- 1Department of Neurology, Shenzhen Hospital, Southern Medical University, Shenzhen, China
- 2School of Physical Education, Southwest University, Chongqing, China
- 3Department of Neurology, Affiliated Xiaolan Hospital, Southern Medical University, Xiaolan People's Hospital, Zhongshan, China
- 4Department of Neurology, Shenzhen University General Hospital, Shenzhen University Clinical Medical Academy, Shenzhen, China
The N6-methyladenosine (m6A) modification acts as a dynamic regulatory factor in diseases by regulating the metabolism and function of the transcriptome, especially mRNAs. However, little is known regarding the functional effects of m6A modifications on circRNAs. In this research, we established a distal middle cerebral artery occlusion (MCAO) model in adult C57BL/6J mice. The mice were divided into three groups: sham surgery, 3 days after MCAO (3d), and 7 days after MCAO (7d). Reverse transcription quantitative polymerase chain reaction (RT-qPCR) demonstrated that the mRNA expression levels of m6A-related methyltransferases (METTL3, METTL14), demethylases (FTO, ALKBH5), and reading proteins (YTHDF1, YTHDF3) altered compared to the sham group. Furthermore, the translation level of ALKBH5 and YTHDF3 was significantly decreased in the 3d group while increased in 7d group. Methylated RNA immunoprecipitation (MeRIP) and circRNA microarray indicated 85 hypermethylated and 1621 hypomethylated circRNAs in the 3d group. In the 7d group, the methylation level increased in 57 and decreased in 66 circRNAs. Subsequently, our results were verified by MeRIP-qPCR. Bioinformatics analysis was performed to analyze the functions of differentially m6A-modified circRNAs. We found some m6A modified-circRNAs associated with cerebral infarction, providing a new direction for the molecular mechanism of stroke.
Introduction
Cerebral infarction causes transcriptional and post-transcriptional alterations including RNA expression levels and epigenetic modifications, resulting in a series of complicated post-stroke pathophysiological processes, such as glutamate excitotoxicity, oxidative stress, apoptosis, nerve regeneration, and angiogenesis (Mehta et al., 2007).
Studies of RNA modification are known as epigenetic transcriptomics. Recent studies have indicated that epigenetic changes occur during the pathophysiological course of diverse diseases as regulators of RNAs (Niu et al., 2013). At present, more than 170 classes of RNA modifications have been identified, among which N6-methyladenosine (m6A) is the most abundant and common modification in Eukaryotes, particularly in the mammalian brains (Dominissini et al., 2012). The m6A modification refers to the methylation of the sixth nitrogen atom in adenosine, which is catalyzed by methyltransferase complexes, such as methyltransferase-like (METTL) 3 and 14 (METTL3 and METTL14), and Wilms' tumor 1-associated protein (WTAP); eliminated by demethylases, such as fat mass and obesity-associated protein (FTO) and alkylation repair homolog protein 5 (ALKBH5); and recognized by m6A readers, such as YTH domain family (YTHDF) 1, 2, and 3 (YTHDF1, YTHDF2, and YTHDF3) (Shi et al., 2019). Thus, the m6A modification is a dynamic and reversible process. The majority of m6A modifications occur near the 3′UTR and the mRNA stop codon, and regulate various molecular functions in mRNAs related to transcription, alternative splicing, nuclear export, translation, stability, and degradation (Niu et al., 2013). It has been researched that m6A-related enzymes play a critical role in ischemic diseases. FTO has been reported to promote angiogenesis and reduce myocardial fibrosis after myocardial ischemia, however, in the brain, it is predominantly expressed in neurons, whereas downregulation of FTO and ALKBH5 after stroke can induce secondary brain injury by destroying neurons (Chokkalla et al., 2019; Mathiyalagan et al., 2019; Xu et al., 2020). METTL3, METTL14, and YTHDF1 have been shown to regulate axon regeneration, neuronal apoptosis, neurogenesis, and inflammation after cerebral ischemia (Weng et al., 2018; Si et al., 2020; Zhang et al., 2020), which indicates that the m6A modification may be involved in a series of post-stroke molecular events.
Non-coding RNAs, especially circRNAs, show tissue specificity during development and are highly expressed in the brain (Rybak-Wolf et al., 2015). Studies have shown that circRNAs are specifically enriched in the subcellular compartments of neurons, including dendrites, axons, and synapses (Rybak-Wolf et al., 2015; You et al., 2015; Shigeoka et al., 2016). Therefore, tracking changes in circRNAs and their m6A modification levels may provide new insights into the pathogenesis of cerebral ischemia from the perspective of epigenetics, thereby improving intervention and prognosis. CircRNAs can regulate secondary brain injury and repair by binding with proteins and acting as “sponges” for miRNA (Wu et al., 2019; Yang et al., 2020). However, the upstream regulatory mechanism for circRNAs remains unknown. Recently, a study focused on the differences in m6A modifications of mRNAs lncRNAs after transient cerebral ischemia (Chokkalla et al., 2019), but the m6A modification of circRNA is not yet known. Therefore, in this study, we screened the entire genome for m6A-modified circRNAs around the site of cerebral infarction to explore the potential significance of m6A-modified circRNAs in stroke.
Methods and Materials
Cerebral Infarction in Mice
All experimental procedures were approved by the Institutional Animal Use Committee of the Shenzhen Hospital of Southern Medical University. Animals were raised in a SPF room with free access to food and water, under a controlled temperature (25 ± 2°C) and a 12/12 h day/night cycle The surgery was performed in accordance with the guidelines for the care and use of laboratory animals. C57BL/6J mice weighing 23–26 g were randomized to three groups: the 3d after MCAO, 7d after MCAO, and sham groups. After anesthetizing the mice with isoflurane, we made a 1 cm incision between the left ear and eye, dissected the temporal muscle, and exposed the left side of the skull under an operating microscope. Next, a small hole was drilled, and a distal middle cerebral artery occlusion (MCAO) was performed with a bipolar electrosurgical knife, leading to a permanent infarction of the local cerebral cortex. And then the bilateral common carotid arteries were immediately occluded for 7 minutes. The sham group underwent the same surgical procedure without the MCAO. During surgery and recovery, the body temperature of the mice were maintained at 37°C.
TTC Staining
Three mice were used for histological staining at 24 h after MCAO. Briefly, the mice were killed by cervical dislocation, and brain tissues were isolated and frozen at −20°C in refrigerator for 5 mins. Coronal sections were sliced into 1 mm and maintained at 37°C in 2% 2,3,5-triphenyltetrazolium chloride (TTC) solution (Sigma Aldrich, St. Louis, MO, USA) for 30 min. TTC staining can effectively detect the location of the primary cerebral infarction after MCAO.
H&E Staining
One week after cerebral infarction operation, the brain tissue was collected and fixed with 4% paraformaldehyde for 1 week, and then embedded in paraffin. Following alcohol dehydration and xylene dewaxing, coronal sections were stained with hematoxylin for 3 min, washed, and then eosin for 2 min. Finally, sections were sealed with neutral gum after being dehydrated.
m6A Immunoprecipitation (MeRIP)
Total RNA (1–3 μg) and m6A spike-in control mixture were added to immunoprecipitation (IP) buffer (50 mmol/L Tris-HCl, pH 7.4; 150 mmol/L NaCl; 0.1% NP40; and 40 U/μL RNase inhibitor), incubated with 2 μg anti-m6A rabbit polyclonal antibody (Synaptic Systems, 202003) at 4°C for 2 h, washed, resuspended in 20 μL Dynabeads™ M-280 sheep anti-rabbit IgG suspension (Invitrogen, 11203D), and incubated at 4°C for 2 h. The beads were then washed three times with 500 μL of IP buffer and twice with 500 μL of wash buffer [50 mmol/L Tris-HCl, pH 7.4; 50 mmol/L NaCl; 0.1% NP 40; and 40 U/μL RNase inhibitor (Enzymatics, Y9240L)]. The enriched RNA was eluted with 200 μL elution buffer (10 mmol/L Tris-HCl, pH 7.4; 1 mmol/L EDTA; 0.05% SDS; and 40 U proteinase K) and extracted using acid phenol-chloroform and ethanol precipitation. MeRIP assays were performed in three replicates in each group (n = 3).
Microarray Hybridization of Methylated CircRNAs
The immunoprecipitate was digested with RNase R to remove linear RNA, amplified, and labeled with super RNA Labeling Kit (Cy3 [for “sup”, supernatant] and Cy5 [for “IP”]) in accordance with the manufacturer's instructions, and then purified using the RNeasy Mini Kit. After fragmentation, the circRNAs were hybridized to an m6A-circRNA epi-transcriptomic microarray slide (Arraystar) and incubated at 65°C for 17 h in a hybridization oven (Agilent, Santa Clara, CA, USA). The hybridized arrays were washed, fixed, and scanned using an Agilent Scanner G2505C.
Analysis of Microarray Data
Acquired array images were analyzed using Agilent Feature Extraction software (version 11.0.1.1). Raw intensities of IP (immunoprecipitated, Cy5-labeled) were normalized at a criterion of log2-scaled intensities in spike-in RNA. The percentage of the IP (Cy5-labeled) normalized intensities was considered as m6A methylation levels. Differentially m6A-modified circRNAs were identified between the two comparison groups by filtering with the fold change and statistical significance (P-value) thresholds. Expression of circRNAs were calculated by the normalized signal value of “IP” plus “sup”, the differentially expressed circRNAs were screened at the criterion of with fold change >1.5 and P < 0.05.
