
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 20 December 2023
Sec. Sleep and Circadian Rhythms
Volume 17 - 2023 | https://doi.org/10.3389/fnins.2023.1323565
This article is part of the Research Topic Signaling Pathways and Brain Circuitry Underlying Circadian Rhythms and Sleep View all 7 articles
The suprachiasmatic nucleus (SCN) of the hypothalamus is the master circadian clock in mammals. SCN neurons exhibit circadian Ca2+ rhythms in the cytosol, which is thought to act as a messenger linking the transcriptional/translational feedback loop (TTFL) and physiological activities. Transcriptional regulation occurs in the nucleus in the TTFL model, and Ca2+-dependent kinase regulates the clock gene transcription. However, the Ca2+ regulatory mechanisms between cytosol and nucleus as well as the ionic origin of Ca2+ rhythms remain unclear. In the present study, we monitored circadian-timescale Ca2+ dynamics in the nucleus and cytosol of SCN neurons at the single-cell and network levels. We observed robust nuclear Ca2+ rhythm in the same phase as the cytosolic rhythm in single SCN neurons and entire regions. Neuronal firing inhibition reduced the amplitude of both nuclear and cytosolic Ca2+ rhythms, whereas blocking of Ca2+ release from the endoplasmic reticulum (ER) via ryanodine and inositol 1,4,5-trisphosphate (IP3) receptors had a minor effect on either Ca2+ rhythms. We conclude that the in-phasic circadian Ca2+ rhythms in the cytosol and nucleus are mainly driven by Ca2+ influx from the extracellular space, likely through the nuclear pore. It also raises the possibility that nuclear Ca2+ rhythms directly regulate transcription in situ.
Almost all living organisms on Earth are regulated by a circadian clock, which anticipates environmental changes for their own survival. In mammals, the master circadian pacemaker is located in the suprachiasmatic nucleus (SCN) of the hypothalamus in the brain (Moore and Eichler, 1972; Stephan and Zucker, 1972), which controls various physiological functions (e.g., sleep–wakefulness cycles, hormone secretion) and animal behavior (Welsh et al., 2010; Mohawk et al., 2012). Rodent SCN is composed of approximately 20,000 neurons in the bilateral side of the hypothalamus, which express a wide range of neuropeptides and their receptors (Van den Pol, 1980; Wen et al., 2020) and constitute a hierarchical multi-oscillator system (Welsh et al., 2010). In individual SCN neurons, circadian rhythms are observed in clock gene transcription (Yoo et al., 2004), neuronal firing (Welsh et al., 1995), hormone secretion (e.g., vasoactive intestinal peptide) (Shinohara et al., 1995), and intracellular Ca2+ concentration (Colwell, 2000; Ikeda et al., 2003; Enoki et al., 2012a). The molecular background of the circadian rhythms of the biological rhythms is based on the transcriptional/translational feedback loop (TTFL), which is composed of clock genes and their products. In the main loop of this model, circadian locomotor output cycles kaput (CLOCK) and brain and muscle ARNT-like1 (BMAL1), as positive regulators, activate the transcription of the Period (Per) and Cryptochrome (Cry) genes via the E-boxes. The expressed CRYs and PERs form their own complex and translocate into the nucleus. Then, they act as negative regulators to repress their own expression and generate rhythms (Mohawk et al., 2012; Takahashi, 2017).
The PER2 expression rhythms were abolished when intracellular Ca2+ was chelated in SCN tissue culture (Lundkvist et al., 2005). Furthermore, the behavioral rhythm in Drosophila was prolonged when parvalbumin, a Ca2+-binding protein, was specifically expressed in clock neurons, suggesting that Ca2+ is an important component of the TTFL (Harrisingh et al., 2007). As circadian Ca2+ rhythms are attenuated by Bmal1 knockdown (Ikeda and Ikeda, 2014) and Cry1/2 knockout (Enoki et al., 2017b), Ca2+ and TTFL are likely to be mutually coupled. A hypothesized model of the molecular mechanism of TTFL regulation by the intracellular Ca2+ is the phosphorylation of CLOCK through the Ca2+/calmodulin-dependent protein kinase (CaMK) II (Kon et al., 2014). In the signaling pathway of light entrainment, glutamate input from the retina triggers Ca2+ signaling via NMDA receptors to activate CaMKII and initiate PER transcription via CREB-CRE signaling (Golombek and Rosenstein, 2010). TTFL may regulate Ca2+ influx by controlling membrane potential via circadian regulation of Na+ and K+ conductance (Hastings et al., 2018); however, tetrodotoxin (TTX), a Na+ channel blocker, does not suspend cytosolic circadian Ca2+ rhythms. It has been reported that Ca2+ release from ryanodine receptors on the endoplasmic reticulum (ER) contributes to cytosolic Ca2+ rhythm, but the results are either supportive (Ikeda et al., 2003; Plante et al., 2021), negative (Noguchi et al., 2017), or mixed (Aguilar-Roblero et al., 2016).
Despite decades of research on circadian Ca2+ rhythms showing the importance of Ca2+ in the TTLF model, the ionic origin of circadian Ca2+ rhythms remains to be elucidated. Ikeda et al. (2003) and Wood et al. (2001) previously examined nuclear and cytosolic Ca2+ dynamics in mouse SCN and in genetically modified tobacco seedlings, and they detected the Ca2+ rhythms in the cytosol but not in the nucleus. In general, the nuclear envelope has a continuous structure with the ER, and ryanodine and inositol 1,4,5-trisphosphate (IP3) receptors are expressed on the inner membrane of the nucleus (Bootman et al., 2009; Secondo et al., 2020). Based on these observations, it can be hypothesized that nuclear Ca2+ is regulated independently of the cytosol. However, it is difficult to assume the independence of nuclear Ca2+ on a circadian timescale as the nucleus has nuclear pore complexes through which molecules smaller than ∼40 kDa can passively diffuse (Lin and Hoelz, 2019) and the diffusion rate of Ca2+ ions in cells is orders of magnitude faster than the circadian timescale (Villarruel et al., 2021). To re-evaluate the mechanism of nuclear Ca2+ regulation in the SCN neuron on a circadian timescale, we performed dual-color Ca2+ imaging in the nucleus and cytosol using highly sensitive and genetically encoded Ca2+ sensors, GCaMP6s (Chen et al., 2013) and jRGECO1a (Dana et al., 2016). To further investigate the regulatory mechanisms of nuclear Ca2+, we administered drugs that inhibit Ca2+ influx via the ER and neuronal firing and also obtained results indicating a regulatory mechanism for nuclear and cytosolic Ca2+ concentration in the SCN neurons.
