
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Neurosci. , 25 March 2022
Sec. Neurodevelopment
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.848648
This article is part of the Research Topic Changes in Metabolic Processes Affecting Brain Development View all 5 articles
Decades of research have unequivocally demonstrated that fetal exposure to both recreational and prescription drugs in utero negatively impacts the developing brain. More recently, the application of cutting-edge techniques in neurodevelopmental research has attempted to identify how the fetal brain responds to specific environmental stimuli. Meanwhile, human fetal brain studies still encounter ethical considerations and technical limitations in tissue collection. Human-induced pluripotent stem cell (iPSC)-derived brain organoid technology has emerged as a powerful alternative to examine fetal neurobiology. In fact, human 3D organoid tissues recapitulate cerebral development during the first trimester of pregnancy. In this review, we aim to provide a comprehensive summary of fetal brain metabolic studies related to drug abuse in animal and human models. Additionally, we will discuss the current challenges and prospects of using brain organoids for large-scale metabolomics. Incorporating cutting-edge techniques in human brain organoids may lead to uncovering novel molecular and cellular mechanisms of neurodevelopment, direct novel therapeutic approaches, and raise new exciting questions.
Recent data indicates that in the last year nearly 275 million people used illicit drugs and over 36 million people are suffering from drug abuse worldwide (United Nations Office on Drugs and Crime, 2021). As the number of drug users increases globally, in the United States, nearly 53 million Americans (19.4% of people over the age of 12) have consumed an illicit drug in the last year (National Center for Drug Abuse Statistics, 2022). Illicit drugs include marijuana/hashish, crack/cocaine, heroin, hallucinogens, inhalants, or illegally obtained prescription psychotherapeutics (Substance Abuse and Mental Health Services Administration, 2009). Among pregnant women, the use of non-illicit drugs such as alcohol and tobacco appeared to stabilize at ∼5.9% reporting use of both of these drugs in 1 year, 8.5% consumed alcohol, and 15.9% smoked cigarettes (Smith, 2021). Taken together, this resulted in over one million offspring that were exposed to an illicit drug, alcohol, or tobacco in utero (Smith, 2021). Additionally, over 40% of the women enrolled in the Infant Development, Environment, and Lifestyle (IDEAL) study reported continuous drug consumption into the third trimester, half of whom did not change their personal use during the entire pregnancy (Della Grotta et al., 2010). Illicit substance abuse has been linked to fetal growth restriction, stillbirth, preterm birth, and neonatal intensive care unit admission (Ross et al., 2015). However, identifying the specific molecular and cellular neurodevelopmental mechanisms underlying these outcomes is challenging. In addition, prenatal drug exposure studies are often confounded by multiple variables including multidrug usage, poverty, and a small number of participants. Thus, most neurodevelopmental outcomes are studied in adult/teenage participants, animal models, and 2D cell cultures. While many of the factors expressed in an adult brain or a neuronal culture are also present in progenitor cells, precise molecular mechanisms by which different illicit drugs shape the neurodevelopmental landscape of a fetal brain are extremely difficult to track with current approaches.
Historically, animal models have provided great insights into the neurodevelopmental consequences of prenatal drug exposure. Marijuana has become the most commonly abused illicit drug during pregnancy in the United States (Hurd et al., 2019). Previous studies in animal models demonstrated that exposure to marijuana during development may predispose children to psychiatric disorders by altering the endocannabinoid (eCB) system, which is key for the proper wiring of the brain (Harkany et al., 2007; Wu et al., 2011; Torii et al., 2017). Additionally, in utero exposure to TΔ-9-tetrahydrocannabinol (THC, the active ingredient in marijuana) can alter the general responsiveness of the mesolimbic dopamine system. Rats exposed to THC in utero exhibited changes in the ratio of excitatory and inhibitory inputs onto dopamine neurons (Frau et al., 2019; Hurd et al., 2019).
Meanwhile, the effects of in utero exposure to opioids on the developing brain largely depend on the type of opioid, dose, and timing of the exposure. Most commonly studied opioids include: morphine, methadone, buprenorphine, and oxycodone (Byrnes and Vassoler, 2018). Rat models of opioid exposure exhibited increased spine density and dendritic length during early development in response to morphine (Ricalde and Hammer, 1990) while changes in myelination and axon length were observed under buprenorphine and methadone treatment (Sanchez et al., 2008; Wu et al., 2014). Similarly, a series of studies reported that prenatal opioid exposure in rats caused significant changes in neurotransmitter content and post-synaptic activity (Robinson et al., 1996; Robinson, 2000, 2002).
Non-illicit drugs such as nicotine and alcohol are also commonly abused and have profound effects on the developing brain. Nicotine binds to nicotinic acetylcholine receptors (nAChR) acting as a neurotransmitter and powerful neuromodulator. Prenatal mice exposed to nicotine showed reductions in both cingulate cortex volume and dopamine turnover (Zhu et al., 2012). Meanwhile, in developing rats, nicotine led to deficits in dendritic branching, dendritic length, and spine density (Muhammad et al., 2012). Similarly, Drosophila melanogaster larvae developmentally exposed to nicotine displayed alterations in the dopaminergic system and brain size (Morris et al., 2018).
Animal model systems have also been critical in establishing the causal relationship between ethanol exposure and developmental brain changes. Perhaps surprisingly, prenatal ethanol exposure is not only affecting the offspring but bears a transgenerational neurodevelopmental impact (Abbott et al., 2018). Prenatal exposure to ethanol in mice also resulted in ectopic intracortical connectivity, decreased global DNA methylation levels (Badanich et al., 2013), and enhanced intrinsic excitability, glutamatergic synaptic signaling, neural progenitor cell survival, and proliferation (Nimitvilai et al., 2016). Rats prenatally exposed to ethanol showed reduced spine density, altered dendritic branching, and increased soma size in the medial prefrontal cortex (King et al., 1988), a brain region known to exert the highest baseline metabolic activity and implicated in the regulation of attention, habit formation, and working memory.