Reverse Transcription Quantitative Polymerase Chain Reaction
After collecting samples at 3d and 7d after MCAO, 1 μg of total RNA was reverse transcribed to cDNA. RT-qPCR was performed to determine the mRNA levels of m6A-related enzymes (METTL3, METTL14, FTO, WTAP, ALKBH5, YTHDF1, and YTHDF3) by the Applied Biosystems 7300/7500 Real-Time PCR System (Foster City, CA, USA). Additionally, to further confirm the results from the m6A immunoprecipitation and microarray, we randomly selected four differentially m6A-modified circRNAs (mmu_circRNA_27268, mmu_circRNA_20673, mmu_circRNA_32905, and mmu_circRNA_29864) in samples of 3d after MCAO and detected their level by MeRIP-qPCR analysis. In brief, equal quantity of immunoprecipitated RNA samples were reverse-transcribed to cDNA using SuperScriptTM III Reverse Transcriptase (Invitrogen, 18080-044), and amplified with gene-specific primers for the circRNAs (Table 1). PCR assays were performed in six replicates in each group (n = 6). All results were analyzed by 2ΔΔCT method.
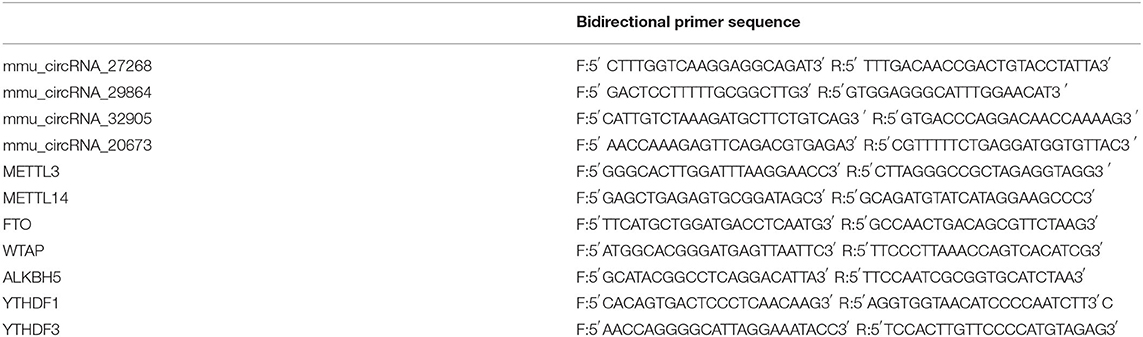
Table 1. The primer sequence of the four differentially m6A-modified circRNAs and m6A-related enzymes.
Western Blots Analysis
Around 50 μg of protein samples extract was resolved in 10% sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) after denaturation and transferred to PVDF membranes. The membranes were blocked in 5% non-fat milk in tris-buffered saline solution (TBS) for 1.5 h at room temperature and then probed with the primary antibodies: rabbit anti-ALKBH5 (Abcam, ab195377, 1:1,000) and rabbit anti-YTHDF3 (Abcam, ab220161, 1:1,000). After overnight incubation at 4°C, the membranes were washed three times with TBS-T and incubated for 1 h at room temperature with an appropriate secondary antibody: horseradish peroxidase-labeled anti-rabbit IgG (1:5,000). The protein bands were visualized using the enhanced chemiluminescence method, and the intensity of band was quantified using Image J software (Version 1.51j8, NIH, Bethesda, MD, USA).
Function and Pathways Analyses
The functions of differentially m6A-modified circRNAs' parent genes and three verified ones' target genes were analyzed by Gene Ontology (GO) analysis (http://www.geneontology.org), and their significant pathways were explored by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (http://www.genome.jp/kegg/) using the DAVID bioinformatics database (https://david.ncifcrf.gov).
Prediction of Network of CircRNA-miRNA-mRNA
Three verified m6A-circRNAs were used to predicted miRNAs by Arraystar's homemade software based on TargetScan and miRanda, and top ten possible miRNAs were screened. Additionally, mRNAs of these ten miRNAs were predicted by miRanda. The network of circRNA-miRNA-mRNA was visualized by Cytoscape.
Statistical Analysis
Our results were analyzed using Statistical Program for Social Sciences (SPSS) software (version 22.0; SPSS, Chicago, IL, USA) and are presented as means ± standard deviations. The student t test was used to detect the significant differences between two groups, one-way ANOVA was used for comparisons between multiple groups, and P < 0.05 was taken as statistically significant.
Results
Establishment and Validation of the Cerebral Infarction Model
TTC and H&E staining were used to confirm the infarction region of the brain after MCAO in mice (Figures 1A,B). TTC staining showed that the infarcted brain tissue was white, and the non-infarct region was red. Additionally, with H&E staining, the infarct area was pale, and the number of neurons was decreased, whereas the opposite was observed in normal tissue.
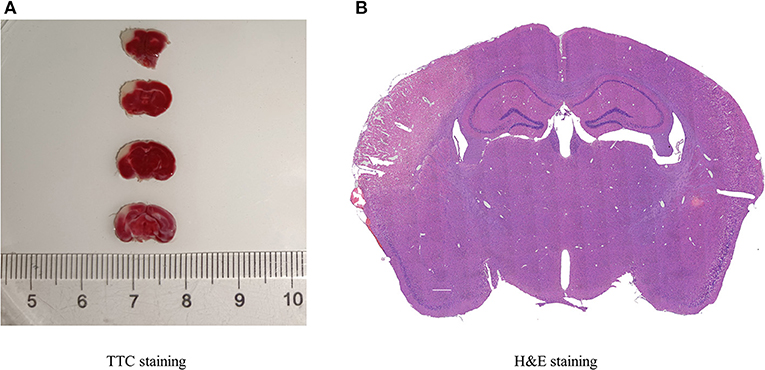
Figure 1. Histopathology of cerebral infarction. (A) TTC staining displayed the infarcted areas in white, and the normal areas in red, (n = 3). (B) H&E staining presented the destruction of morphology in the infarct lateral cortex compared with the Contralateral cortex (n = 3).
The Transcriptional Levels of Methylation-Related Enzymes After Stroke in Mice
To explore the changes in m6A methylation on circRNA transcripts, we first determined transcriptional levels of m6A-related enzymes in the peri-infarct region of the cerebral cerebral cortex (Figure 2A). We observed that mRNA expression of all enzymes tested was significantly downregulated at 3 days after MCAO compared with the sham group. At 7 days after MCAO, the transcriptional levels of METTL3, METTL4, ALKBH5, and YTHDF3 were decreased, whereas increased in FTO significantly. Interestingly, compared with the 3d group, except for METTL3, METTL14, and ALKBH5, significant increases in transcription of other genes occurred in the 7d group.
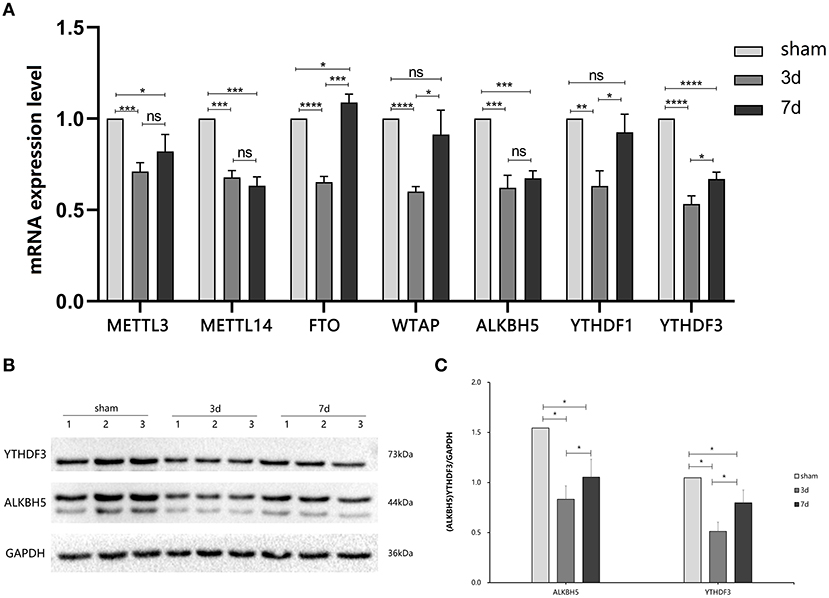
Figure 2. Expression of m6A related enzymes between MCAO and sham groups. (A) The mRNA expression level of m6A writers (METTL3, METTL14, and WTAP), m6A erasers (FTO, ALBKH5), and m6A readers (YTHDF1, YTHDF3) were changed at 3d or 7d after MCAO compared with the sham, (n = 6/group). (B,C) The protein YTHDF3 were significantly decreased in 3d group, while increased in 7d group, (n = 6/group). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ns, not significant.
ALKBH5 and YTHDF3 Showed Dynamic Alterations in Stroke
We further measured the translation level of m6A related enzymes in the area around cerebral cortex infarction (Figures 2B,C). We observed that ALKBH5 and YTHDF3 were significantly decreased at 3 and 7 days after MCAO compared with the sham group. Interestingly, ALKBH5 and YTHDF3 were increased at 7 days in comparison to 3 days after MCAO.
