
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Neurosci. , 01 December 2022
Sec. Neural Technology
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.906290
Deep neural networks (DNNs) have transformed the field of computer vision and currently constitute some of the best models for representations learned via hierarchical processing in the human brain. In medical imaging, these models have shown human-level performance and even higher in the early diagnosis of a wide range of diseases. However, the goal is often not only to accurately predict group membership or diagnose but also to provide explanations that support the model decision in a context that a human can readily interpret. The limited transparency has hindered the adoption of DNN algorithms across many domains. Numerous explainable artificial intelligence (XAI) techniques have been developed to peer inside the “black box” and make sense of DNN models, taking somewhat divergent approaches. Here, we suggest that these methods may be considered in light of the interpretation goal, including functional or mechanistic interpretations, developing archetypal class instances, or assessing the relevance of certain features or mappings on a trained model in a post-hoc capacity. We then focus on reviewing recent applications of post-hoc relevance techniques as applied to neuroimaging data. Moreover, this article suggests a method for comparing the reliability of XAI methods, especially in deep neural networks, along with their advantages and pitfalls.
Machine learning (ML) and deep learning (DL; also known as hierarchical learning or deep structured learning) models have revolutionized computational analysis (Bengio et al., 2013; LeCun et al., 2015; Schmidhuber, 2015) across a variety of fields such as text parsing, facial reconstruction, recommender systems, and self-driving cars (Cheng et al., 2016; Richardson et al., 2017; Young et al., 2018; Grigorescu et al., 2020). These models are particularly successful when applied to images, achieving human-level performance on visual recognition tasks (Kriegeskorte, 2015; LeCun et al., 2015). Among the demanding domains confronting ML/DL researchers are healthcare and medicine (Litjens et al., 2017; Miotto et al., 2017; Shen et al., 2017; Kermany et al., 2018). In medical imaging and neuroimaging, in particular, deep learning has been used to make new discoveries in various domains. For example, conventional wisdom for radiologists was that little or no prognostic information is contained within a tumor, and therefore one should examine its borders. However, a recent deep learning approach coupled to texture analysis found predictive information within the tumor itself (Alex et al., 2017). As another example, Esteva et al. (2017) demonstrated that a single convolutional neural network could classify skin cancer with high predictive performance, on par with the performance of a dermatologist. Numerous other studies have been conducted on various intelligent medical imaging fields, from diabetic retinopathy (Ting et al., 2017) up to lung cancer (Farahani et al., 2018) and Alzheimer's disease (Tang et al., 2019), all of which demonstrate good predictive performance.
However, in practice, data artifacts might compromise the high performance of ML/DL models, making it difficult to find a suitable problem representation (Leek et al., 2010). Ideally, though, these algorithms could be leveraged for both prediction and explanation, where the latter may drive human discovery of improved ways to solve problems (Silver et al., 2016; Hölldobler et al., 2017). Thus, strategies for comprehending and explaining what the model has learned are crucial to deliver a robust validation scheme (Došilović et al., 2018; Lipton, 2018; Montavon et al., 2018), particularly in medicine (Caruana et al., 2015) and neuroscience (Sturm et al., 2016), which must be modeled based on correct features. For example, brain tumor resection requires an interpretation in a feature space that humans can readily understand, such as image or text, to leverage that information in an actionable capacity (Mirchi et al., 2020; Pfeifer et al., 2021).
Conventionally, to compare a new ML/DL technique to the existing gold standard in medicine (i.e., the human in most applications), the sensitivity, specificity, and predictive values are first calculated for each modality. Then, the confusion matrices could be constructed for both the new technique and the clinician (e.g., radiologist) and ultimately compared with each other. However, a significant weakness of this comparison is that it ignores the similarity between the support features of the ML/DL model (e.g., voxels, pixels, edges, etc.) and the features examined by the radiologist (e.g., hand-drawn or eye-tracking features). Accordingly, it is impossible to determine whether the model has learned from the embedded signals or from the artifacts or didactic noise that covary with the target (Goodfellow I. J. et al., 2014; Montavon et al., 2017; Douglas and Farahani, 2020). In other words, the presence of adversarial noise, which could be simply introduced due to instrumentation, prevents achieving a robust explanation of model decisions.
It is widely accepted that the different architectures of DL methods, e.g., recurrent neural network (RNN), long short term memory (LSTM), deep belief network (DBN), convolutional neural network (CNN), and generative adversarial network (GAN), which are well-known for their high predictive performance, are effectively considered to be black boxes, with internal inference engines that users cannot interpret (Guidotti et al., 2018b). Therefore, the limited transparency and explainability in such non-linear methods has prevented their adoption throughout the sciences; as a result, simpler models with higher interpretability (e.g., shallow decision trees, linear regression, or non-negative matrix factorization) remain more popular than complex models in many applications, including bioinformatics and neuroscience though these choices often reduce predictivity (Ma et al., 2007; Devarajan, 2008; Allen et al., 2012; Haufe et al., 2014; Bologna and Hayashi, 2017). Traditionally, some researchers believe that there is a trade-off between prediction performance and explainability for commonly used ML/DL models (Gunning and Aha, 2019). In this respect, decision trees presumably exhibit the highest explainability but are the least likely to deliver accurate results, whereas DL methods represent the best predictive performance and the worst model explainability. However, it is essential to underline that this notion has no proven linear relationship, and it can be bent for specific models/methods and sophisticated setups (Yeom et al., 2021), increasing both prediction performance and explainability.
Recently, this notion has been strongly challenged by novel explainable AI studies, in which well-designed interpretation techniques have shed light on many deep non-linear machine learning models (Simonyan et al., 2013; Zeiler and Fergus, 2014; Bach et al., 2015; Nguyen et al., 2016; Ribeiro et al., 2016; Selvaraju et al., 2017; Montavon et al., 2018; Hall and Gill, 2019; Holzinger et al., 2019). Remarkably, in healthcare and medicine, there is a growing demand for building AI approaches that must perform well and guarantee transparency and interpretability to medical experts (Douglas et al., 2011; Holzinger et al., 2019). Additionally, researchers suggest keeping humans in the loop—considering expert knowledge in interpreting the ML/DL results—leads to user trust and identifying points of model failure (Holzinger, 2016; Magister et al., 2021). In recognition of the importance of transparency in models defined for the medical imaging data, dedicated datasets and XAI exploration environments were recently proposed (Holzinger et al., 2021). Due to the nascent nature of the neuroimaging filed and its extensive use in deep learning studies, techniques such as magnetic resonance imaging (MRI), functional MRI (fMRI), computerized tomography (CT), and ultrasound, have considerably piqued the interest of XAI researchers (Zhu et al., 2019; van der Velden et al., 2022).
The present work provides a systematic review of recent neuroimaging studies that have introduced, discussed, or applied the post-hoc explainable AI methods. The post-hoc methods take a fitted and trained model and extract information about the relationships between the model input and model decision (with no effect on model performance). In contrast, model-based approaches alter the model to allow for mechanistic (functional) or archetypal explanations. In this work, we focused on post-hoc methods of their importance for practitioners and researchers who deal with deep neural networks and neuroimaging techniques. Hence, standard data analysis methods can be utilized to evaluate the extracted information and provide tangible outcomes to the end users. The remaining sections are organized as follows. Section Background: Approaches for interpreting DNNs summarizes the existing techniques for interpreting deep neural networks, categorized into saliency (e.g., gradients, signal, and decomposition) and perturbation methods. Section Methodology discusses our search strategy for identifying relevant publications and their inclusion criterion and validity risk assessment. Section Results provides the results of a literature search, study characteristics, reliability analysis of XAI methods, and quality assessment of the included studies. Finally, section Discussion discusses the significant limitations of XAI-based techniques in medical domains and highlights several challenging issues and future perspectives in this emerging field of research.
A variety of explainable AI (XAI) techniques have been developed in recent years that have taken various approaches. For example, some XAI methods are model agnostic, and some take a local as opposed to a global approach. Some have rendered heatmaps based on “digital staining” or combining weights from feature maps in the last hidden layer (Cruz-Roa et al., 2013; Xu et al., 2017; Hägele et al., 2020). Here we suggest that these methods should be distinguished based on the goal of the explanation: functional, archetypal, or post-hoc (relevance) approximation (Figure 1A).
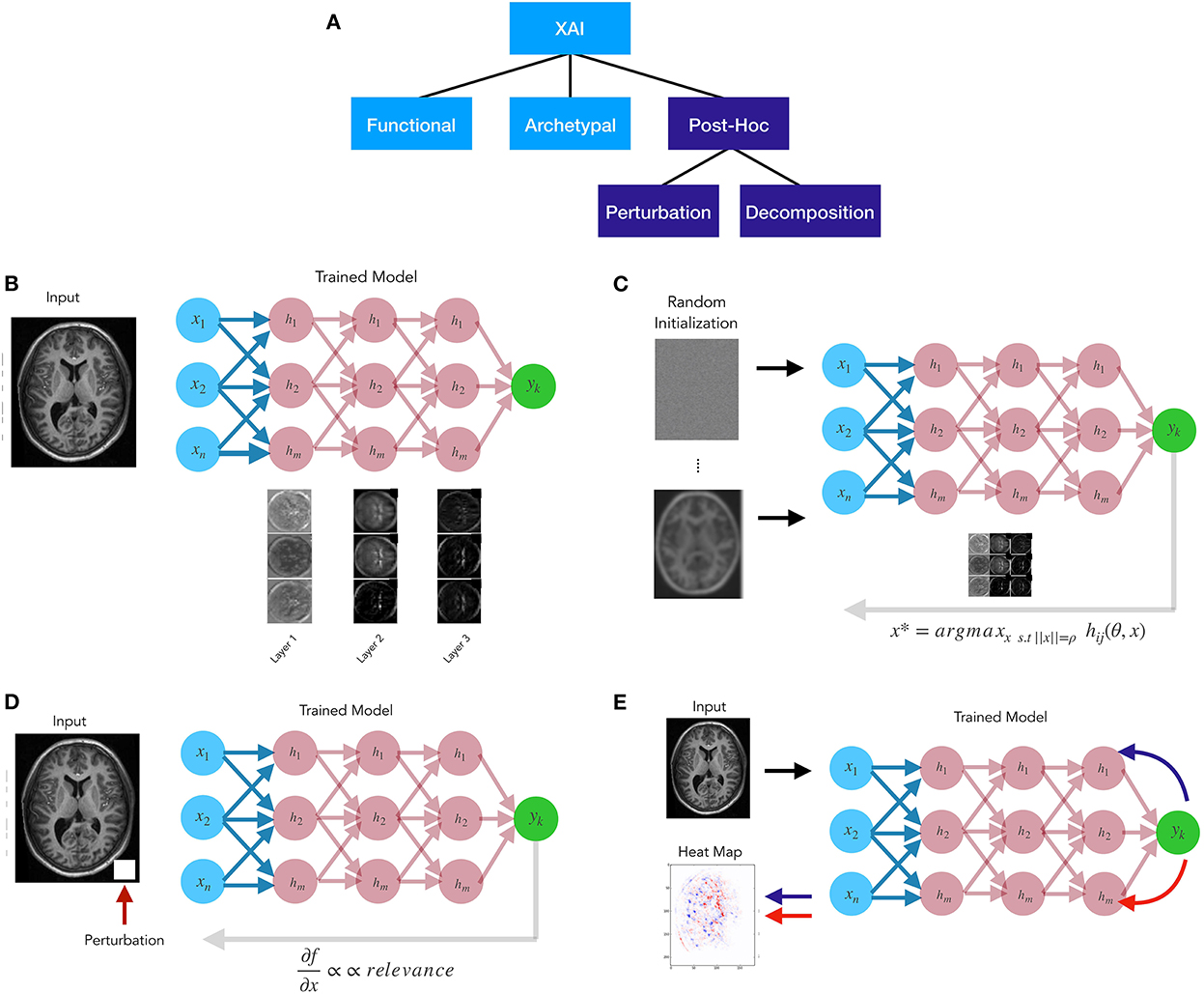
Figure 1. (A) Explainable AI methods taxonomy. (B) Functional approaches attempt to disclose the algorithm's mechanistic aspects. (C) Archetypal approaches, like generative methods, seek to uncover input patterns that yield the best model response. (D) Post-hoc perturbation relevance approaches generally change the inputs or the model's components and then attributing relevance proportionally to the amount of the change in model output. (E) Post-hoc decomposition relevance approaches are propagation-based techniques explaining an algorithm's decisions by redistributing the function value (i.e., the neural network's output) to the input variables, often in a layer-by-layer fashion.