In addition to structural and functional changes observed in animal models of prenatal drug exposure, a growing number of studies explored the brain metabolome. In rats, alteration in one-carbon (C1) metabolism was detected after fetal ethanol treatment (Ngai et al., 2015). C1 metabolism was specifically enhanced by increasing homocysteine and methionine concentrations in offspring plasma. C1 metabolism activity is necessary for healthy brain development during pregnancy and is supported by micronutrients such as methionine, choline, vitamin B12, betaine, and folate (Chen et al., 2020). The precise role of biochemical reactions that support the methylation of molecules involved in metabolic activity in brain development requires further investigation. This is an important point given that gene expression is deeply influenced by DNA methylation. In adult rats exposed to alcohol in utero, genes involved in the hypothalamus pituitary adrenal (HPA)-axis displayed increased gene or promoter methylation as compared to untreated rats (Ngai et al., 2015).
In other animal models, monkeys exposed to ethanol in utero exhibited elevated adrenocorticotropic hormone (ACTH) and cortisol (CORT), leading to increased neonatal irritability (Kraemer et al., 2008). It was also demonstrated that monkeys exposed to alcohol in middle-to-late gestation had lower primary serotonin levels and dopamine metabolites -5-hydroxyindoleacetic acid (5-HIAA) and homovanillic acid (HVA) (Schneider et al., 2011). These findings raise the question of whether abnormal serotonin biological pathways could be responsible for some of the psychiatric conditions associated with fetal alcohol spectrum disorder.
Prenatal exposure to nicotine has also been reported to cause alterations in developing brain metabolism (McCarthy et al., 2018). In a mouse model, chronic cigarette smoke exposure was linked to significant differences in amino acid, purine, lipid, fatty acid, and steroid metabolite levels as compared to unexposed offspring (Cruickshank-Quinn et al., 2014). Additionally, whereas 60% of the metabolite changes were reversible, 40% of metabolites remained altered (including nicotine metabolites) despite 2 months of smoking cessation. Taken together, these animal studies highlight the need to explore the role of dysregulated metabolites in human brain development during fetal drug exposure (Byrnes and Vassoler, 2018). Efforts in this research direction may lead to the identification of novel biomarkers or underappreciated pathogenic pathways.
Although animal models have provided important insights into our understanding of fetal neurodevelopment, amid the similarities of rodent and human brain development, significant differences exist. Humans and rodents share substantial stages of brain development including initial neocortical development, excitatory and inhibitory neuron development, and circuit formation (Robinson et al., 1996; Zhang et al., 2020). Although the developing nervous system is highly conserved, the trajectory and timing of differentiation, as well as maturation of transmitters and receptors, differs between humans and rodents. Additionally, while neurogenesis in humans occurs around 4.5 months of pregnancy (Silbereis et al., 2016), neurogenesis in mice is seen between embryonic days 9–18 (La Manno et al., 2020). As for prenatal substance exposure, the effects of maternal nicotine treatment on fetal neurogenesis was characterized and well described using mouse models (Zhang et al., 2020). However, in contrast to the human brain, the mouse brain exhibits lissencephaly, lacks the large pool of neural stem cells (NSCs) seen in humans, and mouse NSCs exhibit lower number of divisions before differentiation (Sarieva and Mayer, 2021).
A shift from animal to human models of in utero drug exposure has been previously attempted, however, these studies encountered several limitations. Primarily, human studies relied on imaging techniques such as MRI and fMRI (Roussotte et al., 2010; Nygaard et al., 2018), which capture gross brain changes. Additionally, due to the association of socioeconomic status and drug use, studies involving human subjects suffer from participant retention that possibly confounds results (Peterson et al., 2020). An alternative approach to understanding human brain disorders has been the use of human post-mortem tissue. However, post-mortem studies have offered limited insight into the time course of human brain development as well as how prenatal substance exposure impacts gross anatomical and molecular-cellular changes. More importantly, the role of metabolism in human brain development under specific conditions remains enigmatic. One key study demonstrated prenatal cannabis exposure decreased levels of the opioid peptide precursor preproenkephalin (PENK) in the caudal putamen (Wang et al., 2006). Intriguingly, nicotine and alcohol exposure had no effects on opioid peptide precursors (Wang et al., 2006). The main limitations human post-mortem drug studies encounter are referral, ascertainment bias (Peterson et al., 2020), and circumstances of death that often mask the molecular effects of treatment. Furthermore, in fetal development investigations, the primary method of tissue collection is from voluntary saline abortions which involve varying protocols that likely influence the metabolomic end results and highlight the need for more standardized models.
Recent advances in genomics, genetics, and imaging fields, support the importance of shifting from animal and post-mortem studies to more human-descriptive models of brain development research (Linsley et al., 2019). To do so, scientists have employed human embryonic stem cells (ESCs), human induced pluripotent stem cells (iPSCs) and brain organoids, which have proven as models that most faithfully and uniformly recapitulate human specific cerebral development and disease mechanisms. Stem cell-derived brain organoids demonstrate excellent potential to investigate development, disease, signaling pathways, drug toxicity, and the efficacy of various therapeutic agents. It is known that these organoids recapture organ microanatomy and mimic multicellular function at least to a certain extent. However, the field of metabolism in brain organoids is in its infancy, and understanding the role of brain metabolism in neurodevelopment and disease will require thorough evaluation.
To date, studying human brain development using fetal tissues has posed ethical considerations and experimental difficulties. One prominent technical challenge in studying fetal brain development is that fetal neocortical organotypic slice cultures can only be maintained for up to 3 weeks, thus precluding long-term interventions (Eiraku et al., 2008). Ethically, obtaining human fetal tissue for research is allowed in a very small number of cases (Sarieva and Mayer, 2021). The generation of human iPSCs from somatic cells has provided the unique opportunity to study human brain development as well as opened new avenues for precision medicine using personalized cell therapy. iPSCs retain the donor genome, may regenerate indefinitely, and undergo differentiation into virtually any cell type of interest using well-established protocols. Hence, iPSCs are an ideal system for human disease modeling. Three-dimensional (3D) culturing methods, such as the cerebral organoid technique, further enhance the utility of iPSCs to study human brain development and neurodevelopment disease (Figure 1). Using fine-tuned culturing conditions, human iPSCs spontaneously differentiate, self-assemble, and exhibit characteristics of the primitive neurepithelium at the organoid stage (Eiraku et al., 2008; Kadoshima et al., 2013).
