
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 04 October 2022
Sec. Perception Science
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.965871
This article is part of the Research TopicPsychological and Cognitive Evaluation and Intervention Based on Physiological and Behavioral ComputingView all 9 articles
 Peiji Chen1
Peiji Chen1 Bochao Zou2*
Bochao Zou2* Abdelkader Nasreddine Belkacem3
Abdelkader Nasreddine Belkacem3 Xiangwen Lyu4
Xiangwen Lyu4 Xixi Zhao5
Xixi Zhao5 Weibo Yi6
Weibo Yi6 Zhaoyang Huang7
Zhaoyang Huang7 Jun Liang8
Jun Liang8 Chao Chen1,9*
Chao Chen1,9*Current decoding algorithms based on a one-dimensional (1D) convolutional neural network (CNN) have shown effectiveness in the automatic recognition of emotional tasks using physiological signals. However, these recognition models usually take a single modal of physiological signal as input, and the inter-correlates between different modalities of physiological signals are completely ignored, which could be an important source of information for emotion recognition. Therefore, a complete end-to-end multi-input deep convolutional neural network (MI-DCNN) structure was designed in this study. The newly designed 1D-CNN structure can take full advantage of multi-modal physiological signals and automatically complete the process from feature extraction to emotion classification simultaneously. To evaluate the effectiveness of the proposed model, we designed an emotion elicitation experiment and collected a total of 52 participants' physiological signals including electrocardiography (ECG), electrodermal activity (EDA), and respiratory activity (RSP) while watching emotion elicitation videos. Subsequently, traditional machine learning methods were applied as baseline comparisons; for arousal, the baseline accuracy and f1-score of our dataset were 62.9 ± 0.9% and 0.628 ± 0.01, respectively; for valence, the baseline accuracy and f1-score of our dataset were 60.3 ± 0.8% and 0.600 ± 0.01, respectively. Differences between the MI-DCNN and single-input DCNN were also compared, and the proposed method was verified on two public datasets (DEAP and DREAMER) as well as our dataset. The computing results in our dataset showed a significant improvement in both tasks compared to traditional machine learning methods (t-test, arousal: p = 9.7E-03 < 0.01, valence: 6.5E-03 < 0.01), which demonstrated the strength of introducing a multi-input convolutional neural network for emotion recognition based on multi-modal physiological signals.
As an important component of artificial intelligence, affective computing plays an essential role in human–computer interaction (HCI). Emotion is the attitude and experience of people toward objective things; it integrates the state of human feelings, thoughts, and behaviors. When external stimuli cause changes in people's emotions, their physiological responses will change accordingly. The purpose of emotion recognition is to build a harmonious HCI environment by empowering the computer with human emotion capabilities and making the HCI process more intelligent. Research in the field of emotions can be traced back to the nineteenth century when American psychologist William James and Danish physiologist Carl Lange independently proposed the oldest theories of emotion (James, 1922), which supposed that emotional experience was the result of peripheral physiological changes caused by external stimuli. In the current stage, there are many emotion recognition tasks based on facial images (Akhand et al., 2021) and human speech signals (Chourasia et al., 2021; Mustaqeem and Kwon, 2021), which have achieved great development. Simultaneously, according to the maturity of wearable sensor technology, people have paid more and more attention to emotion recognition based on physiological signals, different from facial expressions and speech signals, and most changes in physiological signals cannot be voluntarily controlled, cannot be hidden, and can better reflect people's true emotional state. Furthermore, recording physiological signals can be continuous without interruption, which leads to feasible continuous emotional assessment. In this study, we collected subjects' multi-modal physiological signals, including ECG, EDA, and RSP through an emotion elicitation experiment. ECG reflects small electrical changes on the surface of the skin each time myocardial cells depolarize in the heartbeat, Agrafioti et al. (2012) found that when the induction method is active for subjects, ECG has a higher chance of responding to emotion, which proves the feasibility of using ECG for emotion recognition. EDA is a change in the electrical characteristics of the skin caused by the activity of sweat glands, which can reflect the change in arousal (Lang et al., 1993; Cacioppo et al., 2007). Respiration activity can reflect a person's stress states. When people are in a relaxed or pleasant state, their breathing rate will be relatively gentle, and when in a stressful or frightening state, their breathing rate could change rapidly or even stop. It has been proven in previous studies that the use of multi-modal physiological signals makes up for the limitation of the single modal in emotion recognition (Verma and Tiwary, 2014); applications based on multi-modal physiological signals are also increasing, such as emotion recognition (Zhang X. et al., 2021) and wearable health device (Cosoli et al., 2021). However, there are also disadvantages of physiological signal-based emotion recognition mainly because the difficulty of acquiring physiological signals is much higher than that of facial expressions and speech signals, which also limits the development of this field.
In recent years, the advancement of deep learning promoted the study of emotion recognition (Abbaschian et al., 2021; Makowski et al., 2021; He et al., 2022; Liu et al., 2022). In comparison with traditional machine learning, the use of deep learning for emotion recognition has a high improvement in accuracy and other aspects (Tang et al., 2017; Santamaria-Granados et al., 2019). Research shows that both convolutional neural networks and recurrent neural networks can achieve good results on emotion recognition tasks. Long short-term memory (LSTM) recurrent neural networks can extract context information from input sequences and avoid the long-term dependency problem. Zhang et al. (2017) used a deep learning framework to perform classification tasks on valence and arousal and confirmed its great potential in emotion recognition. Song et al. (2019) weakened the weight of certain useless sequences in the process of emotion classification by adding an attention mechanism to the LSTM network. Research on emotion recognition based on convolutional neural networks (CNNs) using physiological signals has the following two paths: the first is to transform physiological signals into spectrograms and then two-dimensional (2D) convolution is used for feature extraction. 2D convolutional neural networks have a very mature structure, which is beneficial for extracting physiological signal features from 2D images. For instance, Siddharth et al. (2022) used a pre-trained VGG-16 network to extract features from the spectrogram, and then they used a long short-term memory (LSTM) network for classification. The second one is to use a one-dimensional (1D) convolutional neural network to directly classify raw physiological signals. For example, in the research of Zhang Y. et al. (2021), they used a 1D convolutional neural network to extract efficient features from biological signals; this structure can automatically complete the feature extraction and classification of physiological signals. Unlike using 2D convolutional neural networks, the 1D-CNN does not require many preprocesses and can be deployed in an end-to-end manner. However, previous research on emotion recognition based on 1D convolutional neural networks usually utilizes a single modal physiological signal, and even if it is based on multi-modal, it is only a decision-level fusion, not a feature-level fusion. Therefore, we proposed an MI-DCNN, which enables the feature-level fusion of multi-modal physiological signals for emotion recognition.
