- 1Wu Tsai Neurosciences Institute, Stanford University, Stanford, CA, United States
- 2Neurosciences IDP Graduate Program, Stanford University School of Medicine, Stanford, CA, United States
- 3Nevada ENDURE Program, University of Nevada, Reno, Reno, NV, United States
- 4Department of Neurosurgery, Stanford University School of Medicine, Stanford, CA, United States
Gastrointestinal (GI) symptoms are highly prevalent among individuals with autism spectrum disorder (ASD), but the molecular link between ASD and GI dysfunction remains poorly understood. The enteric nervous system (ENS) is critical for normal GI motility and has been shown to be altered in mouse models of ASD and other neurological disorders. Contactin-associated protein-like 2 (Cntnap2) is an ASD-related synaptic cell-adhesion molecule important for sensory processing. In this study, we examine the role of Cntnap2 in GI motility by characterizing Cntnap2’s expression in the ENS and assessing GI function in Cntnap2 mutant mice. We find Cntnap2 expression predominately in enteric sensory neurons. We further assess in vivo and ex vivo GI motility in Cntnap2 mutants and show altered transit time and colonic motility patterns. The overall organization of the ENS appears undisturbed. Our results suggest that Cntnap2 plays a role in GI function and may provide a molecular link between ASD and GI dysfunction.
1. Introduction
Autism spectrum disorder (ASD) is a neurodevelopmental disorder affecting approximately 1 in 36 children in the United States (Maenner et al., 2023). Individuals with ASD often report gastrointestinal (GI) issues, which can lead to irritability and social withdrawal, ultimately affecting quality of life (Valicenti-McDermott et al., 2006; Chaidez et al., 2014; Restrepo et al., 2020). GI issues, including constipation, diarrhea, and abdominal pain (Chaidez et al., 2014; Holingue et al., 2018), have been correlated with sensory over-responsivity in the central and peripheral nervous system in children with ASD (Mazurek et al., 2013). Whether sensory functions of the GI tract are also affected in ASD, and thus potentially contribute to ASD-related GI dysfunction, has not yet been extensively explored.
The enteric nervous system (ENS) is a quasi-autonomous neuronal network that populates the length of the GI tract and can regulate GI function and motility independent of the central nervous system (CNS) (Furness et al., 1994). Enteric neurons and glia cluster into ganglia that reside within the myenteric and submucosal plexuses within the gut wall (Sasselli et al., 2012). GI motility initiates when intrinsic enteric sensory neurons, known as intrinsic primary afferent neurons (IPANs), are activated by chemical or mechanical stimuli. IPANs signal to enteric interneurons that stimulate excitatory or inhibitory motor neurons, resulting in repetitive contractions and propulsive motility (Furness et al., 1994; Rao and Gershon, 2016; Fung and Berghe, 2020). Alterations in ENS activity, organization or gene expression are known to affect digestive function (Avetisyan et al., 2015; Rao and Gershon, 2016). We hypothesize that genes known to be risk factors for ASD are expressed in the ENS and influence enteric neuron activity, and thus could provide a link between ASD and associated GI dysfunction.
ASD-related genes have previously been linked to GI function (Niesler and Rappold, 2021). Mutations in the zebrafish shank3 gene, which encodes a synaptic scaffolding protein critical for synaptic transmission, result in reduced serotonin-expressing enteroendocrine cells and serotonin-filled ENS boutons, and prolonged GI transit (James et al., 2019). In mice, a global deletion of Nlgn3, an ASD-related synaptic cell adhesion molecule, results in increased colonic diameter and faster colonic migrating motor complexes (Leembruggen et al., 2019).
Here we study Contactin-associated protein-like 2 (Cntnap2; also known as Caspr2), an ASD-related cell-adhesion molecule that aids in the formation and function of the central and peripheral nervous system (Poliak et al., 1999; Anderson et al., 2012; Peñagarikano and Geschwind, 2012; Gordon et al., 2016). CNTNAP2 gene mutations have been detected in individuals diagnosed with ASD (Peñagarikano and Geschwind, 2012), and Cntnap2−/− mice show social deficits, communication impairment, and repetitive behaviors, three hallmark characteristics of ASD (Peñagarikano et al., 2011). Additionally, Cntnap2−/− mice have altered neural circuitry in the somatosensory cortex and exhibit hypersensitivity to mechanical stimuli due to enhanced excitability of primary dorsal root afferents (Peñagarikano et al., 2011; Dawes et al., 2018). In the GI tract, CNTNAP2 has been associated with inflammatory bowel disease, and Cntnap2−/− mice have increased intestinal permeability (Buniello et al., 2019; Graf et al., 2019). Whether GI motility, which relies on sensing luminal stimuli, is affected in Cntnap2−/− mice has not been previously investigated.
In this study, we assess Cntnap2 expression in the adult mouse GI tract and ask whether ENS organization and GI motility are altered in Cntnap2−/− mice. We find that Cntnap2 is predominantly expressed in IPANs, being nearly exclusive to IPANs in the colon. We assess GI motility in vivo and focus on colonic motor function in an ex vivo motility monitor in the absence and presence of an artificial stimulus. We find that lack of Cntnap2 results in altered colonic motility. The overall organization of the ENS appears undisturbed.
2. Materials and methods
2.1. Animals
All procedures conformed to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Stanford University Administrative Panel on Laboratory Animal Care. C57BL/6, B6.129(Cg)-Cntnap2tm1Pele/J (Strain #:017482, hereafter Cntnap2− ), and B6.129(Cg)-Cntnap2tm2Pele/J (Strain #:028635, hereafter Cntnap2tlacZ) mice were purchased from The Jackson Laboratory. Mice were maintained on a 12:12 LD cycle and fed a standard rodent diet, containing 18% Protein and 6% Fat (Envigo Teklad). Food and water were provided ad libitum and mice were group housed with a maximum of five adults per cage. Both male and female 8–12 week-old adult mice were used in this study.
2.2. Histology
Mice were euthanized by CO2 followed by cervical dislocation. Segments of SI and colon were dissected, flushed with cold PBS, and cut longitudinally along the mesenteric border. Segments were opened flat, placed between sheets of filter paper, and immersed in 4% PFA at 4°C for 90 min. Tissue was rinsed three times in PBS for 10 min and immersed in a 30% sucrose solution overnight at 4°C. Tissue sections were rolled into a “Swiss-roll” preparation as described in Williams et al. (2016), embedded in OCT (Tissue-Tek), and frozen until use. 14 μm slices were sectioned using a Leica Cryostat (Leica CM3050 S) and mounted on Superfrost glass slides. Slides were stained with hematoxylin and eosin (H&E). Brightfield images were taken by the Human Pathology/Histology Service Center at Stanford School of Medicine and analyzed for villus height, crypt depth, colonic fold thickness, and circular muscle thickness using Leica ImageScope software. Villus height was measured when full lacteal was visible and crypt depth was measured when both villus/crypt junctions were present in the jejunum. Colonic fold thickness was measured from cross sections of mid and distal colon. 10 measurements were taken per animal. To determine muscle thickness, 20 measurements were taken at random points along the length of the jejunum and distal colon.
2.3. Tissue dissection and processing
Dissection and tissue processing of the intestines was performed as previously described in Hamnett et al. (2022). Wholemount muscle-myenteric plexus preparations were made by peeling away the muscularis (longitudinal and circular muscle with myenteric plexus). The tissue was stored in PBS with 0.1% sodium azide at 4°C for up to 3 weeks. Jejunum samples were taken from the middle 1/3 length of the SI. The final 1/3 of the colon was considered distal colon.
2.4. Immunohistochemistry
Segments of the jejunum (≥1 cm in length) and distal colon (≥0.5 cm in length) were used for immunohistochemistry studies. Staining was performed as previously described in Hamnett et al. (2022), with modifications. For cell body labeling with anti-Cntnap2 antibody, PBT contained 0.01% Triton X-100; for all other labeling, PBT contained 0.1% Triton X-100 (Supplementary Figures S1C,D). Primary antibodies used included rabbit anti-Cntnap2 (1:1000; Alomone Labs, APZ-005), rabbit anti-ß-galactosidase (1:1000; gift from J. Sanes), human anti-HuC/D (ANNA1) (1:100,000; gift from V. Lennon), goat anti-Sox10 (1:2,000; R&D Systems, AF2864) and fluorophore-conjugated secondary antibodies (Jackson Labs and Molecular Probes).
2.5. Rnascope in situ hybridization with protein co-detection
Tissue was dissected and prepared for fixation as outlined in Tissue Dissection and Processing. Flat segments of the jejunum and distal colon were fixed overnight in 4% PFA. Segments were rinsed with PBS, and wholemount muscle-myenteric plexus preparations were made by peeling away the muscularis. RNAscope in situ with protein co-detection was performed using Advanced Cell Diagnostics (ACD) RNAscope Multiplex Fluorescent Reagent Kit v2 (Cat# 323100) and ACD RNA-protein Co-detection ancillary kit (Cat# 323180) as described Guyer et al. (2023). The following RNAscope probes were used: Mm-Nmu-C1 (Cat# 446831) and Mm-Cntnap2-C1 (Cat# 449381).
