
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Neurosci., 28 July 2022
Sec. Brain Imaging Methods
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.954237
This article is part of the Research TopicImage Processing Methods in Animal MRI and their Application to Evaluate Brain FunctionView all 12 articles
Aims: To construct an automatic method for individual parcellation of manganese-enhanced magnetic resonance imaging (MEMRI) of rat brain with high accuracy, which could preserve the inherent voxel intensity and Regions of interest (ROI) morphological characteristics simultaneously.
Methods and results: The transformation relationship from standardized space to individual space was obtained by firstly normalizing individual image to the Paxinos space and then inversely transformed. On the other hand, all the regions defined in the atlas image were separated and resaved as binary mask images. Then, transforming the mask images into individual space via the inverse transformations and reslicing using the 4th B-spline interpolation algorithm. The boundary of these transformed regions was further refined by image erosion and expansion operator, and finally combined together to generate the individual parcellations. Moreover, two groups of MEMRI images were used for evaluation. We found that the individual parcellations were satisfied, and the inherent image intensity was preserved. The statistical significance of case-control comparisons was further optimized.
Conclusions: We have constructed a new automatic method for individual parcellation of rat brain MEMRI images, which could preserve the inherent voxel intensity and further be beneficial in case-control statistical analyses. This method could also be extended to other imaging modalities, even other experiments species. It would facilitate the accuracy and significance of ROI-based imaging analyses.
Numerous studies have demonstrated the usefulness of magnetic resonance imaging (MRI) techniques in animal experiments, which is important for pathogenesis researches, drug developments, and so on (Bible et al., 2012; Kim et al., 2012; Wang et al., 2012, 2018; Li et al., 2015; Liang et al., 2020; Tu et al., 2020). Manganese-enhanced MRI (MEMRI), a new MRI technique, could detect the active neurons and trace neuronal pathway by monitoring manganese ions (Mn2+) in vivo (Aoki et al., 2004; Koretsky and Silva, 2004). By tracing the deposition of Mn2+, the active neurons would have high voxel intensity in MEMRI images, while other voxels have low intensity. Recently, the MEMRI technique has been widely used in various studies of the rodent brain (Ho et al., 2018; Perez et al., 2018; Gimenes et al., 2019; Yang et al., 2020; Bearer et al., 2022).
Imaging analysis is a prerequisite in MEMRI studies for quantitative interpretations of neuronal dysfunctions. Regions of interests (ROIs)-based analysis is one of the most frequently used methods in rat brain MEMRI studies (Spurny et al., 2019). Similar to other modalities of MRI, by spatially normalizing individual MEMRI images into a standard space, ROIs could be extracted via a corresponding atlas image automatically (Nie et al., 2013; Barriere et al., 2019). However, it has been found in functional studies that the brain sub-regions vary across individuals (Chong et al., 2017; Salehi et al., 2020; Reijonen et al., 2021). Moreover, because of image interpolations and transformations in spatial normalization, the inherent MEMRI intensity would be confused by adjacent voxels inevitably (Ashburner and Friston, 1999; Zhilkin and Alexander, 2004). Especially, the voxel intensity of active neurons would be affected, even decreased, by adjacent un-activated neurons, and the statistical significance of case-control studies would further be affected (Lv et al., 2021).
Alternatively, manually tracing ROIs in individual images is another important method in MEMRI studies, which could avoid the influences of image normalization and preserve the inherent voxel intensity (Jackson et al., 2011). However, this method has many disadvantages, such as low accuracy, labor-intensive, subjective and poor robustness, and so on. Therefore, it would be highly desirable to develop an automatic method for ROI parcellations in individual space of MEMRI images of rat brain.
In addition, more than a decade ago, our lab had constructed an automatic atlas-based method for tracing ROIs in individual space by registering the template images to individuals (Nie et al., 2010). However, as the resolution of individual images was often lower than the template, the accuracy of this image registration was always lower than spatial normalization, so that the subtle ROIs were hard to trace out precisely.
Therefore, the current study was aimed to introduce a new automatic method for individual parcellation of MEMRI images of rat brain with high accuracy, and both the inherent voxel intensity and ROI morphological characteristics could be preserved simultaneously. For evaluation, individual parcellations were performed on two groups of T2-weighted (T2WI) and MEMRI images, and further comparing with the parcellations in standardized space. Moreover, the method constructed in this study was packaged and available by contacting the corresponding author at bmllYmJAaWhlcC5hYy5jbg==.
Parcellation of rat brain images in individual space could preserve the inherent voxel intensity and ROI morphological characteristics, which would further improve the statistical sensitivity in ROI-based analyses. The automatic individual parcellation method was performed based on the stereotaxic template sets of rat brain in Paxinos space (Liang et al., 2017). The flow chart was shown in Figure 1 and detailed as follows.
