- 1Department of Applied Chemistry and Life Science, Toyohashi University of Technology, Toyohashi, Aichi, Japan
- 2TechnoPro, Inc., Tokyo, Japan
- 3Institute for Research on Next-Generation Semiconductor and Sensing Science, Toyohashi University of Technology, Toyohashi, Aichi, Japan
- 4Health and Medical Research, National Institute of Advanced Industrial Science and Technology (AIST), Takamatsu, Kagawa, Japan
DNA aptamers can bind specifically to biomolecules to modify their function, potentially making them ideal oligonucleotide therapeutics. Herein, we screened for DNA aptamer of melanopsin (OPN4), a blue-light photopigment in the retina, which plays a key role using light signals to reset the phase of circadian rhythms in the central clock. Firstly, 15 DNA aptamers of melanopsin (Melapts) were identified following eight rounds of Cell-SELEX using cells expressing melanopsin on the cell membrane. Subsequent functional analysis of each Melapt was performed in a fibroblast cell line stably expressing both Period2:ELuc and melanopsin by determining the degree to which they reset the phase of mammalian circadian rhythms in response to blue-light stimulation. Period2 rhythmic expression over a 24-h period was monitored in Period2:ELuc stable cell line fibroblasts expressing melanopsin. At subjective dawn, four Melapts were observed to advance phase by >1.5 h, while seven Melapts delayed phase by >2 h. Some Melapts caused a phase shift of approximately 2 h, even in the absence of photostimulation, presumably because Melapts can only partially affect input signaling for phase shift. Additionally, some Melaps were able to induce phase shifts in Per1::luc transgenic (Tg) mice, suggesting that these DNA aptamers may have the capacity to affect melanopsin in vivo. In summary, Melapts can successfully regulate the input signal and shifting phase (both phase advance and phase delay) of mammalian circadian rhythms in vitro and in vivo.
Introduction
Most living organisms exposed to sunlight have evolved a ~ 24-h internal clock, known as a circadian rhythm, and synchronize the phases of this autonomous clock according to environmental clues (Ralph et al., 1990; Pittendrigh, 1993; Davidson et al., 2003; Yoo et al., 2004). Therefore, each organism resets the phase of its circadian rhythm to match the environmental light and dark cycle each morning. In mammals, circadian rhythms are influenced primarily by light, an input signal perceived through the eyes, which is capable of resetting the phase of transcriptional oscillations of clock genes (Takahashi, 1995; Menaker, 2003). The central pacemaker of mammalian circadian rhythms is located in the suprachiasmatic nucleus (SCN). The retinohypothalamic tract (RHT) immediately transmits information about blue light in the early morning to the SCN via the photoreceptor melanopsin (OPN4) in the retina and resets the phase of the circadian clock along the environmental cycle. Melanopsin mainly transfers environmental blue-light signals to the central clock early in the morning to reset the phase of the clock in the SCN (Hattar et al., 2003; Lucas et al., 2003; Foster, 2005; Pulivarthy et al., 2007; Ukai et al., 2007; Guler et al., 2008; LeGates et al., 2012).
Melanopsin is a photoreceptor protein expressed in retinal ganglion cells that absorbs blue light with a maximum absorbance of 477 nm. Although the exact function of melanopsin was unclear until melanopsin-knockout (KO) mice were generated in 2003, melanopsin was known to play an important role in resetting the phase of the mammalian circadian clock by blue light. In the first step, the melanopsin receptor is activated by binding trans-isomerized retinal ligands under blue light through the Gq family. The photosignal affects SCN neurons through Gq family-mediated transient receptor potential polycystin (TRPP) control of cell firing, glutamate transmitters, and pituitary adenylate cyclase-activating polypeptide (PACAP). The transmitted stimuli eventually increase calcium ion flux in the cytoplasm of cells in the SCN to activate protein kinase A (PKA) and PKC. These kinases phosphorylate cAMP response element-binding protein (CREB) and activate the connected to the cAMP response element (CRE) consensus sequence in the Period 1 (Per1) promoter to transiently induce Per1 transcription and reset the phase of circadian rhythms (King et al., 1997; Sun et al., 1997; Tei et al., 1997; Hida et al., 2000; Yamazaki et al., 2000).
The phase of the molecular circadian clock is reset by, and depends on, the timing of light stimulation and transient induction of Per1 by the melanopsin photoreceptor. Autonomous circadian clocks in individual cells of both the SCN and peripheral clocks are constituted by transcription and translation feedback loops involving clock genes, such as Clock, Bmal1, Per1-3, and Cryptochromes, as well as their protein products (Albrecht et al., 1997; Antoch et al., 1997; King et al., 1997; Shearman et al., 1997; Takumi et al., 1998; Zylka et al., 1998; Kume et al., 1999; Bunger et al., 2000; Numano et al., 2006). The photosignal by melanopsin transiently induces transcription of photoreactive clock genes such as Per1 and Per2 in the SCN from 30 min to 2 h following delivery of a light pulse to the retina. Induction of transient expression of these input genes influences the transcriptional and translational feedback loop within a 24-h period as well as the phase of the autonomous clock (Ellington and Szostak, 1990; Albrecht et al., 1997; Shearman et al., 1997; Shigeyoshi et al., 1997; Giebultowicz, 2004; Yan and Silver, 2004).
DNA aptamers are short, single-stranded RNA/DNA molecules that can bind selectively to specific targets, proteins, peptides, and other molecules and can be used clinically to switch the function of target molecules. The main advantages of these aptamers are high target specificity, lack of immunogenicity, and ease of synthesis.
The function of the melanopsin protein is easily modified by the DNA aptamer because it is located on the cell membrane. DNA aptamers are powerful pharmaceutical agents because, unlike antibodies, they can be stored stably and duplicated easily in large quantities using PCR.
People with sleep–wake phase disorder and shift workers who only sleep for 1–4 h may have difficulty falling asleep and waking up immediately in the morning (Takahashi et al., 2008; Gentry et al., 2021). Thus, it would be socially and economically advantageous to improve the sleep–wake cycle indirectly by manipulating the ability of melanopsin to input signals into a central clock. Antagonists of melanopsin acquired via chemical screening of chemical libraries primarily contribute to delaying the rhythm phase (Jones et al., 2013).
In this study, we used the cell systematic evolution of ligands by exponential enrichment (Cell-SELEX) method to identify DNA aptamers (single-stranded DNA; ssDNA) that caused melanopsin (expressed on the cell membrane) to shift the phase of circadian rhythms (Tuerk and Gold, 1990; Hida et al., 2000; Yamazaki et al., 2000; Umekage and Kikuchi, 2006; Dua et al., 2011). In total, 15 types of melanopsin aptamers were analyzed to assess their ability to shift the phase of Per2::ELuc bioluminescent oscillations in Per2:ELuc:TK:Mel stable cells, in which a bioluminescent reporter follows the Per2 promoter region controlling an enhanced green-emitting luciferase from Pyrearinus termitilluminans, with melanopsin overexpressed under the control of the thymidine kinase (TK) promotor (Nakajima et al., 2010). In these stable fibroblast cell lines, the signal pathway is incorporated into a fibroblast cell mimicking the signal pathway from the retina to the SCN by melanopsin.
Among these 15 DNA aptamers of melanopsin (Melapts), four Melapts induced phase advance and seven Melapts induced delay of circadian rhythms (by >1.5 h and > 2 h, respectively) in the Per2::ELuc cell line. Some Melapts induced phase shifts of ~2 h even in the absence of photostimulation in vitro. As the results from Per1::luc transgenic (Tg) mice were similar to the in vitro results from the Per2::ELuc cell line, melanopsin was used to induce phase Shifts in vivo. In summary, Melapts were able to regulate input signals and phase shifts to achieve both phase advance and phase delay of mammalian circadian rhythms in vitro and in vivo.
Materials and methods
Screening of DNA aptamers by the cell-SELEX method
A random ssDNA library for screening of DNA aptamers by the Cell-SELEX method was designed based on a random DNA aptamer (5‘-AAAGGGGAATTCGGATCC-N-40-CTGCAGAAGCTTCCGAAAA-3′) with regions of 40 bases, and 1017 types of DNA aptamers with a fixed leading and trailing region were prepared by Hokkaido System Science (Hokkaido, Japan). A floating PC12 cell (obtained from the Cell Bank, RIKEN BRC, Ibaraki, Japan) was used to perform the Cell-SELEX method, and cells were cultured in low-glucose DMEM (FUJIFILM Wako, Tokyo, Japan) containing 10% fetal bovine serum (FBS; Wako, Tokyo, Japan), 5% donor horse serum (DHS; Wako, Tokyo, Japan), and 1% penicillin–streptomycin (Wako, Tokyo, Japan). The melanopsin expression plasmid (provided by Professor Ueda of the University of Tokyo) was transfected into PC12 cells using Lipofectamine 3000 (Invitrogen, Massachusetts, United States) to prepare (+)melanopsin-transfected PC12 cells ([+] melanopsin cells) in which melanopsin was overexpressed on the cell membrane, whereas in (−) melanopsin-free PC12-negative control cells (other [−] melanopsin cells without transfecting melanopsin: negative selection), melanopsin was not expressed. First, DNA aptamers were mixed and incubated with (−) melanopsin cells, and only unbound DNA aptamers were recovered. Second, the recovered DNA aptamers in solution were mixed and incubated with (+) melanopsin cells, and only DNA aptamer bound to cells was recovered. Then, the recovered DNA aptamers specifically bound to (+) melanopsin cells but not bound to (−) melanopsin cells were amplified by asymmetric PCR amplofication (Figure 1A).
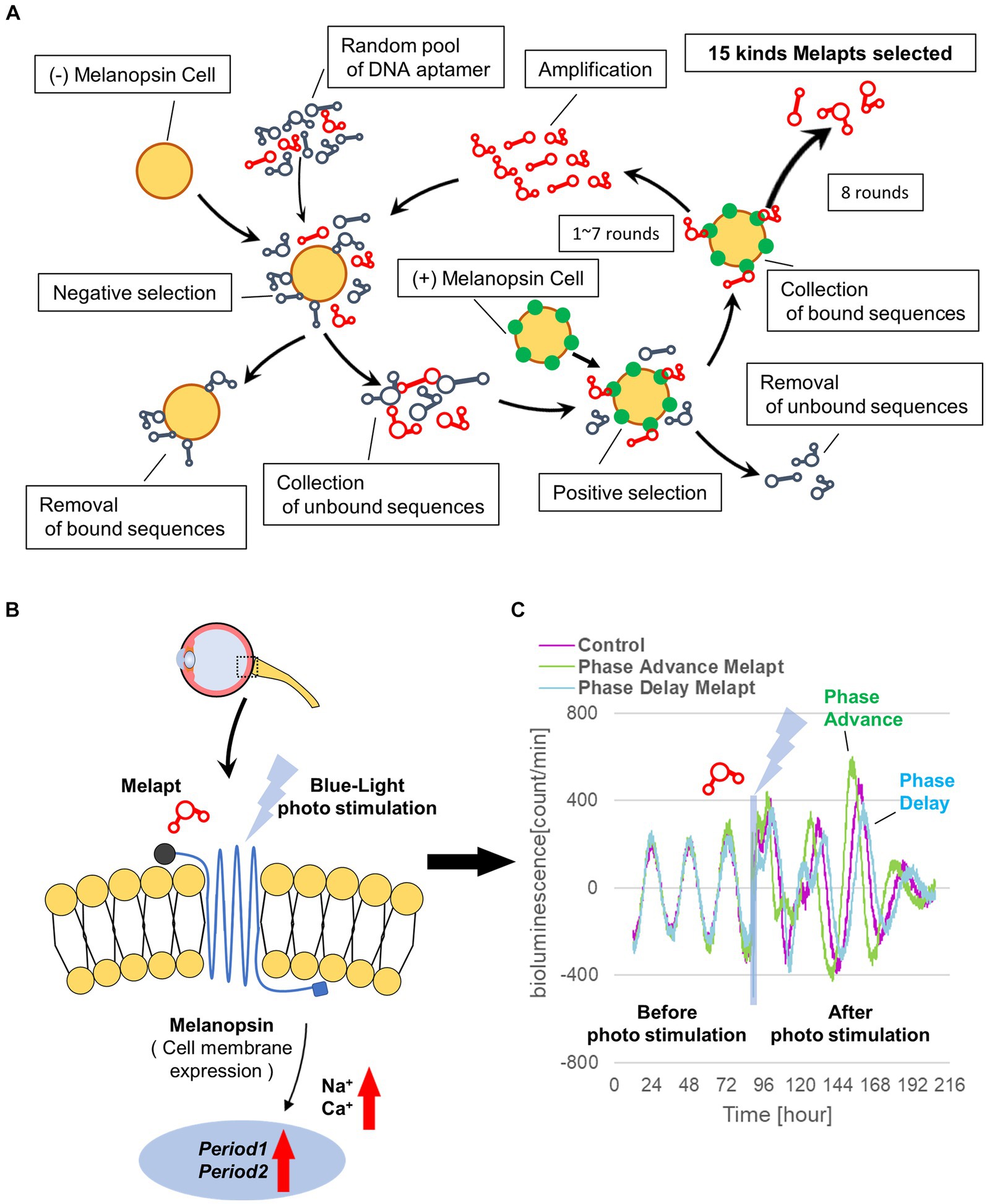
Figure 1. Cell-SELEX method for screening Melapts. Melapts that specifically bind melanopsin were selected by Cell-SELEX because melanopsin is a membrane protein bound and accessible from outside cells. (A) 1017 DNA aptamer were selected from a random ssDNA library. DNA aptamers were mixed and bound to (−) melanopsin cells (negative selection). When unbound DNA aptamers were retrieved, they were combined with (+) melanopsin cells to promote binding to melanopsin protein (positive selection). The Cell-SELEX process was performed over 8 rounds, and 15 types of Melapt were screened. Fluorescence intensity in the wells was measured (at λex 495 nm and λem 517 nm) using a microplate reader (Infinite 1,000; TECAN). (B) Conceptual diagram of Melapt binding to melanopsin followed by induction of the expression of the clock gene Per2. When melanopsin expressed on the surface of the cell membrane is photostimulated by a Melapt, the concentration of Na+ and Ca+ in the cytoplasm increases transiently, inducing the activation of transcriptional factors and the upregulation of Per2 in the cell nucleus. (C) Phase shift of luciferase emission rhythms in Per2:ELuc:TK:Mel cells stably expressing melanopsin. When Per2:ELuc cells expressing melanopsin were photostimulated post-addition of Melapts, Per2:ELuc emission rhythms showed phase advance (green curve) or phase delay (blue curve) relative to controls (purple curve); these shifts were Melapt-dependent. PBS buffer alone was added to control cells.
