
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Neurosci., 14 June 2024
Sec. Brain Imaging Methods
Volume 18 - 2024 | https://doi.org/10.3389/fnins.2024.1401329
This article is part of the Research TopicNew Developments in Artificial Intelligence for Stroke Image AnalysisView all 5 articles
Introduction: Brain medical image segmentation is a critical task in medical image processing, playing a significant role in the prediction and diagnosis of diseases such as stroke, Alzheimer's disease, and brain tumors. However, substantial distribution discrepancies among datasets from different sources arise due to the large inter-site discrepancy among different scanners, imaging protocols, and populations. This leads to cross-domain problems in practical applications. In recent years, numerous studies have been conducted to address the cross-domain problem in brain image segmentation.
Methods: This review adheres to the standards of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for data processing and analysis. We retrieved relevant papers from PubMed, Web of Science, and IEEE databases from January 2018 to December 2023, extracting information about the medical domain, imaging modalities, methods for addressing cross-domain issues, experimental designs, and datasets from the selected papers. Moreover, we compared the performance of methods in stroke lesion segmentation, white matter segmentation and brain tumor segmentation.
Results: A total of 71 studies were included and analyzed in this review. The methods for tackling the cross-domain problem include Transfer Learning, Normalization, Unsupervised Learning, Transformer models, and Convolutional Neural Networks (CNNs). On the ATLAS dataset, domain-adaptive methods showed an overall improvement of ~3 percent in stroke lesion segmentation tasks compared to non-adaptive methods. However, given the diversity of datasets and experimental methodologies in current studies based on the methods for white matter segmentation tasks in MICCAI 2017 and those for brain tumor segmentation tasks in BraTS, it is challenging to intuitively compare the strengths and weaknesses of these methods.
Conclusion: Although various techniques have been applied to address the cross-domain problem in brain image segmentation, there is currently a lack of unified dataset collections and experimental standards. For instance, many studies are still based on n-fold cross-validation, while methods directly based on cross-validation across sites or datasets are relatively scarce. Furthermore, due to the diverse types of medical images in the field of brain segmentation, it is not straightforward to make simple and intuitive comparisons of performance. These challenges need to be addressed in future research.
Medical image segmentation, particularly for the brain, is a crucial and challenging task in the field of medical imaging analysis, with a wide range of applications from disease diagnosis to treatment planning. The complexity of this task is further compounded when considering the cross-domain nature of the data, arising from variations in scanners, imaging protocols, and patient populations among different sites (Dolz et al., 2018; Ravnik et al., 2018). This review aims to provide an overview of the progress made in the domain of cross-domain brain medical image segmentation. As depicted in Figure 1, the brain images and the corresponding segmented lesion areas are illustrated.
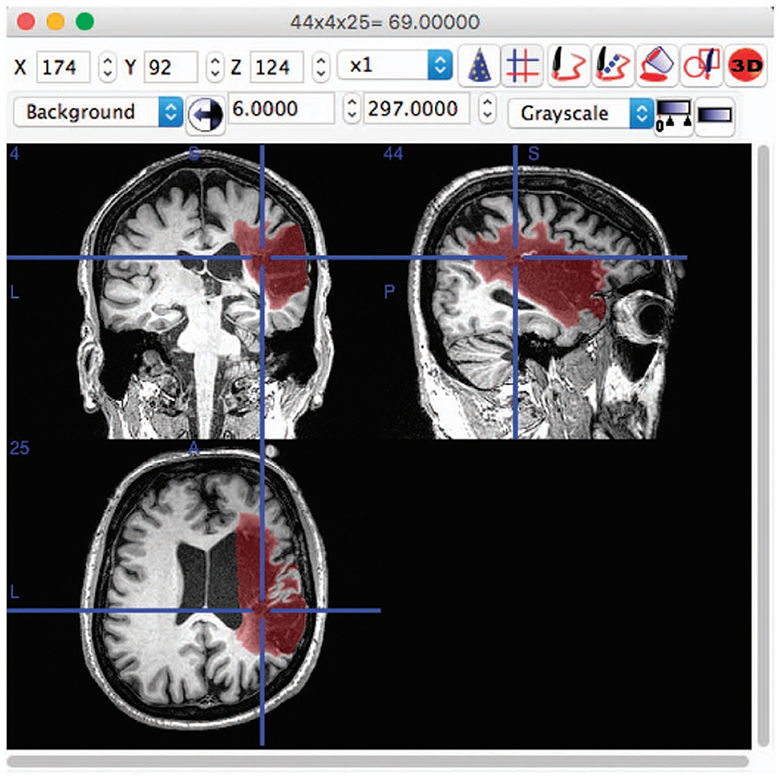
Figure 1. An example of lesion segmentation in brain (Liew et al., 2017).
Domain-adaptive methods are designed to adapt a model that has been trained on one domain (the source domain) to perform well on a different, but related domain (the target domain). This is useful in situations where we have a lot of labeled data in the source domain but little to no labeled data in the target domain. Domain adaptation techniques attempt to learn the shift or differences between the source and target domains and adjust the model accordingly. Techniques can include feature-level adaptation, instance-level adaptation, and parameter-level adaptation, among others.
Non-adaptive methods, on the other hand, do not make any adjustments to account for differences between the source and target domains. They are trained on one domain and then directly applied to another. This approach can work well if the source and target domains are very similar, but performance can degrade if there are significant differences between the two domains. Non-adaptive methods do not leverage any domain adaptation techniques and hence, can suffer from a problem known as domain shift or dataset shift, where the distribution of data in the target domain differs from the distribution in the source domain.
The advent of deep learning methods, especially Convolutional Neural Networks (CNNs) (LeCun et al., 1998) and their variants, has significantly improved the performance of image segmentation tasks (Dolz et al., 2018; Ravnik et al., 2018; Huang et al., 2020; Liu Y. et al., 2020). However, these models often suffer from limited generalization capability when applied to unseen data from different domains (Knight et al., 2018; Bermudez and Blaber, 2020; Zhou et al., 2022). To address this, various domain adaptation techniques have been proposed, including transfer learning, unsupervised learning, and self-supervised learning (Knight et al., 2018; Atlason et al., 2019; Ntiri et al., 2021; Tomar et al., 2022).
Transfer learning has emerged as a popular approach to leverage pre-trained models on new data, demonstrating success in various studies (Knight et al., 2018; Bermudez and Blaber, 2020; Zhou et al., 2022; Liu D. et al., 2023; Torbati et al., 2023). Unsupervised learning methods, which do not require labeled data from the target domain, have also shown promising results in cross-domain brain image segmentation (Atlason et al., 2019; Rao et al., 2022). Recently, self-supervised learning, where models are pre-trained on auxiliary tasks before being fine-tuned on the main task, has been increasingly adopted (Ntiri et al., 2021; Liu et al., 2022a; Tomar et al., 2022).
