- 1Department of Basic Medicine, Zhengzhou Shuqing Medical College, Zhengzhou, Henan, China
- 2Department of Biochemistry and Molecular Biology, Medical School, Henan University, Kaifeng, Henan, China
Post-translational modifications play crucial roles in regulating protein functions and stabilities. PTEN is a critical tumor suppressor involved in regulating cellular proliferation, survival, and migration processes. However, dysregulation of PTEN is common in various human cancers. PTEN stability and activation/suppression have been extensively studied in the context of tumorigenesis through inhibition of the PI3K/AKT signaling pathway. PTEN undergoes various post-translational modifications, primarily including phosphorylation, acetylation, ubiquitination, SUMOylation, neddylation, and oxidation, which finely tune its activity and stability. Generally, phosphorylation modulates PTEN activity through its lipid phosphatase function, leading to altered power of the signaling pathways. Acetylation influences PTEN protein stability and degradation rate. SUMOylation has been implicated in PTEN localization and interactions with other proteins, affecting its overall function. Neddylation, as a novel modification of PTEN, is a key regulatory mechanism in the loss of tumor suppressor function of PTEN. Although current therapeutic approaches focus primarily on inhibiting PI3 kinase, understanding the post-translational modifications of PTEN could help provide new therapeutic strategies that can restore PTEN’s role in PIP3-dependent tumors. The present review summarizes the major recent developments in the regulation of PTEN protein level and activity. We expect that these insights will contribute to better understanding of this critical tumor suppressor and its potential implications for cancer therapy in the future.
Introduction
Post-translational modifications (PTMs) such as phosphorylation, acetylation, ubiquitination, SUMOylation (small ubiquitin-like modifier), neddylation, and oxidation are key regulatory mechanisms in the fine control of protein functions, stabilities, and cell localizations [1]. The phosphatase and tensin-homolog in chromosome 10 (PTEN) is a crucial tumor suppressor protein that negatively regulates the phosphoinositide 3-kinase (PI3K) signaling pathway. Upon activation of PI3K, PTEN converts phosphatidylinositol (3,4,5)-triphosphate (PIP3) to phosphatidylinositol (4,5)-biphosphate (PIP2) and attenuates the downstream AKT activity. Dysregulation of PTEN could lead to overactivation of PI3K/AKT signaling and promotion of tumorigenesis [2]. The activity and stability of PTEN are regulated by various PTMs. Deletion of PTEN activity is also involved in tumorigenesis and progression in many types of cancers [3].
Phosphorylation is one of the most extensively studied PTMs that modulates PTEN activity. Several phosphorylation sites have been identified on PTEN, including serine, threonine, and tyrosine residues. Phosphorylation can either activate or inhibit PTEN functions depending on the specific site and context.
Acetylation is another PTM that influences PTEN protein stability, activity, or subcellular location. Acetylation of specific lysine residues on PTEN have been shown to enhance its stability and tumor-suppressive functions. Protein acetylation is regulated by lysine acetyltransferase and deacetylase. PTEN is associated with acetyltransferase in a growth-factor-dependent manner; this association leads to acetylation of the PTEN catalytic domains K125 and K128, resulting in functional inactivation of PTEN. Understanding the regulatory mechanisms of PTEN acetylation is therefore critical for developing effective cancer therapies [4, 5].
The ubiquitination of PTEN is influenced by multiple factors, including its phosphorylation status and interactions with specific regulatory proteins. Ubiquitination and proteasomal degradation are critical processes involved in PTEN stability and/or translocation. PTEN can be targeted by ubiquitin ligases, which attach ubiquitin molecules to PTEN, marking it for proteasomal degradation [6]. Dysregulation of PTEN ubiquitination disrupts its tumor-suppressive function. When PTEN is monoubiquitinated on the conservative residues K13 and K289, it can localize in the nucleus more easily and retain its tumor suppressor activity. However, when it is polyubiquitinated, it is targeted for degradation in the cytosol, leading to loss of its tumor suppressor activity [7]. Interestingly, the conformational state of PTEN also impacts its susceptibility to ubiquitination, as the open and unphosphorylated conformation is more easily ubiquitinated and degraded [8].
SUMOylation is another covalent modification that attaches small ubiquitin-like modifier proteins to the PTEN proteins. SUMOylation influences PTEN stability, cellular localization, and protein–protein interactions [9]. The process of SUMOylation necessitates the involvement of an E1-activating enzyme, an E2-conjugating enzyme, and an E3 SUMO ligase, although it can occur even in the absence of the latter [10]. SUMOylation of PTEN has been shown to promote its nuclear localization and enhance its interactions with specific transcription factors, leading to altered gene expression patterns [11]. The process by which neural-precursor-cell-expressed developmentally downregulated 8 (NEDD8) is specifically covalently attached to the substrate protein is called neddylation, and PTEN is a new substrate for neddylation modification [12]. PTEN contains cysteine residues in the active sites, which can be regulated in response to oxidative stress.
Loss of PTEN function leads to upregulation of PI3K signaling, which is widely regarded as one of the most common factors in cancer development. In addition, PTEN is involved in the tolerances of cancer cells to chemotherapy drugs and tumor metastasis, together with various other conditions [13]. Recent studies have suggested that PTEN protein levels, activities, and stabilities are tightly regulated by a broad array of PTMs; disruption of these modifications can perturb PTEN functions and contribute to the development and progression of cancers [14]. Further investigation is therefore required to elucidate the precise molecular mechanisms underlying these modifications to develop potential therapeutic strategies for PTEN-related cancers.
Diversity Modifications in the PTEN Protein
PTEN undergoes various PTMs, including phosphorylation, acetylation, ubiquitination, SUMOylation, neddylation, and oxidation. These modifications dynamically regulate PTEN’s subcellular localization, stability, enzymatic activity, and protein–protein interactions [1]. Protein modifications in PTEN can fine-tune its signaling pathways and impact diverse cellular processes (Figure 1).
