
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Neurosci. , 06 October 2022
Sec. Neuropharmacology
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.986874
This article is part of the Research Topic Innovative Treatments for Neuro-Psychiatric Diseases View all 5 articles
Epilepsy is a common and disastrous neurological disorder characterized by abnormal firing of neurons in the brain, affecting about 70 million people worldwide. Long non-coding RNAs (LncRNAs) are a class of RNAs longer than 200 nucleotides without the capacity of protein coding, but they participate in a wide variety of pathophysiological processes. Alternated abundance and diversity of LncRNAs have been found in epilepsy patients and animal or cell models, suggesting a potential role of LncRNAs in epileptogenesis. This review will introduce the structure and function of LncRNAs, summarize the role of LncRNAs in the pathogenesis of epilepsy, especially its linkage with neuroinflammation, apoptosis, and transmitter balance, which will throw light on the molecular mechanism of epileptogenesis, and accelerate the clinical implementation of LncRNAs as a potential therapeutic target for treatment of epilepsy.
Epilepsy (recurrent unprovoked seizures) is a chronic disorder of transient brain dysfunction caused by sudden, abnormal firing of neurons, characterized by unpredictably recurrent seizures involve part or the entire body, leading to severe developmental and functional effects (World Health Organization., 2022). There are about 70 million epilepsy patients worldwide, and infants and the elderly accounting for the largest number of cases (Thijs et al., 2019). Once diagnosed, the first-line treatments for epilepsy patients are antiepileptic drugs and anticonvulsants, which aim to control seizures via regulating neuronal excitability through balancing the abundance of transmitters, especially γ-aminobutyric acid (GABA) and glutamate (Glu). However, there are many adverse drug effects, and one-third of patients have drug-refractory epilepsy. Moreover, lack of professional medical care is still the leading cause of the increased risk of epilepsy (Singh and Trevick, 2016). About 130,000 people die of epilepsy each year, with an annual direct cost of 1736–5848 US dollars and indirect cost of 2037–8587 US dollars for each epilepsy patient (Allers et al., 2015; Singh and Sander, 2020). Thus, epilepsy has been a great public health issue worldwide, and there is a critical need for a greater understanding of the pathogenesis and therapeutic targets of epilepsy, based on which more effective and non-invasive therapies should be explored and developed.
Long non-coding RNAs (LncRNAs) are RNAs with a length greater than 200 nucleotides whose transcripts are not involved in coding proteins. Changes of LncRNAs can be measured by RNA sequencing technology. Up till now only a small amount of lncRNAs have been characterized thoroughly, however, their changes including overexpression, deficiency or mutation have been implicated in numerous human diseases (Esteller, 2011; Ang et al., 2020). Recently, the role of LncRNAs in epileptogenesis has been illustrated gradually, involving mainly with the pathophysiological process of neuroinflammation, neuronal apoptosis, and balance of transmitters. In this paper, we will provide a short but comprehensive description of the research progress about the role of LncRNAs in the pathogenesis of epilepsy.
Epilepsy is one of the world’s oldest recognized conditions, the written records of which could be dated back to 4000 BCE (World Health Organization., 2022). In 2017, according to the onset characteristics, the international league against epilepsy (ILAE) classified epilepsy into four categories including focal, generalized, generalized combined with focal, and epilepsy of unknown classification (Scheffer et al., 2017).
Epilepsy is a multifactorial disease with a strong genetic predisposition, and comorbidities such as depression and cognitive impairments are increasingly taken as important etiological and prognostic markers of epilepsy (Manford, 2017). Evidence from clinical and experimental studies indicate that GABA is substantially involved in the mechanism and treatment of epilepsy, and the widely accepted “GABA-hypothesis” implies that a reduction of GABA-ergic inhibition would lead to the onset of epilepsy while an enhancement of GABA-ergic inhibition could result in an antiepileptic effect (De Deyn et al., 1990). Consistently, abnormal GABAergic function and complex changes in Glu signaling have been demonstrated in the brain of epilepsy patients and animal models, with a confirmed suppression effect of GABA agonists and an inducible effect of GABA antagonists on epilepsy (Pfisterer et al., 2020). Moreover, benzodiazepines and barbiturates, which are widely used antiepileptic drugs, exert their effect via enhancing GABA-mediated inhibition (Treiman, 2001). More recently, it has been reported that neuroinflammation and neuronal apoptosis are also the main causes of epilepsy (Henshall and Simon, 2005; Vezzani et al., 2019; Kumar et al., 2022).
Affecting ion channels or neurotransmitters, the antiseizure medication might suppress seizures in up to two-thirds of all individuals (Manford, 2017). However, apart from the poor tolerability, they could not alter the long-term prognosis, and long-term use of traditional antiepileptic drugs can induce many adverse effects, including central nervous system problems, idiosyncratic reactions, neurocognitive and psychiatric symptoms, and long-term complications (Alsfouk et al., 2020). On February 19, 2016 and June 25, 2018, the FDA approved two new drugs, Briviact and Epidiolex, for treatment of epilepsy, but there are still insurmountable side effects including liver damage, anorexia, and allergic reactions (U.S. Food and Drug Administration, 2022a,b). Thanks to the advances in brain imaging, it is more possible for doctors and clinical investigators to identify the structural and functional causes and consequences of the epilepsies (Duncan, 2022). Based on these, epilepsy surgery has been one of the most effective ways to achieve long-term seizure freedom in special individuals with drug-resistant focal epilepsy (Manford, 2017; Duncan, 2022), but it is not popularized widely due to the limitations of indications and techniques. However, with the improved understanding of the biological mechanisms of epileptogenesis including epigenetic determinants and pharmacogenomics, appears a great hope for better and precisive pharmacological and non-pharmacological strategies to forecast, modify, or even cure epilepsy (Manford, 2017; Proix et al., 2021; Duncan, 2022).
Long non-coding RNAs are transcribed by RNA polymerase II or III and have no structures of appreciable open reading frame (ORF) and Kozak consensus sequence. Besides the traits of diverse and numerous, LncRNAs are involved in numerous important biological phenomena, and their unique characteristics and functional roles have been reviewed detailed in other studies (Quinn and Chang, 2016; Gao et al., 2020; Nojima and Proudfoot, 2022). Similar with that of the messenger RNAs (mRNAs), a maturation process is needed for LncRNAs, and mature LncRNA molecules could be found in the nucleus, cytosol, and organelles including the mitochondria (Villa et al., 2019). Generally, LncRNAs in the nucleus are mainly involved in epigenetic and transcriptional regulation, while LncRNAs in the cytoplasm are often participated in the regulation of post-transcription, through which regulating the mRNA stability, protein translation, and the competitive endogenous RNA (ceRNA) network (Gao et al., 2020).