Besides, different strategies have been proposed to handle specific challenges in cross-domain brain image segmentation. For instance, normalization techniques have been used to reduce the scanner-related variability (Ou et al., 2018; Goubran et al., 2020; Dinsdale et al., 2021). Generative Adversarial Networks (GANs) (Goodfellow et al., 2014) have been employed to generate synthetic images that share the same distribution as the target domain, thus improving the model's generalizability (Zhao et al., 2019; Cerri et al., 2021; Tomar et al., 2022). Model ensembling and federated learning approaches have also been explored to leverage the strengths of multiple models or to perform decentralized learning (Reiche et al., 2019).
Moreover, the application of advanced architectures, such as 3D-CNNs (Ji et al., 2013), Transformers (Vaswani et al., 2017), and UNets, has further enhanced the performance of brain image segmentation across different domains (Dolz et al., 2018; Goubran et al., 2020; Huang et al., 2020; Liu Y. et al., 2020; Basak et al., 2021; Li et al., 2021; Meyer et al., 2021; Sun et al., 2021; Zhao et al., 2021). These models have been applied to various brain structures and conditions, including white matter, brain tumors, multiple sclerosis, and stroke (Erus et al., 2018; Knight et al., 2018; Ravnik et al., 2018; Reiche et al., 2019; Basak et al., 2021; Jiang et al., 2021; Kruger et al., 2021; Li et al., 2021; Sun et al., 2021; Kaffenberger et al., 2022; Zhou et al., 2022; Liu D. et al., 2023; Yu et al., 2023b; Zhang et al., 2023).
Despite the significant progress, cross-domain brain image segmentation remains a challenging problem. Future research directions may include the development of more robust and generalizable models, the exploration of novel domain adaptation techniques, and the incorporation of multimodal imaging data to improve segmentation performance. The studies reviewed herein provide valuable insights into these potential avenues for future advancement (Liu Y. et al., 2020; Jiang et al., 2021; Liu et al., 2022a; Rao et al., 2022; Torbati et al., 2023).
The search process for this study adheres to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (Moher et al., 2009) guidelines. In order to gather relevant research on cross-domain issues in brain medical image segmentation, we have designated three main categories of keywords: Medical Imaging, Segmentation, and Domain. Specific keywords for each category are shown in Table 1. It's worth noting that we use the Boolean operator “OR” to connect keywords within the same category, while “AND” is used to connect different categories. This way, we can construct complex search queries. Because the focus of the research is on cross-domain issues in brain medical image segmentation, these articles will be included in our review.
We used three search engines for literature retrieval: PubMed, IEEE, and Web of Science, with the search time frame being from January 2018 to December 2023 for journal or conference articles. In compliance with the PRISMA guidelines, the first stage of the screening process is to merge duplicate articles from different search engines. In the second stage, we screen based on the title and abstract of the articles, discarding those not relevant to our discussion topic, such as those that do not include keywords like “brain medical imaging,” “segmentation,” or “domain” in the title and abstract. In the third stage, we filter out eligible articles through a full-text review. Reasons for exclusion may include: inability to access the full text; non-English articles; survey studies or literature reviews; non-original research; not focusing on cross-domain issues; not describing experiments or validation studies; not using multi-site or multi-scanner datasets.
From the screened articles, we extracted the following information: author names, publication year, dataset name, dataset size, parts included in the dataset, cross-domain type, solution method, and evaluation metrics. For more detailed information about solution method, please refer to Tables 2, 3.
Enhancements based upon the UNet model continue to represent a prevalent research direction in medical image segmentation. Subsequent models, such as 3D-CNN, exhibit commendable performance in many 3D data scenarios, albeit at the cost of requiring substantial computational resources. In comparison, newer network structures like Transformer are gradually gaining traction in the field of medical segmentation, and it is anticipated that a plethora of innovations will be spawned from this methodology.
Methods grounded in different learning types are somewhat niche in comparison. On the whole, the outcomes of unsupervised and semi-supervised learning methods are not as effective as their supervised counterparts. This discrepancy is likely attributable to the relatively smaller datasets available in the field of medical imaging, unlike the voluminous data present in natural language processing and computer vision.
Mathematically-based methods are currently often amalgamated with deep learning models to enhance their interpretability. This area of work is particularly meaningful and holds significant potential.
There is a broad spectrum of data preprocessing techniques available, including Generative Adversarial Networks (GANs), which can be employed for data augmentation to enhance data diversity.
The array of tools available for medical image segmentation is continually expanding, and the barriers to their utilization are concurrently lowering.
In addition to extracting key data from cross-domain research in the field of brain image segmentation, we have also conducted a focused comparative analysis of cross-domain algorithms for three important branches of brain image segmentation: stroke lesion segmentation, white matter segmentation and brain tumor segmentation.
Due to the variety of datasets employed in the selected articles, it is challenging to compare the merits and demerits of each algorithm on a holistic basis. To compare the effectiveness of these algorithms, it becomes necessary to delve into more specific areas of segmentation. The ATLAS, MICCAI 2017 and BraTS datasets, each employed five times, stand out as the most frequently used. They correspond respectively to stroke lesion segmentation, white matter segmentation and brain tumor segmentation.
Figure 2 presents the PRISMA flow diagram for this task. The number of articles from the three databases (PubMed, IEEE, Web of Science) were 487, 332, and 890 respectively. An additional seven articles were identified through the references of confirmed papers. After merging duplicate studies, 1,286 articles were obtained. Following the title and abstract screening, 364 articles remained. Finally, after full-text review, 71 articles were included for publication. Table 4 documents the details of the finally collected articles.
As illustrated in the Figure 3, the number of papers addressing cross-domain segmentation in brain imaging has been increasing annually from 2018 to the present, with a peak of 15 papers in 2021. This trend indicates that there are still many challenges to overcome in this field, affirming its status as an active area of research.
As can be seen from Table 4 and Figure 4, in the 71 articles reviewed, 41 utilized public datasets, encompassing 56 different types. Among these, from Figure 5, the most frequently used datasets were ATLAS, MICCAI 2017 and BraTS, only five times. The remaining datasets were used less, with the majority being used only once. Thus, within the field of brain image segmentation, many articles addressing cross-domain issues still rely on proprietary datasets, and those that do use public datasets draw from a wide variety.
For a more specific analysis, we have included the disease type or brain region that is segmented' in our data extraction. This addition will enable us to gain a deeper understanding of which diseases are related to brain image segmentation and which regions require segmentation. This detailed approach will significantly contribute to our comprehensive review of cross-domain segmentation in brain medical imaging. Figure 6 shows the disease categories and regions extracted from the reviewed papers. Among them, whole-brain segmentation accounts for the largest proportion.
Based on the data collected, we have identified several types of cross-domain variations present in the field of brain medical image segmentation in Figure 7. The most common type of variation is “multi-site,” with 37 articles addressing this particular challenge. This is followed by “multi-scanner,” which is the focus of 18 articles. Both “multi-center” and “multi-modal” variations were discussed in 10 and six articles each. These findings highlight the diverse range of cross-domain challenges encountered in the segmentation of brain medical images, underscoring the need for further research and method development in this area.