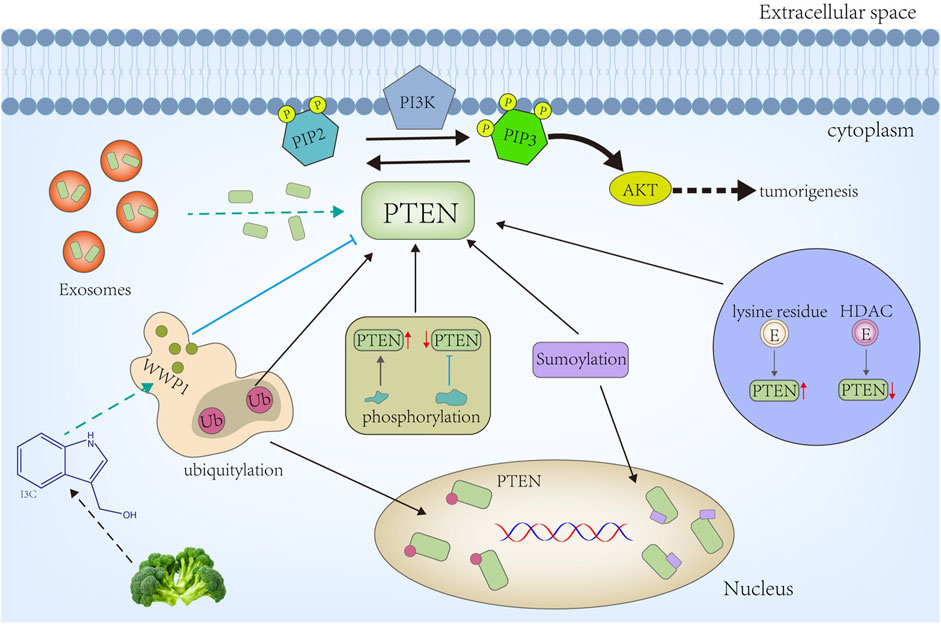
Figure 1. PTEN is recruited into the membrane from the cytoplasm, where it primarily dephosphorylates PIP3 (IP3) to PIP2 and represses the IP3→AKT signaling activity to facilitate tumorigenesis. Diverse modifications occur on the PTEN protein layer, from altering the protein amount to its subcellular location and activity. Among these, phosphorylation and acetylation can have different effects on PTEN activity depending on the modified residues. Ubiquitylation and SUMOylation can alter PTEN’s subcellular location or degradation. Conversely, deubiquitylation stabilizes the PTEN protein. Furthermore, recent studies have revealed that exosomes can transport PTEN proteins directly to other cells, including PTEN-deficient cells.
Phosphorylation
Phosphorylation plays a key role in the regulation of PTEN, and such phosphorylation at multiple residues regulates the activity, localization, stability, and protein interactions [15]. One of the most extensively studied phosphorylation sites in PTEN is serine 380 (Ser380). Glycogen synthetase kinase 3β mediated phosphorylation of PTEN at Ser380 promotes PTEN association with the plasma membrane, resulting in increased dephosphorylation of PIP3 and activation of PI3K/AKT signaling [16]. Conversely, Sema4D promotes PTEN dephosphorylation at Ser380 in the hippocampus and enhances PTEN phosphatase activity [17]. In addition to Ser380, other phosphorylation sites in PTEN, such as T366, T240, and Ser229, have been identified and implicated in modulating its activities and interactions with other proteins [18]. The fibroblast growth factor receptor 2 mediated phosphorylation of PTEN at T240 (pY240-PTEN) protects cells from DNA damage by promoting DNA repair. The nuclear localization and chromatin binding of pY240-PTEN has been shown to increase after treatment with ionizing radiation and promote DNA repair, indicating that pY240-PTEN downregulates PTEN antitumor activity [19].
PTEN protein phosphatase activity can dephosphorylate phosphoglycerate kinase 1 and inhibit its phosphorylation, thereby inhibiting glycolysis [20]. Cellular exposure to recombinant transcription factors or protease activated receptor 2 agonist peptide (PAR2-AP) can activate PTEN. PAR2 activation releases PTEN from the membrane-associated guanylate kinase with inverted configuration (MAGI complex) [21]. At the protein level, melanin II increases PTEN through the melanocortin 1 receptor, mediating reduction of PTEN phosphorylation and inducing PTEN expression upregulation. As a result, upregulation of PTEN inhibits the AKT/nuclear factor κ B signaling pathway, repressing migration, invasion, and colony-forming capability in the melanoma cells [22]. Phosphorylation regulation of PTEN is a complex process that impacts its tumor suppressor functions and cellular processes associated with growth, survival, and proliferation. The balance between phosphorylation and dephosphorylation events at specific residues determines PTEN’s activity and its crucial role in maintaining cellular homeostasis.
Acetylation
Acetylation is a common PTM that regulates PTEN; it occurs at specific lysine residues and influences the activity, stability, and cellular localization of PTEN [23]. One of the extensively studied acetylation sites in PTEN is K125. PTEN interacts with the acetyltransferase p300/CBP-related factor (PCAF) in a manner dependent on growth factors. The acetylation of K125 by PCAF has been demonstrated to augment both PTEN stability and phosphatase activity [5].
Additionally, some histone deacetylase (HDAC) inhibitors have been shown to enhance the translocation of PTEN membrane or promote the activation and expression of PTEN. The translocation of the PTEN membrane is enhanced by trichostatin A or octylaniline hydroxamic acid. PTEN’s C-tail interacts with another part of the protein, blocking its membrane translocation and activation. Acetylation of PTEN at K163 induces conformational changes, facilitating activation and membrane translocation [24]. In addition, valproic acid inhibitor can enhance PTEN expression and block the AKT/mTOR pathway [25]. A negative correlation also exists between the transfer-related protein 1 (MTA1) and PTEN, where MTA1 knockdown could enhance the acetylation of PTEN. Resveratrol, a dietary compound found in grapes and wine, promotes acetylation and reactivation of PTEN by inhibiting the MTA1/HDAC complex [26].
