
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Oncol. , 31 January 2012
Sec. Molecular and Cellular Oncology
Volume 2 - 2012 | https://doi.org/10.3389/fonc.2012.00005
This article is part of the Research Topic The key role of ubiquitination and sumoylation in signaling and cancer View all 11 articles
Ubiquitination has been demonstrated to play a pivotal role in multiple biological functions, which include cell growth, proliferation, apoptosis, DNA damage response, innate immune response, and neuronal degeneration. Although the role of ubiquitination in targeting proteins for proteasome-dependent degradation have been extensively studied and well-characterized, the critical non-proteolytic functions of ubiquitination, such as protein trafficking and kinase activation, involved in cell survival and cancer development, just start to emerge, In this review, we will summarize recent progresses in elucidating the non-proteolytic function of ubiquitination signaling in protein kinase activation and its implications in human cancers. The advancement in the understanding of the novel functions of ubiquitination in signal transduction pathways downstream of growth factor receptors may provide novel paradigms for the treatment of human cancers.
Posttranslational modification of cellular proteins, such as phosphorylation, ubiquitination, SUMOylation, PARylation, to name a few, have been shown to play a pivotal role in multiple cellular processes such as proliferation, apoptosis, senescence, autophagy, DNA damage repair, innate immune response, and neuron degeneration (Kim et al., 2008b; Kirkin et al., 2009; Kraft et al., 2010; Gao et al., 2011). Ubiquitination is originally described as a process by which ubiquitin is covalently attached to the lysine residue of a target protein for proteasome-mediated degradation by three enzymes, namely E1 (ubiquitin-activating enzyme), one of many E2s (ubiquitin-conjugating enzymes, UBCs), and one of many E3s (ubiquitin ligases; Bhoj and Chen, 2009; Figure 1). As ubiquitination is a reversible process, ubiquitin covalently attached to a target protein can be removed by deubiquitinating enzymes (DUBs; Figure 1). Thus the balance between the activities of ubiquitin enzymes and DUBs determines the level and activity of a target protein.
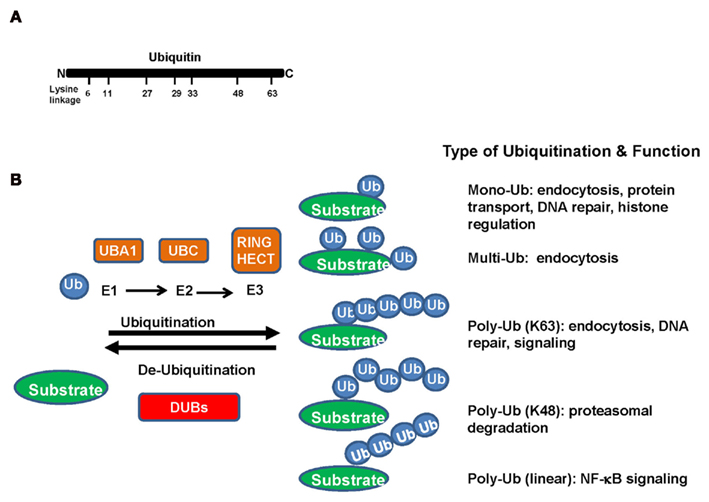
Figure 1. (A) Ubiquitin, a 76-amino acid protein, is highly evolutionarily conserved across species. It contains seven lysine (K) residues (K6, K11, K27, K29, K33, K48, and K63) in the ubiquitin, which have been identified to be utilized for ubiquitination chains. (B) In the ubiquitination process, ubiquitin is covalently conjugated to lysine residues of substrate proteins through a three-step enzymatic reaction. Ubiquitin is first activated by ubiquitin-activating enzyme (E1). The activated ubiquitin is then delivered to the ubiquitin-conjugating enzyme (E2), which will be transferred from the E2 to a lysine residue of a target protein by ubiquitin ligase (E3). The function usefulness of ubiquitin conjugation is not limited to the ubiquitin–proteasome pathway. Mono- and polyubiquitins are used as signals in various pathways including endocytosis, DNA repair, apoptosis, and transcriptional regulation.
Although ubiquitination most often lead to proteasome-mediated degradation of the target protein (Pickart, 2001; Weissman et al., 2011), recent evidence suggests non-proteolytic functions for ubiquitination in the regulation of protein subcellular localization and activity (Giasson and Lee, 2003; Bhoj and Chen, 2009; Chen and Sun, 2009; Hoeller and Dikic, 2009; Raiborg and Stenmark, 2009; de Bie and Ciechanover, 2011). These novel findings suggest that posttranslational modification of protein by ubiquitin and ubiquitin-like proteins is involved in a much wider range of cellular processes and signal transduction pathways important for numerous biological functions.
In this review, we will focus on the recent advances in the novel role of ubiquitination in the NF-κB activation and PKB/Akt kinase activation and their implications in human cancers. In addition, we will discuss some potential therapeutic strategies for targeting the ubiquitination pathways in the treatment of human cancers.
A large number of proteins are posttranslational modified by ubiquitin. Ubiquitin, a 76-amino acid protein, is highly evolutionarily conserved across species. In the ubiquitination process, ubiquitin is covalently conjugated to lysine residues of substrate proteins through a three-step enzymatic reaction. Ubiquitin is first activated by ubiquitin-activating enzyme (E1). The activated ubiquitin is then delivered to the ubiquitin-conjugating enzyme (E2), and will be transferred from the E2 to a lysine residue of a target protein by ubiquitin ligase (E3; Figure 1B).
As a general principle, the ubiquitin-conjugating enzyme E2 determines the type of ubiquitination modification to occur and the ubiquitin ligase E3 confers substrate specificity. Currently, there are two E1s, 50 E2s, and 600 E3s identified in human genome (Bhoj and Chen, 2009). There are seven lysine (K) residues (K6, K11, K27, K29, K33, K48, and K63) in the ubiquitin, which have been identified to be utilized for ubiquitination chains (Figure 1A). Most E2s trigger the K48-linked ubiquitination, in turn leading to proteasome-dependent degradation. However, the functions of ubiquitin conjugation are not limited to the ubiquitin–proteasome pathway. Mono and polyubiquitins are used as signals in various pathways including endocytosis, DNA repair, apoptosis, and transcriptional regulation. Polyubiquitin chains can be formed using lysine residues other than K48, the linkage required for proteasomal degradation (Figure 1B).
Ubiquitin-conjugating enzyme 13 is a major E2 that induces K63-linked ubiquitination with the assistance of its cofactor UEV1A. In stark contrast to the role of K48-linked ubiquitination in targeting its substrate protein for proteasome-degradation, the K63-linked ubiquitination does not induce proteasome-dependent degradation, but serves as a molecular platform for protein/protein interaction important for kinase signaling activation, receptor endocytosis, protein trafficking, and DNA damage repair. Although other types of ubiquitination modifications are also observed, the exact role that they play is unclear. Recent studies suggest that they can be involved in protein degradation or non-proteolytic functions (Figure 1B; Adhikari and Chen, 2009; Heemers and Tindall, 2009; Xu et al., 2009).
The E3s fall into two major families: E3s with HECT (homologous to the E6-AP carboxyl terminus) domain and E3s with RING (a really interesting new gene) or RING-like domain (e.g., U-Box and PHD domain; Bhoj and Chen, 2009). There are ∼600 RING finger and ∼30 HECT ligases in the human genome. Most of E3s recognize substrate proteins for K48-linked ubiquitination. However, several E3s can target proteins for K63-linked ubiquitination, such as HectH9, Mdm2, TNF receptor-associated factor 6 (TRAF6; tumor necrosis factor receptor-associated factor 6), cIAP1/2 (cellular inhibitor of apoptosis protein 1/2), CHIP, Parkin, UCHL1, TRAF2, ITCH, and NEDD4-2 (Adhikary et al., 2005; Huen et al., 2007; Mailand et al., 2007; Bertrand et al., 2008; Bhoj and Chen, 2009; Lim and Lim, 2011). Interestingly, HectH9, Mdm2, RNF8 (ring finger protein 8), and cIAP1/2 can trigger both K63- and K48-linked ubiquitination. For example, HectH9 not only targets Myc for K63-linked ubiquitination, which is critical for Myc transcriptional activation and the expression of a subset of Myc target genes, but also target MCL-1 and p19Arf for ubiquitination-dependent degradation (Adhikary et al., 2005; Chen et al., 2005; Zhong et al., 2005). So far, TRAF6 and RNF168 are the only two known E3s that selectively target substrate proteins for K63-linked ubiquitination, and thus play an important role in the innate immune response, DNA damage response (DDR), and tumorigenesis (Huen et al., 2007; Mailand et al., 2007; Bhoj and Chen, 2009).
Protein ubiquitination is a reversible process, and it is tightly controlled at both the levels of ubiquitin-conjugation and ubiquitin-deconjugation. Removal of ubiquitin on proteins is carried out by the DUBs. There are at least 98 DUBs encoded in the human genome, which can subdivided into six subfamilies based on sequence and structural similarity: ubiquitin-specific proteases (USP), ubiquitin carboxyl-terminal hydrolases (UCH), ovarian tumor-like proteases (OTU), JAMM/MPN metalloproteases, Machado–Jakob-disease (MJD) proteases, and the recently discovered monocyte chemotactic protein-induced protein (MCPIP) family (Sun, 2008; Bhoj and Chen, 2009; Hoeller and Dikic, 2009; Liang et al., 2010; Fraile et al., 2011). One mechanism to control the activities of DUB is the substrate specificity, the type of ubiquitin-chain linkages that are processed. Structural and functional studies have suggested that USPs and OTUs recognize and remove either Lys48- or Lys63-linked polyubiquitin chains. For example, USP14 removes Lys48-linked chains (Hu et al., 2005), whereas CYLD only efficiently processes Lys63 linkages and linear ubiquitin chains (Komander et al., 2009). Some OTU family components such as OTUB1 and A20 hydrolyze Lys48-linked chains, whereas TRABID and OTUD5 have preference for Lys63 linkages (Virdee et al., 2010). Furthermore, Cezanne, suppressesing NF-κB activation by targeting RIP1 for deubiquitination (Enesa et al., 2008), preferentially cleaves Lys11 over Lys48 and Lys63 linkages (Bremm et al., 2010), whereas JAMMs share the specificity for Lys63-linked polyubiquitin (McCullough et al., 2004; Sato et al., 2008; Cooper et al., 2009). Finally, the Josephin ATXN3-editing activity shows a restricted specificity for K63-linked chains (Winborn et al., 2008). The activity of DUBs regulates the turnover rate, activation, recycling, and localization of multiple proteins, and thus plays an essential role in cell homeostasis, protein stability, and a wide range of signaling pathways (Table 1) (Cohn et al., 2007; Shao et al., 2009; Lee et al., 2010; Nakada et al., 2010; Wiltshire et al., 2010). Importantly, deregulated DUB function has been linked to several diseases, including cancer (Table 1) (Nicassio et al., 2007; Bhoj and Chen, 2009; Novak et al., 2009; Fraile et al., 2011; Luise et al., 2011; Metzig et al., 2011).