According to the position in the genome and their orientation to the adjacent protein-coding genes, LncRNAs can be subclassified into major groups including sense, antisense, intronic, intergenic, and bidirectional (Esteller, 2011). Generally, the LncRNA mechanisms of action can be divided into four categories: signal, decoy, guide, and scaffold, which has been described in detail in previous reviews (Gao et al., 2020; Nojima and Proudfoot, 2022). In brief, LncRNAs could regulate the transcription of downstream genes as signal molecules, or affect the expression of target genes by sponging miRNAs (Esteller, 2011). Moreover, LncRNAs could block a certain molecular pathway and act as a decoy molecule. Furthermore, LncRNAs can guide specific proteins to reach target location, or facilitate the interaction of numerous molecules and proteins, promoting the convergence and integration of information among different signaling pathways (Esteller, 2011; Quinn and Chang, 2016; Gao et al., 2020). For example, LncRNA H19 has been reported to inhibit glial cell activation in the hippocampus of epileptic rats by targeting STAT3 signaling way (Han et al., 2020), while LncRNA MEG3 could reduce neuronal apoptosis in the hippocampus of epileptic rats through the PI3K/AKT/mTOR pathway (Zhang et al., 2020). The common models of LncRNAs action are summarized in Figure 1.
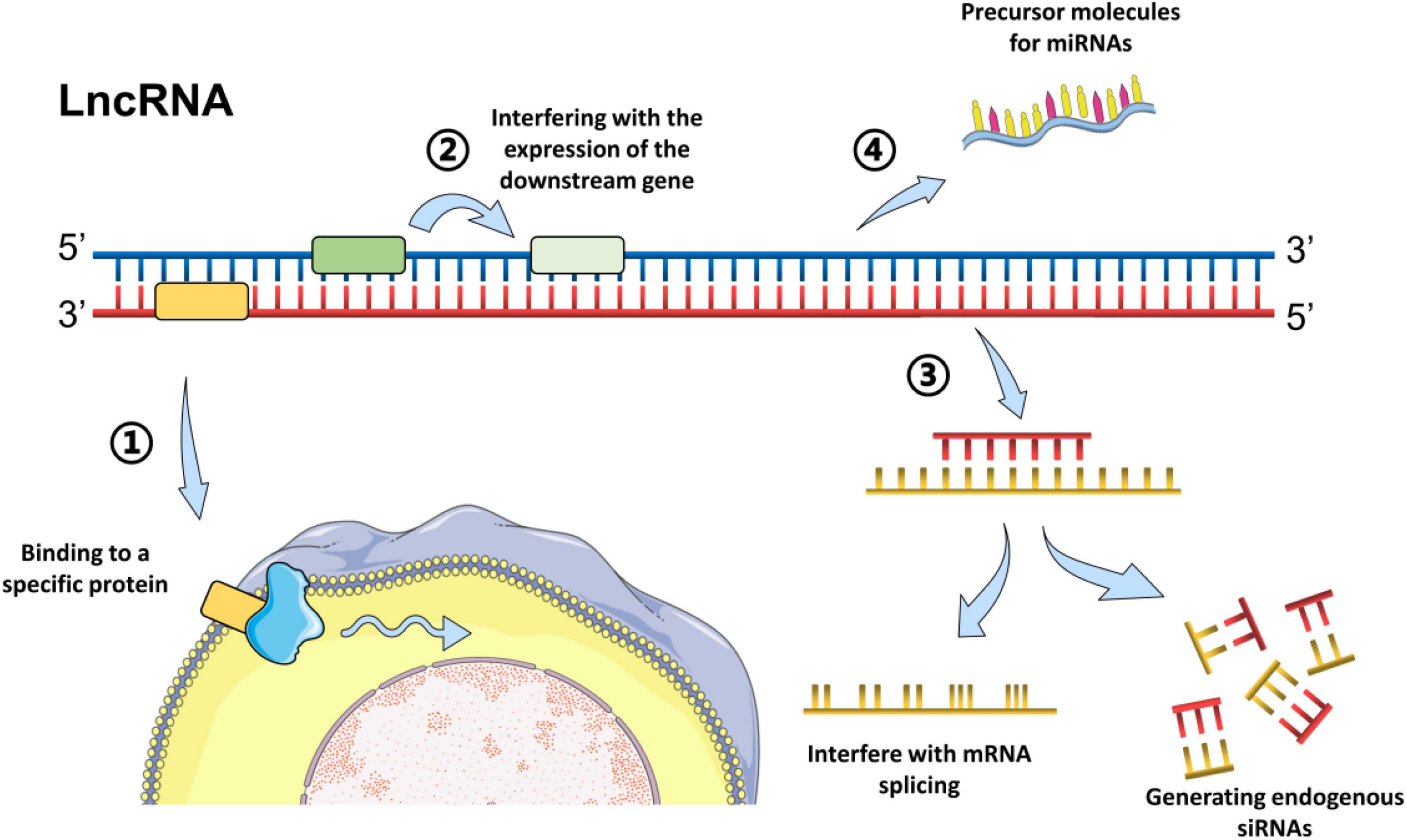
Figure 1. Common models of action of LncRNAs. Long non-coding RNAs can act through different biological mechanisms, mainly via methods as followings: (1) Bind to a specific protein and regulate its activity directly or indirectly by change the cellular localization via forming nucleic acid-protein complexes; (2) Encode the transcriptional process of the upstream promoter region of a protein gene, inhibit RNA polymerase II or mediate chromatin remodeling and histone modification, or interfere with the expression of the downstream gene. (3) Form complementary double strands with the transcription of the protein-encoding gene and generate endogenous siRNAs under the action of dicer enzyme, or interfere with mRNA splicing, and form different cleavage. (4) Serve as the precursor molecules for miRNAs (Grelet et al., 2017; Ali and Grote, 2020; Han et al., 2020; Zhang et al., 2020).
It is worth to mention that apart from regulating the target gene expression at the transcriptional and post-transcriptional level, some LncRNAs could manipulate the expression of the target gene in an epigenetic way (Tsai et al., 2010; Peschansky and Wahlestedt, 2014). Serving as scaffolds, LncRNA could recruit chromatin remodeling complexes to a special combining site, or assemble select histone modification enzymes, thereby specifying the pattern of DNA methylation or histone status (Tsai et al., 2010; Chen and Xue, 2016). Through binding to the enhancer of zester homolog 2 (EZH2), which is a subunit of polycomb repressive complex 2 (PRC2), LncRNA GAS5 has been reported to guide EZH2 to the promoter of interferon response factor 4 (IRF4) and suppress its transcription, eventually leading to an inhibition of microglial M2 polarization (Sun et al., 2017). Acting as a modular scaffold of WDR5 and KAT2A complexes, LncRNA GClnc1 could promote gastric carcinogenesis via altering the pattern of histone modification (Sun et al., 2016). Moreover, it has also been reported that the HOX transcript antisense RNA (HOTAIR) could bind to the Polycomb Repressive Complex 2 (PRC2) and recruit it to target genes such as HOXD, then suppress the transcription of target gene via its histone methylase activity (Tsai et al., 2010). Otherwise, HOTAIR could bind to LSD1, which is a histone demethylase enzyme and part of the CoREST/REST repressor complex. Via the effect of LSD1, HOTAIR could remove the transcription-activating histone marks and reinforce the repressive activity of the PRC2 complex eventually (Tsai et al., 2010).