Functional approaches (Figure 1B) examine the learned representations in the graph to reveal mechanistic aspects of the algorithm (Khaligh-Razavi and Kriegeskorte, 2014; Kriegeskorte and Douglas, 2018). One of the goals of these approaches is to shed light onto how the feature maps or filters learned through the layers help the model achieve its goal, or support its global decision structure. Archetypal methods (Figure 1C) attempt to find an input pattern x that is a prototypical exemplar of y. A simple example of this is activation maximization, whereby an initial input is randomized and the algorithm searches for input patterns that produce maximal response from the model (Erhan et al., 2009). A variety of generative methods have been developed for archetypal purposes, such as generative adversarial network models (Goodfellow I. et al., 2014). Post-hoc (or relevance) methods (Figures 1D,E) attempt to determine which aspects of input x make it likely to take on a group membership or provide supporting evidence for a particular class. In general relevance methods fall into three classes: feature ranking, perturbation methods, and decomposition methods. Feature ranking as well as feature selection methods have existed for many years (e.g., Guyon and Elisseeff, 2003), though their importance or lack thereof in high dimensional medical imaging data sets has been debated (Chu et al., 2012; Kerr et al., 2014). Perturbation relevance methods (Figure 1D) provide a local estimate of the importance of an image region or feature. This class of techniques involve altering some aspect of the inputs or the model, and subsequently assigning relevance as proportional to the magnitude of the alteration in the model output. An initial example of this is the classic pixel flip (Bach et al., 2015; Samek et al., 2017a), whereby small local regions of the image are altered, and the ensuing changes to the output are mapped back in the form of a relevance score to those altered pixels. Alternatively, perturbation methods may alter the weights, and see how this effects the output f(x). Perturbations methods can be model agnostic or not, depending upon their implementation. Lastly, decomposition or redistribution methods (Figure 1E) for relevance assignment attempt to determine the share of relevance through the model layers by examining the model structure, and are thus, typically model dependent. These attribute methods involve message passing, and propose relevance backward through the model, viewing prediction as the output of a computational graph.
The earliest work in XAI can be traced to 1960–1980, when some expert systems were equipped with rules that could interpret their results (McCarthy, 1960; Shortliffe and Buchanan, 1975; Scott et al., 1977). Although the logical inference of such systems was easily readable by humans, many of these systems were never used in practice for poor predictive performance, lack of generalizability, and the high cost of their knowledge base maintenance (Holzinger et al., 2019). The emergence of ML techniques, especially those based on deep neural networks, has overcome many traditional limitations, although their interpretability to users remains their primary challenge (Lake et al., 2017).
Accordingly, in recent years, AI researchers have afforded considerable focus to peering inside the black-box of DNNs and enhancing the system's transparency (Baehrens et al., 2009; Anderson et al., 2012; Haufe et al., 2014; Simonyan and Zisserman, 2014; Springenberg et al., 2014; Zeiler and Fergus, 2014; Bach et al., 2015; Yosinski et al., 2015; Nguyen et al., 2016; Kindermans et al., 2017; Montavon et al., 2017; Smilkov et al., 2017; Sundararajan et al., 2017; Zintgraf et al., 2017). In the following, we review the latest methods used to interpret deep learning models, including perturbation and decomposition (or redistribution) approaches. While unified in purpose, i.e., revealing the relationship between inputs and outputs (or higher levels) of the underlying model, these methods are highly divergent in outcome and explanation mechanism. The post-hoc XAI methods we found in our review are listed in Table 1 and discussed in the following subsections.
The perturbation-based approach is broadly divided into model-specific (e.g., gradients and signal) or model-agnostic methods. Gradients/sensitivity-based methods examine how a slight shift to the input affects the classification score for the output of interest, such as the techniques introduced by Baehrens et al. (2009) and Simonyan et al. (2013), as well as Class Activation Mapping (CAM; Zhou et al., 2016), Gradient-weighted CAM (Grad-CAM; Selvaraju et al., 2017), SmoothGrad (SG; Smilkov et al., 2017), and (Multiscale CAM; Hu et al., 2020). These techniques are easily implemented in DNNs because the gradient is generally computed by backpropagation (Rumelhart et al., 1986; Swartout et al., 1991). Signal methods typically visualize input patterns by stimulating neuron activation in higher layers, resulting in so-called feature maps. DeConvNet (Zeiler and Fergus, 2014), Guided BackProp (Springenberg et al., 2014), PatternNet (Kindermans et al., 2017), and inversion-based techniques (Mahendran and Vedaldi, 2015; Dosovitskiy and Brox, 2016) are some examples of this group. Mahendran and Vedaldi (2015) showed that by moving from the shallower layers to the deeper layers, the feature maps reveal more complex patterns of input [e.g., in human face explanation: from (1) line and edges to (2) eyes, nose, and ears, then to (3) complex facial structures].
On the other hand, model agnostic methods explore the prediction of interest to infer the relevance of the input features toward the output (Ribeiro et al., 2016; Alvarez-Melis and Jaakkola, 2018). Two of the most popular techniques in this category are Local Interpretable Model-agnostic Explanations (LIME; Ribeiro et al., 2016) and SHapley Additive exPlanations (SHAP; Lundberg and Lee, 2017).
Decomposition-based methods seek to identify important features (pixels) in a particular input by decomposing the network classification decision into contributions of the input elements. The earliest study in this class goes back to Bach et al. (2015), who introduced the Layer-Wise Relevance Propagation (LRP) technique, which interprets the DNN decisions using heatmaps (or relevance-maps). Using a set of propagation rules, LRP performs a separate backward pass for each possible target class, satisfying a layer-wise conservation principle (Landecker et al., 2013; Bach et al., 2015). As a result, each intermediate layer up to the input layer is assigned relevance scores. The sum of the scores in each layer equals the prediction output for the class under consideration. The conservation principle is one of the significant differences between the decomposition and gradients methods.
In another technique, Montavon et al. (2017) demonstrated how the propagation rules derived from deep Taylor decomposition relate to those heuristically defined by Bach et al. (2015). Recently, several studies have used LRP to interpret and visualize their network decisions in various applications such as text analysis (Arras et al., 2017a), speech recognition (Becker et al., 2018), action recognition (Srinivasan et al., 2017), and neuroimaging (Thomas et al., 2019). Other recently-developed decomposition-based methods include DeepLIFT (Shrikumar et al., 2017), Integrated Gradients (Sundararajan et al., 2017), Gradient Input (Shrikumar et al., 2017), and Prediction Difference Analysis (PDA; Zintgraf et al., 2017). In recent years, various studies have attempted to test the reliability of explanation techniques compared to each other by introducing several properties such as fidelity (or sensitivity), consistency, stability and completeness (Bach et al., 2015; Kindermans et al., 2017; Sundararajan et al., 2017; Alvarez-Melis and Jaakkola, 2018). In the following chapters, we address this issue.
This systematic review was conducted according to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement guidelines (Moher et al., 2009). To reduce the effect of research expectations on the review, we first identified research questions and search strategies. Moreover, this systematic review adhered to the Cochrane Collaboration methodology (Higgins et al., 2011), to mitigate the risk of bias and error.
Based on the objectives outlined in the abstract, the following research questions were derived and form the cornerstone of our study:
• What are the main challenges in AI that have limited their implementation in medical imaging applications, particularly in neuroimaging, despite their high prediction performance?
• How can we overcome the black-box property of complex and deep neural networks for the user in critical areas such as healthcare and medicine?
• How have recent advances in explainable AI affected machine/deep learning in medical imaging and neuroimaging?
• How can one assess the reliability and generalizability of interpretation techniques?
The current and seminal studies in the realm of XAI with a focus on healthcare and medicine were considered critical sources for this systematic review. A bibliographic search for this work was carried out across the following scientific databases and search engines: PubMed, Scopus, Web of Science, Google Scholar, ScienceDirect, IEEE Xplore, SpringerLink, and arXiv, using the following keyword combinations in the title, keywords, or abstract: (“explainable AI” or “XAI” or “explainability” or “interpretability”) and (“artificial intelligence” or “machine learning” or “deep learning” or “deep neural networks”) and (“medical imaging” or “neuroimaging” or “MRI” or “fMRI” or “CT”). Moreover, the reference lists of the retrieved studies were also screened to find relevant published works.
Published original articles with the following features were included in the current study: (a) be written in English; AND [(b) introduce, identify, or describe XAI-based techniques for visualizing and/or interpreting ML/DL decisions; OR (c) be related to the application of XAI in healthcare and medicine]. Other exclusion criteria were: (a) book chapters; (b) papers that upon review were not related to the research questions; (c) opinions, viewpoints, anecdotes, letters, and editorials. The eligibility criteria were independently assessed by two authors (FF and KF), who screened the titles and abstracts to establish the relevant articles based on the selection criteria. Any discrepancies were resolved through discussion or referral to a third reviewer (BL or WK).
We developed a data extraction sheet, pilot-tested this sheet on randomly selected studies, and refined the sheet appropriately. During a full-text review process, one review author (KF) extracted the following data from the selected studies, and a second review author (FF) crosschecked the collected data, which included: taxonomic topic, first author (year of publication), key contributions, XAI model used, and sample size (if applicable). Disagreements were resolved by discussion between the two review authors, and if necessary, a third reviewer was invoked (BL or WK).
We performed a co-occurrence analysis to analyze text relationships between the shared components of the reviewed studies, including XAI methods, imaging modalities, diseases, and frequently used ML/DL terms. Creating a co-occurrence network entails finding keywords in the text, computing the frequency of co-occurrences, and analyzing the networks to identify word clusters and locate central terms (Segev, 2020). Furthermore, to provide a critical view of the extracted XAI techniques, we carried out an additional subjective examination of articles that have proposed quality tests for evaluating the reliability of these methods.