Adult female mice with newborn pups were purchased from an animal breeder (Japan SLC, Inc., Hamamatsu, Japan). The animals were housed and provided with food and water ad libitum under controlled conditions (temperature, 22°C ± 2°C; humidity, 40% ± 20%; 12-h light/12-h dark cycle, with lights on from 8:00 AM to 8:00 PM). Light intensity was adjusted to approximately 100–200 lx at the cage surface. The animals were fed commercial chow (Labo MR Standard; Nosan Corporation, Yokohama, Japan) and tap water. All animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee of the National Institutes of Natural Sciences and performed according to the National Institute for Physiological Sciences guidelines (approval no. Nos.22A044 and 23A080).
The brains of neonate mice (4–6 days old, both male and female) were rapidly removed and dipped in an ice-cold balanced salt solution comprising (in mM) 87 NaCl, 2.5 KCl, 8 MgCl2, 0.5 CaCl2, 1.2 NaH2PO4, 26 NaHCO3, 25 glucose, 10 HEPES, and 75 sucrose. A 200-μm coronal brain slice containing the mid-rostro-caudal region of the SCN was prepared using a vibratome (VT 1200; Leica Microsystems GmbH, Wetzlar, Germany). The bilateral SCNs were cut out from the slice using a surgical knife and explanted onto a culture membrane (Millicell CM; pore size, 0.4 μ m; Millipore; Merck KGaA, Darmstadt, Germany) in a 35-mm Petri dish containing 1.0-mL DMEM (Invitrogen; Thermo Fisher Scientific Inc.) and 5% FBS (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
The pAAV-hSyn-NLS-GCaMP6s-WPRE plasmid was constructed by VectorBuilder Inc. The AAV vectors were produced by a procedure as described previously (Sano et al., 2020). AAV1-Syn1-nes-jRGECO1a was purchased from Addgene (Dana et al., 2016). AAV aliquots (0.8–1.0 μL) were inoculated onto the surface of the SCN slices on days 3–4 of culture. The infected slice was cultured for a further 10–14 days before imaging. The titer of all AAV was over 1.0 × 1013 genome copies/mL.
For confocal recording, 3–4 days before the recordings, membranes with cultured SCN slices were cut out, flipped over, and transferred to glass-based dishes (3971–035 IWAKI; AGC TECHNO GLASS Co., Ltd., Yoshida, Japan), which were coated with collagen (Cellmatrix Type 1-C, Nitta Gelatin Inc., Yao, Japan). For wide-field recording, culture membrane inserts with cultured SCN slices were transferred to glass-based dishes. Then, the dishes were filled with DMEM (180–200 μL for confocal, 1.2 mL for wide-field) containing 5% FBS and sealed with O2-permeable filters (membrane kit, High Sens; YSI Inc.). The specimen was observed using a time-lapse imaging system as previously described (Enoki et al., 2012b). The system consisted of a Nipkow spinning disk confocal unit (X-Light; CrestOptics S.p.A., Roma, Italy), a sCMOS camera Neo (2,560 × 2,160 pixels, Andor Technology, Oxford Instruments, Belfast, UK), an EM-CCD camera Evolve (512 × 512 pixels, Teledyne Photometrics) or an EM-CCD camera iXon3 (1,024 × 1,024 pixels, Andor Technology), a TIXHB box incubator (Tokai Hit., Co, Ltd., Fujinomiya, Japan), and a Ti-E inverted microscope (Nikon Corporation, Tokyo, Japan). Confocal images were acquired with a 20× objective (NA0.75 PlanApo, Nikon Corporation) at 0.975-μm or 0.8-μm resolution. Wide-field images were acquired with a 10× objective (NA0.45 PlanApo, Nikon Corporation) at 2.6-μm resolution. For acquisition to GCaMP6s fluorescence, the specimen was illuminated with LED light (Light Engine; Lumencor) at cyan light (475/28 nm) and fluorescence was visualized with 495-nm dichroic mirrors and 520/35-nm emission filters. For jRGECO1a, 542/27-nm excitation light, 593-nm dichroic mirrors, and 630/92-nm emission filters were used. All experiments were performed at 36.5°C and 5% CO2. The intensity of the excitation light and the exposure time of the sCMOS and EM-CCD camera were adjusted to obtain optimal images of each sample.
Wide-field time-lapse imaging was performed for 4 days, the medium was switched to DMEM containing 0.1% DMSO or inhibitors [10 μM dantrolene (Supelco; Merck KGaA, Darmstadt, Germany), 100 μM ryanodine (Sigma-Aldrich), 1 μM TTX (Abcam plc., Cambridge, UK), 10 μM Xestospongin C (Abcam plc.)], and imaging was continued for another 4 days.
The SCN slices expressing NLS-GCaMP6s or NES-jRGECO1a were fixed with 4% paraformaldehyde in 0.1 M PBS for 60 min at room temperature. The slices were mounted on a glass bottom dish with ProLong™ Diamond Antifade Mountant with DAPI (Invitrogen). Fluorescence was visualized using a confocal microscope (TCS SP8, Leica Microsystems GmbH) equipped with 63 × objective lens (NA 1.20, HC PL APO, Leica Microsystems GmbH) with the following excitation/emission spectra: DAPI (405 nm/410–483 nm), NLS-GCaMP6s (488 nm/493–592 nm), and NES-jRGECO1a (587 nm/592–779 nm).
For image acquisition, MetaMorph (Molecular Devices, LLC) and LasX (Leica Microsystems GmbH) were used. Fiji (Wayne Rasband, National Institutes of Health) was used to analyze the obtained image data. The neuronal condition was monitored by bright-field imaging at the same time as fluorescence imaging, and the data in which significant cell death, the slice shape change, and the network desynchronization occurred during the time-lapse measurement were excluded from the analysis as they were assumed to be cytotoxic due to light irradiation. To analyze the circadian Ca2+ rhythms in the SCN network, we used a rhythm-fitting program (ImageJ Plugin). Time-lapse images were subjected to 7 × 7 or 8 × 8 binning for analyzing phase differences and 2 × 2 binning for other analysis, noise-filtered (median filter, rad = 1.0 pixel), and converted from 16- to 8-bit images. For each pixel data, the following cosine functions were fitted using the least-squares method in 48-h intervals.
where Yp (ti) denotes the signal intensity at time ti (h); Mp (ti), average fluorescence; Ap (ti), amplitude; Wp (ti), angular velocity; and Φp (ti), phase. To evaluate each fitting rate, an index of the goodness of fit of the fitted values to the measured data was calculated and an uncorrelated test was conducted at a significance level of p < 0.001 for making the phase-difference map and p < 0.00001 for another analysis, such as calculating the percentages of amplitude and trough changes. Only pixels that were rejected were extracted to generate images. In the phase map, phase differences are indicated by pseudo colors, and pixels rejected by the fitting test are shown in white. The acrophase map of the entire SCN region was normalized to zero and displayed.