With the development of reliable technologies and useful tools, our understanding of LncRNAs implicated in epilepsy has made substantial progress. Involved with the fundamental cellular processes via regulating gene expression at epigenetics, transcription, and post-transcription, LncRNAs attribute substantially to the molecular mechanisms of epilepsy development. Results of a genome-wide LncRNA profiling study have demonstrated a differential regulation of LncRNAs in pilocarpine or kainate acid-induced temporal lobe epilepsy animal models (Lee et al., 2015; Jang et al., 2018). Using methylated DNA immunoprecipitation and hybridization technique, Miller-Delaney SF et al. detected and analyzed the methylation status of all annotated C-phosphate-G islands and promoter regions in the human genome, and established a hippocampal methylation profile of patients with temporal lobe epilepsy (Miller-Delaney et al., 2015). They found a total of 146 protein-coding genes exhibited altered DNA methylation in the hippocampus of temporal lobe epilepsy patients, with a close relation with the development processes, neuron maturation and remodeling (Miller-Delaney et al., 2015). Besides these findings, methylation-sensitive microRNAs were also identified in their study, including MiR27A, miR-193a-5p (MIR193A), and miR-876-3p (MIR876). More importantly, they reported the differential methylation of LncRNA in the hippocampus of epilepsy patients for the first time, including urothelial cancer associated 1 (UCA1), adenosine deaminase, RNA-specific, B2, antisense RNA 1 (ADARB2-AS1), long intergenic non-protein coding RNA 324 (LINC324) and mitogen-activated protein kinase 14, and antisense RNA 1 (MAP3K14-AS1) (Miller-Delaney et al., 2015).
Importantly, LncRNAs might be potential drug targets for treatment of epilepsy (Luo et al., 2019), which was supported by the results that administration of an antagoNAT (natural antisense transcript) could mediate an upregulation of the sodium channel protein SCN1A in a mice model of the epilepsy-related disorder, leading to significant improvements in seizure phenotype and excitability of hippocampal interneurons (Hsiao et al., 2016).
Neuroinflammation is a contributing factor for many neuropsychiatric diseases including Alzheimer’s disease and depression (Fang et al., 2020; Qi et al., 2021; Liu S. et al., 2022), and increasing evidence have demonstrated its role in the pathogenesis of epilepsy (Vezzani et al., 2019; Kumar et al., 2022). Using single-cell cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) techniques, Kumar et al. (2022) uncovered a pro-inflammatory microenvironment in the brain lesions of epileptic patients, including an extensive activation of microglia and an infiltration of other pro-inflammatory immune cells, together with a direct interaction between T cells and microglia. It is well known that activated microglia and astrocytes could interact with surrounding cells by secreting inflammatory factors, promoting neuroinflammatory responses into a vicious circle (Ginhoux et al., 2010; Tian et al., 2012; Santiago et al., 2017; Fang et al., 2020; Liu S. et al., 2022). Through microarray screening, Sun et al. (2017) identified that LncRNA GAS5 could regulate the polarization of microglia, acting as an epigenetic regulator. In a bilateral chronic constriction injury rat model, the expression levels of LncRNA 00311 and Lnc-AK141205 were upregulated in the microglia of dorsal spinal. Regulated by the STAT3 pathway, they were closely associated with the expression of IL-1β, IL-6, and TNF-α (Pang et al., 2020). Moreover, results of clinical investigation disclosed a relationship between cerebrospinal fluid IL1-β levels and an allelic variant of the IL1-β gene to the risk of developing epilepsy (Diamond et al., 2014). Furthermore, targeting at the inflammatory mediators in epilepsy, anti-IL-1, anti-IL-6, and anti-CD20 agents have been used in patients with drug-resistant epilepsy and refractory status epilepticus, with promising results and a good safety profile (Costagliola et al., 2022).
Through interacting with proteins, RNA, and DNA, LncRNA could serve as regulators of gene expression pathways involved with the regulation of both the pro-inflammatory and the anti-inflammatory process in the central nervous system (Tripathi et al., 2021). Acting as regulators of inflammation and neuronal differentiation pathways in the epileptic brain, LncRNAs are reported to be involved in the development of epilepsy (Zhao et al., 2019). Results of pre-clinical studies showed that the expressions of LncRNA H19 and LncRNA X inactive specific transcript (XIST) were up-regulated, while the expressions of LncRNA PVT1, LncRNA CACS2, and LncRNA UCA1 were down-regulated in epilepsy animal or cell models (Han et al., 2018a; Wang et al., 2020; Zhang et al., 2021). LncRNA H19 could promote an abnormal activation of rat hippocampal glial cells by targeting STAT3 signaling pathway via binding to miRNA-let-7b (Han et al., 2020), while LncRNA XIST has been reported to promote astrocyte activation through negatively regulating the expression of NEAT5 by binding to miR-29c-3p (Zhang et al., 2021). The down-regulated LncRNA PVT1, LncRNA CACS2, and LncRNA UCA1 could, respectively, regulate the downstream proteins including BDNF, PTEN, or JAK to promote the activation of astrocytes and the progression of epilepsy (Zhao et al., 2019; Wang et al., 2020; Zhu et al., 2020). Moreover, LncRNA UCA1 could inhibit the inflammation via regulating miR-203 mediated regulation of MEF2C/NF-κB signaling in epilepsy (Yu et al., 2020).
Long non-coding RNA growth arrest-specific 5 (GAS5) was markedly upregulated in epileptic cell and animal models, together with a down-regulated expression of miR-219. Moreover, GAS5 could epigenetically suppress the transcriptional miR-219 expression, and knockdown of GAS5 could dramatically promote the cell proliferation of epileptic cells and suppress the inflammation and apoptosis processes, which could be reversed by inhibition of miR-219, suggesting a regulation role of LncRNA GAS5 in the inflammatory response of epilepsy via its effect on miR-219 (Zhao et al., 2022). Cai et al. (2020) reported an elevated expression of LncRNA ILF3-AS1 in the serum and hippocampus tissue of epilepsy patients, which promoted the abnormal activation of astrocytes by specifically targeting at miR-212 and up-regulating the expression of inflammatory molecular such as tumor necrosis factor-α (TNF-α), IL-1β, and IL-6. Serum levels of LncRNA ZFAS1 were also reported higher in temporal lobe epilepsy patients, and ZFAS1 could exacerbate epilepsy development via promoting inflammation (He et al., 2021). Moreover, it has been demonstrated that transfection of pcDNA3.1- ZFAS1 could boost NF-κB activation and elevate the release of cytokines including TNF-α, IL-1, IL-6, and intercellular adhesion molecule-1 in lipopolysaccharide-challenged neuron (He et al., 2021). In another study, the expression of LncRNA NEAT1 was significantly increased in the temporal lobe tissue of epilepsy patients, and it could combine with miR-129-5p, resulting in an increased expression of key molecular in Notch signaling pathway and inflammatory cytokines such as IL-6 and TNF-α (Wan and Yang, 2020; see Table 1 for details).