This study focuses on using multi-modal physiological signals for automatic emotion recognition, and the difference between single-input and multi-input in the CNN will also be discussed. Simultaneously, we evaluate our proposed method on two public datasets and our collected data. This article consists of five sections. Section Related research provides an overview of related research. Section Introduction of our emotional dataset introduces the experimental process and the preprocess methods used in our dataset. Our proposed method is presented in Section Methods. We also give a baseline result of our dataset and evaluation of our proposed method in Section Evaluation, and we then draw conclusions in the last section.
The development of emotion recognition research based on multi-modal physiological signals mainly benefits from the establishment of physiological emotion datasets and the rise of deep learning techniques. The research of Gross and Levenson (1997) shows that audiovisual stimuli can better induce participants' emotions; we, therefore, summarized some popular physiological emotion datasets and related research in Table 1, where all datasets use audiovisual stimuli as elicitation materials.
Previous research studies (Kolodyazhniy et al., 2011; He et al., 2020) have demonstrated the potential of fusion of multi-modality for emotion recognition tasks. Subramanian et al. (2018) recorded electroencephalogram (EEG), electrocardiogram (ECG), and galvanic skin response (GSR) of 58 subjects during watching the video and used physiological features for emotion and personality trait recognition. Abadi et al. (2015) presented a dataset to estimate subjects' physiological responses to elicitation materials using brain signals and several physiological signals from 30 participants. Katsigiannis and Ramzan (2018) recorded EEG and ECG signals of 23 subjects using wearable, low-cost devices and showed the broad prospects of using these devices for emotion recognition. Soleymani et al. published MAHNOB-HCI (Soleymani et al., 2012) and DEAP (Koelstra et al., 2012) to explore the relationship between human physiological responses and emotional states. Miranda-Correa et al. (2021) recorded 40 subjects' EEG, ECG, and GSR signals elicited by visual stimuli for multi-modal research of emotion states on individuals and groups. MPED (Song et al., 2019) consists of EEG, GSR, RSP, and ECG signals of 23 participants while watching videos, and an attention-long short-term memory (A-LSTM) network was designed for emotion recognition. Siddharth et al. (2022) used deep learning methods for physiological signal-based emotion recognition on four public datasets (Koelstra et al., 2012; Soleymani et al., 2012; Katsigiannis and Ramzan, 2018; Miranda-Correa et al., 2021) and demonstrated the superiority of deep learning methods.
As a structure of a deep learning network, the convolutional neural network is mostly used in image classification and other related fields and has made great achievements. However, the 1D convolutional neural network, with its special structure, is widely used to process time series signals. Previous studies (Acharya et al., 2018; Yildirim et al., 2018) had proved the superiority of using the 1D convolutional neural network to classify time series signals. Yildirim et al. (2018) designed a 16-layer CNN model to detect 17 types of arrhythmia diseases and reached an overall classification accuracy of 91.33%. Acharya et al. (2018) used a 13-layer DCNN with an EEG signal to classify normal, preictal, and seizures, which achieved an accuracy of 88.76%. In the field of physiological signal-based automatic emotion recognition, the 1D convolutional neural network also achieved better results. Santamaria-Granados et al. (2019) used 1D deep convolutional neural networks for emotion recognition on the AMIGOS dataset and achieved a great improvement compared with previous research. Several models based on 1D convolutional neural networks are used for automatic emotion recognition, and end-to-end characteristics and better recognition results also prove the advantages of using such models. However, current research on 1D-CNN-based emotion recognition usually used a single-modal physiological signal as input, which neglects the inter-correlates between different modalities of physiological signals; therefore, in this article, we proposed an MI-DCNN for linking the relationship between multi-modalities.
A total of 52 subjects were recruited to participate in our experiment. We elaborately chose 12 videos as elicitation materials, which are shown in Table 2. All of these video materials run 34–201 s and contain different emotional states, such as fear, disgust, sadness, anger, and happiness. Before the formal experiment, the participants need to complete the Patient Health Questionnaire-9 (PHQ-9; Kroenke and Spitzer, 2002) to evaluate their recent emotional state. The participants who do not reach the specified score are not allowed to participate in the experiment, and those who reach the specified score were given MP150 physiological signal recorders in the shielded room to randomly watch the videos, and then three physiological signals (ECG, EDA, and RSP) were collected. There is a piece of 30-s music between every two videos to calm their moods, and the emotion evaluation scheme was the Self-Assessment Manikin (SAM; Morris, 1995). After watching each video, the participants filled in the evaluation according to their current feelings, and the valuation options mainly include the arousal level (discrete value of 1-9), valence level (discrete value of 1-9), and familiar level (-1, 0, 1). Because the nine-point scale was used, a threshold was placed in the middle, which leads to unbalanced classes, also reported by the studies of the first two public datasets (Koelstra et al., 2012; Katsigiannis and Ramzan, 2018).
In addition, each subject will first collect baseline data in the initial state of 60 s, which are used to compensate for data differences caused by individual differences (Wagner et al., 2005). After the trial, the participants watched their recorded video and marked the stimulus segments. Finally, two psychologists evaluated the videos of all participants and cut out the needed parts.