The obtained Melapts were amplified with forward and reverse primers in a 10:1 ratio by asymmetric PCR with reactions containing 10 pmol Melapt, 20 pmol forward primer (5′-AAAGGGGAATTCGGATCC-3′, FASMAC), 2 pmol reverse primer (5′-AAACGGAAGCTTCTGCAG-3′, FASMAC), 5 units Go Taq DNA polymerase (Promega; Wisconsin, USA), 30 nmol Mg2+ (Promega), 2.5 nmol dNTPs (Promega), and PCR buffer (Promega) in a final volume of 20 μL. The PCR amplification profile for Melapts involved preliminary denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 4 min.
Only those amplified as a significant sense band in gel electrophoresis were used in the next round of Cell-SELEX. These steps were repeated for eight rounds to concentrate DNA aptamers by asymmetric PCR amplification. Then, DNA aptamers were cloned into a T vector (Invitrogen), and aptamer sequences were confirmed by Fasmac Corporation (Kanagawa, Japan; Table 1). Finally, 15 DNA aptamers for melanopsin were selected and named Melapt1-Melapt15 (Supplementary Figure S1). Melapts obtained by the Cell-SELEX method appeared to bind specifically to melanopsin alone. Melanopsin-KO cells and melanopsin-KO mice were not used in this study.
Binding assay of Melapts to (+) melanopsin cells
Melapts were amplified and labeled with a 6-FAM-forward primer. Briefly, 10 pmol Melapt, 20 pmol 6-FAM-forward primer (5′-AAAGGGGAATTCGGATCC-′3, Hokkaido System Science), 2 pmol reverse primer (5′-AAACGGAAGCTTCTGCAG-3′, Fasmac Corporation), 1 unit Go Taq DNA polymerase (Promega), 125 pmol dNTPs (Promega), 5× Go Taq Reaction Buffer (Promega), and nuclease-free water were combined in a 20 μL reaction volume using the Cell-SELEX amplification profile (Figure 2A).
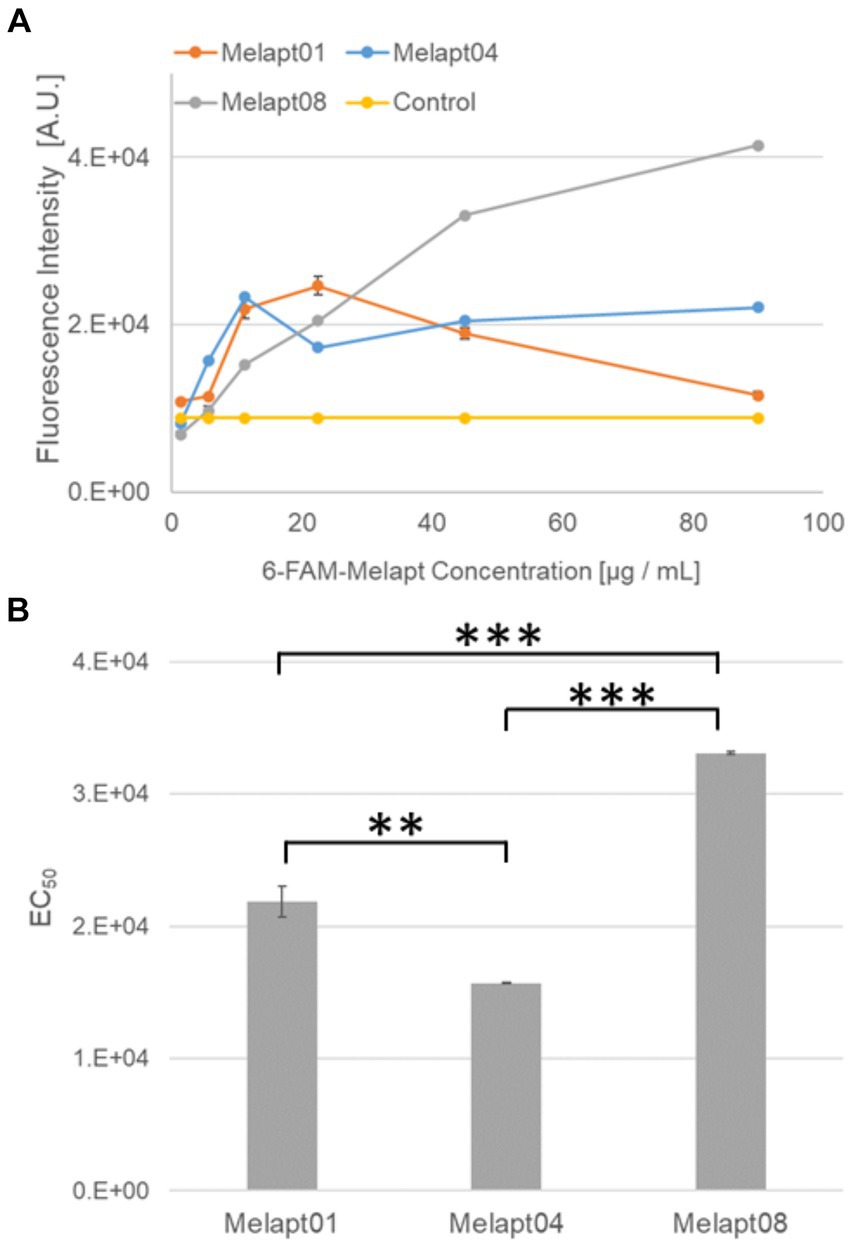
Figure 2. Binding of 6-FAM-Melapts to Per2:ELuc:TK:Mel stable cells expressing melanopsin. (A) 6-FAM-Melapts 01, 04, and 08 were added to cells at 1.405 μg/mL, 5.625 μg/mL, 11.25 μg/mL, 22.5 μg/mL, 45.0 μg/mL, and 90.0 μg/mL, whereas only PBS was added to control cells (n = 3). The 6-FAM fluorescence values of cells were plotted against the concentration of 6-FAM-Melapt. (B) The concentration resulting in EC50 was calculated from Figure 2A and plotted. Melapt01, 11.25 μg/mL; Melapt04, 5.625 μg/mL; Melapt08, 45.0 μg/mL. n = 3, **p < 0.01, ***p < 0.001, Tukey–Kramer test.
Mouse fibroblast cells were cultured in 96-well plates (1.5 × 105 cells/well) using 200 μL DMEM, transfected with melanopsin, and incubated at 37°C and 5% CO2 for 24 h. Then, 6-FAM-Melapt was added to the culture medium at six different concentrations (1.405 μg/mL, 5.625 μg/mL, 11.25 μg/mL, 22.5 μg/mL, 45.0 μg/mL, and 90.0 μg/mL) and incubated for 15 min. After washing, the binding capacity of 6-FAM-Melapts was estimated by measuring the fluorescence signal in (+) melanopsin cells (λex 495 nm, λem 517 nm) using an Infinite 1,000 microplate reader (Tecan, Zürich, Switzerland; Figure 1). The most suitable concentration of Melapts was 22.5 μg/mL. Before performing each measurement, 6-FAM-Melapt was added to three different wells (n = 3). A stable mouse fibroblast cell line (Per2:ELuc:TK:Mel) with Melapts was established to monitor the phase shift of Per2:ELuc bioluminescent emission rhythms using photo-responsive ELuc.
The 5′ flanking region of Per2 (from −2,858 to +144, where +1 indicates the putative transcription start site) was PCR-amplified from the C57BL/6 J mouse genome and cloned into the XhoI and BglII sites of pELuc (PEST)-test (Toyobo, Osaka, Japan). Expression cassettes containing early poly-A (pA) signal, Per2 promoter, ELuc-PEST, and late pA signal were amplified by PCR and cloned into pENTR-D-TOPO (Thermo Fisher Scientific Inc., Waltham, MA, United States), with the attL1 and attL2 sites flanked by the upstream and downstream regions of early and late pA signals, respectively, resulting in pmPer2-ELuc-PEST-pENTR. The expression cassette was recombined into the pBsd-R4 attB vector (a gift from Dr. T. Ohbayashi) by the LR reaction using LR Clonase II Plus Enzyme Mix (Thermo Fisher Scientific, Inc.), yielding pR4-Bsd-mPer2-ELuc-PEST.
Furthermore, mOpn4-Flag, wherein the Flag-tag was fused in-frame to the C-terminus of mouse melanopsin cDNA, was synthesized as double-stranded DNA (GenScript, Tokyo, Japan) and cloned into pUC57. The mOpn4-Flag was excised using NcoI and XbaI and ligated into the NcoI and XbaI site of pTK-SLG-pENTR-D-Topo (Tabei et al., 2017), from which the SLG cDNA was removed, yielding pTK-mOpn4-Flag-pENTR. The expression cassette containing TK promoter, mOpn4-Flag, and late pA signal was recombined into the pNeo-ϕC31 attB vector (Yamaguchi et al., 2011; a gift from Dr. T. Ohbayashi) by the LR reaction, resulting in pϕC31-Neo-mOpn4-Flag.
Mouse fibroblast A9 cells harboring the multi-integrase mouse artificial chromosome (MI-MAC) vector (Takiguchi et al., 2014; kindly provided by Dr. M. Oshimura and Dr. Y. Kazuki) seeded into six-well plates were co-transfected a day later with pR4-Bsd-mPer2-ELuc-PEST and the R4 integrase expression plasmid pCMV-R4 (Yamaguchi et al., 2011; kindly provided by Dr. T. Ohbayashi) and subcultured for selection with 6 μg/mL Blasticidin S (Thermo Fisher Scientific, Inc.). Selected cells were further co-transfected with pϕC31-Neo-mOpn4-Flag and ϕC31 integrase expression plasmid pCMV-ϕC31 (Yamaguchi et al., 2011; kindly provided by Dr. T. Ohbayashi) and subcultured for selection with 800 μg/mL G418 (Nacalai Tesque, Kyoto, Japan). Genomic PCR confirmed the integration of the transgenes into the corresponding sites in the MI-MAC vector. The established cell line was named Per2:ELuc:TK:Mel.
The photo-responsive fibroblast stable cell line for functional analysis of DNA aptamers (ELuc:Per2:ELuc:TK:Mel) was stably transfected into Per2-enhanced green-emitting luciferase cells (Per2:ELuc) with melanopsin (mOPN4) expression under the control of the TK promoter to generate photo-responsive A9 fibroblast cells (Nakajima et al., 2010). Screening of DNA aptamers was performed using blue light-responsive and bioluminescence real-time imaging of circadian rhythms (Supplementary Figure S2). Per2:ELuc stably expresses ELuc under the control of the Per2 promoter because the phase due to the transcriptional activity rhythm of Per2 can be monitored from the emission rhythms of ELuc (Nakajima et al., 2010). Per2:ELuc:TK:Mel stable cells constitutively and stably express melanopsin on the cell surface under the control of the TK promoter. Melanopsin expressed on the surface of the cell membrane transmits external photo-stimuli into the cell and transiently induces Per2 transcription in the cell nucleus. Thus, Per2:ELuc:TK:Mel stable cells are suitable for the phase shift of circadian rhythms in response to blue-light photo-stimuli.
Binding assay for Melapts
A total of 1.5 × 105 cells/well were cultured in 96-well plates for 24 h using 200 μL of DMEM and incubated at 37°C and 5% CO2. 6-FAM-Melapt was added to (+) melanopsin cells at six different concentrations (1.405 μg/mL, 5.625 μg/mL, 11.25 μg/mL, 22.5 μg/mL, 45.0 μg/mL, and 90.0 μg/mL) and incubated for 15 min. After washing, the binding capacity of 6-FAM-Melapt in (+) melanopsin cells was estimated by measuring the 6-FAM signal using a microplate reader (λex 495 nm, λem 517 nm). Before performing each measurement, 6-FAM-Melapt was added to three different wells (n = 3).