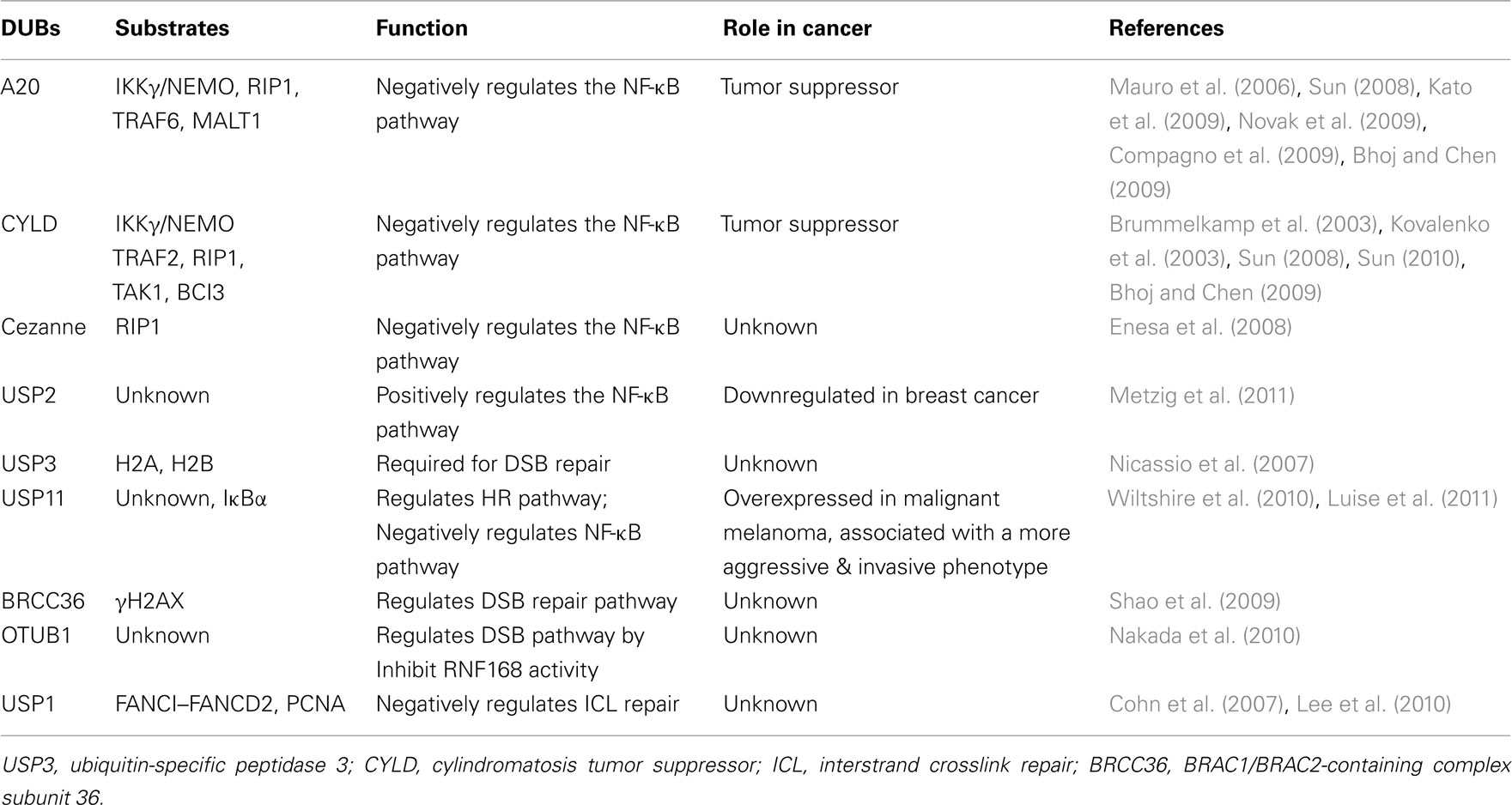
Table 1. Deubiquitinating enzymes (DUBs) play important roles in NF-κB activation, DNA damage response, and cancer.
Emerging evidence suggests that K63-linked ubiquitination also plays a pivotal role in the regulation of pathways that have been implicated in inflammatory response and cancer development, such as NF-κB (Skaug et al., 2009). The canonical NF-κB activation pathway is mediated through IKK, an upstream kinase responsible for IκB phosphorylation and subsequent degradation (Figure 2). IKK is consisted of IKKα kinase, IKKβ kinase, and IKKγ regulatory subunit (also known as NEMO; Chen, 2005; Bhoj and Chen, 2009; Skaug et al., 2009), which phosphorylates IκB and promotes SCF-βTrCP-mediated K48-linked ubiquitination and subsequent degradation by proteasome. Degradation of IκB results in NF-κB nuclear translocation and activation. In the non-canonical pathway, NIK-mediated-phosphorylation of IKKα results in its activation and subsequent phosphorylation of p100, which recruits a K48 E3 ligase to targetp100 for partial proteasomal degradation (Martinez-Forero et al., 2009; Skaug et al., 2009). A stop signal within the p100 sequence will terminate the proteasomal degradation process and the p52 transcription factor is then released and forms heterodimer NF-κB2 transcription factor with REL-B, which translocates to the nucleus and regulates gene expression. K63-linked ubiquitination has been shown to play an important role in both canonical and non-canonical NF-κB activation pathways (Figure 2).
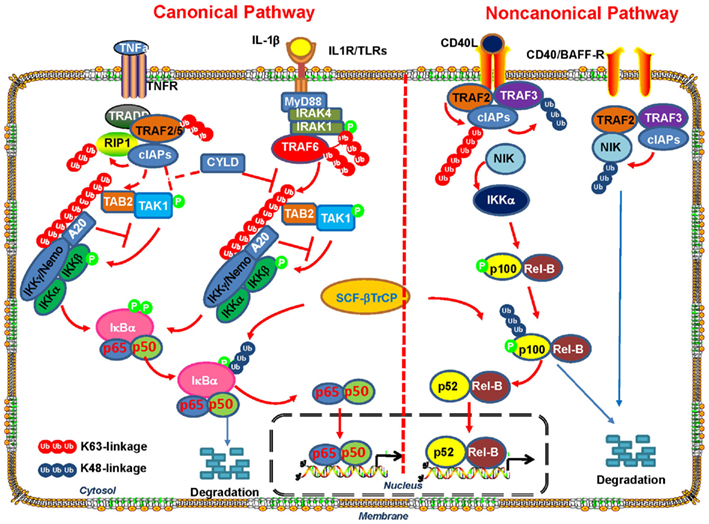
Figure 2. The role of K63-linked polyubiquitination in the activation of canonical and non-canonical NF-κB signaling pathway. In both canonical and non-canonical NF-κB pathways, ligand binding to receptors results in the recruitment of the TNF receptor-associated factors (TRAFs) to the receptors. In the canonical pathway, the recruitment of E3 ligase TRAF6promotesits auto-polyubiquitinating and other neighboring target proteins via K63-linked polyubiquitin chains. Unanchored free K63-linked polyubiquitin then directly activate TAK1 kinase, which then phosphorylates and activates IKK-β to induce NF- κB activation. In the non-canonical pathway, TRAF3, TRAF2, and cIAP promote rapid proteasomal degradation of NIK by the ubiquitin–proteasome system under un-stimulated condition. Various stimuli release NIK from TRAF3 inhibition by triggering cIAP activation by TRAF2-mediated K63-polyubiquitination and subsequent degradation of TRAF3. NIK-mediated-phosphorylation of p100 and its subsequent processing promotes RelB/p52 nuclear translocation and NF- κB activation.
In both canonical and non-canonical NF-κB pathways, the engagement of cell surface receptors, such as TNFR, IL-1R, Toll-like receptor (TLR), and CD40 by ligand stimulation, leads to the recruitment of the TRAFs such as TRAF2 and TRAF6, to the receptors (Skaug et al., 2009; Figure 2). TRAF proteins are ubiquitin ligases containing a highly conserved N-terminal RING domain except for TRAF1. TRAF6 has been demonstrated to play an important role in innate immune response and apoptosis by regulating TLR and TGF-β signaling involved in NF-κB activation and p38 activation, respectively (Chen, 2005; Sorrentino et al., 2008; Yamashita et al., 2008; Heldin et al., 2009; Skaug et al., 2009).
In the canonical pathways, after ligand binding to IL-1R or TLRs, the adaptor protein MyD88 is recruited to the cytoplasmic tail of the receptor and facilitates its interaction with IRAK4 and IRAK1 serine/threonine protein kinases. IRAK4 in turn phosphorylates and activate IRAK1 (Ordureau et al., 2008), which then recruits E3 ligase TRAF6 that functions together with dimeric ubiquitin-conjugating enzyme complex UBC13/UEV1A to induce its auto-polyubiquitinating and other neighboring target proteins via K63-linked polyubiquitin chains (Sorrentino et al., 2008; Yamashita et al., 2008; Martinez-Forero et al., 2009; Skaug et al., 2009). It has been proposed that K63-linked ubiquitination chains induced by TRAF6 serve as docking sites for TAB2, which contains ubiquitin-binding domains (UBD; Kanayama et al., 2004; Kulathu et al., 2009). TAB2 and TAB1 then promote the recruitment of the serine–threonine kinase TAK1 into the complex (Kanayama et al., 2004; Lynch and Gadina, 2004; Sun et al., 2004; Shambharkar et al., 2007) and its subsequent activation. Activated TAK1 then phosphorylates and activates IKK-β to promote IκB proteasomal degradation and translocation of NF-κB members p65 and p50 into the nucleus, in turn resulting in NF-κB activation (Figure 2). K63-linked polyubiquitination is critical in this process, because deletion of the UBAN domain of NEMO (IKKγ) abrogates IKK signaling. Although it was previously proposed that K63-linked polyubiquitination of several proteins in the IL-1LR/TLR pathways, such as TRAF6, IRAK1, and IKKγ/NEMO, are critical for the activation of TAK1 and IKK, a recent report provides compelling evidence that none of these polyubiquitin chains on these proteins are required for TAK1 activation (Xia et al., 2009). Instead, unanchored free K63-linked polyubiquitin chains, which are not conjugated to any target protein, directly activate TAK1 kinase through binding to TAB2 (Xia et al., 2009). Two possible mechanisms have been proposed for the unanchored polyubiquitin chains-mediated activation of TAK1 (Xia et al., 2009). One possible explanation is that TAK1 complex is brought into close proximity by the binding of TAB2 to the polyubiquitin chain, which will result in TAK1 autophosphorylation at Thr-187 and its activation. Another possibility is that polyubiquitin binding induces conformational change of TAK1 and thus allosterically activates the TAK1 complex. Free K63-linked polyubiquitin chains also directly bind to IKKγ/NEMO and activate IKK, which will lead to NF-κB activation (Xia et al., 2009).