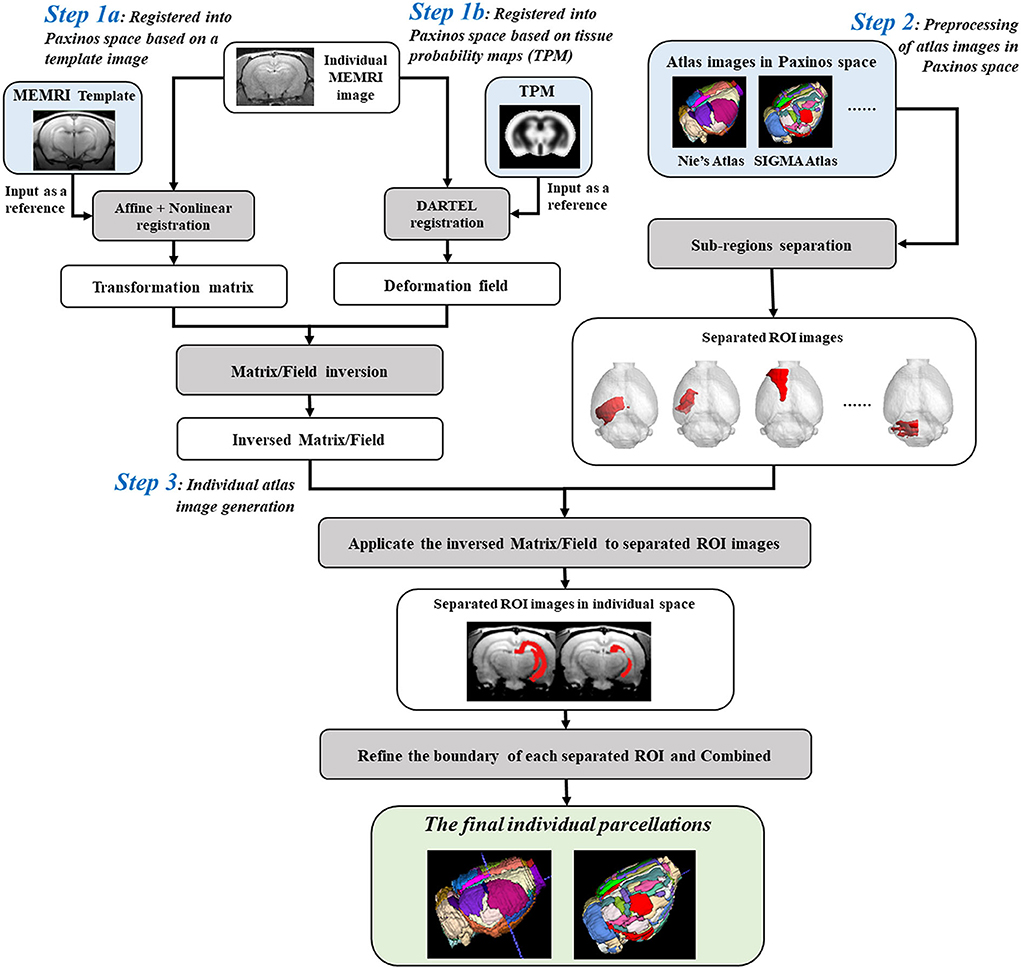
Figure 1. Flow chart for creating individual parcellations of manganese-enhanced magnetic resonance imaging (MEMRI) image of rat brain. Step 1: The individual MEMRI image was registered into Paxinos space either by affine/nonlinear transformations based on the MEMRI template image (Step 1a), or by DARTEL algorithm based on the tissue probability maps (TPM) (Step 1b), and the transformation matrix/deformation field was obtained, named as Matrixsn/Deformy. Inverse transformation of the Matrixsn/Deformy was then calculated, named InvMatrixsn/InvDeformy. Step 2: All the regions of interests (ROIs) defined in atlas images in Paxinos space were firstly separated and resaved as single-ROI mask images. Step 3: The separated ROI mask images were transformed into individual space via the InvMatrixsn/InvDeformy using 4th B-spline interpolation algorithm. The contour of each transformed ROI was identified and further refined by image erosion and expansion operator. Then, each refined ROI was given a unique integer as an index and then combined into a new atlas image. This combined atlas image was the final individual parcellations of MEMRI images.
Firstly, the spatial transformation from individual space to Paxinos space was calculated by registering the MEMRI image to the stereotaxic template. This registration could be performed either by affine/nonlinear transformations based on the MEMRI template image (Figure 1-Step 1a) (Ashburner and Friston, 1999; Zhilkin and Alexander, 2004), or by DARTEL (diffeomorphic anatomical registration through exponentiated lie) algorithm (Mak et al., 2011) based on the tissue probability maps (TPM) (Figure 1-Step 1b). A transformation matrix was obtained via template way, named Matrixsn, while a deformation field was obtained via TPM way, named Deformy. The inverse transformation of Matrixsn or Deformy was then calculated to obtain the spatial transformation from Paxinos space to individual space (Ashburner et al., 2000), and saved as InvMatrixsn or InvDeformy.