The risk of bias in individual studies was ascertained independently by two reviewers (FF and KF) following the Cochrane Collaboration's tool (Higgins et al., 2011). The Cochrane Collaboration's tool assesses random sequence generation, allocation concealment, blinding of participants, blinding of outcome assessment, incomplete outcome data, and selective outcome reporting, and ultimately rates the overall quality of the studies as weak, fair, or good. To appraise the quality of evidence across studies, we examined for lack of completeness (publication bias) and missing data from the included studies (selective reporting within studies). The risk of missing studies is highly dependent on the chosen keywords and the limitations of the search engines. A set of highly-cited articles was used to create the keyword search list in an iterative process to alleviate this risk. Disagreements were resolved by discussion between the study authors.
Following the PRISMA guidelines (Moher et al., 2009), a summary of the process used to identify, screen, and select studies for inclusion in this review is illustrated in Figure 2. First, 357 papers were identified through the initial search, followed by the removal of duplicate articles, which resulted in 263 unique articles. Only ~5% were published before 2010, indicating the novelty of the terminology and the research area. Afterwards, the more relevant studies were identified from the remaining papers by incorporating inclusion and exclusion criteria. The inclusion criteria at this step required the research to (a) be written in English; AND [(b) introduce, identify, or describe post-hoc XAI techniques for visualizing and/or interpreting ML/DL decisions; OR (c) be related to the application of post-hoc XAI in neuroimaging]. Other exclusion criteria included: (a) book chapters; (b) papers which upon review were not related to the research questions; (c) opinions, viewpoints, anecdotes, letters, and editorials. As a result, the implementation of these criteria yielded 126 eligible studies (~48% of the original articles). Subsequently, the full text of these 126 papers was scrutinized in detail to reaffirm the criteria described in the previous step. Eventually, 78 publications remained for systematic review.
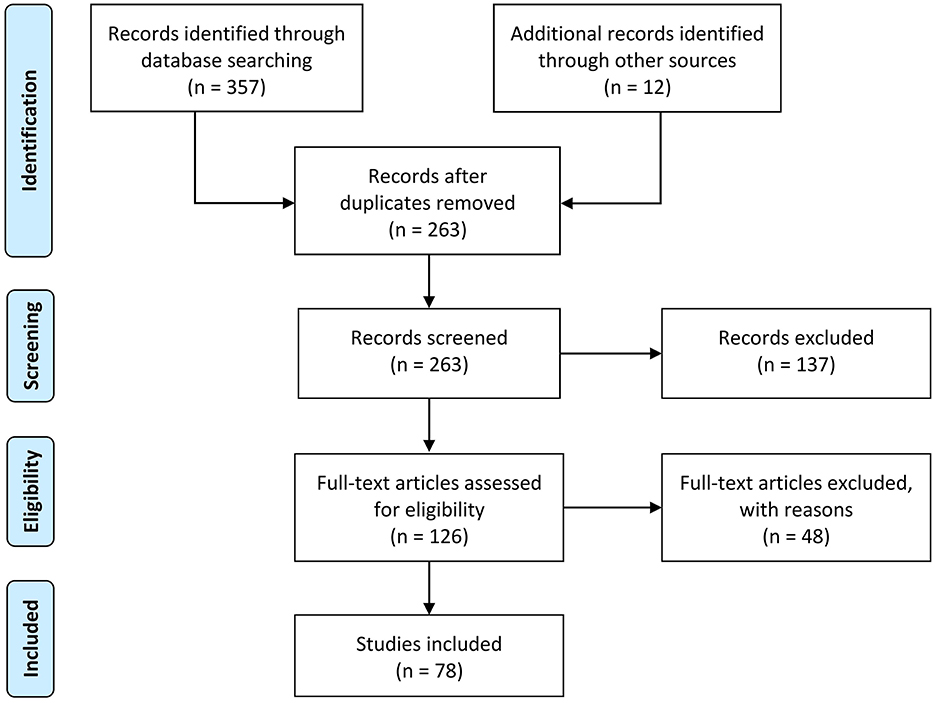
Figure 2. The flow diagram of the methodology and selection processes used in this systematic review follows the PRISMA statement (Moher et al., 2009).
The included studies—published from 2005 to 2021—were binned into three major taxonomies (Figure 3A). The first category was focused on the researchers' efforts to introduce post-hoc XAI techniques and theoretical concepts for the visualization and interpretation of deep neural network predictions (blue slice) and accounted for 37% of the selected articles. In the second category, articles discussing the neuroimaging applications of XAI were collected and reviewed (green slice), accounting for 42% of the selected papers. Finally, the last group consisted of perspective and review studies in the field (yellow slice), either methodologically or medically, which accounted for 21% of the selected articles. Figure 3B, in particular, illustrates the classification of XAI applications in neuroimaging in terms of the post-hoc method they used (along with their percentage). As mentioned in the literature, these methods can be divided into decomposition-based and perturbation-based approaches; the latter can be classified into gradients, signal and model agnostic ones.
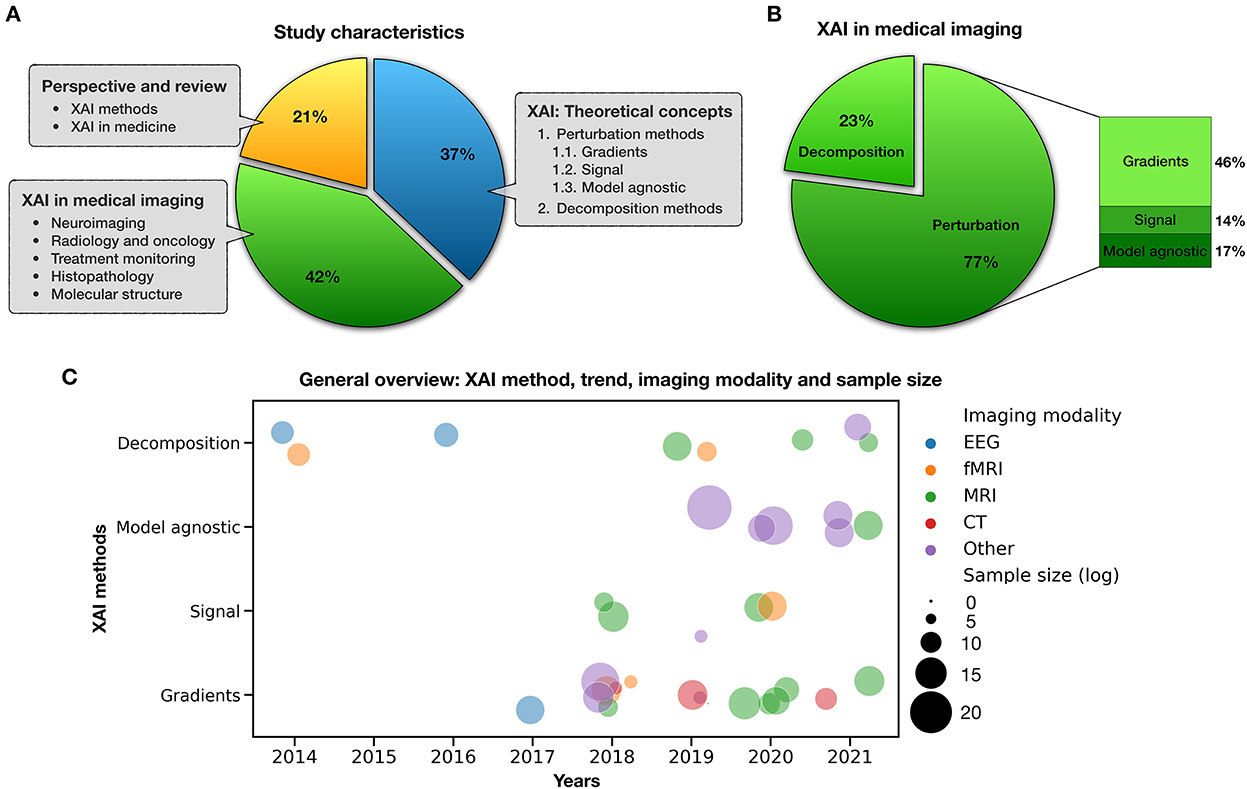
Figure 3. Study characteristics. (A) Categorization of included studies, (B) XAI in medical imaging, and (C) a bubble plot that shows mentioned studies by type of XAI method, imaging modality, sample size, and publication trend in recent years.
Moreover, Figure 3C distinguishes applied XAI studies in neuroimaging from various aspects such as method, imaging modality, sample size, and their publication trend in recent years. In this plot, each circle represents a study whose color determines the type of imaging modality such as EEG, fMRI, MRI, CT, and other (PET, ultrasound, histopathological scans, blood film, electron cryotomography, Aβ plaques, tissue microarrays, etc.), and its size is logarithmically related to the number of people/scans/images used in that study. By focusing on each feature, compelling general information can be extracted from this figure. For example, it can be noted that gradient-based methods cover most imaging modalities well, or those model agnostic methods are suitable for studies with large sample sizes.
A summary of the reviewed articles is provided in Table 2, which contains the author's name, publication year, XAI model examined, and key contributions, respectively, ordered by taxonomy and article date. The table is constructed to provide the reader with a complete picture of the framework and nature of components contributing to XAI in medical applications.
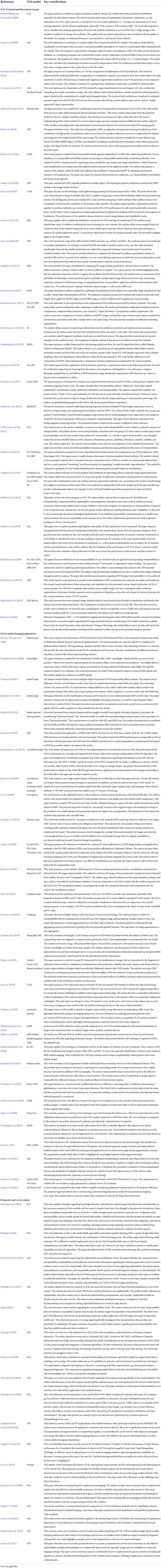
Table 2. Summary of included articles by taxonomy, authors (year), XAI model, and key contributions.
Counting of matched data within a collection unit is what co-occurrence analysis is all about. Figure 4 visualizes the co-occurrences of the key vocabularies of XAI/AI concepts, XAI methodologies, imaging modalities, and diseases from our reviewed papers. In this figure, word clusters are represented by different colors. Also, the bubble size denotes the number of publications, while the connection width reflects the frequency of co-occurrence. We only considered the abstracts in our co-occurrence analysis due to the large number of words within the full texts and the curse of overlapping labels.
XAI methods are likely be unreliable against some factors that do not affect the model outcome. The output of the XAI methods, for instance, could be significantly altered by a slight transformation in the input data, even though the model remains robust to these changes (Kindermans et al., 2019). Accordingly, we conducted a subjective examination on articles that have introduced properties such as completeness, implementation invariance, input invariance, and sensitivity (Bach et al., 2015; Kindermans et al., 2017; Sundararajan et al., 2017; Alvarez-Melis and Jaakkola, 2018) to evaluate the reliability of these methods (Table 3).