Cerebral Infarction Changed the Level of m6A Modification and Expression Profile in CircRNA
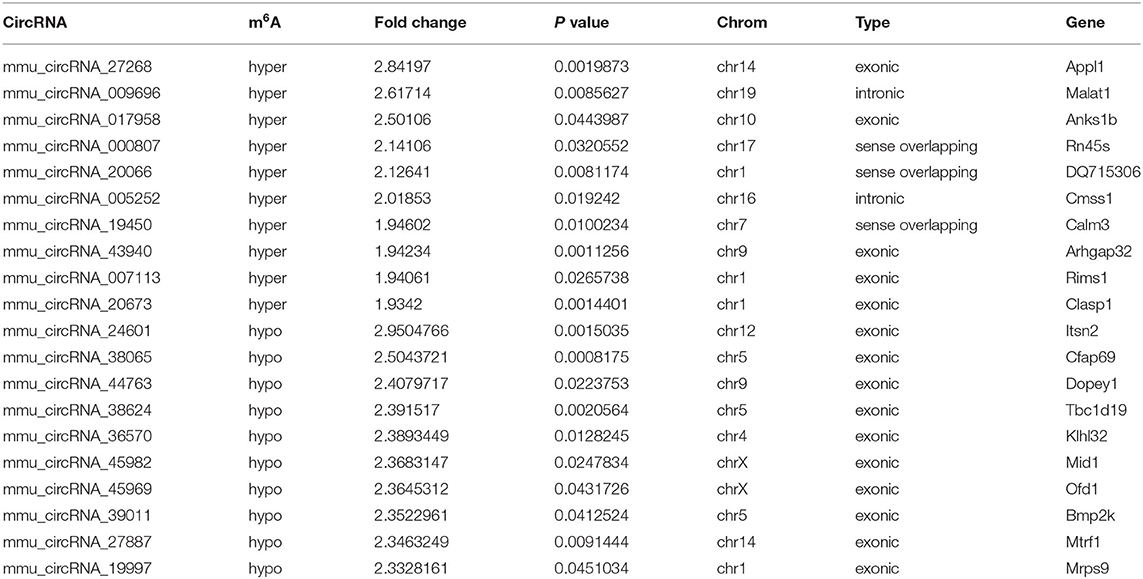
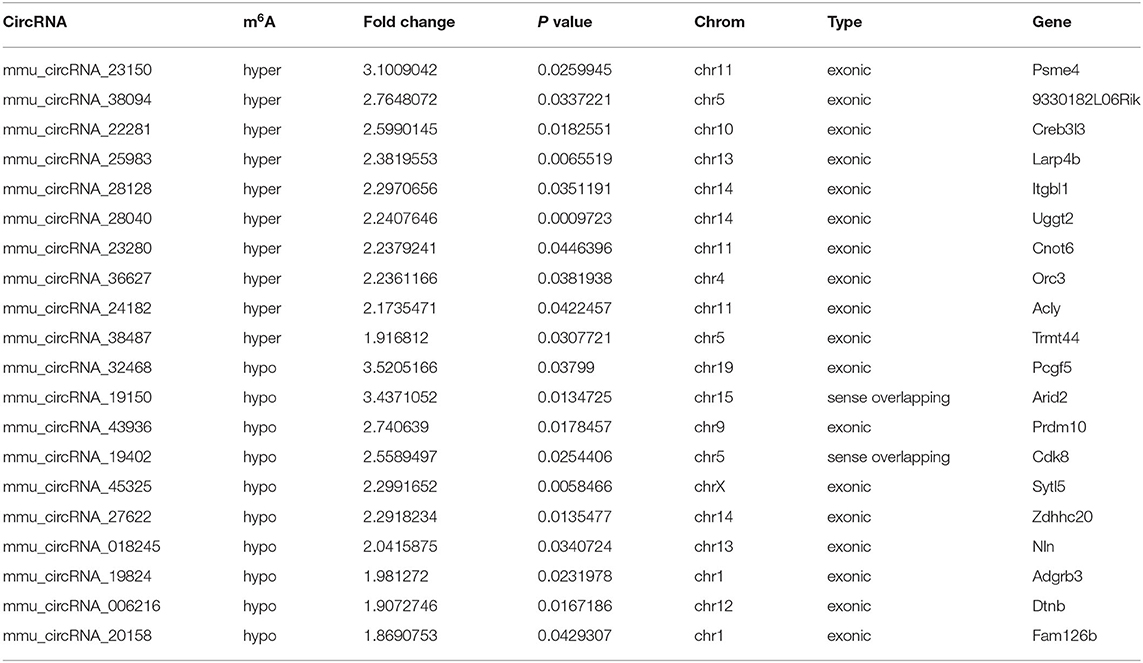
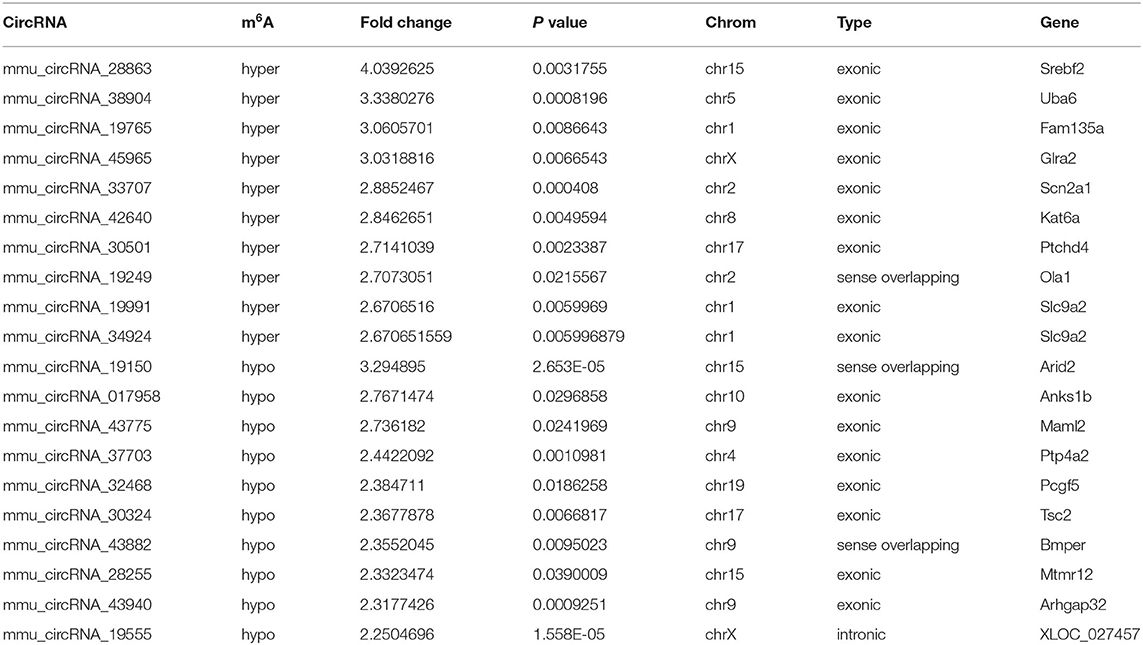
The mRNA expression of m6A-related enzymes in the peri-infarct cortex was altered, suggesting that cerebral ischemia might change m6A modification. We speculated that the methylation modification of circRNAs around cerebral infarction was changed. We performed m6A immunoprecipitation and microarray analysis of circRNAs from cortex tissue surrounding the infarcted area after 3 and 7 days of permanent cerebral ischemia in mice. Heatmaps and volcano graphs showed differences in m6A methylation in circRNAs between the infarct groups and the sham group (fold change > 1.5, P < 0.05, n = 3/group) (Figures 3A–F). A total of 12,345 methylated circRNAs were found in our research. In comparison with the sham group, the m6A modification was significantly changed in 1,706 circRNAs, including 85 hypermethylated and 1,621 hypomethylated circRNAs at 3d after MCAO. However, the methylation levels increased in 57 circRNAs and decreased in 66 circRNAs at 7d after MCAO. In general, at 3d after operation, the hypomethylated transcripts reached to 95% in the m6A-circRNAs, and only 5% were hypermethylated, while the distribution of these two kinds of transcripts was nearly equal at 7d after MCAO compared to the sham group. Additionally, compared to the 3d group, 2632 circRNAs were hypermethylated, while 186 ones were hypomethylated in the 7d group. The top 10 hyper and hypo-methylated circRNAs with different treatments were listed as Tables 2–4.