On the other hand, the atlas images in Paxinos space were preprocessed prior to registering with an individual image (Figure 1-Step 2). To avoid the influences between adjacent regions, all the ROIs defined in the atlas image were separated and resaved as single-ROI mask images. In another word, each ROI was resaved as a 3D binary image, and the image intensity of voxels inside the ROI was assigned 1 and outside was 0.
Finally, transforming the atlas images into individual space to create the parcellation of individual image (Figure 1-Step 3). In detail, all the separated ROI mask images were registered with individual image via the InvMatrixsn/InvDeformy using the 4th B-spline interpolation algorithm. Then, the ROI contour of each registered mask image was identified, and further refined by image erosion and dilation using a disc operator with one voxel radius. Next, each refined ROI was given a unique integer as an index. At last, all the reassigned ROIs were combined together to generate the final individual parcellation image.
Five healthy adult Sprague–Dawley (SD) rats (male, 5; age, 10–11 week old; weight, 250–300 g) and ten modal rats with smell damage (male, 10; age, 10–11 week old; and weight, 250–300 g) were used for method evaluation.
During MRI scan, all rats were anesthetized using inhaled isoflurane/O2 (3% for induction and 1.5–2% for maintenance), and prostrated on a custom-made holder to minimize head motion while respiration was monitored at a rate of 50 breaths per min. For MEMRI study, all these rats were injected with manganese chloride (MnCl2·4H2O, Bio Basic Inc., Canada) dissolved in bicine [di(hydroxyethyl)glycine, Sigma-Ulrich, UK] buffer pH 7.4 in doses of 13.95 mg/kg for continuous 7 days (Tang et al., 2016).
All datasets were acquired on a 7.0T animal MRI scanner (70/16 PharmaScan, Bruker Biospin GmbH, Germany) in Nanjing, using a 38-mm birdcage rat brain quadrature resonator for radiofrequency transmission and receiving. T2-weighted (T2WI) data were obtained with a RARE sequence (RARE factor = 8, TR = 10493 ms, TE = 36 ms, matrix size 256*256*90, voxel size 0.14*0.14*0.3 mm, no slice gap). MEMRI data were obtained right after the last injection with a RAREVTR sequence (TR = 5500 ms, TE = 8 ms, matrix size 256*192*22, voxel size 0.14*0.18*1.2 mm3, no slice gap) for six repetitions. All the Bruker original images were converted to DICOM format with programs (Paravision 5.0) in the scanner. All experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Jiang Su Animal Care and Use Committee.
All the fifteen T2WI images were inspected and found equally of high quality in terms of the image contrast, noise level, and resolution. Firstly, all the individual T2WI images were registered into Paxinos space using DARTEL algorithm, and the corresponding Deformy of each rat was obtained. Then, all the Deformy were inversely transformed, and the corresponding InvDeformy of each rat was obtained. On the other hand, 66 functional ROIs defined in the 3D atlas image in Paxinos space were separated and resaved into 66 binary mask images. Next, all the 66 mask images were registered with each T2WI image via the corresponding InvDeformy repetitively using the 4th B-spline interpolation algorithm. Each registered ROI contour was identified and further refined. Finally, each refined ROI was given a unique integer as an index and combined together to generate the individual parcellations of each T2WI image.
All the fifteen MEMRI images were also inspected and found equally of high quality in terms of the image contrast, noise level, and resolution. Firstly, the MEMRI image series were realigned to remove the head movement, and a mean MEMRI image was created over the six realigned volumes. Next, the mean MEMRI image was registered into Paxinos space via the MEMRI template image of rat brain, and the corresponding Matrixsn of each rat was obtained. Then, all the Matrixsn were inversely transformed, and the corresponding InvMatrixsn of each rat was obtained. The preprocessing of the atlas image was the same with Section Generation of individual parcellations of T2WI images. Then, all the preprocessed ROI mask images were registered with individual mean MEMRI images via InvMatrixsn, and the contour was refined. Finally, each refined ROI was given a unique integer as an index and combined together to generate the individual parcellations of each MEMRI image.
For qualitative evaluation of the individual parcellations, the mean MEMRI values in dentate gyrus (DG) and hippocampus (Hip) of all the fifteen rats were extracted in individual space, respectively, and the time courses of each rat were plotted out. For qualitative comparison, all the fifteen MEMRI images were also normalized into the Paxinos space via the corresponding Matrixsn using the 4th B-spline interpolation algorithm, and resliced to 1*1*1.5 mm3 (after zooming). The mean values in DG and Hip of normalized MEMRI images were also extracted and plotted out.
For quantitative evaluation of the individual parcellations, the mean values of the first MEMRI volume in DG and Hip were analyzed using SPSS software, version 19.0 (SPSS Inc. IBM, Armonk, NY, USA). The mean MEMRI values in ten modal and five healthy rats were compared by two-sample t-tests. Statistical significance was defined as a p < 0.05.