Estimation of phase shifts by Per2: Eluc:TK:Mel stable cells
Screening of DNA aptamers was performed using blue light-responsive and real-time bioluminescence recording of circadian rhythms (Supplementary Figure S2). Per2:ELuc cells were expressed in a stable cell line transfected to express ELuc under the control of the Per2 promoter (Nakajima et al., 2010) because the phase due to the transcriptional activity rhythm of Per2 can be monitored from the emission rhythms of ELuc. Per2:ELuc:TK:Mel stable cells expressed melanopsin stably under the control of the melanopsin under the control of the TK promoter. Melanopsin transmits external photo-stimuli into the cell via signal transduction and transiently induces Per2 transcription in the cell nucleus. Thus, Per2:ELuc:TK:Mel stable cells were deemed suitable for the phase shift of circadian rhythms in response to blue-light photostimulation because of recombinant gene Per2:ELuc and TK:Mel.
Per2: ELuc: TK: Mel stable cells (1.5 × 105) were passaged in a 35-mm dish, forskolin (Invitrogen) was added at a final concentration of 10 μM at 24 h after passaging, and cells were incubated for 30 min at 37°C before replacing the medium. This process synchronizes the circadian phase of Per2 rhythmic expression in all cells to amplify the amplitude of the emission rhythm, monitored by Phot multiple tubes (PMTs). Following forskolin shock, the cells were washed with phosphate-buffered saline (PBS) and exposed to luciferin (Beetle Luciferin, Potassium Salt; Promega) at a final concentration of 0.1 mM in a new medium in a Kronos illuminometer (ATTO, Tokyo, Japan). This was equipped with a PMT to detect luminescence and measure the cell emission signal in dishes in real time for 10 s every 3 min, while the cells were incubated at 37°C with 5% CO2. The phase of Per2:Eluc:TK:Mel stable cells was determined as the middle point between the peak and trough of bioluminescent rhythms as CT12. Then, Melapts were added to the culture medium at 22.5 μg/mL (final concentration) at subjective dawn (CT22) and subjective afternoon (CT8). Photostimulation was then performed in culture dishes in the Kronos for 15 min using a Blue LED (Amon Industry Co., Ltd., Tokyo, Japan). Subsequently, rhythmic bioluminescence in Per2:ELuc:TK:Mel stable cells was observed for 3 days.
The obtained rhythmic emission data were analyzed by cosine fitting over a 12-36-h period after forskolin shock, and after photostimulation, using NINJA software (CHURITSU Electric, Aichi, Japan).
Animals
Per1::luc Tg C57BL/N strain mice with a firefly luciferase gene linked to the downstream region of the 6.7-kb Per1 promoter (provided by Professor Tei, Kanazawa University; Yamazaki et al., 2000) were used to monitor mammalian the circadian rhythm of the central clock in the SCN. Experiments were performed using Per1::luc Tg heterozygous mice. Mice (all male and aged 6 to 12 months) were maintained under SPF conditions at 22°C with a 12:12 h light:dark cycle (from 08:00 to 20:00). Mice were fed standard pellets (CLEA Rodent Diet CE-2, CLEA, Tokyo, Japan, Inc.), and water and food were freely available. All animals were handled according to the guidelines for the use of laboratory animals, Toyohashi University of Technology (DO2021-1).
Injection of Melapts into bulbus oculi of Per1::luc Tg mice
Mice were injected with abatin anesthetic (1.9% w/v, 0.45 mL/20 g body weight). Response reflexes were assessed by pinching the tail and legs 5 min after anesthesia. For the mydriasis tests, one to two drops of Midorin-P ophthalmic solution (tropicamide phenylephrine hydrochloride ophthalmic solution; Santen Pharmaceutical, Osaka, Japan) were injected into the eyes of three Tg mice (CT22) following Melapt injection. Then, 1 μL of Melapt solution (100 ng-300 ng/μL) was injected into both bulbus oculi using a microsyringe (Hamilton, Nevada, United States) from the border between the cornea and sclera into both eyes. After injection, the mice were illuminated with an LED (Yazawa Corporation, Tokyo, Japan) at 1000 LUX for 30 min. Bulbus oculi were removed to prevent them from responding to light. All animal experiments were conducted under dim red LED lights.
Observing circadian bioluminescence rhythms of SCN cultured slices
We prepared 300-μm coronal section slices of SCN using a micro-slicer (Dosaka E.M., Kyoto, Japan) in cooled PBS on ice from whole brains of three Per1::luc Tg mice following Melapt injection. Under a stereomicroscope, only the SCN on the Chiasma was removed as a 1.5-mm triangular tissue section from coronal section slices in 1.2 mL of media. The SCN tissue slices were grown on Millicell membranes (PICM03050; Millipore, Massachusetts, USA) in serum-free DMEM (Invitrogen) containing 10 mM HEPES (pH 7.2; Invitrogen), 2% B27 (Life Technologies, Carlsbad, CA, United States), 25 unit/mL penicillin, and 25 μg/mL streptomycin (Invitrogen) in a 35-mm dish. Beatle luciferin potassium salt (Promega) was added at 0.2 mM where required. Circadian bioluminescence rhythms were continuously monitored using a PMT (Hamamatsu Photonics) in an incubator at 36°C. Emission values of SCN slices were integrated for 1 min over ~5 days and plotted onto graphs. Circadian bioluminescence cycle data were examined using NINJA software.
Statistical analysis
The Tukey–Kramer method in R software v.4.1.0. (R Foundation for Statistical Computing, Vienna, Austria) was used to analyze data. Statistical significance was set at p ≤ 0.05. Error bars on graphs show mean ± standard deviation (SD). All analyses were performed at least three times.
Results
Selecting DNA aptamers for melanopsin: Melapts
From the 1017 different DNA fragments, we used the Cell-SELEX approach to identify DNA aptamers that bind specifically to melanopsin in PC12 cells; eight rounds of Cell-SELEX were performed using 1017 DNA fragments, with (−) melanopsin cells as negative selection and (+) melanopsin cells as positive selection, respectively, by PCR to select 15 Melapts that bind specifically to melanopsin (Figure 1A). Functional screening of the 15 Melapts was performed to identify those able to induce a phase shift in circadian rhythms (Figure 1B). Photostimulation at subjective dawn (CT22) caused phase advance of Per2 expression rhythms in Per2:ELuc:TK:Mel stable fibroblasts expressing melanopsin (Figure 1C, Tables 2, 3). In addition, some Melapts caused a more significant phase advance of circadian rhythms following photostimulation (Figure 1B). These findings suggest that Melapts influence the phase of circadian rhythms by binding to melanopsin, thereby triggering signal transmission into cells and affecting transcription of the clock gene Per2.
Estimating the binding capacity of Melapts
We selected three Melapts labeled with 6-FAM (Melapt01, Melapt04, and Melapt08), Melapt01 (group1) induced both phase advance of Per2::luc bioluminescence rhythms and rhythm phase delay at CT22 (Figure 2).
That was because15 screened Melapts were divided into three groups, group 1: phase advance or delay in Per2:ELuc bioluminescence rhythms at both CT22 and CT8 like Melapt04, group 2: phase advance or delay at CT22 and delay or advance at CT8, respectively, like Melapt01, and group 3: non-phase shifts in Per2:ELuc bioluminescence rhythms like Melapt08. Melapt01, Melapt04, and Melapt08 were selected as representatives of each group and were FAM-labeled to determine a final concentration of Melapt for adding on cells. At 22.5 μg/mL, all three Melapts could bind to melanopsin on the cell membrane sufficiently.
Three Melapts labeled with 6-FAM fluorescence using 6-FAM primers were compared in terms of binding capacity to Per2:ELuc:TK:Mel stable cells. The higher binding capacity of Melapt04 and Melapt01 to cells was saturated at a concentration of 11.25 μg/mL, whereas that of Melapt08 could not be saturated at a concentration of 90 μg/mL (Figure 2A). Thus, in subsequent experiments, the applied concentration of Melapts was 22.5 μg/mL, sufficient to bind to melanopsin on the cell membrane because the half-maximal effective concentration (EC50) of Melapt08 with even lower binding capacity was 22.5 μg/mL (Figure 2B). These results not only confirmed the successful screening of selective aptamers capable of binding to melanopsin cell membranes but also determined the appropriate concentrations of Melapts in the culture medium for phase-shift experiments.
Screening of Melapts using Per2: Eluc:TK:Mel stable cells
Melapt with blue-light photostimulation at CT22
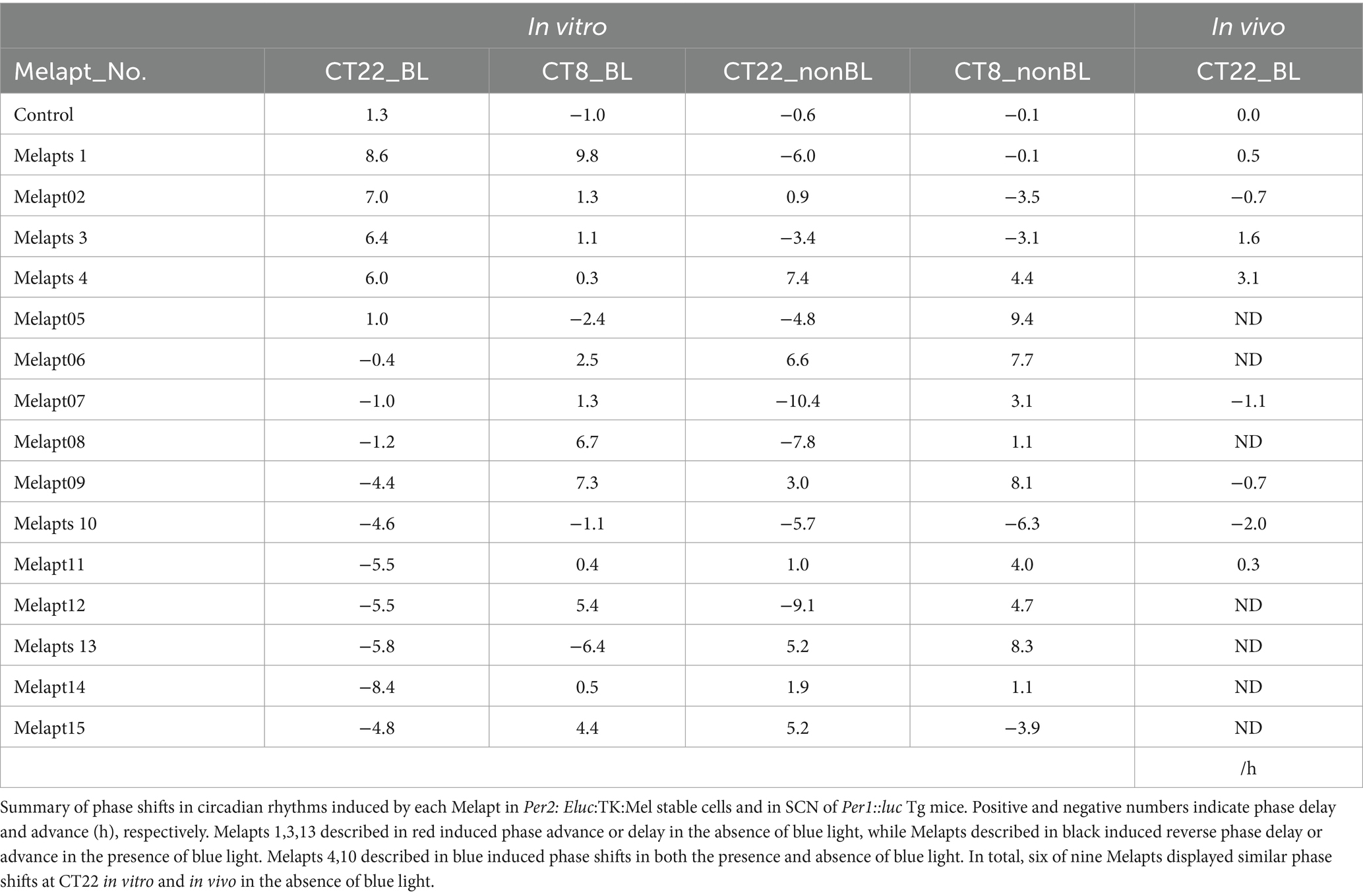
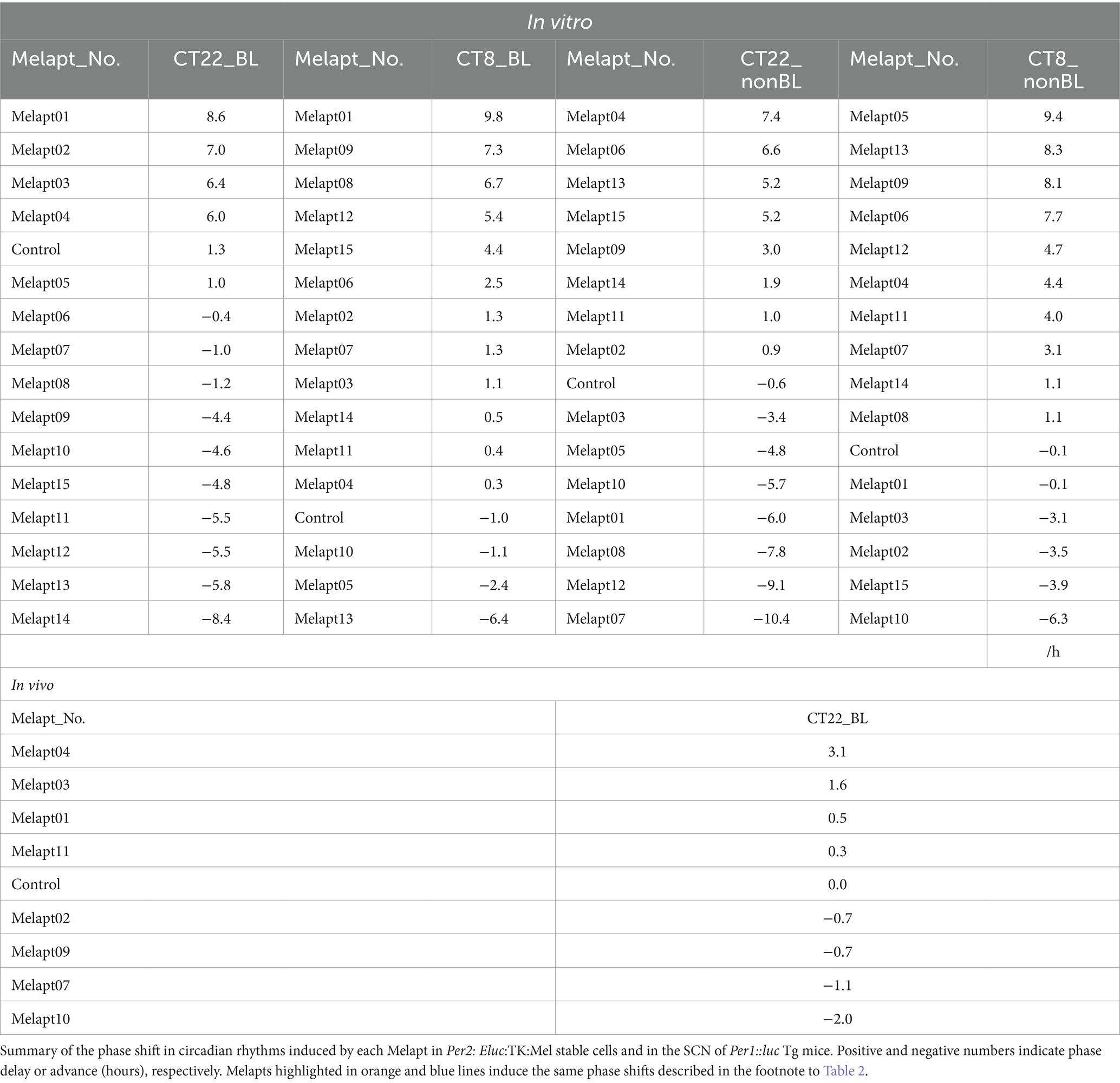
The 15 Melapts were categorized according to their ability to trigger a phase shift in circadian rhythms (Table 2). In addition, Table 3 shows Melapts rearranged from phase advance (positive number) to phase delay (negative number). The phase shift in bioluminescence rhythm depends on the timing of Melapt addition and the presence of photostimulation.