The physiological signal has weak amplitudes and high susceptibility to noise and magneto- and electro-interference. Different source signals come from different parts of the body. Therefore, different preprocesses of raw signals are needed. We use the Neurokit2 Toolbox (Makowski et al., 2021) to process the raw biological signals. ECG signals are mostly concentrated between 0 and 50 HZ, and a finite impulse response (FIR) band-pass filter between 3 and 45 Hz is used to filter the raw ECG signal, as shown in Figure 1A where the gray line represents the raw ECG signal and the pink line represents the filtered ECG signal. Then, filtered data are passed through Hamiltonian segmentation (Hamilton, 2002) to detect the QRS complex of each heartbeat, the heartbeat changes while watching the elicitation materials are recorded as Figure 1B shown, and individual heartbeats during this period are shown in Figure 1C. Finally, the R-peaks are identified as the orange points, as shown in Figure 1A. Extraction of electrodermal activity (EDA) is based on convex optimization (Greco et al., 2016). For respiration, statistical characteristics were calculated, such as mean, maximum, and variance value of the intensity of each breathing cycle. From the three physiological signals collected in our emotion elicitation experiment, 58 features were extracted for traditional machine learning methods, mainly including 47 ECG features (Table 3), four EDA features, and seven RSP features.
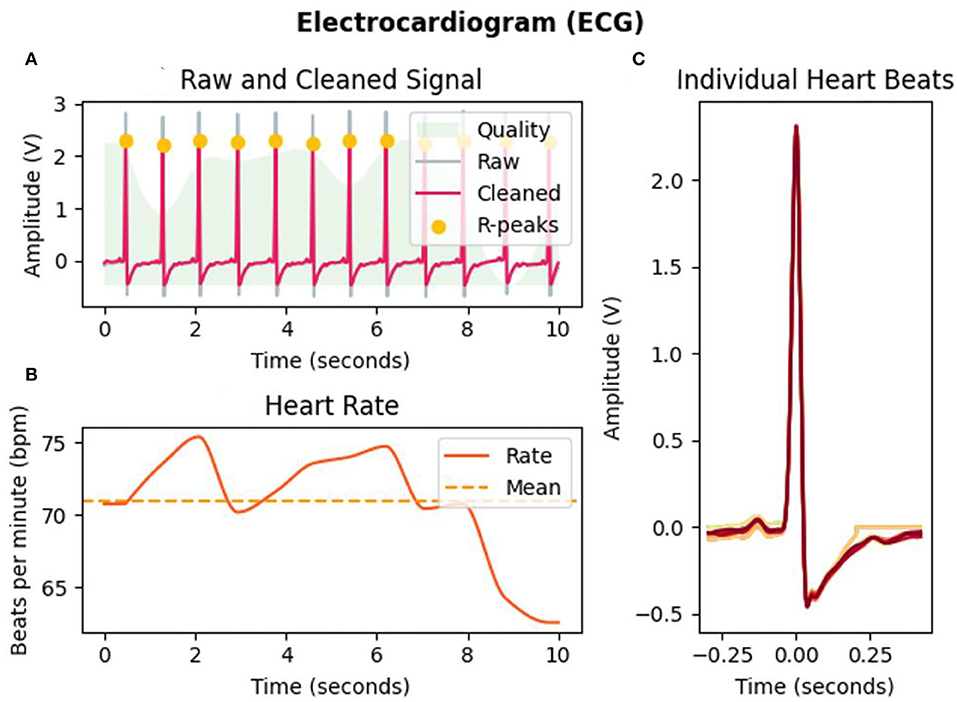
Figure 1. ECG analysis of subject 1 in the selected interval (10 s) of video b. (A) Filtering and R-peaks detection of ECG signals. (B) Heart rate changes during the selected interval. (C) Individual heartbeats.
A multi-input deep convolutional neural network for emotion recognition based on multi-modal physiological signals is designed and compared with the previous DCNN model. Our proposed multi-input deep convolutional neural network can extract the features of different input signals separately, and simultaneously, the filters in different channels are not shared, which guarantees that each input can have a different convolutional setting for the different input signals. The multi-input deep convolutional neural network proposed in this research can be regarded as a data fusion method at the feature level, and each branch in this structure can rely on the 1D-CNN to extract features automatically from the input signals without the need for manual feature extraction and selection, and the feature extracted from each input will be concatenated together through the concatenate layer. Then, the feature vector will be sent to the fully connected network for feature fusion and selection. This efficient feature-level fusion method does not base on any prior knowledge or complex feature engineering, and it can automatically fuse the most distinguishing features from each channel and eliminate redundant features between each channel. The whole process is end-to-end, which is more conducive to real-time subsequent decision-making. In our designed multi-input deep convolutional neural network, each channel inputs one modal of physiological signal; therefore, by adding input channels, the model can take multi-modal physiological signals, instead of a single modal, which may have potential benefit for emotion recognition.
The structure of the model is mainly composed of convolution layers, pooling layers, and fully connected layers. Figure 2 illustrates the overall structure of the multi-input deep convolutional neural network designed in this study, and Table 4 details the parameter setting of the automatic feature extraction structure used in our model.