2.6. Neuron quantification
Images were acquired on a Leica SP8 confocal microscope using 20x and 63x oil objectives. All images were adjusted for brightness and contrast using ImageJ/FIJI. For Cntnap2 quantification, three 20x ROIs (1,000 μm × 1,000 μm) per mouse were randomly selected in both the jejunum and distal colon. HuC/D+ and Cntnap2+ neurons were counted manually using the cell counter tool FIJI (Schindelin et al., 2012). For each region, neurons per ROI were averaged per animal.
For quantification of Cntnap2 co-expression with Nmu transcript, five images (138 μm × 138 μm) were taken at 63x magnification per region per mouse. Regions of interest (ROIs) were created around every HuC/D+ neuron for each image and manually scored as either positive or negative for Cntnap2 or Nmu transcript. Neurons with ≥20 Nmu or Cntnap2 fluorescent transcript dots were considered positive. For each region, the average percentage of co-expression was calculated per mouse.
Quantification of ganglia was performed using COUNTEN (Kobayashi et al., 2021), with = 4.5. For each region, the average of three maximum projection images (1,000 μm × 1,000 μm) were analyzed per mouse.
2.7. Functional behavior
2.7.1. Whole GI transit time
Whole GI transit assay was performed as previously described in Spear et al. (2018). In brief, mice were gavaged with a carmine red-methylcellulose mixture and observed until a red pellet was expelled.
2.7.2. Gastric emptying and SI transit
Gastric emptying and SI transit were determined as previously described in De Lisle (2007) and Spear et al. (2018). In brief, mice were fasted for 12 h and water was removed 3 h before the start of the assay. Mice were gavaged with a 2% methylcellulose mixture containing 2.5 mg/mL Rhodamine B Dextran (Invitrogen, D1841, MW: 70,000). 15 min after gavage, mice were euthanized with CO2 and the stomach and SI were removed. The SI was divided into 10 equal segments that were homogenized in saline. The fluorescence in the stomach and each SI segment was measured. The percentage of gastric emptying and the geometric center were determined as previously described in De Lisle (2007).
2.7.3. Bead expulsion assay
Bead expulsion assay was performed as previously described in Spear et al. (2018). In brief, mice were lightly anesthetized by isoflurane and a 2 mm glass bead was inserted 2 cm into the colon through the anus using a gavage needle. Expulsion time was determined as the time from bead insertion to when the bead was fully expelled.
2.7.4. Fecal water content and pellet length
Fecal water content was assessed as previously described in (Spear et al., 2018) with modifications to allow for measurement of pellet lengths. Mice were housed individually for 1 h during which all fecal pellets were collected immediately after expulsion, photographed, and stored in a pre-weighed tube (1 tube/mouse). After 1 h of collection, tubes were weighed again, incubated for 48 h at 50°C, and weighed a final time to determine the percentage of water content. Pellet length was measured using FIJI (Swaminathan et al., 2019).
2.8. Ex vivo colonic motility assay
Ex vivo motility monitor assay was adapted from Hennig et al. (1999), Swaminathan et al. (2016), and Spear et al. (2018). Colons with cecum attached were removed and placed in warmed Kreb’s solution. The mesentery was cut away, and colons were placed in an organ bath, pinned down at the cecum and distal colon end with care to not impede expulsion of contents. The organ bath was kept at 37°C and filled with circulating warmed Kreb’s solution (NaCl, 120.9 mM; KCl, 5.9 mM; NaHCO3, 25.0 mM; Monobasic NaH2PO4, 1.2 mM; CaCl2, 3.3 mM; MgCl2•6H20, 1.2 mM; D-Glucose, 11.1 mM) saturated with carbogen (95% O2 and 5% CO2). Colons were allowed to acclimate for 10 min in the bath. Colonic motility was recorded ex vivo using a high-resolution monochromatic firewire industrial camera (The Imaging Source, DMK41AF02) mounted directly above the organ bath as previously described in Swaminathan et al. (2016) and Spear et al. (2018).
2.8.1. Motility monitor–natural colonic behavior
After a 10-min acclimation period and additional 20-min to allow for clearing of natural fecal pellets, motility of the empty colon was recorded for a 10-min period. Recorded videos were converted to spatiotemporal maps (STMs) using Scribble 2.0 and Matlab (2012a) plugin Analyze 2.0 (Swaminathan et al., 2016) and annotated to determine characteristics of CMCs, which we considered neurogenic repetitive contractions (Corsetti et al., 2019). Intervals between CMCs were measured from start of one contraction to the start of the next contraction (Fida et al., 2000).
2.8.2. Motility monitor–artificial pellet assay
Dissection was performed as described in “Ex vivo Colonic Motility Assay,” with cecum removed. Artificial pellet assay was adapted from Costa et al. (2021). The colon was flushed of endogenous fecal matter using warmed Kreb’s solution. After 10 min of acclimation, a lubricated (KY jelly) 2 mm 3D-printed pellet was inserted through the proximal colon and gently pushed to the proximal-mid colon junction using a blunt-ended gavage needle. Colonic activity was recorded until the pellet was fully expelled from the distal end of the colon. After at least three successful trials in which the artificial pellet traveled through the colon independently and was fully expelled, 10 additional minutes of empty colonic activity were recorded to ensure normal function. Time to expulsion was determined and the pellet’s path was traced using FIJI plug-in TrackMate (v7.6.1), from which pellet velocity and max speed were determined (Tinevez et al., 2017; Ershov et al., 2021). STMs were generated as described above.
2.9. Statistical analysis
Statistical analyzes were performed using GraphPad Prism (Version 9.4.1) with a 95% confidence limit (p < 0.05). Data are presented as mean ± SEM and checked for normal distribution. Unless otherwise noted, an unpaired t-test was used for comparison between two groups. For comparison between more than two groups, one-way or two-way analysis of variance (ANOVA) was used with Tukey’s multiple comparisons test. To ensure sufficient animals were used for the studies, we performed power analyzes based on early pilot data using a value of p (alpha) of 0.05 and a power (beta) of 0.8. Experimenter and analyzer were blinded to the genotype when feasible and appropriate. “n” refers to the number of animals tested, unless otherwise stated.
3. Results
To define the distribution of Cntnap2 in the mouse intestines, we used an antibody against Cntnap2, which we validated using Cntnap2 transcript co-expression (Figure 1A) and Cntnap2−/− mice (Figure 1B; Poliak et al., 2003). As we were interested in querying the role of Cntnap2 in GI motility, we focused our analysis on the myenteric plexus, which harbors the intrinsic neuronal circuitry required for motility (Spencer and Hu, 2020). We examined Cntnap2tlacz/+ mice (Gordon et al., 2016), and found β-gal-expressing neurons and projections throughout the SI and colon (Figure 1C and data not shown). Given that the expression of neurotransmitters and neuromodulators can differ between intestinal regions (Hamnett et al., 2022), we assessed Cntnap2 expression in distinct regions of the SI (duodenum, jejunum, and ileum) and colon (proximal, mid, and distal) (Figure 1D). Cntnap2 is expressed in 10–25% of HuC/D+ enteric neurons, depending on the region analyzed (Figure 1G). We further observed Cntnap2 expression in a small subset of Sox10+ progenitor/glial cells in the small intestine and colon (Supplementary Figure S1A and data not shown), in agreement with previous reports in mouse and human (Drokhlyansky et al., 2020; Morarach et al., 2021; Progatzky et al., 2021). Additionally, we observed Cntnap2 expression in 5-HT+ intestinal epithelial cells, suggesting that Cntnap2 is present in a subset of enterochromaffin cells along the epithelial layer of small intestine and colon (Supplementary Figure S1B and data not shown).