Per2:ELuc:TK:Mel cells stably expressing melanopsin were exposed to each Melapt under blue-light photostimulation at CT22 as a model for exposure of the retina to intense blue light at dawn (Figure 3; Tables 2, 3).
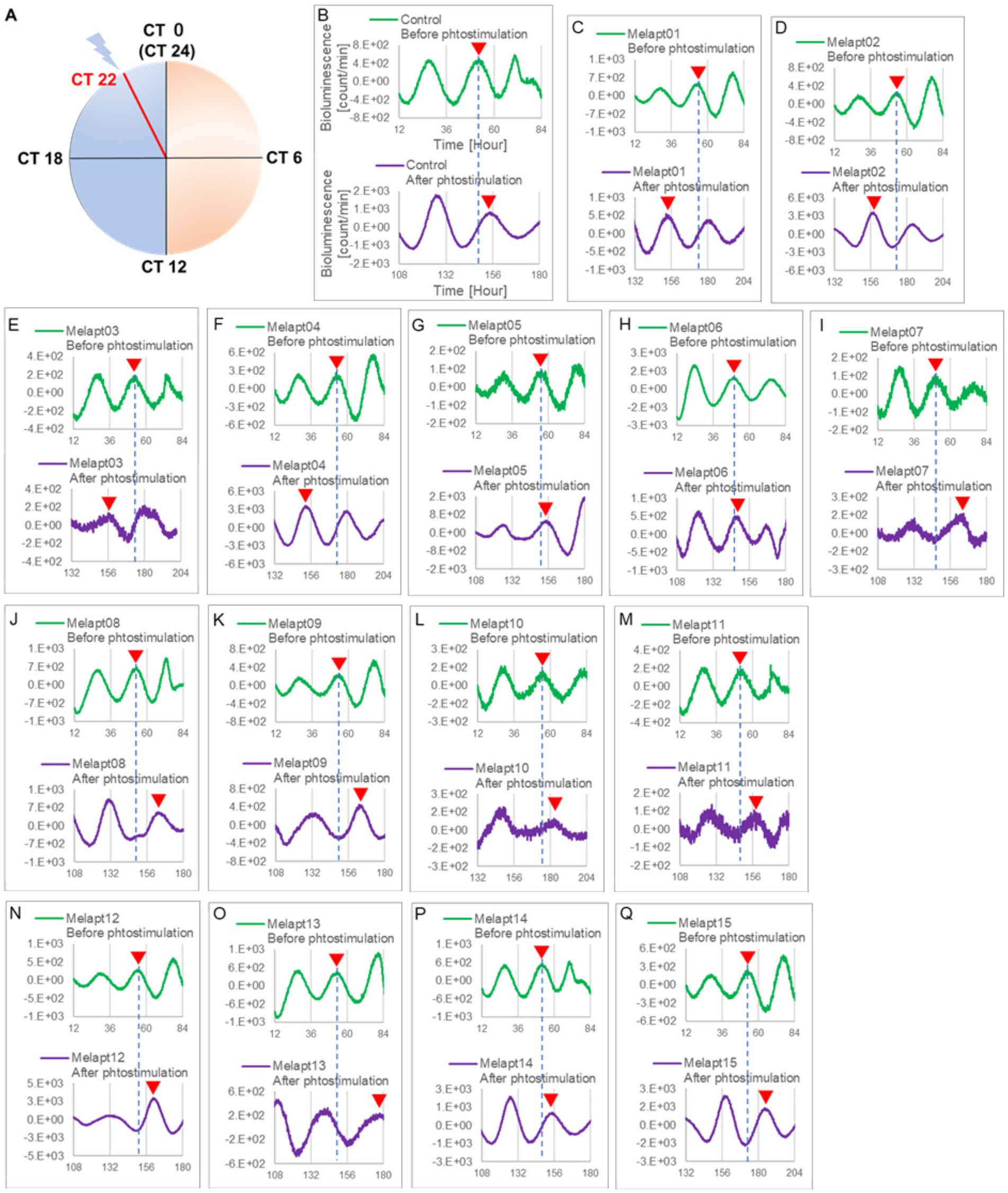
Figure 3. Changes in Per2:ELuc emission rhythms before and after the addition of each Melapt and blue-light photostimulation at CT22. (A) Subjective light period from CT0-12, subjective dark period from CT12-0, and a blue light indicate photostimulation at CT22 of the 24-h clock. (B) Per2:ELuc emission upon adding PBS and blue-light photostimulation as a control. (C–Q). Per2:ELuc emission upon adding Melapt01-Melapt15 and blue-light photostimulation. Upper row (green), Per2:ELuc emission before the addition of Melapt observed for 3 days. Lower row (purple), Per2:ELuc emission after addition of Melapt and blue-light photostimulation observed for 3 days. A red triangle indicates the peak. Bioluminescence traces in Figure 3 were estimated from an individual sample. The peak bioluminescent emission rhythms monitored over 3 days before the addition of Melapts and photostimulation (green) are denoted by a dotted line to allow comparison with the peak rhythms in the lower graphs (plotted post-stimulation and colored purple).
Phase advance of Per2:ELuc rhythmic emission rhythms by ~1.3 h was observed (using a Kronos bio luminometer) in controls following simple blue-light photostimulation in the absence of Melapts at CT22 (Figure 4; Tables 2, 3). Cells treated with Melapt01 showed a phase advance of Per2:ELuc emission rhythms by ~8.6 h (Figure 4A). The group treated with Melapt02 showed a phase advance of Per2:ELuc emission rhythms by ~6.9 h (Figures 4B–E). Melapt05 induced a phase advance of Per2:ELuc emission rhythms by ~1.0 h (Figure 4E). Conversely, Melapt06 induced a phase delay of Per2:ELuc emission rhythms of ~0.4 h (Figure 4F). Groups treated with Melapt07, Melapt08, Melapt09, Melapt10, Melapt11, Melapt12, Melapt13, Melapt14, and Melapt15 showed phase delays of Per2:ELuc emission rhythms by approximately 1.0, 1.2, 4.3, 4.6, 5.4, 5.5, 5.8, 8.4, and 4.7 h, respectively (Figures 4G–O).
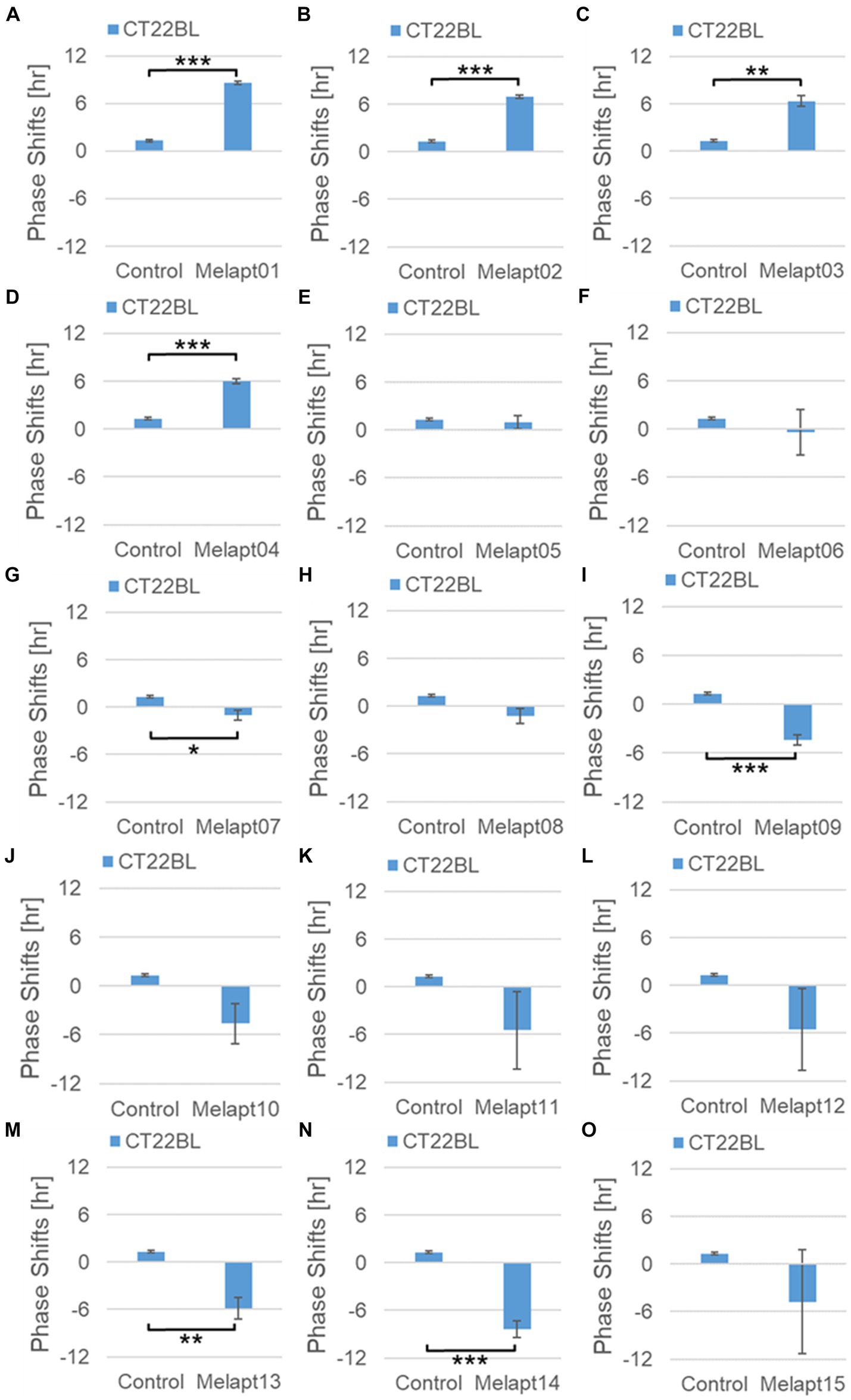
Figure 4. Phase shift due to binding of Melapts plus photostimulation at subjective dawn (CT22). (A–O) Phase-shifts comparison between Melapts and controls with photo-stimulus at CT22. PBS alone was added to control cells. The phase shift was calculated from the cosine-fitting curve in Figure 3 using the NINJA program and plotted on a graph. n = 3 (individual samples); *p < 0.05, **p < 0.01, ***p < 0.001, Tukey–Kramer test.