TAK1 not only plays a role in NF-κB activation, but also in MAPK activation in response to numerous inflammatory stimuli (Wang et al., 2001; Skaug et al., 2009). TAK1 directly phosphorylates and activates MKK6 in an ubiquitin-dependent manner. In addition, knockdown of TAK1 by siRNA or TAK1 inhibitors suppresses IL-1β- and TNFα-induced IKK and c-Jun N-terminal kinase (JNK) activation (Ninomiya-Tsuji et al., 2003; Takaesu et al., 2003; Kanayama et al., 2004). Furthermore, inactivation of Tak1 in Drosophila melanogaster (Vidal et al., 2001; Chen et al., 2002; Silverman et al., 2003), or deletion of Tak1 in multiple mouse cell types abolishes IKK and MAPK activation by a variety of NF-κB agonists (Sato et al., 2005; Shim et al., 2005; Liu et al., 2006; Wan et al., 2006). Another study has further demonstrated a role for ubiquitination of TAK1 as a scaffold to recruit mitogen-activated MEKK3 (protein kinase kinase kinase 3) and subsequently to activate the MAP kinase pathway (Yamazaki et al., 2009).
In addition to TAK1, other kinases also play a role in NF-κB activation. RICK (also known as RIP2), a caspase-recruitment domain-containing kinase, plays a critical role in the NF-κB activation in response to Nod1 and Nod2 stimulation (Inohara et al., 2000; Park et al., 2007). RICK also undergoes K63-linked ubiquitination upon Nod1 and Nod2 stimulation at K209 located within its kinase domain. This ubiquitination triggers the oligomerization of RICK and the recruitment of TAK1 kinase, in turn facilitating IKK activation and secretion of cytokine and chemokine (Hasegawa et al., 2008). IRAK1, involved in TLR and IL-1R-mediated NF-κB activation, undergoes K63-linked ubiquitination upon IL-1 treatment, which is presumably ubiquitinated by E3 ligase TRAF6 (Conze et al., 2008). Ubiquitinated IRAK1 then recruits IKKγ and activates NF-κB signaling (Conze et al., 2008).
Stimulation of TNFR by the binding of TNFα triggers the recruitment of TRADD, TRAF2, and RIP1 to the receptor and also results in the activation of the canonical NF-κB pathway (Ermolaeva et al., 2008; Ha et al., 2009; Skaug et al., 2009; Figure 2). Similar to TRAF6, overexpressed TRAF2 is polyubiquitinated via K63-linked ubiquitination and is sufficient to induce NF-κB activation. However, there is functional redundancy between TRAF2 and TRAF5, as TRAF2-deficient mouse embryonic fibroblasts (MEFs) still activate NF-κB in response to TNFα, whereas TRAF2/5 compound mutant MEFs is compromised in IKK activation (Yeh et al., 1997; Grech et al., 2004; Skaug et al., 2009; Zhang et al., 2009). RIP1 is also polyubiquitinated via K63-linked chains by TRAF2/5 and cIAP1/2 complex, which is required for TNFα-induced NF-κB activation (Varfolomeev et al., 2008; Bertrand et al., 2011). The polyubiquitin chains on RIP1 serve as docking sites to recruit the TAK1 and IKK complexes to TNFR because the RIP1-K377R mutant failed to recruit TAK1 and IKK to the receptor, while retained the ability to recruit the adaptor protein TRADD or TRAF2 (Ea et al., 2006). However, the identification of free and unanchored K63-linked polyubiquitin chains directly activating TAK1 and IKK in the IL-1R/TLR pathway (Xia et al., 2009) suggests that such a mechanism may also exist in the TNFR pathway.
Interestingly, a recent report identified a novel mechanism by which enteropathogenic Escherichia coli NleE, a conserved bacterial type-III-secreted effector, regulates ubiquitin-chain signaling and its specificity in NF-κB activation during infection (Zhang et al., 2012). Specifically, NleE was found to possess S-adenosyl-L-methionine-dependent methyltransferase activity and modified a zinc-coordinating cysteine in the Npl4 zinc finger (NZF) domains in TAB2 and TAB3, which led to the loss of zinc ion and their ubiquitin-chain binding activity and subsequent inactivation of host NF-κB signaling activation.
On the other hand, the non-canonical NF-κB pathway regulation depends on tightly controlled degradation of NIK. TRAF3, TRAF2, and cIAP promote rapid proteasomal degradation of NIK by the ubiquitin–proteasome system (UPS). Therefore, under basal conditions, NIK protein is almost undetectable (Sasaki et al., 2008; Vallabhapurapu et al., 2008; Zarnegar et al., 2008a,b; Razani et al., 2010). However, various stimuli, such as CD40L and BAFF, trigger TRAF2-mediated K63-polyubiquitination of cIAP, which promotes K48-linked ubiquitination of TRAF3 and targets it to the proteasome (Vallabhapurapu et al., 2008; Zarnegar et al., 2008b). NIK is released from TRAF3 inhibition and phosphorylates and activates IKKα, which then induces phosphorylation of p100 and its subsequent processing by proteasome (Skaug et al., 2009; Sun, 2011). The processing of p100 is an important step in the activation of non-canonical NF-κB pathway, which not only generates p52 transcription factor, but also induces the nuclear translocation of RelB/p52 by releasing from its cytoplasmic localizing sequence present in the digested fragment (Skaug et al., 2009; Sun, 2011).
Interestingly, DDR can results in NF-κB activation via K63-linked ubiquitination as well as a novel linear ubiquitination. It has been shown that the E3 ligase linear ubiquitin-chain assembly complex (LUBAC), regulates NF-κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO (Niu et al., 2011). In addition, ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress (Wu et al., 2010). Furthermore, a cytoplasmic ATM–TRAF6–cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation (Hinz et al., 2010).
Since the process of ubiquitination of target proteins is tightly regulated by the activities of DUBs, it is conceivable that DUBs also play critical role in the regulation of NF-κB signaling. CYLD, a tumor suppressor which is found to be mutated in patients with familial cylindromatosis in which affected individuals develop multiple benign tumors of the skin appendages (Bignell et al., 2000), shows specific DUB activity toward K63-linked polyubiquitin chains, negatively regulating IKK activation. CYLD has been shown to target TRAF6, TRAF2, and NEMO for deubiquitination (Brummelkamp et al., 2003; Kovalenko et al., 2003; Mauro et al., 2006; Sun, 2008, 2010; Bhoj and Chen, 2009), thus presumably reduced their activities in NF-κB activation. Furthermore, CYLD also cleaves free unanchored K63-linked polyubiquitin chains, thus inhibiting TAK1 and IKK activation (Xia et al., 2009). Importantly, the tumor suppressive function of CYLD has been demonstrated in vivo using genetically engineered mouse (GEMs) model. Cyld-deficient mice are highly susceptible to chemical-induced skin tumors, chemical-induced colitis, and colon-tumorigenesis (Massoumi et al., 2006; Reiley et al., 2006, 2007; Zhang et al., 2006).
Another DUB, A20, which contains an N-terminal OTU-type DUB domain and seven C-terminal zinc fingers, has been suggested to play a critical role in the negative regulation of NF-κB activation, and A20-deficient mice have hyperactivation of NF-κB pathway and multi-organ inflammation (Brummelkamp et al., 2003; Kovalenko et al., 2003; Mauro et al., 2006; Sun, 2008, 2010; Bhoj and Chen, 2009). Recent evidence also implicates that A20 also plays an essential role in human diseases (Hymowitz and Wertz, 2010). For example, polymorphisms in the A20 locus are associated with multiple autoimmune diseases including systemic lupus erythematosis. In addition, A20 acts as a tumor suppressor in B cell lymphoma (Compagno et al., 2009; Kato et al., 2009) and mutated in marginal zone lymphomas (MZLs) (Novak et al., 2009). A20 can deubiquitinate RIP1 via its OTU domain and also act as an E3 ligase to add K48 polyubiquitin chains to RIP1 via the ZnF region to promote its proteasomal degradation (Wertz et al., 2004). In addition, A20 has been shown to promote disassembly of ubiquitination complexes in the IL-1R and TNFR pathways, including TRAF6–UBC13, cIAP1/2–UBC13, and cIAP1/2–UBCH5, as well as proteasomal degradation of UBC13 and UBCH5 (Shembade et al., 2009, 2010). Furthermore, A20 has also been reported to block recruitment of the adaptor proteins TRADD and RIP1 to TNFR (He and Ting, 2002), and to promote lysosomal degradation of TRAF2 (Li et al., 2009). Interestingly, A20 is recently shown to inhibit IKK activation through direct and non-catalytic mechanism (Figure 2). Specifically, A20 can bind to the unanchored polyubiquitin chain via its ZnF7 domain and interacts with IKKγ/NEMO, which results in an impairment of TAK1-mediated phosphorylation of IKK. Importantly, the catalytic Cys 103 residue of A20 is not required for the inhibition of IKK activation (Bosanac et al., 2010; Skaug et al., 2011).
Recently, it has been demonstrated that E3 ligase ITCH and CYLD formed a complex which can sequentially cleave K63-linked ubiquitin-chain and catalyzes K48-linked ubiquitination to deactivate TAK1 and terminate NF-κB signaling (Ahmed et al., 2011), providing an example of how K48 and K63-linked ubiquitinations are closely linked and can be differentially utilized to control kinase activation and deactivation.