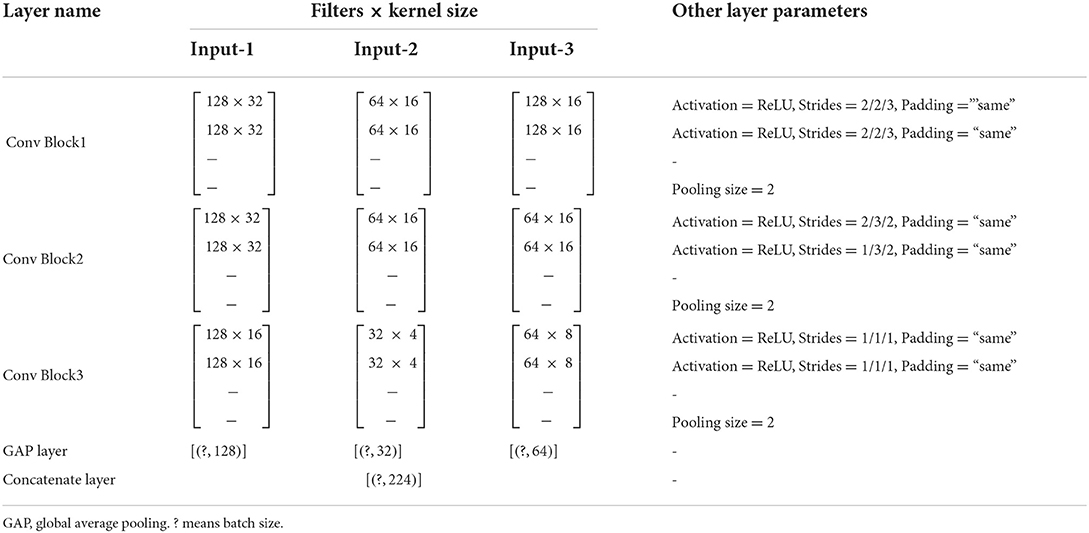
Table 4. Detailed feature extraction structure of the multi-input deep convolutional neural network.
The model is divided into three parts according to its functionality (convolutional, concatenate, and fully connected layers). Convolutional layers play the role of feature extraction, concatenate layer will concatenate the features of each channel from deep convolution layers, and fully connected layers get emotion class by these features. As shown in Figure 2, in each processing channel, multiple convolution blocks are stacked to form a deep convolution structure. The composition structure of the convolutional blocks is shown in Figure 3, which consists of four parts: (1) 1D convolutional layer, (2) activation function, (3) batch normalization layer, and (4) max-pooling layer.
(1) One-dimensional convolutional layer: It consists of fixed-number and fixed-size filters (convolution kernels). Each filter can be regarded as a fuzzy filter (Zhang Y. et al., 2021), and it slides on the input signal or is a result of the previous layer, and performs the convolution operation according to Equation 1:
Where represents the input vector of this convolutional layer, Mj indicates the size of the receptive field, and , are two trainable parameters, where denotes the kernel weight between the ith neuron in the k−1 layer and the jth neuron in the k layer; denotes the bias coefficient of the jth neuron in the k layer; and represents the output vector corresponding to the jth convolution kernel in the k layer.
(2) Activation function: We add an activation function after each convolutional layer, and the activation function can increase the non-linearity of the model and improve the expression ability of the model. There are many types of activation functions. In order to prevent the vanishing gradient problem, we choose the rectified linear unit (ReLU) as the activation function. Equation 2 shows the ReLU function.
(3) Batch normalization: This method has been proposed by Ioffe and Szegedy (2015), which is based on adjusting the activation value distribution of each layer to have the appropriate breadth, eliminating the problem of internal covariate shift. Simultaneously, the addition of the batch normalization layer can also accelerate the learning process and suppress overfitting. This can be expressed in a mathematical formula as follows:
where μB and represent the mean and variance of the data input to the batch normalization layer. Equation 5 indicates that the input data are regularized with a mean value of 0 and a variance of 1. In order to prevent the situation of dividing by 0, a minimum value ε is added. Equation 6 represents the scaling and translation transformation of regularized data; , γ, and β are parameters, which will be adjusted to appropriate values by the training of the model.
(4) Max-pooling layer: A max-pooling layer is added at the end of each convolution block. The max-pooling layer removes redundant information from the input vector and extracts important features by taking the maximum value of the input vector within the set range. Simultaneously, the complexity and calculation of the model are reduced, and overfitting is prevented.
A global average pooling layer is added at the end of the deep convolution structure of each channel. It is an important step to achieve multiple inputs. Through this step, the features extracted from different channels can be concatenated. We also evaluated that the global average pooling layer was added behind the concatenate layer, but in this case, it is necessary to ensure that the shape of the feature map output by each channel must be consistent, which also limits the structure of the model. Finally, the prediction results are output through two fully connected layers. During the model training process, the Adam (Kingma and Ba, 2015) optimizer is selected for parameter update, and cross-entropy is used as the loss function. The used framework is TensorFlow (Abadi et al., 2016). The network is trained on a GTX2080Ti GPU. The dataset is divided into two parts (train dataset and test dataset), with a division ratio of 80/20, and a 5-fold cross-validation was performed.
In this article, we will use three traditional machine learning methods (support vector machines, random forests, K-nearest neighbors) and deep learning methods to analyze our dataset and public datasets, including the classification of valence and arousal.
The results obtained by the three traditional machine learning methods are used as the baseline data of our dataset, which can be used for follow-up research. The machine learning method used in this study is from scikit-learn (Pedregosa et al., 2018). Simultaneously, a single-input DCNN is used to evaluate the three modalities together and separately, and the evaluation results, along with the results from our MI-DCNN, were used to investigate when the input signals are significantly different, and whether the multi-input DCNN model performs better than the single-input DCNN model. Figure 4 shows the emotion recognition framework based on traditional machine learning methods and our proposed deep learning method, where the blue arrow represents the emotion recognition process based on traditional machine learning and the red arrow represents the emotion recognition process based on deep learning. For the traditional machine learning process, the following steps are required: (a) data collection from the subjects, (b) signal preprocessing and feature extraction for raw signals, (c) feature engineering and construction of balanced subsets, and (d) training the ML models and predicting the emotion class. For our proposed deep learning methods, the following steps are necessary: (a) data collection from the subjects, (b) construction of balanced subsets, (c) training the DL models, and predicting the emotion class. Compared with traditional machine learning methods, our proposed deep learning emotion recognition method eliminates the process of signal processing, feature extraction, and feature engineering, which is a complete end-to-end model.