As show in Figure 8, in the landscape of cross-domain segmentation in brain medical imaging, a diverse range of techniques are employed. The most prevalent methods include UNet, CNN, 3D-CNN, and Transfer Learning, indicating a strong reliance on convolutional architectures and leveraging pre-existing models. Other techniques such as Normalization, Self-Supervised learning, and GANs are also being utilized, albeit less frequently. A handful of studies explore alternative approaches including Unsupervised learning, Data Augmentation, and Transformer-based methods. This diversity of methodologies underscores the complexity of the challenge and the ongoing innovation in the field.
Due to the diversity in datasets and experimental methods, it is not feasible to compare the performance of all algorithms. However, it is possible to compare the algorithms that have utilized the ATLAS, MICCAI 2017 and BraTS datasets.
To begin with, we introduce the dataset used, ATLAS. The MR modality of the Anatomical Tracings of Lesions After Stroke (ATLAS) dataset is T1. It has two versions: ATLAS v1.2 (Liew et al., 2017), released in 2018, includes 304 cases from 11 research centers worldwide; and ATLAS v2.0 (Liew et al., 2022), released in 2022, includes 12,71 cases. Although v2.0 contains more data, its relatively recent release means that fewer articles have used it for cross-domain image segmentation to date. Therefore, we have chosen ATLAS v1.2 as our comparison dataset. As shown in Table 5, ATLAS v1.2 includes nine sites.
Cross-domain algorithms, as the name suggests, are designed to generalize and perform well across multiple, diverse datasets. A notable example from 2023, the Fan-Net (Yu et al., 2023b), utilizes Fourier-based adaptive normalization for stroke lesion segmentation. In 2021, the Unlearning algorithm (Dinsdale et al., 2020) was proposed to unlearn dataset biases for MRI harmonization and confound removal. Similarly, SAN-Net (Yu et al., 2023a) in 2023 and RAM-DSIR (Zhou et al., 2022) in 2022 showcased learning generalization to unseen sites and generalizable medical image segmentation via random amplitude mixup, respectively.
On the other hand, for performance comparison, we have also selected some non-cross-domain algorithms that are optimized for specific tasks or datasets. For instance, U-Net (Ronneberger et al., 2015), proposed in 2015, is an early example of convolutional networks for biomedical image segmentation. In 2018, DeepLab v3+ (Chen et al., 2018) introduced atrous separable convolution for semantic image segmentation. More recently, in 2020, nnU-Net (Isensee et al., 2021) presented a self-configuring method for deep learning-based biomedical image segmentation.
In the realm of cross-domain segmentation in brain medical imaging, specifically for stroke lesion segmentation, the performance of various methods demonstrates a compelling trend toward the adoption of cross-domain algorithms.
As can be seen from Table 6, Among the non-cross-domain algorithms, CLCI-Net exhibits the highest Dice and F1-score, demonstrating superior performance in segmentation accuracy. However, nnU-Net, despite having a slightly lower Dice score, presents the least Floating Point Operations Per Second (FLOPs), indicating a more efficient use of computational resources.
Shifting focus to cross-domain algorithms, SAN-Net outperforms the rest in all three performance metrics—Dice, Recall, and F1-score, highlighting its robustness in handling cross-domain segmentation tasks. Notably, RAM-DSIR, despite having the least number of parameters, delivers competitive results, suggesting an efficient model with less complexity.
In conclusion, while non-cross-domain algorithms such as CLCI-Net and nnU-Net exhibit commendable performance, cross-domain algorithms, particularly SAN-Net and RAM-DSIR, demonstrate superior performance and efficiency in stroke lesion segmentation. This underscores the potential and advantages of cross-domain approaches in this field, prompting further exploration and development in this direction.
In order to benchmark stroke lesion segmentation algorithms under non-domain adaptation scenarios, we refer to the dataset collated in this study (Malik et al., 2024). As shown in Table 7, eight stroke lesion segmentation algorithms from the ATLAS project were employed. Many of these algorithms achieved a Dice Similarity Coefficient (DSC) of up to 0.7, with the highest-performing algorithm, the seventh one, reaching 0.844. This significantly surpasses the maximum DSC of 0.597 achieved when conducting domain adaptation testing. Therefore, it is currently challenging for domain adaptation algorithms to achieve performance levels comparable to those of algorithms tested without domain adaptation, due to the necessity of conducting domain adaptation testing.
As shown in Table 8, the dataset MICCAI 2017 is derived from the WMH MICCAI 2017 challenge (Kuijf et al., 2019). This dataset encompasses MRI scans from multiple sites, including the University Medical Center Utrecht (UMC Utrecht), the National University Health System Singapore (NUHS Singapore), the VU University Medical Center Amsterdam (VU Amsterdam), and two undisclosed locations.
The MRI scans in the dataset are obtained from a variety of scanners, including 3T Philips Achieva, 3T Siemens TrioTim, 3T GE Signa HDxt, 1.5T GE Signa HDxt, and 3T Philips Ingenuity. The T1 voxel sizes and FLAIR scan sizes captured by these scanners vary, ranging from 0.87*0.87*1.00 mm3 to 1.21*1.21*1.30 mm3.
In total, 60 samples are utilized for training, while the testing set comprises 110 samples. The diversity and scale of this dataset allow us to evaluate the performance of our methods in a comprehensive and accurate manner. The training data can be downloaded at https://wmh.isi.uu.nl.
In the context of white matter medical imaging, several notable papers stand out. The Voxel-Wise Logistic Regression (VLR) (Knight et al., 2018) algorithm, introduced in 2018, leveraged voxel-wise logistic regression for FLAIR-based white matter hyperintensity segmentation. An innovative approach was presented in 2019 with the Skip Connection U-net (SC U-net) (Zhang et al., 2019), which added skip connections to the classic U-net architecture. In 2021, the MixDANN (Kushibar et al., 2021) algorithm tackled the challenging scenario of domain generalization (DG), i.e., training a model without any knowledge about the test distribution. The same year, an Ensemble U-net (Park et al., 2021) with multi-scale highlighted foreground (HF) was introduced for white matter hyperintensity segmentation, demonstrating its effectiveness in cross-domain segmentation in the 2017 MICCAI white matter hyperintensity segmentation challenge. A Transductive Transfer Learning Approach (TDA) (Kruger et al., 2021) was proposed in 2021 for domain adaptation, aiming to reduce the domain shift effect in brain MRI segmentation.
Table 9 presents the results of five different methods, all of which focus on the cross-domain segmentation problem in white matter imaging. In the table, – means there is no valid data. However, it is important to note that, with the exception of the second and third methods, the experimental datasets and experimental procedures used in each method are distinct from each other.