Ubiquitination
Ubiquitination plays a crucial role in the regulation of PTEN; when PTEN is subjected to ubiquitination by several E3 ubiquitin ligases, such as NEDD4-1, WW-domain-containing E3 ubiquitin protein ligase 2 (WWP2), BRCA1-associated protein 1 (BAP1), ovarian tumor protease deubiquitination enzyme 5 (OTUD5), and ovarian-tumor-domain-containing protein 3 (OTUD3). Ubiquitination of PTEN can have dual effects on its stability and activity. Inhibiting PTEN ubiquitination mediated by E3 ligases through drugs or gene targeting can effectively unleash PTEN’s anticancer activity and inhibit tumorigenesis [27].
NEDD4-1 was the first identified PTEN E3 ubiquitin ligase; it recognizes specific motifs in PTEN and catalyzes the addition of ubiquitin chains to PTEN, marking it for proteasomal degradation [28]. WWP2-mediated ubiquitination of PTEN produces different outcomes [29]; instead of promoting degradation, WWP2-mediated ubiquitination has been reported to stabilize PTEN [30]. Studies have shown that deubiquitination is related to the PTEN protein level. The BAP1 deubiquitinating enzyme plays a crucial role in deubiquitination of PTEN; overexpression of BAP1 can upregulate PTEN protein levels. BAP1 can physically combine with and deubiquitinate PTEN, thereby inhibiting the ubiquitin-mediated degradation of PTEN and stabilizing the PTEN protein [31].
USP10 significantly increases the phosphatase activity of PTEN via deubiquitination and stabilization of PTEN [32]. Differentially, Gao et al. found that once USP25 was knocked down, the cell proliferation increased, apoptosis decreased, and PI3K/AKT signaling pathway activated by the PTEN protein level decreased [33].
OTUD5 can deubiquitinate and stabilize PTEN. However, miR-652-3p can target and repress OTUD5 expression, thereby inhibiting its deubiquitination effect on PTEN and downregulating PTEN protein levels [34]. Moreover, OTUD3 depletion results in cell transformation, cancer metastasis induction, and AKT signaling pathway activation. Interestingly, OTUD3 transgenic mice showed elevated levels of PTEN and were less prone to developing tumors. Thus, the OTUD3-PTEN axis could be a promising target for cancer prevention and treatment [35, 36].
The ubiquitination and protein levels of PTEN are influenced by the isoleucine levels in the cytoplasm. Isoleucine promotes PTEN entry into the nucleus, increases PTEN protein level, and inhibits PTEN ubiquitination to maintain PTEN stability in the nucleus, where PTEN stabilizes the genome. Therefore, isoleucine may achieve antitumor activity by influencing the level of PTEN accumulation in the nucleus instead of transcriptional activation [37].
The balance between ubiquitination and deubiquitination thus determines the stability and activity of PTEN.
Focus on WWP1 and PTEN Activities
As an E3 ubiquitin ligase, WWP1 recognizes and binds to PTEN, facilitating transfer of ubiquitinated PTEN to the proteasome for degradation [38].
The ubiquitination and degradation of PTEN by WWP1 reduces the PTEN protein levels, impairing its tumor suppressor function. WWP1 variants have been shown to cause cancer in many patients without PTEN deficiency. In a recent study, Jiang et al. identified two functional gain variants of WWP1, namely K740N and N745S, which have the ability to activate WWP1 E3 ligase while promoting ubiquitination and degradation of PTEN to activate the PI3K pathway [39]. WWP1 blocked PTEN activity by polyubiquitination, blocking of PTEN dimerization, and membrane translocation. Indole-3-methanol (I3C) derived from cruciferous vegetables was discovered to inhibit WWP1 activity effectively [40]. Similarly, Wang et al. found that the expressions of WWP1 mRNA and proteins increased in thyroid papillary carcinoma cells; knocking down WWP1 could upregulate PTEN expression and inhibit cancer [41]. PTEN membrane localization can be restored by genetic or chemical inhibition of WWP1, and tumor growth can be inhibited in vitro and in vivo in cooperation with PI3K inhibitors [42].
SUMOylation
SUMOylation governs an array of processes including transcriptional regulation, subcellular localization, protein stability, protein–protein interactions, and protein–DNA interactions [43].
Recent research has revealed that PTEN is modified by SUMO1 at the K254 and K266 residues, especially the latter. This modification plays a key role in preventing activation of the PI3K/AKT signaling pathway and anchoring PTEN to the plasma membrane [44]. The conjugation of SUMO to K254 has been suggested to enhance its nuclear localization. This finding highlights the importance of the interactions between PTEN and SUMO, revealing how PTEN localization is controlled at the molecular level [45]. Recently, new discoveries have been reported on the SUMOylation of PTEN, revealing that PTEN’s K266 and K289 are required for its antiviral activity against the vesicular stomatitis virus [46]. In addition, K254 SUMOylation does not affect PTEN’s ability against PI3K but is involved in the DNA damage response [45].
SUMOylation of PTEN modulates its protein–protein interactions. SUMOylation has been shown to enhance the interactions between PTEN and other proteins, such as p53, resulting in synergistic tumor suppressor effects [47]. By activating PTEN SUMO, the protein inhibitor of the activated STAT xα (PIASxα) reduces PTEN ubiquitination to promote PTEN protein stability and upregulate the protein level. However, the mutation of PTEN’s SUMO1 binding site can neutralize the effects of PIASxα on the half-life of the PTEN protein. These findings provide important insights into the regulation of PTEN protein stability and role of PIASxα in this process [48].
Dysregulation of PTEN SUMOylation has been observed in various cancers, leading to altered PTEN stability, subcellular distribution, and functional interactions. Investigating the regulation of PTEN SUMOylation provides insights into the intricate mechanisms controlling PTEN’s tumor suppressor activities and involvement in various cellular processes, particularly in cancer.
Neddylation
NEDD8 is the molecule that is most similar to ubiquitin and can bind to PTEN. Xie et al. showed that under high glucose conditions, neddylation promotes PTEN entry into the nucleus and enhances the PI3K/AKT signaling pathway to promote tumorigenesis; neddylation also controls PTEN subcellular localization when glucose is excessive or lacking [49].