The Cochrane Collaboration's tool (Higgins et al., 2011) was used to assess the risk of bias in each trial (Figure 5). The articles were categorized as: (a) low risk of bias, (b) high risk of bias, or (c) unclear risk of bias for each domain. We judged most domains to be unclear or not reported using the Cochrane Collaboration. Finally, the overall quality of the studies was classified into weak, fair, or good, if < 3, 3, or ≥4 domains were rated as low risk, respectively. Among the 78 studies included in the systematic review, 22 were categorized as good quality, 50 were fair quality, and 6 were low quality.
The current study provides an overview of applications of post-hoc XAI techniques in neuroimaging analysis. We focused on post-hoc approaches since interpreting weight vectors has historically been the standard practice when applying encoding and decoding models to functional imaging and neuroimaging data. However, it is generally challenging to interpret decoding and encoding models (Kriegeskorte and Douglas, 2019). In light of this, post-hoc procedures provide a novel strategy that would make it possible to use these techniques for predictions and gain scientific and/or neuroscientific knowledge during the interpretation step.
For many years, ML and DL algorithms have established a strong presence in various medical imaging research with examples of performance at least equaling that of radiologists (Khan et al., 2001; Hosny et al., 2018; Li et al., 2018; Lee et al., 2019; Lundervold and Lundervold, 2019; Tang et al., 2019). In contrast to linear models, many practitioners regard DNNs as a “black box,” and this lack of transparency has hindered the adoption of deep learning methods in certain domains where explanations are crucial (Guidotti et al., 2018b). Transparency builds trust, subtends the evaluation of fairness, and helps identify points of model failure (Kindermans et al., 2017; Rajkomar et al., 2018; Vayena et al., 2018; Wilson et al., 2019). In many cases, trustworthy models may be essential to verify that the model is not exploiting artifacts in the data, or operating on spurious attributes that covary with meaningful support features (Leek et al., 2010; Lapuschkin et al., 2016; Montavon et al., 2018).
The need for interpreting the black-box decisions of DNNs (Holzinger, 2014; Biran and Cotton, 2017; Doshi-Velez and Kim, 2017; Lake et al., 2017; Lipton, 2018) was answered by leveraging a variety of post-hoc explanation techniques in recent years. These models can assign relevance to inputs for the predictions carried out by trained deep learning models either for each instance separately (Robnik-Šikonja and Kononenko, 2008; Zeiler and Fergus, 2014; Ribeiro et al., 2016; Sundararajan et al., 2017) or on the class or model level (Datta et al., 2016; Guidotti et al., 2018a; Staniak and Biecek, 2018; Ghorbani et al., 2019b). Because of the successful applications of CNNs in image analysis, particularly in the medical domain, several XAI methods were proposed solely for explaining predictions of 2D (Springenberg et al., 2014; Bach et al., 2015; Smilkov et al., 2017) and 3D images (Yang et al., 2018; Zhao et al., 2018; Thomas et al., 2019).
This systematic review also reveals the need to involve medical personnel in developing ML, DL, and XAI for the medical domains. Without feedback from clinicians' active participation, it will be unlikely to create ML models dedicated solely to the medical fields (Ustun and Rudin, 2016; Lamy et al., 2019). Familiarizing AI researchers with the original needs and point-of-view of specialists from the medical domain and its subdomains (Tonekaboni et al., 2019) would be beneficial because it would allow focusing on the detailed shortcomings of the state-of-the-art XAI methods, followed by their significant improvement.
From the perspective of health stakeholders (e.g., patients, physicians, pharmaceutical firms and government), interpretability is an integral part of choosing the optimal model. As shown in Figure 6, interpretability could also be used to ensure other significant desiderata of medical intelligent systems such as transparency, causality, privacy, fairness, trust, usability, and reliability (Doshi-Velez and Kim, 2017). In this sense, transparency indicates how a model reached a given result; causality examines the relationships between model variables; privacy assesses the possibility of original training data leaking out of the system; fairness shows whether there is bias aversion in a learning model; trust indicates how assured a model is in the face of trouble; usability is an indicator of how efficient the interaction between the user and the system is; and reliability is about the stability of the outcomes under similar settings (Doshi-Velez and Kim, 2017; Miller, 2019; Barredo Arrieta et al., 2020; Jiménez-Luna et al., 2020; Fiok et al., 2022).
Even though AI methods have successfully been utilized in medical research and neuroimaging studies, these methods still have not advanced into everyday real-life applications. Researchers name several reasons for this fact: (1) Lack of interpretability, transparency, trust, and clear causality of the black-box AI models continues to be a vital issue (Holzinger et al., 2017; Došilović et al., 2018; Hoffman et al., 2018) despite the research already carried out on XAI. (2) Speeding up model convergence while maintaining predictive performance is important in scenarios where data is naturally homogeneous or spatially normalized (e.g., fMRI, MRI sequences, PET, CT). This is crucial for neuroimaging research since the data are relatively homogeneous, unlike natural images, because of their uniform structure and spatial normalization (Eitel et al., 2021). (3) Despite improvements in medical data availability for AI training (Hosny et al., 2018; Lundervold and Lundervold, 2019), an insufficient amount/quality of data for training ML and DL solutions remains a significant limitation, with the result that many studies are carried out on small sample sizes of subjects (13 in Blankertz et al., 2011; 10 in Sturm et al., 2016; 10 in Tonekaboni et al., 2019). (4) It is believed that trained AI models that achieve super-human performance on data from a distribution (e.g., a specific hospital) cannot adapt appropriately to unseen data drawn from other medical units since it comes from a different distribution (Yasaka and Abe, 2018). (5) Compliance with legislation that calls for the “right for explanation” is also considered (Holzinger et al., 2017; Samek et al., 2017b) to be a limiting factor regarding the use of ML and DL without the ability to provide explanations for each use case.
Understanding the need for XAI in the DNNs community seems to spread rapidly or can be already considered widespread. However, the importance of XAI, particularly for the medical domain, is still underestimated. When human health and life is at stake, it is insufficient to decide based solely on a “black box” prediction even when obtained from a superhuman model. It is far not enough to classify; instead, interpretation is the key to achieving if XAI manages to deliver a complete and exhaustive description of voxels that constitute a part of a tumor. The potential for XAI in medicine is exceptional as it can answer why we should believe that the diagnosis is correct.
In recent years, various computational techniques have been proposed (Table 3) to objectively evaluate explainers based on accuracy, fidelity, consistency, stability and completeness (Robnik-Šikonja and Kononenko, 2008; Sundararajan et al., 2017; Molnar, 2020; Mohseni et al., 2021). Accuracy and fidelity (sensitivity or correctness in the literature) are closely related; the former refers to how well an explainer predicts unseen data, and the latter indicates how well an explainer detect relevant components of the input that the black box model operates upon (notably, in case of high model accuracy and high explainer fidelity, the explainer also has high accuracy). Consistency refers to the explainer's ability to capture the common components under different trained models on the same task with similar predictions. However, high consistency is not desirable when models' architectures are not functionally equivalent, but their decisions are the same (due to the “Rashomon effect”). While consistency compares explanations between models, stability compares explanations under various transformations or adversaries to a fixed model's input. Stability examines how slight variations in the input affect the explanation (assuming the model predictions are the same for both the original and transformed inputs). Eventually, completeness reveals a complete picture of features essential for decisions, so how well humans understand the explanations. It looks like the elephant in the room, that somewhat abstruse to measure, but very important to get right in future research. Particularly in medicine, we need a holistic picture of the disease, such as a complete and exhaustive description of voxels that are part of a tumor. What if altered medial temporal lobe shape covaries with a brain tumor (because the tumor moves it somehow)? Should we then resect the temporal lobe? Thus, further research is needed on this property.
In post-hoc explanation, fidelity has been studied more than accuracy (in fact, high accuracy is solely important when an explanation is used for predictions). In this respect, Bach et al. (2015) and Samek et al. (2017a) suggested a framework to evaluate saliency explanation techniques by pixel-flipping in an image input repeatedly (based on their relevance importance), then quantifying the effect of this perturbation on the classifier prediction. Their framework inspired many other studies (Ancona et al., 2017; Lundberg and Lee, 2017; Sundararajan et al., 2017; Chen et al., 2018; Morcos et al., 2018). The common denominator of fidelity metrics is that the greater the change in prediction performance, the more accurate the relevance. However, this approach may lead to unreliable predictions when the model receives out-of-distribution input images (Osman et al., 2020). To solve this problem, Osman et al. (2020) developed a synthetic dataset with explanation ground truth masks and two relevance accuracy measures for evaluating explanations. Their approach provides an unbiased and transparent comparison of XAI techniques, and it uses data with a similar distribution to those during model training.
Another possible way for appraising explanations is to leverage the saliency maps for object detection, e.g., by setting a threshold on the relevance and then calculating the Jaccard index (also known as Intersection over Union) concerning bounding box annotations as a measure of relevance accuracy (Simonyan et al., 2013; Zhang et al., 2018). However, since the classifier's decision is based solely on the object and not the background (contradictory to the real world) in this approach, the evaluation could be misleading. In many other cases, comparing a new explainer with those state-of-the-art techniques is utilized to measure explanation quality (Lundberg and Lee, 2017; Ross et al., 2017; Shrikumar et al., 2017; Chu et al., 2018).
On the other hand, Kindermans et al. (2019) proposed an input invariance property. They revealed that explainers might have instabilities in their results after slight image transformations, and consequently, their saliency maps could be misleading and unreliable. They assessed the quality of interpretation methods such as Gradients, GI, SG, DeConvNets, GBP, Taylor decomposition, and IG. Only Taylor decomposition and IG passed this property, subject to the choice of reference and type of transformation. In another study, Sundararajan et al. (2017) introduced two measures for evaluating the reliability of XAI methods, one called sensitivity (or fidelity) and the other as implementation invariance (i.e., a requirement that models with different architectures that achieve the same results should also provide the same explanations). In their paper, the sensitivity test was failed by Gradients, DeConvNets, and GBP, while DeepLift, LRP, and IG passed the test; in contrast, the implementation invariance was failed by DeepLift and LRP, while IG passed. To extend a similar idea, Adebayo et al. (2018) proposed another evaluation approach (to test Gradients, GI, IG, GBP, Guided Grad-CAM, and SG methods) by applying randomizations tests on the model parameters and input data, to confirm that the explanation relies on both these factors. Here, Gradients and Grad-CAM methods succeeded in the former and GI and IG in the latter. While these assessments can serve as a first sanity check for explanations, they cannot directly evaluate the explanation's adequacy.