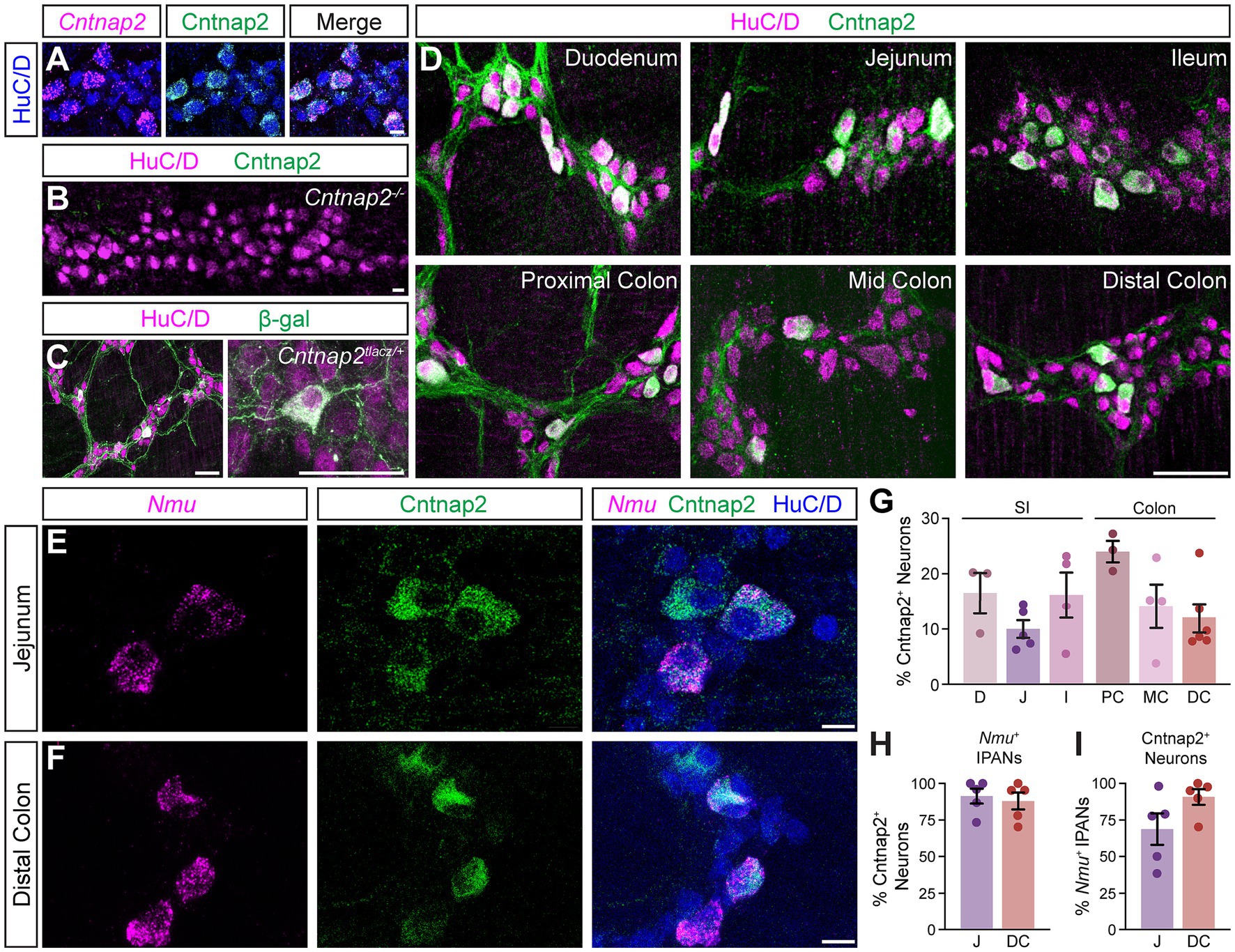
Figure 1. Cntnap2 expression in enteric sensory neurons. (A) Cntnap2 (green) colocalizes with Cntnap2 transcript (magenta) in adult jejunum myenteric plexus. Enteric neurons labeled with HuC/D (blue). (B) Cntnap2 (green) is absent from HuC/D+ (magenta) enteric neurons in Cntnap2−/− myenteric plexus of adult jejunum. (C) β-gal (green) expression in HuC/D+ (magenta) neurons of adult Cntnap2tlacZ/+ jejunum. (D) Cntnap2 (green) is expressed in a subset of HuC/D+ (magenta) neurons throughout the small intestine and colon. (E,F) A subset of Nmu+ (magenta) sensory neurons expresses Cntnap2 (green) in the jejunum (E) and distal colon (F). (G) Quantification of Cntnap2+ HuC/D+ neurons in the small intestine [D: 16.4 ± 3.6% (n = 3); J: 10.0 ± 1.6% (n = 5); I: 16.1 ± 4.0% (n = 4)] and colon [PC: 23.9 ± 1.9% (n = 3); MC: 14.2 ± 3.9% (n = 4); DC: 11.9 ± 2.5% (n = 6)]. (H) The majority of Nmu+ IPANs express Cntnap2 in the jejunum [91.2 ± 5.1% (n = 5)] and distal colon [87.8 ± 5.7% (n = 5)]. (I) The majority of Cntnap2+ neurons express Nmu in the jejunum [68.6 ± 10.7% (n = 5)] and distal colon [90.5 ± 5.3% (n = 5)]. Scale bars (A–F) 10 μm, (C,D) 50 μm. D: Duodenum, J: Jejunum, I: Ileum, PC: Proximal colon, MC: Mid colon, DC: Distal colon.
We next asked whether Cntnap2 expression in the myenteric ENS was confined to a particular neuronal subtype. We focused this and all future analyzes on the distal region of the colon due to its association with the propulsion of formed fecal pellets, and for comparison, chose the jejunum as a representative region within the SI. scRNA-sequencing studies of the mouse ENS have reported high Cntnap2 expression in putative sensory neuron populations in both the SI and colon (Zeisel et al., 2018; Drokhlyansky et al., 2020; Morarach et al., 2021). IPANs make up approximately 26% of enteric neurons in the SI and have Dogiel Type II morphology, based on their large and smooth cell bodies and two or more long axons (Qu et al., 2008). We observed that the majority of Cntnap2+ neurons were large in shape with smooth cell bodies (Supplementary Figure S1A). We further assessed Cntnap2 expression in IPANs, using Nmu transcript as a sensory neuron marker (Morarach et al., 2021; Figures 1E,F). Over 80% of Nmu+ neurons in both SI and colon co-expressed Cntnap2 (Figure 1H) and over half of Cntnap2+ neurons in the SI and over 80% of Cntnap2+ neurons in the colon colocalized with Nmu (Figure 1I). Taken together, these results suggest that Cntnap2 has a subtype and region-specific expression profile, and that the majority of Cntnap2+ myenteric plexus neurons in the colon are putative sensory neurons.
We next asked whether the absence of Cntnap2 affects GI morphology and function. Cntnap2−/− mice survive (Poliak et al., 2003), have normal body weight and SI and colon length (Figures 2A–C). We found no changes in SI villi height, crypt depth, and circular muscle thickness (Figures 2D,E,G,H). Also, colonic fold depth and circular muscle thickness was the same in Cntnap2−/− as compared to Cntnap2WT mice (Figures 2F,H). To assess whole GI transit time, we measured the length of time needed for a carmine red mixture gavaged into the stomach to be expelled as a red fecal pellet (Spear et al., 2018). We observed a 23% increase in whole GI transit time when comparing Cntnap2−/− to Cntnap2WT mice (Figure 2I). We found no changes in fecal water content and pellet length (Figures 2J,K). Thus, whole gut transit is prolonged in the absence of Cntnap2.
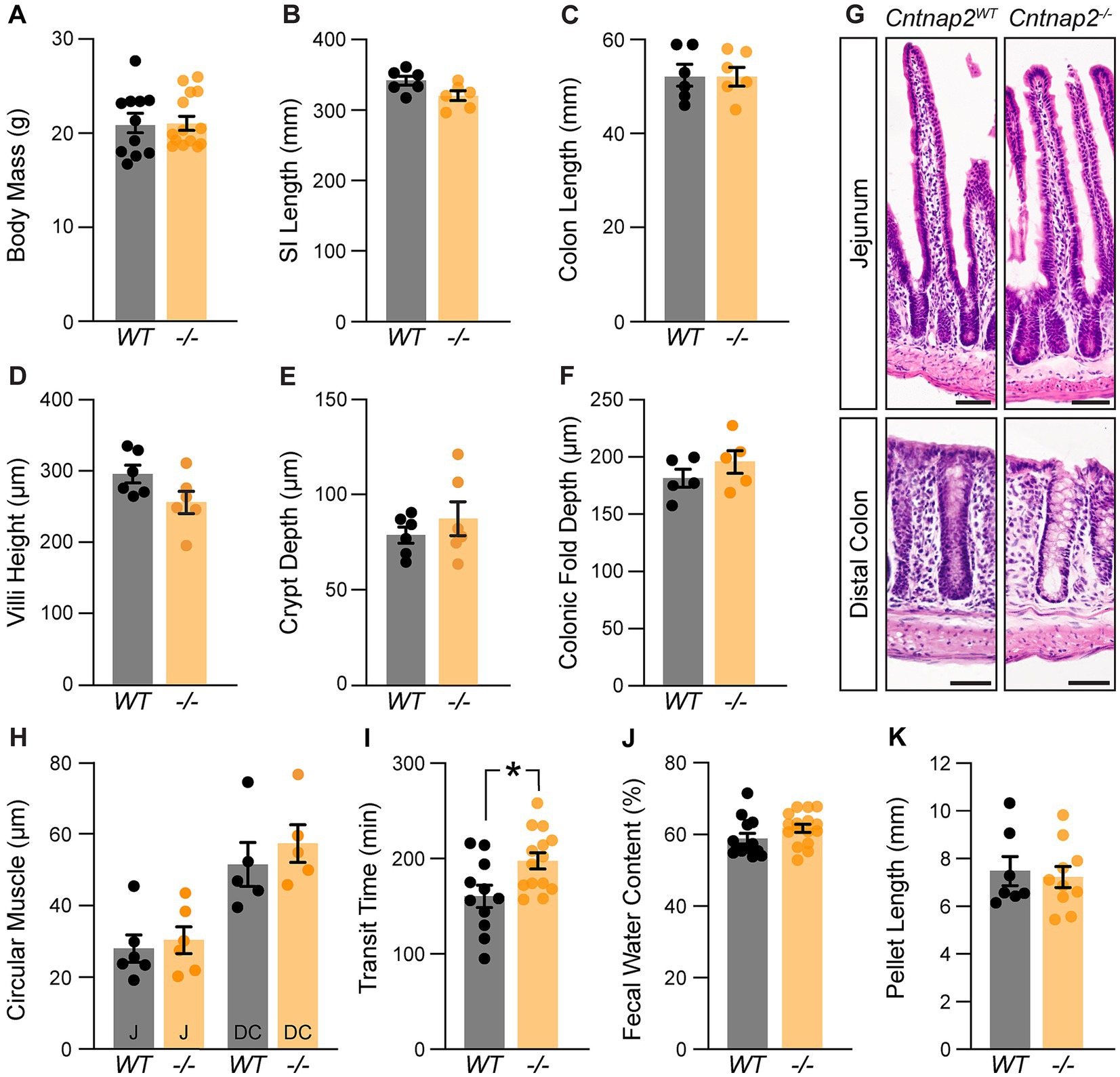