Nod-like receptor protein 3 (NLRP3) inflammasome-mediated inflammation has emerged as a contributor to epileptogenesis (Yue et al., 2020). Using mouse and cell epilepsy models, Zhang et al. (2022) found that NLRP3 inflammasome activation could enhance the expression of adenosine kinase and accelerate epilepsy via regulating CREB/REST/SP1 signaling pathway (Zhang et al., 2022). In line with these findings, Gong et al. reported that NLRP3 was increasingly expressed in epileptic neurons and rats, together with a downregulated expression of miR-138-5p and an upregulated expression of LncRNA ZNF883 and ubiquitin-specific peptidase 47 (USP47), and concurrent with aggravated inflammation and apoptosis (Gong et al., 2022). These findings suggested that inhibition of NLRP3 inflammasome might be a target for treatment of epilepsy. Furthermore, Gong et al. demonstrated a close relationship between LncRNA ZNF883 and NLPR3, via the regulation of USP47 and miR-138-5p in epilepsy models in vivo and in vitro (Gong et al., 2022). Moreover, four LncRNAs including Gm26917, Gm42418, Gm26767, and Neat1 were found to be enriched in NLRP3 inflammasomes (Zhang et al., 2019). Neat1, which was normally resided in the paraspeckles of macrophages, has been demonstrated to disassociate from these nuclear bodies and translocate to the cytoplasm under hypoxic conditions or stimulated with LPS, and modulate inflammasome activation via regulating caspase-1 activation, cytokine production, and pyroptotic cell death, or in a HIF-2α-dependent manner. Besides, LincRNA Cox2 has been reported to regulate NLRP3 inflammasome and autophagy mediated neuroinflammation through binding to NF-κB p65 and promoting its nuclear translocation and transcription (Xue et al., 2019).
Abnormal neuronal function and viability are the most important features of epilepsy. Increased neuronal apoptosis is strongly associated with epilepsy progression (Henshall and Simon, 2005). In a non-Mg2+ treated epileptic neuronal model and rat models induced by LiCl or pilocarpine, the expressions of LncRNA SNHG1 and LncRNA MALAT1 were significantly increased, while the expressions of LncRNA FTX and LncRNA MEG3 were significantly decreased (Wu and Yi, 2018; Li et al., 2019; Cai et al., 2020; Zhang et al., 2020).
Apart from promoting neuroinflammation, LncRNA ZFAS1 could also exacerbate epilepsy development via accelerating neuronal apoptosis. ZFAS1 expression in neurons was raised by pcDNA3.1- ZFAS1 and declined after silencing of ZFAS1 (He et al., 2021). Transfection of pcDNA- ZFAS1 could promote apoptosis (He et al., 2021), while knockdown of ZFAS1 could inhibit the apoptosis and autophagy of hippocampal neurons by activating the PI3K/AKT pathway via up-regulating miR-421 in epilepsy (Hu et al., 2020). LncRNA TUG1 could also regulate the cell activity and apoptosis of hippocampal neuron via sponging miR-199a-3p (Li et al., 2021). Besides the regulation effect of LncRNA 17A in autophagy and apoptosis, there were approximately 100 dysregulated LncRNA transcripts found in amyloid β peptide treated SH-SY5Y cells (Wang et al., 2019). LncRNA UCA1 could suppress pilocarpine-induced epilepsy by inhibiting apoptosis of hippocampal neurons via miR-495/Nrf2-ARE pathway (Geng et al., 2018). Furthermore, LncRNA H19 was highly expressed in the latent period of epilepsy, contributing to the apoptosis process of hippocampal neurons by targeting let-7b (Han et al., 2018b). Moreover, results of clinical observation indicated that the rs6478974 TT genotype could inhibit the susceptibility to epilepsy by reducing the levels of transforming growth factor beta receptor 1 (TGFBR1) mRNA, which is a target of let-7b (Zheng et al., 2021). In addition, the altered LncRNAs could involve with neuronal apoptosis by participating in the regulation of downstream oxidative stress and the expression of apoptosis-related proteins such as Nrf2, BCL2, and Caspase3 (Feng et al., 2020, 2021; Gong et al., 2020; Zhao et al., 2020; see Table 1 for details).
Accumulating evidences from postmortem studies, brain imaging, and animal models demonstrate dysfunction of inhibitory GABAergic interneuron and excitatory Glu neurons contribute to epileptogenesis (De Deyn et al., 1990; Treiman, 2001; Pfisterer et al., 2020). The main clinical anticonvulsants, such as lacosamide, levetiracetam, phenobarbital, phenytoin, and valproate, are mainly targeting at improving the GABAergic activity or decreasing Glu-ergic activity (De Deyn et al., 1990). More importantly, vigabatrin, a second-generation antiepileptic drug with a remarkable effect against infantile spasms and focal seizures, is a structural analog of GABA that irreversibly inhibits GABA-transaminase (GABA-T) and increase brain levels of GABA (O’Callaghan et al., 2017; Knupp et al., 2022).
