kim TS, Lee EH, Park SY, Zhou M, et al. Bimodal interaction of mammalian polo-like kinase 1 and a centrosomal scaffold, Cep192, in the regulation of bipolar spindle formation. Mol Cell Biol (2015) 35:2626–40. doi:10.1128/MCB.00068-15
43. Lindqvist A, Rodríguez-Bravo V, Medema RH. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol (2009) 185:193–202. doi:10.1083/jcb.200812045
44. Tavernier N, Noatynska A, Panbianco C, Martino L, Van Hove L, Schwager F. Cdk1 phosphorylates SPAT-1/Bora to trigger PLK-1activation and drive mitotic entry in C. elegans embryos. J Cell Biol (2015) 6:661–9. doi:10.1083/jcb.201408064
45. Feine O, Hukasova E, Bruinsma W, Freire R, Fainsod A, Gannon J, et al. Phosphorylation-mediated stabilization of Bora in mitosis coordinates Plx1/Plk1 and Cdk1 oscillations. Cell Cycle (2014) 13:1727–36. doi:10.4161/cc.28630
46. Seki A, Coppinger JA, Du H, Jang CY, Yates JR III, Fang G. Plk1- and beta -TrCP - dependent degradation of Bora controls mitotic progression. J Cell Biol (2008) 181:65–78. doi:10.1083/jcb.200712027
47. Bruinsma W, Aprelia M, Kool J, Macurek L, Lindqvist A, Medema RH. Spatial separation of Plk1 phosphorylation and activity. Front Oncol (2015) 5:e132. doi:10.3389/fonc.2015.00132
48. Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell (2010) 18:533–43. doi:10.1016/j.devcel.2010.02.013
49. Lindqvist A. Cyclin B-Cdk1 activates its own pump to get into the nucleus. J Cell Biol (2010) 189:197–9. doi:10.1083/jcb.201003032
50. Lee YC, Que J, Chen YC, Lin JT, Liou YC, Liao PC, et al. Pin1 acts as a negative regulator of the G2/M transition by interacting with the Aurora-A-Bora complex. J Cell Sci (2013) 126:4862–72. doi:10.1242/jcs.121368
51. Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C, et al. Polo-like Kinase 1-mediated phosphorylation stabilizes pin1 by inhibiting its ubiquitination in human cells. J Biol Chem (2005) 280:36575–83. doi:10.1074/jbc.M504548200
52. Alvarez-Fernández M, Malumbres M. Preparing a cell for nuclear envelope breakdown: Spatio-temporal control of phosphorylation during mitotic entry. Bioessays (2014) 36:757–65. doi:10.1002/bies.201400040
53. Jeong AL, Yang Y. PP2A function toward mitotic kinases and substrates during the cell cycle. BMB Rep (2013) 46:289–94. doi:10.5483/BMBRep.2013.46.6.041
54. Rebelo S, Santos M, Martins F, da Cruz E, Silva EF, da Cruz E, et al. Protein phosphatase 1 is a key player in nuclear events. Cell Signal (2015). pii: S0898–6568(15)00238-7. doi:10.1016/j.cellsig.2015.08.007
55. Álvarez-Fernández M, Sánchez-Martínez R, Sanz-Castillo B, Gan PP, Sanz-Flores M, Trakala M, et al. Greatwall is essential to prevent mitotic collapse after nuclear envelope breakdown in mammals. Proc Natl Acad Sci U S A (2013) 110:17374–9. doi:10.1073/pnas.1310745110
56. Wang P, Galan JA, Normandin K, Bonneil É, Hickson GR, Roux PP, et al. Cell cycle regulation of Greatwall kinase nuclear localization facilitates mitotic progression. J Cell Biol (2013) 202:277–93. doi:10.1083/jcb.201211141
57. Taniguchi E, Toyoshima-Morimoto F, Nishida E. Nuclear translocation of plk1 mediated by its bipartite nuclear localization signal. J Biol Chem (2002) 277:48884–8. doi:10.1074/jbc.M206307200
58. Lee MS, Huang YH, Huang SP, Lin RI, Wu SF, Li C. Identification of a nuclear localization signal in the polo box domain of Plk1. Biochim Biophys Acta (2009) 1793:1571–8. doi:10.1016/j.bbamcr.2009.07.005
59. van de Weerdt BC, van Vugt MA, Lindon C, Kauw JJ, Rozendaal MJ, Klompmaker R, et al. Uncoupling anaphase-promoting complex/cyclosome activity from spindle assembly checkpoint control by deregulating polo-like kinase 1. Mol Cell Biol (2005) 25:2031–44. doi:10.1128/MCB.25.5.2031-2044.2005
60. Hanisch A, Wehner A, Nigg EA, Silljé HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell (2006) 17:448–59. doi:10.1091/mbc.E05-08-0801
61. Lindqvist A, van Zon W, Karlsson Rosenthal C, Wolthuis RM. Cyclin B1-Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol (2007) 5:e123. doi:10.1371/journal.pbio.0050123
62. Bruinsma W, Macurek LM, Freire R, Lindqvist A, Medema RH. Bora and Aurora-A continue to activate Plk1 in mitosis. J Cell Sci (2014) 127:801–11. doi:10.1242/jcs.137216