Figure 2. A role for Cntnap2 in whole GI transit. (A) Body mass is the same in Cntnap2WT and Cntnap2−/− mice [WT: 21.1 ± 1.0 g (n = 11); −/−: 21.3 ± 0.8 g (n = 14)]. Unpaired t-test, p = 0.89. (B) Length of small intestine is the same in Cntnap2WT and Cntnap2−/− mice [WT: 341.5 ± 6.5 mm (n = 6); −/−: 319.8 ± 7.0 mm (n = 6)]. Unpaired t-test, p = 0.05. (C) Length of colon is the same in Cntnap2WT and Cntnap2−/− mice [WT: 52.5 ± 2.3 mm (n = 6); −/−: 52.4 ± 1.9 mm (n = 6)]. Unpaired t-test, p = 0.98. (D) Villi height is the same in Cntnap2WT [294.8 ± 12.5 μm (n = 6)] and Cntnap2−/− [256.1 ± 15.6 μm (n = 6)] jejunum. Unpaired t-test, p = 0.08. (E) Crypt depth is the same in Cntnap2WT [78.6 ± 4.2 μm (n = 6)] and Cntnap2−/− [87.6 ± 8.8 μm (n = 6)] mice. Unpaired t-test, p = 0.38. (F) Depth of colonic folds is the same in Cntnap2WT [181.4 ± 7.8 μm (n = 5)] and Cntnap2−/− [195.9 ± 10.3 μm (n = 5)] mice. Unpaired t-test, p = 0.29. (G) H&E stained cross sections of jejunum and distal colon from Cntnap2WT and Cntnap2−/− mice. (H) Circular muscle thickness is the same in Cntnap2WT and Cntnap2−/− jejunum [WT: 27.9 ± 3.8 μm (n = 6); −/−: 30.2 ± 3.7 μm (n = 5)] and distal colon [WT: 51.5 ± 6.1 μm (n = 5); −/−: 57.4 ± 5.3 μm (n = 5)]. Two-way ANOVA: genotype, F(1,18) = 0.77, p = 0.39; region, F(1,18) = 29.12, p < 0.001; interaction, F(1,18) = 0.14, p = 0.71. (I) Whole GI transit time is increased in Cntnap2−/− [197.6 ± 8.5 min, (n = 14)] compared to Cntnap2WT [160.5 ± 11.6 min (n = 11)] mice. Unpaired t-test, p = 0.01. (J) Fecal water content is the same in 9 week old Cntnap2WT [58.8 ± 1.5% (n = 13)] and Cntnap2−/− [61.8 ± 1.1% (n = 16)] mice. Unpaired t-test, p = 0.12. (K) Fecal pellet length is the same in 9-week-old Cntnap2WT [7.5 ± 0.6 mm (n = 7)] and Cntnap2−/− [7.2 ± 0.4 mm (n = 10)] mice. Unpaired t-test, p = 0.73. All mice were 11 weeks old unless stated otherwise. Tukey’s multiple comparison test: *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar, 50 μm. WT: Cntnap2WT; −/−: Cntnap2−/−; SI: Small Intestine; J: Jejunum; DC: Distal Colon.
Whole gut transit provides information about stomach, small intestine and colon transit combined. To specifically focus on the colon, we next recorded and analyzed colonic motility using an ex vivo motility monitor (Swaminathan et al., 2016). In this setup, the colon is isolated from extrinsic innervation and allows us to assess loss of GI tract intrinsic Cntnap2. We generated spatiotemporal maps (STMs) of the empty colon (Figure 3A) and observed that CMCs were 31% shorter-lasting in Cntnap2−/− compared to Cntnap2WT mice (Figures 3F,G). CMC intervals, number, velocity, and length remained unchanged (Figures 3B–E,G). Thus, repetitive contractions are shortened in isolated empty Cntnap2 mutant colons.
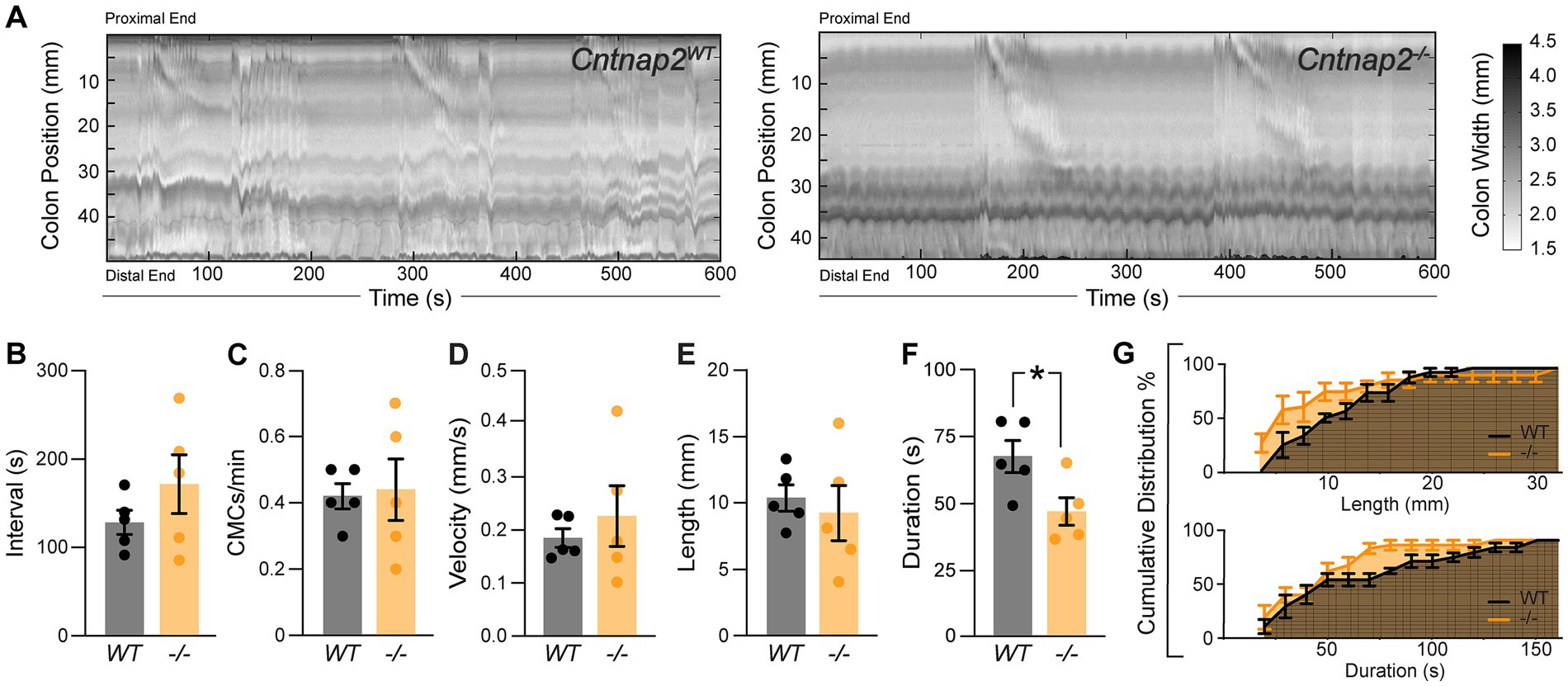
Figure 3. Altered ex-vivo motility in empty Cntnap2−/− colons. (A) Representative spatiotemporal maps of 10 min video recordings from Cntnap2WT and Cntnap2−/− empty colons. Gray scale indicates colonic diameter. (B) Intervals between CMC onsets are unchanged in Cntnap2−/− [171.7 ± 33.2 s (n = 5)] compared to Cntnap2WT [128.8 ± 13.7 s (n = 5)] mice. Unpaired t-test, p = 0.27. (C) Number of CMCs per minute are the same in Cntnap2WT [0.42 ± 0.04 (n = 5)] and Cntnap2−/− [0.44 ± 0.09 (n = 5)] mice. Unpaired t-test, p = 0.84. (D) CMC velocity is the same in Cntnap2WT [0.18 ± 0.02 mm/s (n = 5)] and Cntnap2−/− [0.23 ± 0.06 mm/s (n = 5)] mice. Unpaired t-test, p = 0.49. (E) Length of CMCs is the same in Cntnap2WT [10.3 ± 1.0 mm, (n = 5)] and Cntnap2−/− [9.3 ± 2.1 mm (n = 5)] mice. Unpaired t-test, p = 0.66. (F) CMCs are shorter-lasting in Cntnap2−/− [47.6 ± 5.2 s (n = 5)] compared to Cntnap2WT [68.5 ± 6.0 (n = 5)] mice. Unpaired t-test, p = 0.03. (G) Cumulative distributions for CMC length (mm) and duration (s) in Cntnap2WT and Cntnap2−/− mice.