Protein kinases play a pivotal role in signaling transduction cascade, and they act as signaling mediators to convey the extracellular clues, such as growth factors and cytokines, from the outside of the cells to the nucleus by inducing protein phosphorylation (Hynes and MacDonald, 2009; Klein and Levitzki, 2009). The level of phosphorylation of a target protein is counteracted by the activity of phosphatase. Protein kinases are also subjected to phosphorylation, which regulates their activities and expressions. Recently, emerging evidence suggests a critical role for K63-linked ubiquitination in the regulation of kinase activation (Yang et al., 2009, 2010a,b). Here we will focus on the regulatory function of K63-linked polyubiquitination in kinase activation using AKT as an example. AKT (also known as PKB) is a serine/threonine protein kinase, which is a keystone in growth factor and cytokine-mediated signaling cascade (Datta et al., 1999; Brazil et al., 2002; Cantley, 2002; Gonzalez and McGraw, 2009; Yang et al., 2010a). Giving that Akt regulate numerous biological functions, such as cell growth, survival, cell migration, and metabolism, by phosphorylating several downstream effectors, the activity of Akt must be stringently controlled. Aberrant Akt activation is observed in various human cancers, and importantly, Akt1, Akt2, and Akt3 isoforms are found to be overexpressed in human cancers (Staal, 1987; Cheng et al., 1992, 1996; Bellacosa et al., 1995; Nakatani et al., 1999; Stahl et al., 2004). Recent studies show that Akt1 mutations are observed in a subset of human cancers and are associated with Akt hyperactivation (Carpten et al., 2007; Kim et al., 2008a; Malanga et al., 2008; Mohamedali et al., 2008; Askham et al., 2010; Shoji et al., 2009; Zilberman et al., 2009). The role of Akt in cancer development has been supported by numerous animal tumor models. For example, Pten+/− mice with aberrant Akt activation develop multiple tumors, which can be inhibited by Akt1 deficiency (Di Cristofano et al., 2001; Chen et al., 2006). In addition, the prostate-specific expression of constitutively active Akt1 in mice leads to prostate intraepithelial neoplasia (Majumder et al., 2003, 2004). Accordingly, these results highlight the critical role of the PI3K/Akt pathway in cancer development.
Ubiquitination has been shown to regulate Akt protein degradation. Several E3 ligases, such as CHIP (carboxyl terminus of Hsc-70 interacting protein), BRCA1, and TTC3, have been shown to target AKT for ubiquitination and subsequent proteasome degradation (Dickey et al., 2008; Xiang et al., 2008; Suizu et al., 2009; Toker, 2009). In addition, mTORC2-mediated phosphorylation of Akt at T450 also regulates Akt stability as Sin1 deficiency or mTOR knockdown results in reduced T450 phosphorylation and increased Akt ubiquitination and degradation (Facchinetti et al., 2008).
Recently, TRAF6 has been identified as a unique E3 ligase for Akt and induces K63-linked Akt ubiquitination, in turn facilitating Akt membrane recruitment and subsequent Akt T308 phosphorylation and activation (Yang et al., 2009, 2010a). The notion is supported by the fact that primary MEFs deficient for Traf6 display profound defects in Akt membrane recruitment and activation in response to various stimuli such as growth factors and cytokines. The ubiquitination of Akt occurs on the K8 and K14 residues within the PH (pleckstrin homology) domain of Akt. Notably, Akt K8R and Akt K14R mutants display defects in Akt ubiquitination, correlated with the impairment of Akt membrane recruitment and T308 phosphorylation, suggesting that Akt ubiquitination plays an important role in Akt membrane recruitment and activation.
Analysis of K8R mutant of Akt reveals that the regulation of Akt membrane recruitment by Akt ubiquitination does not result from its impact on the PIP3 binding. It is known that Akt membrane localization is essentially dependent on its binding to phosphoinositol (3,4,5) trisphosphate (PIP3) in the plasma membrane (Brazil et al., 2002; Varnai et al., 2005). The K14R Akt mutant fails to bind to PIP3, as the K14 residue is within the PIP3 binding pocket. However, the K8 residue is not within the PIP3 binding pocket (Bellacosa et al., 1998; Rong et al., 2001; Carpten et al., 2007) and there is no difference in the binding to PIP3 between the Akt K8R mutant and the wild-type (Wt) Akt (Yang et al., 2009).
One Akt mutant (E17K), found in a subset of human cancers, displays higher PIP3 binding, membrane localization, and activation (Brazil et al., 2002; Varnai et al., 2005). Interestingly, this cancer-associated Akt mutant also displays a hyperubiquitination of Akt, and the blocking of this hyperubiquitination profoundly impairs Akt membrane localization, phosphorylation, and activation (Carpten et al., 2007; Yang et al., 2009). These results suggest that mutation of E17 residue to lysine residue in Akt results in not only hyperubiquitination but also hyper-PIP3 binding, and both of these events appear to contribute to the constitutive membrane localization and activation of Akt. Collectively, these results demonstrate that TRAF6-mediated Akt ubiquitination is relevant and important for human cancers. Along this line, another Akt mutation E49K has been recently identified in bladder cancer, which also displays Akt hyperphosphorylation and activation compared to WtAkt (Askham et al., 2010). Given this Akt mutation is located in the PH domain of Akt but not in the PIP3 binding pocket, we speculate that this mutation may not affect Akt binding to the PIP3. Since this Akt mutant gains an additional lysine residue, it is very likely that such mutation may also results in hyperubiquitination of Akt, in turn contributing to its hyperactivation and hyper-oncogenic potential.
Currently, it remains unclear how the K63-linked ubiquitination of Akt results in its recruitment to plasma membrane and subsequent activation. One possibility is that TRAF6-mediated polyubiquitination of Akt may increase its binding to PIP3 on the membrane (Figure 3). However, it may be not the case, since the Akt K8R mutant, which shows an impairment in Akt ubiquitination, membrane recruitment and activation, displays similar binding affinity to PIP3 as WtAkt does (Yang et al., 2009), suggesting that other mechanisms contribute to ubiquitination-mediated Akt membrane recruitment and activation. It has been shown that TCL1 family proteins form heterodimer with Akt and promote Akt dimerization, in turn augmenting Akt activation (Teitell, 2005). Therefore, another possibility is that K63-polyubiquitination of Akt may enhance the formation of TCL1–Akt complex and Akt dimerization, thus promoting Akt membrane localization and Akt activation (Figure 3). Furthermore, since K63-linked polyubiquitin chains have been proposed to serve as docking sites for many signaling molecules, it is highly possible that K63-ubiquitination of Akt enhances its binding to membrane bound adaptor protein containing ubiquitin-binding property, thereby facilitating Akt membrane localization and activation (Figure 3). More works will be required to further dissect these possibilities.
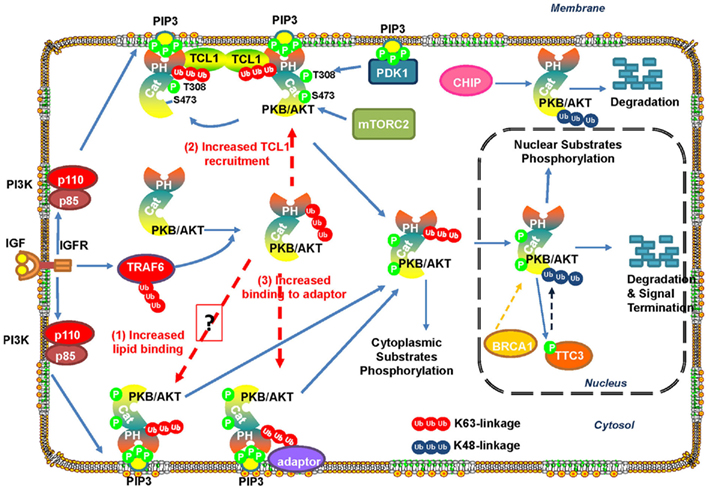
Figure 3. Ubiquitination regulates Akt stability and activation. IGF-1-induced activation of IGF-1R promotes TRAF6 activation, which then triggers K63-linked ubiquitination of Akt and promotes Akt membrane recruitment and subsequent phosphorylation by PDK1 and mTORC2. Active Akt then phosphorylates its substrates in the cytoplasm and nucleus to exert its biological functions. E3 ligase TTC3 has been suggested to play a role in the termination of active Akt signaling in the nucleus by promoting Akt ubiquitination and degradation by the proteasome. TTC3 activity is regulated by Akt-mediated phosphorylation at S378, thus providing a negative feedback loop for Akt inactivation. In addition, Akt ubiquitination is promoted by CHIP or BRCA1 and leads to Akt degradation.
Ubiquitination and deubiquitination mediated through the E3 ligases and DUBs regulate the activities and functions of a large amount of protein, including tumor suppressors and oncogenes important for cell proliferation, apoptosis, and tumorigenesis. Importantly, the important roles of these proteins in tumorigenesis are supported by their frequent deregulations in human cancer and by mouse tumor models to reveal their important roles in cancer development. Therefore, targeting these proteins may have important therapeutic implications for the treatment of human cancer. Efforts have been invested to target the proteasome subunits (the proteolytic 20S subunit and the regulatory 19S subunit), and E3 ubiquitin ligase, and DUBs.
Bortezomib, an inhibitor proteasome 20S subunit, has been approved by the FDA for the treatment of multiple myeloma (Adams and Kauffman, 2004; Bold, 2004). Recently, the small molecule b-AP15, which inhibits the activity of two 19S regulatory-particle-associated DUBs, ubiquitin C-terminal hydrolase 5 (UCHL5), and ubiquitin-specific peptidase 14 (USP14), can induce tumor cell apoptosis that is insensitive to TP53 status and overexpression of the apoptosis inhibitor BCL2. Furthermore, b-AP15 inhibits tumor progression in four different in vivo solid tumor xenograft models, including FaDu squamous carcinoma, HCT-116 colon carcinoma, Lewis lung carcinomas (LLCs), orthotopic breast carcinoma (4T1), and also inhibits organ infiltration in one acute myeloid leukemia model (C1498 leukemia; D’Arcy et al., 2011).
Many E3 ligases have been shown to play an oncogenic role in tumorigenesis, therefore, E3 ubiquitin ligases, which confer substrate specificity, are attractive targets for human cancers. For example, efforts have been invested on targeting HDM2, the E3 ligase for tumor suppressor p53, aiming to elevate the protein level of p53. Nutlin 3/R7112, an inhibitor that disrupts the interaction between HDM2 and its substrate p53, has entered clinical trials (Cohen and Tcherpakov, 2010). In addition, several inhibitors that target inhibitors of apoptosis proteins (IAPs) have also entered clinical trials (Cohen and Tcherpakov, 2010). These inhibitors induce the autoubiquitination and degradation of the IAPs and promote cancer death by stimulating the TNF-α pathway (Wu et al., 2007).
Given the critical role of TRAF6 in the regulation of Akt ubiquitination, membrane localization, and activation (Yang et al., 2009), small molecules targeting TRAF6 can suppress Akt signaling and may be considered as potential or adjuvant agents for cancer therapy. In line with this notion, TRAF6 inhibition by shRNA knockdown reduces Akt activation in prostate cancer cells, suppresses tumor formation in the xenograft tumor model, and potentiates apoptosis induced by chemotherapy agents (Yang et al., 2009). Therefore, TRAF6 can be considered as a potential therapeutic target for human cancer. In addition to small molecules inhibitors for TRAF6, miR-145, and miR-146a have shown to repress TRAF6 protein translation and display a potent tumor suppressive effect on restricting the formation of primary and metastatic breast cancer (Hou et al., 2009; Hurst et al., 2009; Starczynowski et al., 2010; Yuan et al., 2010). Therefore, miR-145 and miR-146a can be applied as another strategy to target TRAF6 for cancer treatment.
