- 1Institute of Anatomy and Cell Biology, National Yang Ming Chiao Tung University, Taipei, Taiwan
- 2Institute of Brain Science, College of Medicine, National Yang Ming Chiao Tung University, Taipei, Taiwan
- 3Brain Research Center, National Yang Ming Chiao Tung University, Taipei, Taiwan
Expansion of the neocortex is thought to pave the way toward acquisition of higher cognitive functions in mammals. The highly conserved Notch signaling pathway plays a crucial role in this process by regulating the size of the cortical progenitor pool, in part by controlling the balance between self-renewal and differentiation. In this review, we introduce the components of Notch signaling pathway as well as the different mode of molecular mechanisms, including trans- and cis-regulatory processes. We focused on the recent findings with regard to the expression pattern and levels in regulating neocortical formation in mammals and its interactions with other known signaling pathways, including Slit–Robo signaling and Shh signaling. Finally, we review the functions of Notch signaling pathway in different species as well as other developmental process, mainly somitogenesis, to discuss how modifications to the Notch signaling pathway can drive the evolution of the neocortex.
General Introduction of Notch Signaling
Over a century ago, Morgan and Dexter identified hereditary mutant flies having wings with serrated edges (Morgan, 1911; Dexter, 1914) because of Notch deficiency (Morgan, 1917). Subsequently, studies have revealed that Notch and the corresponding signal pathways are highly conserved among species including Drosophila melanogaster (Go et al., 1998), Caenorhabditis elegans (Chen and Greenwald, 2004), Lytechinus variegatus (Sherwood and McClay, 1997), Danio rerio (Liao et al., 2016), and Mus musculus (Shimojo et al., 2008; Borrell et al., 2012; Cárdenas et al., 2018). Notch is involved in the regulation of cell fates in variable lineages (Artavanis-Tsakonas et al., 1999), cell survival, proliferation (Purow et al., 2005), and differentiation (Apelqvist et al., 1999) in a juxtacrine manner through the crosstalk between corresponding ligands and receptors.
Notch signaling, also known as the canonical Notch signaling pathway, is initiated through the interaction of a ligand on a signal-sending cell with a receptor on a signal-receiving cell (Figure 1A). The majority of Notch ligands and their receptors are single-pass type I transmembrane proteins with an intracellular C terminus and an extracellular N terminus (Figure 1B). Notch ligands contain the extracellular delta, serrate, and lag2 (DSL) domain that selects the corresponding receptors to mediate Notch activities (Kopan and Ilagan, 2009). Notch receptors contain extracellular epidermal growth factor (EGF)–like repeats that interact with the DSL domain of Notch ligands. The interaction triggers the cleavage of the intracellular Notch receptor to release the Notch intracellular domain (NICD) fragment. Subsequently, the NICD fragment is translocated into the nucleus to activate the downstream gene cascade by interacting with DNA-binding transcription factors such as CBF1, SU(H), and LAG1 (CSL) in vertebrates (Figure 1A). In addition, another non-canonical Notch signaling pathway has been uncovered in the recent two decades (Shawber et al., 1996; Nofziger et al., 1999; Bush et al., 2001). Unlike the canonical Notch signaling pathway, the non-canonical Notch signaling pathway activates Notch receptors independent of the DSL domain of Notch ligands or regulates downstream genes independent of CSL transcription factors (Andersen et al., 2012).