63. Ikeda M, Chiba S, Ohashi K, Mizuno K. Furry protein promotes Aurora A-mediated polo-like kinase 1 activation. J Biol Chem (2012) 33:27670–81. doi:10.1074/jbc.M112.378968
64. Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, et al. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol (2008) 18:136–41. doi:10.1016/j.cub.2007.12.055
65. Joukov V, De Nicolo A, Rodriguez A, Walter JC, Livingston DM. Centrosomal protein of 192 kDa (Cep192) promotes centrosome-driven spindle assembly by engaging in organelle-specific Aurora A activation. Proc Natl Acad Sci U S A (2010) 107:21022–7. doi:10.1073/pnas.1014664107
66. Joukov V, Walter JC, De Nicolo A. The Cep192-organized Aurora A–Plk1 cascade is essential for centrosome cycle and bipolar spindle assembly. Mol Cell (2014) 55:578–91. doi:10.1016/j.molcel.2014.06.016
67. Haren L, Stearns T, Lüders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS One (2009) 4:e5976. doi:10.1371/journal.pone.0005976
68. Lee K, Rhee K. PLK1 phosphorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J Cell Biol (2011) 195:1093–101. doi:10.1083/jcb.201106093
69. Bertran MT, Sdelci S, Regue L, Avruch J, Caelles C, Roig J. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J (2011) 30:2634–47. doi:10.1038/emboj.2011.179
70. Mardin BR, Agircan FG, Lange C, Schiebel E. Plk1 controls the Nek2A-PP1γ antagonism in centrosome disjunction. Current Biol (2011) 21:1145–51. doi:10.1016/j.cub.2011.05.047
71. Smith E, Hégarat N, Vesely C, Roseboom I, Larch C, Streicher H, et al. Differential control of Eg5-dependent centrosome separation by Plk1 and Cdk1. EMBO J (2011) 30:2233–45. doi:10.1038/emboj.2011.120
72. Nam HJ, van Deursen JM. Cyclin B2 and p53 control proper timing of centrosome separation. Nat Cell Biol (2014) 16:538–49. doi:10.1038/ncb2952
73. Tsai MY, Zheng Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr Biol (2005) 15:2156–63. doi:10.1016/j.cub.2005.10.054
74. Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell (2001) 104:83–93. doi:10.1016/S0092-8674(01)00193-3
75. Ma N, Titus J, Gable A, Ross JL, Wadsworth P. TPX2 regulates the localization and activity of Eg5 in the mammalian mitotic spindle. J Cell Biol (2011) 195:87–98. doi:10.1083/jcb.201106149
76. Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell (2003) 12:851–62. doi:10.1016/S1097-2765(03)00392-7
77. Giubettini M, Asteriti IA, Scrofani J, De Luca M, Lindon C, Lavia P, et al. Control of Aurora-A stability via interaction with TPX2. J Cell Sci (2011) 124:113–22. doi:10.1242/jcs.075457
78. Eckerdt F, Pascreau G, Phistry ML, Lewellyn A, DePaoli-Roach AA, Maller JL. Phosphorylation of TPX2 by Plx1 enhances activation of Aurora A. Cell Cycle (2009) 8:2413–9. doi:10.4161/cc.8.15.9086
79. Kettenbach AN, Schweppe DK, Faherty BK, Pechenick D, Pletnev AA, Gerber SA. Quantitative phosphoproteomics identifies substrates and functional modules of aurora and polo-like kinase activities in mitotic cells. Sci Signal (2011) 4:rs5. doi:10.1126/scisignal.2001497
80. Santamaria A, Wang B, Elowe S, Malik R, Zhang F, Bauer M, et al. The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol Cell Proteomics (2011) 10:M110.004457. doi:10.1074/mcp.M110.004457
81. Shim SY, Perez de Castro I, Neumayer G, Wang J, Park SK, Sanada K, et al. Phosphorylation of targeting protein for Xenopus kinesin-like protein 2 (TPX2) at threonine 72 in spindle assembly. J Biol Chem (2015) 290:9122–34. doi:10.1074/jbc.M114.591545
82. Du J, Jablonski S, Yen TJ, Hannon GJ. Astrin regulates Aurora-A localization. Biochem Biophys Res Commun (2008) 370:213–9. doi:10.1016/j.bbrc.2008.03.072
83. Asteriti IA, Giubettini M, Lavia P, Guarguaglini G. Aurora-A inactivation causes mitotic spindle pole fragmentation by unbalancing microtubule-generatedforces. Mol Cancer (2011) 10:131. doi:10.1186/1476-4598-10-131
84. Dunsch AK, Linnane E, Barr FA, Gruneberg U. The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J Cell Biol (2011) 192:959–68. doi:10.1083/jcb.201008023