Deubiquitinating enzymes are another class of potential drug targets for cancer treatment. DUBs have been shown to play an important role in the regulation of the stability of many oncoproteins by counteracting the activity of E3 ligase. USP7 (ubiquitin-specific protease 7), which deubiquitinates HDM2, can lead to increased levels of HDM2 and decreased levels of p53. Therefore, inhibitors for USP7 will promote HDM2 degradation and p53 accumulation. Interestingly, as USP7 silencing stabilizes p53 and enhances its activity, small molecular inhibitors for USP7 are expected to suppress cancer cell growth. As expected, a novel USP7 inhibitor indeed induces p53-dependent apoptosis by stabilizing p53 (Colland et al., 2009). USP20, also called VDU2 (von Hippel–Lindau deubiquitinating enzyme 2) stabilizes hypoxia-inducible factor 1α (HIF-1α; Li et al., 2005), which is overexpressed in many human cancers. Therefore, targeting USP20 can induce degradation of HIF-1α and be a potential drug target for human cancer. Furthermore, many transcription factors have been demonstrated to be overexpressed and play potent oncogenic functions in tumorigenesis, such as Myc (Pelengaris et al., 2002), CyclinD1 (Musgrove et al., 2011) and inhibitors of DNA binding (ID; Hasskarl and Munger, 2002). However, it has been well-accepted that it is difficult to target transcription factors with small molecules since they lack enzymatic activity, although there is a report that small molecule has been designed to target Bcl-6 in mouse model for cancer therapy (Cerchietti et al., 2010). Targeting Myc is recently demonstrated to be very effective in the treatment of human cancers (Adhikary and Eilers, 2005; Soucek et al., 2008; Herold et al., 2009). In addition, inactivation of Cyclin D1 can abolish breast cancer development driven by oncogenic Ras or Neu (Lee et al., 2000; Yu et al., 2001). Further, IDs overexpression is observed in a broad range of dedifferentiated primary human malignancies, such as pancreatic carcinoma and neuroblastoma (Perk et al., 2005). Since many of these oncogenic transcription factors are short-lived oncoproteins undergoing fast turnover, therapeutic strategies by targeting their corresponding DUBs will be an alternative approach, which will lead to rapid degradation of these oncoproteins. USP28, USP2, USP1, Dub3, and USP9x have been shown to play an important role in stabilizing oncogenic signals, such as Myc (Popov et al., 2007), Cyclin D1 (Shan et al., 2009), and IDs (Williams et al., 2011) and CDC25A (Pereg et al., 2010), MCL-1 (Opferman and Green, 2010; Schwickart et al., 2010) respectively. Therefore, small molecules inhibitors to target these DUBs are theoretically possible to design and to target these oncogenic transcription factors indirectly. Future experiments using small molecule inhibitors to target these DUBs in preclinical mouse tumor models may provide in vivo evidence that targeting the DUBs can be ideal therapeutic strategies for the treatment of human cancers.
A large amount of recent studies in the area of ubiquitination study have revealed several novel roles of ubiquitination in signaling transduction pathways involved in a broad range of biological functions, such as DDR, NF-κB activation, and kinase activation. Ubiquitination can play a distinct role in the regulation of protein fate, depending on which type of the polyubiquitin chains formed within the proteins (Figure 1). Mono-ubiquitination plays a role in endocytosis, protein transport, DNA repair, and histone regulation. K48-linked polyubiquitination signals mainly target proteins for proteasome-dependent protein degradation, whereas K63-linked ubiquitination serves as a scaffold for protein/protein interaction, in turn regulating the localization and activity of protein kinase. In addition, a novel type of linear polyubiquitination plays a role in the NF-κB activation. It is anticipated that other novel functions on this posttranslational modification will be uncovered in the near future.
As strong evidence has demonstrated an important role for ubiquitination in cancers, efforts to specifically target this pathway with small molecules inhibitors have now been actively investigated. Proteasome inhibitor Bortezomib has been approved to be used in the treatment of multiple myeloma. Several small molecule inhibitors targeting E3 ligases have entered clinical trials, such as inhibitors for HDM2 and IAPs. In addition, efforts have been invested to target DUBs and small molecule inhibitors have been developed and demonstrated efficacy in the treatment of human cancer. We anticipate to see more and more small molecule inhibitors targeting this pathway will be developed and eventually utilized for the treatment of human cancers.
However, there still remains several outstanding questions and warrant for future investigations. Currently only a few E3 ligases are able to trigger K63-linked ubiquitination. Will more such E3 ligases be discovered in the genome? As many proteins can undergo both K48- and K63-linked ubiquitination, it remains to be determined how the E3 ligases and DUBs coordinate to regulate signaling activation for protein kinases. A recent report on ITVH–CYLD sheds light on how K48- and K63-link ubiquitination/deubiquitination are closely coupled to activate and inactivate TAK1 to tightly control the activation of NF-κB signaling pathway. In addition, it remains largely unknown for the roles of the novel linear polyubiquitin chains in the regulation of target protein function. Moreover, much work is needed to determine the mechanisms for the regulation of linear vs. branch polyubiquitination and how the ubiquitination regulates the localization and activation of protein kinase. Furthermore, a deeper mechanistic understanding of ubiquitination and deubiquitination and kinase regulation by ubiquitin remains as an important challenge for this field. In addition, we need to understand how substrate binding and activity of ubiquitination and DUBs are regulated. Addressing all these important questions will lead to comprehensive understanding of the complicated mode of ubiquitination and deubiquitination in regulating signaling transduction pathways.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the members of the Lin’s lab for their critical reading and comments on our manuscript. This work is supported by M. D. Anderson Research Trust Scholar Fund, NIH RO1 grants, CPRIT grant, and New Investigator Award from Department of Defense to H. K. Lin.
Adams, J., and Kauffman, M. (2004). Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 22, 304–311.
Adhikary, S., and Eilers, M. (2005). Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 6, 635–645.
Adhikary, S., Marinoni, F., Hock, A., Hulleman, E., Popov, N., Beier, R., Bernard, S., Quarto, M., Capra, M., Goettig, S., Kogel, U., Scheffner, M., Helin, K., and Eilers, M. (2005). The ubiquitin ligase HectH9 regulates transcriptional activation by Myc and is essential for tumor cell proliferation. Cell 123, 409–421.
Ahmed, N., Zeng, M., Sinha, I., Polin, L., Wei, W. Z., Rathinam, C., Flavell, R., Massoumi, R., and Venuprasad, K. (2011). The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat. Immunol. 12, 1176–1183.
Askham, J. M., Platt, F., Chambers, P. A., Snowden, H., Taylor, C. F., and Knowles, M. A. (2010). AKT1 mutations in bladder cancer: identification of a novel oncogenic mutation that can co-operate with E17K. Oncogene 29, 150–155.
Bellacosa, A., Chan, T. O., Ahmed, N. N., Datta, K., Malstrom, S., Stokoe, D., McCormick, F., Feng, J., and Tsichlis, P. (1998). Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene 17, 313–325.
Bellacosa, A., de Feo, D., Godwin, A. K., Bell, D. W., Cheng, J. Q., Altomare, D. A., Wan, M., Dubeau, L., Scambia, G., Masciullo, V., Ferrandina, G., Benedetti Panici, P., Mancuso, S., Neri, G., and Testa, J. R. (1995). Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int. J. Cancer 64, 280–285.
Bertrand, M. J., Lippens, S., Staes, A., Gilbert, B., Roelandt, R., De Medts, J., Gevaert, K., Declercq, W., and Vandenabeele, P. (2011). cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1-4). PLoS ONE 6, e22356. doi:10.1371/journal.pone.0022356
Bertrand, M. J., Milutinovic, S., Dickson, K. M., Ho, W. C., Boudreault, A., Durkin, J., Gillard, J. W., Jaquith, J. B., Morris, S. J., and Barker, P. A. (2008). cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 30, 689–700.
Bhoj, V. G., and Chen, Z. J. (2009). Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437.
Bignell, G. R., Warren, W., Seal, S., Takahashi, M., Rapley, E., Barfoot, R., Green, H., Brown, C., Biggs, P. J., Lakhani, S. R., Jones, C., Hansen, J., Blair, E., Hofmann, B., Siebert, R., Turner, G., Evans, D. G., Schrander-Stumpel, C., Beemer, F. A., van Den Ouweland, A., Halley, D., Delpech, B., Cleveland, M. G., Leigh, I., Leisti, J., and Rasmussen, S. (2000). Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25, 160–165.
Bold, R. (2004). Development of the proteasome inhibitor Velcade (Bortezomib) by Julian Adams, Ph.D., and Michael Kauffman, M.D., Ph.D. Cancer Invest. 22, 328–329.
Bosanac, I., Wertz, I. E., Pan, B., Yu, C., Kusam, S., Lam, C., Phu, L., Phung, Q., Maurer, B., Arnott, D., Kirkpatrick, D. S., Dixit, V. M., and Hymowitz, S. G. (2010). Ubiquitin binding to A20 ZnF4 is required for modulation of NF-kappaB signaling. Mol. Cell 40, 548–557.
Brazil, D. P., Park, J., and Hemmings, B. A. (2002). PKB binding proteins. Getting in on the Akt. Cell 111, 293–303.
Bremm, A., Freund, S. M., and Komander, D.. (2010). Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 17, 939–947.
Brummelkamp, T. R., Nijman, S. M., Dirac, A. M., and Bernards, R. (2003). Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424, 797–801.
Carpten, J. D., Faber, A. L., Horn, C., Donoho, G. P., Briggs, S. L., Robbins, C. M., Hostetter, G., Boguslawski, S., Moses, T. Y., Savage, S., Uhlik, M., Lin, A., Du, J., Qian, Y. W., Zeckner, D. J., Tucker-Kellogg, G., Touchman, J., Patel, K., Mousses, S., Bittner, M., Schevitz, R., Lai, M. H., Blanchard, K. L., and Thomas, J. E. (2007). A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444.
Cerchietti, L. C., Ghetu, A. F., Zhu, X., Da Silva, G. F., Zhong, S., Matthews, M., Bunting, K. L., Polo, J. M., Farès, C., Arrowsmith, C. H., Yang, S. N., Garcia, M., Coop, A., Mackerell, A. D. Jr., Privé, G. G., and Melnick, A. (2010). A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell 17, 400–411.
Chen, D., Kon, N., Li, M., Zhang, W., Qin, J., and Gu, W. (2005). ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell 121, 1071–1083.