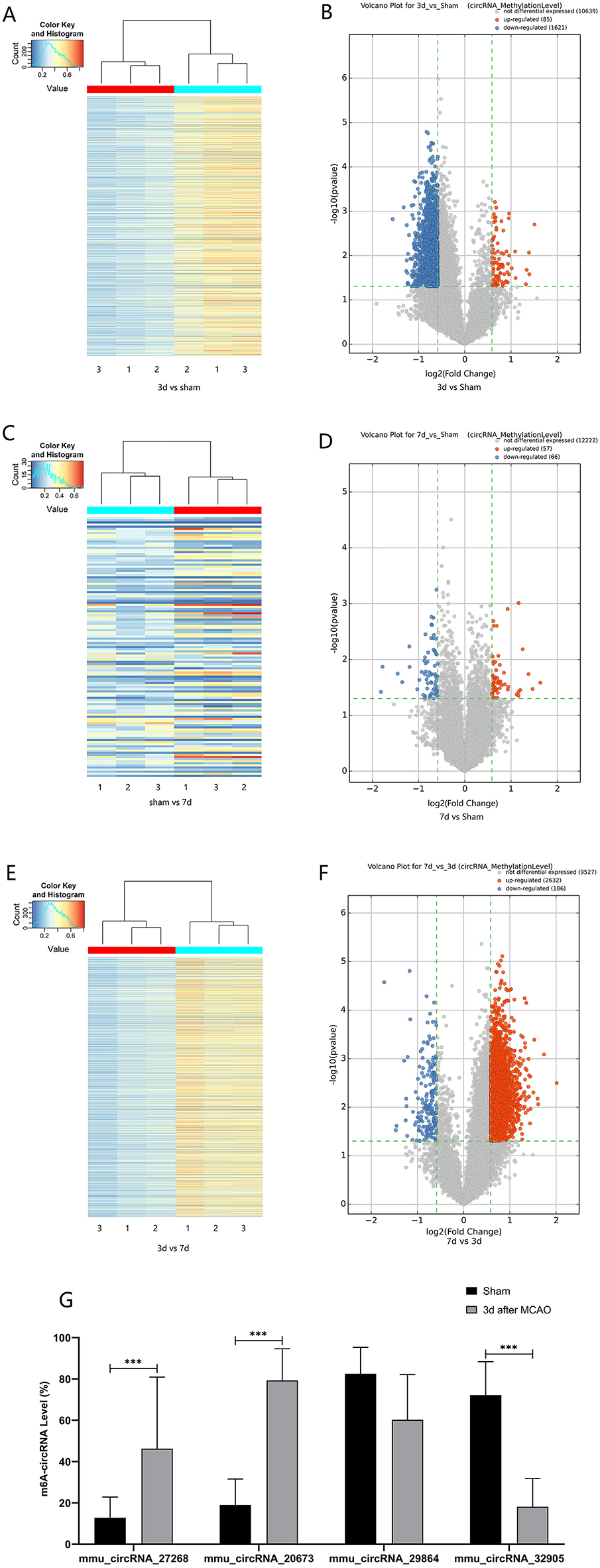
Figure 3. Alteration of m6A methylated-circRNAs. Different methylation modification in circRNAs in 3d (A) or 7d (C) vs. sham and 7d vs. 3d (E) are shown in heatmaps. The volcano plots display differentially m6A-methylated circRNAs in 3d (B) or 7d (D) vs. sham and 7d vs. 3d (F): the green line means the screening criteria with fold change >1.5, red dots represent the hypermethylated circRNAs while green dots represent the hypomethylated ones. (G) The verified results of mmu_circRNA_27268, mmu_circRNA_20673, mmu_circRNA_32905, and mmu_ circRNA_29864 by MeRIP-qPCR in MCAO and sham groups (MeRIP assays, n = 3/group; MeRIP-qPCR, n = 6/group). *P < 0.05, ***P < 0.001; ns, not significant.
As for circRNA expression level, compared with the sham group, numerous circRNAs were differentially expressed, including 163 up-regulated and 289 were down-regulated in the 3d group, while 502 up-regulated and 512 down-regulated in the 7d group. In comparison to the 3d group, 697 circRNAs were significantly increased and 1,012 were decreased in 7 days after MCAO. Besides, we further explored the relation between methylation and expression (Figures 4A–C). We found that in 3d group, 24, 0, 3, and 72 circRNAs were hypomethylated-up expressed, hypermethylated-up expressed, hypermethylated-down expressed, and hypomethylated-down expressed respectively, while turned to 8, 0, 5, 2 ones in 7d group. Compared with the 3d group, there were 63 hypomethylated-up expressed, 14 hypermethylated-up expressed, 448 hypermethylated-down expressed and 3 hypomethylated-down expressed circRNAs in 7d group.
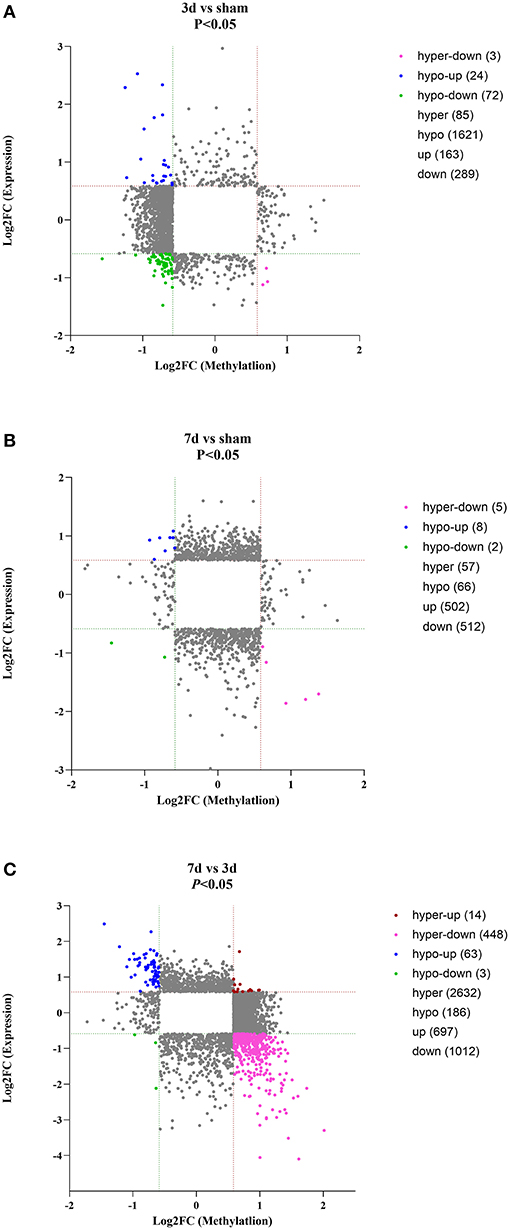
Figure 4. Conjoint analysis for expression and m6A methylation profiles of circRNAs around the infarct cortex in 3d vs. sham (A), 7d vs. sham (B), and 7d vs. 3d (C).
Verification of the Change in m6A Levels of CircRNAs After Cerebral Infarction
Post-stroke alterations of m6A modifications demonstrated by m6A immunoprecipitation and microarray analysis were confirmed by MeRIP-qPCR. In comparison with the sham group, the methylation levels of mmu_circRNA_27268 and mmu_circRNA_20673 were significantly up-regulated in the 3d group, while mmu_circRNA_32905 was hypomethylated. Although the difference in methylation of mmu_ circRNA_29864 was not statistically significant, there was a downward trend. In general, the validation data was consistent with the m6A immunoprecipitation and circRNA microarray analysis (Figure 3G).
Functional Prediction for Parental Genes of Differentially m6A-Modified CircRNAs
To understand the significance of the m6A modification in the pathology of cerebral ischemia, GO and KEGG analyses were performed to compare the parental genes of differentially m6A-modified circRNAs at 3d or 7d after cerebral infarction. The top 10 most significant enrichment scores in biological processes (BP), cellular components (CC), and molecular functions (MF) are displayed in the histograms (Figures 5A–F). With the development of cerebral ischemia, the BP for hypermethylated circRNAs was established for localization in cells and cellular localization at 3 days after ischemia onset, which displayed a transition to meiotic telomere clustering and establishment of the blood-brain barrier at 7 days after MCAO. Analogously, CC showed enrichment of major synapses and cellular junctions at 3d group, which shifted to integral components of the postsynaptic density membrane at 7d group. We observed the involvement of the glutamate receptor, GTPase activator, single-stranded telomeric DNA binding, and phosphotransferase activity in MF. As for the hypomethylated circRNAs, BP, CC, and MF enrichment concentrated mainly on cellular processes, intracellular, and binding at 3 days after MCAO, which changed to protein ubiquitination, ubiquitin ligase complexes, and ubiquitin-like protein ligase activity at 7 days after MCAO.