Highly expressed in the brain, LncRNAs are involved in important neurobiological process, including neurotransmitter synthesis and transmission, neurogenesis, and neural plasticity (Bond et al., 2009). It has been reported that the LncRNAs that associated with PICK1, GADL1, and PMD6 genes are abundant in the pathways involved with the ionotropic Glu receptor and GABA synthesis (Muniz et al., 2022). LncRNA Evf2 has been reported to play a crucial role in regulation of homeodomain transcription factors and the formation of GABA-dependent neuronal circuitry in the developing mouse forebrain, and LncRNA MALAT1 can modulate synaptic plasticity and neuronal regeneration via promoting the density of dendritic spines (Wu et al., 2013). Apart from abrogating the activation of astrocyte, over-expression of LncRNA UCA1 could suppresses the expression of astrocyte glutamate aspartate transporter (GLAST) via JAK/STAT signaling pathway, result in a reduced frequency of the of epilepsy seizures and a promotion of learning and memory in temporal lobe epilepsy rats (Wang et al., 2020). Besides, the regulation effect of LncRNA GAS5 in epilepsy was also involved in the regulation of Calmodulin-dependent protein kinase II (CaMKII)γ/N-methyl-D-aspartate receptor (NMDAR) pathway (Zhao et al., 2022). Moreover, non-coding RNA-dependent balanced gene regulation in embryonic brain has been reported to be critical for proper formation of GABA-dependent neuronal circuitry in adult brain (Bond et al., 2009). Furthermore, studies targeting at neuroblastoma have found that LINC00622 could promote GABBR1 expression via mediating the activity of transcriptional factor androgen receptor (Guo et al., 2022). Additionally, a dysfunction of LncRNAs/miRNAs/GABA regulatory axis has also been reported in depression animal model (Xue et al., 2022), and Glu decarboxylase-67 (Gad-67) and vesicular GABA transporter (VGAT) have been demonstrated to be directly regulated by miRNA-144-3p, miRNA-15b-5p, and miR-879-5p. Although LncRNA-mediated sponge regulatory network underlying GABA deficit in epilepsy remains unclear, a bidirectional relationship of epilepsy and depression or dementia has been demonstrated, involving both biological and psychosocial factors (De Deyn et al., 1990; Lothe et al., 2008; Sen and Romoli, 2021), sharing abnormalities in lots of neurotransmitters including Glu, with an antiepileptic activity of NMDA receptor antagonists (Lothe et al., 2008). Moreover, Valproic acid (VPA), a common anticonvulsant, has been reported to display an antitumor effect through regulating non-coding RNAs controlling gene expression via H19→EZH2→p21/PTEN pathway (Sajadpoor et al., 2018). Furthermore, it has been demonstrated that loss of LncRNA maternally expressed gene 3 (MEG3) could alleviate the impairment of learning and memory abilities of autism spectrum disorder rats induced by VPA, via promoting neuronal viability but inhibiting apoptosis (Liu X. et al., 2022). Although not based on epilepsy models, these findings indicated a close linkage between changes of LncRNAs and the GABA/Glu balance in epilepsy and the effect of anticonvulsants.
Although a lot of energy and funds were invested in the research and development of antiepileptic drugs every year, the effective treatment for epilepsy are still unsatisfactory. Given the close linkage with neuroinflammation, apoptosis, and balance of transmitters especially GABA and Glu, LncRNAs might be taken as potential therapeutic targets for treatment of epilepsy. Though numerous LncRNAs have been identified, the mechanistic and functional roles for most LncRNAs are still unknown. Moreover, there is no reports about the dynamic changes of LncRNAs and their function in the progress of epilepsy, especially in different types of attacks. Thus, detailed investigation should be carried out not only about the changes of the abundance and diversity of LncRNAs in epilepsy, but the exact neurobiological mechanism as well, especially the mechanism about how they regulate the activity of transcription factors, through which how their regulations affect the downstream target genes expression.
F-HC and J-FG proposed the conception of this work and supervised. SL and J-FG wrote the draft of the manuscript. SL, J-FG, and F-HC revised the manuscript. SL and MF searched literatures and analyzed the data. M-DM helped modifying the figure and table. All authors have read and approved the final manuscript.
This study was supported by the Key Scientific Research Platform for Collaborative Tackling of Universities and Collaborative Innovation of Public Health in Anhui Province (GXXT-2020-062), National Natural Science Foundation of China (81870403), and The Comprehensive Reform Pilot Project of “San Quan Education” of Anhui Medical University (2021xsqyr03).
We would like to acknowledge Dr. Qiao Jinping for his help in revising the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ali, T., and Grote, P. (2020). Beyond the RNA-dependent function of LncRNA genes. eLife 9:e60583. doi: 10.7554/eLife.60583
Allers, K., Essue, B. M., Hackett, M. L., Muhunthan, J., Anderson, C. S., Pickles, K., et al. (2015). The economic impact of epilepsy: a systematic review. BMC Neurol. 15:245. doi: 10.1186/s12883-015-0494-y
Alsfouk, B. A. A., Brodie, M. J., Walters, M., Kwan, P., and Chen, Z. (2020). Tolerability of antiseizure medications in individuals with newly diagnosed epilepsy. JAMA Neurol. 77, 574–581. doi: 10.1001/jamaneurol.2020.0032
Ang, C. E., Trevino, A. E., and Hy, C. (2020). Diverse lncRNA mechanisms in brain development and disease. Curr. Opin. Genet. Dev. 65, 42–46. doi: 10.1016/j.gde.2020.05.006
Bond, A. M., VanGompel, M. J. W., Sametsky, E. A., Clark, M. F., Savage, J. C., Disterhoft, J. F., et al. (2009). Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat. Neurosci. 12, 1020–1027. doi: 10.1038/nn.2371
Cai, X., Long, L., Zeng, C., Ni, G., Meng, Y., Guo, Q., et al. (2020). LncRNA ILF3-AS1 mediated the occurrence of epilepsy through suppressing hippocampal miR-212 expression. Aging 12, 8413–8422. doi: 10.18632/aging.103148
Chen, J., and Xue, Y. (2016). Emerging roles of non-coding RNAs in epigenetic regulation. Sci. China Life Sci. 59, 227–235. doi: 10.1007/s11427-016-5010-0
Costagliola, G., Depietri, G., Michev, A., Riva, A., Foiadelli, T., Savasta, S., et al. (2022). Targeting inflammatory mediators in epilepsy: a systematic review of its molecular basis and clinical applications. Front. Neurol. 13:741244. doi: 10.3389/fneur.2022.741244
De Deyn, P. P., Marescau, B., and Rl, M. (1990). Epilepsy and the GABA-hypothesis a brief review and some examples. Acta Neurol. Belg. 2, 65–81.
Diamond, M. L., Ritter, A. C., Failla, M. D., Boles, J. A., Conley, Y. P., Kochanek, P. M., et al. (2014). IL-1b associations with posttraumatic epilepsy development: a genetics and biomarker cohort study. Epilepsia 7, 1109–1119. doi: 10.1111/epi.12628
Duncan, J. S. (2022). Epilepsy in the 21st century. Lancet Neurol. 21, 501–503. doi: 10.1016/S1474-4422(22)00175-2
Esteller, M. (2011). Non-coding RNAs in human disease. Nat. Rev. Genet. 12, 861–874. doi: 10.1038/nrg3074
Fang, K., Li, H., Chen, X., Gao, X., Huang, L., Du, A., et al. (2020). Quercetin alleviates LPS-induced depression-like behavior in rats via regulating BDNF-related imbalance of copine 6 and TREM1/2 in the Hippocampus and PFC. Front. Pharmacol. 11:518. doi: 10.3389/fphar.2020.00518
Feng, H., Gui, Q., Wu, G., Zhu, W., Dong, X., Shen, M., et al. (2021). Long noncoding RNA Nespas inhibits apoptosis of epileptiform hippocampal neurons by inhibiting the PI3K/Akt/mTOR pathway. Exp. Cell Res. 398:112384. doi: 10.1016/j.yexcr.2020.112384
Feng, H., Gui, Q., Zhu, W., Wu, G., Dong, X., Shen, M., et al. (2020). Long-noncoding RNA Peg13 alleviates epilepsy progression in mice via the miR-490-3p/Psmd11 axis to inactivate the Wnt/beta-catenin pathway. Am. J. Transl. Res. 12, 7968–7981.