Chen, M. L., Xu, P. Z., Peng, X. D., Chen, W. S., Guzman, G., Yang, X., Di Cristofano, A., Pandolfi, P. P., and Hay, N. (2006). The deficiency of Akt1 is sufficient to suppress tumor development in Pten+/− mice. Genes Dev. 20, 1569–1574.
Chen, W., White, M. A., and Cobb, M. H. (2002). Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J. Biol. Chem. 277, 49105–49110.
Chen, Z. J., and Sun, L. J. (2009). Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286.
Cheng, J. Q., Godwin, A. K., Bellacosa, A., Taguchi, T., Franke, T. F., Hamilton, T. C., Tsichlis, P. N., and Testa, J. R. (1992). AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc. Natl. Acad. Sci. U.S.A. 89, 9267–9271.
Cheng, J. Q., Ruggeri, B., Klein, W. M., Sonoda, G., Altomare, D. A., Watson, D. K., and Testa, J. R. (1996). Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. U.S.A. 93, 3636–3641.
Cohen, P., and Tcherpakov, M. (2010). Will the ubiquitin system furnish as many drug targets as protein kinases? Cell 143, 686–693.
Cohn, M. A., Kowal, P., Yang, K., Haas, W., Huang, T. T., Gygi, S. P., and D’Andrea, A. D. (2007). A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 28, 786–797.
Colland, F., Formstecher, E., Jacq, X., Reverdy, C., Planquette, C., Conrath, S., Trouplin, V., Bianchi, J., Aushev, V. N., Camonis, J., Calabrese, A., Borg-Capra, C., Sippl, W., Collura, V., Boissy, G., Rain, J. C., Guedat, P., Delansorne, R., and Daviet, L. (2009). Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Mol. Cancer Ther. 8, 2286–2295.
Compagno, M., Lim, W. K., Grunn, A., Nandula, S. V., Brahmachary, M., Shen, Q., Bertoni, F., Ponzoni, M., Scandurra, M., Califano, A., Bhagat, G., Chadburn, A., Dalla-Favera, R., and Pasqualucci, L. (2009). Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 459, 717–721.
Conze, D. B., Wu, C. J., Thomas, J. A., Landstrom, A., and Ashwell, J. D. (2008). Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol. Cell. Biol. 28, 3538–3547.
Cooper, E. M., Cutcliffe, C., Kristiansen, T. Z., Pandey, A., Pickart, C. M., and Cohen, R. E. (2009). K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631.
D’Arcy, P., Brnjic, S., Olofsson, M. H., Fryknäs, M., Lindsten, K., De Cesare, M., Perego, P., Sadeghi, B., Hassan, M., Larsson, R., and Linder, S. (2011). Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat. Med. 17, 1636–1640.
Datta, S. R., Brunet, A., and Greenberg, M. E. (1999). Cellular survival: a play in three Akts. Genes Dev. 13, 2905–2927.
de Bie, P., and Ciechanover, A. (2011). Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death Differ. 18, 1393–1402.
Di Cristofano, A., De Acetis, M., Koff, A., Cordon-Cardo, C., and Pandolfi, P. P. (2001). Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat. Genet. 27, 222–224.
Dickey, C. A., Koren, J., Zhang, Y. J., Xu, Y. F., Jinwal, U. K., Birnbaum, M. J., Monks, B., Sun, M., Cheng, J. Q., Patterson, C., Bailey, R. M., Dunmore, J., Soresh, S., Leon, C., Morgan, D., and Petrucelli, L. (2008). Akt and CHIP coregulate tau degradation through coordinated interactions. Proc. Natl. Acad. Sci. U.S.A. 105, 3622–3627.
Ea, C. K., Deng, L., Xia, Z. P., Pineda, G., and Chen, Z. J. (2006). Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22, 245–257.
Enesa, K., Zakkar, M., Chaudhury, H., Luong le, A., Rawlinson, L., Mason, J. C., Haskard, D. O., Dean, J. L., and Evans, P. C. (2008). NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J. Biol. Chem. 283, 7036–7045.
Ermolaeva, M. A., Michallet, M. C., Papadopoulou, N., Utermöhlen, O., Kranidioti, K., Kollias, G., Tschopp, J., and Pasparakis, M. (2008). Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat. Immunol. 9, 1037–1046.
Facchinetti, V., Ouyang, W., Wei, H., Soto, N., Lazorchak, A., Gould, C., Lowry, C., Newton, A. C., Mao, Y., Miao, R. Q., Sessa, W. C., Qin, J., Zhang, P., Su, B., and Jacinto, E. (2008). The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943.
Fraile, J. M., Quesada, V., Rodríguez, D., Freije, J. M., and López-Otín, C. (2011). Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. doi: 10.1038/onc.2011.443. [Epub ahead of print].
Gao, Z., Xu, M. S., Barnett, T. L., and Xu, C. W. (2011). Resveratrol induces cellular senescence with attenuated mono-ubiquitination of histone H2B in glioma cells. Biochem. Biophys. Res. Commun. 407, 271–276.
Giasson, B. I., and Lee, V. M. (2003). Are ubiquitination pathways central to Parkinson’s disease? Cell 114, 1–8.
Gonzalez, E., and McGraw, T. E. (2009). The Akt kinases: isoform specificity in metabolism and cancer. Cell Cycle 8, 2502–2508.
Grech, A. P., Amesbury, M., Chan, T., Gardam, S., Basten, A., and Brink, R. (2004). TRAF2 differentially regulates the canonical and noncanonical pathways of NF-kappaB activation in mature B cells. Immunity 21, 629–642.
Ha, H., Han, D., and Choi, Y. (2009). TRAF-mediated TNFR-family signaling. Curr. Protoc. Immunol. Chapter 11, Unit11 19D.
Hasegawa, M., Fujimoto, Y., Lucas, P. C., Nakano, H., Fukase, K., Núñez, G., and Inohara, N. (2008). A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 27, 373–383.
Hasskarl, J., and Munger, K. (2002). Id proteins – tumor markers or oncogenes? Cancer Biol. Ther. 1, 91–96.
He, K. L., and Ting, A. T. (2002). A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol. Cell. Biol. 22, 6034–6045.
Heemers, H. V., and Tindall, D. J. (2009). Unraveling the complexities of androgen receptor signaling in prostate cancer cells. Cancer Cell 15, 245–247.
Heldin, C. H., Landstrom, M., and Moustakas, A. (2009). Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr. Opin. Cell Biol. 21, 166–176.
Herold, S., Herkert, B., and Eilers, M. (2009). Facilitating replication under stress: an oncogenic function of MYC? Nat. Rev. Cancer 9, 441–444.
Hinz, M., Stilmann, M., Arslan, S. Ç., Khanna, K. K., Dittmar, G., and Scheidereit, C. (2010). A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-kappaB activation. Mol. Cell 40, 63–74.
Hoeller, D., and Dikic, I. (2009). Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444.
Hou, J., Wang, P., Lin, L., Liu, X., Ma, F., An, H., Wang, Z., and Cao, X. (2009). MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183, 2150–2158.
Hu, M., Li, P., Song, L., Jeffrey, P. D., Chenova, T. A., Wilkinson, K. D., Cohen, R. E., and Shi, Y. (2005). Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756.
Huen, M. S., Grant, R., Manke, I., Minn, K., Yu, X., Yaffe, M. B., and Chen, J. (2007). RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914.
Hurst, D. R., Edmonds, M. D., Scott, G. K., Benz, C. C., Vaidya, K. S., and Welch, D. R. (2009). Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 69, 1279–1283.
Hymowitz, S. G., and Wertz, I. E. (2010). A20: from ubiquitin editing to tumour suppression. Nat. Rev. Cancer 10, 332–341.
Hynes, N. E., and MacDonald, G. (2009). ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184.
Inohara, N., Koseki, T., Lin, J., del Peso, L., Lucas, P. C., Chen, F. F., Ogura, Y., and Núñez, G. (2000). An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 275, 27823–27831.
Kanayama, A., Seth, R. B., Sun, L., Ea, C. K., Hong, M., Shaito, A., Chiu, Y. H., Deng, L., and Chen, Z. J. (2004). TAB2 and TAB3 activate the NF-kappaB pathway through binding to polyubiquitin chains. Mol. Cell 15, 535–548.
Kato, M., Sanada, M., Kato, I., Sato, Y., Takita, J., Takeuchi, K., Niwa, A., Chen, Y., Nakazaki, K., Nomoto, J., Asakura, Y., Muto, S., Tamura, A., Iio, M., Akatsuka, Y., Hayashi, Y., Mori, H., Igarashi, T., Kurokawa, M., Chiba, S., Mori, S., Ishikawa, Y., Okamoto, K., Tobinai, K., Nakagama, H., Nakahata, T., Yoshino, T., Kobayashi, Y., and Ogawa, S. (2009). Frequent inactivation of A20 in B-cell lymphomas. Nature 459, 712–716.
Kim, M. S., Jeong, E. G., Yoo, N. J., and Lee, S. H. (2008a). Mutational analysis of oncogenic AKT E17K mutation in common solid cancers and acute leukaemias. Br. J. Cancer 98, 1533–1535.
Kim, P. K., Hailey, D. W., Mullen, R. T., and Lippincott-Schwartz, J. (2008b). Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. U.S.A. 105, 20567–20574.
Kirkin, V., McEwan, D. G., Novak, I., and Dikic, I. (2009). A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269.
Klein, S., and Levitzki, A. (2009). Targeting the EGFR and the PKB pathway in cancer. Curr. Opin. Cell Biol. 21, 185–193.
Komander, D., Reyes-Turcu, F., Licchesi, J. D., Odenwaelder, P., Wilkinson, K. D., and Barford, D. (2009). Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 10, 466–473.
Kovalenko, A., Chable-Bessia, C., Cantarella, G., Israël, A., Wallach, D., and Courtois, G. (2003). The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424, 801–805.
Kraft, C., Peter, M., and Hofmann, K. (2010). Selective autophagy: ubiquitin-mediated recognition and beyond. Nat. Cell Biol. 12, 836–841.
Kulathu, Y., Akutsu, M., Bremm, A., Hofmann, K., and Komander, D. (2009). Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16, 1328–1330.
Lee, K. Y., Yang, K., Cohn, M. A., Sikdar, N., D’Andrea, A. D., and Myung, K. (2010). Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J. Biol. Chem. 285, 10362–10369.
Lee, R. J., Albanese, C., Fu, M., D’Amico, M., Lin, B., Watanabe, G., Haines, G. K. III, Siegel, P. M., Hung, M. C., Yarden, Y., Horowitz, J. M., Muller, W. J., and Pestell, R. G. (2000). Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20, 672–683.
Li, L., Soetandyo, N., Wang, Q., and Ye, Y. (2009). The zinc finger protein A20 targets TRAF2 to the lysosomes for degradation. Biochim. Biophys. Acta 1793, 346–353.