One more known approach for evaluating visual explanations is to expose the input data to adversaries, unintentional or malicious purposes, which are generally unrecognizable to the human eyes (Paschali et al., 2018; Douglas and Farahani, 2020). For example, Douglas and Farahani (2020) developed a structural similarity analysis and compared the reliability of explanation techniques by adding small amounts of Rician noise to the structural MRI data (in the real world, this kind of adversary can be caused by the physical and temporal variability across instrumentation). In this study, while not significantly changing CNN's prediction performance for both the original and attacked images, the obtained relevance heatmaps showed the superiority of decomposition-based algorithms such as LRP over the others. In another study, Alvarez-Melis and Jaakkola (2018) proposed a Lipschitz estimate to evaluate explainers' stability by adding Gaussian noise to the input data. The authors showed SHAP was more stable than LIME when a random forest was considered. They also assessed explanations provided by LIME, IG, GI, Occlusion sensitivity, Saliency Maps, and LRP over CNNs. They reported acceptable results by all methods (IG was the most stable) but LIME. Finally, Ghorbani et al. (2019a) proposed to measure fragility, i.e., given an adversarial input image (perturbed original), the degree of behavioral change of the XAI method. In their work, Gradients and DeepLIFT were found to be more fragile than IG.
While there is an ongoing discussion regarding the virtues that XAI should exhibit, so far, no consensus has been reached, even regarding fundamental notions such as interpretability (Lipton, 2018). Terms such as completeness, trust, causality, explainability, robustness, fairness, and many others are actively brought up and discussed by different authors (Biran and Cotton, 2017; Doshi-Velez and Kim, 2017; Lake et al., 2017), as researchers now struggle to achieve common definitions of most important XAI nomenclature (Doshi-Velez and Kim, 2017). Given the research community's activity in this field, it is very likely that additional requirements and test proposals will be formulated shortly. Moreover, new XAI methods will undoubtedly emerge. We also note that not a single XAI method passed all proposed tests, and not all tests were conducted with all available algorithms. The abovementioned reasons force us to infer that the currently available XAI methods (Holzinger et al., 2022a,b), exhibit significant potential, although they remain immature. Therefore, we agree with Lipton (2018), which clearly warns about blindly trusting XAI interpretations because they can potentially be misleading.
AI has already inevitably changed medical research perspectives, but without explaining the rationale for undertaking decisions, it could not provide a high level of trust required in medical applications. With current developments of XAI techniques, this is about to change. Research on fighting cardiovascular disease (Weng et al., 2017), hypoxemia during surgery (Lundberg and Lee, 2017), Alzheimer's disease (Tang et al., 2019), breast cancer (Lamy et al., 2019), acute intracranial hemorrhage (Lee et al., 2019) and coronavirus disease (Wang et al., 2019), can serve as examples of developing successful AI+XAI systems that managed to adequately explain their decisions and pave the way to many other medical applications, notably neuroimaging studies. However, the XAI in this research field is still immature and young. If we expect to overcome XAI's current imperfections, great effort is still needed to foster XAI research. Finally, medical AI and XAI's needs cannot be achieved without keeping medical practitioners in the loop.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
FF and KF conducted the literature search and prepared the initial draft of the paper. FF, KF, BL, and PD were involved in study conception and contributed to intellectual content. WK and PD supervised all aspects of manuscript preparations, revisions, editing, and final intellectual content. FF, KF, and BL edited the final draft of the paper. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adebayo, J., Gilmer, J., Muelly, M., Goodfellow, I., Hardt, M., and Kim, B. (2018). “Sanity checks for saliency maps,” in Advances in Neural Information Processing Systems, Vol. 31.
Alex, V., KP, M. S., Chennamsetty, S. S., and Krishnamurthi, G. (2017). “Generative adversarial networks for brain lesion detection,” in Proc.SPIE.
Allen, J. D., Xie, Y., Chen, M., Girard, L., and Xiao, G. (2012). Comparing statistical methods for constructing large scale gene networks. PLoS ONE 7, e29348. doi: 10.1371/journal.pone.0029348
Alvarez-Melis, D., and Jaakkola, T. S. (2018). On the robustness of interpretability methods. arXiv [Preprint]. arXiv: 1806.08049.
Ancona, M., Ceolini, E., Öztireli, C., and Gross, M. (2017). Towards better understanding of gradient-based attribution methods for deep neural networks. arXiv [Preprint]. arXiv: 1711.06104.
Anderson, A., Han, D., Douglas, P. K., Bramen, J., and Cohen, M. S. (2012). Real-time functional MRI classification of brain states using Markov-SVM hybrid models: peering inside the rt-fMRI Black Box BT - machine learning and interpretation in neuroimaging,” in eds G. Langs, I. Rish, M. Grosse-Wentrup, and B. Murphy, B (Berlin, Heidelberg: Springer Berlin Heidelberg), 242–255.
Arras, L., Horn, F., Montavon, G., Müller, K.-R., and Samek, W. (2017a). “What is relevant in a text document?”: An interpretable machine learning approach. PLoS ONE 12, e0181142. doi: 10.1371/journal.pone.0181142
Arras, L., Montavon, G., Müller, K. R., and Samek, W. (2017b). Explaining recurrent neural network predictions in sentiment analysis. arXiv [Preprint]. arXiv: 1706.07206.
Bach, S., Binder, A., Montavon, G., Klauschen, F., Müller, K. R., and Samek, W. (2015). On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 10, 1–46. doi: 10.1371/journal.pone.0130140
Baehrens, D., Schroeter, T., Harmeling, S., Kawanabe, M., Hansen, K., and Müller, K. R. (2009). How to explain individual classification decisions. arXiv [Preprint]. arXiv: 0912.1128.
Barredo Arrieta, A., Díaz-Rodríguez, N., Del Ser, J., Bennetot, A., Tabik, S., Barbado, A., et al. (2020). Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 58, 82–115. doi: 10.1016/j.inffus.2019.12.012
Becker, S., Ackermann, M., Lapuschkin, S., Müller, K.-R., and Samek, W. (2018). Interpreting and explaining deep neural networks for classification of audio signals. arXiv [Preprint]. arXiv: 1807.03418.
Bengio, Y., Courville, A., and Vincent, P. (2013). Representation learning: a review and new perspectives. IEEE Trans. Pattern Anal. Mach. Intell. 35, 1798–1828. doi: 10.1109/TPAMI.2013.50
Binder, A., Bockmayr, M., Hägele, M., Wienert, S., Heim, D., Hellweg, K., et al. (2021). Morphological and molecular breast cancer profiling through explainable machine learning. Nat. Mach. Intell. 3, 355–366. doi: 10.1038/s42256-021-00303-4
Biran, O., and Cotton, C. (2017). “Explanation and justification in machine learning: A survey,” in IJCAI-17 workshop on explainable AI (XAI), Vol. 8. p. 8–13.
Blankertz, B., Lemm, S., Treder, M., Haufe, S., and Müller, K. R. (2011). Single-trial analysis and classification of ERP components - A tutorial. Neuroimage 56, 814–825. doi: 10.1016/j.neuroimage.2010.06.048
Böhle, M., Eitel, F., Weygandt, M., and Ritter, K. (2019). Layer-wise relevance propagation for explaining deep neural network decisions in MRI-Based alzheimer's disease classification. Front. Aging Neurosci. 11, 194. doi: 10.3389/fnagi.2019.00194
Bologna, G., and Hayashi, Y. (2017). Characterization of symbolic rules embedded in deep DIMLP networks: A challenge to transparency of deep learning. J. Artif. Intell. Soft Comput. Res. 7, 265–286. doi: 10.1515/jaiscr-2017-0019
Bosse, S., Wiegand, T., Samek, W., and Lapuschkin, S. (2022). From “where” to “what”: Towards human-understandable explanations through concept relevance propagation. arXiv [Preprint]. arXiv: 2206.03208.
Caruana, R., Lou, Y., Gehrke, J., Koch, P., Sturm, M., and Elhadad, N. (2015). “Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission,” in Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD '15. (New York, NY: Association for Computing Machinery), 1721–1730.
Chen, J., Song, L., Wainwright, M. J., and Jordan, M. I. (2018). “Learning to explain: An information-theoretic perspective on model interpretation,” in International Conference on Machine Learning (PMLR), 883–892.
Cheng, H.-T., Koc, L., Harmsen, J., Shaked, T., Chandra, T., Aradhye, H., et al. (2016). “Wide & deep learning for recommender systems,” in Proceedings of the 1st Workshop on Deep Learning for Recommender Systems, DLRS 2016 (New York, NY: Association for Computing Machinery), 7–10.
Chereda, H., Bleckmann, A., Menck, K., Perera-Bel, J., Stegmaier, P., Auer, F., et al. (2021). Explaining decisions of graph convolutional neural networks: patient-specific molecular subnetworks responsible for metastasis prediction in breast cancer. Genome Med. 13, 42. doi: 10.1186/s13073-021-00845-7
Chu, C., Hsu, A.-L., Chou, K.-H., Bandettini, P., and Lin, C. (2012). Does feature selection improve classification accuracy? Impact of sample size and feature selection on classification using anatomical magnetic resonance images. Neuroimage 60, 59–70. doi: 10.1016/j.neuroimage.2011.11.066
Chu, L., Hu, X., Hu, J., Wang, L., and Pei, J. (2018). “Exact and consistent interpretation for piecewise linear neural networks: a closed form solution,” in Proceedings of the 24th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, KDD '18 (New York, NY: Association for Computing Machinery), 1244–1253.
Couture, H. D., Marron, J. S., Perou, C. M., Troester, M. A., and Niethammer, M. (2018). “Multiple instance learning for heterogeneous images: training a CNN for histopathology,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, eds A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer International Publishing), 254–262.
Cruz-Roa, A. A., Arevalo Ovalle, J. E., Madabhushi, A., and González Osorio, F. A. (2013). “A deep learning architecture for image representation, visual interpretability and automated basal-cell carcinoma cancer detection BT,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013, eds K. Mori, I. Sakuma, Y. Sato, C. Barillot, and N. Navab (Berlin, Heidelberg: Springer Berlin Heidelberg), 403–410.
Datta, A., Sen, S., and Zick, Y. (2016). “Algorithmic transparency via quantitative input influence: theory and experiments with learning systems,' in Proc. - 2016 IEEE Symp. Secur. Privacy, SP 2016, 598–617.
Devarajan, K. (2008). Nonnegative matrix factorization: an analytical and interpretive tool in computational biology. PLoS Comput. Biol. 4, e1000029. doi: 10.1371/journal.pcbi.1000029
Doshi-Velez, F., and Kim, B. (2017). Towards a rigorous science of interpretable machine learning. arXiv [Preprint]. arXiv: 1702.08608.
Došilović, F. K., Brčić, M., and Hlupić, N. (2018). “Explainable artificial intelligence: a survey,” in 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), 210–215.
Dosovitskiy, A., and Brox, T. (2016). “Inverting visual representations with convolutional networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4829–4837.
Douglas, P. K., and Farahani, F. V. (2020). On the similarity of deep learning representations across didactic and adversarial examples. arXiv [Preprint]. arXiv: 2002.06816.