Melapt with blue-light photostimulation at CT8
Then, we examined the effects on Per2:ELuc emission rhythms of applying Melapts and blue-light photostimulation in the subjective afternoon (CT8) to mimic exposure to intense afternoon light. Kronos bio luminometer results revealed that controls receiving only blue-light photostimulation without any Melapts at CT8 showed a phase delay of Per2:ELuc emission rhythms by ~0.6 h (Figure 5; Tables 2, 3).
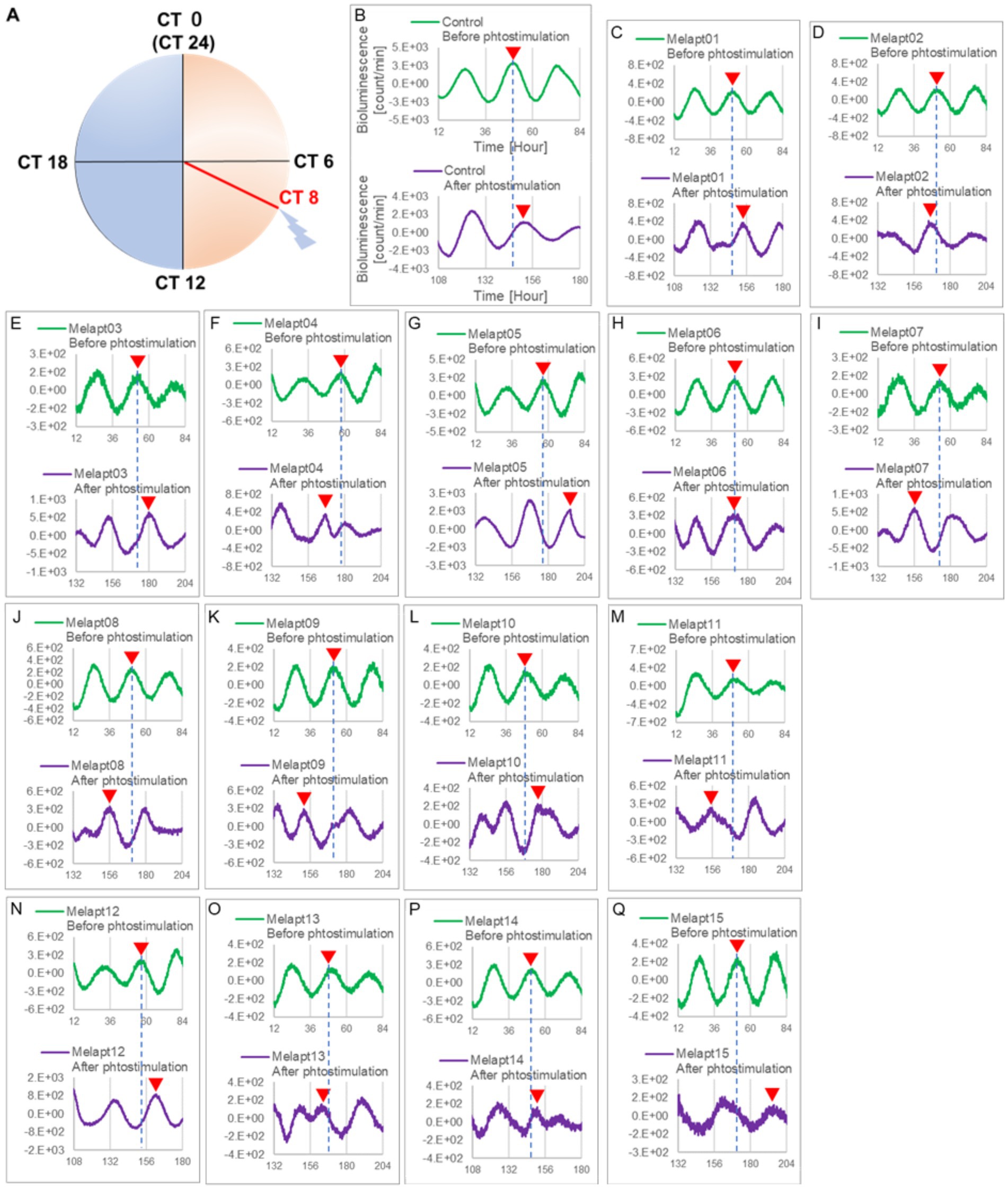
Figure 5. Changes in Per2:ELuc emission rhythms before and after applying Melapts and blue-light photostimulation at CT8. (A) Subjective light period from CT0-12, subjective dark period from CT12-0, and blue light indicate photostimulation at CT8 of the 24-h clock. (B) Per2:ELuc emission upon applying PBS and blue-light photostimulation to controls. (C–Q) Per2:ELuc emission upon applying Melapt01- Melapt15 and blue-light photostimulation. Upper row (green), Per2:ELuc emission before the addition of Melapt observed for 3 days. Lower row (purple), Per2:ELuc emission after addition of Melapt and blue-light photostimulation observed for 3 days. Red triangles indicate the peak. Bioluminescence traces in Figure 5 were estimated from an individual sample. The peak of bioluminescent emission rhythms measured over the 3 days prior to the addition of Melapts and photostimulation (green) is denoted by a dotted line to allow comparison with peak rhythms in the lower graphs (potted post-stimulation).
Cells treated with Melapt01 showed a phase delay of Per2:ELuc emission rhythms by ~5.9 h (Figure 6A). The Melapt03, Melapt05, Melapt07, Melapt08, Melapt10, and Melapt12 groups showed a phase delay of Per2:ELuc emission rhythms by approximately 3.4, 4.8, 10.4, 7.8, 5.6, and 9.1 h, respectively (Figures 6C,E,G,H,J,L). Melapt09 triggered a phase advance of Per2:ELuc emission rhythms by approximately 3.0 h (Figure 6I). By contrast, Melapt06 induced a phase delay of Per2:ELuc emission rhythms by ~6.6 h (Figure 6F). Groups treated with Melapt02, Melapt04, Melapt11, Melapt13, and Melapt14 displayed a phase advance of Per2:ELuc emission rhythms by approximately 0.9, 7.3, 1.0, 8.3, and 1.9 h, respectively (Figures 6B,D,K,M,N). Melapt15 induced a phase advance of Per2:ELuc emission rhythms by ~5.1 h (Figure 6O).
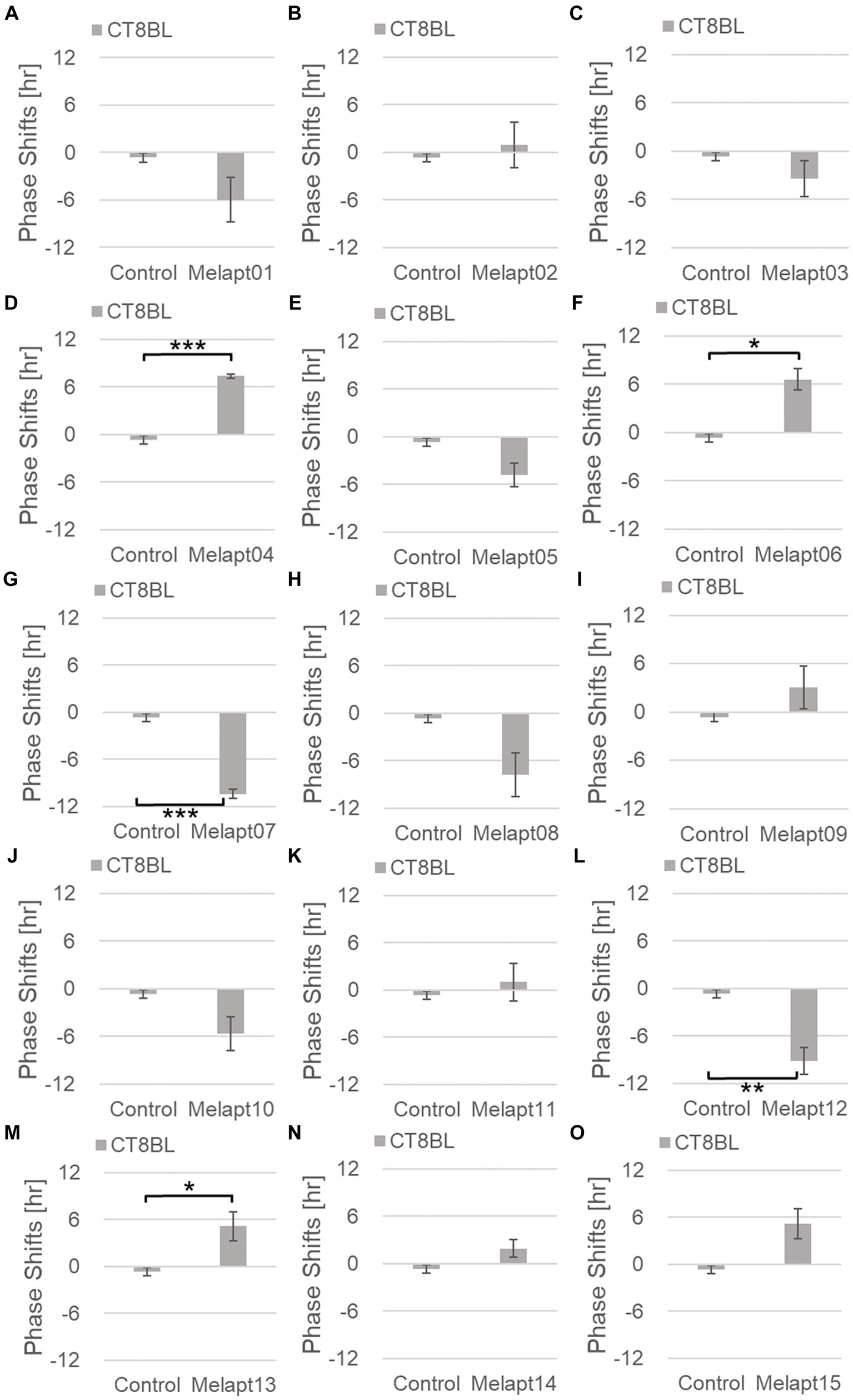
Figure 6. Phase shifts upon binding of Melapts plus photostimulation at subjective afternoon (CT8). (A–O) Phase-shifts comparison between Melapt-treated groups and controls with photo-stimulus at CT8. PBS alone was added to the controls. n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, Tukey–Kramer test.
Melapt with and without blue-light photostimulation at CT22 and CT8
At CT22, Melapt01, Melapt03, and Melapt04 plus photostimulation induced a phase advance, while Melapt10 and Melapt13 at CT22 induced a phase delay (Figures 7A, 8A). We picked up Melapt4 and Melapt10 as representatives of Melapts in group1, which induced a phase advance and phase delay of Per2:ELuc bioluminescence rhythms in the same direction at both CT22 and CT8 (Figure 7). Melapt1, Melapt3, and Melapt13 were selected as representatives of Melapts in group2, which induced those of Per2:ELuc rhythms in opposite direction at CT22 and CT8 (Figure 8), for example, phase advance at both CT22 and CT8 by Melapt4, while phase advance at CT22 and phase delay at CT8 by Melapt1.