Li, Z., Wang, D., Messing, E. M., and Wu, G. (2005). VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 6, 373–378.
Liang, J., Saad, Y., Lei, T., Wang, J., Qi, D., Yang, Q., Kolattukudy, P. E., and Fu, M. (2010). MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 207, 2959–2973.
Lim, K. L., and Lim, G. G. (2011). K63-linked ubiquitination and neurodegeneration. Neurobiol. Dis. 43, 9–16.
Liu, H. H., Xie, M., Schneider, M. D., and Chen, Z. J. (2006). Essential role of TAK1 in thymocyte development and activation. Proc. Natl. Acad. Sci. U.S.A. 103, 11677–11682.
Luise, C., Capra, M., Donzelli, M., Mazzarol, G., Jodice, M. G., Nuciforo, P., Viale, G., Di Fiore, P. P., and Confalonieri, S. (2011). An atlas of altered expression of deubiquitinating enzymes in human cancer. PLoS One 6, e15891.
Lynch, O. T., and Gadina, M. (2004). Ubiquitination for activation: new directions in the NF-kappaB roadmap. Mol. Interv. 4, 144–146.
Mailand, N., Bekker-Jensen, S., Faustrup, H., Melander, F., Bartek, J., Lukas, C., and Lukas, J. (2007). RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900.
Majumder, P. K., Febbo, P. G., Bikoff, R., Berger, R., Xue, Q., McMahon, L. M., Manola, J., Brugarolas, J., McDonnell, T. J., Golub, T. R., Loda, M., Lane, H. A., and Sellers, W. R. (2004). mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 10, 594–601.
Majumder, P. K., Yeh, J. J., George, D. J., Febbo, P. G., Kum, J., Xue, Q., Bikoff, R., Ma, H., Kantoff, P. W., Golub, T. R., Loda, M., and Sellers, W. R. (2003). Prostate intraepithelial neoplasia induced by prostate restricted Akt activation: the MPAKT model. Proc. Natl. Acad. Sci. U.S.A. 100, 7841–7846.
Malanga, D., Scrima, M., De Marco, C., Fabiani, F., De Rosa, N., De Gisi, S., Malara, N., Savino, R., Rocco, G., Chiappetta, G., Franco, R., Tirino, V., Pirozzi, G., and Viglietto, G. (2008). Activating E17K mutation in the gene encoding the protein kinase AKT1 in a subset of squamous cell carcinoma of the lung. Cell Cycle 7, 665–669.
Martinez-Forero, I., Rouzaut, A., Palazon, A., Dubrot, J., and Melero, I. (2009). Lysine 63 polyubiquitination in immunotherapy and in cancer-promoting inflammation. Clin. Cancer Res. 15, 6751–6757.
Massoumi, R., Chmielarska, K., Hennecke, K., Pfeifer, A., and Fässler, R. (2006). Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125, 665–677.
Mauro, C., Pacifico, F., Lavorgna, A., Mellone, S., Iannetti, A., Acquaviva, R., Formisano, S., Vito, P., and Leonardi, A. (2006). ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J. Biol. Chem. 281, 18482–18488.
McCullough, J., Clague, M. J., and Urbé S. (2004). AMSH is an endosome-associated ubiquitin isopeptidase. J. Cell Biol. 166, 487–492.
Metzig, M., Nickles, D., Falschlehner, C., Lehmann-Koch, J., Straub, B. K., Roth, W., and Boutros, M. (2011). An RNAi screen identifies USP2 as a factor required for TNF-alpha-induced NF-kappaB signaling. Int. J. Cancer 129, 607–618.
Mohamedali, A., Lea, N. C., Feakins, R. M., Raj, K., Mufti, G. J., and Kocher, H. M. (2008). AKT1 (E17K) mutation in pancreatic cancer. Technol. Cancer Res. Treat. 7, 407–408.
Musgrove, E. A., Caldon, C. E., Barraclough, J., Stone, A., and Sutherland, R. L. (2011). Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572.
Nakada, S., Tai, I., Panier, S., Al-Hakim, A., Iemura, S., Juang, Y. C., O’Donnell, L., Kumakubo, A., Munro, M., Sicheri, F., Gingras, A. C., Natsume, T., Suda, T., and Durocher, D. (2010). Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946.
Nakatani, K., Thompson, D. A., Barthel, A., Sakaue, H., Liu, W., Weigel, R. J., and Roth, R. A. (1999). Up-regulation of Akt3 in estrogen receptor-deficient breast cancers and androgen-independent prostate cancer lines. J. Biol. Chem. 274, 21528–21532.
Nicassio, F., Corrado, N., Vissers, J. H., Areces, L. B., Bergink, S., Marteijn, J. A., Geverts, B., Houtsmuller, A. B., Vermeulen, W., Di Fiore, P. P., and Citterio, E. (2007). Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17, 1972–1977.
Ninomiya-Tsuji, J., Kajino, T., Ono, K., Ohtomo, T., Matsumoto, M., Shiina, M., Mihara, M., Tsuchiya, M., and Matsumoto, K. (2003). A resorcylic acid lactone, 5Z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J. Biol. Chem. 278, 18485–18490.
Niu, J., Shi, Y., Iwai, K., and Wu, Z. H. (2011). LUBAC regulates NF-kappaB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. EMBO J. 30, 3741–3753.
Novak, U., Rinaldi, A., Kwee, I., Nandula, S. V., Rancoita, P. M., Compagno, M., Cerri, M., Rossi, D., Murty, V. V., Zucca, E., Gaidano, G., Dalla-Favera, R., Pasqualucci, L., Bhagat, G., and Bertoni, F. (2009). The NF-{kappa}B negative regulator TNFAIP3 (A20) is inactivated by somatic mutations and genomic deletions in marginal zone lymphomas. Blood 113, 4918–4921.
Opferman, J. T., and Green, D. R. (2010). DUB-le trouble for cell survival. Cancer Cell 17, 117–119.
Ordureau, A., Smith, H., Windheim, M., Peggie, M., Carrick, E., Morrice, N., and Cohen, P. (2008). The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochem. J. 409, 43–52.
Park, J. H., Kim, Y. G., McDonald, C., Kanneganti, T. D., Hasegawa, M., Body-Malapel, M., Inohara, N., and Núñez, G. (2007). RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 178, 2380–2386.
Pelengaris, S., Khan, M., and Evan, G. (2002). c-MYC: more than just a matter of life and death. Nat. Rev. Cancer 2, 764–776.
Pereg, Y., Liu, B. Y., O’Rourke, K. M., Sagolla, M., Dey, A., Komuves, L., French, D. M., and Dixit, V. M. (2010). Ubiquitin hydrolase Dub3 promotes oncogenic transformation by stabilizing Cdc25A. Nat. Cell Biol. 12, 400–406.
Perk, J., Iavarone, A., and Benezra, R. (2005). Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5, 603–614.
Popov, N., Wanzel, M., Madiredjo, M., Zhang, D., Beijersbergen, R., Bernards, R., Moll, R., Elledge, S. J., and Eilers, M. (2007). The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774.
Raiborg, C., and Stenmark, H. (2009). The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452.
Razani, B., Zarnegar, B., Ytterberg, A. J., Shiba, T., Dempsey, P. W., Ware, C. F., Loo, J. A., and Cheng, G. (2010). Negative feedback in noncanonical NF-kappaB signaling modulates NIK stability through IKKalpha-mediated phosphorylation. Sci. Signal. 3, ra41.
Reiley, W. W., Jin, W., Lee, A. J., Wright, A., Wu, X., Tewalt, E. F., Leonard, T. O., Norbury, C. C., Fitzpatrick, L., Zhang, M., and Sun, S. C. (2007). Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. J. Exp. Med. 204, 1475–1485.
Reiley, W. W., Zhang, M., Jin, W., Losiewicz, M., Donohue, K. B., Norbury, C. C., and Sun, S. C. (2006). Regulation of T cell development by the deubiquitinating enzyme CYLD. Nat. Immunol. 7, 411–417.
Rong, S. B., Hu, Y., Enyedy, I., Powis, G., Meuillet, E. J., Wu, X., Wang, R., Wang, S., and Kozikowski, A. P. (2001). Molecular modeling studies of the Akt PH domain and its interaction with phosphoinositides. J. Med. Chem. 44, 898–908.
Sasaki, Y., Calado, D. P., Derudder, E., Zhang, B., Shimizu, Y., Mackay, F., Nishikawa, S., Rajewsky, K., and Schmidt-Supprian, M. (2008). NIK overexpression amplifies, whereas ablation of its TRAF3-binding domain replaces BAFF:BAFF-R-mediated survival signals in B cells. Proc. Natl. Acad. Sci. U.S.A. 105, 10883–10888.
Sato, S., Sanjo, H., Takeda, K., Ninomiya-Tsuji, J., Yamamoto, M., Kawai, T., Matsumoto, K., Takeuchi, O., and Akira, S. (2005). Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 6, 1087–1095.
Sato, Y., Yoshikawa, A., Yamagata, A., Mimura, H., Yamashita, M., Ookata, K., Nureki, O., Iwai, K., Komada, M., and Fukai, S. (2008). Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455, 358–362.
Schwickart, M., Huang, X., Lill, J. R., Liu, J., Ferrando, R., French, D. M., Maecker, H., O’Rourke, K., Bazan, F., Eastham-Anderson, J., Yue, P., Dornan, D., Huang, D. C., and Dixit, V. M. (2010). Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107.
Shambharkar, P. B., Blonska, M., Pappu, B. P., Li, H., You, Y., Sakurai, H., Darnay, B. G., Hara, H., Penninger, J., and Lin, X. (2007). Phosphorylation and ubiquitination of the IkappaB kinase complex by two distinct signaling pathways. EMBO J. 26, 1794–1805.
Shan, J., Zhao, W., and Gu, W. (2009). Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 36, 469–476.
Shao, G., Lilli, D. R., Patterson-Fortin, J., Coleman, K. A., Morrissey, D. E., and Greenberg, R. A. (2009). The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl. Acad. Sci. U. S. A. 106, 3166–3171.
Shembade, N., Ma, A., and Harhaj, E. W. (2010). Inhibition of NF-kappaB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139.
Shembade, N., Parvatiyar, K., Harhaj, N. S., and Harhaj, E. W. (2009). The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-kappaB signalling. EMBO J. 28, 513–522.
Shim, J. H., Xiao, C., Paschal, A. E., Bailey, S. T., Rao, P., Hayden, M. S., Lee, K. Y., Bussey, C., Steckel, M., Tanaka, N., Yamada, G., Akira, S., Matsumoto, K., and Ghosh, S. (2005). TAK1, but not TAB1 or TAB2, plays an essential role in multiple signaling pathways in vivo. Genes Dev. 19, 2668–2681.