Considering our dataset is not a balanced dataset, we randomly created five fully balanced datasets on the current dataset according to the least type of categories to make our results more convincing. For each sub-dataset, 80% of the samples are used as the training set, the remaining 20% of the samples are used as the test set, and five-fold cross-validation was performed. The average of the results of the five sub-datasets is regarded as the result of the sentiment dataset.
We used single-modal physiological signals (ECG, EDA, and RSP) and multi-modal physiological signals for emotion recognition, respectively. While using traditional machine learning classifiers, we also used random classifiers and majority classifiers as baseline comparisons. Random classifier makes random predictions on samples, and the majority classifier is a model that makes predictions based on most samples. For the training process of the three traditional machine learning algorithms, the grid search was used to search and compare the inherent parameters of each machine learning model to select the best combination of parameters. The specific results of using traditional machine learning methods are shown in Table 5.
We analyzed the baseline results of the dataset and discussed the performance differences of emotion recognition based on single-modality and multi-modality physiological signals on different traditional machine learning classifiers. For arousal, the results of using the multi-modal had a significant improvement than single modal (t-test, ECG: 6.5E-04 < 0.01, EDA: 7.5E-04 < 0.01, RSP: 3.6E-05 < 0.01), and for valence, the results of using the multi-modal also showed a significant improvement than the single modal (t-test, ECG: 5.2E-06 < 0.01, EDA: 1.2E-04 < 0.01, RSP: 8.9E-04 < 0.01). The support vector machine algorithm achieved the best classification results in arousal classification tasks, with an accuracy of 62.9 ± 0.9% and an f1-score of 62.8 ± 1.0%. Compared with the results of arousal, the results of valence were relatively inferior at the level of the classifier or at the level of different modalities. The support vector machine algorithm also achieved the best results on the valence task, with an accuracy of 60.3 ± 0.8% and an f1-score of 60.0 ± 1.0%.
To evaluate the quality of our dataset, we compared the baseline results of our dataset with those of three public datasets. The comparison results are shown in Table 6. First, a brief introduction to the three public datasets was performed.
A total of 32 participants were recorded with EEG data, peripheral physiological data, frontal facial videos, and participants' self-assessment reports while watching emotion elicitation video stimuli. Considering the relevance of the study, only peripheral physiological signals and participants' self-assessment reports were analyzed in this study. In total, 40 emotional videos were selected as stimuli, and through an effective highlighting algorithm, the 60-s segment with maximum emotional content is detected. The emotional evaluation model uses the Self-Assessment Manikin (SAM; Morris, 1995), including valence, arousal, and dominance, in which the dominance scale ranges from submissive to dominant. The values of all three options are continuous values from 1 to 9; in addition, the division of valence/arousal degree is also strictly in accordance with the study mentioned in Koelstra et al. (2012).
Instead of medical-grade devices, the DREAMER dataset is used with low-cost, wearable devices. The stimuli materials were obtained from a dataset consisting of 18 videos evaluated by Gabert-Quillen et al. (2015). EEG, ECG, and self-assessment reports of 23 participants were recorded through the emotional eliciting experiment, and the length of stimuli videos ranges from 65 to 393 s. Only the data corresponding to the last 60 s of each video are clipped for research. Similar to DEAP, the emotional evaluation model also uses the Self-Assessment Manikin, but the scope scale is different.
It is a multi-modal physiological signal dataset for personality characteristics and emotion recognition research of individuals and groups, using long and short video-inducted materials, collecting EEG signals, ECG signals, GSR signals, and facial videos of 40 participants. The emotional evaluation model is also a Self-Assessment Manikin.
Different from the other three datasets, we did not collect EEG data in our experiment, so we will not discuss EEG signals in detail. For peripheral physiological signals, the baseline results of arousal and valence in the DEAP dataset are lower than those in our dataset, which had an accuracy of 57 and 62.7%, and an f1-score of 0.533 and 0.608, respectively. In addition, the results in the DEAP dataset were not obtained by using balanced data, and from the majority class classifier, it can be seen that when the model predicts the test samples according to the proportion, the accuracy is even higher than that of the trained machine learning model, but the f1-score is very low. This is because the majority class classifier predicts the test set into the same class (the class with more samples in the training set). The DREAMER dataset uses EEG and ECG to perform the emotion recognition task, which had accuracies of arousal and valence of 62.4 and 62.4%, and f1-scores of arousal and valence of 0.580 and 0.531. The AMIGOS dataset only calculated the results of the f1-score, which were slightly lower than our results when using fusion peripheral data. In our dataset, we first construct a fully balanced dataset and then judge on the balanced dataset so that the results obtained will not be falsely high, from the majority class classification used in our baseline results. It can be seen that when the test set is predicted according to the class with more samples in the training set, the result is 50.0%, which is a completely random result for a binary classification task. In addition, the f1-score is only 0.33. It is because all the samples in the test set are predicted to be of a certain category, and the proportions of the two categories in the test set are exactly the same. Compared with the public datasets, our research obtained a better result based on the use of a balanced dataset, which also shows that our dataset has better quality.
This section discusses the results of our proposed methods. The convolutional neural network is a network structure widely used for image processing. It has been well-applied in computer vision and related fields, but it is still in the exploration stage in the field of emotion recognition using physiological signals. Unlike the three-channel picture information, each type of physiological signal is one unique channel signal; they are independent and represent different functions, so if stacked different physiological signals into one input for convolution like the process for a 2D image, we may not be able to get the effective features of each physiological signal, or even get some meaningless features because in the generation process of each feature map, all the input channels should be considered. Therefore, we introduced the concept of multiple inputs and verified our idea on our collected dataset. Table 7 shows the comparison results of using single-input convolution and multi-input convolution. The first three schemes are based on single-input single-modal, the fourth scheme is single-input multi-modal, and the fifth scheme is multi-input multi-modal. The results show that when the network structure is a single-input DCNN, the classification result of fusion data is worse than that of ECG, which proved that if the single input DCNN is used for different input data, the performance of the classifier may be degraded. However, when using an MI-DCNN, the best results were obtained and compared with the former schemes, and the classification results on valence and arousal were significantly improved, which verified the effectiveness of MI-DCNN. Simultaneously, the fifth scheme also confirmed the superiority of using multi-modality for emotion recognition. We compared the classification performance between traditional machine learning methods and our proposed MI-CNN, and the detailed results are shown in Figure 5. Compared with traditional machine learning methods, our proposed MI-CNN showed a significant improvement in both tasks (t-test, arousal: p = 9.7E-03 < 0.01, valence: 6.5E-03 < 0.01).