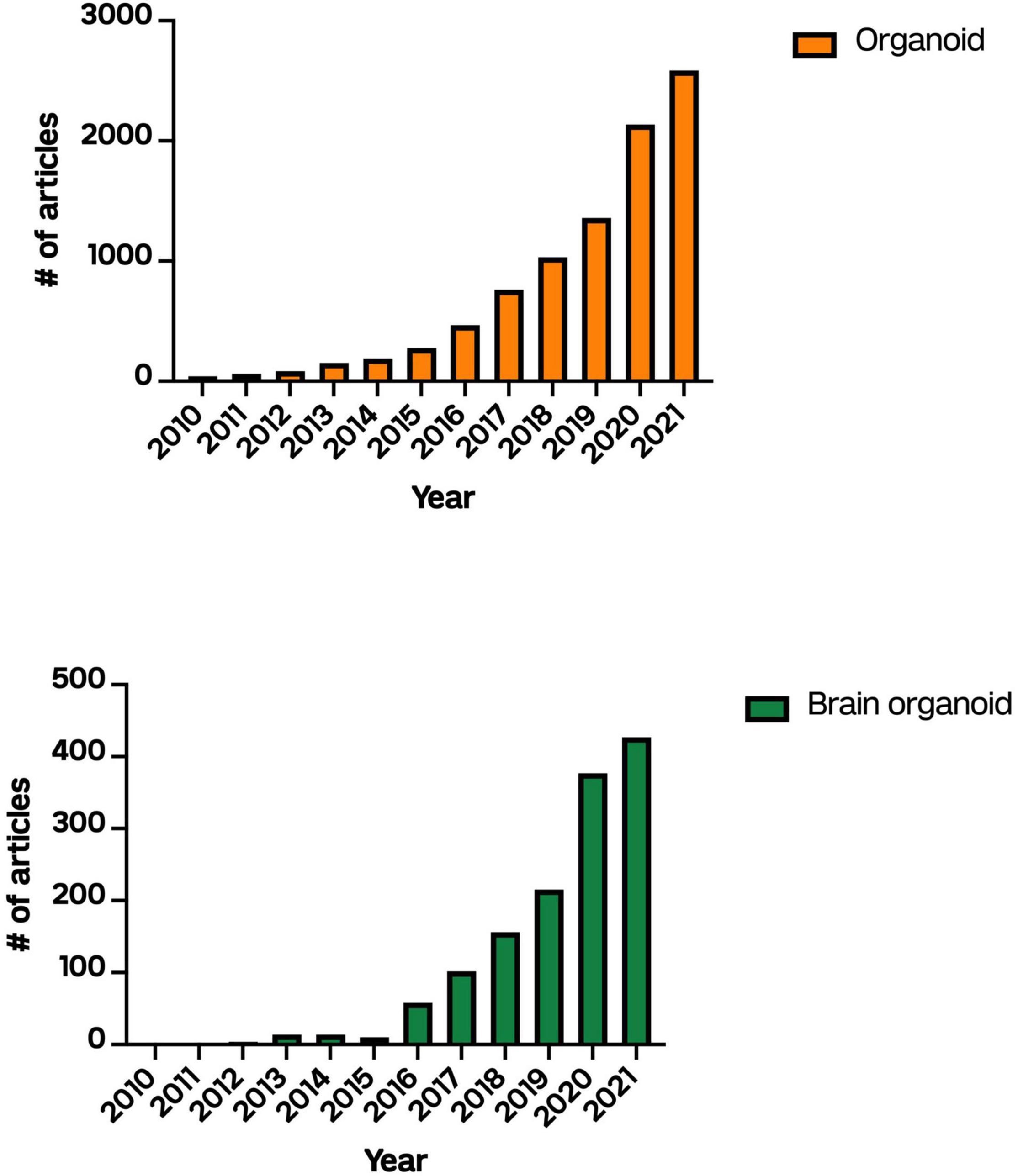
Figure 1. Number of articles published describing “organoids” or “brain organoids” on PubMed between 2010 and 2021.
Previous studies have shown that human iPSC-derived brain organoids mimic the developing human cerebral cortex, as they share transcriptomic, proteomic, and cellular similarities, making human brain organoids an excellent model for studying the early stages of human brain development, e.g., the first trimester of pregnancy (Lancaster et al., 2013; Lancaster and Knoblich, 2014). Of note, numerous studies have shown that 3D organoid models contain both neuronal progenitor and early-born neuron populations, as seen during early cortical development (Velasco et al., 2019; Tanaka et al., 2020), and mature neurons and astrocytes (Pasca et al., 2015; Quadrato and Arlotta, 2017). They also showcase cellular architecture observed during early corticogenesis and exhibit calcium activity (Qian et al., 2020). In general, generating 3D brain organoids relies on unguided or guided methods dictated by the scientific question of interest. Unguided differentiation of brain organoids relies on intrinsic morphogens and signaling of human iPSCs to generate brain organoids with a variety of cell lineages seen in the dorsal and ventral forebrain, midbrain, hindbrain, hippocampus, retina, choroid plexus and others (Lancaster et al., 2013; Lancaster and Knoblich, 2014; Quadrato and Arlotta, 2017). Guided differentiation utilizes small molecules and patterning factors that promote specific cell lineages to form structures resembling discrete brain regions such as cerebral cortex, cerebellum, midbrain, thalamus, hypothalamus, and others (Kadoshima et al., 2013; Yoon et al., 2019; Xu and Wen, 2021). While unguided differentiation provides opportunities to study brain region interactions, it displays high variability across organoid batches. On the other hand, guided differentiation benefits from less variation in batches and cell lines at the cost of limiting the heterogeneity observed in the self-organizing developing brain.