Furthermore, the accuracy of the individual parcellations was evaluated by a spatial index, named Dice similarity coefficient (Dice). It describes the similarity of the volume and position between the MEMRI image (MEMRI) and its corresponding individual parcellation image (Parcellation), named DiceMePa, as shown in Equation (1).
The derivation of the Dice is detailed previously (Gutierrez and Zaidi, 2012). The excellent agreement value of Dice is more than 80%. For comparison, the Dice was calculated between the template image and normalized MEMRI image, which was regarded as the golden standard, named DiceTempMe. Moreover, the previous method proposed by our group was also executed (Nie et al., 2010). In detail, each individual MEMRI image was selected as the reference, and the template image was transformed to register with it. And the Dice between the MEMRI image and registered template image was also calculated, named DiceMeTemp.
The individual parcellations of MRI images were shown in Figure 2. Two rats of T2WI images (Figure 2A) or MEMRI images (Figure 2B) were randomly selected. The individual parcellations were shown in color scale, while the MRI images were in gray scale. A 3D surface of individual parcellations was shown to the left, while three coronal slices were shown to the right. The individual parcellations were superimposed on the corresponding T2WI image (Figure 2A), or mean intracranial MEMRI image (Figure 2B). As illustrated in Figure 2, the parcellations were registered into the individual space accurately.
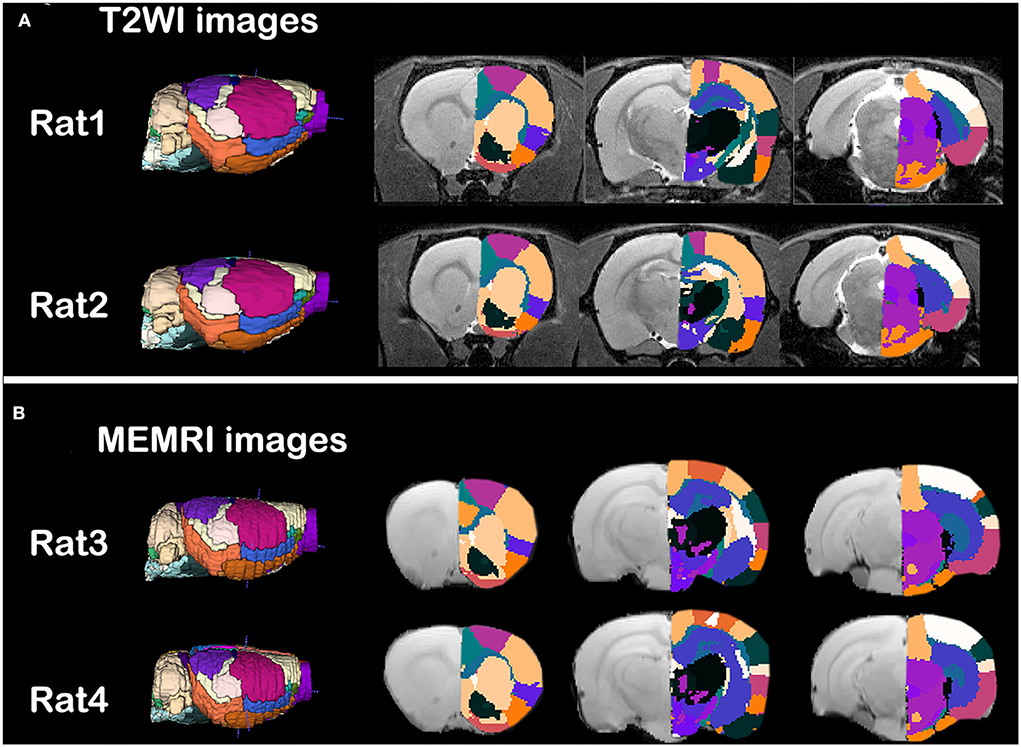
Figure 2. Qualitative evaluation of the individual parcellations of (A) T2WI images and (B) intercranial mean MEMRI images of rat brain. Four rats were randomly selected. Right side of the individual parcellations was shown in color scaled and superimposed on the corresponding MRI image. The T2WI/MEMRI MRI images were shown in gray scaled as background.
The evaluation of individual parcellations of two randomly selected MEMRI images was shown in Figure 3. Unilateral DG and Hip were superimposed on the first volume of MEMRI images (Figures 3A,B). The MEMRI time course in individual space was plotted out in blue color, while it in Paxinos space was in green (Figures 3C,D). As illustrated in Figure 3, although the downtrend of time courses was similar, the MEMRI voxel intensity in individual space was always higher than it in standardized space. It could be speculated that the inherent MEMRI signal could be persevered by individual parcellations.