Statistical analyses were conducted using Microsoft Excel and RStudio (RStudio, PBC). Paired or unpaired t-tests were used when comparing two dependent and independent group means.
To reveal the circadian-timescale Ca2+ dynamics in the nucleus and cytosol in the SCN neurons at the network and single-cell level, we expressed spectrally distinct and genetically encoded Ca2+ probes, GCaMP6s (green) and jRGECO1a (red), in the cultured SCN slices from mice. Nuclear localization signal-tagged GCaMP6s (NLS-GCaMP6s) and nuclear export signal-tagged jRGECO1a (NES-jRGECO1a) were expressed in the SCN neurons in slices using (AAV) transfection under the control of the neuron-specific promoter, human synapsin I. The expressions of NLS-GCaMP6s and NES-jRGECO1a were colocalized or exclusively localized with the nuclear marker (DAPI) at the single-cell level, and both expressions were widely detectable in the entire area, including the dorsal and ventral SCN subregions (Figures 1A,B). These results ensure the origin of the optical signals in the subsequent experiments.
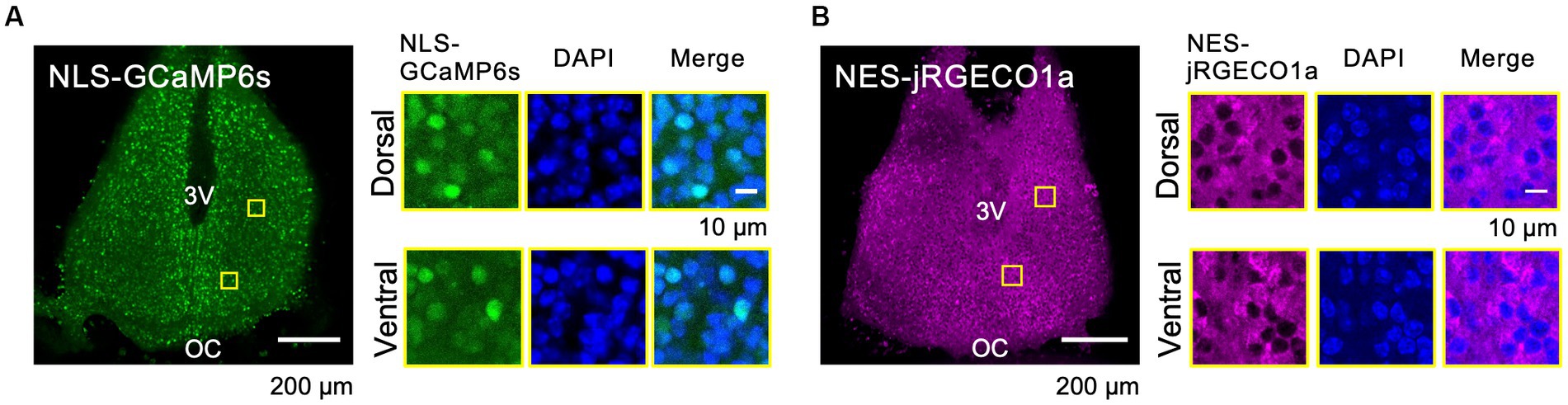
Figure 1. Expression Patterns of the Ca2+ Probes in the SCN Slices. (A–B) Expression patterns of the probes of NLS-GCaMP6s (A) and NES-jRGECO1a (B) in the whole SCN (left) and in dorsal and ventral SCN subregions (right). ROIs are indicated by yellow squares in the SCN images. Images are NLS-GCaMP6s or NES-jRGECO1a (left), DAPI (middle), and the merged one (right), respectively. 3 V, third ventricle; OC, optic chiasm.
Next, we performed time-lapse confocal Ca2+ imaging in the SCN slices expressing NLS-GCaMP6s and NES-jRGECO1a over a period of several days (Figure 2; Supplementary Video 1). We detected circadian Ca2+ rhythms in the cytosol and nucleus in entire SCN subregions (Figure 2B). The nuclear and cytosolic Ca2+ rhythms were in-phasic at the single-cell level in the dorsal and ventral SCN neurons (Figure 2C). Then, we constructed and compared acrophase maps (peak phase maps) to quantitatively analyze the topological patterns of the nuclear and cytosolic Ca2+ rhythms in the SCN network. We found that the spatial patterns of nuclear and cytosolic Ca2+ rhythms were almost identical in the entire SCN, with the dorsal subregion phase-leading the ventral subregion (Figure 2D). The phase-difference map (difference of two acrophase maps) confirmed that the two rhythms were in-phasic in the entire SCN regions (Figure 2E). This observation was held true for all the SCN slices examined (−0.19 ± 0.18 h, n = 5 slices) (Figure 2F). We further evaluated the phase difference of the two Ca2+ rhythms in the dorsal and ventral SCN subregions, normalized to the mean phase of the entire SCN, and found that the phase of nuclear Ca2+ rhythm was −1.35 ± 0.77 h in the dorsal and 1.30 ± 1.01 h in the ventral subregions and that of cytosolic Ca2+ rhythm was −1.16 ± 0.41 h in the dorsal and 1.22 ± 1.19 h in the ventral subregions, respectively (Supplementary Figure S1C). Significant differences were observed between the dorsal and ventral subregions in both nuclear and cytosolic Ca2+ rhythms (p = 0.0013 in the nucleus, p = 0.0012 in the cytosol) but not between the nuclear and cytosolic Ca2+ rhythms within subregions (p = 0.26 in the dorsal region, p = 0.33 in the ventral region). Single-cell analysis further confirmed our observation that the cytosolic and nuclear Ca2+ rhythms were in-phase in individual SCN neurons (0.08 ± 0.33 h, n = 30 cells in 5 slices) (Supplementary Figure S1D).