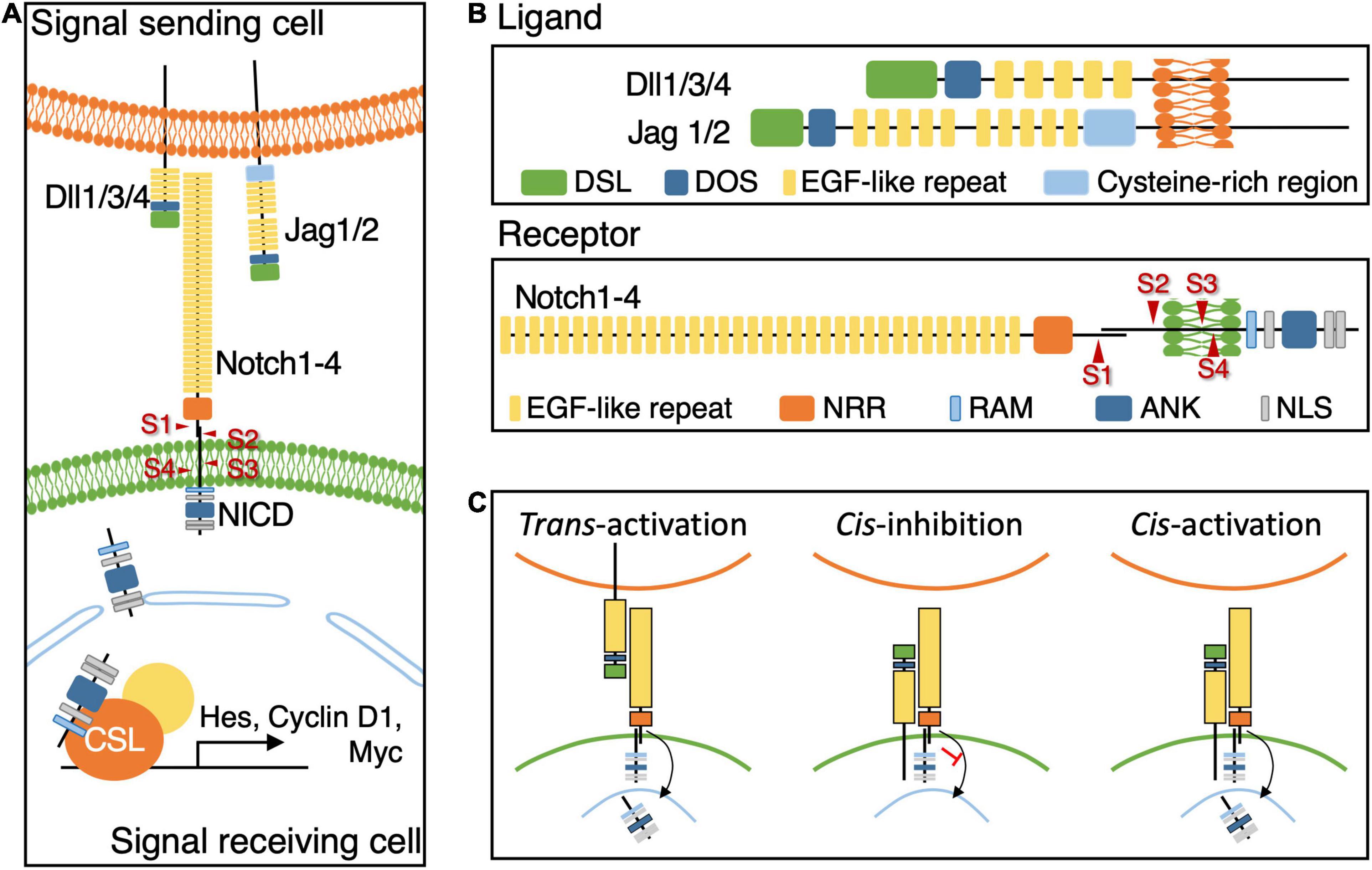
Figure 1. The components and Core Notch Signaling Pathway. (A) The illustration diagram of canonical Notch signal pathway showing the crosstalk between a ligand on the signal sending cells and a receptor on the signal receiving cell triggers the cleavage of Notch receptor to release NICD fragment. Nuclear-translocated NICD fragment interacts with CSL transcription factors to activate the downstream genes. (B) The schematic diagram showing the composition of Notch ligands, Dll1/3/4 and Jag1/2, and receptors, Notch 1–4 in mammals. Arrows indicate four protease cleavage cites on Notch receptors, S1–4. (C) The schematic diagram of different regulatory processes of Notch receptor-ligand interaction. The cell membrane marked in green represent the signal receiving cell.
The structure of Notch ligands is critical in regulating the activity of canonical Notch signaling (Figure 1B). The extracellular N terminus contains several conserved domains including the DSL domain and EGF-like repeats. The DSL domain selects the corresponding subtype receptors, and EGF-like repeats determine the binding affinity to Notch receptors. Most of the Notch ligands possess a transmembrane domain at the C terminus, while some of them are not observed in C. elegans. On the basis of the absence or presence of cysteine-rich regions located between EGF-like repeats and the transmembrane domain, drosophila Notch ligands can be classified into two groups: Delta and Serrate. The vertebrate orthologs of Delta and Serrate are known as Delta-like and Jagged, respectively (Fleming, 1998). Mutagenesis analysis of subunits of Notch ligands revealed their roles in mediating Notch signaling, such as DSL domain (Henderson et al., 1994, 1997) or EGF-like repeats (Tax et al., 1994). In addition to the drosophila studies, missense mutant of Jagged1 induces Nodder (Hansson et al., 2010) and Slalom (Tsai et al., 2001) in mice and, in Human, mutations in DSL and EGF-like repeats domains of JAG1 cause Alagille syndrome and mutations in only EGF-like repeats cause familial tetralogy of Fallot (Eldadah et al., 2001). Another highly conserved DOS domain (Delta and OSM-11-like proteins) sitting between the DSL and EGF-like repeats domains is known to cooperate with the DSL domain to facilitate Notch signaling (Komatsu et al., 2008), although it is missing in the majority of Notch ligand subtypes in C. elegans. Komatsu et al. (2008) found an OSM-11 protein carrying the DOS domain supports Notch ligands to activate Notch signaling during vulval development in C. elegans. Moreover, they demonstrated that the mammalian non-canonical Notch ligand Deltalike1 (Dlk1) can replace OSM-11 during the development of C. elegans, suggesting the presence of another mechanism that activates Notch signaling by using non-canonical ligands with the DOS domain, such as Dlk1/2, in invertebrates and vertebrates (Komatsu et al., 2008).