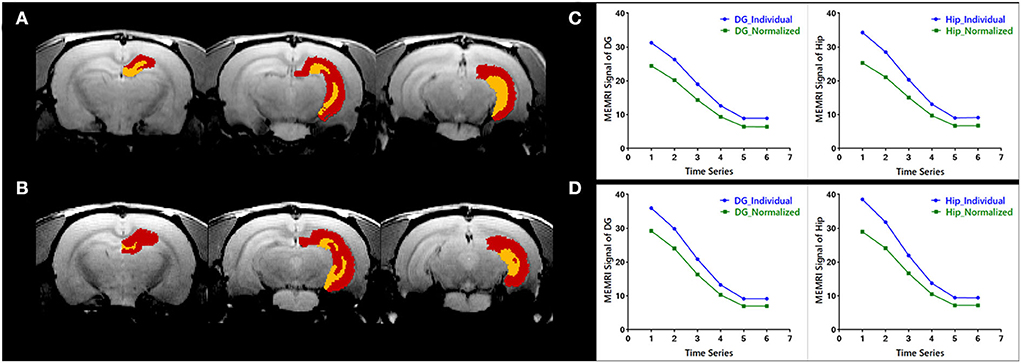
Figure 3. Quantitative evaluation of the individual parcellations of MEMRI images of (A,C) a modal rat and (B,D) a healthy rat. The individual parcellations were shown in color scaled in which the right dentate gyrus was yellow and the right hippocampus was red. The MEMRI images were shown in gray scaled as background. Based on the individual parcellations, the mean MEMRI signal in each ROI was extracted and the curve is shown in blue color (C,D). As a comparison, the MEMRI images of these two rats were also standardized into Paxinos space, and the mean signal in each ROI was then extracted based on the atlas in Paxinos space. The signal curve extracted in traditional way was shown in green color (C,D).
The Dice similarity coefficients between the reference images and transformed images were listed in Table 1. The DiceTempMe between the template image and normalized MEMRI image was selected as the golden standard. As illustrated in Table 1, all the Dice coefficients were bigger than 80%. Compared with the DiceMeTemp, the accuracy proposed in this study was higher.
The individual parcellations of MEMRI images were further applicate in ROI-based analyses of case-control studies. The statistical results were shown in Figure 4. The MEMRI signal of each group was presented as mean ± SD (standard deviation). The MEMRI signal in modal rats is significantly lower than the healthy rats in both DG (p < 0.001) and Hip (p < 0.05). Further studies of neural mechanisms would report in our coming article.
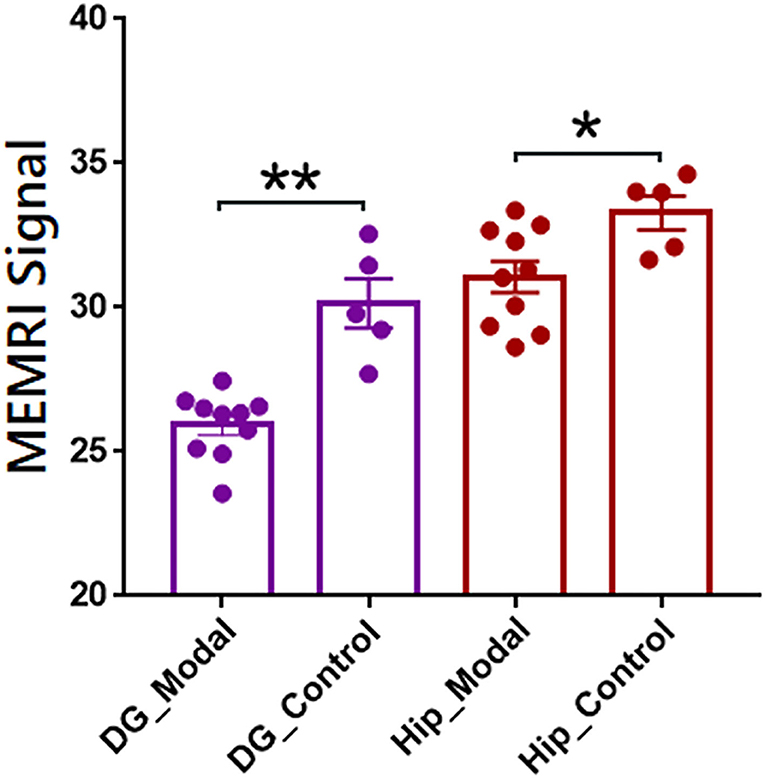
Figure 4. Regions of interest-based quantitative analysis results of MEMRI images between modal and healthy rats. The mean MEMRI signal in bilateral dentate gyrus (purple) and hippocampus (red) were shown as mean ± SE. **p < 0.001; *p < 0.05; DG, dentate gyrus; Hip, hippocampus.
In this study, the new automatic method for individual parcellation of rat brain MEMRI images was constructed. In our proposed procedure, the individual MEMRI images were remained untransformed, so the inherent voxel intensity could be preserved. It would further be beneficial to case-control ROI-based statistical analyses.