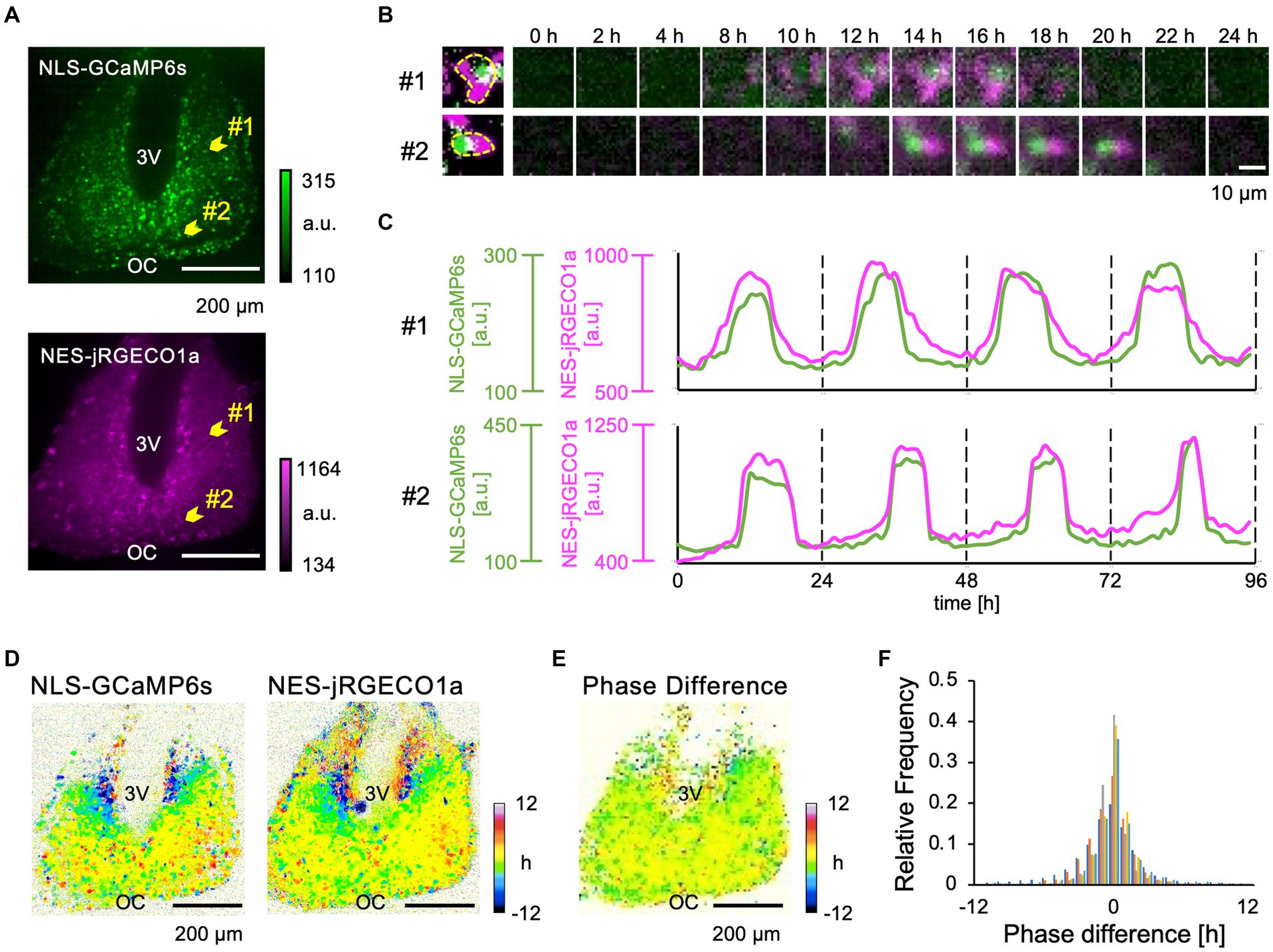
Figure 2. Dual-color imaging of the Ca2+ Dynamics in the nucleus and cytosol of SCN Neurons. (A) Expression patterns of NLS-GCaMP6s (upper) and NES-jRGECO1a (lower) in the SCN. 3 V, third ventricle; OC, optic chiasm. (B) Two-hourly montage images of two representative individual SCN neurons, as indicated by arrowheads in panel (A). The leftmost images show the edge of single SCN neuron. (C) Raw traces of the Ca2+ dynamics of the nucleus (green) and the cytosol (magenta) of two representative SCN neurons. (D) Acrophase maps of the Ca2+ rhythms of the nucleus (left) and the cytosol (right). The mean acrophase of the entire SCN region was normalized to zero. Color bars indicate the relative time of day (hours). (E) Phase-difference map between the nuclear and cytosolic Ca2+ rhythms. (F) Histogram of the pixel distribution of the phase-difference maps (normalized to the total pixels). The frequency (y-axis) represents the value relative to the total number of pixels. Individual SCN data are represented by different colors (n = 5 slices).
Ca2+ release from the ER has been reported to contribute to the Ca2+ rhythm in the cytosol (Ikeda et al., 2003; Plante et al., 2021). Type 2 ryanodine receptors have been reported to be expressed in the SCN (Díaz-Muñoz et al., 1999). Furthermore, it has been demonstrated that the ER and nuclear envelope are structurally interconnected, and the Ca2+ release channels, the ryanodine and IP3 receptor, are present in the inner membrane of the nucleus (Bootman et al., 2009; Secondo et al., 2020). Thus, it is plausible that nuclear Ca2+ may be amplified by Ca2+ release from the nuclear envelope as internal stores. To test this hypothesis, we applied inhibitors for ryanodine and IP3 receptors, major Ca2+ release channels in ER, and examined the nuclear and cytosolic Ca2+ rhythms. Dantrolene, a type 1 and 2 ryanodine receptor antagonist (Oo et al., 2015), had no significant effect on the amplitude and trough level of both nuclear and cytosolic Ca2+ rhythms (amplitude: 101% ± 21% in the nucleus, 92% ± 22% in the cytosol; trough level: 105% ± 3% in the nucleus and 102% ± 3% in the cytosol) (n = 5 slices). To further confirm this observation, we applied another ryanodine receptor antagonist, a high concentration of ryanodine (100 μM), in the SCN slices. High ryanodine concentrations bind to the receptor sites and subsequently cause a decrease in Ca2+ conductance (Thomas and Williams, 2012). We found that the amplitude and trough of both nuclear and cytosolic Ca2+ rhythms were slightly but significantly increased (amplitude: 126% ± 21% in the nucleus, 115% ± 27% in the cytosol; trough: 110% ± 3% in the nucleus and 105% ± 2% in the cytosol) (n = 6 slices). These results indicate that ryanodine receptor-mediated Ca2+ release is not a major determinant of Ca2+ rhythm generation but rather contributes to the control of baseline levels of Ca2+ concentration in SCN neurons.
To further investigate the contribution of Ca2+ release from the ER, we applied the IP3 receptor antagonist, Xestospongin C (10 μM), and statistically analyzed the effect on the Ca2+ rhythms. We found that neither the amplitude nor the trough level of both nuclear and cytosolic Ca2+ rhythms was affected (amplitude: 117% ± 12% in the nucleus, 110% ± 14% in the cytosol, trough: 104% ± 1% in the nucleus and 103% ± 2% in the cytosol) (n = 6 slices) (Figures 3C,E,F).