Oxidation
Oxidation of PTEN results in its dissociation from the plasma membrane; this suggests that PTEN oxidation not only affects its activity but also regulates its cellular localization, effectively removing it from the main site of lipid phosphatase activity [50]. Peroxidoredoxin can prevent the oxidation of PTEN under benign oxidative stress through direct interactions, thereby maintaining and promoting the tumor suppressor function of PTEN [51]. In streptozotocin (STZ)-induced diabetes, STZ toxicity is mediated by cytokine and oxidative stress, leading to β cell damage. Under mechanical conditions, these factors inhibit AKT activation by upregulating PTEN expression and p-PTEN levels, thereby inducing β cell apoptosis [52].
Regulation of PTEN via Exosomes
Exosomes are small extracellular vesicles released by various cell types [53]; they have emerged as important regulators of intercellular communications since they can transfer functional biomolecules, including proteins, lipids, and nucleic acids, between cells to influence cellular processes [54]. Recent studies have shown that exosomes can also modulate PTEN activity through different mechanisms.
It has been identified that exosomes regulate PTEN functions through the transfer of miRNAs. For example, miR-152-3p in the mesenchymal stem cell (MSC)-derived exosomes has been shown to upregulate PTEN expressions in the recipient cells, promoting wound healing in diabetic foot ulcers [55]. Other studies have shown that miR-144-5p directly targets PTEN and promotes PTEN expression, leading to apoptosis of vascular endothelial cells in intracranial aneurysms [56]. Interestingly, exosomes from human umbilical cord MSCs were found to contain lncRNA PTENP1, which stabilizes PTEN expression by competitively binding to miR-10a-5p in the U87 cells [57].
In contrast, some miRNAs in the exosomes have been shown to downregulate PTEN expressions in recipient cells, leading to activation of the PI3K/AKT pathway. Some examples of these are miR-18b-5p [58], miR-148-3p [59], miR-21 [60], miR-181b-5p [61], miR-205 [62], and miR-934 [63].
Moreover, exosomes can directly modulate PTEN activity through transfer of the PTEN protein itself. The NEDD4 family interacting protein 1 (Ndfip1) is a molecular regulator of PTEN secretion and impacts non-cellular autonomous PTEN activity. PTEN secretion in the exocrine system requires Ndfip1. K13 in PTEN is required for transport of the PTEN exosomes [64]. Similarly, exosomes secreted by cells with PTEN mutations or loss may contribute to PTEN inactivation in the recipient cells. These exosomes may contain altered miRNA profiles or lack functional PTEN proteins, promoting PTEN deficiencies in the recipient cells and oncogenic signaling thereof.
Protein–Protein Interactions
The activities and subcellular localizations of the PTEN proteins are regulated through interactions with specific proteins [65]. In addition to its interactions with AKT, PTEN can form complexes with other proteins like the discoid homologous region (PDZ-domain) proteins, which help stabilize PTEN and regulate its cellular localization. Scaffold proteins like MAGI-1b, MAGI-2, and MAGI-3 interact with PTEN’s PDZ binding domain and recruit PTEN to the membrane complex, where PTEN dephosphorylates PIP3 [66–68]. A recent study revealed that the β-suppressor scaffold protein is the upstream signal regulator of PTEN; here, PTEN membrane localization and lipid phosphatase activity increase through direct contact [69].
The maintenance of chromosome stability is ensured by the nuclear PTEN through its interactions with the centromere protein-C [70]. The interactions between PTEN and specific partners also affect PTEN’s subcellular localizations. For instance, ubiquitin-conjugating enzymes interact with PTEN and modulate its translocation to the nucleus [71]. Moreover, ataxia telangiectasia mutated gene phosphorylates PTEN and transfers it to the cytoplasm, where it interacts with p53 to regulate cell-cycle progression [72].
Perspectives
PTEN is the second most-mutated tumor suppressor after p53 in cancer and is associated with many human diseases. Understanding and studying the PTMs of PTEN can help in the discovery of new therapeutic strategies to restore PTEN activities and protein levels in PIP3-dependent tumors (Table 1). PIK3CA is the catalytic subunit of the PI3KsIA type; mutations in the PIK3CA gene promote cell carcinogenesis through the PI3K/AKT pathway [73]. G protein signal transduction 12 binds to PTEN through the PDZ domain, upregulating the phosphorylation and SUMOylation of PTEN as well as inhibiting oral cancer development [74]. PTEN deficiency promotes gemcitabine efficacy in cancer by modulating the phosphorylation of protein phosphatase 2A [75]. PTEN inactivation mediated by overexpression of E3 ubiquitin ligase causes glioblastoma cells to adapt to the PTEN deficiency, thereby developing resistance to epidermal growth factor receptor tyrosine kinase inhibitors and immunotherapies [76].
Conclusion
As an important tumor suppressor, PTEN effectively inhibits the activity of the PI3K/AKT carcinogenic signaling pathway through its lipid phosphatase activity. In addition, PTEN achieves inhibitory effects on tumor cells through other mechanisms without depending on the PI3K/AKT pathway. It is well known that PTEN dysregulation is extensively involved in tumor occurrence and development. Therefore, the regulatory mechanisms of PTEN capacity have become the focus in the field of tumor biology research. A comprehensive understanding of the molecular and cellular mechanisms underlying the regulation of PTEN activity and protein level is crucial for clinical practitioners and researchers. In addition, this provides new research directions and therapeutic avenues for the application of PTEN in anticancer therapies.