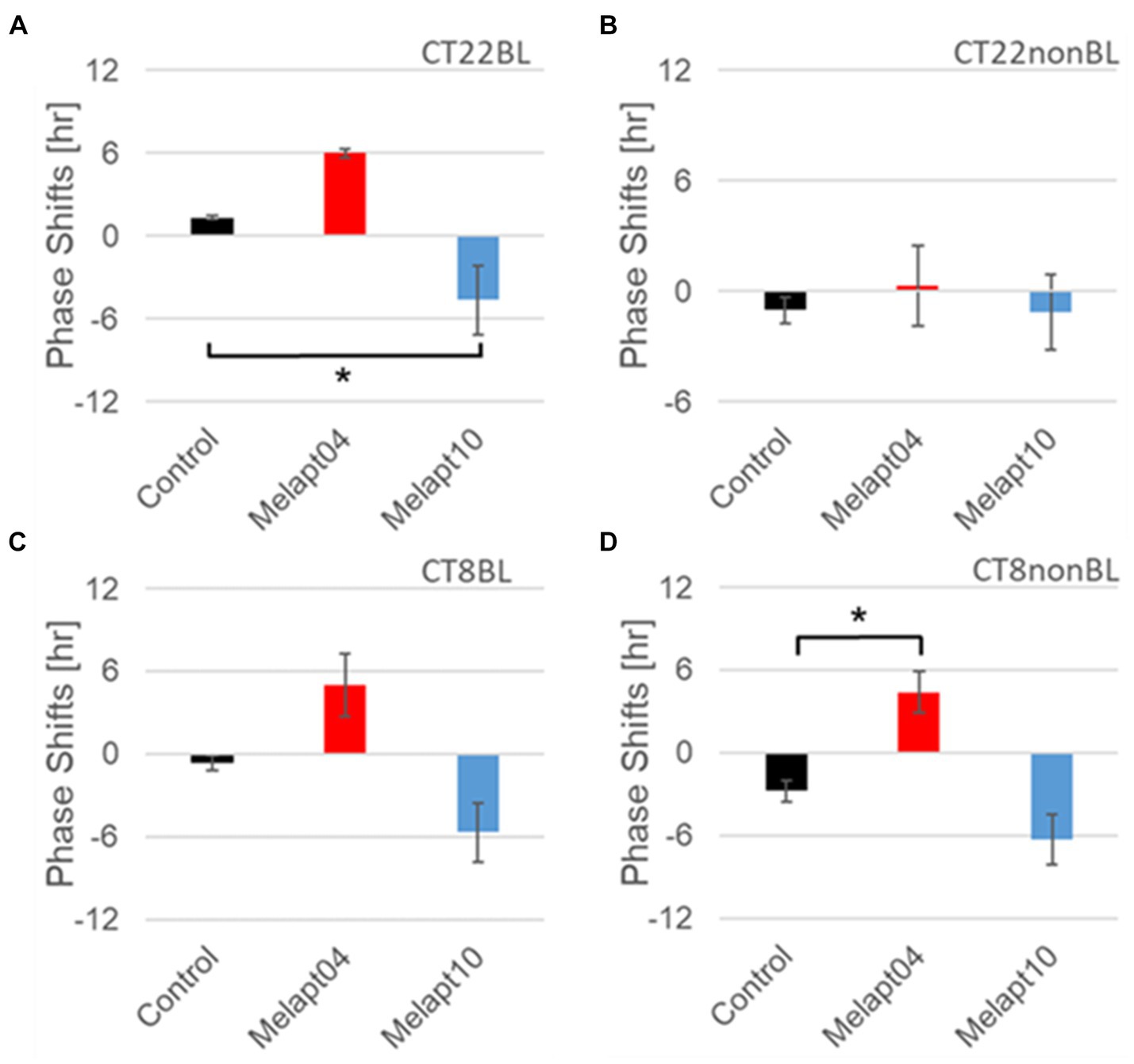
Figure 7. Similar phase-shift abilities of representative Melapts at both CT22 and CT8. (A) Melapt binding-mediated phase shift induced via photostimulation at subjective dawn (CT22BL). (B) Melapt-mediated phase shift at subjective dawn (CT22nonBL). (C) Melapt binding-mediated phase shifts induced via photostimulation in the afternoon (CT8BL). (D) Melapt-mediated phase shifts in the afternoon (CT8nonBL). Controls showed phase shifts in the absence of Melapts under all conditions. The upper direction shows phase advance, whereas the lower direction shows phase delay. Red: phase advance. Blue: phase delay. *p < 0.05, Tukey–Kramer test.
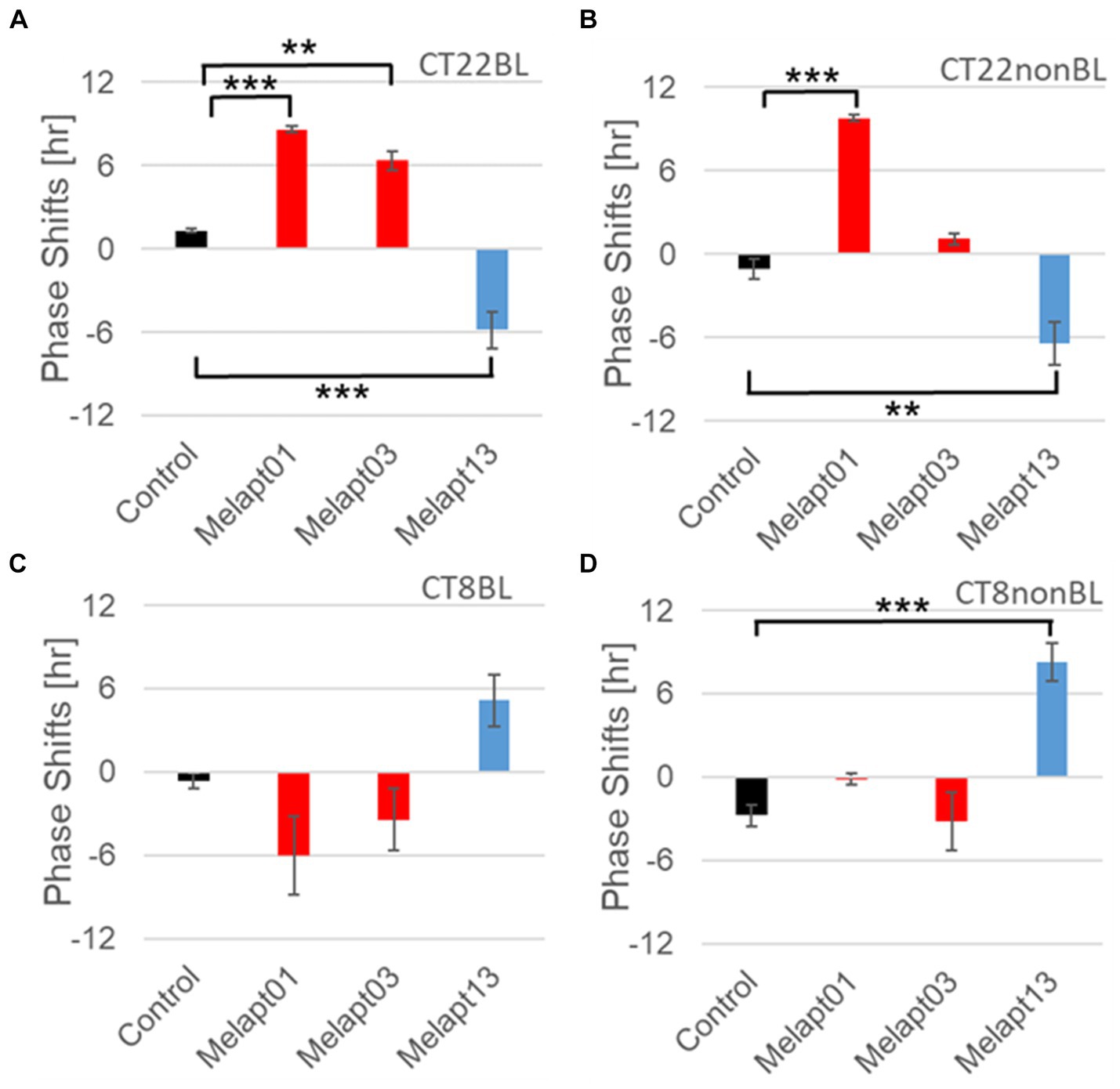
Figure 8. Reverse-phase shift abilities of representative Melapts at CT22 and CT8. (A) Melapt-mediated phase shift induced by photostimulation at subjective dawn (CT22BL). (B) Melapt-mediated phase shift at subjective dawn (CT22nonBL). (C) Melapt-mediated phase shift induced by photostimulation in the afternoon (CT8BL). (D) Melapt-mediated phase shift in the afternoon (CT8nonBL). Controls were phase shifts without any Melapts under all conditions. The upper direction shows phase advance, whereas the lower direction shows phase delay. Red: phase advance. Blue: phase delay. **p < 0.01, ***p < 0.001, Tukey–Kramer Test.
Although these phase shifts could be observed even without photostimulation when adding Melapts, Melapt04 and Melapt10 enhanced the phase shift compared with controls without photostimulation at CT22 (Figures 7B, 8B).
The phase-shift abilities of Melapts without photostimulation were evaluated (Figures 7B, 8B).
Melapts plus intense photostimulation in the subjective afternoon (CT8) caused phase shifts of Per2 emission rhythms in the same direction (advance or delay) as at dawn (CT22; Figures 7A,C). In addition, at CT22, the degree of phase shift was changed more than at CT8 in both advanced and delayed directions. For Melapt01 and Melapt13, a more significant phase advance or delay at CT22 was observed in the absence of photostimulation. By contrast, for Melapt03, Melapt04, Melapt10, and Melapt13, a more significant phase advance or delay at CT8 was observed in the absence of photostimulation.
Furthermore, Melapt01 and Melapt03 advanced the phase at CT22, and Melapt13 delayed the Phase, while the opposite phase shifts were observed at CT8 (Figures 8A,C). Like Melapt04, Melapt01 and Melapt03 induced expression of Per2 by binding to melanopsin at CT22, probably resulting in a phase advance, whereas they caused a phase delay at CT8.
Melapt03 induced a phase advance and delay at CT22 and CT8, while Melapt04 induced a phase advance at both CT22 and CT8. Nevertheless, the phase shifts of Melapt03 and Melapt04 without blue-light photostimulation were less than those with photostimulation at CT22, when Per2 transcription was slightly upregulated (Figures 7A,B, 8A,B). However, phase shifts of Melapt03 and Melapt04 were the same with and without blue-light photostimulation at CT8, when Per2 transcription was shown to be high (Figures 7A,B, 8A,B). Melapt04 and Melapt10 displayed little difference in phase-shift ability between CT22 and CT8 upon photostimulation (Figures 7A,C).
Functional analysis of Melapts in Per1::Luc Tg mice
We performed in vivo experiments similar to the in vitro experiments to investigate whether Melapts binding to melanopsin in the retina projecting to the SCN affected the phase shifts of the central clock in the SCN. Eight types of Melapt causing phase-shift responses in Per2 expression rhythms in in vitro experiments were injected into the bulbs of eyes of Per1::luc Tg mice at CT22 (Figure 9; Tables 2, 3; Supplementary Figure S3). Melapt01, Melapt03, Melapt04, Melapt07, Melapt09, and Melapt10 displayed similar phase-shift abilities in the in vivo and in vitro experiments. Figures 4, 9 show that Melapt01, Melapt03, and Melapt04 induce phase advance and that Melapt07, Melapt09, and Melapt10 induce phase delay in CT22 both in vivo and in vitro as Melapts effect on phase shift by in vivo experiments can be predicted from in vitro experiments. On the other hand, total phase shifts were limited to 3 h in intact animals, regardless of how much advance or delay by Melapts in Per2:Eluk:TK:Mel cells.
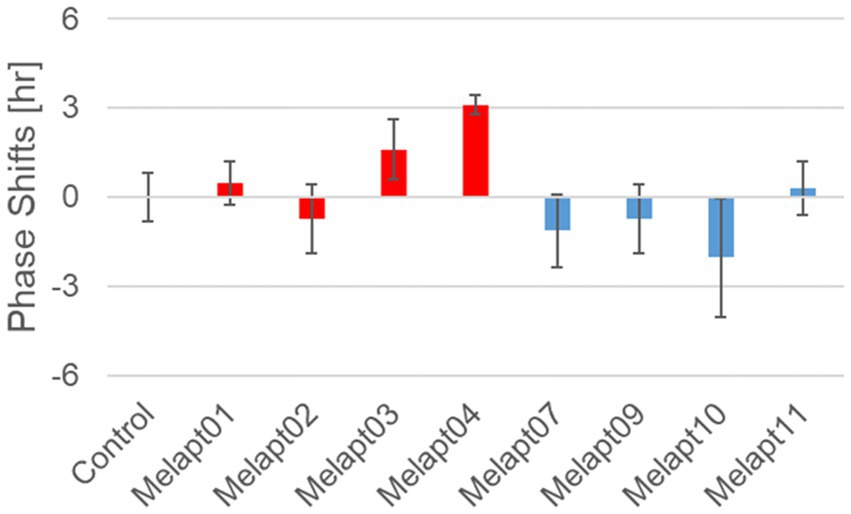
Figure 9. Phase shift of Per1::luc expressional rhythms in SCN slices following injection of Melapts into both bulbus oculi of Per1::luc Tg mice. Phase shifts following injection of Melapts into bulbus oculi with LED light stimulus at subjective dawn (CT22). SCN slices were obtained from mice injected with Melapts, and bioluminescence was observed for approximately 5 days. The NINJA program was used to calculate phase shifts. The upper direction shows phase advance, whereas the lower direction shows phase delay. Red: phase advance. Blue: phase delay. n = 3, *p < 0.05, Tukey–Kramer test.
For example, Melapt04 caused a maximum phase advance of 3 h, and Melapt10 caused a maximum phase delay at 3 h, in experiments in Per2:ELuc:TK:Mel stable cells and in SCN of Per1::luc Tg mice similar to the restriction of phase shifts to a maximum of 3–4 h in mammals (Gillette and Mitchell, 2002; Guler et al., 2008; Iyer et al., 2014; Figure 9; Tables 2, 3).
In total, two/three out of the nine Melapts showed similar phase shifts at CT22 in vitro and in vivo in the absence of blue light (Tables 2, 3). This suggests that each Melapt induced a similar phase shift in both mice and Per2:ELuc:TK:Mel stable cells.