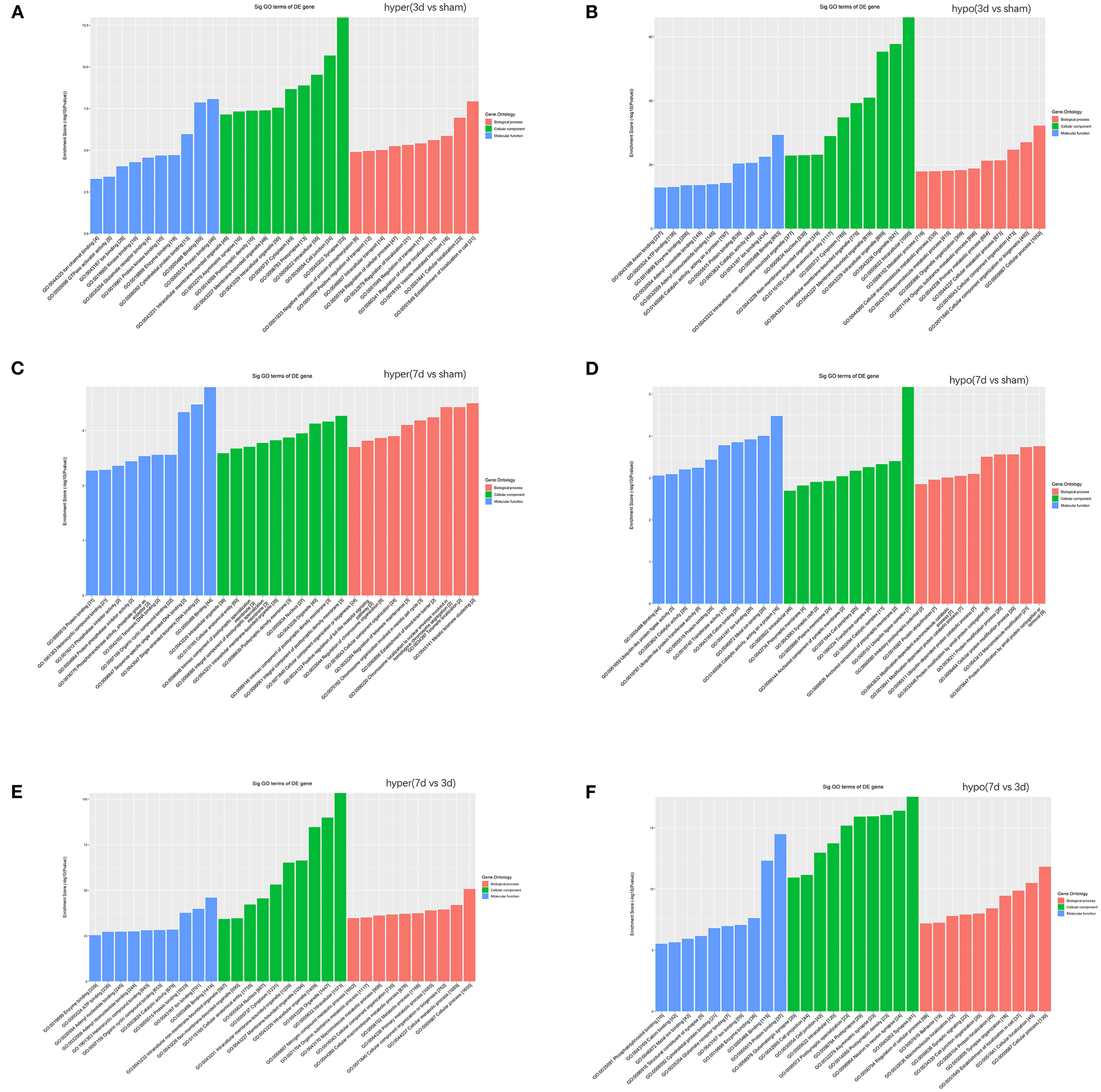
Figure 5. Function analyses of the parent genes of differentially m6A-modified circRNAs. Function analyses of the parent genes of significantly hypermethylated and hypomethylated circRNAs around the infarct cortex in 3d vs. sham (A,B), 7d vs. sham (C,D), and 7d vs. 3d (E,F).
Pathway Prediction for Parental Genes of Differentially m6A-Modified CircRNAs
KEGG analysis demonstrated the most enrichment pathways (Figures 6A–C). Compared with the sham group, various pathways including Hippo, neurotrophin, ubiquitin-mediated proteolysis, and axon guidance signaling were the major enrichment pathways at 3 days after MCAO, then turned into phosphorylated inositol signaling, ubiquitin-mediated proteolysis, and the glutamatergic synapse pathway at 7 days after MCAO. As for differentially m6A-modified circRNAs between 7d and 3d groups, the pathways mainly enriched in adherens junction, phosphatidylinositol signaling system, AMPK signaling pathway, and glutamatergic synapse.
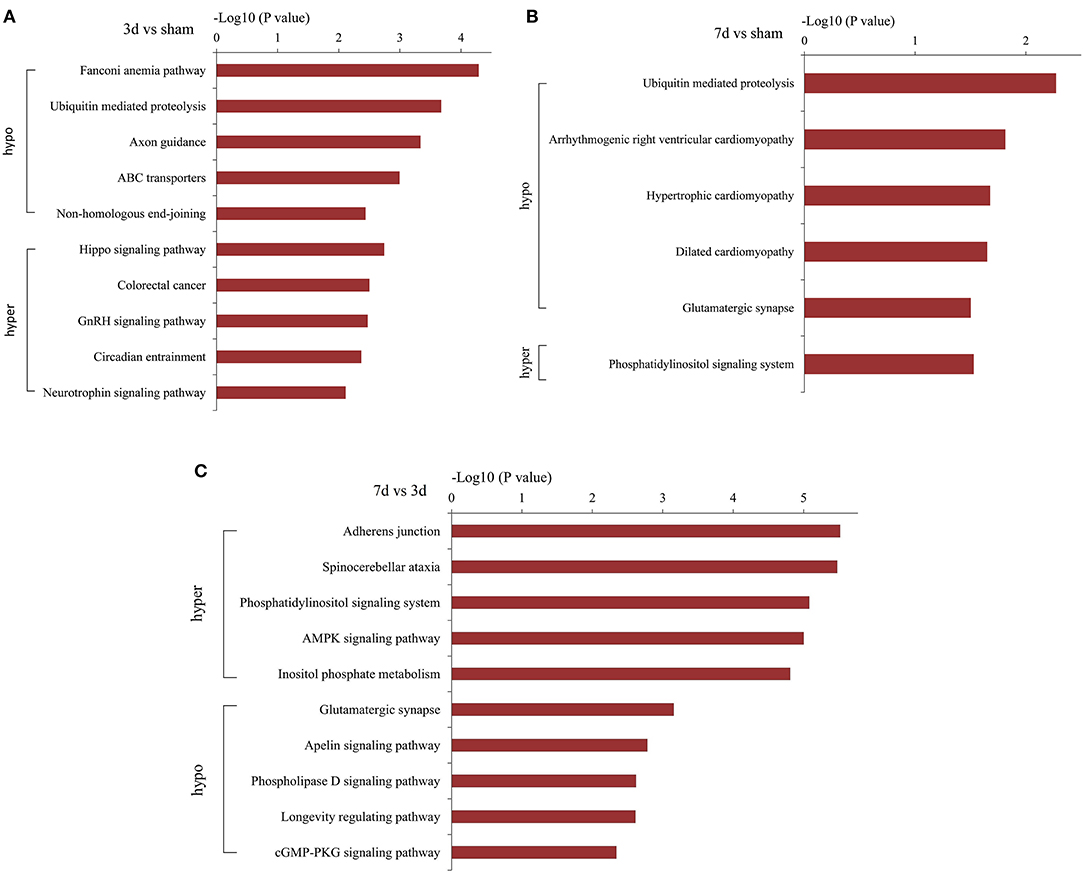
Figure 6. Pathway analyses of the parent genes of differentially m6A-modified circRNAs. Pathway analyses of the parent genes of significantly hypermethylated and hypomethylated circRNAs around the infarct cortex in 3d vs. sham (A), 7d vs. sham (B), and 7d vs. 3d (C).
The Interaction of CircRNA-miRNA-mRNA
Recent evidences have demonstrated that circular RNAs play a crucial role in regulating the level of miRNA-mediated gene expression by sequestering the miRNAs. Their interaction with disease-associated miRNAs indicates that circular RNAs are important for disease regulation. Ten most significant miRNAs were predicted for three verified m6A-circRNAs, and the binding sites were shown in Figure 7. At the same time, we predicted 273 target genes of miRNA and constructed the circRNA-miRNA-mRNA network (Figure 8A). GO analysis indicated that the top ten results of the BP, CC, and MF of target genes were almost related to calcium ion, and the most significant enrichment functions, respectively, were calcium ion transmembrane transport, voltage gated calcium channel complex, and voltage-gated calcium channel activity (Figure 8B). KEGG of target genes revealed the MAPK signaling pathway, focal adhesion, axon guidance, glutamatergic synapse, long-term potentiation, neurotrophin signaling pathway, and PI3K-Akt signaling pathway were mainly enriched pathways (Figure 8C).
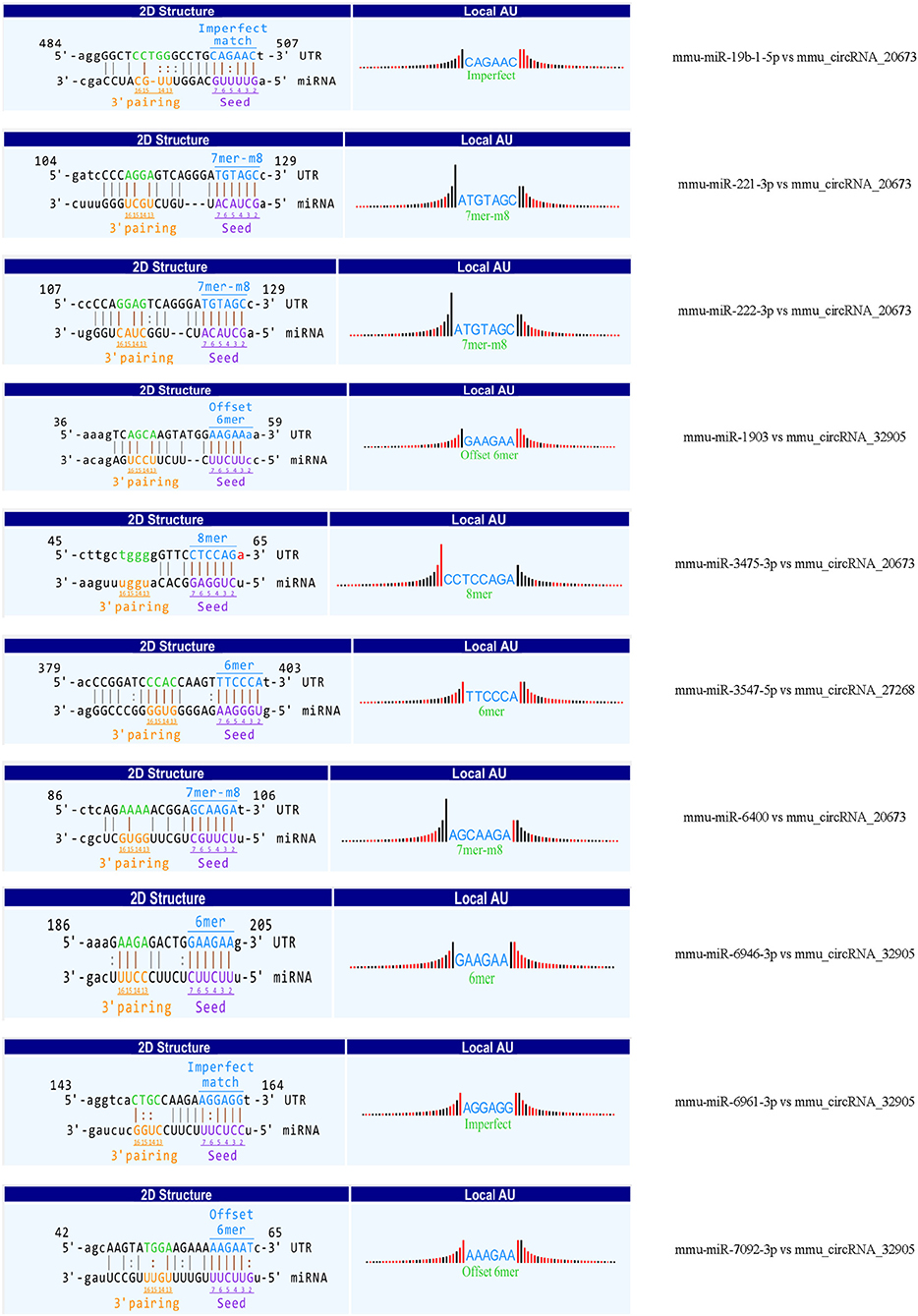
Figure 7. The predicted binding sites between three verified differentially m6A-modified circRNAs and ten miRNAs.