Author Contributions
XL: Data curation, Formal analysis, Investigation, Methodology, Resources, Writing–original draft. PY: Conceptualization, Data curation, Resources, Validation, Writing–original draft. XH: Data curation, Investigation, Validation, Writing–original draft. SJ: Conceptualization, Formal analysis, Funding acquisition, Supervision, Validation, Writing–review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 31371386 SJ) and Natural Science Foundation of Shaanxi Province (No. 2021JZ-27). The funding agencies were not involved in the study design, collection, analysis, interpretation of data, writing of this article, or decision to submit it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. González-García, A, Garrido, A, and Carrera, AC. Targeting PTEN Regulation by Post Translational Modifications. Cancers (Basel) (2022) 14(22):5613. doi:10.3390/cancers14225613
2. Liu, A, Zhu, Y, Chen, W, Merlino, G, and Yu, Y. PTEN Dual Lipid- and Protein-Phosphatase Function in Tumor Progression. Cancers (Basel) (2022) 14(15):3666. doi:10.3390/cancers14153666
3. Kotelevets, L, Trifault, B, Chastre, E, and Scott, MGH. Posttranslational Regulation and Conformational Plasticity of PTEN. Cold Spring Harb Perspect Med (2020) 10(7):a036095. doi:10.1101/cshperspect.a036095
4. Drazic, A, Myklebust, LM, Ree, R, and Arnesen, T. The World of Protein Acetylation. Biochim Biophys Acta (Bba) - Proteins Proteomics (2016) 1864(10):1372–401. doi:10.1016/j.bbapap.2016.06.007
5. Okumura, K, Mendoza, M, Bachoo, RM, DePinho, RA, Cavenee, WK, and Furnari, FB. PCAF Modulates PTEN Activity. J Biol Chem (2006) 281(36):26562–8. doi:10.1074/jbc.m605391200
6. Xu, W, Yang, Z, Zhou, SF, and Lu, N. Posttranslational Regulation of Phosphatase and Tensin Homolog (PTEN) and its Functional Impact on Cancer Behaviors. Drug Des Develop Ther (2014) 8:1745–51. doi:10.2147/dddt.s71061
7. Trotman, LC, Wang, X, Alimonti, A, Chen, Z, Teruya-Feldstein, J, Yang, H, et al. Ubiquitination Regulates PTEN Nuclear Import and Tumor Suppression. Cell (2007) 128(1):141–56. doi:10.1016/j.cell.2006.11.040
8. Song, MS, Carracedo, A, Salmena, L, Song, SJ, Egia, A, Malumbres, M, et al. Nuclear PTEN Regulates the APC-CDH1 Tumor-Suppressive Complex in a Phosphatase-independent Manner. Cell (2011) 144(2):187–99. doi:10.1016/j.cell.2010.12.020
9. González-Santamaría, J, Campagna, M, Ortega-Molina, A, Marcos-Villar, L, de la Cruz-Herrera, CF, González, D, et al. Regulation of the Tumor Suppressor PTEN by SUMO. Cell Death Dis (2012) 3(9):e393. doi:10.1038/cddis.2012.135
10. Gareau, JR, and Lima, CD. The SUMO Pathway: Emerging Mechanisms that Shape Specificity, Conjugation and Recognition. Nat Rev Mol Cel Biol (2010) 11(12):861–71. doi:10.1038/nrm3011
11. Yeh, ET, Gong, L, and Kamitani, T. Ubiquitin-like Proteins: New Wines in New Bottles. Gene (2000) 248(1-2):1–14. doi:10.1016/s0378-1119(00)00139-6
12. Du, MG, Peng, ZQ, Gai, WB, Liu, F, Liu, W, Chen, YJ, et al. The Absence of PTEN in Breast Cancer Is a Driver of MLN4924 Resistance. Front Cel Dev Biol (2021) 9:667435. doi:10.3389/fcell.2021.667435
13. Zheng, F, Zhong, J, Chen, K, Shi, Y, Wang, F, Wang, S, et al. PINK1-PTEN axis Promotes Metastasis and Chemoresistance in Ovarian Cancer via Non-canonical Pathway. J Exp Clin Cancer Res (2023) 42(1):295. doi:10.1186/s13046-023-02823-w
14. Ngeow, J, and Eng, C. PTEN in Hereditary and Sporadic Cancer. Cold Spring Harb Perspect Med (2020) 10(4):a036087. doi:10.1101/cshperspect.a036087
15. Misra, S, Chowdhury, SG, Ghosh, G, Mukherjee, A, and Karmakar, P. Both Phosphorylation and Phosphatase Activity of PTEN Are Required to Prevent Replication fork Progression during Stress by Inducing Heterochromatin. Mutat Research/Fundamental Mol Mech Mutagenesis (2022) 825:111800. doi:10.1016/j.mrfmmm.2022.111800
16. Lin, J, Song, T, Li, C, and Mao, W. GSK-3β in DNA Repair, Apoptosis, and Resistance of Chemotherapy, Radiotherapy of Cancer. Biochim Biophys Acta (Bba) - Mol Cel Res (2020) 1867(5):118659. doi:10.1016/j.bbamcr.2020.118659
17. Oinuma, I, Ito, Y, Katoh, H, and Negishi, M. Semaphorin 4D/Plexin-B1 Stimulates PTEN Activity through R-Ras GTPase-Activating Protein Activity, Inducing Growth Cone Collapse in Hippocampal Neurons. J Biol Chem (2010) 285(36):28200–9. doi:10.1074/jbc.m110.147546
18. Dempsey, DR, Viennet, T, Iwase, R, Park, E, Henriquez, S, Chen, Z, et al. The Structural Basis of PTEN Regulation by Multi-Site Phosphorylation. Nat Struct Mol Biol (2021) 28(10):858–68. doi:10.1038/s41594-021-00668-5