Notch receptors are type-1 transmembrane proteins (Figure 1B). In mammals, four paralogs of Notch receptors (Notch1–4) have been identified with similar structures but distinct corresponding ligands and functions. Mase et al. (2021) reported that the Notch1 receptor substantially maintains the radial glia (RG) pool during the early neurogenic stage of forebrain development, whereas Notch1 and 2 receptors contribute during the late stage. The extracellular domain of Notch receptors contains multiple EGF-like repeats that interact with Notch ligands and control the binding affinity. The negative regulatory region (NRR) adjacent to EGF-like repeats prevents the activation of the Notch receptor without binding to ligands. Intracellular Notch receptors contain a RBP-Jκ associated molecule (RAM), multiple ankyrin (ANK), and one-to-two nuclear location signal (NLS) domains. One of the NLS domain is located between the RAM and ANK domain and the other, if there is, is after the ANK domain (Lubman et al., 2007). RAM and ANK domains recruit transcription factors, and the NLS domain helps in their transportation into the nucleus. In addition, four proteolytic sites (S1–S4) are present between the intracellular and extracellular domains. S1 is cleaved by furin convertase to form the complete structure of the Notch receptor (Bray, 2006). S2 is located near the transmembrane domain on the extracellular side and is cleaved by ADAM metalloproteases. S3 and S4 are located in the transmembrane domain and would be cleaved by γ-secretase. Once Notch signaling is activated by the ligand–receptor interaction, S2 is first cleaved, followed by S3 and S4 (Figure 1). The cleavage releases NICD fragments containing RAM and ANK domains that translocate into the nucleus to control downstream target gene expression (Kopan and Ilagan, 2009).
Downstream target genes of Notch signaling include genes encoding the hairy and enhancer of split (Hes) protein family such as E(spl) genes in drosophila, her1 and hey1 in zebrafish, and Hes1 and Hes5 genes in mice (Jarriault et al., 1998). The cluster of Hes proteins belongs to the basic helix-loop-helix family. They function as transcriptional repressors to suppress differentiation genes, such as Ngn2, to retain the abilities of self-renewal and differentiation capacity (Tomita et al., 1999; Borrell et al., 2012). Moreover, Hes proteins may upregulate downstream genes such as the cell cycle regulator Cyclin D1, the upregulation of which would maintain cells in the cell cycle (Ronchini and Capobianco, 2001) and the protooncogene cMyc in cancer cells (Weng et al., 2006; Figure 1A).
Given the delicate and complex structure of Notch receptors and their ligands, Notch signaling is involved in various regulatory mechanisms. The extracellular calcium concentration affects Notch activity during left-right determination in vertebrates (Raya et al., 2004). This effect can be attributable to EGF-like domains in Notch receptors and their ligands that interact with calcium ions, which affect the ligand–receptor binding affinity (Rao et al., 1995; Cordle et al., 2008a,b). In support of this, the NRR in Notch receptors contains Ca2+-binding sites observed in the X-ray structure (Gordon et al., 2007). In contrast to the activation of Notch signaling by Notch ligands and receptors in adjacent cells (trans-activation), the interaction between Notch ligands and receptors within the same cell can inhibit Notch signaling (cis-inhibition) (Figure 1C). Although conflicts may occur in the binding sites of Notch receptors and their ligands during trans-activation and cis-inhibition, trans-activation and cis-inhibition can compete with each other (Cordle et al., 2008a). del Álamo et al. (2011) proposed that proteolytic sites responsible for generating NICD fragments are shed and that Notch signaling cannot be initiated when Notch ligands and their receptors are concurrently bound in the same cell through cis-inhibition (Figure 1). Because Notch ligands contain multiple proteolytic sites that can be either cleaved by ADAM metalloproteases or γ-secretases near the transmembrane domain (Zolkiewska, 2008), some Notch ligands appear to be soluble, even though they contain transmembrane domains, such as DeltaC in zebrafish and Dll3 in mammals (Geffers et al., 2007). Soluble Notch ligands may not be able to activate Notch signaling and instead act as an antagonist (Ladi et al., 2005; Chapman et al., 2011). This phenomenon might be induced by the binding of soluble Notch ligands to their corresponding Notch receptors in a cis-inhibitory conformation (D’souza et al., 2008); however, this regulatory mechanism is still under debate (Geffers et al., 2007). Beyond the classical concept of trans-activation and cis-inhibition (Sprinzak et al., 2010; LeBon et al., 2014); Nandagopal et al. (2019) demonstrated that cis-activation of Notch signaling can occur when the cell density was rigorously controlled