Gao, N., Li, Y., Li, J., Gao, Z., Yang, Z., Li, Y., et al. (2020). Long Non-Coding RNAs: the regulatory mechanisms, research strategies, and future directions in cancers. Front. Oncol. 10:598817. doi: 10.3389/fonc.2020.598817
Geng, J. F., Liu, X., Zhao, H. B., Fan, W. F., Geng, J. J., and Xz, L. (2018). LncRNA UCA1 inhibits epilepsy and seizure-induced brain injury by regulating miR-495/Nrf2-ARE signal pathway. Int. J. Biochem. Cell Biol. 99, 133–139. doi: 10.1016/j.biocel.2018.03.021
Ginhoux, F., Greter, M., Leboeuf, M., Nandi, S., See, P., Gokhan, S., et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845. doi: 10.1126/science.1194637
Gong, L., Han, Y., Chen, R., Yang, P., and Zhang, C. (2022). LncRNA ZNF883-mediated NLRP3 inflammasome activation and epilepsy development involve USP47 upregulation. Mol. Neurobiol. 59, 5207–5221. doi: 10.1007/s12035-022-02902-7
Gong, L., Yang, P., Hu, L., and Zhang, C. (2020). MiR-181b suppresses the progression of epilepsy by regulation of lncRNA ZNF883. Am. J. Transl. Res. 12, 2769–2780.
Grelet, S., Link, L. A., Howley, B., Obellianne, C., Palanisamy, V., Gangaraju, V. K., et al. (2017). A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 19, 1105–1115. doi: 10.1038/ncb3595
Guo, M., Li, D., Feng, Y., Li, M., and Yang, B. (2022). Adipose-derived stem cell-derived extracellular vesicles inhibit neuroblastoma growth by regulating GABBR1 activity through LINC00622-mediated transcription factor AR. J. Leukocyte Biol. 111, 19–32. doi: 10.1002/JLB.1MIA0321-164R
Han, C., Ge, M., Liu, Y., Zhao, X., Wang, K., Chen, N., et al. (2018a). LncRNA H19 contributes to hippocampal glial cell activation via JAK/STAT signaling in a rat model of temporal lobe epilepsy. J. Neuroinflamm. 15:103. doi: 10.1186/s12974-018-1139-z
Han, C., Ge, M., Liu, Y., Zhao, X., Wang, K., Chen, N., et al. (2018b). Long non-coding RNA H19 contributes to apoptosis of hippocampal neurons by inhibiting let-7b in a rat model of temporal lobe epilepsy. Cell Death Dis. 9:617. doi: 10.1038/s41419-018-0496-y
Han, C. L., Liu, Y. P., Guo, C. J., Du, T. T., Jiang, Y., Wang, K. L., et al. (2020). The lncRNA H19 binding to let-7b promotes hippocampal glial cell activation and epileptic seizures by targeting Stat3 in a rat model of temporal lobe epilepsy. Cell Proliferat. 53:e12856. doi: 10.1111/cpr.12856
He, C., Su, C., Zhang, W., Zhou, Q., Shen, X., Yang, J., et al. (2021). Modulatory potential of LncRNA Zfas1 for inflammation and neuronal apoptosis in temporal lobe epilepsy. Yonsei Med. J. 62:215. doi: 10.3349/ymj.2021.62.3.215
Henshall, D. C., and Simon, R. P. (2005). Epilepsy and apoptosis pathways. J. Cereb. Blood Flow Metab. 25, 1557–1572. doi: 10.1038/sj.jcbfm.9600149
Hsiao, J., Yuan, T. Y., Tsai, M. S., Lu, C. Y., Lin, Y. C., Lee, M. L., et al. (2016). Upregulation of haploin sufficient gene expression in the brain by targeting a long non-coding RNA improves seizure phenotype in a model of dravet syndrome. EBioMedicine 9, 257–277. doi: 10.1016/j.ebiom.2016.05.011
Hu, F., Shao, L., Zhang, J., Zhang, H., Wen, A., and Zhang, P. (2020). Knockdown of ZFAS1 Inhibits hippocampal neurons apoptosis and autophagy by activating the PI3K/AKT Pathway via Up-regulating miR-421 in epilepsy. Neurochem. Res. 45, 2433–2441. doi: 10.1007/s11064-020-03103-1
Jang, Y., Moon, J., Lee, S. T., Jun, J. S., Kim, T. J., Lim, J. A., et al. (2018). Dysregulated long non-coding RNAs in the temporal lobe epilepsy mouse model. Seizure 58, 110–119. doi: 10.1016/j.seizure.2018.04.010
Knupp, K. G., Coryell, J., Singh, R. K., Gaillard, W. D., Shellhaas, R. A., Koh, S., et al. (2022). Comparison of cosyntropin, vigabatrin, and combination therapy in new-onset infantile spasms in a prospective randomized trial. J. Child Neurol. 37, 186–193. doi: 10.1177/08830738211073400
Kumar, P., Lim, A., Hazirah, S. N., Chua, C. J. H., Ngoh, A., Poh, S. L., et al. (2022). Single-cell transcriptomics and surface epitope detection in human brain epileptic lesions identifies pro-inflammatory signaling. Nat. Neurosci. 25, 956–966. doi: 10.1038/s41593-022-01095-5
Lee, D. Y., Moon, J., Lee, S. T., Jung, K. H., Park, D. K., Yoo, J. S., et al. (2015). Dysregulation of long non-coding RNAs in mouse models of localization-related epilepsy. Biochem. Biophys. Res. Commun. 462, 433–440. doi: 10.1016/j.bbrc.2015.04.149
Li, C., Zheng, X., Liu, P., and Li, M. (2021). Clinical value of lncRNA TUG1 in temporal lobe epilepsy and its role in the proliferation of hippocampus neuron via sponging miR-199a-3p. Bioengineered 12, 10666–10673. doi: 10.1080/21655979.2021.2001904
Li, X., Giri, V., Cui, Y., Yin, M., Xian, Z., and Li, J. (2019). LncRNA FTX inhibits hippocampal neuron apoptosis by regulating miR-21-5p/SOX7 axis in a rat model of temporal lobe epilepsy. Biochem. Biophys. Res. Commun. 512, 79–86. doi: 10.1016/j.bbrc.2019.03.019
Liu, S., Fan, M., Xu, J., Yang, L., Qi, C., Xia, Q., et al. (2022). Exosomes derived from bone-marrow mesenchymal stem cells alleviate cognitive decline in AD-like mice by improving BDNF-related neuropathology. J. Neuroinflamm. 19:35. doi: 10.1186/s12974-022-02393-2
Liu, X., Wang, Z., Zhang, X., Zhang, D., Yang, Q., and Hu, P. F. L. (2022). LncRNA MEG3 activates CDH2 expression by recruitment of EP300 in valproic acid-induced autism spectrum disorder. Neurosci. Lett. 783:136726. doi: 10.1016/j.neulet.2022.136726
Lothe, A., Didelot, A., Hammers, A., Costes, N., Saoud, M., Gilliam, F., et al. (2008). Comorbidity between temporal lobe epilepsy and depression: a [18 F]MPPF PET study. Brain 131, 2765–2782. doi: 10.1093/brain/awn194