Discussion
Melapts were identified to affect the phase of the circadian rhythms
This study focused on controlling the ability of melanopsin to manipulate the phase resetting of circadian rhythms caused by blue-light input from the eyes using Melapts identified by screening and Per2:ELuc:TK:Mel stable cells for functional analysis of phase-shift ability in molecular clock rhythms. Because we used cells exhibiting stable expression of melanopsin via the TK promoter, whereas previous studies (Pulivarthy et al., 2007; Ukai et al., 2007) used cells transiently transfected with melanopsin, we were able to perform more quantitative functional analyses of the phase response to aptamer concentration and light intensity.
Among the 15 Melapts obtained by Cell-SELEX, two showed characteristic responses: Melapt04 caused a phase advance and Melapt10 caused a phase delay at both subjective dawn (CT22) and in the afternoon (CT8). At dawn (CT22), when transcript levels of clock gene Per2 began to increase, Melapt04, Melapt01, and Melapt03 increased signaling into cells to upregulate Per2 transcription and induce a phase advance, while Melapt10 and Melapt13 repressed intracellular signaling to downregulate Per2 expression and induce a phase delay. In the afternoon (CT8), Per2 transcription was almost at its peak. It appeared that Melapt04 and Melapt10 with photostimulation caused the same phase shift as at CT22 and that Melapt1, Melapt3, and Melapt13 with photostimulation caused the opposite phase shift to that at CT22. At CT8, the transition between stopping the upregulation of Per2 transcription and beginning its downregulation could account for the instability effect of the phase shift upon adding Melapt. Furthermore, we found that in both the presence/absence of blue light, the phase of the molecular clock can be shifted (either advanced or delayed) simply by the addition of a Melapt in contrast with previous reports (Takumi et al., 1998; Pulivarthy et al., 2007; Ukai et al., 2007).
Photostimulation by blue-light exposure of the retina of mice before dawn (CT22) resulted in a phase advance of activity rhythms, similar to previous reports in rats (Honma et al., 1985). However, adding a particular Melapt (01, 03, 04) at CT22 resulted in a more significant phase advance, whereas other Melapts (10 and 13) resulted in a phase delay.
At CT22 (subjective dawn), as Per2 expression levels were increasing, the melanopsin signal bound to Melapt01, Melapt03, and Melapt04 with photostimulation had a complementary effect and enhanced Per2 expression, although the detailed underlying mechanism is not known. The extent of the phase advance was higher than in controls only receiving photostimulation at the same time.
By contrast, among the phase delay group of Melapts, Melapt10 and Melapt13 at CT22 could be binding to melanopsin, leading to upregulation of Per2 expression and repression of signal transduction in cells, and induction of Per2 gene expression in cells. Therefore, the delay of PER2 protein accumulation in cells could lead to a phase delay of intracellular circadian rhythms.
Melapts binding to melanopsin could directly or indirectly modify the structure of the domain in melanopsin that binds to the retinal, altering the phase shifts. Melanopsin loses its photosensitivity when its structure, function, and retinoid cycle are altered (Dua et al., 2011; Harrison et al., 2021). For example, cis-retinal is present in cells bound to melanopsin without post-stimulation, which mediates photostimulation signal transduction into cells, as Melapts bound to melanopsin may influence intracellular signaling and phase shifts without photostimulation.
The secondary structure of Melapts was assessed (not shown) using GENETYX Ver. 9 (GENETYX CORPORATION). Melapt02 and Melapt04 in the phase advance group have an outwardly protruding C-A-G-A-G sequence within the secondary structure of the stem and loop. The stem–loop conformation may enhance the binding of Melapts to melanopsin. However, it is unclear whether Melapts directly or indirectly alter their binding domain to affect signal transfer or their ability to phase-shift circadian rhythms.
Both per1 and per2 are clock genes involved in an input pathway, and their transcription is induced transiently by photosignals, resulting in a phase shift in rhythmic transcription (Shigeyoshi et al., 1997; Yan et al., 1999; Ikegami et al., 2020). Therefore, the phase shift of per1::luc and per2::luc rhythm induced by Melapts is synchronized. We confirmed whether the aptamers obtained by Selex screening and functionally analyzed using a per2::luc reporter cells in vitro could trigger the same phase shift in the SCN of per1::luc Tg mice in vivo. We identified an agonist DNA aptamer that caused a maximum 8-h shift in vitro and a 3-h shift in vivo. There was a 3-h phase-shift limit in vivo because the photic input signal by melanopsin is transferred to the SCN projected from retinal ganglion cells by synaptic transmission (Hanna et al., 2017).
The 11 cis-retinal bound to melanopsin is isomerized to trans-retinal using blue light (485 nm), causing its dissociation from melanopsin (Schwartz and Zimmerman, 1990). Compared with photostimulation, Melapts that induced a phase advance (Melapt03 and Melapt04) and a phase delay (Melapt10) displayed lower phase-shift abilities, possibly because most retinal molecules remained in the cis isomeric form and therefore did not dissociate from melanopsin and transmit signals into cells, thereby repressing the phase shift. This signal of melanopsin is transmitted inside the cell via G protein-coupled receptors, eventually upregulating transcription of the clock gene Per2 in cells (Iyer et al., 2014; Krzysztynska-Kuleta et al., 2021), resulting in a phase shift of circadian rhythms. Further studies are needed to understand how the downregulation/upregulation of Per2 upon Melapt–melanopsin binding induces phase shifts of circadian rhythms via CREB phosphorylation.
Future research on Melapt based regulation of the intracellular clock
Melapt04 and Melapt10 induced phase advance or delay of the circadian clock by ~3 h, respectively, at both CT22 and CT8 during the photo-signal input process. This suggests that Melapt04 regulates the phase of circadian rhythms, and facilitates falling asleep and waking, mainly via Phase Advance. Considering the social desire to advance the phase of the circadian clock to facilitate going to bed earlier at night and waking up earlier in the morning, Melapts could be used to trigger phase advance in humans. We chose to use DNA aptamers because they can be synthesized easily by PCR and are stable in the bloodstream. However, delivering aptamers to the retina is very difficult in humans (Hosoya and Tomi, 2005; Del Amo et al., 2017; Ikegami et al., 2020; Ham et al., 2023); here, we injected aptamers intraocularly into mice. Therefore, we need to develop a method for introducing aptamers efficiently and stably into the retina, possibly using a cell-permeable drug delivery with functions like ribosomes administered via drops into the eye (Wang et al., 2010; An et al., 2019; Yan et al., 2019; Tawfik et al., 2022).
Melapts could be used to regulate the function of membrane proteins by inducing conformational changes, thereby allowing more complex functional switching of the target (i.e., melanopsin) in the presence or absence of external photo-stimuli (Faria and Ulrich, 2002; Ulrich, 2005; Wang et al., 2010; Jones et al., 2013). However, the functional stability of Melapt is uncertain because the 3D structure adopted by the Melapt aptamer when bound to melanopsin is difficult to ascertain. In addition, it can be difficult for DNA aptamers to reach the retina, especially if injected into the eye bulb intravenously (Kwak and D'Amico, 1992; Hosoya and Tomi, 2005; Repetto et al., 2010; Patane et al., 2011); also, the stability of aptamers in blood and other body fluids will need to be assessed (Ma et al., 2021). Despite these limitations, Melapts could contribute to future research focused on resetting the phase of circadian clocks. Melapts could help us to better adapt to modern social life cycles, allow crops and domestic animals to be improved for greater productivity, and help shift workers in overcoming social jet lag by adjusting the phases of the circadian clock (Davidson et al., 2006; Saksvik et al., 2011; Barclay et al., 2012; Vetter et al., 2015; Li et al., 2021). These Melapts could contribute to resetting the phase of circadian clocks in photic input pathways.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, AF147789.1.
Ethics statement
The animal studies were approved by Animal care and use committee of Toyohashi Tech University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
RN and KN conducted the experiments, analyzed the data, performed the statistical analyses, prepared the figures, and contributed to the manuscript writing. MM assisted the experiments. YN established the photo-responsible cell line, while YK and YN conducted experiments with these cells. We are grateful to YK and YN for their supervision and helpful discussion. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by research funding from TechnoPro, Inc. TechnoPro R&D Company and the Program to Foster Young Researchers in Cutting-Edge Interdisciplinary Research (to RN). Funding for Kiban Scientists (to RN 24590350, 20H00614) was obtained from the Japan Society for the Promotion of Science (JSPS), the Mitsubishi Science Foundation (to RN), and a Research Grant for Science & Technology Innovation at Toyohashi University of Technology (to RN). This study was also supported by the Ministry of Education, Culture, Sports, Science, and Technology in Japan (to YN 21H02083). KN was supported by a scheme for employees to obtain a Ph.D. from TechnoPro, Inc. TechnoPro R&D Company (0007077240 to KN). TechnoPro, Inc was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Acknowledgments
We thank H. Tei, Y. Sakaki, S. Yamazaki, and H. Ueda for providing plasmid DNA and Tg mice. We also thank S. Umekage, S. Yamashita and N. Danno for helpful discussion and technical assistance, and N. Kimura and A. Ohtake (Toyohashi University of Technology) for animal care. We are grateful to T. Ohbayashi, Y. Kazuki, and M. Oshimura (Tottori University) for providing A9 cells harboring the MI-MAC vector and recombinase expression plasmids. We acknowledge the excellent technical assistance from T. Iwaki (Advanced Industrial Science and Technology).