Given that IPANs are sensitive to stretch (Furness et al., 2004), we next assessed ex vivo colonic motility of Cntnap2−/− mice in response to a stimulus. We inserted a natural-shaped 3D-printed artificial fecal pellet through the proximal colon and recorded colonic behavior until complete pellet expulsion (Costa et al., 2021). The artificial pellet served as a normalized stimulus that was able to travel the entire length of the mid and distal colon (Figures 4A,B). The time to pellet expulsion was shortened by 51% in Cntnap2−/− mice compared to Cntnap2WT littermate controls (Figure 4C). Using TrackMate (v7.6.1) (Tinevez et al., 2017; Ershov et al., 2021) to create a trace of the pellet’s movement (Figure 4A’), we observed a 42% reduction in the number of pellet movements in Cntnap2−/− compared to Cntnap2WT mice (Figure 4D), which was also visible when plotting mean pellet speed along the length of the colon (Figure 4E). The mean pellet speed per trial trended higher in Cntnap2−/− mice (Figure 4F), but was not statistically significant. Maximum pellet speed was unchanged (Figure 4G). Thus, artificial fecal pellets move more continuously, and colonic transit is accelerated in isolated Cntnap2 mutant colons.
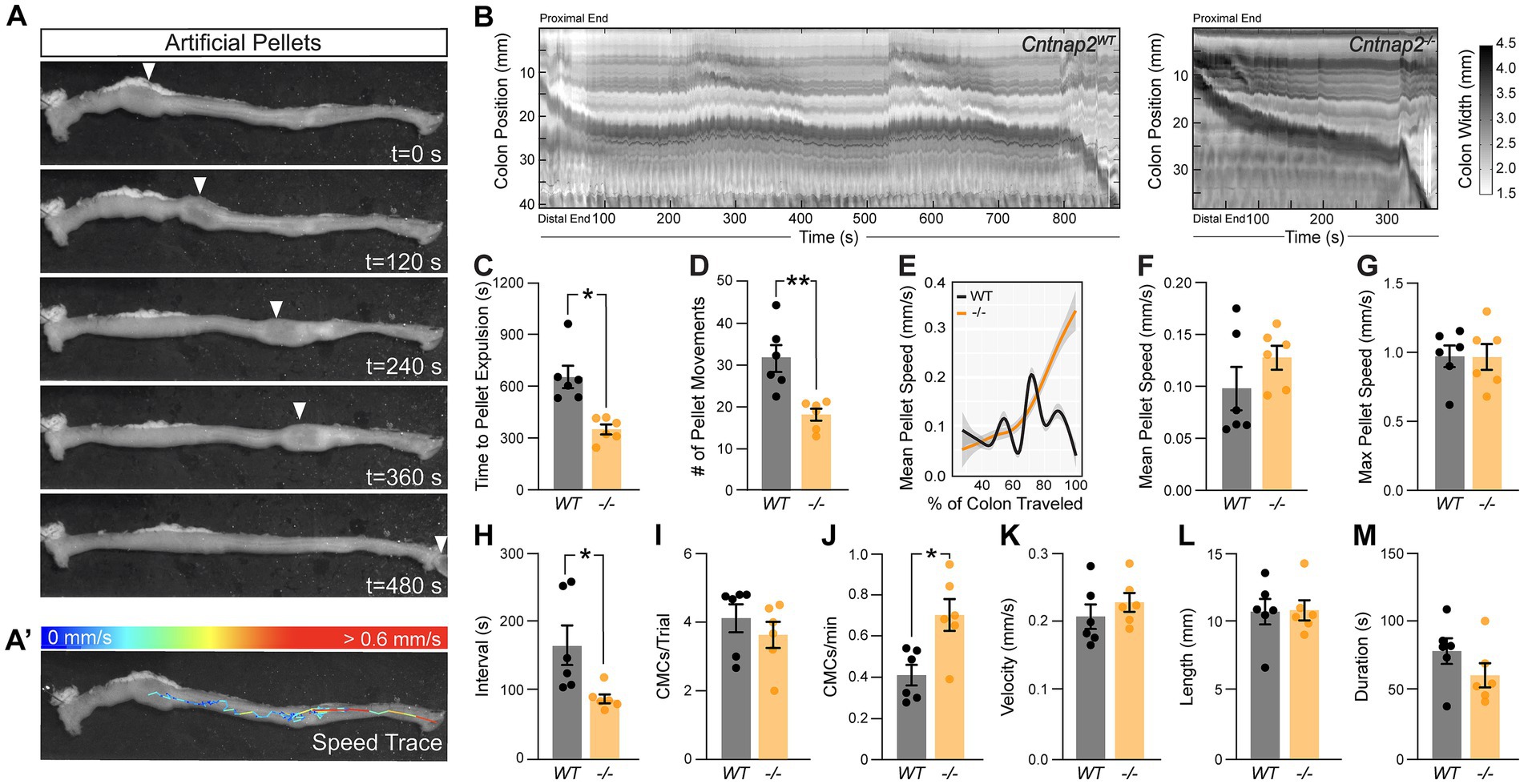
Figure 4. Accelerated ex-vivo pellet expulsion in Cntnap2−/− colons. (A) Time series images of artificial pellet traversing the colon over time. White arrowheads indicate center of artificial pellet. (A’) Trace of pellet path with speed of pellet represented by color scale. (B) Representative spatiotemporal maps of full-length artificial pellet trials from Cntnap2WT and Cntnap2−/− mice. Gray scale indicates colonic diameter. (C) Time to pellet expulsion is shorter in Cntnap2−/− mice [350.2 ± 28.2 s (n = 6)] compared to Cntnap2WT [657.9 ± 65.0 s (n = 6)] mice. Unpaired t-test, p = 0.002. (D) Number of pellet movement intervals is significantly reduced in Cntnap2−/− [18.2 ± 1.4 (n = 6)] when compared to Cntnap2WT [31.5 ± 3.2 (n = 6)] mice. Unpaired t-test, p = 0.003. (E) Local regression (LOESS) of mean pellet speed for each genotype as a function of the % of colon length traveled. 95% confidence interval shown in gray. (F) Mean speed of artificial pellet per trial is the same in Cntnap2WT [0.10 ± 0.02 mm/s (n = 6)] and Cntnap2−/− [0.13 ± 0.01 mm/s (n = 6)] mice. Unpaired t-test, p = 0.24. (G) Max speed of pellet is the same in Cntnap2WT [0.97 ± 0.08 mm/s (n = 6)] and Cntnap2−/− [0.97 ± 0.09 mm/s (n = 6)] mice. Unpaired t-test, p = 0.96. (H) Intervals between CMCs are significantly reduced in Cntnap2−/− [87.1 ± 6.6 s (n = 6)] compared to Cntnap2WT [165.3 ± 29.0 s (n = 6)] mice. Unpaired t-test, p = 0.03. (I) Number of CMCs during trial period are similar between Cntnap2WT [4.1 ± 0.4 (n = 6)] and Cntnap2−/− [3.6 ± 0.4 min (n = 6)] mice. Unpaired t-test, p = 0.4. (J) Number of CMCs per minute are increased in Cntnap2−/− [0.71 ± 0.08 (n = 6)] compared to Cntnap2WT [0.41 ± 0.05 (n = 6)] mice. Unpaired t-test, p = 0.0099. (K) Velocity of CMCs is the same in Cntnap2WT [0.21 ± 0.02 mm/s (n = 6)] and Cntnap2−/− [0.23 ± 0.01 mm/s (n = 6)] mice. Unpaired t-test, p = 0.38. (L) Length of CMCs is the same in Cntnap2WT [10.8 ± 0.9 mm (n = 6)] and Cntnap2−/− [10.9 ± 0.7 mm (n = 6)] mice. Unpaired t-test, p = 0.93. (M) Duration of CMCs is the same in Cntnap2WT [79.1 ± 9.4 s (n = 6)] and Cntnap2−/− [61.5 ± 8.8 s (n = 6)] mice. Unpaired t-test, p = 0.2. Tukey’s multiple comparison test: *p < 0.05, **p < 0.01, ***p < 0.001. WT: Cntnap2WT; −/−: Cntnap2−/−.
We next performed STM analysis of these ex vivo colonic motility data in the presence of a stimulus. The interval between CMC onsets was shortened by 47% in Cntnap2−/− compared to Cntnap2WT mice (Figure 4H), while the number of CMCs per trial remained the same (Figure 4I). As a result, CMC frequency was 73% increased in Cntnap2−/− mice (Figure 4J). CMC velocity, length, and duration remained the same (Figures 4K–M). These findings suggest that in the presence of a luminal stimulus, CMC frequency is increased.
Given the predominant expression of Cntnap2 in IPANs, we next asked whether lack of Cntnap2 impacts IPANs. The total number and distribution of HuC/D+ myenteric neurons in the distal colon were unchanged in Cntnap2−/− mice (Figures 5A–E). Also, the number of Nmu+ neurons were unchanged (Figures 5F,G). Thus, the number of myenteric plexus-resident sensory neurons is unchanged.