in vitro (Figure 1C). They found Notch signaling can be activated in a cell which expressed both Notch ligands and receptors in the absence of surrounding cells. While this finding of cis-activation expends the possibility of regulatory mechanisms of Notch signaling, the related biological functions as well as the interaction with the conventional ways of trans-activation and cis-inhibition remained to be elucidated (Nandagopal et al., 2019). Thus, the phenotype induced by mutant Notch ligands lacking the C-terminus, including intracellular and transmembrane domains, might not be due to haploinsufficiency but dominant negative effects (Bulman et al., 2000; Warthen et al., 2006; Fischer-Zirnsak et al., 2019). However, Notch signaling mediates cell fate determination in variable cell types. Restricted combinations of ligand and receptors in canonical Notch signaling pathway may not be sufficient for all Notch-mediated developmental processes, suggesting an alternative pathway may be involved in. That might be the non-canonical Notch signaling pathway as conserved receptors are utilized, although detailed functions remain unclear (D’souza et al., 2010).
Notch Signaling in Neocortex Formation in Mammals
At the early beginning of embryo development, the telencephalon originates from the most anterior part of the neural tube arising from a single layer of epithelial cells. On the basis of the anatomical position and composition of cell types, the telencephalon can be categorized into dorsal and ventral compartments. The neocortex, which is believed to be responsible for higher cognitive functions, is a major part of the dorsal telencephalon. The neocortex is formed by a six-layer laminated structure composed of glutamatergic excitatory neurons. Here, we focus on the involvement of the Notch signaling pathway in the formation of the laminated structure.
Transition From Neuroepithelial Cells to Radial Glias
Distinct types of neurons in the neocortex are all derived from neural progenitor cells. Hence, the number of neural progenitor cells is critical to determine the size of the brain. The development of the neocortex begins with the generation and expansion of neural progenitor cells. In mammals, at least three types of neural progenitor cells are involved in the development of the neocortex: neuroepithelial cells (NECs), RGs, and intermediate progenitor cells (IPCs). NECs are the earliest type of neural progenitor cells that are highly polarized in a pseudostratified pattern (His, 1889; Ramon y Cajal and Azoulay, 1955). Because NECs are believed to generate all other types of cells in the neocortex, the size of the NEC pool is crucial to determine the numbers of progenitor cells and even the final number of cortical neurons (Malatesta et al., 2000; Noctor et al., 2001, 2002). To amplify their pool, NECs keep dividing symmetrically and exponentially before the onset of neurogenesis. NECs gradually transform into RGs for the onset of neurogenesis. Although RGs still maintain some NEC characteristics, such as bipolar morphology and apical–basal polarity (Rakic, 1972), they begin to lose tight junctions (Aaku-Saraste et al., 1996) and express specific RG proteins (Levitt and Rakic, 1980), such as glutamate/aspartate transporter (Shibata et al., 1997) and brain lipid-binding proteins (Feng et al., 1994). Although RGs could symmetrically divide to expand its pool as NECs, they can undergo asymmetrical division to produce neurons. In addition to their self-renewal and differentiation functions, the radial fiber of RGs guides neuronal migration. In this process, the overexpression of cleaved NICD fragments promote progenitor cells to express RG-specific markers (Gaiano et al., 2000). No differences in the number of NECs in the neural tube were observed between Hes1/5 double-knockout mice and control mice at the NEC stage E8.5, whereas the number of RGs decreased due to prematuration at later stages (E9.5–10.5) when NECs begin to transform to RGs (Hatakeyama et al., 2004). These studies suggest that the transition of NECs to RGs is dependent on Notch signaling, whereas the formation and expansion of NECs is independent of Notch signaling.
Generating Intermediate Progenitors or Neurons From Radial Glia
Neurogenesis from RGs to neurons can occur in a direct or an indirect manner. Direct neurogenesis is one RG divides to generate an RG and a neuron in the ventricular zone (VZ), and indirect neurogenesis is one RG may generate two RGs or two other types of progenitor cells, such as IPCs. Subsequently, IPCs symmetrically divide to generate two neurons. Indirect neurogenesis is beneficial for the increase in the final neuron pool and is more common in the mammalian neocortex compared with direct neurogenesis, which is the predominant neurogenesis manner in the developing cortex of other vertebrates, such as birds and reptiles (Englund et al., 2005; Guillemot, 2005; Cárdenas et al., 2018).