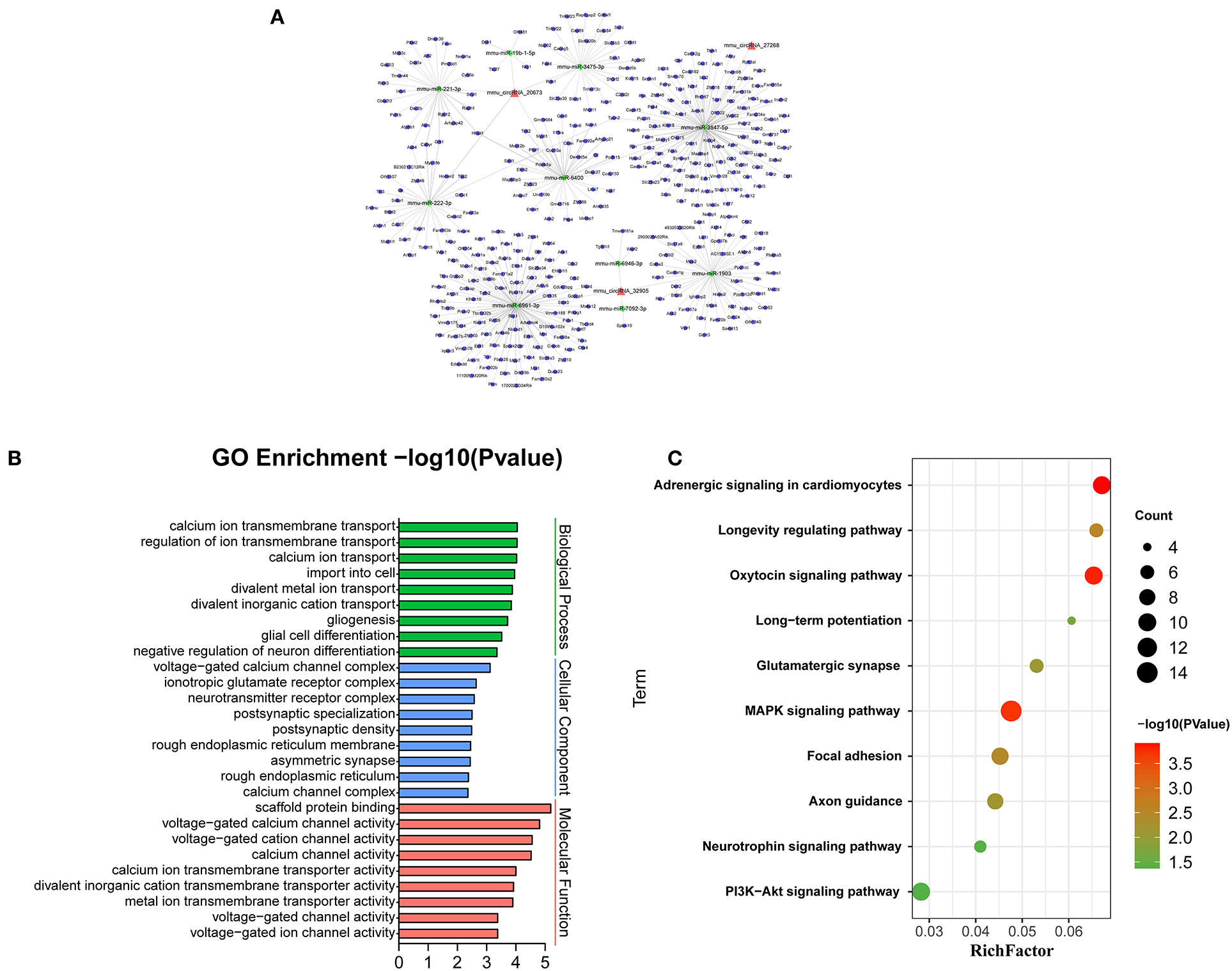
Figure 8. Bioinformatics analysis for target genes of three verified differentially m6A-modified circRNAs. (A) The circRNA-miRNA-mRNA network contains 3 circRNAs, 10 miRNAs, and 273 mRNAs. (B) The top ten most enriched functions of the target genes. (C) The top ten most enriched pathways of target genes.
Discussion
It's reported that different pathophysiological processes occur after 3d and 7d of cerebral infarction (F. Zhang et al., 2019), and we took 3d and 7d as time points to explore the dynamic change of m6A level after cerebral infarction. Our research suggests that stroke alters the methylation status of circRNAs dynamically. To our knowledge, this is the first study to explore methylation of circRNAs around the cerebral infarct region after MCAO in mice. And we found that the mRNA levels of m6A-related enzymes were significantly altered, which were supposed to be a contributing factor of the alteration in methylation of circRNAs. Additionally, we further showed that ALKBH5 and YTHDF3 proteins were changed at different times after stroke. The bioinformatics analysis showed that the differentially methylated circRNAs may act in some pathological molecular events after cerebral ischemia.
Notably, the methylation of circRNAs acts as a regulator of other diseases. It has been reported that decreased m6A abundance of methylated circRNAs can be observed in senile cataract and hypoxia-induced pulmonary hypertension (Li et al., 2020; Su et al., 2020), while the abundance is increased in some cancers (He et al., 2021). Therefore, the pathophysiological process of diseases may be related to an imbalance in the N6-methylation of adenosine in circRNAs. Recent studies have shown that stroke significantly changes the global level of m6A in peri-infarct tissue, and enzymes associated with the m6A modification are believed to mediate post-stroke secondary brain damage by participating in apoptosis and inflammation (Chokkalla et al., 2019; Si et al., 2020; Zhang et al., 2020).
Our results also indicated that the transcripts of these enzymes were differentially expressed between the sham and the MCAO groups, which implies that the homeostasis of m6A may change after stroke. Interestingly, the transcription level of the demethylase FTO decreased significantly at 3 days after MCAO compared with the sham group, while it increased significantly at 7d after MCAO compared to the 3d group. This indicates that FTO is decreased in the short term after stroke, which was confirmed by a recent report (Chokkalla et al., 2019). In the brain, FTO and ALKBH5 are predominantly expressed in neurons and increase dynamically during neural development, leading to the proliferation and differentiation of neural stem cells (Li et al., 2017; Chokkalla et al., 2019; Du et al., 2020). Studies have shown the protective role of FTO in proving brain damage and myocardial inflammation by regulating the m6A level (Chokkalla et al., 2019; Dubey et al., 2022). Another study suggested that FTO can increase lipid levels, which leads to a decrease in adenosine, an important factor that promotes neuronal apoptosis. Therefore, FTO deficiency can lead to adenosine accumulation, resulting in neuronal apoptosis (Gao et al., 2020). Although ALKBH5 has been reported to selectively demethylate BCL2 transcripts, which can prevent the degradation of BCL2 mRNA, enhance the expression of the anti-apoptotic BCL2 protein, and inhibit neuronal apoptosis (Xu et al., 2020), the role of ALKBH5 is less to know. Our study showed that ALKBH5 protein decreased in 3d group but increased in 7d group, suggesting that it could be one of the potential targets for treatment of stroke, and increases in demethylase may improve cerebral infarction by changing the m6A modification. Additionally, YTHDF3 also showed the same trend as ALKBH5, and the latest study showed that YTHDF3 and ALKBH5 were involved in synaptic plasticity (Martinez De La Cruz et al., 2021). However, the specific mechanisms require further study.