Regardless of the method of differentiation, recent studies have reported that spontaneous excitatory post-synaptic currents can be detected in organoid cultures starting at 4 months in vitro (Li et al., 2017). Moreover, organoid models are also suitable to investigate neuronal network formation at early or late stages of brain development and exhibit spontaneous network oscillations similar to those observed in pre-term human electroencephalography (Trujillo et al., 2019).
In the last 2 years, brain organoids have revolutionized how we study the developmental effects of prenatal drug exposure. To date, several reports (Wang et al., 2018; Ao et al., 2020; Dang et al., 2021; Notaras et al., 2021) have been published establishing human brain organoids as a valuable model system to identify the impact of prenatal drugs on human brain development.
Notaras et al. (2021) aimed to determine prenatal signatures that arise from narcotic use and environmental risk factors in human iPSC-derived forebrain organoids. Using this human-derived model, they studied exposure to opiates (μ-opioid agonist endomorphin), cannabinoids (WIN 55,212-2), alcohol (ethanol), smoking (nicotine), chronic stress (human cortisol), and maternal immune activation (human IL17a). Of the mimetics tested, the cannabinoid agonist WIN 55,212-2 was shown to significantly impact normal cortical development by inducing DNA fragmentation and increasing cell death in organoids. At a metabolic level, all treatment groups showed differential expression of L-phenylalanine. This is of note, as previous studies in rats have shown that phenylalanine can induce neuronal death (Li et al., 2010; Zhu et al., 2017) as well as contribute to oxidative stress in the developing brain (Fernandes et al., 2010). Additionally, organoids that were treated with ethanol, endomorphin, and nicotine exhibited differential expression of GTP. Hydrolysis of GTP into 7,8-DHNP-3′-TP is required for the biosynthesis of numerous monoamine neurotransmitters (Notaras et al., 2021) and GTPase activators are ubiquitously expressed during corticogenesis. One can postulate that differential expression of GTP upon drug enviromimetic treatment is likely to hold important implications for cortical development and synaptic plasticity later in life. Taken together, this study is pivotal for both uncovering novel cellular mechanisms of prenatal drug action and establishing 3D brain organoids as a system for evaluating the effects of illicit and non-illicit drugs on the developing brain.
Two other studies linked novel pathways to methamphetamine (METH) and ethanol prenatal exposure. Dang et al. (2021) combined 10-month-old ESC-derived organoids with single-cell RNA sequencing technologies to evaluate the effects of prenatal METH exposure on the developing brain. A robust transcriptional response within astroglial cell types in response to METH treatment was observed. Metabolically, METH treatment disrupted cAMP signaling and glutamate regulation in organoids. Intriguingly, enhanced IL-8, IL-6, GFAP, and NLRP levels were detected that ultimately elicited gliosis, neuroinflammation, and neurotoxicity in cerebral organoids (Dang et al., 2021). To our knowledge, this study was the first report describing how METH treatment elicits neuroinflammation in a human-derived model of the developing brain. Similarly, Zhu et al. (2017) modeled prenatal ethanol exposure (PAE) in vitro using brain organoids. Zhu et al. observed that upon ethanol exposure brain organoids displayed attenuated neurite outgrowth, disrupted neural maturation, and altered gene expression involved in organogenesis, synaptic plasticity, neural transmission, stem cell proliferation, and differentiation. This study was also the first to link GSX2, RSPO2, and the Hippo signaling pathway to ethanol-induced impaired neurogenesis (Zhu et al., 2017).
Another novel approach is combining brain organoid techniques with microfluidic device technology to evaluate the effects of prenatal drugs on the developing brain. Wang et al. (2018) took this approach to study the effects of nicotine exposure on brain development, while Ao et al. (2020) perfused brain organoids with THC to evaluate the impact of cannabinoids on corticogenesis. In a brain organoid model of nicotine exposure, another group observed abnormal neuronal differentiation and migration suggesting nicotine elicits impaired neurogenesis during early fetal brain development in humans. Meanwhile, in the presence of THC, cerebral organoids exhibited reduced neuronal maturation, downregulation of cannabinoid receptor type 1 (CB1) receptors, impaired neurite outgrowth, and decreased spontaneous firing (Wang et al., 2018).
Taken together, these studies pioneered the approach of using brain organoids for screening commonly abused drugs during pregnancy. While the cellular mechanisms, signaling pathways, and metabolites identified in developing brain models are toxin-specific, drug screens show an effect on neurons, astrocytes, and other glial cells. Neurons often displayed impaired neurogenesis, maturation, and migration, while studies evaluating other cell types described cellular toxicity and inflammation (Figure 2).