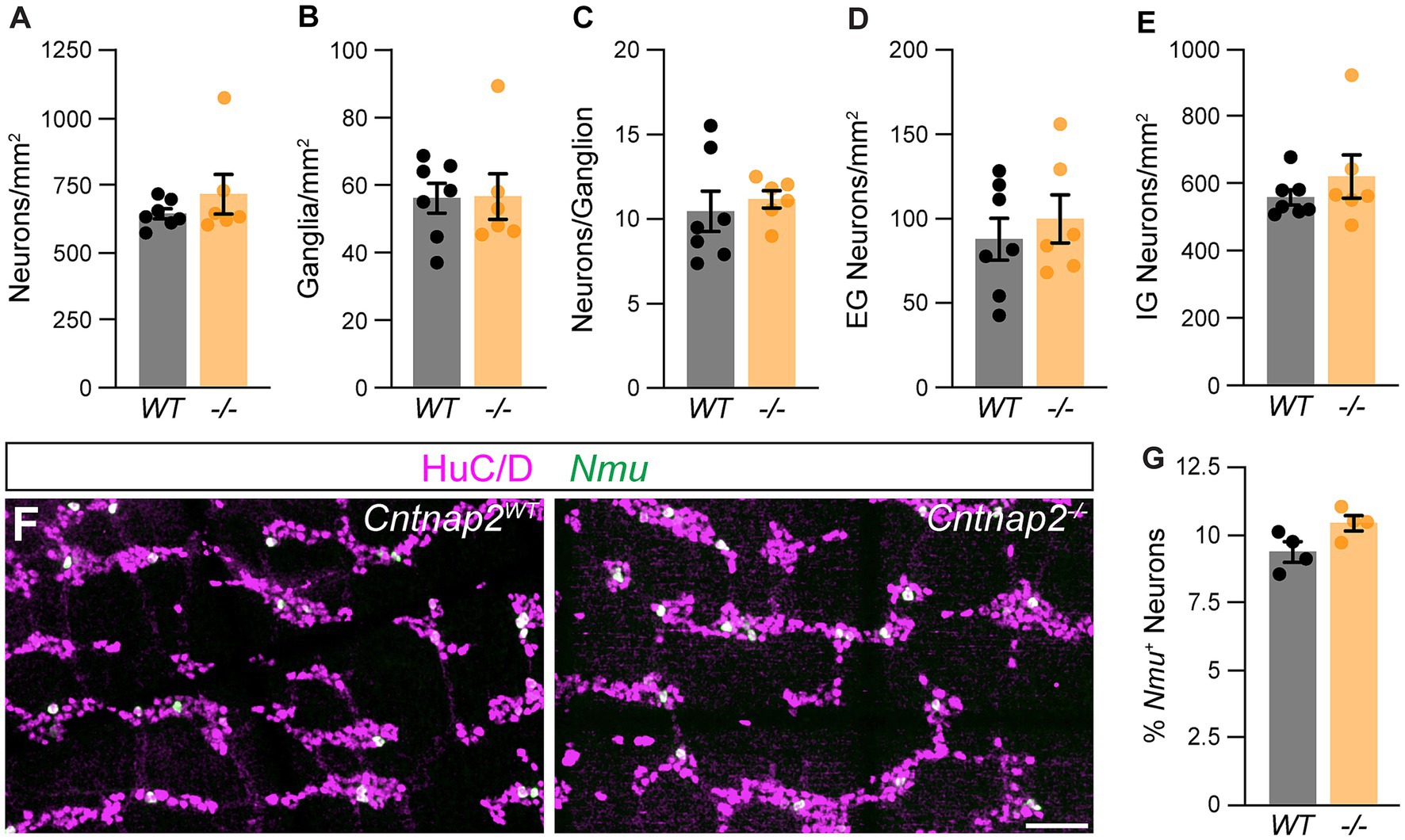
Figure 5. Normal ENS organization in Cntnap2mutant distal colon. (A) Number of HuC/D+ neurons is the same in Cntnap2WT [647.7 ± 18.8 (n = 7)] and Cntnap2−/− [720.9 ± 73.6 (n = 6)] mice. Unpaired t-test, p = 0.32. (B) Number of enteric ganglia is the same in Cntnap2−/− [56.7 ± 6.8 (n = 6)] compared to Cntnap2WT [56.2 ± 4.4 (n = 7)] mice. Unpaired t-test, p = 0.95. (C) Number of neurons per ganglion is unchanged in Cntnap2−/− [11.2 ± 0.5 (n = 6)] compared to Cntnap2WT [10.5 ± 1.2 (n = 7)] mice. Unpaired t-test, p = 0.62. (D,E) Number of extra-(D) and intra-ganglionic (E) neurons are similar in Cntnap2WT [extra: 88.7 ± 12.6; intra: 559.0 ± 22.7 (n = 7)] and Cntnap2−/− mice [extra: 100.8 ± 14.4; intra: 620.1 ± 23.8 (n = 7)]. Unpaired t-test, P (extra, intra) = 0.54, 0.36. (F) Nmu (green) is expressed in a subset of HuC/D+ (magenta) neurons in Cntnap2WT and Cntnap2−/− distal colon. (G) Percent of Nmu+ HuC/D+ neurons is unchanged in Cntnap2−/− [10.5 ± 0.3 (n = 4)] compared to Cntnap2WT [9.4 ± 0.3 (n = 4)] mice. Unpaired t-test, p = 0.05. Scale bar, 50 μm.
4. Discussion
GI dysfunction is a prevalent symptom in individuals with ASD (Holingue et al., 2018). In this study, we aimed to determine whether the ASD-related gene, Cntnap2, plays a role in mouse GI function by characterizing Cntnap2’s expression in the intestines and assessing colonic function and ENS organization in Cntnap2−/− mice. Our findings reveal that Cntnap2 is expressed in colonic sensory neurons, and a subset of progenitor/glial cells and intestinal epithelial cells. Whole gut transit is slowed in Cntnap2−/− mice and repetitive contractions are shortened in isolated empty Cntnap2 mutant colons. In the presence of a luminal stimulus, CMC frequency is increased and colonic transit is accelerated in isolated Cntnap2 mutant colons. The overall organization of the ENS appears unchanged.
Sensory over-responsivity has been correlated with the presence of GI issues in children diagnosed with ASD (Mazurek et al., 2013) and Cntnap2 has been linked to sensory processing deficits and hypersensitivity in the mouse CNS and PNS (Peñagarikano et al., 2011; Dawes et al., 2018; Fernández et al., 2021). Our finding that Cntnap2 is expressed in the majority of Nmu+ IPANs is consistent with scRNA-seq studies reporting a high expression of Cntnap2 in putative enteric sensory neuron classes in mice (Zeisel et al., 2018; Drokhlyansky et al., 2020; Morarach et al., 2021). RNA-seq data from human colon also show CNTNAP2 expression in a subset of enteric sensory neurons, in addition to pronounced expression in subsets interneurons and excitatory motor neurons (Drokhlyansky et al., 2020). IPANs are thought to be critical for initiating propulsive CMCs and downstream motility patterns (Spencer et al., 2018; Fung et al., 2021; Nestor-Kalinoski et al., 2021; Feng et al., 2022) and the observed changes to CMCs in ex vivo Cntnap2−/− colon preparations suggest a role for Cntnap2 in enteric sensory function.
One caveat of this study is that the germline Cntnap2 deletion model deletes Cntnap2 not only in intrinsic ENS cells, but also from a small number of progenitor/glial cells and EECs that might contribute to the observed phenotypes (Rao et al., 2017; Servin-Vences et al., 2022). Further, in the SI, Cntnap2 might be additionally expressed in other ENS subsets, such as cholinergic interneurons (Zeisel et al., 2018; Morarach et al., 2021).
Recent studies have provided growing evidence for the role of ASD-related genes in GI function. Other ASD mouse models that have been used to investigate ENS organization include Slc6a4−/− (SERTKO) mice, SERT Ala56 mice (common SERT variant), Nlgn3−/− mice, and NL3R451C mice (human neuroligin-3 R451C missense mutation; Margolis et al., 2016;Hosie et al., 2019; Leembruggen et al., 2019). Three out of these four mouse models show changes to the ENS and all mutants demonstrate altered GI function. SERT Ala56 mice have a hypoplastic ENS and SERTKO mice have a hyperplastic ENS (Margolis et al., 2016), both resulting in slower colonic motility. NL3R451C mice have an increased number of neurons in the SI (Hosie et al., 2019; Leembruggen et al., 2019), while Nlgn3−/− mice have overall normal numbers of enteric neurons. We find no changes in the number and organization of neurons within the myenteric plexus, but given the function of Caspr2 as a cell-adhesion molecule in neural circuit assembly (Anderson et al., 2012) further investigation is needed to assess whether the connectivity and function, particularly of colonic IPANs, is altered in Cntnap2−/− mice.
While Cntnap2 was first identified as playing a role in the localization of potassium channels to the juxtaparanodal regions of myelinated axons (Poliak et al., 1999, 2003), considering Cntnap2’s described roles and functions in other systems may shed light on its mechanisms of action in the unmyelinated ENS. Cntnap2 regulates the excitability of DRG sensory neurons by altering Kv1 channel function (Dawes et al., 2018). The absence of Cntnap2 leads to a reduction in overall expression of Kv1.2 channels at the soma membrane of DRG neurons, resulting in altered electrical properties and increased neuronal excitability (Dawes et al., 2018). The gene encoding Kv1.2 (Kcna2) is also highly expressed in colonic sensory neurons (Drokhlyansky et al., 2020). Furthermore, similar to the altered cerebellar response to somatosensory stimuli previously reported in Cntnap2−/− mice (Fernández et al., 2021), IPANs may become hyperexcitable when activated in the absence of Cntnap2, contributing to increased CMC frequency and a shorter time to expulsion during the artificial pellet assay. In contrast, repetitive contractions in isolated empty Cntnap2 mutant colons were shortened and we attribute this difference in ex vivo colonic motility to the differential activation of sensory neurons (empty colon versus artificial fecal pellet). IPAN-specific manipulations will be instrumental in further investigating the role of Cntnap2 in gut-intrinsic sensory neurons.