Douglas, P. K., Harris, S., Yuille, A., and Cohen, M. S. (2011). Performance comparison of machine learning algorithms and number of independent components used in fMRI decoding of belief vs. disbelief. Neuroimage 56, 544–553. doi: 10.1016/j.neuroimage.2010.11.002
Eitel, F., Albrecht, J. P., Weygandt, M., Paul, F., and Ritter, K. (2021). Patch individual filter layers in CNNs to harness the spatial homogeneity of neuroimaging data. Sci. Rep. 11, 24447. doi: 10.1038/s41598-021-03785-9
El-Sappagh, S., Alonso, J. M., Islam, S. M. R., Sultan, A. M., and Kwak, K. S. (2021). A multilayer multimodal detection and prediction model based on explainable artificial intelligence for Alzheimer's disease. Sci. Rep. 11, 2660. doi: 10.1038/s41598-021-82098-3
Erhan, D., Bengio, Y., Courville, A., and Vincent, P. (2009). Visualizing Higher-Layer Features of a Deep Network. Montreal, QC: University of Montreal.
Essemlali, A., St-Onge, E., Descoteaux, M., and Jodoin, P.-M. (2020). “Understanding Alzheimer disease's structural connectivity through explainable AI,” in Proceedings of the Third Conference on Medical Imaging with Deep Learning, Proceedings of Machine Learning Research, eds T. Arbel, I. Ben Ayed, M. de Bruijne, M. Descoteaux, H. Lombaert, and C. Pal (PMLR), 217–229.
Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., et al. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118. doi: 10.1038/nature21056
Farahani, F. V., Ahmadi, A., and Zarandi, M. H. F. (2018). Hybrid intelligent approach for diagnosis of the lung nodule from CT images using spatial kernelized fuzzy c-means and ensemble learning. Math. Comput. Simul. 149, 48–68. doi: 10.1016/j.matcom.2018.02.001
Fiok, K., Farahani, F. V., Karwowski, W., and Ahram, T. (2022). Explainable artificial intelligence for education and training. J. Def. Model. Simul. 19, 133–144.
Gaonkar, B., and Davatzikos, C. (2013). Analytic estimation of statistical significance maps for support vector machine based multi-variate image analysis and classification. Neuroimage 78, 270–283. doi: 10.1016/j.neuroimage.2013.03.066
Ghorbani, A., Abid, A., and Zou, J. (2019a). “Interpretation of neural networks is fragile,” in Proceedings of the AAAI Conference on Artificial Intelligence, 3681–3688.
Ghorbani, A., Zou, J., Wexler, J., and Kim, B. (2019b). Towards Automatic Concept-based Explanations.
Goodfellow, I., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. (2014). “Generative adversarial nets,” in Advances in Neural Information Processing Systems 27, eds Z. Ghahramani, M. Welling, C. Cortes, N. D. Lawrence, and K. Q. Weinberger (Curran Associates, Inc.), 2672–2680.
Goodfellow, I. J., Shlens, J., and Szegedy, C. (2014). Explaining and harnessing adversarial examples. arXiv [Preprint]. arXiv: 1412.6572.
Grigorescu, S., Trasnea, B., Cocias, T., and Macesanu, G. (2020). A survey of deep learning techniques for autonomous driving. J. F. Robot. 37, 362–386. doi: 10.1002/rob.21918
Guidotti, R., Monreale, A., Ruggieri, S., Pedreschi, D., Turini, F., and Giannotti, F. (2018a). Local rule-based explanations of black box decision systems. arXiv [Preprint]. arXiv: 1805.10820.
Guidotti, R., Monreale, A., Ruggieri, S., Turini, F., Giannotti, F., and Pedreschi, D. (2018b). A survey of methods for explaining black box models. ACM Comput. Surv. 51, 1–42. doi: 10.1145/3236009
Gunning, D., and Aha, D. (2019). DARPA's explainable artificial intelligence (XAI) Program. AI Mag. 40, 44–58. doi: 10.1145/3301275.3308446
Guyon, I., and Elisseeff, A. (2003). An introduction to variable and feature selection. J. Mach. Learn. Res. 3, 1157–1182.
Hägele, M., Seegerer, P., Lapuschkin, S., Bockmayr, M., Samek, W., Klauschen, F., et al. (2020). Resolving challenges in deep learning-based analyses of histopathological images using explanation methods. Sci. Rep. 10, 6423. doi: 10.1038/s41598-020-62724-2
Hall, P., and Gill, N. (2019). An Introduction to Machine Learning Interpretability. O'Reilly Media, Incorporated.
Haufe, S., Meinecke, F., Görgen, K., Dähne, S., Haynes, J.-D., Blankertz, B., et al. (2014). On the interpretation of weight vectors of linear models in multivariate neuroimaging. Neuroimage 87, 96–110. doi: 10.1016/j.neuroimage.2013.10.067
Herent, P., Jegou, S., Wainrib, G., and Clozel, T. (2018). Brain age prediction of healthy subjects on anatomic MRI with deep learning: going beyond with an “explainable AI” mindset. bioRxiv. 413302. doi: 10.1101/413302
Higgins, J. P. T., Altman, D. G., Gøtzsche, P. C., Jüni, P., Moher, D., Oxman, A. D., et al. (2011). The cochrane collaboration's tool for assessing risk of bias in randomised trials. BMJ 343. doi: 10.1136/bmj.d5928
Hoffman, R. R., Mueller, S. T., Klein, G., and Litman, J. (2018). Metrics for explainable AI: Challenges and prospects. arXiv [Preprint]. arXiv:1812.04608.
Hölldobler, S., Möhle, S., and Tigunova, A. (2017). “Lessons learned from alphago,” in YSIP. p. 92–101.
Holzinger, A. (2014). Trends in interactive knowledge discovery for personalized medicine: cognitive science meets machine learning. IEEE Intell. Inform. Bull. 15, 6–14.
Holzinger, A. (2016). Interactive machine learning for health informatics: when do we need the human-in-the-loop? Brain Informat. 3, 119–131. doi: 10.1007/s40708-016-0042-6
Holzinger, A. (2018). “From machine learning to explainable AI,” in DISA 2018 - IEEE World Symp. Digit. Intell. Syst. Mach. Proc., 55–66.
Holzinger, A., Biemann, C., Pattichis, C. S., and Kell, D. B. (2017). What do we need to build explainable AI systems for the medical domain?. arXiv [Preprint]. arXiv: 1712.09923.
Holzinger, A., Dehmer, M., and Jurisica, I. (2014). Knowledge discovery and interactive data mining in bioinformatics - state-of-the-art, future challenges and research directions. BMC Bioinformat. 15, I1. doi: 10.1186/1471-2105-15-S6-I1
Holzinger, A., Goebel, R., Fong, R., Moon, T., Müller, K.-R., and Samek, W. (2022a). “xxAI-beyond explainable artificial intelligence,” in International Workshop on Extending Explainable AI Beyond Deep Models and Classifiers (Springer), p. 3–10.
Holzinger, A., Langs, G., Denk, H., Zatloukal, K., and Müller, H. (2019). Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 9, 1–13. doi: 10.1002/widm.1312
Holzinger, A., Saranti, A., Molnar, C., Biecek, P., and Samek, W. (2022b). “Explainable AI methods-a brief overview,” in International Workshop on Extending Explainable AI Beyond Deep Models and Classifiers, eds A. Holzinger, R. Goebel, R. Fong, T. Moon, K.-R. Müller, and W. Samek (Cham: Springer International Publishing), 13–38.
Holzinger, A., Saranti, A., and Mueller, H. (2021). KANDINSKYPatterns–An experimental exploration environment for pattern analysis and machine intelligence. arXiv [Preprint]. arXiv: 2103.00519.
Hosny, A., Parmar, C., Quackenbush, J., Schwartz, L. H., and Aerts, H. J. W. L. (2018). Artificial intelligence in radiology. Nat. Rev. Cancer 18, 500–510. doi: 10.1038/s41568-018-0016-5
Hryniewska, W., Bombiński, P., Szatkowski, P., Tomaszewska, P., Przelaskowski, A., and Biecek, P. (2021). Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognit. 118, 108035. doi: 10.1016/j.patcog.2021.108035
Hu, S., Gao, Y., Niu, Z., Jiang, Y., Li, L., Xiao, X., et al. (2020). Weakly supervised deep learning for COVID-19 infection detection and classification from CT images. IEEE Access 8, 118869–118883. doi: 10.1109/ACCESS.2020.3005510
Jiménez-Luna, J., Grisoni, F., and Schneider, G. (2020). Drug discovery with explainable artificial intelligence. Nat. Mach. Intell. 2, 573–584. doi: 10.1038/s42256-020-00236-4
Joshi, G., Walambe, R., and Kotecha, K. (2021). A review on explainability in multimodal deep neural nets. IEEE Access. 9, 59800–59821.
Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C. S., Liang, H., Baxter, S. L., et al. (2018). Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 172, 1122–1131.e9. doi: 10.1016/j.cell.2018.02.010
Kerr, W. T., Douglas, P. K., Anderson, A., and Cohen, M. S. (2014). The utility of data-driven feature selection: Re: Chu et al. 2012. Neuroimage 84, 1107–1110. doi: 10.1016/j.neuroimage.2013.07.050
Khaligh-Razavi, S.-M., and Kriegeskorte, N. (2014). Deep supervised, but not unsupervised, models may explain IT cortical representation. PLoS Comput. Biol. 10, e1003915. doi: 10.1371/journal.pcbi.1003915
Khan, J., Wei, J. S., Ringnér, M., Saal, L. H., Ladanyi, M., Westermann, F., et al. (2001). Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat. Med. 7, 673–679. doi: 10.1038/89044
Kim, B., Wattenberg, M., Gilmer, J., Cai, C., Wexler, J., and Viegas, F. (2018). “Interpretability beyond feature attribution: Quantitative testing with concept activation vectors (tcav),” in International Conference on Machine Learning (PMLR), 2668–2677.
Kindermans, P.-J., Hooker, S., Adebayo, J., Alber, M., Schütt, K. T., Dähne, S., et al. (2019). “The (un)reliability of saliency methods,” in Explainable AI: Interpreting, Explaining and Visualizing Deep Learning, eds W. Samek, G. Montavon, A. Vedaldi, L. K. Hansen, and K.-R. Müller (Cham: Springer International Publishing), 267–280.
Kindermans, P. J., Schütt, K. T., Alber, M., Müller, K. R., Erhan, D., Kim, B., et al. (2017). Learning how to explain neural networks: Patternnet and patternattribution. arXiv [Preprint]. arXiv: 1705.05598.