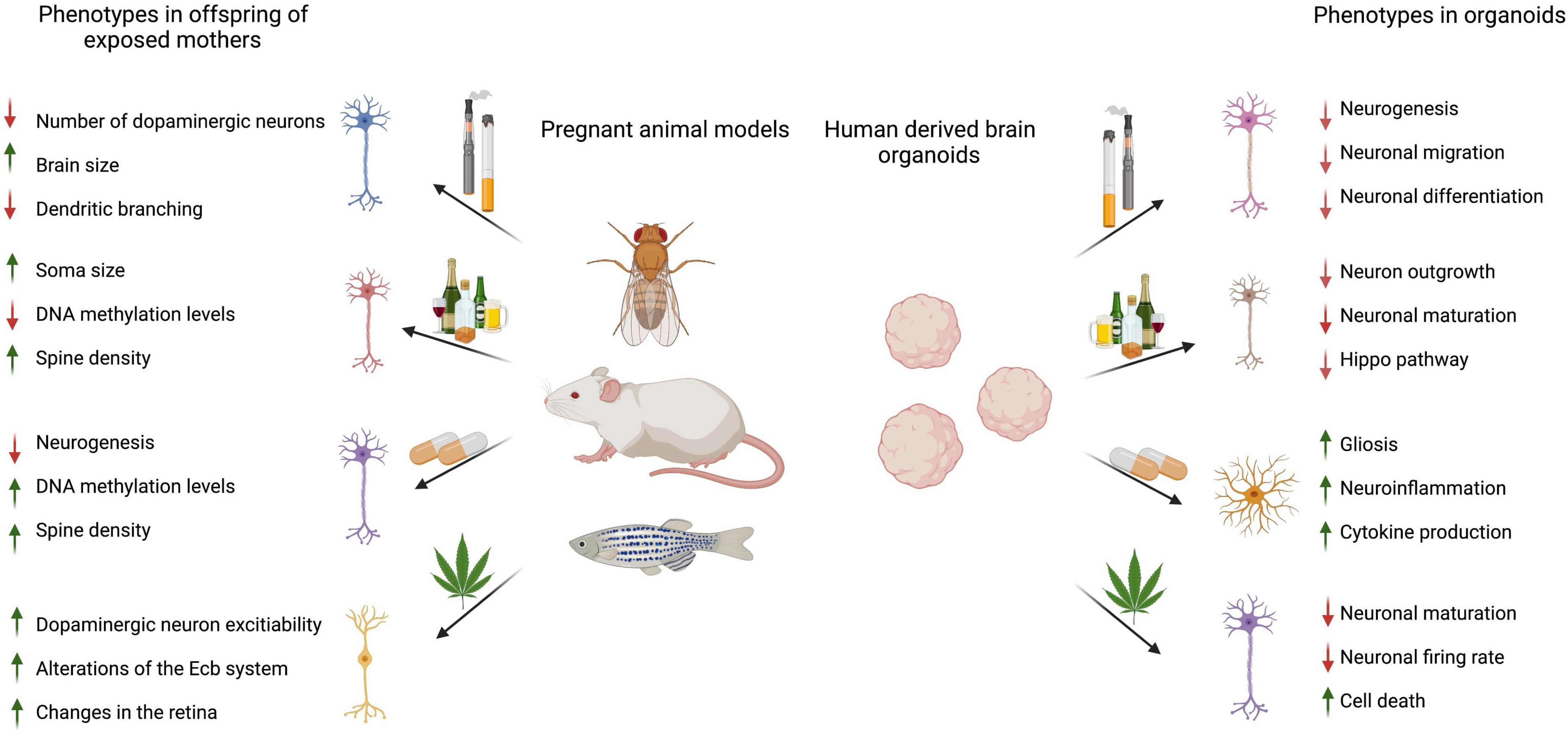
Figure 2. Summary of referenced animal and brain organoid studies describing the effects of prenatal drugs on offspring brains or 3D brain organoids. Image created using BioRender.com.
Three-dimensional brain organoids are slowly emerging as the leading model for studying human neurodevelopment and hold promise in advancing our understanding of fetal neurobiology in health and disease. Due to their small size and short generation time, researchers can produce hundreds to thousands of organoids from relatively small batches of stem cells. Using human-derived tissue allows for rapid drug screening of candidate therapeutics that may have been incorrectly discarded in animal trials. Importantly, the broader application of organoid technology outside of the academic setting has several challenges and limitations that will need to be resolved (Chen et al., 2019). One limitation is the batch-to-batch variability of cerebral organoids, despite derivation from cells of the same donor. Methods to enhance cellular diversity in organoids will also need to be developed as the technology advances. In this regard, groups have managed to grow organoids for long periods of time to promote the generation of various cell types (Stratoulias et al., 2019; Sun et al., 2021). However, certain brain-specific cell types such as microglia remain challenging to populate in cerebral organoids (Stratoulias et al., 2019). Brain organoids also cannot fully reproduce the spatial organization of neurons observed in the human brain. Nevertheless, neurons in organoids display various organizational structures that resemble the cortical plate (Lancaster et al., 2013; Lancaster and Knoblich, 2014). In addition, recapitulating the interaction of neurons and the blood-brain barrier in the organoid system requires further exploration. Thus, we can postulate that brain organoids are better suited to model specific neurodevelopmental diseases and facilitate the testing of compounds that do not depend on distinct cortical plate organization.
As novel technologies continue to emerge, interdisciplinary approaches that combine brain organoids with microfluidic chip devices are in sight. Such organoid-on-a-chip model has provided a promising platform to interrogate neurodevelopmental disorders under environmental exposure (Wang et al., 2018; Ao et al., 2020). Moreover, the organ-on-a-chip strategy offers advantages including relatively low cost, precise control of the microenvironment (i.e., external stimuli and mechanical fluidic cues), and tissue longevity. Successful applications of organ-on-a-chip models have been highlighted in liver, lung, and hear tissues (Huh et al., 2010; Ribas et al., 2016; Moradi et al., 2020). Meanwhile, the extension of organ-on-a-chip in neurodevelopmental disease and drug screening is on the horizon. Additionally, with the explosion of omics techniques, solving a decade-long question on the exact molecular alterations caused by drug exposure during fetal brain development is becoming attainable. 3D brain organoids represent the most faithful and ethical model of human development to date, and combining this approach with omics techniques will only begin to scrape the surface of human brain development in health and disease. In this regard, evaluating how distinct metabolites regulate gene expression programs to ultimately influence trajectories of the developing brain is within our grasp.