Luo, Z. H., Walid, A. A., Xie, Y., Long, H., Xiao, W., Xu, L., et al. (2019). Construction and analysis of a dysregulated lncRNA-associated ceRNA network in a rat model of temporal lobe epilepsy. Seizure 69, 105–114. doi: 10.1016/j.seizure.2019.04.010
Manford, M. (2017). Recent advances in epilepsy. J. Neurol. 264, 1811–1824. doi: 10.1007/s00415-017-8394-2
Miller-Delaney, S. F., Bryan, K., Das, S., McKiernan, R. C., Bray, I. M., Reynolds, J. P., et al. (2015). Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain 138, 616–631. doi: 10.1093/brain/awu373
Muniz, M. M. M., Simielli Fonseca, L. F., Scalez, D. C. B., Vega, A. S., Silva, D. B. D. S., Ferro, J. A., et al. (2022). Characterization of novel lncRNA muscle expression profiles associated with meat quality in beef cattle. Evol. Appl. 15, 706–718. doi: 10.1111/eva.13365
Nojima, T., and Proudfoot, N. J. (2022). Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 23, 389–406. doi: 10.1038/s41580-021-00447-6
O’Callaghan, F. J. K., Edwards, S. W., Alber, F. D., Hancock, E., Johnson, A. L., Kennedy, C. R., et al. (2017). Safety and effectiveness of hormonal treatment versus hormonal treatment with vigabatrin for infantile spasms (ICISS): a randomised, multicentre, open-label trial. Lancet Neurol. 16, 33–42. doi: 10.1016/S1474-4422(16)30294-0
Pang, H., Ren, Y., Li, H., Chen, C., and Zheng, X. (2020). LncRNAs linc00311 and AK141205 are identified as new regulators in STAT3-mediated neuropathic pain in bCCI rats. Eur. J. Pharmacol. 868:172880. doi: 10.1016/j.ejphar.2019.172880
Peschansky, V. J., and Wahlestedt, C. (2014). Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 9, 3–12. doi: 10.4161/epi.27473
Pfisterer, U., Petukhov, V., Demharter, S., Meichsner, J., Thompson, J. J., Batiuk, M. Y., et al. (2020). Identification of epilepsy-associated neuronal subtypes and gene expression underlying epileptogenesis. Nat. Commun. 11:5038. doi: 10.1038/s41467-020-18752-7
Proix, T., Truccolo, W., Leguia, M. G., Tcheng, T. K., King-Stephens, D., Rao, V. R., et al. (2021). Forecasting seizure risk in adults with focal epilepsy: a development and validation study. Lancet Neurol. 20, 127–135. doi: 10.1016/S1474-4422(20)30396-3
Qi, C., Chen, X., Gao, X., Xu, J., Liu, S., and Ge, J. (2021). Impaired learning and memory ability induced by a bilaterally hippocampal injection of streptozotocin in mice: involved with the adaptive changes of synaptic plasticity. Front. Aging Neurosci. 13:633495. doi: 10.3389/fnagi.2021.633495
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Sajadpoor, Z., Amini-Farsani, Z., Teimori, H., Shamsara, M., Sangtarash, M. H., Ghasemi-Dehkordi, P., et al. (2018). Valproic acid promotes apoptosis and cisplatin sensitivity through downregulation of H19 Noncoding RNA in Ovarian A2780 Cells. Appl. Biochem. Biotech. 185, 1132–1144. doi: 10.1007/s12010-017-2684-0
Santiago, A. R., Bernardino, L., Agudo-Barriuso, M., and Gonçalves, J. (2017). Microglia in Health and Disease: A Double-Edged Sword. Mediat. Inflamm. 2017:7034143. doi: 10.1155/2017/7034143
Scheffer, I. E., Berkovic, S., Capovilla, G., Connolly, M. B., French, J., Guilhoto, L., et al. (2017). ILAE classification of the epilepsies: position paper of the ILAE commission for classification and terminology. Epilepsia 58, 512–521. doi: 10.1111/epi.13709
Sen, A., and Romoli, M. (2021). Pathological brain ageing in epilepsy and dementia: two sides of the same coin? Brain 144, 9–11. doi: 10.1093/brain/awaa441
Singh, A., and Trevick, S. (2016). The epidemiology of global epilepsy. Neurol. Clin. 34, 837–847. doi: 10.1016/j.ncl.2016.06.015
Singh, G., and Sander, J. W. (2020). The global burden of epilepsy report: Implications for low- and middle-income countries. Epilepsy. Behav. 105:106949. doi: 10.1016/j.yebeh.2020.106949
Sun, D., Yu, Z., Fang, X., Liu, M., Pu, Y., Shao, Q., et al. (2017). LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 18, 1801–1816. doi: 10.15252/embr.201643668
Sun, T. T., He, J., Liang, Q., Ren, L. L., Yan, T. T., Yu, T. C., et al. (2016). LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 6, 784–801. doi: 10.1158/2159-8290.CD-15-0921
Thijs, R. D., Surges, R., O’Brien, T. J., and Sander, J. W. (2019). Epilepsy in adults. Lancet 393, 689–701. doi: 10.1016/S0140-6736(18)32596-0
Tian, L., Ma, L., Kaarela, T., and Li, Z. (2012). Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. J. Neuroinflammation 9:155. doi: 10.1186/1742-2094-9-155
Treiman, D. M. (2001). GABAergic mechanisms in epilepsy. Epilepsia 42, 8–12. doi: 10.1046/j.1528-1157.2001.042suppl.3008.x
Tripathi, S., Shree, B., Mohapatra, S., Swati, Basu, A., and Sharma, V. (2021). the expanding regulatory mechanisms and cellular functions of Long Non-coding RNAs (lncRNAs) in neuroinflammation. Mol. Neurobiol. 58, 2916–2939. doi: 10.1007/s12035-020-02268-8
Tsai, M. C., Manor, O., Wan, Y., Mosammaparast, N., Wang, J. K., Lan, F., et al. (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. doi: 10.1126/science.1192002
U.S. Food and Drug Administration (2022a). FDA approves Briviact to treat partial onset seizures. Available online at: https://www.fda.gov/news-events/press-announcements/fda-approves-briviact-treat-partial-onset-seizures (accessed July 1, 2022).
U.S. Food and Drug Administration (2022b). FDA Approves First Drug Comprised of an Active Ingredient Derived from Marijuana to Treat Rare, Severe Forms of Epilepsy. Available online at: https://www.fda.gov/news-events/press-announcements/fda-approves-first-drug-comprised-active-ingredient-derived-marijuana-treat-rare-severe-forms (accessed July 1, 2022).