Recently, research on m6A has focused on the transcription and post-transcriptional processes of mRNAs. Although great progress has been made with mRNAs (Niu et al., 2013), the mechanisms involved in the m6A methylation in non-coding RNAs, especially circRNAs, are still poorly understood. Limited research has shown that the m6A modification regulates the biogenesis, degradation, stability, translation, and immunogenicity of circRNAs (Chen et al., 2017, 2021; Knuckles and Buhler, 2018; Park et al., 2019; Di Timoteo et al., 2020). CircRNA is produced by selective reverse splicing of primary transcripts, interestingly, the deposition of specific exon regions with m6A was suggested to facilitate this process, resulting in the formation of some circRNAs (Di Timoteo et al., 2020). As an m6A reading protein, YTHDF2 is known as a degradation tool of mRNAs. YTHDF2 was later found to combine with RNase P/MRP to recognize the degradation of circRNAs with the m6A modification (Park et al., 2019). One study showed that circRNAs containing m6A motifs were translated in a cap-independent manner, and translation efficiency was related to m6A levels levels (Knuckles and Buhler, 2018; Di Timoteo et al., 2020). These pieces of evidence show that the m6A modification may act not only on post-transcriptional regulation of mRNAs, but also on circRNAs. Another special regulatory mechanism of m6A modification of circRNAs has been demonstrated. Exogenous circRNAs that activate immune responses are thought to be “circFOREIGN”; however, the m6A modification masks them as “circSELF”, which greatly reduces their immunogenicity modifications can directly change the abundance of circRNAs by regulating their biogenesis, degradation, and stability, as well as alter the characteristics of circRNAs, such as their immunogenicity. These studies have demonstrated the potential functions of m6A-modified circRNAs in disease regulation.
MeRIP combined with microarray analysis revealed that the m6A modification was altered in a considerable number of circRNAs at 3 and 7 days post-cerebral infarction compared with the sham group. In the early stage after cerebral infarction, the majority of circRNAs were hypomethylated, while the circRNAs were hyper-and hypomethylated to same extent in the later stage. Our results indicated that m6A methylation was a dynamic process after stroke. Abnormal methylation of circRNAs was supposed to affect multiple molecular events that lead to secondary injury after ischemia. Numerous circRNAs, such as circTTC3, circCCDC9, and circTLK, have been found to regulate autophagy, nerve regeneration, axonal plasticity, and the blood-brain barrier after cerebral infarction (Wu et al., 2019, 2020; Chen et al., 2020; Yang et al., 2020, 2021), and correcting their dysregulation improved the brain damage.
However, the upstream regulatory mechanisms of these circRNAs are not well-known. Interestingly, N6-methylation of adenosine alters the abundance of circRNAs by regulating their generation and degradation (Park et al., 2019; Di Timoteo et al., 2020). Therefore, changing the methylation status may correct the level of circRNAs and affect their functions in cerebral ischemia. Our conjoint analysis of circRNAs' expression and m6A methylation profiles screened all circRNAs changed in both methylation and expression levels. We found that the differentially expressed circRNAs mainly shows hypomethylation at 3 days after MCAO. However, the trend of methylation and expression seems to be opposite at 7 days after MCAO. The hypermethylated circRNAs were mainly down-expressed, while the hypomethylated ones were up-expressed. Our study indicated that the methylation level and abundance of circRNAs are not the only relationship. Other research also reported that increasing methylation levels can both increase or decrease the expression of circRNAs (Park et al., 2019; Su et al., 2020; Chen et al., 2021). It will be necessary to investigate the specific mechanisms that alter m6A-modified circRNAs in the pathological process after stroke.
CircRNA is derived from the precursor of mRNA. Thus, it can make great significance in pathophysiological process of diseases by regulating its parental gene. To further determine the function of differentially methylated circRNAs around cerebral infarction, we conducted the bioinformatics analysis. Our genomic analysis showed that BP and CC terms were involved in cell localization and intracellular transition at 3 days after MCAO, and shifted to the establishment of the blood-brain barrier and protein ubiquitination at 7 days after MCAO. Moreover, MF term happened the transition of glutamate receptor to phosphotransferase activity. Our results show that the secondary changes after stroke mainly occur from early nerve injury to late brain repair, indicating that differentially methylated circRNAs may regulate these pathological processes. In addition, previous research displayed that cerebral ischemia caused oxidative stress, neurovascular regeneration, and synaptic formation (Mehta et al., 2007). In our study, numerous signaling pathways, including ubiquitin-mediated proteolysis, axon guidance, hippo, neurotrophin, glutamatergic synapse, adherens junction, and phosphatidylinositol signaling, were significantly enriched. These pathways regulate the processes of oxidative stress, nerve regeneration, angiogenesis, synaptic formation, and plasticity after cerebral ischemia (Reichardt, 2006; Ding et al., 2014; Zhao et al., 2016). Glutamate is an excitatory neurotransmitter that mediates excitatory synaptic transmission. During cerebral ischemia and hypoxia, glutamate is released in large quantities, which over-activates glutamate receptors, leads to excessive Ca2+ influx, and causes brain damage and neuronal death (Mayor and Tymianski, 2018), a major cause of secondary brain injury. Therefore, understanding the regulation mechanism of m6A-modified circRNAs in the glutamate signaling pathway might be helpful for better intervention and prognosis and reduces secondary brain injury after stroke.
CircRNA can act as a sponge of miRNA to reduce the inhibitory effect of miRNA on mRNA to promote the expression of mRNA (Wu et al., 2019). Therefore, we selected the verified circRNAs to predict the network of circRNA-miRNA-mRNA. BP, CC, and MF show the predicted mRNAs mainly involve calcium ion transmembrane transport, voltage-gated calcium channel complex, voltage-gated calcium channel activity. Research shows calcium overload occurs in damaged neurons to mediate mitochondrial swelling injury and neuronal necrosis in the peri-ischemic cortex (Pivovarova and Andrews, 2010). It is reported that target genes cacna1g, tpcn2, panx1, and Fyn are involved in pathophysiological processes such as neuroexcitatory conduction, autophagy, inflammation, and synaptic plasticity in cerebral infarction (Shin et al., 2008; Shestopalov and Slepak, 2014; Guo et al., 2020; Rajani et al., 2021). In addition, 10 miRNAs have been predicted in our circRNA-miRNA-mRNA network. At present, little research has focused on these miRNAs. Interestingly, mmu-mir-3547-5 has been reported to play an important role in anti-atherosclerosis, and atherosclerosis is related to the pathogenesis of cerebral infarction (Zhang et al., 2018). In this network, mir-3547-5p and tpcn2 were the downstream targets of mmu_circRNA 27268 and mmu-mir-3547-5p, respectively. Therefore, we speculated that the mmu_circRNA 27268, mir-3547-5p, and tpcn2 may form a network axis, and participate in the pathological process of cerebral infarction by regulating calcium overload.
Conclusion
In summary, this study is the first to show changes of m6A methylation in circRNAs in cerebral infarction. The bioinformatics analysis predicted the possible functions and related pathways of m6A modified-circRNAs in secondary injury and the repair of cerebral infarction. However, understanding how the methylated circRNAs regulate the post-stroke pathophysiological mechanism will require further investigation. Our results provide new insights into the molecular mechanism of ischemia stroke.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE196448.
Ethics Statement
The animal study was reviewed and approved by Institutional Animal Use Committee of the Shenzhen Hospital of Southern Medical University.
Author Contributions
FL and LL conceived and designed the study and interpreted experiments. YL performed the experiments and wrote the manuscript. HL performed the experiments. YL, XL, ZC, and WZ analyzed the data. All authors read and approved the final submission.
Funding
This study was supported by the grants from the Science and Technology Project of Shenzhen (JCYJ20190814112201888), Sanming Project of Medicine in Shenzhen (SZSM201812047), Shenzhen Key Medical Discipline Construction Fund (SZXK074), and Guangdong Basic and Applied Basic Research Foundation (2018A030313586).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Editage for their work.