IS and DC came up with the topic of the review and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbott, C. W., Rohac, D. J., Bottom, R. T., Patadia, S., and Huffman, K. J. (2018). Prenatal ethanol exposure and neocortical development: a transgenerational model of FASD. Cereb. Cortex. 28, 2908–2921. doi: 10.1093/cercor/bhx168
Ao, Z., Cai, H., Havert, D. J., Wu, Z., Gong, Z., Beggs, J. M., et al. (2020). One-stop microfluidic assembly of human brain organoids to model prenatal cannabis exposure. Anal. Chem. 92, 4630–4638. doi: 10.1021/acs.analchem.0c00205
Badanich, K. A., Mulholland, P. J., Beckley, J. T., Trantham-Davidson, H., and Woodward, J. J. (2013). Ethanol reduces neuronal excitability of lateral orbitofrontal cortex neurons via a glycine receptor dependent mechanism. Neuropsychopharmacology 38, 1176–1188. doi: 10.1038/npp.2013.12
Byrnes, E. M., and Vassoler, F. M. (2018). Modeling prenatal opioid exposure in animals: current findings and future directions. Front. Neuroendocrinol. 51, 1–13. doi: 10.1016/j.yfrne.2017.09.001
Chen, H. I., Wolf, J. A., Blue, R., Song, M. M., Moreno, J. D., Ming, G. L., et al. (2019). Transplantation of human brain organoids: revisiting the science and ethics of brain chimeras. Cell Stem Cell 25, 462–472. doi: 10.1016/j.stem.2019.09.002
Chen, S., Alhassen, W., Yoshimura, R., De Silva, A., Abbott, G. W., Baldi, P., et al. (2020). Metabolomic and transcriptomic signatures of prenatal excessive methionine support nature rather than nurture in schizophrenia pathogenesis. Commun. Biol. 3:409. doi: 10.1038/s42003-020-01124-8
Cruickshank-Quinn, C. I., Mahaffey, S., Justice, M. J., Hughes, G., Armstrong, M., Bowler, R. P., et al. (2014). Transient and persistent metabolomic changes in plasma following chronic cigarette smoke exposure in a mouse model. PLoS One 9:e101855. doi: 10.1371/journal.pone.0101855
Dang, J., Tiwari, S. K., Agrawal, K., Hui, H., Qin, Y., Rana, T. M., et al. (2021). Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol. Psychiatry 26, 1194–1207. doi: 10.1038/s41380-020-0676-x
Della Grotta, S., LaGasse, L. L., Arria, A. M., Derauf, C., Grant, P., Smith, L. M., et al. (2010). Patterns of methamphetamine use during pregnancy: results from the Infant Development, Environment, and Lifestyle (IDEAL) study. Matern. Child Health J. 14, 519–527. doi: 10.1007/s10995-009-0491-0
Eiraku, M., Watanabe, K., Matsuo-Takasaki, M., Kawada, M., Yonemura, S., Matsumura, M., et al. (2008). Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 3, 519–532. doi: 10.1016/j.stem.2008.09.002
Fernandes, C. G., Leipnitz, G., Seminotti, B., Amaral, A. U., Zanatta, Â, Vargas, C. R., et al. (2010). Experimental evidence that phenylalanine provokes oxidative stress in hippocampus and cerebral cortex of developing rats. Cell Mol. Neurobiol. 30, 317–326. doi: 10.1007/s10571-009-9455-6
Frau, R., Miczán, V., Traccis, F., Aroni, S., Pongor, C. I., Saba, P., et al. (2019). Prenatal THC produces a hyperdopaminergic phenotype accompanied by maladaptive behavioral susceptibility. Nat. Neurosci. 22:1975–1985. doi: 10.1038/s41593-019-0512-2
Harkany, T., Guzmán, M., Galve-Roperh, I., Berghuis, P., Devi, L. A., and Mackie, K. (2007). The emerging functions of endocannabinoid signaling during CNS development. Trends Pharmacol. Sci. 28, 83–92. doi: 10.1016/j.tips.2006.12.004
Huh, D., Matthews, B. D., Mammoto, A., Montoya-Zavala, M., Hsin, H. Y., and Ingber, D. E. (2010). Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668. doi: 10.1126/science.1188302
Hurd, Y. L., Manzoni, O. J., Pletnikov, M. V., Lee, F. S., Bhattacharyya, S., and Melis, M. (2019). Cannabis and the developing brain: insights into its long-lasting effects. J. Neurosci. 39, 8250–8258. doi: 10.1523/JNEUROSCI.1165-19.2019
Kadoshima, T., Sakaguchi, H., Nakano, T., Soen, M., Ando, S., Eiraku, M., et al. (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U.S.A. 110, 20284–20289. doi: 10.1073/pnas.1315710110
King, M. A., Hunter, B. E., and Walker, D. W. (1988). Alterations and recovery of dendritic spine density in rat hippocampus following long-term ethanol ingestion. Brain Res. 459, 381–385. doi: 10.1016/0006-8993(88)90656-7
Kraemer, G. W., Moore, C. F., Newman, T. K., Barr, C. S., and Schneider, M. L. (2008). Moderate level fetal alcohol exposure and serotonin transporter gene promoter polymorphism affect neonatal temperament and limbic-hypothalamic-pituitary-adrenal axis regulation in monkeys. Biol. Psychiatry 63, 317–324. doi: 10.1016/j.biopsych.2007.07.017
La Manno, G., Siletti, K., Furlan, A., Gyllborg, D., Vinsland, E., Langseth, C. M., et al. (2020). Molecular architecture of the developing mouse brain. bioRxiv [Preprint]. doi: 10.1101/2020.07.02.184051
Lancaster, M. A., and Knoblich, J. A. (2014). Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340. doi: 10.1038/nprot.2014.158
Lancaster, M. A., Renner, M., Martin, C.-A., Wenzel, D., Bicknell, L. S., Hurles, M. E., et al. (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. doi: 10.1038/nature12517
Li, D., Gu, X., Lu, L., and Liang, L. (2010). Effects of phenylalanine on the survival and neurite outgrowth of rat cortical neurons in primary cultures: possible involvement of brain-derived neurotrophic factor. Mol. Cell Biochem. 339, 1–7. doi: 10.1007/s11010-009-0364-2
Li, R., Sun, L., Fang, A., Li, P., Wu, Q., and Wang, X. (2017). Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 8, 823–833. doi: 10.1007/s13238-017-0479-2
Linsley, J. W., Tripathi, A., Epstein, I., Schmunk, G., Mount, E., Campioni, M., et al. (2019). Automated four-dimensional long term imaging enables single cell tracking within organotypic brain slices to study neurodevelopment and degeneration. Commun. Biol. 2:115. doi: 10.1038/s42003-019-0411-9
McCarthy, D. M., Morgan, T. J. Jr., Lowe, S. E., Williamson, M. J., Spencer, T. J., Biederman, J., et al. (2018). Nicotine exposure of male mice produces behavioral impairment in multiple generations of descendants. PLoS Biol. 16:e2006497. doi: 10.1371/journal.pbio.2006497
Moradi, E., Jalili-Firoozinezhad, S., and Solati-Hashjin, M. (2020). Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomaterial. 116, 67–83. doi: 10.1016/j.actbio.2020.08.041
Morris, M., Shaw, A., Lambert, M., Perry, H. H., Lowenstein, E., Valenzuela, D., et al. (2018). Developmental nicotine exposure affects larval brain size and the adult dopaminergic system of Drosophila melanogaster. BMC Dev. Biol. 18:13. doi: 10.1186/s12861-018-0172-6
Muhammad, A., Mychasiuk, R., Nakahashi, A., Hossain, S. R., Gibb, R., and Kolb, B. (2012). Prenatal nicotine exposure alters neuroanatomical organization of the developing brain. Synapse 66, 950–954. doi: 10.1002/syn.21589
National Center for Drug Abuse Statistics (2022). Drug Abuse Statistics. National Institute on Drug Abuse (n.d.). Rockville, MD: National Center for Drug Abuse Statistics.