Prematuration is observed in animal models with a Notch signaling deficiency. The aforementioned studies have indicated that defects in the activation of Notch signaling inhibited the transition from NECs to RGs. Because Mind bomb 1 (Mib1), a RING-type E3 ubiquitin ligase, promotes the endocytosis of canonical Notch ligands, knocking out the Mib1 gene can impair Notch signaling. Conventional Mib1 knockout mice exhibited deficient Notch signaling that led to prematuration at E9.0–E9.5, resulting in the death of embryos before E12.5 (Koo et al., 2005). Furthermore, in animal models with Notch signaling deficiency, RGs transformed into IPCs early before differentiating into neurons. In Nestin-driven Mib1 knockout mice, the numbers of IPCs and mitotic cells outside the VZ region were increased at E13.5 (Yoon et al., 2008), resulting in an increase in the number of neurons from E14.0. Those findings suggest that Notch signaling activity is high in RGs but low in IPCs and neurons. To determine the activity of Notch signaling in RGs and IPCs separately, overexpression of NICD together with CBF1-EGFP, a reporter of Notch signaling, was utilized. The results revealed that NICD activated the CBF1-binding site in RGs but not in IPCs. Because NICD cannot activate Notch signaling in IPCs, Hes proteins can be overexpressed as an alternative method to activate Notch signaling. However, the numbers of IPCs decreased when Hes proteins were overexpressed (Mizutani et al., 2007; Ohtsuka and Kageyama, 2021b); this finding is in contrast to that of knockout experiments that indicated the attenuation of Notch activity. However, the reason underlying the inactivation of Notch signaling in IPCs remains to be elucidated. Because IPCs mediate indirect neurogenesis to effectively increase cell numbers and emergence of IPCs is crucial in the evolution of the mammalian neocortex (Cárdenas et al., 2018), the evolution of the mammalian neocortex should be examined by investigating the functional roles and molecular mechanisms of IPCs.
In gyrencephalic species, such as ferret and primates, a large population of proliferative cells can be noted in the basal region of the VZ. They are a subtype of RGs, called basal RGs (bRGs). These bRGs, unlike IPCs, have radial fibers but lose the apical attachment to the ventricular surface, unlike their apical cohorts, apical RGs (aRGs). bRGs can undergo self-renewal to expand the progenitor pool in the SVZ region. In the developing primate neocortex, the majority of bRGs are positioned in the outer SVZ (OSVZ), which is separated from the inner SVZ (ISVZ) by an inner fiber layer. During neocortical expansion, the thickness of the OSVZ gradually increases with the expansion of the VZ (Rakic, 1974; Smart et al., 2002; Lukaszewicz et al., 2005; Lui et al., 2011). Except for the similarity in morphological characteristics between bRG and aRGs, bRGs express some aRG genes, such as SOX2, PAX6, nestin, and GFAP, and undergo a Notch signaling–dependent pathway to self-renew or generate IPCs in the OSVZ (Fietz et al., 2010; Hansen et al., 2010). The induction of radial glial fiber divergence in the superficial neocortex by a large number of bRGs produced through the basal process combined with neuronal migration along the newly formed fibers can cause lateral dispersion and promote cortical folding in gyrencephalic species (Reillo et al., 2011; Gertz and Kriegstein, 2015; Llinares-Benadero and Borrell, 2019). Moreover, because of the abundant generation of bRGs and their daughter cells, the OSVZ was determined to be the predominant neurogenic zone at the mid-gestational stage that caused marked cortical neuronal expansion and an increase in brain size in humans, thus leading to the evolution of the cerebral cortex (Hansen et al., 2010; Lui et al., 2011; Llinares-Benadero and Borrell, 2019).