Kohoutová, L., Heo, J., Cha, S., Lee, S., Moon, T., Wager, T. D., et al. (2020). Toward a unified framework for interpreting machine-learning models in neuroimaging. Nat. Protoc. 15, 1399–1435. doi: 10.1038/s41596-019-0289-5
Kriegeskorte, N. (2015). Deep neural networks: a new framework for modeling biological vision and brain information processing. Annu. Rev. Vis. Sci. 1, 417–446. doi: 10.1146/annurev-vision-082114-035447
Kriegeskorte, N., and Douglas, P. K. (2018). Cognitive computational neuroscience. Nat. Neurosci. 21, 1148–1160. doi: 10.1038/s41593-018-0210-5
Kriegeskorte, N., and Douglas, P. K. (2019). Interpreting encoding and decoding models. Curr. Opin. Neurobiol. 55, 167–179. doi: 10.1016/j.conb.2019.04.002
Kriegeskorte, N., Goebel, R., and Bandettini, P. (2006). Information-based functional brain mapping. Proc. Natl. Acad. Sci. U. S. A. 103, 3863–3868. doi: 10.1073/pnas.0600244103
Lake, B. M., Ullman, T. D., Tenenbaum, J. B., and Gershman, S. J. (2017). Building machines that learn and think like people. Behav. Brain Sci. 40, e253. doi: 10.1017/S0140525X16001837
Lamy, J.-B., Sekar, B., Guezennec, G., Bouaud, J., and Séroussi, B. (2019). Explainable artificial intelligence for breast cancer: a visual case-based reasoning approach. Artif. Intell. Med. 94, 42–53. doi: 10.1016/j.artmed.2019.01.001
Landecker, W., Thomure, M. D., Bettencourt, L. M. A., Mitchell, M., Kenyon, G. T., and Brumby, S. P. (2013). “Interpreting individual classifications of hierarchical networks,” in Proc. 2013 IEEE Symp. Comput. Intell. Data Mining, CIDM 2013 - 2013 IEEE Symp. Ser. Comput. Intell. SSCI 2013, 32–38.
Langlotz, C. P., Allen, B., Erickson, B. J., Kalpathy-Cramer, J., Bigelow, K., Cook, T. S., et al. (2019). A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 291, 781–791. doi: 10.1148/radiol.2019190613
Lapuschkin, S., Binder, A., Montavon, G., Muller, K.-R., and Samek, W. (2016). “Analyzing classifiers: fisher vectors and deep neural networks,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2912–2920.
Lapuschkin, S., Wäldchen, S., Binder, A., Montavon, G., Samek, W., and Müller, K. R. (2019). Unmasking Clever Hans predictors and assessing what machines really learn. Nat. Commun. 10, 1–8. doi: 10.1038/s41467-019-08987-4
LeCun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521, 436–444. doi: 10.1038/nature14539
Lee, H., Yune, S., Mansouri, M., Kim, M., Tajmir, S. H., Guerrier, C. E., et al. (2019). An explainable deep-learning algorithm for the detection of acute intracranial haemorrhage from small datasets. Nat. Biomed. Eng. 3, 173–182. doi: 10.1038/s41551-018-0324-9
Leek, J. T., Scharpf, R. B., Bravo, H. C., Simcha, D., Langmead, B., Johnson, W. E., et al. (2010). Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739. doi: 10.1038/nrg2825
Li, X., Dvornek, N. C., Zhuang, J., Ventola, P., and Duncan, J. S. (2018). “Brain biomarker interpretation in ASD using deep learning and fMRI,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, eds A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer International Publishing), 206–214.
Lipton, Z. C. (2018). The mythos of model interpretability. Commun. ACM 61, 36–43. doi: 10.1145/3233231
Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., et al. (2017). A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88. doi: 10.1016/j.media.2017.07.005
Lucieri, A., Dengel, A., and Ahmed, S. (2021). Deep learning based decision support for medicine–a case study on skin cancer diagnosis. arXiv [Preprint]. arXiv: 2103.05112.
Lundberg, S. M., and Lee, S. I. (2017). “A unified approach to interpreting model predictions,” in Advances in Neural Information Processing Systems, Vol. 30.
Lundervold, A. S., and Lundervold, A. (2019). An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 29, 102–127. doi: 10.1016/j.zemedi.2018.11.002
Ma, S., Song, X., and Huang, J. (2007). Supervised group Lasso with applications to microarray data analysis. BMC Bioinformat. 8, 60. doi: 10.1186/1471-2105-8-60
Magister, L. C., Kazhdan, D., Singh, V., and Liò, P. (2021). GCExplainer: Human-in-the-loop concept-based explanations for graph neural networks. arXiv [Preprint]. arXiv: 2107.11889.
Mahendran, A., and Vedaldi, A. (2015). “Understanding deep image representations by inverting them,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 5188–5196.
Meske, C., and Bunde, E. (2020). “Transparency and trust in human-AI-interaction: the role of model-agnostic explanations in computer vision-based decision support BT,” in Artificial Intelligence in HCI, eds H. Degen, and L. Reinerman-Jones (Cham: Springer International Publishing), 54–69.
Miller, T. (2019). Explanation in artificial intelligence: insights from the social sciences. Artif. Intell. 267, 1–38. doi: 10.1016/j.artint.2018.07.007
Miotto, R., Wang, F., Wang, S., Jiang, X., and Dudley, J. T. (2017). Deep learning for healthcare: review, opportunities and challenges. Brief. Bioinformat. 19, 1236–1246. doi: 10.1093/bib/bbx044
Mirchi, N., Bissonnette, V., Yilmaz, R., Ledwos, N., Winkler-Schwartz, A., and Del Maestro, R. F. (2020). The virtual operative assistant: an explainable artificial intelligence tool for simulation-based training in surgery and medicine. PLoS ONE 15, e0229596. doi: 10.1371/journal.pone.0229596
Moher, D., Liberati, A., Tetzlaff, J., and Altman, D. G.. (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA Statement. Ann. Intern. Med. 151, 264–269. doi: 10.7326/0003-4819-151-4-200908180-00135
Mohseni, S., Zarei, N., and Ragan, E. D. (2021). “A multidisciplinary survey and framework for design and evaluation of explainable AI systems,” in ACM Transactions on Interactive Intelligent Systems, Vol. 11(ACM), 1–45.
Montavon, G., Lapuschkin, S., Binder, A., Samek, W., and Müller, K. R. (2017). Explaining nonlinear classification decisions with deep Taylor decomposition. Pattern Recognit. 65, 211–222. doi: 10.1016/j.patcog.2016.11.008
Montavon, G., Samek, W., and Müller, K. R. (2018). Methods for interpreting and understanding deep neural networks. Digit. Signal Process. A Rev. J. 73, 1–15. doi: 10.1016/j.dsp.2017.10.011
Morcos, A. S., Barrett, D. G., Rabinowitz, N. C., and Botvinick, M. (2018). On the importance of single directions for generalization. arXiv [Preprint]. arXiv: 1803.06959.
Mourão-Miranda, J., Bokde, A. L. W., Born, C., Hampel, H., and Stetter, M. (2005). Classifying brain states and determining the discriminating activation patterns: support vector machine on functional MRI data. Neuroimage 28, 980–995. doi: 10.1016/j.neuroimage.2005.06.070
Nguyen, A., Dosovitskiy, A., Yosinski, J., Brox, T., and Clune, J. (2016). “Synthesizing the preferred inputs for neurons in neural networks via deep generator networks,” in Advances in Neural Information Processing Systems 29, eds D. D. Lee, M. Sugiyama, U. V. Luxburg, I. Guyon, and R. Garnett (Curran Associates, Inc.), 3387–3395.
Nigri, E., Ziviani, N., Cappabianco, F., Antunes, A., and Veloso, A. (2020). “Explainable deep CNNs for MRI-based diagnosis of Alzheimer's Disease,” in 2020 International Joint Conference on Neural Networks (IJCNN), 1–8.
Osman, A., Arras, L., and Samek, W. (2020). Towards ground truth evaluation of visual explanations. arXiv e-prints, arXiv-2003.
Palatnik de Sousa, I., Maria Bernardes Rebuzzi Vellasco, M., and Costa da Silva, E. (2019). Local interpretable model-agnostic explanations for classification of lymph node metastases. Sensors. 19, 2969. doi: 10.3390/s19132969
Papanastasopoulos, Z., Samala, R. K., Chan, H.-P., Hadjiiski, L., Paramagul, C., Helvie, M. A., et al. (2020). “Explainable AI for medical imaging: deep-learning CNN ensemble for classification of estrogen receptor status from breast MRI,” in Proc.SPIE.
Paschali, M., Conjeti, S., Navarro, F., and Navab, N. (2018). “Generalizability vs. robustness: investigating medical imaging networks using adversarial examples,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, eds A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer International Publishing), 493–501.
Pennisi, M., Kavasidis, I., Spampinato, C., Schinina, V., Palazzo, S., Salanitri, F. P., et al. (2021). An explainable AI system for automated COVID-19 assessment and lesion categorization from CT-scans. Artif. Intell. Med. 118, 102114. doi: 10.1016/j.artmed.2021.102114
Pfeifer, B., Baniecki, H., Saranti, A., Biecek, P., and Holzinger, A. (2021). Graph-guided random forest for gene set selection.
Puri, N., Gupta, P., Agarwal, P., Verma, S., and Krishnamurthy, B. (2017). MAGIX: model agnostic globally interpretable explanations. arXiv [Preprint]. arXiv:1 706.07160.
Qin, Y., Kamnitsas, K., Ancha, S., Nanavati, J., Cottrell, G., Criminisi, A., et al. (2018). “Autofocus layer for semantic segmentation,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, eds A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer International Publishing), 603–611.
Rajkomar, A., Oren, E., Chen, K., Dai, A. M., Hajaj, N., Hardt, M., et al. (2018). Scalable and accurate deep learning with electronic health records. npj Digit. Med. 1, 18. doi: 10.1038/s41746-018-0029-1
Ribeiro, M. T., Singh, S., and Guestrin, C. (2016). Nothing else matters: Model-agnostic explanations by identifying prediction invariance. arXiv [Preprint]. arXiv: 1611.05817.
Richardson, E., Sela, M., Or-El, R., and Kimmel, R. (2017). “Learning detailed face reconstruction from a single image,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1259–1268.
Robnik-Šikonja, M., and Kononenko, I. (2008). Explaining classifications for individual instances. IEEE Trans. Knowl. Data Eng. 20, 589–600. doi: 10.1109/TKDE.2007.190734
Ross, A. S., Hughes, M. C., and Doshi-Velez, F. (2017). Right for the right reasons: Training differentiable models by constraining their explanations. arXiv [Preprint]. arXiv: 1703.03717.
Rumelhart, D. E., Hinton, G. E., and Williams, R. J. (1986). Learning representations by back-propagating errors. Nature 323, 533–536. doi: 10.1038/323533a0
Samek, W., Binder, A., Montavon, G., Lapuschkin, S., and Müller, K. (2017a). Evaluating the visualization of what a deep neural network has learned. IEEE Trans. Neural Networks Learn. Syst. 28, 2660–2673. doi: 10.1109/TNNLS.2016.2599820
Samek, W., Montavon, G., Binder, A., Lapuschkin, S., and Müller, K.-R. (2016). Interpreting the predictions of complex ML models by layer-wise relevance propagation. arXiv [Preprint]. arXiv: 1611.08191.
Samek, W., Wiegand, T., and Müller, K.-R. (2017b). Explainable artificial intelligence: understanding, visualizing and interpreting deep learning models. arXiv [Preprint]. arXiv: 1708.08296.