19. Ma, J, Benitez, JA, Li, J, Miki, S, Ponte de Albuquerque, C, Galatro, T, et al. Inhibition of Nuclear PTEN Tyrosine Phosphorylation Enhances Glioma Radiation Sensitivity through Attenuated DNA Repair. Cancer Cell (2019) 35(3):816–518.e7. doi:10.1016/j.ccell.2019.04.011
20. Qian, X, Li, X, Shi, Z, Xia, Y, Cai, Q, Xu, D, et al. PTEN Suppresses Glycolysis by Dephosphorylating and Inhibiting Autophosphorylated PGK1. Mol Cel (2019) 76(3):516–27.e7. doi:10.1016/j.molcel.2019.08.006
21. Mohammad, MA, Greenman, J, Maraveyas, A, and Ettelaie, C. Activation of PAR2 by Tissue Factor Induces the Release of the PTEN from MAGI Proteins and Regulates PTEN and Akt Activities. Sci Rep (2020) 10(1):20908. doi:10.1038/s41598-020-77963-6
22. Wu, JC, Tsai, HE, Hsiao, YH, Wu, JS, Wu, CS, and Tai, MH. Topical MTII Therapy Suppresses Melanoma through PTEN Upregulation and Cyclooxygenase II Inhibition. Int J Mol Sci (2020) 21(2):681. doi:10.3390/ijms21020681
23. Chen, H, Zhang, Y, Zou, M, Sun, X, Huang, X, and Xu, S. Dibutyl Phthalate-Induced Oxidative Stress and Apoptosis in Swine Testis Cells and Therapy of Naringenin via PTEN/PI3K/AKT Signaling Pathway. Environ Toxicol (2022) 37(8):1840–52. doi:10.1002/tox.23531
24. Meng, Z, Jia, LF, and Gan, YH. PTEN Activation through K163 Acetylation by Inhibiting HDAC6 Contributes to Tumour Inhibition. Oncogene (2016) 35(18):2333–44. doi:10.1038/onc.2015.293
25. Shi, X, Liu, Y, Zhang, D, and Xiao, D. Valproic Acid Attenuates Sepsis-Induced Myocardial Dysfunction in Rats by Accelerating Autophagy through the PTEN/AKT/mTOR Pathway. Life Sci (2019) 232:116613. doi:10.1016/j.lfs.2019.116613
26. Dhar, S, Kumar, A, Li, K, Tzivion, G, and Levenson, AS. Resveratrol Regulates PTEN/Akt Pathway through Inhibition of MTA1/HDAC Unit of the NuRD Complex in Prostate Cancer. Biochim Biophys Acta (Bba) - Mol Cel Res (2015) 1853(2):265–75. doi:10.1016/j.bbamcr.2014.11.004
27. Wang, K, Liu, J, Li, YL, Li, JP, and Zhang, R. Ubiquitination/de-ubiquitination: A Promising Therapeutic Target for PTEN Reactivation in Cancer. Biochim Biophys Acta (Bba) - Rev Cancer (2022) 1877(3):188723. doi:10.1016/j.bbcan.2022.188723
28. Wang, K, Yu, Y, Wang, W, Jiang, Y, Li, Y, Jiang, X, et al. Targeting the E3 Ligase NEDD4 as a Novel Therapeutic Strategy for IGF1 Signal Pathway-Driven Gastric Cancer. Oncogene (2023) 42(14):1072–87. doi:10.1038/s41388-023-02619-4
29. Zhang, R, Zhang, J, Luo, W, Luo, Z, and Shi, S. WWP2 Is One Promising Novel Oncogene. Pathol Oncol Res (2019) 25(2):443–6. doi:10.1007/s12253-018-0506-5
30. Chen, W, Jiang, X, and Luo, Z. WWP2: A Multifunctional Ubiquitin Ligase Gene. Pathol Oncol Res (2014) 20(4):799–803. doi:10.1007/s12253-014-9838-y
31. Deng, R, Guo, Y, Li, L, He, J, Qiang, Z, Zhang, H, et al. BAP1 Suppresses Prostate Cancer Progression by Deubiquitinating and Stabilizing PTEN. Mol Oncol (2021) 15(1):279–98. doi:10.1002/1878-0261.12844
32. He, Y, Jiang, S, Mao, C, Zheng, H, Cao, B, Zhang, Z, et al. The Deubiquitinase USP10 Restores PTEN Activity and Inhibits Non-small Cell Lung Cancer Cell Proliferation. J Biol Chem (2021) 297(3):101088. doi:10.1016/j.jbc.2021.101088
33. Gao, Y, Chen, J, Ji, R, Ding, J, Zhang, Y, and Yang, J. USP25 Regulates the Proliferation and Apoptosis of Ovarian Granulosa Cells in Polycystic Ovary Syndrome by Modulating the PI3K/AKT Pathway via Deubiquitinating PTEN. Front Cel Dev Biol (2021) 9:779718. doi:10.3389/fcell.2021.779718
34. Li, X, Lu, B, Zhang, L, Yang, J, Cheng, Y, and Yan, D. Mechanism of OTUD5 in Non-small Cell Lung Cancer Cell Proliferation, Invasion, and Migration. Bosnian J Basic Med Sci (2022) 22(6):901–11. doi:10.17305/bjbms.2022.7206
35. Yuan, L, Lv, Y, Li, H, Gao, H, Song, S, Zhang, Y, et al. Deubiquitylase OTUD3 Regulates PTEN Stability and Suppresses Tumorigenesis. Nat Cel Biol (2015) 17(9):1169–81. doi:10.1038/ncb3218
36. Liu, YZ, Du, XX, Zhao, QQ, Jiao, Q, and Jiang, H. The Expression Change of OTUD3-PTEN Signaling axis in Glioma Cells. Ann Transl Med (2020) 8(7):490. doi:10.21037/atm.2020.03.51
37. Wang, H, Chen, S, Kang, W, Ding, B, Cui, S, Zhou, L, et al. High Dose Isoleucine Stabilizes Nuclear PTEN to Suppress the Proliferation of Lung Cancer. Discover Oncol (2023) 14(1):25. doi:10.1007/s12672-023-00634-1
38. Lee, YR, Yehia, L, Kishikawa, T, Ni, Y, Leach, B, Zhang, J, et al. WWP1 Gain-Of-Function Inactivation of PTEN in Cancer Predisposition. N Engl J Med (2020) 382(22):2103–16. doi:10.1056/nejmoa1914919
39. Jiang, H, Dempsey, DR, and Cole, PA. Ubiquitin Ligase Activities of WWP1 Germline Variants K740N and N745S. Biochemistry (2021) 60(5):357–64. doi:10.1021/acs.biochem.0c00869