Figure 6 shows confusion matrixes using fusion data of our proposed method, where the row represents the actual label and the column represents the predicted label. Through the confusion matrix, we can observe the specific classification results of our proposed method in each category, including precision and recall. For arousal, the precision of low arousal is 77.57% and recall is 83.84%, and the precision of high arousal is 82.42% and recall is 75.76%. For valence, the precision of low valence is 64.38% and recall is 76.42%, and the precision of high valence is 71.00% and recall is 57.72%. Table 8 shows the comparison of results on two public datasets. The results of our proposed methods were higher than the baseline results (Koelstra et al., 2012; Katsigiannis and Ramzan, 2018). Simultaneously, compared with other end-to-end research (Wang and Shang, 2013), our method also had improvement, which shows a high potential for using the 1D convolutional neural network for automatic emotion recognition tasks.
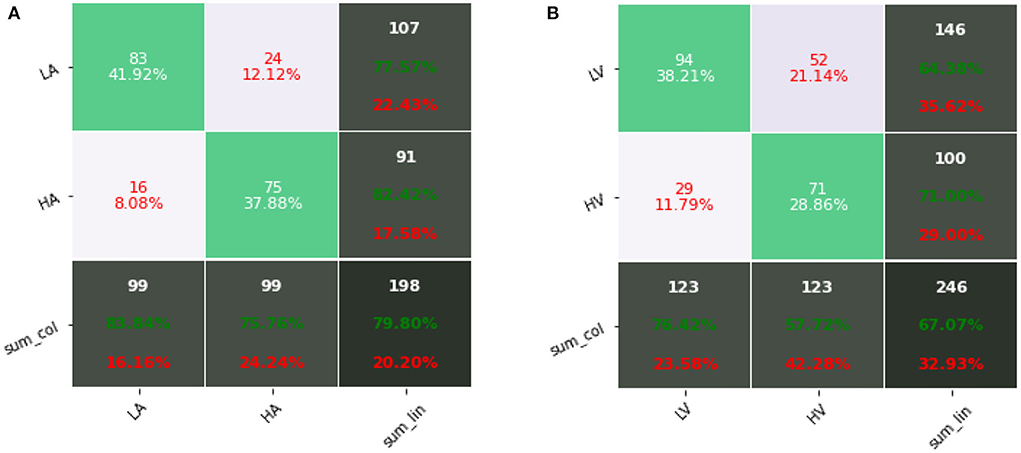
Figure 6. Confusion matrixes using fusion data (ECG&EDA&RSP) of our proposed method: (A) arousal; (B) valence.
This work studied the response patterns of physiological signals in different emotions. By designing an emotional induction paradigm, a physiological emotional dataset was constructed, and a complete end-to-end MI-DCNN structure was designed. The results of the MI-DCNN in emotion recognition tasks showed better performance than traditional machine learning methods, and less preprocessing and automatic feature extraction progress showed the MI-DCNN more suitable for automatic emotion recognition. We also studied physiological signal-based emotion recognition from a new perspective, and physiological signals are different from picture information. Considering the application scenario, we propose a multi-input deep convolutional neural network that implements end-to-end automatic emotion recognition based on multi-modal physiological signals. By adding input channels, we solved the problem of interference between channels and achieved the automatic feature extraction step. Simultaneously, by comparing the difference between a single-input CNN and a multi-input CNN, we elaborated on the necessity of using a multi-input CNN when the channel information is quite different. However, some limitations should be noted. First, to obtain a model with stronger generalization ability, the individual difference and temporal difference of biological signals should be considered. Second, we only discussed the benefits of adding input channels to the classification results, ignoring the increase in complexity and parameters they bring to the model. In our future research, we will seek to develop lightweight neural networks that can learn individual and temporal differences from biological signals for automatic emotion recognition.
The datasets presented in this article are not readily available because the experiment data is not available online for further research, but available on reasonable request according to the policy of Tianjin University, Capital Medical University, and Tianjin University of Technology. Requests to access the datasets should be directed to CC, Y2Njb3ZiQGhvdG1haWwuY29t.
The studies involving human participants were reviewed and approved by Beijing Anding Hospital, Capital Medical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
BZ and CC designed coordinated this research and helped perform the analysis through constructive discussions. PC carried out the experiments and the data analysis and wrote the manuscript. AB, XL, and XZ conceived the study and participated in the research coordination. WY, ZH, and JL contributed significantly to the analysis and manuscript preparation. All authors contributed to the article and approved the submitted version.
This research was supported in part by the National Natural Science Foundation of China (61806146, 82101448, 62006014, and U19B2032), the Beijing Municipal Hospital Research and Development Project (PX2021068), the Advanced Innovation Center for Human Brain Protection Project (3500-12020137), the Guangdong Basic and Applied Basic Research Foundation (2021A1515110249), the Scientific and Technological Innovation 2030 (Grant 2021ZD0204300 and 2021Z D0204303), Tianjin Enterprise Science and Technology Commissioner Project under (Grant No. 20YDTPJC00790), and Beijing Key Laboratory of Mental Disorders (Code: 2020JSJB04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abadi, M., Barham, P., Chen, J., Chen, Z., Davis, A., Dean, J., et al. (2016). “TensorFlow: a system for large-scale machine learning,” in Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI '16) (Savannah, GA), 265–283.