For instance, the VLR method employed three datasets, which included seven sites, and performed a leave-one-out cross-validation with respect to these sites. The SC U-net and MixDANN methods, on the other hand, only employed three sites from the MICCAI 2017 training data for cross-validation. The Ensemble U-net method used all of the training data from MICCAI 2017 for training and the test data for testing. Lastly, the TDA method utilized both the MICCAI 2017 and VH datasets, performing cross-validation between these datasets. In addition, VH is a private dataset.
Therefore, while there are numerous studies addressing the cross-domain problem in the field of white matter segmentation, direct comparisons between them are challenging. This is due to the variations in the experimental data and procedures used, even when the same dataset is utilized in different studies. The differences in experimental procedures are manifested in whether cross-validation is performed between sites or between datasets.
Although it is challenging to make a direct comparison between each algorithm, an overall observation can be made in the field of white matter segmentation. Specifically, the Dice Similarity Coefficient (DSC) is above 0.7 when cross-validation is conducted between sites, while the DSC is only around 0.5 when cross-validation is carried out between datasets. This observation suggests that cross-validation between datasets is more challenging, yet it is also closer to real-world scenarios.
In Table 10, the BraTS datasets comprises three dataset: BraTS 2015, BraTS 2018, and BraTS 2019, each with varying numbers of cases. The datasets are categorized into two major classes: High-Grade Gliomas (HGG) and Low-Grade Gliomas (LGG). Each case consists of four modalities (T1, T2, FLAIR, T1ce) and requires segmentation into three parts: Whole Tumor (WT), Enhancing Tumor (ET), and Tumor Core (TC). The BraTS 2019 can be downloaded at https://www.med.upenn.edu/cbica/brats-2019/.
In 2021, a learnable Self-Attentive Spatial Adaptive Normalization (SASAN) (Tomar et al., 2021) method was introduced, utilizing adversarial training to address the domain gap in radiological images. In 2022, two algorithms were presented. One algorithm is grounded in a knowledge distillation scheme incorporating exponential mixup decay (EMD) (Liu et al., 2022b) to progressively acquire target-specific representations, while the other algorithm is the Unsupervised Domain Adaptation (UDA) method based on Self-Semantic Contour Adaptation (SSCA) (Liu et al., 2022a). In 2023, another UDA (Qin et al., 2023) method, based on semi-supervised learning, was proposed. Additionally, in the same year, the Multimodal Contrastive Domain Sharing (Multi-ConDoS) (Zhang et al., 2023) generative adversarial networks were introduced.
As shown in Table 11, Whole, Core, and Enh represent the Dice Similarity Coefficient (DSC) for whole tumor, core tumor, and enhanced tumor, respectively. While all five articles conducted cross-domain studies on brain tumor segmentation using the BraTS datasets, each article employed different source and target domains. As a result, direct comparisons of algorithm performance across the experimental results are challenging.
The field of brain medical image segmentation has seen significant advancements with the widespread application of deep learning technologies. However, the challenge of domain adaptation continues to be a crucial issue. In our review, we have identified a variety of methods proposed to address this issue, including transfer learning, normalization, unsupervised learning, Transformer models, and convolutional neural networks, among others. Each of these methods has its strengths but also comes with certain limitations.
Transfer learning is a common approach to addressing domain adaptation issues, with the main idea being to apply knowledge learned in one domain (source domain) to another domain (target domain). However, the effectiveness of this method is influenced by the distribution difference between the source and target domains. If the distribution difference is too large, the effectiveness of transfer learning may be compromised.
Normalization is another common method for addressing domain adaptation issues, with the main idea being to reduce the differences between different datasets by adjusting the brightness and contrast of images. However, this method may result in the loss of some important image information, thereby affecting the accuracy of segmentation results.
Unsupervised learning and Transformer models have also been used in some studies to address domain adaptation issues. The advantage of unsupervised learning is that it does not require labeled data, but its performance is usually not as good as supervised learning. The advantage of Transformer models is that they can handle long-distance dependencies, but they have a high computational complexity and require a large amount of computational resources.
Furthermore, we have observed that despite the application of various techniques to address domain adaptation issues in brain medical imaging, there currently exists a lack of unified dataset collections and experimental standards.
For instance, as illustrated in Figure 4, 42.3% of the papers only use private data, while 8.5% of the papers use both public and private data. As shown in Figure 7, even when public datasets are used, there is significant diversity amongst them. As indicated in Tables 9, 11, even when a single identical dataset is used, if the experimental data and methods differ, it remains challenging to make comparisons among various algorithms. Moreover, the vast majority of current algorithms are not open-source, making it nearly impossible to reproduce the algorithms in the papers and design similar experiments for comparison.
Consequently, this makes it difficult to compare the performance of different studies and accurately assess the effectiveness of new methods. Therefore, future research needs to further develop more effective domain adaptation methods and establish unified dataset collections and experimental standards.
In conclusion, domain adaptation in brain medical image segmentation is a challenging research field that necessitates further exploration and development. Although numerous methods have been proposed to tackle this issue, each possesses its own strengths and limitations. Future research needs to delve deeper into novel methods to enhance the performance of domain adaptation in brain medical image segmentation.
Moreover, it is imperative to establish unified dataset collections and experimental standards for a more accurate evaluation of the performance of different methods. Only through this approach can we gain a better understanding of the strengths and weaknesses of various methods and develop more effective solutions.
Finally, we anticipate further advancements in deep learning technologies to address the domain adaptation problem in brain medical image segmentation. This progress will improve the accuracy of medical image analysis and, ultimately, enhance patient diagnosis and treatment.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
MY: Writing – original draft, Writing – review & editing, Investigation, Methodology, Validation. CS: Data curation, Investigation, Methodology, Software, Writing – review & editing. LW: Data curation, Investigation, Validation, Visualization, Writing – review & editing. YZ: Formal analysis, Investigation, Validation, Writing – review & editing. AW: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the project supported by the Special Fund of Advantageous and Characteristic Disciplines (Group) of Hubei Province and the Scientific Research Plan Project of Hubei Province Department of Education 2021.B 2021312. This work was partially funded by the Health Research Council of New Zealand's project 21/144, the MBIE Catalyst: Strategic Fund NZ-Singapore Data Science Re-search Programme UOAX2001, the Marsden Fund Project 22-UOA-120, and the Royal Society Catalyst: Seeding General Project 23-UOA-055-CSG.
We extend our heartfelt appreciation to Wuhan Technology and Business University, School of Artificial Intelligence Academy for their invaluable and generous support.
CS was employed by Wuhan Dobest Information Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
MS, multiple sclerosis; ICC, intra-class correlations; HD, Hausdorff distance; TPR, true positive rate; FPR, false positive rate; NMI, normalized mutual information; ARI, adjusted rand index; MHD, modified Hausdorff distance; ASD, average surface distance; AP, average precision; H95, Housdorff distance; AVD, absolute volume difference; MAE, mean absolute error; PSNR, signal-noise ratio; ASSD, the average symmetric surface distance; DSC, dice similarity coefficient; PPV, positive predictive value; LTPR, lesion-wise TPR; LFPR, lesion-wise false positive rate; Acc, accuracy rate; IoU, intersection over union; FLAIR, fluid-attenuated inversion recovery; LST, lesion segmentation tool algorithms; LVD, lesion volume difference; SSD, sym metric surface distance; CV, coefcient of variation; TRV, test-retest variability; ROI, regions of interest; OSM, OATS and Sydney MAS; CNSR, Chinese National Stroke Registry; TDA, transductive domain adaptation; MSD, medical segmentation decathlon; RM, repeated measure; CND, Chinese normative data.