By demonstrating altered GI motility, the Cntnap2−/− mouse model contributes to our understanding of the relationship between ASD and GI dysfunction. Our findings show that, in addition to previously described phenotypes in the CNS and PNS, Cntnap2−/− mice display changes in colonic motility. Cntnap2’s expression in enteric sensory neurons suggest that sensory dysfunction might contribute to disrupted GI motility. Our findings therefore may have important implications for the diagnosis and treatment of GI symptoms in individuals with ASD.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Stanford University Administrative Panel on Laboratory Animal Care. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BR: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft, Writing – review & editing. BO: Writing – review & editing, Formal analysis, Methodology. KR: Formal analysis, Methodology, Writing – review & editing. JK: Conceptualization, Data curation, Funding acquisition, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by an National Institutes of Health (NIH) NIMH T32 Stanford Neurosciences Program Training Grant, T32MH020016 (BR and KR); a Bertarelli Foundation Fellowship, the Stanford Master of Science in Medicine Program (BR); a BP-ENDURE grant from NIH NINDS awarded to the University of Nevada, Reno, R25NS119709 (BO); the Wu Tsai Neurosciences Institute, the Stanford University Department of Neurosurgery, and research grants from The Shurl and Kay Curci Foundation, and The Firmenich Foundation (JK).
Acknowledgments
We thank J. Gomez-Frittelli, R. Hamnett, and C. Plant for feedback on the manuscript. We thank V. Lennon (Mayo Clinic) for the HuC/D antibody, J. Sanes (Harvard) for the β-galactosidase antibody, and G. Hennig for the Volumetry software. We thank the Human Pathology/Histology Service Center at Stanford School of Medicine for their slide scanning services. We acknowledge the usage of generative AI technology (ChatGPT May 24 version) for proofreading parts of the discussion section. We have followed all Frontiers guidelines and policies, including verifying the factual accuracy and ensuring plagiarism-free content, and made every reasonable attempt to correct obvious errors.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2023.1287057/full#supplementary-material
References
Anderson, G. R., Galfin, T., Xu, W., Aoto, J., Malenka, R. C., and Südhof, T. C. (2012). Candidate autism gene screen identifies critical role for cell-adhesion molecule CASPR2 in dendritic arborization and spine development. Proc. Natl. Acad. Sci. 109, 18120–18125. doi: 10.1073/pnas.1216398109
Avetisyan, M., Schill, E. M., and Heuckeroth, R. O. (2015). Building a second brain in the bowel. J. Clin. Investig. 125, 899–907. doi: 10.1172/JCI76307
Buniello, A., MacArthur, J. A. L., Cerezo, M., Harris, L. W., Hayhurst, J., Malangone, C., et al. (2019). The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012. doi: 10.1093/nar/gky1120
Chaidez, V., Hansen, R. L., and Hertz-Picciotto, I. (2014). Gastrointestinal problems in children with autism, developmental delays or typical development. J. Autism Dev. Disord. 44, 1117–1127. doi: 10.1007/s10803-013-1973-x
Corsetti, M., Costa, M., Bassotti, G., Bharucha, A. E., Borrelli, O., Dinning, P., et al. (2019). First translational consensus on terminology and definitions of colonic motility in animals and humans studied by manometric and other techniques. Nat. Rev. Gastroenterol. Hepatol. 16, 559–579. doi: 10.1038/s41575-019-0167-1
Costa, M., Keightley, L. J., Hibberd, T. J., Wiklendt, L., Dinning, P. G., Brookes, S. J., et al. (2021). Motor patterns in the proximal and distal mouse colon which underlie formation and propulsion of feces. Neurogastroenterol. Motil. 2020, 1–14. doi: 10.1111/nmo.14098
Dawes, J. M., Weir, G. A., Middleton, S. J., Patel, R., Chisholm, K. I., Pettingill, P., et al. (2018). Immune or genetic-mediated disruption of CASPR2 causes pain hypersensitivity due to enhanced primary afferent excitability. Neuron 97, 806–822.e10. doi: 10.1016/j.neuron.2018.01.033
De Lisle, R. C. (2007). Altered transit and bacterial overgrowth in the cystic fibrosis mouse small intestine. American journal of physiology-gastrointestinal and liver. Physiology 293, G104–G111. doi: 10.1152/ajpgi.00548.2006
Drokhlyansky, E., Smillie, C. S., Wittenberghe, N. V., Ericsson, M., Griffin, G. K., Eraslan, G., et al. (2020). The human and mouse enteric nervous system at single-cell resolution. Cells 182, 1606–1622.e23. doi: 10.1016/j.cell.2020.08.003
Ershov, D., Phan, M.-S., Pylvänäinen, J. W., Rigaud, S. U., Le Blanc, L., Charles-Orszag, A., et al. (2021). Bringing TrackMate into the era of machine-learning and deep-learning. BioRxiv 2021:458852. doi: 10.1101/2021.09.03.458852
Feng, J., Hibberd, T. J., Luo, J., Yang, P., Xie, Z., Travis, L., et al. (2022). Modification of neurogenic colonic motor Behaviours by Chemogenetic ablation of Calretinin neurons. Front. Cell. Neurosci. 16:799717. doi: 10.3389/fncel.2022.799717
Fernández, M., Sánchez-León, C. A., Llorente, J., Sierra-Arregui, T., Knafo, S., Márquez-Ruiz, J., et al. (2021). Altered cerebellar response to somatosensory stimuli in the Cntnap2 mouse model of autism. ENeuro 8:333. doi: 10.1523/ENEURO.0333-21.2021
Fida, R., Bywater, R. A. R., Lyster, D. J. K., and Taylor, G. S. (2000). Chronotropic action of 5-hydroxytryptamine (5-HT) on colonic migrating motor complexes (CMMCs) in the isolated mouse colon. J. Auton. Nerv. Syst. 80, 52–63. doi: 10.1016/s0165-1838(00)00074-6
Fung, C., and Berghe, P. V. (2020). Functional circuits and signal processing in the enteric nervous system. Cell. Mol. Life Sci. 77, 4505–4522. doi: 10.1007/s00018-020-03543-6
Fung, C., Hao, M. M., Obata, Y., Tack, J., Pachnis, V., Boesmans, W., et al. (2021). Luminal nutrients activate distinct patterns in submucosal and myenteric neurons in the mouse small intestine. BioRxiv, 1–46. doi: 10.1101/2021.01.19.427232
Furness, J. B., Bornstein, J. C., Pompolo, S., Young, H. M., Kunze, W., and Kelly, H. (1994). The circuitry of the enteric nervous system. Neurogastroenterol. Motil. 6, 241–253. doi: 10.1111/j.1365-2982.1994.tb00190.x
Furness, J. B., Jones, C., Nurgali, K., and Clerc, N. (2004). Intrinsic primary afferent neurons and nerve circuits within the intestine. Prog. Neurobiol. 72, 143–164. doi: 10.1016/j.pneurobio.2003.12.004
Gordon, A., Salomon, D., Barak, N., Pen, Y., Tsoory, M., Kimchi, T., et al. (2016). Expression of Cntnap2 (Caspr2) in multiple levels of sensory systems. Mol. Cell. Neurosci. 70, 42–53. doi: 10.1016/j.mcn.2015.11.012
Graf, R., Campbell, S., Donabedian, D., Preston, G., Hanania, T., Thiede, L., et al. (2019). Characterization of GI barrier integrity and gut microbiome-derived metabolites in BTBR, Shank3 and Cntnap2 mouse models of ASD, and demonstration of AB-2004 as a potential mitigating therapeutic. INSAR: Palais des Congres de Montreal Montreal, Québec, Canada, poster #30563. Available at: https://insar.confex.com/insar/2019/webprogram/Paper30563.html.