Abadi, M. K., Subramanian, R., Kia, S. M., Avesani, P., Patras, I., and Sebe, N. (2015). DECAF: MEG-based multimodal database for decoding affective physiological responses. IEEE Trans. Affect. Comput. 6, 209–222. doi: 10.1109/TAFFC.2015.2392932
Abbaschian, B. J., Sierra-Sosa, D., and Elmaghraby, A. (2021). Deep learning techniques for speech emotion recognition, from databases to models. Sensors 21, 1249. doi: 10.3390/s21041249
Acharya, U. R., Oh, S. L., Hagiwara, Y., Tan, J. H., and Adeli, H. (2018). Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 100, 270–278. doi: 10.1016/j.compbiomed.2017.09.017
Agrafioti, F., Hatzinakos, D., and Anderson, A. K. (2012). ECG pattern analysis for emotion detection. IEEE Trans. Affect. Comput. 3, 102–115. doi: 10.1109/T-AFFC.2011.28
Akhand, M. A. H., Roy, S., Siddique, N., Kamal, M. A. S., and Shimamura, T. (2021). Facial emotion recognition using transfer learning in the deep CNN. Electronics 10, 1036. doi: 10.3390/electronics10091036
Cacioppo, J. T., Tassinary, L. G., and Berntson, G. (2007). Handbook of Psychophysiology. Cambridge: Cambridge University Press.
Chourasia, M., Haral, S., Bhatkar, S., and Kulkarni, S. (2021). “Emotion recognition from speech signal using deep learning,” in Intelligent Data Communication Technologies and Internet of Things Lecture Notes on Data Engineering and Communications Technologies, eds J. Hemanth, R. Bestak, and J. I.-Z. Chen (Singapore: Springer), 471–481. doi: 10.1007/978-981-15-9509-7_39
Cosoli, G., Poli, A., Scalise, L., and Spinsante, S. (2021). Measurement of multimodal physiological signals for stimulation detection by wearable devices. Measurement 184, 109966. doi: 10.1016/j.measurement.2021.109966
Gabert-Quillen, C. A., Bartolini, E. E., Abravanel, B. T., and Sanislow, C. A. (2015). Ratings for emotion film clips. Behav. Res. Methods 47, 773–787. doi: 10.3758/s13428-014-0500-0
Greco, A., Valenza, G., Lanata, A., Scilingo, E. P., and Citi, L. (2016). cvxEDA: a convex optimization approach to electrodermal activity processing. IEEE Trans. Biomed. Eng. 63, 797–804. doi: 10.1109/TBME.2015.2474131
Gross, J. J., and Levenson, R. W. (1997). Hiding feelings: the acute effects of inhibiting negative and positive emotion. J. Abnormal Psychol. 106, 95–103. doi: 10.1037/0021-843X.106.1.95
Hamilton, P. (2002). Open source ECG analysis. Comput. Cardiol. 2002, 101–104. doi: 10.1109/CIC.2002.1166717
He, Z., Li, Z., Yang, F., Wang, L., Li, J., Zhou, C., et al. (2020). Advances in multimodal emotion recognition based on brain–computer interfaces. Brain Sci. 10, 687. doi: 10.3390/brainsci,10100687
He, Z., Zhong, Y., and Pan, J. (2022). An adversarial discriminative temporal convolutional network for EEG-based cross-domain emotion recognition. Comput. Biol. Med. 141, 105048. doi: 10.1016/j.compbiomed.2021.105048
Hjorth, B. (1970). EEG analysis based on time domain properties. Electroencephalogr. Clin. Neurophysiol. 29, 306–310. doi: 10.1016/0013-4694(70)90143-4
Ioffe, S., and Szegedy, C. (2015). “Batch normalization: accelerating deep network training by reducing internal covariate shift,” in Proceedings of the 32nd International Conference on Machine Learning (PMLR) (Lille), 448–456.
Katsigiannis, S., and Ramzan, N. (2018). DREAMER: a database for emotion recognition through EEG and ECG signals from wireless low-cost off-the-shelf devices. IEEE J. Biomed. Health Informat. 22, 98–107. doi: 10.1109/JBHI.2017.2688239
Kingma, D. P., and Ba, L. J. (2015). “Adam: a method for stochastic optimization,” in International Conference on Learning Representations (San Diego, CA).
Koelstra, S., Muhl, C., Soleymani, M., Lee, J.-S., Yazdani, A., Ebrahimi, T., et al. (2012). DEAP: a database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 3, 18–31. doi: 10.1109/T-AFFC.2011.15
Kolodyazhniy, V., Kreibig, S. D., Gross, J. J., Roth, W. T., and Wilhelm, F. H. (2011). An affective computing approach to physiological emotion specificity: toward subject-independent and stimulus-independent classification of film-induced emotions. Psychophysiology 7, 908–922. doi: 10.1111/j.1469-8986.2010.01170.x
Kroenke, K., and Spitzer, R. L. (2002). The PHQ-9: a new depression diagnostic and severity measure. Psychiatr. Ann. 32, 509–515. doi: 10.3928/0048-5713-20020901-06
Lang, P. J., Greenwald, M. K., Bradley, M. M., and Hamm, A. O. (1993). Looking at pictures: affective, facial, visceral, and behavioral reactions. Psychophysiology 30, 261–273. doi: 10.1111/j.1469-8986.1993.tb03352.x
Liu, W., Qiu, J.-L., Zheng, W.-L., and Lu, B.-L. (2022). Comparing recognition performance and robustness of multimodal deep learning models for multimodal emotion recognition. IEEE Trans. Cogn. Dev. Syst. 14, 715–729. doi: 10.1109/TCDS.2021.3071170
Makowski, D., Pham, T., Lau, Z. J., Brammer, J. C., Lespinasse, F., Pham, H., et al. (2021). NeuroKit2: a Python toolbox for neurophysiological signal processing. Behav. Res. 53, 1689–1696. doi: 10.3758/s13428-020-01516-y
Miranda-Correa, J. A., Abadi, M. K., Sebe, N., and Patras, I. (2021). AMIGOS: a dataset for affect, personality and mood research on individuals and groups. IEEE Trans. Affect. Comput. 12, 479–493. doi: 10.1109/TAFFC.2018.2884461
Morris, J. D. (1995). Observations: SAM: the self-assessment manikin: an efficient cross-cultural measurement of emotional response. J. Advert. Res. 35, 63–68.