Conflict of interest
KN was employed by company TechnoPro, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2024.1186677/full#supplementary-material
References
Albrecht, U., Sun, Z. S., Eichele, G., and Lee, C. C. (1997). A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell 91, 1055–1064. doi: 10.1016/s0092-8674(00)80495-x
An, Y., Yan, H., Li, X., Li, Z., Duan, J., and Yang, X.-D. (2019). Selection of a novel DNA aptamer against OFA/iLRP for targeted delivery of doxorubicin to AML cells. Sci. Rep. 9:7343. doi: 10.1038/s41598-019-43910-3
Antoch, M. P., Song, E. J., Chang, A. M., Vitaterna, M. H., Zhao, Y., Wilsbacher, L. D., et al. (1997). Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell 89, 655–667. doi: 10.1016/s0092-8674(00)80246-9
Barclay, J. L., Husse, J., Bode, B., Naujokat, N., Meyer-Kovac, J., Schmid, S. M., et al. (2012). Circadian desynchrony promotes metabolic disruption in a mouse model of shiftwork. PLoS One 7:e37150. doi: 10.1371/journal.pone.0037150
Bunger, M. K., Wilsbacher, L. D., Moran, S. M., Clendenin, C., Radcliffe, L. A., Hogenesch, J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. doi: 10.1016/s0092-8674(00)00205-1
Davidson, A. J., Sellix, M. T., Daniel, J., Yamazaki, S., Menaker, M., and Block, G. D. (2006). Chronic jet-lag increases mortality in aged mice. Curr. Biol. 16, R914–R916. doi: 10.1016/j.cub.2006.09.058
Davidson, A. J., Yamazaki, S., and Menaker, M. (2003). SCN: ringmaster of the circadian circus or conductor of the circadian orchestra? Novartis Found. Symp. 253:110. doi: 10.1002/0470090839.ch9
Del Amo, E. M., Rimpelä, A.-K., Heikkinen, E., Kari, O. K., Ramsay, E., Lajunen, T., et al. (2017). Pharmacokinetic aspects of retinal drug delivery. Prog. Retin. Eye Res. 57, 134–185. doi: 10.1016/j.preteyeres.2016.12.001
Dua, P., Kim, S., and Lee, D. K. (2011). Nucleic acid aptamers targeting cell-surface proteins. Methods 54, 215–225. doi: 10.1016/j.ymeth.2011.02.002
Ellington, A. D., and Szostak, J. W. (1990). In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822. doi: 10.1038/346818a0
Faria, M., and Ulrich, H. (2002). The use of synthetic oligonucleotides as protein inhibitors and anticode drugs in cancer therapy: accomplishments and limitations. Curr. Cancer Drug Targets 2, 355–368. doi: 10.2174/1568009023333827
Gentry, N. W., Ashbrook, L. H., Fu, Y. H., and Ptacek, L. J. (2021). Human circadian variations. J. Clin. Invest. 131:16. doi: 10.1172/JCI148282
Giebultowicz, J. (2004). Chronobiology: biological timekeeping. Integr. Comp. Biol. 44:266. doi: 10.1093/icb/44.3.266
Gillette, M. U., and Mitchell, J. W. (2002). Signaling in the suprachiasmatic nucleus: selectively responsive and integrative. Cell Tissue Res. 309, 99–107. doi: 10.1007/s00441-002-0576-1
Guler, A. D., Ecker, J. L., Lall, G. S., Haq, S., Altimus, C. M., Liao, H. W., et al. (2008). Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature 453, 102–105. doi: 10.1038/nature06829
Ham, Y., Mehta, H., Kang-Mieler, J., Mieler, W. F., and Chang, A. (2023). Novel drug delivery methods and approaches for the treatment of retinal diseases. Asia Pac J Ophthalmol (Phila). 12, 402–413. doi: 10.1097/APO.0000000000000623
Hanna, L., Walmsley, L., Pienaar, A., Howarth, M., and Brown, T. M. (2017). Geniculohypothalamic GABAergic projections gate suprachiasmatic nucleus responses to retinal input. J. Physiol. 595, 3621–3649. doi: 10.1113/JP273850
Harrison, K. R., Reifler, A. N., Chervenak, A. P., and Wong, K. Y. (2021). Prolonged Melanopsin-based Photoresponses depend in part on RPE65 and cellular Retinaldehyde-binding protein (CRALBP). Curr. Eye Res. 46, 515–523. doi: 10.1080/02713683.2020.1815793
Hattar, S., Lucas, R. J., Mrosovsky, N., Thompson, S., Douglas, R. H., Hankins, M. W., et al. (2003). Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature 424, 76–81. doi: 10.1038/nature01761
Hida, A., Koike, N., Hirose, M., Hattori, M., Sakaki, Y., and Tei, H. (2000). The human and mouse Period1 genes: five well-conserved E-boxes additively contribute to the enhancement of mPer1 transcription. Genomics 65, 224–233. doi: 10.1006/geno.2000.6166
Honma, K., Honma, S., and Hiroshige, T. (1985). Response curve, free-running period, and activity time in circadian locomotor rhythm of rats. Jpn. J. Physiol. 35, 643–658. doi: 10.2170/jjphysiol.35.643
Hosoya, K.-i., and Tomi, M. (2005). Advances in the cell biology of transport via the inner blood-retinal barrier: establishment of cell lines and transport functions. Biol. Pharm. Bull. 28, 1–8. doi: 10.1248/bpb.28.1
Ikegami, K., Nakajima, M., Minami, Y., Nagano, M., Masubuchi, S., and Shigeyoshi, Y. (2020). cAMP response element induces Per1 in vivo. Biochem. Biophys. Res. Commun. 531, 515–521. doi: 10.1016/j.bbrc.2020.07.105
Iyer, R., Wang, T. A., and Gillette, M. U. (2014). Circadian gating of neuronal functionality: a basis for iterative metaplasticity. Front. Syst. Neurosci. 8:164. doi: 10.3389/fnsys.2014.00164
Jones, K. A., Hatori, M., Mure, L. S., Bramley, J. R., Artymyshyn, R., Hong, S. P., et al. (2013). Small-molecule antagonists of melanopsin-mediated phototransduction. Nat. Chem. Biol. 9, 630–635. doi: 10.1038/nchembio.1333
King, D. P., Zhao, Y., Sangoram, A. M., Wilsbacher, L. D., Tanaka, M., Antoch, M. P., et al. (1997). Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. doi: 10.1016/s0092-8674(00)80245-7
Krzysztynska-Kuleta, O. I., Olchawa, M. M., and Sarna, T. J. (2021). Melanopsin signaling pathway in HEK293 cell line with stable expression of human Melanopsin: possible participation of phospholipase C beta 4 and diacylglycerol. Photochem. Photobiol. 97, 1136–1144. doi: 10.1111/php.13453
Kume, K., Zylka, M. J., Sriram, S., Shearman, L. P., Weaver, D. R., Jin, X., et al. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. doi: 10.1016/s0092-8674(00)81014-4
Kwak, H. W., and D'Amico, D. J. (1992). Evaluation of the retinal toxicity and pharmacokinetics of dexamethasone after intravitreal injection. Arch. Ophthalmol. 110, 259–266. doi: 10.1001/archopht.1992.01080140115038
LeGates, T. A., Altimus, C. M., Wang, H., Lee, H. K., Yang, S., Zhao, H., et al. (2012). Aberrant light directly impairs mood and learning through melanopsin-expressing neurons. Nature 491, 594–598. doi: 10.1038/nature11673
Li, H., Li, K., Zhang, K., Li, Y., Haotian, G., Liu, H., et al. (2021). The circadian physiology: implications in livestock health. Int. J. Mol. Sci. 22:2111. doi: 10.3390/ijms22042111
Lucas, R. J., Hattar, S., Takao, M., Berson, D. M., Foster, R. G., and Yau, K. W. (2003). Diminished pupillary light reflex at high irradiances in melanopsin-knockout mice. Science 299, 245–247. doi: 10.1126/science.1077293
Ma, W., Zhan, Y., Zhang, Y., Mao, C., Xie, X., and Lin, Y. (2021). The biological applications of DNA nanomaterials: current challenges and future directions. Signal Transduct. Target. Ther. 6:351. doi: 10.1038/s41392-021-00727-9
Menaker, M. (2003). Circadian rhythms. Circadian photoreception. Sci. 299, 213–214. doi: 10.1126/science.1081112
Nakajima, Y., Yamazaki, T., Nishii, S., Noguchi, T., Hoshino, H., Niwa, K., et al. (2010). Enhanced beetle luciferase for high-resolution bioluminescence imaging. PLoS One 5:e10011. doi: 10.1371/journal.pone.0010011
Numano, R., Yamazaki, S., Umeda, N., Samura, T., Sujino, M., Takahashi, R., et al. (2006). Constitutive expression of the Period1 gene impairs behavioral and molecular circadian rhythms. Proc. Natl. Acad. Sci. USA 103, 3716–3721. doi: 10.1073/pnas.0600060103
Patane, M. A., Cohen, A., From, S., Torkildsen, G., Welch, D., and Ousler 3rd, G. W. (2011). Ocular iontophoresis of EGP-437 (dexamethasone phosphate) in dry eye patients: results of a randomized clinical trial. Clin. Ophthalmol. 5, 633–643. doi: 10.2147/OPTH.S19349
Pittendrigh, C. S. (1993). Temporal organization: reflections of a Darwinian clock-watcher. Annu. Rev. Physiol. 55, 16–54. doi: 10.1146/annurev.ph.55.030193.000313
Pulivarthy, S. R., Tanaka, N., Welsh, D. K., De Haro, L., Verma, I. M., and Panda, S. (2007). Reciprocity between phase shifts and amplitude changes in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 104, 20356–20361. doi: 10.1073/pnas.0708877104
Ralph, M. R., Foster, R. G., Davis, F. C., and Menaker, M. (1990). Transplanted suprachiasmatic nucleus determines circadian period. Science 247, 975–978. doi: 10.1126/science.2305266
Repetto, R., Siggers, J. H., and Stocchino, A. (2010). Mathematical model of flow in the vitreous humor induced by saccadic eye rotations: effect of geometry. Biomech. Model. Mechanobiol. 9, 65–76. doi: 10.1007/s10237-009-0159-0
Saksvik, I. B., Bjorvatn, B., Hetland, H., Sandal, G. M., and Pallesen, S. (2011). Individual differences in tolerance to shift work--a systematic review. Sleep Med. Rev. 15, 221–235. doi: 10.1016/j.smrv.2010.07.002
Schwartz, W. J., and Zimmerman, P. (1990). Circadian timekeeping in BALB/c and C57BL/6 inbred mouse strains. J. Neurosci. 10, 3685–3694. doi: 10.1523/JNEUROSCI.10-11-03685.1990
Shearman, L. P., Zylka, M. J., Weaver, D. R., Kolakowski, L. F. Jr., and Reppert, S. M. (1997). Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269. doi: 10.1016/s0896-6273(00)80417-1
Shigeyoshi, Y., Taguchi, K., Yamamoto, S., Takekida, S., Yan, L., Tei, H., et al. (1997). Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053. doi: 10.1016/s0092-8674(00)80494-8
Sun, Z. S., Albrecht, U., Zhuchenko, O., Bailey, J., Eichele, G., and Lee, C. C. (1997). RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003–1011. doi: 10.1016/s0092-8674(00)80366-9
Tabei, Y., Murotomi, K., Umeno, A., Horie, M., Tsujino, Y., Masutani, B., et al. (2017). Antioxidant properties of 5-hydroxy-4-phenyl-butenolide via activation of Nrf2/ARE signaling pathway. Food Chem. Toxicol.: Int. J. Published British Indus. Biolog. Res. Assoc. 107, 129–137. doi: 10.1016/j.fct.2017.06.039
Takahashi, J. S. (1995). Molecular neurobiology and genetics of circadian rhythms in mammals. Annu. Rev. Neurosci. 18, 531–553. doi: 10.1146/annurev.ne.18.030195.002531
Takahashi, J. S., Hong, H. K., Ko, C. H., and McDearmon, E. L. (2008). The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. doi: 10.1038/nrg2430
Takiguchi, M., Kazuki, Y., Hiramatsu, K., Abe, S., Iida, Y., Takehara, S., et al. (2014). A novel and stable mouse artificial chromosome vector. ACS Synth. Biol. 3, 903–914. doi: 10.1021/sb3000723
Takumi, T., Taguchi, K., Miyake, S., Sakakida, Y., Takashima, N., Matsubara, C., et al. (1998). A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 17, 4753–4759. doi: 10.1093/emboj/17.16.4753
Tawfik, M., Chen, F., Goldberg, J. L., and Sabel, B. A. (2022). Nanomedicine and drug delivery to the retina: current status and implications for gene therapy. Naunyn Schmiedeberg's Arch. Pharmacol. 395, 1477–1507. doi: 10.1007/s00210-022-02287-3
Tei, H., Okamura, H., Shigeyoshi, Y., Fukuhara, C., Ozawa, R., Hirose, M., et al. (1997). Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512–516. doi: 10.1038/39086
Tuerk, C., and Gold, L. (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249, 505–510. doi: 10.1126/science.2200121
Ukai, H., Kobayashi, T. J., Nagano, M., Masumoto, K. H., Sujino, M., Kondo, T., et al. (2007). Melanopsin-dependent photo-perturbation reveals desynchronization underlying the singularity of mammalian circadian clocks. Nat. Cell Biol. 9, 1327–1334. doi: 10.1038/ncb1653
Ulrich, H. (2005). DNA and RNA aptamers as modulators of protein function. Med. Chem. 1, 199–208. doi: 10.2174/1573406053175274
Umekage, S., and Kikuchi, Y. (2006). Production of circular form of streptavidin RNA aptamer in vitro. Nucleic Acids Symp Ser (Oxf). 50, 323–324. doi: 10.1093/nass/nrl161
Vetter, C., Fischer, D., Matera, J. L., and Roenneberg, T. (2015). Aligning work and circadian time in shift workers improves sleep and reduces circadian disruption. Curr. Biol. 25, 907–911. doi: 10.1016/j.cub.2015.01.064
Wang, H., Zhao, P., Wenya, S., Wang, S., Liao, Z., Niu, R., et al. (2010). PLGA/polymeric liposome for targeted drug and gene co-delivery. Biomaterials 31, 8741–8748. doi: 10.1016/j.biomaterials.2010.07.082
Yamaguchi, S., Kazuki, Y., Nakayama, Y., Nanba, E., Oshimura, M., and Ohbayashi, T. (2011). A method for producing transgenic cells using a multi-integrase system on a human artificial chromosome vector. PLoS One 6:e17267. doi: 10.1371/journal.pone.0017267
Yamazaki, S., Numano, R., Abe, M., Hida, A., Takahashi, R., Ueda, M., et al. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685. doi: 10.1126/science.288.5466.682
Yan, G., Chen, X., Wang, Y., Zhou, X., Fang, L., and Cao, F. (2019). Multifunctional nanocomposites based on liposomes and layered double hydroxides conjugated with Glycylsarcosine for efficient topical drug delivery to the posterior segment of the eye. Mol. Pharm. 16, 2845–2857. doi: 10.1021/acs.molpharmaceut.8b01136
Yan, L., and Silver, R. (2004). Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur. J. Neurosci. 19, 1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x
Yan, L., Takekida, S., Shigeyoshi, Y., and Okamura, H. (1999). Per1 and Per2 gene expression in the rat suprachiasmatic nucleus: circadian profile and the compartment-specific response to light. Neuroscience 94, 141–150. doi: 10.1016/s0306-4522(99)00223-7
Yoo, S. H., Yamazaki, S., Lowrey, P. L., Shimomura, K., Ko, C. H., Buhr, E. D., et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 101, 5339–5346. doi: 10.1073/pnas.0308709101
Keywords: melanopsin (OPN4), DNA aptamer, Melapt, circadian rhythm, molecular clock, phase shift, period gene
Citation: Nakazawa K, Matsuo M, Kikuchi Y, Nakajima Y and Numano R (2024) Melanopsin DNA aptamers can regulate input signals of mammalian circadian rhythms by altering the phase of the molecular clock. Front. Neurosci. 18:1186677. doi: 10.3389/fnins.2024.1186677
Edited by:
Takahiro J. Nakamura, Meiji University, JapanReviewed by:
Atomu Sawatari, The University of Sydney, AustraliaShigenori Nonaka, Graduate University for Advanced Studies (Sokendai), Japan
Copyright © 2024 Nakazawa, Matsuo, Kikuchi, Nakajima and Numano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rika Numano, bnVtYW5vQGVpaXJpcy50dXQuYWMuanA=
 Kazuo Nakazawa
Kazuo Nakazawa Minako Matsuo3
Minako Matsuo3 Yoshihiro Nakajima
Yoshihiro Nakajima Rika Numano
Rika Numano