Atlason, H. E., Love, A., and Sigurdsson, S. (2019). Segae: unsupervised white matter lesion segmentation from brain MRIs using a CNN autoencoder. Neuroimage Clin. 24:102085. doi: 10.1016/j.nicl.2019.102085
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., et al. (2018). Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv [Preprint]. arXiv:1811.02629. doi: 10.48550/arXiv.1811.02629
Basak, H., Hussain, R., and Rana, A. (2021). Dfenet: a novel dimension fusion edge guided network for brain MRI segmentation. SN Comput. Sci. 2, 1–11. doi: 10.1007/s42979-021-00835-x
Bermudez, C., and Blaber, J. (2020). Generalizing deep whole brain segmentation for pediatric and post- contrast MRI with augmented transfer learning. Image Process. 11313, 111–118. doi: 10.1117/12.2548622
Billast, M., Meyer, M. I., Sima, D. M., and Robben, D. (2019). “Improved inter-scanner ms lesion segmentation by adversarial training on longitudinal data,” in Lecture Notes in Computer Science, Vol. 11992 (Cham: Springer), 98–107. doi: 10.1007/978-3-030-46640-4_10
Borges, P., Sudre, C., Varsavsky, T., and Thomas, D. (2019). “Physics-informed brain MRI segmentation,” in Simulation and Synthesis in Medical Imaging, Vol. 11827 (Cham: Springer), 100–109. doi: 10.1007/978-3-030-32778-1_11
Brown, E., Pierce, M., Clark, D., Fischl, B., Iglesias, J., Milberg, W., et al. (2020). Test-retest reliability of freesurfer automated hippocampal subfield segmentation within and across scanners. Neuroimage 210:116563. doi: 10.1016/j.neuroimage.2020.116563
Bui, T. D., and Wang, L. (2019). “Multi-task learning for neonatal brain segmentation using 3D dense-Unet with dense attention guided by geodesic distance,” in Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data (Cham: Springer), 243–251. doi: 10.1007/978-3-030-33391-1_28
Cerri, S., Puonti, O., and Meier, D. S. (2021). A contrast-adaptive method for simultaneous whole-brain and lesion segmentation in multiple sclerosis. Neuroimage 225:117471. doi: 10.1016/j.neuroimage.2020.117471
Chen, L.-C., Zhu, Y., Papandreou, G., Schroff, F., and Adam, H. (2018). “Encoderdecoder with atrous separable convolution for semantic image segmentation,” in Proceedings of the European Conference on Computer Vision (ECCV) (Cham: Springer), 801–818. doi: 10.1007/978-3-030-01234-2_49
Dewey, B., Zhao, C., Reinhold, J., Carass, A., Fitzgerald, K., Sotirchos, E., et al. (2019). Deepharmony: a deep learning approach to contrast harmonization across scanner changes. Magn. Reson. Imaging 64, 160–170. doi: 10.1016/j.mri.2019.05.041
Dinsdale, N. K., Jenkinson, M., and Namburete, A. I. L. (2020). “Unlearning scanner bias for MRI harmonisation in medical image segmentation,” in Medical Image Understanding and Analysis. MIUA 2020. Communications in Computer and Information Science, Vol. 1248 (Cham: Springer). doi: 10.1007/978-3-030-52791-4_2
Dinsdale, N. K., Jenkinson, M., and Namburete, A. I. L. (2021). Deep learning-based unlearning of dataset bias for MRI harmonisation and confound removal. Neuroimage 228:117689. doi: 10.1016/j.neuroimage.2020.117689
Dolz, J., Desrosiers, C., and Ayed, I. B. (2018). 3D fully convolutional networks for subcortical segmentation in MRI: a large-scale study. Neuroimage 170, 456–470. doi: 10.1016/j.neuroimage.2017.04.039
Doyle, A., Elliott, C., Karimaghaloo, Z., Subbanna, N., Arnold, D. L., and Arbel, T. (2018). “Lesion detection, segmentation and prediction in multiple sclerosis clinical trials,” in BrainLesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2017. Lecture Notes in Computer Science, Vol 10670, eds. A. Crimi, S. Bakas, H. Kuijf, B. Menze, and M. Reyes (Cham: Springer), 14–25. doi: 10.1007/978-3-319-75238-9_2
Erus, G., Doshi, J., and An, Y. (2018). Longitudinally and inter-site consistent multi-atlas based parcellation of brain anatomy using harmonized atlases. Neuroimage 166, 71–78. doi: 10.1016/j.neuroimage.2017.10.026
Fung, Y., Ng, K., Vogrin, S., Meade, C., Ngo, M., Collins, S., et al. (2019). Comparative utility of manual versus automated segmentation of hippocampus and entorhinal cortex volumes in a memory clinic sample. J. Alzheimers Dis. 68, 159–171. doi: 10.3233/JAD-181172
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). Generative adversarial nets. Adv. Neural Inform. Process. Syst. 27, 2672–2680.