Ngai, Y. F., Sulistyoningurm, D. C., O’Neill, R., Innis, S. M., Weinberg, J., and Devlin, A. M. (2015). Prenatal alcohol exposure alters methyl metabolism and programs serotonin transporter and glucocorticoid receptor expression in brain. Am. J. Physiol. Regul. Integr. Comp. Physiol. 309, R613–R622. doi: 10.1152/ajpregu.00075.2015
Nimitvilai, S., Lopez, M. F., Mulholland, P. J., and Woodward, J. J. (2016). Chronic intermittent ethanol exposure enhances the excitability and synaptic plasticity of lateral orbitofrontal cortex neurons and induces a tolerance to the acute inhibitory actions of ethanol. Neuropsychopharmacology 41, 1112–1127. doi: 10.1038/npp.2015.250
Notaras, M., Lodhi, A., Barrio-Alonso, E., Foord, C., Rodrick, T., Jones, D., et al. (2021). Neurodevelopmental signatures of narcotic and neuropsychiatric risk factors in 3D human-derived forebrain organoids. Mol. Psychiatry doi: 10.1038/s41380-021-01189-9 [Epub ahead of print].
Nygaard, E., Slinning, K., Moe, V., Due-Tønnessen, P., Fjell, A., and Walhovd, K. B. (2018). Neuroanatomical characteristics of youths with prenatal opioid and poly-drug exposure. Neurotoxicol. Teratol. 68, 13–26. doi: 10.1016/j.ntt.2018.04.004
Pasca, A. M., Sloan, S. A., Clarke, L. E., Tian, Y., Makinson, C. D., Huber, N., et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 12, 671–678. doi: 10.1038/nmeth.3415
Peterson, B. S., Rosen, T., Dingman, S., Toth, Z. R., Sawardekar, S., Hao, X., et al. (2020). Associations of maternal prenatal drug abuse with measures of newborn brain structure, tissue organization, and metabolite concentrations. JAMA Pediatr. 174, 831–842. doi: 10.1001/jamapediatrics.2020.1622
Qian, X., Su, Y., Adam, C. D., Deutschmann, A. U., Pather, S. R., Goldberg, E. M., et al. (2020). Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell 26, 766.e9–781.e9. doi: 10.1016/j.stem.2020.02.002
Quadrato, G., and Arlotta, P. (2017). Present and future of modeling human brain development in 3D organoids. Curr. Opin. Cell Biol. 49, 47–52. doi: 10.1016/j.ceb.2017.11.010
Ribas, J., Sadeghi, H., Manbachi, A., Leijten, J., Brinegar, K., Zhang, Y. S., et al. (2016). Cardiovascular organ-on-a-chip platforms for drug discovery and development. Appl. In Vitro Toxicol. 2, 82–96. doi: 10.1089/aivt.2016.0002
Ricalde, A. A., and Hammer, R. P. Jr. (1990). Perinatal opiate treatment delays growth of cortical dendrites. Neurosci. Lett. 115, 137–143. doi: 10.1016/0304-3940(90)90444-e
Robinson, S. E. (2000). Effect of prenatal opioid exposure on cholinergic development. J. Biomed. Sci. 7, 253–257. doi: 10.1007/BF02255474
Robinson, S. E. (2002). Effects of perinatal buprenorphine and methadone exposures on striatal cholinergic ontogeny. Neurotoxicol. Teratol. 24, 137–142. doi: 10.1016/s0892-0362(01)00185-4
Robinson, S. E., Guo, H., Maher, J. R., McDowell, K. P., and Kunko, P. M. (1996). Postnatal methadone exposure does not prevent prenatal methadone-induced changes in striatal cholinergic neurons. Brain Res. Dev. Brain Res. 95, 118–121.