Compared with our previous work, the individual parcellation method proposed in this study could perform more precisely. Firstly, in this study, widespread image registration methods were adopted. Especially, by DARTEL algorithm, individual images could be normalized into the template space via a set of tissue probability maps iteratively (Zhang et al., 2022). However, it couldn't be adopted by our previous work, for the lacking of tissue probability maps in individual space. Therefore, although the DARTEL algorithm has been recognized as a more accurate method, it couldn't be used in transforming the standard space to an individual space. On the other hand, the template has also been regarded as a representative image, so transforming the individual image to register with the template would have higher accuracy.
In spatial normalization, the MEMRI images could be firstly normalized into the standardized space via both template way and DARTEL way, that there was also a preferential suggestion. When the individual MRI image has a nearly isotropic resolution, the DARTEL algorithm would perform better than the template way. Otherwise, the template-based method would be more suitable, for its higher arithmetic speed. In the current study, the time course of MEMEI was preferentially considered, so the image resolution was not better enough to use the DARTEL way. Therefore, for demonstration, the MEMRI images were performed in the template way, while the corresponding T2WI structural images were in the DARTEL way.
The parcellation boundary is a problem when transforming the atlas image. Traditionally, in order to protect the ROI index, the nearest neighbor interpolation method was frequently used in atlas image transformations, so that the parcellation boundary was always dentate. To address this issue, we separated all the ROIs into a new binary mask image prior to transformation, and the 4th B-spline interpolation method was chosen. Moreover, the transformed boundary was further refined by erosion and expansion operators after transformation. By this strategy, our parcellation boundary in individual space would be smooth.
The usage and accuracy of a newly developed method are always evaluated by applications in various studies. In this study, a group of modal rats with smell damage was used, and two representative ROIs were selected. As demonstrated in the figures, compared with parcellations in standardized space, the inherent MEMRI intensity could be preserved in individual space, which was in line with the previous studies (Salehi et al., 2020; Reijonen et al., 2021; Wang et al., 2021). Image interpolation is ineluctable in image registration. However, the enhanced voxels are always subtle in MEMRI images, which would be influenced by adjacent un-enhanced voxels in image interpolation. Therefore, the image intensity in standard space is always lower than individual space. Moreover, although the downtrend of MEMRI time courses was similar, the slope is different.
Actually, the individual parcellation method proposed in this study could be applied to other imaging modalities of rat brain studies, such as functional MRI (fMRI), positron emission tomography (PET), diffusion tensor imaging (DTI), and so on. The operation was similar to MEMRI studies, except for choosing the corresponding modality template image (Figure 1-Step 1a). If the DARTEL algorithm was chosen (Figure 1-Step 1b), all the operations were the same with MEMRI studies.
Moreover, this method could also be generalized in data analyses of other species as long as there is a stereotaxic template set, such as human beings, monkeys, mice, tree shrews, and so on. In our released package, these species were all included based on our previous works of stereotaxic templates constructions.
Finally, there are some notes that should be pointed out. Firstly, the individual parcellations would be inaccurate if the corresponding image could not be successfully registered into the standardized space. Secondly, the origin of individual images should be manually set to the same point as template, such as the D3V in rat brain studies (Nie et al., 2013). Thirdly, in rodent brain studies, the image voxel size should be enlarged ten times before spatial normalization. Finally, this individual parcellation method was not suitable in cerebral tumor or infarction imaging studies. Because the sub-anatomical structures are deformed, the traditional template or TPM cannot register the image into standardized space accurately.
In this study, we have constructed a new automatic method for individual parcellation of rat brain MEMRI images, which could preserve the inherent voxel intensity and further be beneficial to case-control statistical analyses. This method could also be extended to other imaging modalities, even other experiments species. It would facilitate the accuracy and significance of ROI-based imaging analyses.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by Jiang Su Animal Care and Use Committee.