Vezzani, A., Balosso, S., and Ravizza, T. (2019). Neuroinflammatory pathways as treatment targets and biomarkers in epilepsy. Nat. Rev. Neurol. 15, 459–472. doi: 10.1038/s41582-019-0217-x
Villa, C., Lavitrano, M., and Combi, R. (2019). Long Non-Coding RNAs and related molecular pathways in the pathogenesis of epilepsy. Int. J. Mol. Sci. 20:4898. doi: 10.3390/ijms20194898
Wan, Y., and Yang, Z. Q. (2020). LncRNA NEAT1 affects inflammatory response by targeting miR-129-5p and regulating Notch signaling pathway in epilepsy. Cell Cycle 19, 419–431. doi: 10.1080/15384101.2020.1711578
Wang, H., Yao, G., Li, L., Ma, Z., Chen, J., and Chen, W. (2020). LncRNA-UCA1 inhibits the astrocyte activation in the temporal lobe epilepsy via regulating the JAK/STAT signaling pathway. J. Cell Biochem. 121, 4261–4270. doi: 10.1002/jcb.29634
Wang, X., Zhang, M., and Liu, H. (2019). LncRNA17A regulates autophagy and apoptosis of SH-SY5Y cell line as an in vitro model for Alzheimer’s disease. Biosci. Biotechnol. Biochem. 83, 609–621. doi: 10.1080/09168451.2018.1562874
World Health Organization. (2022). Epilepsy. Available online at: https://www.who.int/en/news-room/fact-sheets/detail/epilepsy (accessed July 1, 2022).
Wu, P., Zuo, X., Deng, H., Liu, X., and Liu, L. (2013). A J: Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res. Bull. 97, 69–80. doi: 10.1016/j.brainresbull.2013.06.001
Wu, Q., and Yi, X. (2018). Down-regulation of long noncoding RNA MALAT1 protects hippocampal neurons against excessive autophagy and apoptosis via the PI3K/Akt Signaling Pathway in Rats with Epilepsy. J. Mol. Neurosci. 65, 234–245. doi: 10.1007/s12031-018-1093-3
Xue, X., Pan, J., Zhang, H., Lu, Y., Mao, Q., and Ma, K. (2022). Baihe Dihuang (Lilium Henryi Baker and Rehmannia Glutinosa) decoction attenuates somatostatin interneurons deficits in prefrontal cortex of depression via miRNA-144–3p mediated GABA synthesis and release. J. Ethnopharmacol. 292:115218. doi: 10.1016/j.jep.2022.115218
Xue, Z., Zhang, Z., Liu, H., Li, W., Guo, X., Zhang, Z., et al. (2019). lincRNA-Cox2 regulates NLRP3 inflammasome and autophagy mediated neuroinflammation. Cell Death Differ. 26, 130–145. doi: 10.1038/s41418-018-0105-8
Yu, Q., Zhao, M. W., and Yang, P. (2020). LncRNA UCA1 Suppresses the Inflammation Via Modulating miR-203-Mediated Regulation of MEF2C/NF-κB Signaling Pathway in Epilepsy. Neurochem. Res. 45, 783–795. doi: 10.1007/s11064-019-02952-9
Yue, J., Wei, Y. J., Yang, X. L., Liu, S. Y., Yang, H., and Zhang, C. Q. (2020). NLRP3 inflammasome and endoplasmic reticulum stress in the epileptogenic zone in temporal lobe epilepsy: molecular insights into their interdependence. Neuropathol. Appl. Neurobiol. 46, 770–785. doi: 10.1111/nan.12621
Zhang, H., Tao, J., Zhang, S., and Lv, X. (2020). LncRNA MEG3 reduces hippocampal neuron apoptosis via the PI3K/AKT/mTOR pathway in a rat model of temporal lobe epilepsy. Neuropsychiatr. Dis. Treat. 16, 2519–2528. doi: 10.2147/NDT.S270614
Zhang, H., Yu, S., Xia, L., Peng, X., Wang, S., and Yao, B. (2022). NLRP3 inflammasome activation enhances ADK expression to accelerate epilepsy in mice. Neurochem. Res. 47, 713–722. doi: 10.1007/s11064-021-03479-8
Zhang, M., Yang, H., Chen, Z., Hu, X., Wu, T., and Liu, W. (2021). Long noncoding RNA X-inactive-specific transcript promotes the secretion of inflammatory cytokines in LPS stimulated astrocyte cell via sponging miR-29c-3p and regulating nuclear factor of activated T cell 5 expression. Front. Endocrinol. 12:573143. doi: 10.3389/fendo.2021.573143
Zhang, P., Cao, L., Zhou, R., Yang, X., and Wu, M. (2019). The lncRNA Neat1 promotes activation of inflammasomes in macrophages. Nat. Commun. 10:1495. doi: 10.1038/s41467-019-09482-6
Zhao, C. S., Liu, D. X., Fan, Y. H., and Wu, J. K. (2022). LncRNA GAS5 promotes epilepsy progression through the epigenetic repression of miR-219, in turn affecting CaMKIIγ/NMDAR pathway. J. Neurogenet. 36, 32–42. doi: 10.1080/01677063.2022.2067536
Zhao, M., Qiu, W., and Yang, P. (2020). SP1 activated-lncRNA SNHG1 mediates the development of epilepsy via miR-154-5p/TLR5 axis. Epilepsy. Res. 168:106476. doi: 10.1016/j.eplepsyres.2020.106476
Zhao, T., Ding, Y., Li, M., Zhou, C., and Lin, W. (2019). Silencing lncRNA PVT1 inhibits activation of astrocytes and increases BDNF expression in hippocampus tissues of rats with epilepsy by downregulating the Wnt signaling pathway. J. Cell Physiol. 234, 16054–16067. doi: 10.1002/jcp.28264
Zheng, Z., Yan, Y., Guo, Q., Wang, L., Han, X., and Liu, S. (2021). Genetic interaction of H19 and TGFBR1 polymorphisms with risk of epilepsy in a chinese population. Pharmgenomics Pers. Med. 14, 77–86. doi: 10.2147/PGPM.S279664
Keywords: lncRNAs, epilepsy, drug discovery, neuroinflammation, apoptosis, GABA
Citation: Liu S, Fan M, Ma M-D, Ge J-F and Chen F-H (2022) Long non-coding RNAs: Potential therapeutic targets for epilepsy. Front. Neurosci. 16:986874. doi: 10.3389/fnins.2022.986874
Received: 05 July 2022; Accepted: 16 September 2022;
Published: 06 October 2022.
Edited by:
Ying Xu, University at Buffalo, United StatesReviewed by:
Santosh Kumar, National Institute of Technology Rourkela, IndiaCopyright © 2022 Liu, Fan, Ma, Ge and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Fang Ge, Z2VqaW5mYW5nQGFobXUuZWR1LmNu; Fei-Hu Chen, Y2ZoY2hpbmFAc29odS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.