Schirrmeister, R. T., Springenberg, J. T., Fiederer, L. D. J., Glasstetter, M., Eggensperger, K., Tangermann, M., et al. (2017). Deep learning with convolutional neural networks for EEG decoding and visualization. Hum. Brain Mapp. 38, 5391–5420. doi: 10.1002/hbm.23730
Schmidhuber, J. (2015). Deep learning in neural networks: an overview. Neural Netw. 61, 85–117. doi: 10.1016/j.neunet.2014.09.003
Scott, A. C., Clancey, W. J., Davis, R., and Shortliffe, E. H. (1977). Explanation Capabilities of Production-Based Consultation Systems. Stanford Univ CA Dept of Computer Science.
Segev, E. (2020). Textual network analysis: detecting prevailing themes and biases in international news and social media. Sociol. Compass 14, e12779. doi: 10.1111/soc4.12779
Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., and Batra, D. (2017). “Grad-cam: visual explanations from deep networks via gradient-based localization,” in Proceedings of the IEEE International Conference on Computer Vision, 618–626.
Selvaraju, R. R., Das, A., Vedantam, R., Cogswell, M., Parikh, D., and Batra, D. (2016). Grad-CAM: Why did you say that?. arXiv [Preprint]. arXiv: 1611.07450.
Shen, D., Wu, G., and Suk, H.-I. (2017). Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248. doi: 10.1146/annurev-bioeng-071516-044442
Shortliffe, E. H., and Buchanan, B. G. (1975). A model of inexact reasoning in medicine. Math. Biosci. 23, 351–379. doi: 10.1016/0025-5564(75)90047-4
Shrikumar, A., Greenside, P., and Kundaje, A. (2017). “Learning important features through propagating activation differences,” in Proceedings of the 34th International Conference on Machine Learning - Volume 70, ICML'17. (JMLR.org), 3145–3153.
Silver, D., Huang, A., Maddison, C. J., Guez, A., Sifre, L., van den Driessche, G., et al. (2016). Mastering the game of Go with deep neural networks and tree search. Nature 529, 484–489. doi: 10.1038/nature16961
Simonyan, K., Vedaldi, A., and Zisserman, A. (2013). Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv [Preprint]. arXiv: 1312.6034.
Simonyan, K., and Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. arXiv: 1409.1556.
Singh, A., Sengupta, S., Mohammed, A. R., Faruq, I., et al. (2020). “What is the optimal attribution method for explainable ophthalmic disease classification?,” in BT - Ophthalmic Medical Image Analysis, eds H. Fu, M. K. Garvin, T. MacGillivray, Y. Xu, and Y. Zheng (Cham: Springer International Publishing), 21–31.
Smilkov, D., Thorat, N., Kim, B., Viégas, F., and Wattenberg, M. (2017). SmoothGrad: removing noise by adding noise. arXiv [Preprint]. arXiv: 1706.03825.
Springenberg, J. T., Dosovitskiy, A., Brox, T., and Riedmiller, M. (2014). Striving for simplicity: the all convolutional net. arXiv [Preprint]. arXiv: 1412.6806
Srinivasan, V., Lapuschkin, S., Hellge, C., Muller, K. R., and Samek, W. (2017). Interpretable human action recognition in compressed domain. ICASSP, IEEE Int. Conf. Acoust. Speech Signal Process. - Proc., 1692–1696.
Staniak, M., and Biecek, P. (2018). Explanations of model predictions with live and breakDown packages. arXiv [Preprint]. arXiv: 1804.01955.
Sturm, I., Lapuschkin, S., Samek, W., and Müller, K. R. (2016). Interpretable deep neural networks for single-trial EEG classification. J. Neurosci. Methods 274, 141–145. doi: 10.1016/j.jneumeth.2016.10.008
Sundararajan, M., Taly, A., and Yan, Q. (2017). “Axiomatic attribution for deep networks,” in Proceedings of the 34th International Conference on Machine Learning - Volume 70, ICML'17 (JMLR.org), 3319–3328.
Swartout, W., Paris, C., and Moore, J. (1991). Explanations in knowledge systems: design for explainable expert systems. IEEE Expert 6, 58–64. doi: 10.1109/64.87686
Tang, Z., Chuang, K. V., DeCarli, C., Jin, L. W., Beckett, L., Keiser, M. J., et al. (2019). Interpretable classification of Alzheimer's disease pathologies with a convolutional neural network pipeline. Nat. Commun. 10, 1–14. doi: 10.1038/s41467-019-10212-1
Thomas, A. W., Heekeren, H. R., Müller, K. R., and Samek, W. (2019). Analyzing Neuroimaging Data Through Recurrent Deep Learning Models. Front. Neurosci. 13, 1–18. doi: 10.3389/fnins.2019.01321
Ting, D. S. W., Cheung, C. Y.-L., Lim, G., Tan, G. S. W., Quang, N. D., Gan, A., et al. (2017). Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA 318, 2211–2223. doi: 10.1001/jama.2017.18152
Tjoa, E., and Guan, C. (2020). A survey on explainable artificial intelligence (xai): Toward medical xai. IEEE Trans. Nural Netw. Learn. Syst. 32, 4793–4813.
Tonekaboni, S., Joshi, S., McCradden, M. D., and Goldenberg, A. (2019). “What clinicians want: contextualizing explainable machine learning for clinical end use,” in Machine Learning for Healthcare Conference (PMLR), 359–380.
Tulio Ribeiro, M., Singh, S., and Guestrin, C. (2016). Nothing else matters: model-agnostic explanations by identifying prediction invariance.
Ustun, B., and Rudin, C. (2016). Supersparse linear integer models for optimized medical scoring systems. Mach. Learn. 102, 349–391. doi: 10.1007/s10994-015-5528-6
van der Velden, B. H. M., Kuijf, H. J., Gilhuijs, K. G. A., and Viergever, M. A. (2022). Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 79, 102470. doi: 10.1016/j.media.2022.102470
Vayena, E., Blasimme, A., and Cohen, I. G. (2018). Machine learning in medicine: addressing ethical challenges. PLoS Med. 15, e1002689. doi: 10.1371/journal.pmed.1002689
Wang, C. J., Hamm, C. A., Savic, L. J., Ferrante, M., Schobert, I., Schlachter, T., et al. (2019). Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 29, 3348–3357. doi: 10.1007/s00330-019-06214-8
Wang, Z., Childress, A. R., Wang, J., and Detre, J. A. (2007). Support vector machine learning-based fMRI data group analysis. Neuroimage 36, 1139–1151. doi: 10.1016/j.neuroimage.2007.03.072
Weng, S. F., Reps, J., Kai, J., Garibaldi, J. M., and Qureshi, N. (2017). Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE. 12, e0174944.
Wilson, B., Hoffman, J., and Morgenstern, J. (2019). Predictive inequity in object detection. arXiv [Preprint]. arXiv: 1902.11097.
Windisch, P., Weber, P., Fürweger, C., Ehret, F., Kufeld, M., Zwahlen, D., et al. (2020). Implementation of model explainability for a basic brain tumor detection using convolutional neural networks on MRI slices. Neuroradiology 62, 1515–1518. doi: 10.1007/s00234-020-02465-1
Xu, F., Uszkoreit, H., Du, Y., Fan, W., Zhao, D., and Zhu, J. (2019). “Explainable AI: a brief survey on history, research areas, approaches and challenges,” in Natural Language Processing and Chinese Computing, eds J. Tang. M.-Y., Kan, D. Zhao, S. Li„ and H. Zan (Cham: Springer International Publishing), 563–574.
Xu, Y., Jia, Z., Wang, L.-B., Ai, Y., Zhang, F., Lai, M., et al. (2017). Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinforma. 18, 281. doi: 10.1186/s12859-017-1685-x
Yang, C., Rangarajan, A., and Ranka, S. (2018). “Visual explanations from deep 3D convolutional neural networks for Alzheimer's Disease Classification,” in AMIA. Annu. Symp. proceedings. AMIA Symp. 2018, 1571–1580.
Yasaka, K., and Abe, O. (2018). Deep learning and artificial intelligence in radiology: current applications and future directions. PLoS Med. 15, 2–5. doi: 10.1371/journal.pmed.1002707
Yeom, S.-K., Seegerer, P., Lapuschkin, S., Binder, A., Wiedemann, S., Müller, K.-R., et al. (2021). Pruning by explaining: A novel criterion for deep neural network pruning. Pattern Recognit. 115, 107899. doi: 10.1016/j.patcog.2021.107899
Yosinski, J., Clune, J., Nguyen, A., Fuchs, T., and Lipson, H. (2015). Understanding neural networks through deep visualization. arXiv [Preprint]. arXiv: 1506.06579.
Young, T., Hazarika, D., Poria, S., and Cambria, E. (2018). Recent trends in deep learning based natural language processing. IEEE Comput. Intell. Mag. 13, 55–75. doi: 10.1109/MCI.2018.2840738
Zeiler, M. D., and Fergus, R. (2014). “Visualizing and understanding convolutional networks,” inEuropean Conference on Computer Vision, eds D. Fleet, T. Pajdla, B. Schiele, and T. Tuytelaars (Cham: Springer International Publishing), 818–833.
Zhang, J., Bargal, S. A., Lin, Z., Brandt, J., Shen, X., and Sclaroff, S. (2018). Top-down neural attention by excitation backprop. Int. J. Comput. Vis. 126, 1084–1102. doi: 10.1007/s11263-017-1059-x
Zhang, X., Han, L., Zhu, W., Sun, L., and Zhang, D. (2021). An explainable 3D residual self-attention deep neural network for joint atrophy localization and Alzheimer's disease diagnosis using structural MRI. IEEE J. Biomed. Heal. Informatics, 1. doi: 10.1109/JBHI.2021.3066832
Zhao, G., Zhou, B., Wang, K., Jiang, R., and Xu, M. (2018). “Respond-CAM: analyzing deep models for 3D imaging data by visualizations,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018, eds A. F. Frangi, J. A. Schnabel, C. Davatzikos, C. Alberola-López, and G. Fichtinger (Cham: Springer International Publishing), 485–492.
Zhou, B., Khosla, A., Lapedriza, A., Oliva, A., and Torralba, A. (2016). “Learning deep features for discriminative localization,” in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 2921–2929.
Zhu, G., Jiang, B., Tong, L., Xie, Y., Zaharchuk, G., and Wintermark, M. (2019). Applications of deep learning to neuro-imaging techniques. Front. Neurol. 10, 869. doi: 10.3389/fneur.2019.00869
Keywords: explainable AI, interpretability, artificial intelligence (AI), deep learning, neural networks, medical imaging, neuroimaging
Citation: Farahani FV, Fiok K, Lahijanian B, Karwowski W and Douglas PK (2022) Explainable AI: A review of applications to neuroimaging data. Front. Neurosci. 16:906290. doi: 10.3389/fnins.2022.906290
Received: 28 March 2022; Accepted: 14 November 2022;
Published: 01 December 2022.
Edited by:
Takashi Hanakawa, Kyoto University, JapanReviewed by:
Andreas Holzinger, Medical University Graz, AustriaCopyright © 2022 Farahani, Fiok, Lahijanian, Karwowski and Douglas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Farzad V. Farahani, ZmZhcmFoYTJAamh1LmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.