40. Lee, YR, Chen, M, Lee, JD, Zhang, J, Lin, SY, Fu, TM, et al. Reactivation of PTEN Tumor Suppressor for Cancer Treatment through Inhibition of a MYC-WWP1 Inhibitory Pathway. Science (2019) 364(6441):eaau0159. doi:10.1126/science.aau0159
41. Wang, X, Bi, Y, Liu, X, Liu, L, Hao, M, Tian, M, et al. High Expression of WWP1 Associates with Tumor Progression in Papillary Thyroid Cancer. Cancer Biother Radiopharm (2022) 37(4):313–23. doi:10.1089/cbr.2020.4148
42. Kishikawa, T, Higuchi, H, Wang, L, Panch, N, Maymi, V, Best, S, et al. WWP1 Inactivation Enhances Efficacy of PI3K Inhibitors while Suppressing Their Toxicities in Breast Cancer Models. J Clin Invest (2021) 131(24):e140436. doi:10.1172/jci140436
43. Han, ZJ, Feng, YH, Gu, BH, Li, YM, and Chen, H. The post-translational Modification, SUMOylation, and Cancer (Review). Int J Oncol (2018) 52(4):1081–94. doi:10.3892/ijo.2018.4280
44. Huang, J, Yan, J, Zhang, J, Zhu, S, Wang, Y, Shi, T, et al. SUMO1 Modification of PTEN Regulates Tumorigenesis by Controlling its Association with the Plasma Membrane. Nat Commun (2012) 3:911. doi:10.1038/ncomms1919
45. Bassi, C, Ho, J, Srikumar, T, Dowling, RJ, Gorrini, C, Miller, SJ, et al. Nuclear PTEN Controls DNA Repair and Sensitivity to Genotoxic Stress. Science (2013) 341(6144):395–9. doi:10.1126/science.1236188
46. Li, S, Zhu, M, Pan, R, Fang, T, Cao, YY, Chen, S, et al. The Tumor Suppressor PTEN Has a Critical Role in Antiviral Innate Immunity. Nat Immunol (2016) 17(3):241–9. doi:10.1038/ni.3311
47. Rodríguez, JA. Interplay between Nuclear Transport and Ubiquitin/SUMO Modifications in the Regulation of Cancer-Related Proteins. Semin Cancer Biol (2014) 27:11–9. doi:10.1016/j.semcancer.2014.03.005
48. Wang, W, Chen, Y, Wang, S, Hu, N, Cao, Z, Wang, W, et al. PIASxα Ligase Enhances SUMO1 Modification of PTEN Protein as a SUMO E3 Ligase. J Biol Chem (2014) 289(6):3217–30. doi:10.1074/jbc.m113.508515
49. Xie, P, Peng, Z, Chen, Y, Li, H, Du, M, Tan, Y, et al. Neddylation of PTEN Regulates its Nuclear Import and Promotes Tumor Development. Cell Res (2021) 31(3):291–311. doi:10.1038/s41422-020-00443-z
50. Hempel, N, and Melendez, JA. Intracellular Redox Status Controls Membrane Localization of Pro- and Anti-migratory Signaling Molecules. Redox Biol (2014) 2:245–50. doi:10.1016/j.redox.2014.01.005
51. Nguyen Huu, T, Park, J, Zhang, Y, Park, I, Yoon, HJ, Woo, HA, et al. Redox Regulation of PTEN by Peroxiredoxins. Antioxidants (Basel) (2021) 10(2):302. doi:10.3390/antiox10020302
52. Hou, R, Zhang, J, Yin, T, Cao, H, Zhang, N, Li, X, et al. Upregulation of PTEN by Peroxynitrite Contributes to Cytokine-Induced Apoptosis in Pancreatic β-cells. Apoptosis (2010) 15(8):877–86. doi:10.1007/s10495-010-0510-z
53. Kimiz-Gebologlu, I, and Oncel, SS. Exosomes: Large-Scale Production, Isolation, Drug Loading Efficiency, and Biodistribution and Uptake. J Controlled Release (2022) 347:533–43. doi:10.1016/j.jconrel.2022.05.027
54. Liang, Y, Duan, L, Lu, J, and Xia, J. Engineering Exosomes for Targeted Drug Delivery. Theranostics (2021) 11(7):3183–95. doi:10.7150/thno.52570
55. Li, B, Luan, S, Chen, J, Zhou, Y, Wang, T, Li, Z, et al. The MSC-Derived Exosomal lncRNA H19 Promotes Wound Healing in Diabetic Foot Ulcers by Upregulating PTEN via MicroRNA-152-3p. Mol Ther - Nucleic Acids (2020) 19:814–26. doi:10.1016/j.omtn.2019.11.034
56. Yang, G, Qin, H, Liu, B, Zhao, X, and Yin, H. Mesenchymal Stem Cells-Derived Exosomes Modulate Vascular Endothelial Injury via miR-144-5p/PTEN in Intracranial Aneurysm. Hum Cel (2021) 34(5):1346–59. doi:10.1007/s13577-021-00571-7
57. Hao, SC, Ma, H, Niu, ZF, Sun, SY, Zou, YR, and Xia, HC. hUC-MSCs Secreted Exosomes Inhibit the Glioma Cell Progression through PTENP1/miR-10a-5p/PTEN Pathway. Eur Rev Med Pharmacol Sci (2019) 23(22):10013–23. doi:10.26355/eurrev_201911_19568
58. Zhou, Z, Tu, Z, Zhang, J, Tan, C, Shen, X, Wan, B, et al. Follicular Fluid-Derived Exosomal MicroRNA-18b-5p Regulates PTEN-Mediated PI3K/Akt/mTOR Signaling Pathway to Inhibit Polycystic Ovary Syndrome Development. Mol Neurobiol (2022) 59(4):2520–31. doi:10.1007/s12035-021-02714-1
59. Shan, G, Zhou, X, Gu, J, Zhou, D, Cheng, W, Wu, H, et al. Downregulated Exosomal microRNA-148b-3p in Cancer Associated Fibroblasts Enhance Chemosensitivity of Bladder Cancer Cells by Downregulating the Wnt/β-Catenin Pathway and Upregulating PTEN. Cel Oncol (Dordr) (2021) 44(1):45–59. doi:10.1007/s13402-020-00500-0