Shoji, K., Oda, K., Nakagawa, S., Hosokawa, S., Nagae, G., Uehara, Y., Sone, K., Miyamoto, Y., Hiraike, H., Hiraike-Wada, O., Nei, T., Kawana, K., Kuramoto, H., Aburatani, H., Yano, T., and Taketani, Y. (2009). The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br. J. Cancer 101, 145–148.
Silverman, N., Zhou, R., Erlich, R. L., Hunter, M., Bernstein, E., Schneider, D., and Maniatis, T. (2003). Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J. Biol. Chem. 278, 48928–48934.
Skaug, B., Chen, J., Du, F., He, J., Ma, A., and Chen, Z. J. (2011). Direct, noncatalytic mechanism of IKK inhibition by A20. Mol. Cell 44, 559–571.
Skaug, B., Jiang, X., and Chen, Z. J. (2009). The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 78, 769–796.
Sorrentino, A., Thakur, N., Grimsby, S., Marcusson, A., von Bulow, V., Schuster, N., Zhang, S., Heldin, C. H., and Landström, M. (2008). The type I TGF-beta receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207.
Soucek, L., Whitfield, J., Martins, C. P., Finch, A. J., Murphy, D. J., Sodir, N. M., Karnezis, A. N., Swigart, L. B., Nasi, S., and Evan, G. I. (2008). Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683.
Staal, S. P. (1987). Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc. Natl. Acad. Sci. U.S.A. 84, 5034–5037.
Stahl, J. M., Sharma, A., Cheung, M., Zimmerman, M., Cheng, J. Q., Bosenberg, M. W., Kester, M., Sandirasegarane, L., and Robertson, G. P. (2004). Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res. 64, 7002–7010.
Starczynowski, D. T., Kuchenbauer, F., Argiropoulos, B., Sung, S., Morin, R., Muranyi, A., Hirst, M., Hogge, D., Marra, M., Wells, R. A., Buckstein, R., Lam, W., Humphries, R. K., and Karsan, A. (2010). Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 16, 49–58.
Suizu, F., Hiramuki, Y., Okumura, F., Matsuda, M., Okumura, A. J., Hirata, N., Narita, M., Kohno, T., Yokota, J., Bohgaki, M., Obuse, C., Hatakeyama, S., Obata, T., and Noguchi, M. (2009). The E3 ligase TTC3 facilitates ubiquitination and degradation of phosphorylated Akt. Dev. Cell 17, 800–810.
Sun, L., Deng, L., Ea, C. K., Xia, Z. P., and Chen, Z. J. (2004). The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14, 289–301.
Sun, S. C. (2008). Deubiquitylation and regulation of the immune response. Nat. Rev. Immunol. 8, 501–511.
Sun, S. C. (2010). CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 17, 25–34.
Takaesu, G., Surabhi, R. M., Park, K. J., Ninomiya-Tsuji, J., Matsumoto, K., and Gaynor, R. B. (2003). TAK1 is critical for IkappaB kinase-mediated activation of the NF-kappaB pathway. J. Mol. Biol. 326, 105–115.
Teitell, M. A. (2005). The TCL1 family of oncoproteins: co-activators of transformation. Nat. Rev. Cancer 5, 640–648.
Vallabhapurapu, S., Matsuzawa, A., Zhang, W., Tseng, P. H., Keats, J. J., Wang, H., Vignali, D. A., Bergsagel, P. L., and Karin, M. (2008). Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat. Immunol. 9, 1364–1370.
Varfolomeev, E., Goncharov, T., Fedorova, A. V., Dynek, J. N., Zobel, K., Deshayes, K., Fairbrother, W. J., and Vucic, D. (2008). c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J. Biol. Chem. 283, 24295–24299.
Varnai, P., Bondeva, T., Tamás, P., Tóth, B., Buday, L., Hunyady, L., and Balla, T. (2005). Selective cellular effects of overexpressed pleckstrin-homology domains that recognize PtdIns(3,4,5)P3 suggest their interaction with protein binding partners. J. Cell. Sci. 118(Pt 20), 4879–4888.
Vidal, S., Khush, R. S., Leulier, F., Tzou, P., Nakamura, M., and Lemaitre, B. (2001). Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 15, 1900–1912.
Virdee, S., Ye, Y., Nguyen, D. P., Komander, D., and Chin, J. W. (2010). Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757.
Wan, Y. Y., Chi, H., Xie, M., Schneider, M. D., and Flavell, R. A. (2006). The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat. Immunol. 7, 851–858.
Wang, C., Deng, L., Hong, M., Akkaraju, G. R., Inoue, J., and Chen, Z. J. (2001). TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351.
Weissman, A. M., Shabek, N., and Ciechanover, A. (2011). The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620.
Wertz, I. E., O’Rourke, K. M., Zhou, H., Eby, M., Aravind, L., Seshagiri, S., Wu, P., Wiesmann, C., Baker, R., Boone, D. L., Ma, A., Koonin, E. V., and Dixit, V. M. (2004). De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699.
Williams, S. A., Maecker, H. L., French, D. M., Liu, J., Gregg, A., Silverstein, L. B., Cao, T. C., Carano, R. A., and Dixit, V. M. (2011). USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 146, 918–930.
Wiltshire, T. D., Lovejoy, C. A., Wang, T., Xia, F., O’Connor, M. J., and Cortez, D. (2010). Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 285, 14565–14571.
Winborn, B. J., Travis, S. M., Todi, S. V., Scaglione, K. M., Xu, P., Williams, A. J., Cohen, R. E., Peng, J., and Paulson, H. L. (2008). The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443.
Wu, H., Tschopp, J., and Lin, S. C. (2007). Smac mimetics and TNFalpha: a dangerous liaison? Cell 131, 655–658.
Wu, Z. H., Wong, E. T., Shi, Y., Niu, J., Chen, Z., Miyamoto, S., and Tergaonkar, V. (2010). ATM- and NEMO-dependent ELKS ubiquitination coordinates TAK1-mediated IKK activation in response to genotoxic stress. Mol. Cell 40, 75–86.
Xia, Z. P., Sun, L., Chen, X., Pineda, G., Jiang, X., Adhikari, A., Zeng, W., and Chen, Z. J. (2009). Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119.
Xiang, T., Ohashi, A., Huang, Y., Pandita, T. K., Ludwig, T., Powell, S. N., and Yang, Q. (2008). Negative regulation of AKT activation by BRCA1. Cancer Res. 68, 10040–10044.
Xu, K., Shimelis, H., Linn, D. E., Jiang, R., Yang, X., Sun, F., Guo, Z., Chen, H., Li, W., Chen, H., Kong, X., Melamed, J., Fang, S., Xiao, Z., Veenstra, T. D., and Qiu, Y. (2009). Regulation of androgen receptor transcriptional activity and specificity by RNF6-induced ubiquitination. Cancer Cell 15, 270–282.
Yamashita, M., Fatyol, K., Jin, C., Wang, X., Liu, Z., and Zhang, Y. E. (2008). TRAF6 mediates Smad-independent activation of JNK and p38 by TGF-beta. Mol. Cell 31, 918–924.
Yamazaki, K., Gohda, J., Kanayama, A., Miyamoto, Y., Sakurai, H., Yamamoto, M., Akira, S., Hayashi, H., Su, B., and Inoue, J. (2009). Two mechanistically and temporally distinct NF-kappaB activation pathways in IL-1 signaling. Sci. Signal. 2, ra66.
Yang, W. L., Wang, J., Chan, C. H., Lee, S. W., Campos, A. D., Lamothe, B., Hur, L., Grabiner, B. C., Lin, X., Darnay, B. G., and Lin, H. K. (2009). The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138.
Yang, W. L., Wu, C. Y., Wu, J., and Lin, H. K. (2010a). Regulation of Akt signaling activation by ubiquitination. Cell Cycle 9, 487–497.
Yang, W. L., Zhang, X., and Lin, H. K. (2010b). Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene 29, 4493–4503.
Yeh, W. C., Shahinian, A., Speiser, D., Kraunus, J., Billia, F., Wakeham, A., de la Pompa, J. L., Ferrick, D., Hum, B., Iscove, N., Ohashi, P., Rothe, M., Goeddel, D. V., and Mak, T. W. (1997). Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity 7, 715–725.
Yu, Q., Geng, Y., and Sicinski, P. (2001). Specific protection against breast cancers by cyclin D1 ablation. Nature 411, 1017–1021.
Yuan, J., Luo, K., Zhang, L., Cheville, J. C., and Lou, Z. (2010). USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 140, 384–396.
Zarnegar, B., Yamazaki, S., He, J. Q., and Cheng, G. (2008a). Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 3503–3508.
Zarnegar, B. J., Wang, Y., Mahoney, D. J., Dempsey, P. W., Cheung, H. H., He, J., Shiba, T., Yang, X., Yeh, W. C., Mak, T. W., Korneluk, R. G., and Cheng, G. (2008b). Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 9, 1371–1378.
Zhang, J., Stirling, B., Temmerman, S. T., Ma, C. A., Fuss, I. J., Derry, J. M., and Jain, A. (2006). Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. J. Clin. Invest. 116, 3042–3049.
Zhang, L., Blackwell, K., Thomas, G. S., Sun, S., Yeh, W. C., and Habelhah, H. (2009). TRAF2 suppresses basal IKK activity in resting cells and TNFalpha can activate IKK in TRAF2 and TRAF5 double knockout cells. J. Mol. Biol. 389, 495–510.
Zhang, L., Ding, X., Cui, J., Xu, H., Chen, J., Gong, Y. N., Hu, L., Zhou, Y., Ge, J., Lu, Q., Liu, L., Chen, S., and Shao, F. (2012). Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature 481, 204–208.
Zhong, Q., Gao, W., Du, F., and Wang, X. (2005). Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 121, 1085–1095.
Keywords: protein kinase, ubiquitination, phosphorylation, Akt, TRAF6, NF-κB, tumorigenesis
Citation: Wang G, Gao Y, Li L, Jin G, Cai Z, Chao J-I and Lin H-K (2012) K63-linked ubiquitination in kinase activation and cancer. Front. Oncol. 2:5. doi: 10.3389/fonc.2012.00005
Received: 15 December 2011;
Accepted: 10 January 2012;
Published online: 31 January 2012.
Edited by:
Wenyi Wei, Beth Israel Deaconess Medical Center, Harvard Medical School, USAReviewed by:
Paolo Pinton, University of Ferrara, ItalyCopyright: © 2012 Wang, Gao, Li, Jin, Cai, Chao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Hui-Kuan Lin, Department of Molecular and Cellular Oncology, The University of Texas M. D. Anderson Cancer Center, Houston, TX 77030, USA. e-mail:aGtsaW5AbWRhbmRlcnNvbi5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.