Goodkin, O., Prados, F., Vos, S., Pemberton, H., Collorone, S., Hagens, M., et al. (2021). Flair-only joint volumetric analysis of brain lesions and atrophy in clinically isolated syndrome (CIS) suggestive of multiple sclerosis. NeuroImage: Clin. 29:102542. doi: 10.1016/j.nicl.2020.102542
Goubran, M., Ntiri, E. E., and Akhavein, H. (2020). Hippocampal segmentation for brains with extensive atrophy using three-dimensional convolutional neural networks. Hum. Brain Mapp. 41, 291–308. doi: 10.1002/hbm.24811
Han, D., Yu, R., and Li, S. (2023). “MR image harmonization with transformer,” in 2023 IEEE International Conference on Mechatronics and Automation (ICMA) (Harbin: IEEE), 2448–2453. doi: 10.1109/ICMA57826.2023.10215948
Hindsholm, A. M., Andersen, F. L., and Cramer, S. P. (2023). Scanner agnostic large-scale evaluation of MS lesion delineation tool for clinical MRI. Front. Neurosci. 17:1177540. doi: 10.3389/fnins.2023.1177540
Huang, H., Lin, L., Tong, R., and Hu, H. (2020). “Unet 3+: a full-scale connected Unet for medical image segmentation,” in ICASSP 2020 - 2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Barcelona: IEEE), 1055–1059. doi: 10.1109/ICASSP40776.2020.9053405
Hui, H., Zhang, X., Wu, Z., and Li, F. (2021). Dual-path attention compensation U-net for stroke lesion segmentation. Comput. Intell. Neurosci. 2021:7552185. doi: 10.1155/2021/7552185
Isensee, F., Jaeger, P. F., Kohl, S. A. A., Petersen, J., and Maier-Hein, K. H. (2021). nnU-net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211. doi: 10.1038/s41592-020-01008-z
Ji, S., Xu, W., Yang, M., and Yu, K. (2013). 3D convolutional neural networks for human action recognition. IEEE Trans. Pattern Anal. Mach. Intell. 35, 221–231. doi: 10.1109/TPAMI.2012.59
Jiang, Y., Gu, X., and Wu, D. (2021). A novel negative-transfer-resistant fuzzy clustering model with a shared cross-domain transfer latent space and its application to brain CT image segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 18, 40–52. doi: 10.1109/TCBB.2019.2963873
Kaffenberger, T., Venkatraman, V., and Steward, C. (2022). Stroke population-specific neuroanatomical CT-MRI brain atlas. Neuroradiology 64, 1557–1567. doi: 10.1007/s00234-021-02875-9
Kalkhof, J., González, C., and Mukhopadhyay, A. (2022). “Disentanglement enables cross-domain hippocampus segmentation,” in 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) (Kolkata: IEEE), 1–5. doi: 10.1109/ISBI52829.2022.9761560
Kamraoui, R. A., Ta, V.-T., Tourdias, T., Mansencal, B., Manjon, J. V., Coupé, P., et al. (2022). Deeplesionbrain: towards a broader deep-learning generalization for multiple sclerosis lesion segmentation. Med. Image Anal. 76:102312. doi: 10.1016/j.media.2021.102312
Karani, N., Chaitanya, K., Baumgartner, C., and Konukoglu, E. (2018). “A lifelong learning approach to brain mr segmentation across scanners and protocols,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, Volume 11070 of Lecture Notes in Computer Science, eds. A. Frangi, J. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer). doi: 10.1007/978-3-030-00928-1_54
Kazerooni, A. F., Arif, S., and Madhogarhia, R. (2023). Automated tumor segmentation and brain tissue extraction from multiparametric MRI of pediatric brain tumors: a multi-institutional study. Neurooncol. Adv. 5:vdad027. doi: 10.1093/noajnl/vdad027
Khademi, A., Reiche, B., DiGregorio, J., Arezza, G., and Moody, A. (2020). Whole volume brain extraction for multi-centre, multi-disease flair MRI datasets. Magn. Reson. Imaging 66, 116–130. doi: 10.1016/j.mri.2019.08.022
Kim, R., Lee, M., Kang, D., Wang, S., Kim, N., Lee, M., et al. (2020). Deep learning-based segmentation to establish east asian normative volumes using multisite structural MRI. Diagnostics 11:13. doi: 10.3390/diagnostics11010013
Knight, J., Taylor, G. W., and Khademi, A. (2018). Voxel-wise logistic regression and leave-onesource-out cross validation for white matter hyperintensity segmentation. Magn. Reson. Imaging 54, 119–136. doi: 10.1016/j.mri.2018.06.009
Kruger, J., Ostwaldt, A.-C., and Spies, L. (2021). Infratentorial lesions in multiple sclerosis patients: intra- and inter-rater variability in comparison to a fully automated segmentation using 3D convolutional neural networks. Eur. Radiol. 32, 2798–2809. doi: 10.1007/s00330-021-08329-3
Kuijf, H. J., Biesbroek, J. M., De Bresser, J., Heinen, R., Andermatt, S., Bento, A., et al. (2019). Standardized assessment of automatic segmentation of white matter hyperintensities and results of the WMH segmentation challenge. IEEE TMI 38, 2556–2568. doi: 10.1109/TMI.2019.2905770
Kushibar, K., Salem, M., and Valverde, S. (2021). Transductive transfer learning for domain adaptation in brain magnetic resonance image segmentation. Front. Neurosci. 15:608808. doi: 10.3389/fnins.2021.608808
Le, M., Tang, L., Hernández-Torres, E., Jarrett, M., Brosch, T., Metz, L., et al. (2019). Flair2 improves lesiontoads automatic segmentation of multiple sclerosis lesions in non-homogenized, multi-center, 2D clinical magnetic resonance images. Neuroimage Clin. 23:101918. doi: 10.1016/j.nicl.2019.101918
LeCun, Y., Bottou, L., Bengio, Y., and Haffner, P. (1998). Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324. doi: 10.1109/5.726791
Li, B., You, X., and Wang, J. (2021). IAS-NET: joint intraclassly adaptive gan and segmentation network for unsupervised cross-domain in neonatal brain MRI segmentation. Med. Phys. 48, 6962–6975. doi: 10.1002/mp.15212
Li, C. (2021). Stroke lesion segmentation with visual cortex anatomy alike neural nets. arXiv [Preprint]. arXiv:2105.06544. doi: 10.48550/arXiv.2105.06544
Liew, S. L., Anglin, J. M., Banks, N. W., Sondag, M., Ito, K. L., Kim, H., et al. (2017). The anatomical tracings of lesions after stroke (atlas) dataset-release 1.1. bioRxiv 179614. doi: 10.1101/179614
Liew, S. L., Lo, B. P., Donnelly, M. R., Zavaliangos-Petropulu, A., Jeong, J. N., Barisano, G., et al. (2022). A large, curated, open-source stroke neuroimaging dataset to improve lesion segmentation algorithms. Sci. Data 9:320. doi: 10.1038/s41597-022-01401-7
Liu, D., Cabezas, M., and Wang, D. (2023). Multiple sclerosis lesion segmentation: revisiting weighting mechanisms for federated learning. Front. Neurosci. 17:1167612. doi: 10.3389/fnins.2023.1167612
Liu, S., Hou, B., Zhang, Y., Lin, T., Fan, X., You, H., et al. (2020). Inter-scanner reproducibility of brain volumetry: influence of automated brain segmentation software. BMC Neurosci. 21:35. doi: 10.1186/s12868-020-00585-1
Liu, X., Shih, H. A., and Xing, F. (2023). “Incremental learning for heterogeneous structure segmentation in brain tumor MRI,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, Volume 14221 of Lecture Notes in Computer Science, eds. H. Greenspan, A. Madabhushi, P. Mousavi, S. Salcudean, J. Duncan, and T. Syeda-Mahmood (Cham: Springer). doi: 10.1007/978-3-031-43895-0_5
Liu, X., Xing, F., Fakhri, G. E., and Woo, J. (2022a). “Self-semantic contour adaptation for cross modality brain tumor segmentation,” in 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI) (Kolkata: IEEE), 1–5. doi: 10.1109/ISBI52829.2022.9761629
Liu, X., Yoo, C., Xing, F., Kuo, C., Fakhri, G., Woo, J., et al. (2022b). Unsupervised domain adaptation for segmentation with black-box source model. doi: 10.1117/12.2607895
Liu, Y., Nacewicz, B. M., and Zhao, G. (2020). A 3D fully convolutional neural network with top-down attention-guided refinement for accurate and robust automatic segmentation of amygdala and its subnuclei. Front. Neurosci. 14:260. doi: 10.3389/fnins.2020.00260