60. Zhao, S, Li, W, Yu, W, Rao, T, Li, H, Ruan, Y, et al. Exosomal miR-21 from Tubular Cells Contributes to Renal Fibrosis by Activating Fibroblasts via Targeting PTEN in Obstructed Kidneys. Theranostics (2021) 11(18):8660–73. doi:10.7150/thno.62820
61. Lv, PY, Gao, PF, Tian, GJ, Yang, YY, Mo, FF, Wang, ZH, et al. Osteocyte-derived Exosomes Induced by Mechanical Strain Promote Human Periodontal Ligament Stem Cell Proliferation and Osteogenic Differentiation via the miR-181b-5p/PTEN/AKT Signaling Pathway. Stem Cel Res Ther (2020) 11(1):295. doi:10.1186/s13287-020-01815-3
62. He, L, Zhu, W, Chen, Q, Yuan, Y, Wang, Y, Wang, J, et al. Ovarian Cancer Cell-Secreted Exosomal miR-205 Promotes Metastasis by Inducing Angiogenesis. Theranostics (2019) 9(26):8206–20. doi:10.7150/thno.37455
63. Zhao, S, Mi, Y, Guan, B, Zheng, B, Wei, P, Gu, Y, et al. Tumor-derived Exosomal miR-934 Induces Macrophage M2 Polarization to Promote Liver Metastasis of Colorectal Cancer. J Hematol Oncol (2020) 13(1):156. doi:10.1186/s13045-020-00991-2
64. Putz, U, Howitt, J, Doan, A, Goh, CP, Low, LH, Silke, J, et al. The Tumor Suppressor PTEN Is Exported in Exosomes and Has Phosphatase Activity in Recipient Cells. Sci Signal (2012) 5(243):ra70. doi:10.1126/scisignal.2003084
65. Malaney, P, Uversky, VN, and Davé, V. PTEN Proteoforms in Biology and Disease. Cell Mol Life Sci (2017) 74(15):2783–94. doi:10.1007/s00018-017-2500-6
66. Kotelevets, L, van Hengel, J, Bruyneel, E, Mareel, M, van Roy, F, and Chastre, E. Implication of the MAGI-1b/PTEN Signalosome in Stabilization of Adherens Junctions and Suppression of Invasiveness. Faseb j (2005) 19(1):115–7. doi:10.1096/fj.04-1942fje
67. Subauste, MC, Nalbant, P, Adamson, ED, and Hahn, KM. Vinculin Controls PTEN Protein Level by Maintaining the Interaction of the Adherens Junction Protein β-Catenin with the Scaffolding Protein MAGI-2. J Biol Chem (2005) 280(7):5676–81. doi:10.1074/jbc.m405561200
68. Wu, Y, Dowbenko, D, Spencer, S, Laura, R, Lee, J, Gu, Q, et al. Interaction of the Tumor Suppressor PTEN/MMAC with a PDZ Domain of MAGI3, a Novel Membrane-Associated Guanylate Kinase. J Biol Chem (2000) 275(28):21477–85. doi:10.1074/jbc.m909741199
69. Lima-Fernandes, E, Enslen, H, Camand, E, Kotelevets, L, Boularan, C, Achour, L, et al. Distinct Functional Outputs of PTEN Signalling Are Controlled by Dynamic Association with β-arrestins: β-Arrestins Control PTEN Function. EMBO J (2011) 30(13):2557–68. doi:10.1038/emboj.2011.178
70. Shen, WH, Balajee, AS, Wang, J, Wu, H, Eng, C, Pandolfi, PP, et al. Essential Role for Nuclear PTEN in Maintaining Chromosomal Integrity. Cell (2007) 128(1):157–70. doi:10.1016/j.cell.2006.11.042
71. Norris, KL, Hao, R, Chen, LF, Lai, CH, Kapur, M, Shaughnessy, PJ, et al. Convergence of Parkin, PINK1, and α-Synuclein on Stress-Induced Mitochondrial Morphological Remodeling. J Biol Chem (2015) 290(22):13862–74. doi:10.1074/jbc.m114.634063
72. Chen, JH, Zhang, P, Chen, WD, Li, DD, Wu, XQ, Deng, R, et al. ATM-Mediated PTEN Phosphorylation Promotes PTEN Nuclear Translocation and Autophagy in Response to DNA-Damaging Agents in Cancer Cells. Autophagy (2015) 11(2):239–52. doi:10.1080/15548627.2015.1009767
73. Choo, F, Odintsov, I, Nusser, K, Nicholson, KS, Davis, L, Corless, CL, et al. Functional Impact and Targetability of PI3KCA, GNAS, and PTEN Mutations in a Spindle Cell Rhabdomyosarcoma with MYOD1 L122R Mutation. Cold Spring Harb Mol Case Stud (2022) 8(1):a006140. doi:10.1101/mcs.a006140
74. Fu, C, Yuan, G, Yang, ST, Zhang, D, and Yang, S. RGS12 Represses Oral Cancer via the Phosphorylation and SUMOylation of PTEN. J Dent Res (2021) 100(5):522–31. doi:10.1177/0022034520972095
75. Jiang, TY, Cui, XW, Zeng, TM, Pan, YF, Lin, YK, Feng, XF, et al. PTEN Deficiency Facilitates Gemcitabine Efficacy in Cancer by Modulating the Phosphorylation of PP2Ac and DCK. Sci Transl Med (2023) 15(704):eadd7464. doi:10.1126/scitranslmed.add7464
Keywords: PTEN, phosphorylation, acetylation, ubiquitination, SUMOylation
Citation: Li X, Yang P, Hou X and Ji S (2024) Post-Translational Modification of PTEN Protein: Quantity and Activity. Oncol. Rev. 18:1430237. doi: 10.3389/or.2024.1430237
Received: 09 May 2024; Accepted: 04 July 2024;
Published: 31 July 2024.
Edited by:
Deepa Kushwaha, Rare Genomics Institute, United StatesReviewed by:
Constantinos Savva, University of Southampton, United KingdomDanfeng Guo, First Affiliated Hospital of Zhengzhou University, China
Copyright © 2024 Li, Yang, Hou and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoli Hou, aG91eGlhb2xpMjAwNEAxMjYuY29t; Shaoping Ji, c2hhb3BpbmdqaUBoZW51LmVkdS5jbg==
 Xiao Li
Xiao Li Pu Yang1
Pu Yang1 Shaoping Ji
Shaoping Ji