Ross, E. J., Graham, D. L., Money, K. M., and Stanwood, G. D. (2015). Developmental consequences of fetal exposure to drugs: what we know and what we still must learn. Neuropsychopharmacology 40, 61–87. doi: 10.1038/npp.2014.147
Roussotte, F., Soderberg, L., and Sowell, E. (2010). Structural, metabolic, and functional brain abnormalities as a result of prenatal exposure to drugs of abuse: evidence from neuroimaging. Neuropsychol. Rev. 20, 376–397. doi: 10.1007/s11065-010-9150-x
Sanchez, E. S., Bigbee, J. W., Fobbs, W., Robinson, S. E., and Sato-Bigbee, C. (2008). Opioid addiction and pregnancy: perinatal exposure to buprenorphine affects myelination in the developing brain. Glia 56, 1017–1027. doi: 10.1002/glia.20675
Sarieva, K., and Mayer, S. (2021). The effects of environmental adversities on human neocortical neurogenesis modeled in brain organoids. Front. Mol. Biosci. 8:686410. doi: 10.3389/fmolb.2021.686410
Schneider, M. L., Moore, C. F., Barr, C. S., Larson, J. A., and Kraemer, G. W. (2011). Moderate prenatal alcohol exposure and serotonin genotype interact to alter CNS serotonin function in rhesus monkey offspring. Alcoh. Clin. Exp. Res. 35, 912–920. doi: 10.1111/j.1530-0277.2010.01421.x
Silbereis, J. C., Pochareddy, S., Zhu, Y., Li, M., and Sestan, N. (2016). The cellular and molecular landscapes of the developing human central nervous system. Neuron 89, 248–268. doi: 10.1016/j.neuron.2015.12.008
Smith, B. L. (2021). Improving translational relevance: the need for combined exposure models for studying prenatal adversity. Brain Behav. Immun. Health 16:100294. doi: 10.1016/j.bbih.2021.100294
Stratoulias, V., Venero, J. L., Tremblay, M. E., and Joseph, B. (2019). Microglial subtypes: diversity within the microglial community. EMBO J. 38:e101997. doi: 10.15252/embj.2019101997
Substance Abuse and Mental Health Services Administration (2009). The NSDUH Report: Substance use Among Women During Pregnancy and Following Childbirth. Rockville, MD: Office of Applied Studies.
Sun, N., Meng, X., Liu, Y., Song, D., Jiang, C., and Cai, J. (2021). Applications of brain organoids in neurodevelopment and neurological diseases. J. Biomed. Sci. 28:30. doi: 10.1186/s12929-021-00728-4
Tanaka, Y., Cakir, B., Xiang, Y., Sullivan, G. J., and Park, I.-H. (2020). Synthetic analyses of single-cell transcriptomes from multiple brain organoids and fetal brain. Cell Rep. 30, 1682–9.e3. doi: 10.1016/j.celrep.2020.01.038
Torii, M., Sasaki, M., Chang, Y. W., Ishii, S., Waxman, S. G., Kocsis, J. D., et al. (2017). Detection of vulnerable neurons damaged by environmental insults in utero. Proc. Natl. Acad. Sci. U.S.A. 114, 2367–2372. doi: 10.1073/pnas.1620641114
Trujillo, C. A., Gao, R., Negraes, P. D., Gu, J., Buchanan, J., Preissl, S., et al. (2019). Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell 25, 558–569.e7. doi: 10.1016/j.stem.2019.08.002
United Nations Office on Drugs and Crime (2021). UNODC World Drug Report 2021: Pandemic Effects Ramp up Drug Risks, as Youth Underestimate Cannabis Dangers. New York, NY: United Nations Office on Drugs and Crime.
Velasco, S., Kedaigle, A. J., Simmons, S. K., Nash, A., Rocha, M., Quadrato, G., et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527. doi: 10.1038/s41586-019-1289-x
Wang, X., Dow-Edwards, D., Anderson, V., Minkoff, H., Hurd, Y. L., et al. (2006). Discrete opioid gene expression impairment in the human fetal brain associated with maternal marijuana use. Pharmacogenomics J. 6, 255–264. doi: 10.1038/sj.tpj.6500375
Wang, Y., Wang, L., Zhu, Y., and Qin, J. (2018). Human brain organoid-on-a-chip to model prenatal nicotine exposure. Lab. Chip. 18, 851–860. doi: 10.1039/C7LC01084B
Wu, C. C., Hung, C. J., Shen, C. H., Chen, W. Y., Chang, C. Y., Pan, H. C., et al. (2014). Prenatal buprenorphine exposure decreases neurogenesis in rats. Toxicol. Lett. 225, 92–101. doi: 10.1016/j.toxlet.2013.12.001
Wu, C. S., Jew, C. P., and Lu, H. C. (2011). Lasting impacts of prenatal cannabis exposure and the role of endogenous cannabinoids in the developing brain. Future Neurol. 6, 459–480. doi: 10.2217/fnl.11.27
Xu, J., and Wen, Z. (2021). Brain organoids: studying human brain development and diseases in a dish. Stem Cells Int. 2021:5902824. doi: 10.1155/2021/5902824
Yoon, S. J., Elahi, L. S., Paşca, A. M., Marton, R. M., Gordon, A., Revah, O., et al. (2019). Reliability of human cortical organoid generation. Nat. Methods 16, 75–78. doi: 10.1038/s41592-018-0255-0
Zhang, B.-Z., Chu, H., Han, S., Shuai, H., Deng, J., Hu, Y.-F., et al. (2020). SARS-CoV-2 infects human neural progenitor cells and brain organoids. Cell Res. 30, 928–931. doi: 10.1038/s41422-020-0390-x
Zhu, J., Zhang, X., Xu, Y., Spencer, T. J., Biederman, J., and Bhide, P. G. (2012). Prenatal nicotine exposure mouse model showing hyperactivity, reduced cingulate cortex volume, reduced dopamine turnover, and responsiveness to oral methylphenidate treatment. J. Neurosci. 32, 9410–9418. doi: 10.1523/JNEUROSCI.1041-12.2012
Keywords: organoids, metabolism, drugs, brain development, prenatal
Citation: Stankovic IN and Colak D (2022) Prenatal Drugs and Their Effects on the Developing Brain: Insights From Three-Dimensional Human Organoids. Front. Neurosci. 16:848648. doi: 10.3389/fnins.2022.848648
Received: 04 January 2022; Accepted: 01 February 2022;
Published: 25 March 2022.
Edited by:
Gregory Wohl Kirschen, Johns Hopkins Medicine, United StatesReviewed by:
Teresa P. Silva, Universidade de Lisboa, PortugalCopyright © 2022 Stankovic and Colak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isidora N. Stankovic, aXNzNDAwNkBtZWQuY29ybmVsbC5lZHU=; Dilek Colak, ZGljMjAwOUBtZWQuY29ybmVsbC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.