Malik, M., Chong, B., Fernandez, J., Shim, V., Kasabov, N. K., Wang, A., et al. (2024). Stroke lesion segmentation and deep learning: a comprehensive review. Bioengineering 11:86. doi: 10.3390/bioengineering11010086
McClure, P., Rho, N., and Leel, J. A. (2019). Knowing what you know in brain segmentation using bayesian deep neural networks. Front. Neurosci. 13:67. doi: 10.3389/fninf.2019.00067
Memmel, M., Gonzalez, C., and Mukhopadhyay, A. (2021). “Adversarial continual learning for multi-domain hippocampal segmentation,” in Domain Adaptation and Representation Transfer, and Affordable Healthcare and AI for Resource Diverse Global Health, eds. S. Albarqouni, M. J. Cardoso, Q. Dou, K. Kamnitsas, B. Khanal, I. Rekik, et al. (Cham: Springer International Publishing), 35–45. doi: 10.1007/978-3-030-87722-4_4
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., et al. (2015). The multimodal brain tumor image segmentation benchmark (BraTS). IEEE Trans. Med. Imaging 34, 1993–2024. doi: 10.1109/TMI.2014.2377694
Meyer, M. I., de la Rosa, E., and de Barros, N. P. (2021). A contrast augmentation approach to improve multi-scanner. Front. Neurosci. 15:708196. doi: 10.3389/fnins.2021.708196
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 8, 336–341. doi: 10.1016/j.ijsu.2010.02.007
Monteiro, M., Newcombe, V. F., Mathieu, F., Adatia, K., Kamnitsas, K., Ferrante, E., et al. (2020). Multiclass semantic segmentation and quantification of traumatic brain injury lesions on head CT using deep learning: an algorithm development and multicentre validation study. Lancet Digit. Health 2, e314–e322. doi: 10.1016/S2589-7500(20)30085-6
Nair, T., Precup, D., Arnold, D. L., and Arbel, T. (2020). Exploring uncertainty measures in deep networks for multiple sclerosis lesion detection and segmentation. Med. Image Anal. 59:101557. doi: 10.1016/j.media.2019.101557
Niu, K., Li, X., and Zhang, L. (2022). Improving segmentation reliability of multi-scanner brain images using a generative adversarial network. Quant. Imaging Med. Surg. 12, 1775–1786. doi: 10.21037/qims-21-653
Ntiri, E. E., Holmes, M. F., and Forooshani, P. M. (2021). Improved segmentation of the intracranial and ventricular volumes in populations with cerebrovascular lesions and atrophy using 3D CNNs. Neuroinformatics 19, 597–618. doi: 10.1007/s12021-021-09510-1
Opfer, R., Krüger, J., and Spies, L. (2023). Automatic segmentation of the thalamus using a massively trained 3D convolutional neural network: higher sensitivity for the detection of reduced thalamus volume by improved inter-scanner stability. Eur. Radiol. 33, 1852–1861. doi: 10.1007/s00330-022-09170-y
Ou, Y., Zollei, L., and Da, X. (2018). Field of view normalization in multi-site brain MRI. Neuroinform 16, 431–444. doi: 10.1007/s12021-018-9359-z
Park, G., Hong, J., Duffy, B., Lee, J., and Kim, H. (2021). White matter hyperintensities segmentation using the ensemble U-net with multi-scale highlighting foregrounds. Neuroimage 237:118140. doi: 10.1016/j.neuroimage.2021.118140
Qi, K., Yang, H., and Li, C. (2019a). “Xnet: brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Volume 11766 of MICCAI 2019 (Cham: Springer), 247–255. doi: 10.1007/978-3-030-32248-9_28
Qi, K., Yang, H., Li, C., Liu, Z., Wang, M., Liu, Q., et al. (2019b). “X-net: brain stroke lesion segmentation based on depthwise separable convolution and long-range dependencies,” in Medical Image Computing and Computer Assisted Intervention - MICCAI 2019 (Cham: Springer), 247–255. doi: 10.1007/978-3-030-32248-9_28
Qin, C., Li, W., Zheng, B., Zeng, J., Liang, S., Zhang, X., et al. (2023). Dual adversarial models with cross-coordination consistency constraint for domain adaption in brain tumor segmentation. Front. Neurosci. 17:1043533. doi: 10.3389/fnins.2023.1043533
Rao, V. M., Wan, Z., and Arabshahi, S. (2022). Improving across-dataset brain tissue segmentation for MRI imaging using transformer. Front. Neuroimaging 1:1023481. doi: 10.3389/fnimg.2022.1023481
Ravnik, D., Jerman, T., Pernus, F., Likar, B., and Spiclin, Z. (2018). Dataset variability leverages white-matter lesion segmentation performance with convolutional neural network. Proc. SPIE 10574, 388–396. doi: 10.1117/12.2293702
Reiche, B., Moody, A. R., and Khademi, A. (2019). Pathology-preserving intensity standardization framework for multi-institutional flair MRI datasets. Magn. Reson. Imaging 62, 59–69. doi: 10.1016/j.mri.2019.05.001
Ribaldi, F., Altomare, D., Jovicich, J., Ferrari, C., Picco, A., Pizzini, F., et al. (2021). Accuracy and reproducibility of automated white matter hyperintensities segmentation with lesion segmentation tool: a European multi-site 3T study. Magn. Reson. Imaging 76, 108–115. doi: 10.1016/j.mri.2020.11.008
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: convolutional networks for biomedical image segmentation,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, Volume 9351 of Lecture Notes in Computer Science, eds. N. Navab, J. Hornegger, W. Wells, and A. Frangi (Cham: Springer). doi: 10.1007/978-3-319-24574-4_28
Sheng, M., Xu, W., Yang, J., and Chen, Z. (2022). Cross-attention and deep supervision Unet for lesion segmentation of chronic stroke. Front. Neurosci. 16:836412. doi: 10.3389/fnins.2022.836412
Srinivasan, D., Erus, G., Doshi, J., Wolk, D., Shou, H., Habes, M., et al. (2020). A comparison of freesurfer and multi-atlas muse for brain anatomy segmentation: findings about size and age bias, and inter-scanner stability in multi-site aging studies. Neuroimage 223:117248. doi: 10.1016/j.neuroimage.2020.117248
Sun, Y., Gao, K., and Lin, W. (2021). “Multi-scale self-supervised learning for multi-site pediatric brain MR image segmentation with motion/gibbs artifacts,” in Machine Learning in Medical Imaging. MLMI 2021, Volume 12966 of Lecture Notes in Computer Science, eds. C. Lian, X. Cao, I Rekik., X. Xu, and P. Yan (Cham: Springer). doi: 10.1007/978-3-030-87589-3_18
Sundaresan, V., Zamboni, G., Dinsdale, N. K., Rothwell, P. M., Griffanti, L., and Jenkinson, M. (2021). Comparison of domain adaptation techniques for white matter hyperintensity segmentation in brain MR images. Med. Image Anal. 74:102215. doi: 10.1016/j.media.2021.102215
Tomar, D., Bozorgtabar, B., and Lortkipanidze, M. (2022). “Self-supervised generative style transfer for one-shot medical image segmentation,” in 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) (Waikoloa, HI), 1737–1747. doi: 10.1109/WACV51458.2022.00180
Tomar, D., Lortkipanidze, M., Vray, G., Bozorgtabar, B., and Thiran, J.-P. (2021). Self-attentive spatial adaptive normalization for cross-modality domain adaptation. IEEE Trans. Med. Imaging 40, 2926–2938. doi: 10.1109/TMI.2021.3059265
Torbati, M. E., Minhas, D. S., and Laymon, C. M. (2023). Mispel: a supervised deep learning harmonization method for multiscanner neuroimaging data. Med. Image Anal. 89:102926. doi: 10.1016/j.media.2023.102926
Trinh, M.-N., Pham, V.-T., and Tran, T.-T. (2022). “A deep learning-based approach with semi-supervised level set loss for infant brain MRI segmentation,” in Pervasive Computing and Social Networking, Volume 475 of Lecture Notes in Networks and Systems, eds. G. Ranganathan, R. Bestak, and X. Fernando (Singapore: Springer). doi: 10.1007/978-981-19-2840-6_41
van Opbroek, A., Achterberg, H. C., Vernooij, M. W., Ikram, M. A., and de Bruijne, M. (2018). Transfer learning by feature-space transformation: a method for hippocampus segmentation across scanners. Neuroimage Clin. 20, 466–475. doi: 10.1016/j.nicl.2018.08.005
Vaswani, A., Shazeer, N., and Parmar, N. (2017). Attention is all you need. Adv. Neural Inform. Process. Syst. 5998–6008.