Oscillation Pattern of Notch Signaling in Neural Progenitor Cells
In the last decade, a group led by Professor Ryoichiro Kageyama in Japan published a series of discoveries describing several components in the Notch signaling pathway are expressed in a dynamic pattern called oscillation, which has been reported earlier and is essential in somitogenesis (Palmeirim et al., 1997). They found that the oscillation of Hes1 can maintain the pool of neural progenitor cells. Concurrently, the expression of Notch ligand Dll1 and the proneural gene Ngn2 were fluctuated in a manner which is coordinated but opposite to the oscillated expression pattern of Hes1. The fine balance of the oscillating gene expression pattern is orchestrated by several elaborate transcriptional regulatory mechanisms. The oscillating pattern of Hes1 expression can be regulated through a negative feedback loop. After the activation of Hes1 by the Notch ligand–receptor interaction, Hes1 protein cis-represses its own transcription by directly targeting its promoter. Another key is the short half-life of Hes1 mRNA and Hes1 protein. The half-life of Hes1 mRNA and Hes1 protein is as short as 20 min. As both Hes1 mRNA and Hes1 protein are degraded soon after their production, the Hes1 promoter can be released from autoinhibition. Also Hes protein represses proneural genes such as Mash1 (Chen et al., 1997) and the expression of Dll1 is directly regulated by Ngn2 and Mash1 through the regulation of enhancer regulatory elements (Castro et al., 2006), the oscillated pattern of Dll1 and proneuronal genes Ngn2 and Mash1 are similar to and follow that of Hes1 (Shimojo et al., 2008; Imayoshi et al., 2013). Nonetheless, the oscillating Ngn2 expression remains to be validated because previous findings have indicated that most cells, if not all, of Neurogenin2 CreER and R26R-CAG-loxPstop-EGFP mice had left the progenitor pool at 12 h after tamoxifen administration (Miyoshi and Fishell, 2012). Thus, the oscillating Dll1 expression pattern should be the most critical event in orchestrating Hes1 expression and Mash1 may be the upstream activator of Dll1 instead of Ngn2 (Imayoshi et al., 2013; Sueda et al., 2019). Interestingly, while the Hes genes oscillated in multiple tissues across species, the frequency varies. For instance, during somitogenesis when the oscillated Her/Hes expression regulated the formation of new somite, the frequency differs in different species: 30 min in zebrafish, 90 min in chick, 2 h in mouse (Cinquin, 2007), and 4–6 h in humans (Turnpenny et al., 2007; Kageyama et al., 2012; Hubaud and Pourquié, 2014; Matsuda et al., 2020). The period of Hes1 oscillation in mouse neural progenitor cells and fibroblasts is 2 h. However, the period is 3–5 h in mES cells (Kobayashi et al., 2009; Kobayashi and Kageyama, 2011), suggesting that the period may vary among cell types as well as the regulatory machinery. If the oscillation of Hes1 can maintain the pool of neural progenitor cells, the neuronal production step in neurogenesis indicates the escape of the oscillation cycle. Hence, neuronal differentiation can be induced by the sustained Ngn2 expression in the replacement of oscillatory Ngn2 expression (Shimojo et al., 2008). However, in this scheme, how Ngn2 and Dll1 expression escape the negative feedback loop controlled by Hes1 and changes from the oscillatory pattern to a sustained high expression pattern remain unclear.
The oscillatory Hes1 expression can be used to maintain neural progenitors in the cell cycle, whereas sustained Hes1 expression promotes cells to stay in a quiescent state (Sang et al., 2008; Sueda et al., 2019) that may contribute to boundary formation such as the boundary between the dorsal and ventral telencephalon (Baek et al., 2006). The sustained overexpression of Hes1 in mouse neural progenitor cells at E13.5 reduced the expression of Notch ligands (Dll1 and Jag1), proneural genes (Mash1 and Ngn2), and cell cycle regulators (cyclin D1 and cyclin E1) (Shimojo et al., 2008; Sueda et al., 2019). This result suggested that the sustained overexpression of Hes1 repressed both proliferation and differentiation. Thus, cells in the boundaries of the brain were not able to proliferate or differentiate. In Hes1-overexpressing transgenic mice, Pax6+/Hes1+ neural progenitor cells were maintained for a long time in the VZ even after birth. Nonetheless, compared with control mice, Hes1-overexpressing mice exhibited the suppressed proliferation of abnormal neural progenitor cells and a markedly elongated cell cycle length; this finding is in agreement with the previous study indicating that the sustained overexpression of Hes1 reduced the expression of cell cycle–related proteins such as cyclin D1 (Shimojo et al., 2008). Further investigation using transgenic mice to engineered wild-type Hes1 gene into the shortened or elongated form found both amplitude and frequency of oscillated Hes1 expression were impaired which resulted in neural prematuration and reduced brain size (Ochi et al., 2020) similar to the phenotype induced by engineered Dll1 gene (Shimojo et al., 2016). Notably, the shortened or elongated form of Dll1 gene would cause the deficiency in both neural development and somite formation. In addition to manipulating the pattern of oscillation, the basal level of Hes1 expression is also critical to its biological functions. Contrary to the mutant Hes1 mice expressing reduced as well as sustained levels of Hes1, overexpression of Hes1 prevented neural progenitor cells from self-renewal and differentiation, thus leading to a smaller brain size, a thinner cerebral cortex, the enlarged ventricles in Hes1-overexpressing mice and an apparent increase in the number of neural progenitor cells even in the late corticogenesis (Ohtsuka and Kageyama, 2021b). However, another interpretation has been raised by Borrell et al. (2012) proposing that Hes1 expression is crucial to maintaining the progenitor cell pool in the VZ by overexpressing Hes1 cDNA or downregulating Hes1 expression by using the RNA interference (RNAi) technique. Another study showed that activation of Notch signaling maintains the neural progenitor cell pool by overexpressing the NICD fragment (Mizutani et al., 2007). Thus, whether maintaining the neural progenitor pool is controlled by simply activation of Notch signaling or in the combination of the oscillated Hes1 expression remains to be clarified.