85. Reboutier D, Troadec MB, Cremet JY, Fukasawa K, Prigent C. Nucleophosmin/B23 activates Aurora A at the centrosome through phosphorylation of serine 89. J Cell Biol (2012) 197:19–26. doi:10.1083/jcb.201107134
86. Zhang H, Shi X, Paddon H, Hampong M, Dai W, Pelech S. B23/nucleophosmin serine 4 phosphorylation mediates mitotic functions of polo-like kinase 1. J Biol Chem (2004) 279:35726–34. doi:10.1074/jbc.M403264200
87. Chou CH, Loh JK, Yang MC, Lin CC, Hong MC, Cho CL, et al. AIBp regulates mitotic entry and mitotic spindle assembly by controlling activation of both Aurora-A and Plk1. Cell Cycle (2015) 14:2764–76. doi:10.1080/15384101.2015.1066536
88. Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2kinases at the centrosome. Nat Cell Biol (2005) 7:937–46. doi:10.1038/ncb1309
89. Zhao ZS, Lim JP, Ng YW, Lim L, Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates aurora-A. Mol Cell (2005) 20:237–49. doi:10.1016/j.molcel.2005.08.035
90. Maroto B, Ye MB, von Lohneysen K, Schnelzer A, Knaus UG. P21-activated kinase is required for mitotic progression and regulates Plk1. Oncogene (2008) 27:4900–8. doi:10.1038/onc.2008.131
91. Ice RJ, McLaughlin SL, Livengood RH, Culp MV, Eddy ER, Ivanov AV, et al. NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors. Cancer Res (2013) 73:3168–80. doi:10.1158/0008-5472.CAN-12-4008
92. Plotnikova OV, Nikonova AS, Loskutov YV, Kozyulina PY, Pugacheva EN, Golemis EA. Calmodulin activation of Aurora-A kinase (AURKA) is required during ciliary disassembly and in mitosis. Mol Biol Cell (2012) 23:2658–70. doi:10.1091/mbc.E11-12-1056
93. Lee KH, Johmura Y, Yu LR, Park JE, Gao Y, Bang JK, et al. Identification of a novel Wnt5a-CK1ɛ-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J (2012) 31:3104–17. doi:10.1038/emboj.2012.144
94. Cotteret S, Chernoff J. Pak GITs to Aurora-A. Dev Cell (2005) 9:573–4. doi:10.1016/j.devcel.2005.10.005
95. Pugacheva EN, Golemis EA. HEF1-Aurora A interactions points of dialog between the cell cycle and cell attachment signaling networks. Cell Cycle (2006) 5:384–91. doi:10.4161/cc.5.4.2439
96. Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell (2007) 129:1351–63. doi:10.1016/j.cell.2007.04.035
97. Schimizzi GV, Longmore GD. Ajuba proteins. Curr Biol (2015) 25:R445–6. doi:10.1016/j.cub.2015.02.034
98. Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, et al. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell (2003) 114:585–98. doi:10.1016/S0092-8674(03)00642-1
99. Bai M, Ni J, Wu J, Wang B, Shen S, Yu L. A novel mechanism for activation of Aurora-A kinase by Ajuba. Gene (2014) 543:133–9. doi:10.1016/j.gene.2014.03.048
100. Sabino D, Brown NH, Basto R. Drosophila Ajuba is not an Aurora-A activator but is required to maintain Aurora-A at the centrosome. J Cell Sci (2011) 124:1156–66. doi:10.1242/jcs.076711
101. Lioutas A, Vernos I. Aurora A kinase and its substrate TACC3 are required for central spindle assembly. EMBO Rep (2013) 14:829–36. doi:10.1038/embor.2013.109
102. Reboutier D, Troadec MB, Cremet JY, Chauvin L, Guen V, Salaun P, et al. Aurora A is involved in central spindle assembly through phosphorylation of Ser 19 in P150Glued. J Cell Biol (2013) 201:65–79. doi:10.1083/jcb.201210060
103. Chmátal L, Yang K, Schultz RM, Lampson MA. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr Biol (2015) 25:1835–41. doi:10.1016/j.cub.2015.05.013
Keywords: kinases, mitosis, G2/M transition, centrosomes, spindle
Citation: Asteriti IA, De Mattia F and Guarguaglini G (2015) Cross-Talk between AURKA and Plk1 in Mitotic Entry and Spindle Assembly. Front. Oncol. 5:283. doi: 10.3389/fonc.2015.00283
Received: 30 September 2015; Accepted: 30 November 2015;
Published: 23 December 2015
Edited by:
Ignacio Perez De Castro, Spanish National Cancer Research Centre (CNIO), SpainReviewed by:
Wytse Bruinsma, Memorial Sloan Kettering Cancer Center, USADavid Reboutier, Université de Rennes 1, France
Copyright: © 2015 Asteriti, De Mattia and Guarguaglini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giulia Guarguaglini, Z2l1bGlhLmd1YXJndWFnbGluaUB1bmlyb21hMS5pdA==
 Italia Anna Asteriti
Italia Anna Asteriti Fabiola De Mattia
Fabiola De Mattia Giulia Guarguaglini
Giulia Guarguaglini