Mustaqeem, A., and Kwon, S. (2021). MLT-DNet: speech emotion recognition using 1D dilated CNN based on multi-learning trick approach. Expert Syst. Appl. 167, 114177. doi: 10.1016/j.eswa.2020.114177
Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., et al. (2018). Scikit-learn: Machine Learning in Python. arXiv [Preprint]. arXiv: 1201.0490. doi: 10.48550/arXiv.1201.0490
Santamaria-Granados, L., Munoz-Organero, M., Ramirez-González, G., Abdulhay, E., and Arunkumar, N. (2019). Using deep convolutional neural network for emotion detection on a physiological signals dataset (AMIGOS). IEEE Access 7, 57–67. doi: 10.1109/ACCESS.2018.2883213
Siddharth, A., Jung, T.-P., and Sejnowski, T. J. (2022). Utilizing deep learning towards multi-modal bio-sensing and vision-based affective computing. IEEE Trans. Affect. Comput. 13, 96–107. doi: 10.1109/TAFFC.2019.2916015
Soleymani, M., Lichtenauer, J., Pun, T., and Pantic, M. (2012). A multimodal database for affect recognition and implicit tagging. IEEE Trans. Affect. Comput. 3, 42–55. doi: 10.1109/T-AFFC.2011.25
Song, T., Zheng, W., Lu, C., Zong, Y., Zhang, X., and Cui, Z. (2019). MPED: a multi-modal physiological emotion database for discrete emotion recognition. IEEE Access 7, 12177–12191. doi: 10.1109/ACCESS.2019.2891579
Subramanian, R., Wache, J., Abadi, M. K., Vieriu, R. L., Winkler, S., and Sebe, N. (2018). ASCERTAIN: emotion and personality recognition using commercial sensors. IEEE Trans. Affect. Comput. 9, 147–160. doi: 10.1109/TAFFC.2016.2625250
Tang, H., Liu, W., Zheng, W.-L., and Lu, B.-L. (2017). “Multimodal emotion recognition using deep neural networks,” in Neural Information Processing Lecture Notes in Computer Science, eds D. Liu, S. Xie, Y. Li, D. Zhao, and E.-S. M. El-Alfy (Cham: Springer International Publishing), 811–819. doi: 10.1007/978-3-319-70093-9_86
Tripathi, S., Acharya, S., Sharma, R. D., Mittal, S., and Bhattacharya, S. (2017). “Using deep and convolutional neural networks for accurate emotion classification on DEAP dataset,” in Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence AAAI'17 (San Francisco, California, USA: AAAI Press), 4746–4752. doi: 10.1609/aaai.v31i2.19105
Verma, G. K., and Tiwary, U. S. (2014). Multimodal fusion framework: a multiresolution approach for emotion classification and recognition from physiological signals. NeuroImage 102, 162–172. doi: 10.1016/j.neuroimage.2013.11.007
Wagner, J., Kim, J., and Andre, E. (2005). “From physiological signals to emotions: implementing and comparing selected methods for feature extraction and classification,” in 2005 IEEE International Conference on Multimedia and Expo (Amsteram), 940–943. doi: 10.1109/ICME.2005.1521579
Wang, D., and Shang, Y. (2013). Modeling physiological data with deep belief networks. Int. J. Inf. Educ. Technol. 3, 505–511. doi: 10.7763/IJIET.2013.V3.326
Yildirim, Ö., Pławiak, P., Tan, R.-S., and Acharya, U. R. (2018). Arrhythmia detection using deep convolutional neural network with long duration ECG signals. Comput. Biol. Med. 102, 411–420. doi: 10.1016/j.compbiomed.2018.09.009
Zhang, Q., Chen, X., Zhan, Q., Yang, T., and Xia, S. (2017). Respiration-based emotion recognition with deep learning. Comput. Indus. 92–93, 84–90. doi: 10.1016/j.compind.2017.04.005
Zhang, X., Liu, J., Shen, J., Li, S., Hou, K., Hu, B., et al. (2021). Emotion recognition from multimodal physiological signals using a regularized deep fusion of kernel machine. IEEE Trans. Cybernet. 51, 4386–4399. doi: 10.1109/TCYB.2020.2987575
Zhang, Y., Hossain, M. Z., and Rahman, S. (2021). “DeepVANet: a deep end-to-end network for multi-modal emotion recognition,” in Human-Computer Interaction – INTERACT 2021 Lecture Notes in Computer Science, eds C. Ardito, R. Lanzilotti, A. Malizia, H. Petrie, A. Piccinno, G. Desolda, et al. (Cham: Springer International Publishing), 227–237. doi: 10.1007/978-3-030-85613-7_16
Keywords: biological signals, multi-modality, emotion recognition, convolutional neural network, machine learning
Citation: Chen P, Zou B, Belkacem AN, Lyu X, Zhao X, Yi W, Huang Z, Liang J and Chen C (2022) An improved multi-input deep convolutional neural network for automatic emotion recognition. Front. Neurosci. 16:965871. doi: 10.3389/fnins.2022.965871
Received: 10 June 2022; Accepted: 29 August 2022;
Published: 04 October 2022.
Edited by:
Chee-Kong Chui, National University of Singapore, SingaporeReviewed by:
Jiahui Pan, South China Normal University, ChinaCopyright © 2022 Chen, Zou, Belkacem, Lyu, Zhao, Yi, Huang, Liang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bochao Zou, em91Ym9jaGFvQHVzdGIuZWR1LmNu; Chao Chen, Y2Njb3ZiQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.