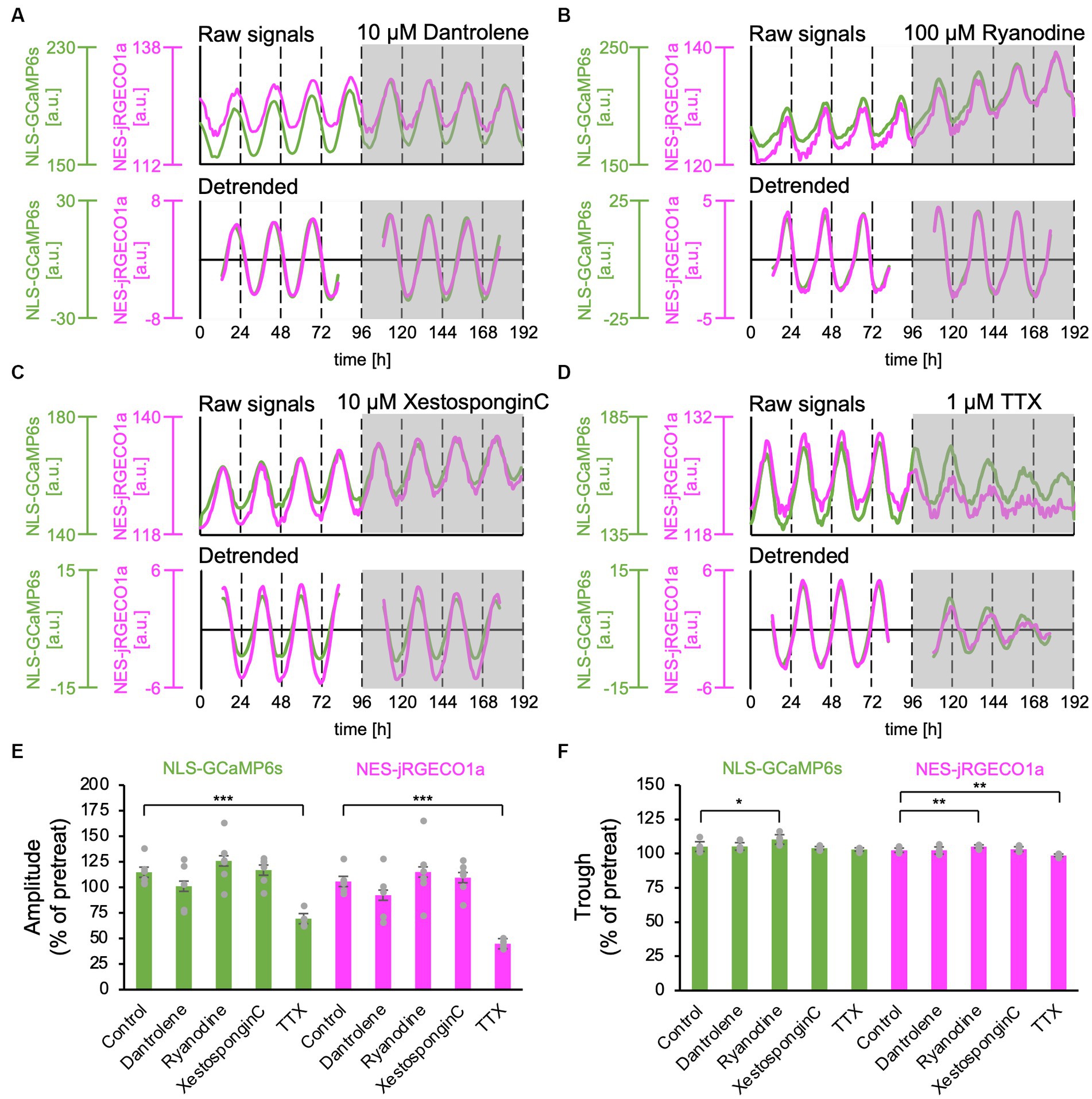
Figure 3. Effect of Ca2+ release inhibitors from the ER on the nuclear and cytosolic Ca2+ Rhythms. (A–D) Representative traces of the Ca2+ rhythms in the nucleus (green) and cytosol (magenta). The upper and lower traces in each panel show raw and 24-h detrended data (smoothed with a 3-h moving average), respectively. (A) Ryanodine receptor blocker, dantrolene (10 μM) (n = 5 slices). (B) Ryanodine receptor blocker, high concentration of ryanodine (100 μM) (n = 6 slices). (C) IP3 receptor blocker, Xestospongin C (10 μM) (n = 6 slices). (D) Sodium channel blocker, TTX (1 μM) (n = 5 slices). (E–F) Summary of blocker effects on the amplitude (E) and trough (F) of the nuclear (green) and cytosolic (magenta) Ca2+ rhythms. Data are expressed as percentage of the pretreatment value. One-sample t-test was employed to evaluate the blocker effects. *p < 0.05; **p < 0.01; ***p < 0.001. All data are expressed as mean ± SD.
Next, we applied 1-μM TTX to block sodium-dependent action potential firing in SCN neurons. As previously reported (Ikeda et al., 2003; Enoki et al., 2012a), after the TTX application, the amplitudes of the Ca2+ rhythm in both the nucleus and cytosol significantly decreased in the entire SCN compared with the pretreat levels (69% ± 7% in the nucleus, 44% ± 4% in the cytosol) (n = 5 slices) (Figures 2D,E). The trough of the Ca2+ rhythm in the cytosol was also significantly reduced (98% ± 9%) but not in the nucleus (103% ± 1%) (Figure 2F).
Taken together, these results indicate that in-phasic Ca2+ rhythms in the nucleus and cytosol are mainly driven from the extracellular space rather than from internal Ca2+ stores in ER or nuclear envelope.
As is common in eukaryotes, the nucleoplasm is generally covered by a nuclear envelope that is contiguous with the ER, and the nuclear envelope contains nuclear pores (Devos et al., 2014). It has been reported that Ca2+ transporters, such as ryanodine and IP3 receptor, are expressed on the inner nuclear membrane (Bootman et al., 2009; Secondo et al., 2020). These structural features suggest that the nuclear envelope functions as a Ca2+ store and that nuclear Ca2+ dynamics are regulated independently of the cytosol. Indeed, nuclear and cytosolic Ca2+ dynamics have been previously reported in mouse SCN (Ikeda et al., 2003) and genetically modified tobacco seedlings (Wood et al., 2001), both of which show the Ca2+ rhythms in the cytosol but not in the nucleus. However, it is difficult to assume the functional independence of nuclear Ca2+ on a circadian time scale because of nuclear pore complexes. In this study, we observed the circadian Ca2+ rhythms in the cytosol and nucleus by expressing highly sensitive, single-molecule, genetically encoded fluorescent Ca2+ sensors, NLS-GCaMP6s and NES-jRGECO1a, in the SCN neurons. We concluded that the nuclear and cytosolic Ca2+ dynamics are synchronized on a circadian time scale.