Wang, L., Wu, Z., Chen, L., Sun, Y., Lin, W., and Li, G. (2023). iBEAT V2.0: a multisite-applicable, deep learning-based pipeline for infant cerebral cortical surface reconstruction. Nat. Protoc. 18, 1488–1509. doi: 10.1038/s41596-023-00806-x
Wang, S., Chen, Z., You, S., Wang, B., Shen, Y., Lei, B., et al. (2022). Brain stroke lesion segmentation using consistent perception generative adversarial network. Neural Comput. Appl. 34, 8657–8669. doi: 10.1007/s00521-021-06816-8
Wang, Y., Haghpanah, F. S., and Zhang, X. (2022). ID-SEG: an infant deep learning-based segmentation framework to improve limbic structure estimates. Brain Inf. 9:12. doi: 10.1186/s40708-022-00161-9
Wu, Z., Zhang, X., Li, F., Wang, S., and Huang, L. (2022). Multi-scale long-range interactive and regional attention network for stroke lesion segmentation. Comput. Electr. Eng. 103:108345. doi: 10.1016/j.compeleceng.2022.108345
Yang, H., Huang, W., and Qi, K. (2019). “CLCI-net: cross-level fusion and context inference networks for lesion segmentation of chronic stroke,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Vol. 11766 (Singapore: Springer), 266–274. doi: 10.1007/978-3-030-32248-9_30
Yu, W., Huang, Z., Zhang, J., and Shan, H. (2023a). SAN-NET: learning generalization to unseen sites for stroke lesion segmentation with self-adaptive normalization. Comput. Biol. Med. 156:106717. doi: 10.1016/j.compbiomed.2023.106717
Yu, W., Lei, Y., and Shan, H. (2023b). “FAN-NET: Fourier-based adaptive normalization for cross-domain stroke lesion segmentation,” in ICASSP 2023 - 2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (Rhodes Island: IEEE), 1–5. doi: 10.1109/ICASSP49357.2023.10096381
Zhang, J., Zhang, S., Shen, X., Lukasiewicz, T., and Xu, Z. (2023). Multicondos: multimodal contrastive domain sharing generative adversarial networks for selfsupervised medical image segmentation. IEEE Trans. Med. Imaging 43, 76–95. doi: 10.1109/TMI.2023.3290356
Zhang, X., Xu, H., Liu, Y., Liao, J., Cai, G., Su, J., et al. (2021). “A multiple encoders network for stroke lesion segmentation,” in Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV) (Cham: Springer International Publishing), 524–535. doi: 10.1007/978-3-030-88010-1_44
Zhang, Y., Wu, J., Chen, W., Liu, Y., Lyu, J., Shi, H., et al. (2019). Fully automatic white matter hyperintensity segmentation using U-net and skip connection. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2019, 974–977. doi: 10.1109/EMBC.2019.8856913
Zhang, Z., Liu, Q., and Wang, Y. (2018). Road extraction by deep residual U-net. IEEE Geosci. Remote Sens. Lett. 15, 749–753. doi: 10.1109/LGRS.2018.2802944
Zhao, F., Wu, Z., and Wang, L. (2019). Harmonization of infant cortical thickness using surface-to-surface cycle-consistent adversarial networks. Med. Image Comput. Comput. Assist. Interv. 11767, 475–483. doi: 10.1007/978-3-030-32251-9_52
Zhao, X., Sicilia, A., and Minhas, D. S. (2021). “Robust white matter hyperintensity segmentation on unseen domain,” in 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) (Nice: IEEE), 1047–1051. doi: 10.1109/ISBI48211.2021.9434034
Zhou, Y., Huang, W., Dong, P., Xia, Y., and Wang, S. (2019). D-Unet: a dimension-fusion u shape network for chronic stroke lesion segmentation. IEEE/ACM Trans. Comput. Biol. Bioinform. 18, 940–950. doi: 10.1109/TCBB.2019.2939522
Zhou, Z., Qi, L., and Shi, Y. (2022). “Generalizable medical image segmentation via random amplitude mixup and domain-specific image restoration,” in European Conference on Computer Vision (Berlin: Springer), 420–436. doi: 10.1007/978-3-031-19803-8_25
Keywords: brain medical image, segmentation, cross-domain, stroke, white matter, brain tumor, normalization
Citation: Yanzhen M, Song C, Wanping L, Zufang Y and Wang A (2024) Exploring approaches to tackle cross-domain challenges in brain medical image segmentation: a systematic review. Front. Neurosci. 18:1401329. doi: 10.3389/fnins.2024.1401329
Received: 15 March 2024; Accepted: 28 May 2024;
Published: 14 June 2024.
Edited by:
Chunliang Wang, Royal Institute of Technology, SwedenReviewed by:
Prasanna Parvathaneni, Flagship Biosciences, Inc, United StatesCopyright © 2024 Yanzhen, Song, Wanping, Zufang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alan Wang, YWxhbi53YW5nQGF1Y2tsYW5kLmFjLm56
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.