References
Chen, C., Yuan, W., Zhou, Q., Shao, B., Guo, Y., Wang, W., et al. (2021). N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics 11, 4298–4315. doi: 10.7150/thno.51342
Chen, W., Wang, H., Zhu, Z., Feng, J., and Chen, L. (2020). Exosome-Shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 axis. Mol. Ther. Nucleic Acids 22, 657–672. doi: 10.1016/j.omtn.2020.09.027
Chen, Y. G., Kim, M. V., Chen, X., Batista, P. J., Aoyama, S., Wilusz, J. E., et al. (2017). Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e225. doi: 10.1016/j.molcel.2017.05.022
Chokkalla, A. K., Mehta, S. L., Kim, T., Chelluboina, B., Kim, J., and Vemuganti, R. (2019). Transient focal ischemia significantly alters the m(6)A epitranscriptomic tagging of RNAs in the brain. Stroke 50, 2912–2921. doi: 10.1161/STROKEAHA.119.026433
Di Timoteo, G., Dattilo, D., Centron-Broco, A., Colantoni, A., Guarnacci, M., Rossi, F., et al. (2020). Modulation of circRNA Metabolism by m(6)A Modification. Cell Rep. 31:107641. doi: 10.1016/j.celrep.2020.107641
Ding, Q., Liao, S. J., and Yu, J. (2014). Axon guidance factor netrin-1 and its receptors regulate angiogenesis after cerebral ischemia. Neurosci. Bull. 30, 683–691. doi: 10.1007/s12264-013-1441-9
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Du, T., Li, G., Yang, J., and Ma, K. (2020). RNA demethylase Alkbh5 is widely expressed in neurons and decreased during brain development. Brain Res Bull. 163, 150–159. doi: 10.1016/j.brainresbull.2020.07.018
Dubey, P. K., Patil, M., Singh, S., Dubey, S., Ahuja, P., Verma, S. K., et al. (2022). Increased m6A-RNA methylation and FTO suppression is associated with myocardial inflammation and dysfunction during endotoxemia in mice. Mol. Cell. Biochem. 477, 129–141. doi: 10.1007/s11010-021-04267-2
Gao, H., Cheng, X., Chen, J., Ji, C., Guo, H., Qu, W., et al. (2020). Fto-modulated lipid niche regulates adult neurogenesis through modulating adenosine metabolism. Hum. Mol. Genet. 29, 2775–2787. doi: 10.1093/hmg/ddaa171
Guo, C., Webb, S. E., Chan, C. M., and Miller, A. L. (2020). TPC2-mediated Ca(2+) signaling is required for axon extension in caudal primary motor neurons in zebrafish embryos. J. Cell Sci. 133:jcs244780. doi: 10.1242/jcs.244780
He, R. Z., Jiang, J., and Luo, D. X. (2021). M6A modification of circNSUN2 promotes colorectal liver metastasis. Genes Dis. 8, 6–7. doi: 10.1016/j.gendis.2019.12.002
Knuckles, P., and Buhler, M. (2018). Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett. 592, 2845–2859. doi: 10.1002/1873-3468.13107
Li, L., Zang, L., Zhang, F., Chen, J., Shen, H., Shu, L., et al. (2017). Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 26, 2398–2411. doi: 10.1093/hmg/ddx128
Li, P., Yu, H., Zhang, G., Kang, L., Qin, B., Cao, Y., et al. (2020). Identification and characterization of N6-methyladenosine CircRNAs and methyltransferases in the lens epithelium cells from age-related cataract. Invest. Ophthalmol. Vis. Sci. 61:13. doi: 10.1167/iovs.61.10.13
Martinez De La Cruz, B., Markus, R., Malla, S., Haig, M. I., Gell, C., Sang, F., et al. (2021). Modifying the m(6)A brain methylome by ALKBH5-mediated demethylation: a new contender for synaptic tagging. Mol. Psychiatry 26, 7141–7153. doi: 10.1038/s41380-021-01282-z
Mathiyalagan, P., Adamiak, M., Mayourian, J., Sassi, Y., Liang, Y., Agarwal, N., et al. (2019). FTO-Dependent N(6)-Methyladenosine regulates cardiac function during remodeling and repair. Circulation 139, 518–532. doi: 10.1161/CIRCULATIONAHA.118.033794
Mayor, D., and Tymianski, M. (2018). Neurotransmitters in the mediation of cerebral ischemic injury. Neuropharmacology. 134, 178–188. doi: 10.1016/j.neuropharm.2017.11.050
Mehta, S. L., Manhas, N., and Raghubir, R. (2007). Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res. Rev. 54, 34–66. doi: 10.1016/j.brainresrev.2006.11.003
Niu, Y., Zhao, X., Wu, Y. S., Li, M. M., Wang, X. J., and Yang, Y. G. (2013). N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genomics Proteom Bioinformat. 11, 8–17. doi: 10.1016/j.gpb.2012.12.002
Park, O. H., Ha, H., Lee, Y., Boo, S. H., Kwon, D. H., Song, H. K., et al. (2019). Endoribonucleolytic cleavage of m(6)A-Containing RNAs by RNase P/MRP complex. Mol. Cell. 74, 494–507.e498. doi: 10.1016/j.molcel.2019.02.034
Pivovarova, N. B., and Andrews, S. B. (2010). Calcium-dependent mitochondrial function and dysfunction in neurons. FEBS J. 277, 3622–3636. doi: 10.1111/j.1742-4658.2010.07754.x
Rajani, V., Sengar, A. S., and Salter, M. W. (2021). Src and Fyn regulation of NMDA receptors in health and disease. Neuropharmacology 193:108615. doi: 10.1016/j.neuropharm.2021.108615
Reichardt, L. F. (2006). Neurotrophin-regulated signalling pathways. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1545–1564. doi: 10.1098/rstb.2006.1894
Rybak-Wolf, A., Stottmeister, C., Glazar, P., Jens, M., Pino, N., Giusti, S., et al. (2015). Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell 58, 870–885. doi: 10.1016/j.molcel.2015.03.027
Shestopalov, V. I., and Slepak, V. Z. (2014). Molecular pathways of pannexin1-mediated neurotoxicity. Front. Physiol. 5:23. doi: 10.3389/fphys.2014.00023
Shi, H., Wei, J., and He, C. (2019). Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650. doi: 10.1016/j.molcel.2019.04.025
Shigeoka, T., Jung, H., Jung, J., Turner-Bridger, B., Ohk, J., Lin, J. Q., et al. (2016). Dynamic axonal translation in developing and mature visual circuits. Cell 166, 181–192. doi: 10.1016/j.cell.2016.05.029
Shin, H. S., Cheong, E. J., Choi, S., Lee, J., and Na, H. S. (2008). T-type Ca2+ channels as therapeutic targets in the nervous system. Curr. Opin. Pharmacol. 8, 33–41. doi: 10.1016/j.coph.2007.12.003
Si, W., Li, Y., Ye, S., Li, Z., Liu, Y., Kuang, W., et al. (2020). Methyltransferase 3 mediated miRNA m6A methylation promotes stress granule formation in the early stage of acute ischemic stroke. Front. Mol. Neurosci. 13:103. doi: 10.3389/fnmol.2020.00103
Su, H., Wang, G., Wu, L., Ma, X., Ying, K., and Zhang, R. (2020). Transcriptome-wide map of m(6)A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics 21:39. doi: 10.1186/s12864-020-6462-y
Weng, Y. L., Wang, X., An, R., Cassin, J., Vissers, C., Liu, Y., et al. (2018). Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron 97, 313–325.e316. doi: 10.1016/j.neuron.2017.12.036
Wu, F., Han, B., Wu, S., Yang, L., Leng, S., Li, M., et al. (2019). Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J. Neurosci. 39, 7369–7393. doi: 10.1523/JNEUROSCI.0299-19.2019
Wu, L., Xu, H., Zhang, W., Chen, Z., Li, W., and Ke, W. (2020). Circular RNA circCCDC9 alleviates ischaemic stroke ischaemia/reperfusion injury via the Notch pathway. J. Cell. Mol. Med. 24, 14152–14159. doi: 10.1111/jcmm.16025
Xu, K., Mo, Y., Li, D., Yu, Q., Wang, L., Lin, F., et al. (2020). N(6)-methyladenosine demethylases Alkbh5/Fto regulate cerebral ischemia-reperfusion injury. Ther. Adv. Chronic Dis. 11:2040622320916024. doi: 10.1177/2040622320916024
Yang, B., Zang, L., Cui, J., and Wei, L. (2021). Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372-3p/TLR4 axis in cerebral infarction. Stem Cell Res. Ther. 12:125. doi: 10.1186/s13287-021-02187-y
Yang, L., Han, B., Zhang, Z., Wang, S., Bai, Y., Zhang, Y., et al. (2020). Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation 142, 556–574. doi: 10.1161/CIRCULATIONAHA.120.045765
You, X., Vlatkovic, I., Babic, A., Will, T., Epstein, I., Tushev, G., et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610. doi: 10.1038/nn.3975
Zhang, F., Zhao, Q., Jiang, Y., Liu, N., Liu, Q., Shi, F. D., et al. (2019). Augmented brain infiltration and activation of leukocytes after cerebral ischemia in type 2 diabetic mice. Front. Immunol. 10:2392. doi: 10.3389/fimmu.2019.02392
Zhang, Y., Ma, X., Li, X., Zhang, T., Qin, M., and Ren, L. (2018). Effects of Icariin on atherosclerosis and predicted function regulatory network in ApoE deficient mice. Biomed. Res. Int. 2018:9424186. doi: 10.1155/2018/9424186
Zhang, Z., Wang, Q., Zhao, X., Shao, L., Liu, G., Zheng, X., et al. (2020). YTHDC1 mitigates ischemic stroke by promoting Akt phosphorylation through destabilizing PTEN mRNA. Cell Death Dis. 11:977. doi: 10.1038/s41419-020-03186-2
Keywords: circRNA, N6-methyladenosine (m6A), m6A-related methyltransferases, middle cerebral artery occlusion, cerebral infarction
Citation: Li Y, Li H, Luo Y, Li X, Chen Z, Zhang W, Li F and Ling L (2022) The Alteration Profiles of m6A-Tagged circRNAs in the Peri-Infarct Cortex After Cerebral Ischemia in Mice. Front. Neurosci. 16:869081. doi: 10.3389/fnins.2022.869081
Received: 03 February 2022; Accepted: 02 May 2022;
Published: 07 June 2022.
Edited by:
Divya Jha, Icahn School of Medicine at Mount Sinai, United StatesReviewed by:
Xiaolin Qu, Shanghai Changzheng Hospital, ChinaPraveen K. Dubey, University of Alabama at Birmingham, United States
Copyright © 2022 Li, Li, Luo, Li, Chen, Zhang, Li and Ling. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Ling, bGluZ2xpcmFiYml0QDE2My5jb20=; Fangming Li, bGlmbHkwNTA0MTNAMTYzLmNvbQ==
 Yudi Li
Yudi Li Hanzhao Li1
Hanzhao Li1 Yang Luo
Yang Luo Li Ling
Li Ling