Among the three isoforms, the type 2 ryanodine receptor has been reported to be expressed in rodent SCN (Díaz-Muñoz et al., 1999). However, the involvement of the ryanodine receptor in Ca2+ rhythm regulation in the SCN neurons remains elusive. In the present study, we administered two types of ryanodine receptor blockers, dantrolene and ryanodine at a high concentration, and found that the amplitudes of Ca2+ rhythms were not significantly altered by either dantrolene or ryanodine, but the trough levels were slightly increased by a high dose of ryanodine. Dantrolene is known to decrease the open probability of ryanodine receptor type 2 in the presence of calmodulin (Oo et al., 2015), whereas ryanodine at a high concentration increases the open probability and decreases conductance (Thomas and Williams, 2012). These different pharmacological actions may account for the different effects on the trough levels of the Ca2+ rhythms.
Previous studies have demonstrated that the high dose of ryanodine reduced the amplitude of Ca2+ rhythms (Ikeda et al., 2003) and dantrolene decreased the Ca2+ levels at both the peak and trough phases in the SCN neurons of tissue cultures (Plante et al., 2021), whereas another study showed no significant effect on Ca2+ rhythms in dissociated SCN neurons (Noguchi et al., 2017). There have also been reports of Ca2+ level measurement when drugs were applied at a specific timing (CT 4–8), but the results were inconsistent; Ca2+ levels have been shown to increase, decrease, or remain unchanged, strongly correlating with the baseline Ca2+ levels measured at day and night in SCN slices (Aguilar-Roblero et al., 2016). The reason for these conflicting results is unknown, but it may be due to different experimental conditions (e.g., dynamic range and sensitivity of Ca2+ probe, gene transfection methods, timing of pharmacological administration) and/or heterogeneity of SCN neurons (Wen et al., 2020).
Regarding the contribution of IP3 receptors on Ca2+ rhythms in the SCN, it has been reported that IP3 receptors are involved in photoentrainment to the SCN (Hamada et al., 1999). Furthermore, because the Gq-mediated plasticity of the SCN network has been reported (Brancaccio et al., 2013), IP3-mediated Ca2+ release may be involved in light-induced resetting or synchronization of the SCN. In the present study, using an IP3 inhibitor, we found that the IP3 receptor is involved neither in the amplitude nor trough level of Ca2+ rhythms. Taken together, we conclude that neither ryanodine nor IP3 receptor-mediated Ca2+ release had a major contribution to the autonomous Ca2+ rhythms in SCN neurons.
A recent study demonstrated that the knockdown of the leucine zipper-EF-hand-containing transmembrane protein 1 (LETM1), a mitochondrial Ca2+/H+ exchanger, dampened the cytosolic Ca2+ rhythms in the SCN (Morioka et al., 2022) and that mitochondria Ca2+ content rhythmically oscillates in SCN astrocytes and U-2 OS cells (Scrima et al., 1867; Burkeen et al., 2011). Therefore, the mitochondria may function as an ion source of Ca2+ rhythms.
Cytosolic circadian Ca2+ rhythms have been reported in cultured SCN from neonate mice (Colwell, 2000; Ikeda et al., 2003; Enoki et al., 2012a, 2017a,b; Brancaccio et al., 2013; Noguchi et al., 2017) as well as in adult mouse SCN (Aguilar-Roblero et al., 2016; Jones et al., 2018). Since the development of the SCN has been reported (Myung et al., 2021), we can not exclude the possibility that the properties of the nuclear circadian Ca2+ rhythms are altered during development.
Ca2+ is a molecule critical for the three essential properties of the circadian clock: autonomous oscillation, entrainment, and temperature compensation (Cavieres-Lepe and Ewer, 2021; Kon et al., 2021). The present study suggests that nuclear and cytosolic Ca2+ rhythms may regulate these circadian properties via Ca2+-dependent signaling in situ.
It has been reported that CaMKII and PKCα (protein kinase Cα), Ca2+-dependent kinases, are expressed in the SCN (Van der Zee and Bult, 1995) and are candidate molecules that can be directly activated by nuclear Ca2+. In general, these protein kinases are thought to translocate from the cytosol to the nucleus upon Ca2+ elevation; however, some isoforms have been reported in the nucleus of mammalian cells (Robles et al., 2010; Mishra et al., 2011). CaMKIV, CBP/p300 and Downstream regulatory element antagonist modulator might be other candidates for nuclear Ca2+-dependent kinases, although their subcellular expression in SCN neurons has not yet been investigated.
The current view of the TTFL model is based on the nuclear translocation of clock proteins (Mohawk et al., 2012; Takahashi, 2017); however, a recent live-cell imaging of endogenous clock proteins shows that clock proteins (e.g., BMAL and CRY1) are predominantly present in the nuclear compartment (Gabriel et al., 2021), challenging the view of circadian gating of nuclear import. It may be plausible that cytoplasmic Ca2+ translocates to the nucleus and regulates transcription of clock genes in situ.
This study highlights the important but long overlooked function of nuclear Ca2+, which may directly activate Ca2+-dependent kinases and regulate the transcriptional regulation of clock genes. The key molecules that could be activated by nuclear Ca2+ signaling remain to be identified in the SCN neurons, but this could be an important target for the Ca2+ signaling of circadian functions in future studies.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by National Institute for Physiological Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
SH: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. KK: Methodology, Resources, Writing – review & editing. TN: Conceptualization, Funding acquisition, Project administration, Writing – review & editing. RE: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT)/Japan Society for the Promotion of Science KAKENHI to RE (20H05769, 20H03425, 22 K19319, and 23H04943), to TN (20H05669, 20H00523), Takeda Science Foundation, the Mochida Memorial Foundation, Joint Research of the ExCELLS (No, 21–205), and Cooperative Study Program (nos, 21–136 and 22–159) of the National Institute for Physiological Sciences, and the NINS program of Promoting Research by Networking among Institutions (No. 01412303).