Guyer, R. A., Stavely, R., Robertson, K., Bhave, S., Mueller, J. L., Picard, N. M., et al. (2023). Single-cell multiome sequencing clarifies enteric glial diversity and identifies an intraganglionic population poised for neurogenesis. Cell Rep. 42:112194. doi: 10.1016/j.celrep.2023.112194
Hamnett, R., Dershowitz, L. B., Sampathkumar, V., Wang, Z., Gomez-Frittelli, J., De Andrade, V., et al. (2022). Regional cytoarchitecture of the adult and developing mouse enteric nervous system. Curr. Biol. 32, 4483–4492.e5. doi: 10.1016/j.cub.2022.08.030
Hennig, G. W., Costa, M., Chen, B. N., and Brookes, S. J. H. (1999). Quantitative analysis of peristalsis in the guinea-pig small intestine using spatio-temporal maps. J. Physiol. 517, 575–590. doi: 10.1111/j.1469-7793.1999.0575t.x
Holingue, C., Newill, C., Lee, L., Pasricha, P. J., and Fallin, M. D. (2018). Gastrointestinal symptoms in autism spectrum disorder: A review of the literature on ascertainment and prevalence. Autism Res. 11, 24–36. doi: 10.1002/aur.1854
Hosie, S., Ellis, M., Swaminathan, M., Ramalhosa, F., Seger, G. O., Balasuriya, G. K., et al. (2019). Gastrointestinal dysfunction in patients and mice expressing the autism-associated R451C mutation in neuroligin-3. Autism Res. 12, 1043–1056. doi: 10.1002/aur.2127
James, D. M., Kozol, R. A., Kajiwara, Y., Wahl, A. L., Storrs, E. C., Buxbaum, J. D., et al. (2019). Intestinal dysmotility in a zebrafish (Danio rerio) shank3a;shank3b mutant model of autism. Mol. Autism. 10:3. doi: 10.1186/s13229-018-0250-4
Kobayashi, Y., Bukowski, A., Das, S., Espenel, C., Gomez-Frittelli, J., Wagle, N., et al. (2021). Counten, an ai-driven tool for rapid and objective structural analyses of the enteric nervous system. ENeuro 8:92. doi: 10.1523/ENEURO.0092-21.2021
Leembruggen, A. J. L., Balasuriya, G. K., Zhang, J., Schokman, S., Swiderski, K., Bornstein, J. C., et al. (2019). Colonic dilation and altered ex vivo gastrointestinal motility in the neuroligin-3 knockout mouse. Autism Res. 82, 691–701. doi: 10.1002/aur.2109
Maenner, M. J., Warren, Z., Williams, A. R., Amoakohene, E., Bakian, A. V., Bilder, D. A., et al. (2023). Prevalence and characteristics of autism Spectrum disorder among children aged 8 years–autism and developmental disabilities monitoring network, 11 sites, United States, 2020. MMWR Surveill. Summ. 72, 1–14. doi: 10.15585/mmwr.ss7202a1
Margolis, K. G., Li, Z., Stevanovic, K., Saurman, V., Israelyan, N., Anderson, G. M., et al. (2016). Serotonin transporter variant drives preventable gastrointestinal abnormalities in development and function. J. Clin. Investig. 126, 2221–2235. doi: 10.1172/JCI84877
Mazurek, M. O., Vasa, R. A., Kalb, L. G., Kanne, S. M., Rosenberg, D., Keefer, A., et al. (2013). Anxiety, sensory over-responsivity, and gastrointestinal problems in children with autism Spectrum disorders. J. Abnorm. Child Psychol. 41, 165–176. doi: 10.1007/s10802-012-9668-x
Morarach, K., Mikhailova, A., Knoflach, V., Memic, F., Kumar, R., Li, W., et al. (2021). Diversification of molecularly defined myenteric neuron classes revealed by single-cell RNA sequencing. Nat. Neurosci. 24, 34–46. doi: 10.1038/s41593-020-00736-x
Nestor-Kalinoski, A., Smith-Edwards, K. M., Meerschaert, K., Margiotta, J. F., Rajwa, B., Davis, B. M., et al. (2021). Unique neural circuit connectivity of mouse proximal, middle, and distal Colon defines regional colonic motor patterns. Cell. Mol. Gastroenterol. Hepatol. 13, 309–337.e3. doi: 10.1016/j.jcmgh.2021.08.016
Niesler, B., and Rappold, G. A. (2021). Emerging evidence for gene mutations driving both brain and gut dysfunction in autism spectrum disorder. Mol. Psychiatry 26, 1442–1444. doi: 10.1038/s41380-020-0778-5
Peñagarikano, O., Abrahams, B. S., Herman, E. I., Winden, K. D., Gdalyahu, A., Dong, H., et al. (2011). Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and Core autism-related deficits. Cells 147, 235–246. doi: 10.1016/j.cell.2011.08.040
Peñagarikano, O., and Geschwind, D. H. (2012). What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol. Med. 18, 156–163. doi: 10.1016/j.molmed.2012.01.003
Poliak, S., Gollan, L., Martinez, R., Custer, A., Einheber, S., Salzer, J. L., et al. (1999). Caspr2, a new member of the Neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron 24, 1037–1047. doi: 10.1016/s0896-6273(00)81049-1
Poliak, S., Salomon, D., Elhanany, H., Sabanay, H., Kiernan, B., Pevny, L., et al. (2003). Juxtaparanodal clustering of shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J. Cell Biol. 162, 1149–1160. doi: 10.1083/jcb.200305018
Progatzky, F., Shapiro, M., Chng, S. H., Garcia-Cassani, B., Classon, C. H., Sevgi, S., et al. (2021). Regulation of intestinal immunity and tissue repair by enteric glia. Nature 599, 125–130. doi: 10.1038/s41586-021-04006-z
Qu, Z.-D. D., Thacker, M., Castelucci, P., Bagyánszki, M., Epstein, M. L., and Furness, J. B. (2008). Immunohistochemical analysis of neuron types in the mouse small intestine. Cell Tissue Res. 334, 147–161. doi: 10.1007/s00441-008-0684-7
Rao, M., and Gershon, M. D. (2016). The bowel and beyond: the enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 13, 517–528. doi: 10.1038/nrgastro.2016.107
Rao, M., Rastelli, D., Dong, L., Chiu, S., Setlik, W., Gershon, M. D., et al. (2017). Enteric glia regulate gastrointestinal motility but are not required for maintenance of the epithelium in mice. Gastroenterology 153, 1068–1081.e7. doi: 10.1053/j.gastro.2017.07.002
Restrepo, B., Angkustsiri, K., Taylor, S. L., Rogers, S. J., Cabral, J., Heath, B., et al. (2020). Developmental–behavioral profiles in children with autism spectrum disorder and co-occurring gastrointestinal symptoms. Autism Res. 13, 1778–1789. doi: 10.1002/aur.2354
Sasselli, V., Pachnis, V., and Burns, A. J. (2012). The enteric nervous system. Dev. Biol. 366, 64–73. doi: 10.1016/j.ydbio.2012.01.012
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Servin-Vences, M. R., Lam, R. M., Koolen, A., Wang, Y., Saade, D. N., Loud, M., et al. (2022). PIEZO2 in somatosensory neurons controls gastrointestinal transit. bioRxiv 2022:518109. doi: 10.1101/2022.11.27.518109
Spear, E. T., Holt, E. A., Joyce, E. J., Haag, M. M., Mawe, S. M., Hennig, G. W., et al. (2018). Altered gastrointestinal motility involving autoantibodies in the experimental autoimmune encephalomyelitis model of multiple sclerosis. Neurogastroenterol. Motil. 30:e13349. doi: 10.1111/nmo.13349
Spencer, N. J., Hibberd, T. J., Travis, L., Wiklendt, L., Costa, M., Hu, H., et al. (2018). Identification of a rhythmic firing pattern in the enteric nervous system that generates rhythmic electrical activity in smooth muscle. J. Neurosci. 38, 5507–5522. doi: 10.1523/JNEUROSCI.3489-17.2018
Spencer, N. J., and Hu, H. (2020). Enteric nervous system: sensory transduction, neural circuits and gastrointestinal motility. Nat. Rev. Gastroenterol. Hepatol. 17, 338–351. doi: 10.1038/s41575-020-0271-2
Swaminathan, M., Fung, C., Finkelstein, D. I., Bornstein, J. C., and Foong, J. P. P. (2019). α-Synuclein regulates development and function of cholinergic enteric neurons in the mouse Colon. Neuroscience 423, 76–85. doi: 10.1016/j.neuroscience.2019.10.029
Swaminathan, M., Hill-Yardin, E., Ellis, M., Zygorodimos, M., Johnston, L. A., Gwynne, R. M., et al. (2016). Video imaging and spatiotemporal maps to analyze gastrointestinal motility in mice. J. Vis. Exp. 108:53828. doi: 10.3791/53828
Tinevez, J.-Y., Perry, N., Schindelin, J., Hoopes, G. M., Reynolds, G. D., Laplantine, E., et al. (2017). TrackMate: an open and extensible platform for single-particle tracking. Methods 115, 80–90. doi: 10.1016/j.ymeth.2016.09.016
Valicenti-McDermott, M., McVicar, K., Rapin, I., Wershil, B. K., Cohen, H., and Shinnar, S. (2006). Frequency of gastrointestinal symptoms in children with autistic spectrum disorders and association with family history of autoimmune disease. J. Dev. Behav. Pediatr. 27, S128–S136. doi: 10.1097/00004703-200604002-00011
Williams, J. M., Duckworth, C. A., Vowell, K., Burkitt, M. D., and Pritchard, D. M. (2016). Intestinal preparation techniques for histological analysis in the mouse. Curr. Protoc. Mouse Biol. 6, 148–168. doi: 10.1002/cpmo.2
Keywords: autism spectrum disorder, gastrointestinal dysmotility, Cntnap2, Caspr2, enteric nervous system, sensory neurons, intrinsic primary afferent neurons
Citation: Robinson BG, Oster BA, Robertson K and Kaltschmidt JA (2023) Loss of ASD-related molecule Cntnap2 affects colonic motility in mice. Front. Neurosci. 17:1287057. doi: 10.3389/fnins.2023.1287057
Edited by:
Lihua Ye, The Ohio State University, United StatesReviewed by:
Howard Sirotkin, Stony Brook University, United StatesFränze Progatzky, University of Oxford, United Kingdom
Copyright © 2023 Robinson, Oster, Robertson and Kaltschmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia A. Kaltschmidt, anVrYWx0c0BzdGFuZm9yZC5lZHU=; @kaltschmidt_lab
 Beatriz G. Robinson1,2
Beatriz G. Robinson1,2 Keiramarie Robertson
Keiramarie Robertson Julia A. Kaltschmidt
Julia A. Kaltschmidt