BN led the project. ZB, TZ, and TP created the figures and wrote the manuscript. TZ established the software. TP carried out the MEMRI data analysis. WZ and SZ made substantial contributions to the manuscript and provided critical comments. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (12175268 and 11975249).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aoki, I., Naruse, S., and Tanaka, C. (2004). Manganese-enhanced magnetic resonance imaging (MEMRI) of brain activity and applications to early detection of brain ischemia. NMR Biomed. 17, 569–580. doi: 10.1002/nbm.941
Ashburner, J., Andersson, J. L., and Friston, K. J. (2000). Image registration using a symmetric prior–in three dimensions. Hum. Brain Mapp. 9, 212–25. doi: 10.1002/(SICI)1097-0193(200004)9:4<212::AID-HBM3>3.0.CO;2-#
Ashburner, J., and Friston, K. J. (1999). Nonlinear spatial normalization using basis functions. Hum. Brain Mapp. 7, 254–66. doi: 10.1002/(SICI)1097-0193(1999)7:4<254::AID-HBM4>3.0.CO;2-G
Barriere, D. A., Magalhaes, R., Novais, A., Marques, P., Selingue, E., Geffroy, F., et al. (2019). The SIGMA rat brain templates and atlases for multimodal MRI data analysis and visualization. Nat. Commun. 10, 5699. doi: 10.1038/s41467-019-13575-7
Bearer, E. L., Zhang, X., and Jacobs, R. E. (2022). Studying axonal transport in the brain by manganese-enhanced magnetic resonance imaging (MEMRI). Methods Mol. Biol. 2431, 111–142. doi: 10.1007/978-1-0716-1990-2_6
Bible, E., Dell'Acqua, F., Solanky, B., Balducci, A., Crapo, P. M., Badylak, S. F., et al. (2012). Non-invasive imaging of transplanted human neural stem cells and ECM scaffold remodeling in the stroke-damaged rat brain by (19)F- and diffusion-MRI. Biomaterials 33, 2858–2871. doi: 10.1016/j.biomaterials.2011.12.033
Chong, M., Bhushan, C., Joshi, A. A., Choi, S., Haldar, J. P., Shattuck, D. W., et al. (2017). Individual parcellation of resting fMRI with a group functional connectivity prior. Neuroimage 156, 87–100. doi: 10.1016/j.neuroimage.2017.04.054
Gimenes, C., Malheiros, J. M., Battapady, H., Tannus, A., Hamani, C., and Covolan, L. (2019). The neural response to deep brain stimulation of the anterior nucleus of the thalamus: A MEMRI and c-Fos study. Brain Res. Bull. 147, 133–139. doi: 10.1016/j.brainresbull.2019.01.011
Gutierrez, D. F., and Zaidi, H. (2012). Automated analysis of small animal PET studies through deformable registration to an atlas. Eur. J. Nucl. Med. Mol. Imaging 39, 1807–1820. doi: 10.1007/s00259-012-2188-7
Ho, Y. J., Weng, J. C., Lin, C. L., Shen, M. S., Li, H. H., Liao, W. C., et al. (2018). Ceftriaxone treatment for neuronal deficits: a histological and MEMRI study in a rat model of dementia with lewy bodies. Behav. Neurol. 2018, 4618716. doi: 10.1155/2018/4618716
Jackson, S. J., Hussey, R., Jansen, M. A., Merrifield, G. D., Marshall, I., MacLullich, A., et al. (2011). Manganese-enhanced magnetic resonance imaging (MEMRI) of rat brain after systemic administration of MnCl(2): hippocampal signal enhancement without disruption of hippocampus-dependent behavior. Behav. Brain Res. 216, 293–300. doi: 10.1016/j.bbr.2010.08.007
Kim, Y. B., Kalthoff, D., Po, C., Wiedermann, D., and Hoehn, M. (2012). Connectivity of thalamo-cortical pathway in rat brain: combined diffusion spectrum imaging and functional MRI at 11.7 T. NMR Biomed. 25, 943–952. doi: 10.1002/nbm.1815
Koretsky, A. P., and Silva, A. C. (2004). Manganese-enhanced magnetic resonance imaging (MEMRI). NMR Biomed. 17, 527–531. doi: 10.1002/nbm.940
Li, R., Liu, X., Sidabras, J. W., Paulson, E. S., Jesmanowicz, A., Nencka, A. S., et al. (2015). Restoring susceptibility induced MRI signal loss in rat brain at 9.4 T: a step towards whole brain functional connectivity imaging. PLoS ONE 10, e0119450. doi: 10.1371/journal.pone.0119450
Liang, S., Wu, S., Huang, Q., Duan, S., Liu, H., Li, Y., et al. (2017). Rat brain digital stereotaxic white matter atlas with fine tract delineation in Paxinos space and its automated applications in DTI data analysis. Magn. Reson. Imaging 43, 122–128. doi: 10.1016/j.mri.2017.07.011
Liang, S., Zhang, J., Zhang, Q., Li, L., Zhang, Y., Jin, T., et al. (2020). Longitudinal tracing of white matter integrity on diffusion tensor imaging in the chronic cerebral ischemia and acute cerebral ischemia. Brain Res. Bull. 154, 135–141. doi: 10.1016/j.brainresbull.2019.10.015
Lv, Q., Yan, M., Shen, X., Wu, J., Yu, W., Yan, S., et al. (2021). Normative analysis of individual brain differences based on a population MRI-based atlas of cynomolgus macaques. Cereb. Cortex 31, 341–355. doi: 10.1093/cercor/bhaa229