We are grateful to the members of the Biophotonics Laboratory (Hirokazu Ishii, Motosuke Tsusumi, Kazushi Yamaguchi, and Joe Sakamoto), the Transformative Research Area “Hibernation Biology,” for the valuable advice and discussion. We also thank the Genetically-Encoded Neuronal Indicator and Effector Project and the Janelia Farm Research Campus of the Howard Hughes Medical Institute for sharing the GCaMP6s and jRGECO1a constructs; Rie Kageyama for packaging AAV; Yuki Watakabe, Maki Watanabe, and Yoshiko Yamada for the excellent technical support on the experiments; Miwa Kawachi, Miyoko Shimomura, and Chiemi Hyodo for their help with animal care and laboratory management. The authors would like to thank enago (www.enago.jp) for the English language review.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2023.1323565/full#supplementary-material
Aguilar-Roblero, R., Quinto, D., Báez-Ruíz, A., Chávez, J. L., Belin, A. C., Díaz-Muñoz, M., et al. (2016). Ryanodine-sensitive intracellular Ca2+ channels are involved in the output from the SCN circadian clock. Eur. J. Neurosci. 44, 2504–2514. doi: 10.1111/ejn.13368
Bootman, M. D., Fearnley, C., Smyrnias, I., MacDonald, F., and Roderick, H. L. (2009). An update on nuclear calcium signalling. J. Cell Sci. 122, 2337–2350. doi: 10.1242/jcs.028100
Brancaccio, M., Maywood, E. S., Chesham, J. E., Loudon, A. S. I., and Hastings, M. H. (2013). A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728. doi: 10.1016/j.neuron.2013.03.011
Burkeen, J. F., Womac, A. D., Earnest, D. J., and Zoran, M. J. (2011). Mitochondrial calcium signaling mediates rhythmic extracellular ATP accumulation in suprachiasmatic nucleus astrocytes. J. Neurosci. 31, 8432–8440. doi: 10.1523/JNEUROSCI.6576-10.2011
Cavieres-Lepe, J., and Ewer, J. (2021). Reciprocal relationship between calcium signaling and circadian clocks: implications for calcium homeostasis, clock function, and therapeutics. Front. Mol. Neurosci. 14:666673. doi: 10.3389/fnmol.2021.666673
Chen, T. W., Wardill, T. J., Sun, Y., Pulver, S. R., Renninger, S. L., Baohan, A., et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. doi: 10.1038/nature12354
Colwell, C. S. (2000). Circadian modulation of calcium levels in cells in the suprachiasmatic nucleus. Eur. J. Neurosci. 12, 571–576. doi: 10.1046/j.1460-9568.2000.00939.x
Dana, H., Mohar, B., Sun, Y., Narayan, S., Gordus, A., Hasseman, J. P., et al. (2016). Sensitive red protein calcium indicators for imaging neural activity. elife 5:e12727. doi: 10.7554/eLife.12727
Devos, D. P., Gräf, R., and Field, M. C. (2014). Evolution of the nucleus. Curr. Opin. Cell Biol. 28, 8–15. doi: 10.1016/j.ceb.2014.01.004
Díaz-Muñoz, M., Dent, M. A. R., Granados-Fuentes, D., Hall, A. C., Hernández-Cruz, A., Harrington, M. E., et al. (1999). Circadian modulation of the ryanodine receptor type 2 in the SCN of rodents. Neuroreport 10, 481–486. doi: 10.1097/00001756-199902250-00007
Enoki, R., Kuroda, S., Ono, D., Hasan, M. T., Ueda, T., Honma, S., et al. (2012a). Topological specificity and hierarchical network of the circadian calcium rhythm in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U. S. A. 109, 21498–21503. doi: 10.1073/pnas.1214415110
Enoki, R., Oda, Y., Mieda, M., Ono, D., Honma, S., and Honma, K. I. (2017a). Synchronous circadian voltage rhythms with asynchronous calcium rhythms in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. U. S. A. 114, E2476–E2485. doi: 10.1073/pnas.1616815114
Enoki, R., Ono, D., Hasan, M. T., Honma, S., and Honma, K. (2012b). Single-cell resolution fluorescence imaging of circadian rhythms detected with a Nipkow spinning disk confocal system. J. Neurosci. Methods 207, 72–79. doi: 10.1016/j.jneumeth.2012.03.004
Enoki, R., Ono, D., Kuroda, S., Honma, S., and Honma, K. I. (2017b). Dual origins of the intracellular circadian calcium rhythm in the suprachiasmatic nucleus. Sci. Rep. 7:41733. doi: 10.1038/srep41733
Gabriel, C. H., Del Olmo, M., Zehtabian, A., Jäger, M., Reischl, S., Van Dijk, H., et al. (2021). Live-cell imaging of circadian clock protein dynamics in CRISPR-generated knock-in cells. Nat. Commun. 12:3796. doi: 10.1038/s41467-021-24086-9
Golombek, D. A., and Rosenstein, R. E. (2010). Physiology of circadian entrainment. Physiol. Rev. 90, 1063–1102. doi: 10.1152/physrev.00009.2009
Hamada, T., Liou, S. Y., Fukushima, T., Maruyama, T., Watanabe, S., Mikoshiba, K., et al. (1999). The role of inositol trisphosphate-induced Ca2+ release from IP3- receptor in the rat suprachiasmatic nucleus on circadian entrainment mechanism. Neurosci. Lett. 263, 125–128. doi: 10.1016/s0304-3940(99)00111-1
Harrisingh, M. C., Wu, Y., Lnenicka, G. A., and Nitabach, M. N. (2007). Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J. Neurosci. 27, 12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007
Hastings, M. H., Maywood, E. S., and Brancaccio, M. (2018). Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469. doi: 10.1038/s41583-018-0026-z
Ikeda, M., and Ikeda, M. (2014). Bmal1 is an essential regulator for circadian cytosolic Ca2+ rhythms in suprachiasmatic nucleus neurons. J. Neurosci. 34, 12029–12038. doi: 10.1523/JNEUROSCI.5158-13.2014
Ikeda, M., Sugiyama, T., Wallace, C. S., Gompf, H. S., Yoshioka, T., Miyawaki, A., et al. (2003). Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38, 253–263. doi: 10.1016/S0896-6273(03)00164-8
Jones, J. R., Simon, T., Lones, L., and Herzog, E. D. (2018). SCN VIP neurons are essential for normal light-mediated resetting of the circadian system. J. Neurosci. 38, 7986–7995. doi: 10.1523/JNEUROSCI.1322-18.2018
Kon, N., Wang, H. T., Kato, Y. S., Uemoto, K., Kawamoto, N., Kawasaki, K., et al. (2021). Na+/Ca2+ exchanger mediates cold Ca2+ signaling conserved for temperature-compensated circadian rhythms. Sci. Adv. 7:eabe8132. doi: 10.1126/sciadv.abe8132
Kon, N., Yoshikawa, T., Honma, S., Yamagata, Y., Yoshitane, H., Shimizu, K., et al. (2014). CaMKII is essential for the cellular clock and coupling between morning and evening behavioral rhythms. Genes Dev. 28, 1101–1110. doi: 10.1101/gad.237511.114