Combinational Effects of Notch Signaling and Other Signaling Pathways in the Developing Brain
Slit–Robo Signaling
Robo signaling is a widely known pathway involved in neural development. Robos and Slits (ligands of Robo receptors) are responsible for regulating axon guidance which contributes to cortical circuits (Brose et al., 1999; Dickson and Gilestro, 2006). Moreover, Robo–slit signaling regulates neurogenesis in the central nervous system (CNS) of drosophila (Mehta and Bhat, 2001) and mice (Andrews et al., 2008). In the neocortex of Robo1/2 knockout mouse, neural progenitor cells in the VZ underwent a premature asymmetric division and increased the generation of IPCs, thus reducing the brain size. This deficiency was found to be mediated by Robo-mediated transcriptional activation of the Notch effector Hes1, which suggested the interplay between Robo and Notch signaling is crucial to regulate neurogenesis precisely (Borrell et al., 2012).
CNS evolution across species has been investigated for decades; however, it still remains largely unclear. The differential regulation of direct and indirect neurogenesis in different species is one of the most prominent hypotheses. Recently, a study examined the switch between Dll1–Notch and Robo–Slit signaling in corticogenesis to determine the predominant mode of indirect or direct neurogenesis along with its effects on the neuron number, brain size, and neural circuit complexity across amniotes. To elucidate the involvement of Notch–Dll1 and Robo–Slit signaling, the expression level in the neural progenitors of each representative species among diverse amniotes (snake, chick, mouse, and human) was analyzed. Data indicated a high Robo expression level and a low Dll1 expression level in brain structures including the dorsal telencephalon of snake, the medial dorsal telencephalon of chick, and the olfactory bulb, hippocampus, and spinal cord of mammals, but a high Dll1 expression level and a low Robo expression level in the advanced brain structures including the lateral dorsal telencephalon of chick and the neocortex of mammals (Cárdenas et al., 2018; Cárdenas and Borrell, 2020). In brief, Robo expression declined in the evolutionary process, whereas Dll1 expression increased during the evolution of amniotes. Furthermore, the combined gain-of-function of Dll1 and loss-of-function of Robo in the evolutionarily old region of the telencephalon in mouse, chick, and snake indicated the promotion of indirect neurogenesis. By contrast, the combined gain-of-function of Robo and loss-of-function of Dll1 in the evolutionarily young region of the telencephalon including the mouse neocortex and human cerebral organoids indicated the promotion of direct neurogenesis. This observation is correlated to the switch between direct and indirect neurogenesis. Progenitors in the snake dorsal cortex exhibit mostly direct neurogenesis with no indirect neurogenesis, as indicated by the absence of IPCs. By contrast, progenitors in the mammalian neocortex exhibit indirect neurogenesis most frequently. These findings were further confirmed in human organoids, indicating that the Robo–Dll reciprocal expression–based balance of direct/indirect neurogenesis is the key factor for evolution among amniotes (Cárdenas et al., 2018; Cárdenas and Borrell, 2020).
Sonic Hedgehog Signaling
Sonic hedgehog (Shh) is a secreted protein encoded by Shh gene. Initially, hedgehog gene was identified from Drosophila melanogaster. Mutations in hedgehog gene lead to abnormal segmental patterning and polarity in flies (Nüsslein-Volhard and Wieschaus, 1980; Mohler, 1988). Shh signaling is essential for embryonic development in two stages. In the early stage, Shh is secreted from the notochord, located ventrally to the neural tube, and controls the neural axis by creating a concentration gradient (Echelard et al., 1993; Roelink et al., 1995). In the later stage, Shh regulates cell proliferation and differentiation during brain development by controlling cell cycle kinetics in various tissues and species such as the mouse neocortex (Bertrand and Dahmane, 2006; Komada et al., 2008; Komada, 2012) and chick spinal cord (Saade et al., 2013). Shh is essential to the development of IPCs (Shikata et al., 2011). Mutations in human SHH gene cause holoprosencephaly (HPE), which is an autosomal dominantly inherited disorder. Patients with HPE have intellectual disability, microcephaly, and epilepsy (Tekendo-Ngongang et al., 1993; Belloni et al., 1996; Roessler et al., 1996). Shh protein initiates signaling by binding to the transmembrane receptor Pathed (Ptch), which inhibits Smoothened (Smo) in the absence of Shh (Murone et al., 1999). When Smo is de-repressed, it causes Gli1-3 to move to the nucleus, thus inducing downstream gene expression (Wickström et al., 2013).