Mak, H. K., Zhang, Z., Yau, K. K., Zhang, L., Chan, Q., Chu, L. W., et al. (2011). Efficacy of voxel-based morphometry with DARTEL and standard registration as imaging biomarkers in Alzheimer's disease patients and cognitively normal older adults at 3.0 Tesla MR imaging. J. Alzheimers Dis. 23, 655–664. doi: 10.3233/JAD-2010-101659
Nie, B., Chen, K., Zhao, S., Liu, J., Gu, X., Yao, Q., et al. (2013). A rat brain MRI template with digital stereotaxic atlas of fine anatomical delineations in paxinos space and its automated application in voxel-wise analysis. Hum. Brain Mapp. 34, 1306–1318. doi: 10.1002/hbm.21511
Nie, B., Hui, J., Wang, L., Chai, P., Gao, J., Liu, S., et al. (2010). Automatic method for tracing regions of interest in rat brain magnetic resonance imaging studies. J. Magn. Reson. Imaging 32, 830–835. doi: 10.1002/jmri.22283
Perez, P. D., Hall, G., Zubcevic, J., and Febo, M. (2018). Cocaine differentially affects synaptic activity in memory and midbrain areas of female and male rats: an in vivo MEMRI study. Brain Imaging Behav. 12, 201–216. doi: 10.1007/s11682-017-9691-1
Reijonen, J., Kononen, M., Tuunanen, P., Maatta, S., and Julkunen, P. (2021). Atlas-informed computational processing pipeline for individual targeting of brain areas for therapeutic navigated transcranial magnetic stimulation. Clin. Neurophysiol. 132, 1612–1621. doi: 10.1016/j.clinph.2021.01.037
Salehi, M., Greene, A. S., Karbasi, A., Shen, X., Scheinost, D., Constable, R. T., et al. (2020). There is no single functional atlas even for a single individual: functional parcel definitions change with task. Neuroimage 208, 116366. doi: 10.1016/j.neuroimage.2019.116366
Spurny, B., Heckova, E., Seiger, R., Moser, P., Klobl, M., Vanicek, T., et al. (2019). Automated ROI-based labeling for multi-voxel magnetic resonance spectroscopy data using FreeSurfer. Front. Mol. Neurosci. 12, 28. doi: 10.3389/fnmol.2019.00028
Tang, X., Wu, D., Gu, L. H., Nie, B. B., Qi, X. Y., Wang, Y. J., et al. (2016). Spatial learning and memory impairments are associated with increased neuronal activity in 5XFAD mouse as measured by manganese-enhanced magnetic resonance imaging. Oncotarget 7, 57556–57570. doi: 10.18632/oncotarget.11353
Tu, T. W., Kovacs, Z. I., Sundby, M., Witko, J. A., Papadakis, G. Z., Reid, W. C., et al. (2020). Diffusion tensor imaging and chemical exchange saturation transfer MRI evaluation on the long-term effects of pulsed focused ultrasound and microbubbles blood brain barrier opening in the rat. Front. Neurosci. 14, 908. doi: 10.3389/fnins.2020.00908
Wang, H., Sun, J., Cui, D., Wang, X., Jin, J., Li, Y., et al. (2021). Quantitative assessment of inter-individual variability in fMRI-based human brain atlas. Quant. Imaging Med. Surg. 11, 810–822. doi: 10.21037/qims-20-404
Wang, J., Hao, Z., and Wang, H. (2018). Generation of individual whole-brain atlases with resting-state fMRI data using simultaneous graph computation and parcellation. Front. Hum. Neurosci. 12, 166. doi: 10.3389/fnhum.2018.00166
Wang, S., Tryggestad, E., Zhou, T., Armour, M., Wen, Z., Fu, D. X., et al. (2012). Assessment of MRI parameters as imaging biomarkers for radiation necrosis in the rat brain. Int. J. Radiat. Oncol. Biol. Phys. 83, e431–e436. doi: 10.1016/j.ijrobp.2011.12.087
Yang, J., Gao, J., Han, D., Li, Q., Liao, C., Li, J., et al. (2020). Hippocampal changes in inflammasomes, apoptosis, and MEMRI after radiation-induced brain injury in juvenile rats. Radiat. Oncol. 15, 78. doi: 10.1186/s13014-020-01525-3
Zhang, T., Nie, B., Liu, H., Shan, B., and Alzheimer's Disease Neuroimaging Initiative (2022). Unified spatial normalization method of brain PET images using adaptive probabilistic brain atlas. Eur. J. Nucl. Med. Mol. Imaging 49, 3073–3085. doi: 10.1007/s00259-022-05752-6
Keywords: individual parcellations, ROI-based analysis, manganese-enhanced magnetic resonance imaging (MEMRI), rat brain, stereotaxic template set
Citation: Bao Z, Zhang T, Pan T, Zhang W, Zhao S, Liu H and Nie B (2022) Automatic method for individual parcellation of manganese-enhanced magnetic resonance imaging of rat brain. Front. Neurosci. 16:954237. doi: 10.3389/fnins.2022.954237
Received: 27 May 2022; Accepted: 05 July 2022;
Published: 28 July 2022.
Edited by:
Shengxiang Liang, Fujian University of Traditional Chinese Medicine, ChinaReviewed by:
Chuanli Cheng, Shenzhen Institutes of Advanced Technology (CAS), ChinaCopyright © 2022 Bao, Zhang, Pan, Zhang, Zhao, Liu and Nie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Binbin Nie, bmllYmJAaWhlcC5hYy5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.