Lin, D. H., and Hoelz, A. (2019). The structure of the nuclear pore complex (an update). Annu. Rev. Biochem. 88, 725–783. doi: 10.1146/annurev-biochem-062917-011901
Lundkvist, G. B., Kwak, Y., Davis, E. K., Tei, H., and Block, G. D. (2005). A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J. Neurosci. 25, 7682–7686. doi: 10.1523/JNEUROSCI.2211-05.2005
Mishra, S., Gray, C. B. B., Miyamoto, S., Bers, D. M., and Brown, J. H. (2011). Location matters: clarifying the concept of nuclear and cytosolic CaMKII subtypes. Circ. Res. 109, 1354–1362. doi: 10.1161/CIRCRESAHA.111.248401
Mohawk, J. A., Green, C. B., and Takahashi, J. S. (2012). Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35, 445–462. doi: 10.1146/annurev-neuro-060909-153128
Moore, R. Y., and Eichler, V. B. (1972). Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 42, 201–206. doi: 10.1016/0006-8993(72)90054-6
Morioka, E., Kasuga, Y., Kanda, Y., Higashida, H., Holmes, T. C., Correspondence, M. I., et al. (2022). Mitochondrial LETM1 drives ionic and molecular clock rhythms in circadian pacemaker neurons. Cell Rep. 39:110787. doi: 10.1016/j.celrep.2022.110787
Myung, J., Nakamura, T. J., Jones, J. R., Silver, R., and Ono, D. (2021). Editorial: development of circadian clock functions. Front. Neurosci. 15:735007. doi: 10.3389/fnins.2021.735007
Noguchi, T., Leise, T. L., Kingsbury, N. J., Diemer, T., Wang, L. L., Henson, M. A., et al. (2017). Calcium circadian rhythmicity in the suprachiasmatic nucleus: cell autonomy and network modulation. eNeuro 4, ENEURO.0160–ENEU17.2017. doi: 10.1523/ENEURO.0160-17.2017
Oo, Y. W., Gomez-Hurtado, N., Walweel, K., Van Helden, D. F., Imtiaz, M. S., Knollmann, B. C., et al. (2015). Essential role of calmodulin in RyR inhibition by dantrolene. Mol. Pharmacol. 88, 57–63. doi: 10.1124/mol.115.097691
Plante, A. E., Rao, V. P., Rizzo, M. A., and Meredith, A. L. (2021). Comparative Ca2+ channel contributions to intracellular Ca2+ levels in the circadian clock. Biophys. Reports 1:100005. doi: 10.1016/j.bpr.2021.100005
Robles, M. S., Boyault, C., Knutti, D., Padmanabhan, K., and Weitz, C. J. (2010). Identification of RACK1 and protein kinase Cα as integral components of the mammalian circadian clock. Science 327, 463–466. doi: 10.1126/science.1180067
Sano, H., Kobayashi, K., Yoshioka, N., Takebayashi, H., and Nambu, A. (2020). Retrograde gene transfer into neural pathways mediated by adeno-associated virus (AAV)-AAV receptor interaction. J. Neurosci. Methods 345:108887. doi: 10.1016/j.jneumeth.2020.108887
Scrima, R., Cela, O., Agriesti, F., Piccoli, C., Tataranni, T., Pacelli, C., et al. (1867). Mitochondrial calcium drives clock gene-dependent activation of pyruvate dehydrogenase and of oxidative phosphorylation. Biochim. Biophys. Acta, Mol. Cell Res. 1867:118815. doi: 10.1016/j.bbamcr.2020.118815
Secondo, A., Petrozziello, T., Tedeschi, V., Boscia, F., Pannaccione, A., Molinaro, P., et al. (2020). Nuclear localization of NCX: role in Ca2+ handling and pathophysiological implications. Cell Calcium 86:102143. doi: 10.1016/j.ceca.2019.102143
Shinohara, K., Honma, S., Katsuno, Y., Abe, H., and Honma, K. (1995). Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc. Natl. Acad. Sci. U. S. A. 92, 7396–7400. doi: 10.1073/pnas.92.16.7396
Stephan, F. K., and Zucker, I. (1972). Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc. Natl. Acad. Sci. U. S. A. 69, 1583–1586. doi: 10.1073/pnas.69.6.1583
Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Thomas, N. L., and Williams, A. J. (2012). Pharmacology of ryanodine receptors and Ca2+-induced Ca2+ release. Wiley Interdiscip. Rev. Membr. Transp. Signal. 1, 383–397. doi: 10.1002/wmts.34
Van den Pol, A. N. (1980). The hypothalamic suprachiasmatic nucleus of rat: intrinsic anatomy. J. Comp. Neurol. 191, 661–702. doi: 10.1002/cne.901910410
Van der Zee, E. A., and Bult, A. (1995). Distribution of AVP and Ca2+-dependent PKC-isozymes in the suprachiasmatic nucleus of the mouse and rabbit. Brain Res. 701, 99–107. doi: 10.1016/0006-8993(95)00968-1
Villarruel, C., Aguilar, P. S., and Ponce Dawson, S. (2021). High rates of calcium-free diffusion in the cytosol of living cells. Biophys. J. 120, 3960–3972. doi: 10.1016/j.bpj.2021.08.019
Welsh, D. K., Logothetis, D. E., Meister, M., and Reppert, S. M. (1995). Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron 14, 697–706. doi: 10.1016/0896-6273(95)90214-7
Welsh, D. K., Takahashi, J. S., and Kay, S. A. (2010). Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577. doi: 10.1146/annurev-physiol-021909-135919
Wen, S., Ma, D., Zhao, M., Xie, L., Wu, Q., Gou, L., et al. (2020). Spatiotemporal single-cell analysis of gene expression in the mouse suprachiasmatic nucleus. Nat. Neurosci. 23, 456–467. doi: 10.1038/s41593-020-0586-x
Wood, N. T., Haley, A., Viry-Moussaïd, M., Johnson, C. H., Van der Luit, A. H., and Trewavas, A. J. (2001). The calcium rhythms of different cell types oscillate with different circadian phases. Plant Physiol. 125, 787–796. doi: 10.1104/pp.125.2.787
Yoo, S. H., Yamazaki, S., Lowrey, P. L., Shimomura, K., Ko, C. H., Buhr, E. D., et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U. S. A. 101, 5339–5346. doi: 10.1073/pnas.0308709101
Keywords: circadian clock, intracellular Ca2+, SCN, imaging, nucleus, organelle
Citation: Hiro S, Kobayashi K, Nemoto T and Enoki R (2023) In-phasic cytosolic-nuclear Ca2+ rhythms in suprachiasmatic nucleus neurons. Front. Neurosci. 17:1323565. doi: 10.3389/fnins.2023.1323565
Received: 18 October 2023; Accepted: 30 November 2023;
Published: 20 December 2023.
Edited by:
Mino Belle, The University of Manchester, United KingdomReviewed by:
Ryan W. Logan, Boston University, United StatesCopyright © 2023 Hiro, Kobayashi, Nemoto and Enoki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryosuke Enoki, ZW5va2lAbmlwcy5hYy5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.