Ohtsuka and Kageyama (2021b) reported that the sustained overexpression of Hes1 in mice retained abnormal neural progenitors with both Pax6 and Hes1 expression in the VZ even after birth but still accompanied by smaller brains, thinner cerebral cortices, and enlarged ventricles due to defects in proliferation and neurogenesis. Later, Ohtsuka and Kageyama (2021a) observed that Hes1-overexpressing mice could be rescued from their defects by crossing them with transgenic mice expressing constitutively active Smo, an effector of Shh signaling. This result suggested that dysfunction in Notch signaling can be complemented by promoting Shh signaling (Ohtsuka and Kageyama, 2021a). However, as both the pathways are crucial during embryonic development, detailed molecular mechanisms through which they work together in parallel or complement remain to be elucidated.
Evolutionary and Comparative Perspectives
Pallial Organization and Evolution in Vertebrates
The cortex of most of the reptiles such as alligators, geckos, and turtles shows a mixed pattern of the layered structure in the dorsal pallium dorsal to the ventricles and nuclear structures in the dorsal ventricular ridge ventral to the ventricles (Goffinet et al., 1986; Suzuki and Hirata, 2014; Briscoe and Ragsdale, 2018a,b; Nomura et al., 2020). In addition to different cytoarchitecture, neurons in the layered structure of reptiles migrate and integrate into the cortex roughly through an outside–in migration pattern (Suzuki and Hirata, 2014; Luzzati, 2015; Tosches et al., 2018; Nomura et al., 2020), opposite to the inside–out migration pattern in the developing mammalian neocortex. The pallium of birds is composed of four major subdivisions: hyperpallium, mesopallium, nidopallium, and arcopallium (Jarvis, 2009). The pallium in birds and some reptiles have a nuclear-type structure, in which neuronal cell bodies aggregate instead of layered laminated structures such as the neocortex in mammals. On the basis of trajectory tracing and in situ hybridization analyses, recent studies have identified that neurons with similar functions and molecular expression across the species have a nuclear or laminar structure, regardless of different cytoarchitectures (Zeier and Karten, 1971; Karten and Shimizu, 1989; Dugas-Ford et al., 2012; Suzuki et al., 2012; Chen et al., 2013). For example, neurons in the L2 field of the cortex in zebra finches receive signals from the thalamus and express genes such as Rorβ, similar to layer IV sensory neurons in the mammalian neocortex; neurons in the mesopallium and nidopallium and neurons in the arcopallium exhibit conserved projections and molecular expression similar to layer II–III and layer V–VI neurons in the mammalian neocortex, respectively (Chen et al., 2013). Considering conserved functions and the phylogenic tree, the laminated structure should be evolved from the nuclear type. A nuclear-to-layered hypothesis proposed by Karten indicated that the laminated pallium of the mammalian neocortex might be transformed from the nuclear type pallium in birds or reptiles (Karten, 1991).
Comparative analysis of neuroanatomical structures, gene expression profiles, and neural circuits is a common approach used to study pallium formation in teleosts (Wullimann and Mueller, 2004; Wullimann, 2009). The structure of the pallium shows distinct morphological features in different teleosts. However, mechanisms underlying the development of the teleost cortex and the gene expression profiles of neuronal connections remain largely unknown. The teleosts are close to land vertebrates such as amphibians and reptiles in evolution, and can be divided into ray-finned fishes and lobe-finned fishes. A ray-finned fish, called zebrafish, is the most common animal model used to study embryonic development, diseases, and neurological behaviors. Accumulating results of in situ hybridization, immunostaining, and neural circuit tracing indicate that molecular profiles and presumptive functions in the pallium and subpallium of the teleostean cortex are similar to those of other vertebrates. For example, Emx genes are enriched in the pallium and Dlx genes are enriched in the subpallium across species (Wullimann and Mueller, 2004; Wullimann, 2009). These genetic studies suggested that pallium formation from the neural tube in ray-finned fishes follows a special method called “eversion,” in which the neural tube bends outward to form two cerebral hemispheres, separated by an unpaired ventricle and covered with a thin roof plate. In contrast to ray-finned fishes, the pallium of other vertebrates, such as lobe-finned fishes, amphibians, reptiles, birds, and mammals, is generated during an evagination process, in which the roof of the neural tube is sunken down to separate two lateral ventricles (Huesa et al., 2009; Yamamoto et al., 2017). The two prominent differences between these processes in terms of morphological changes are the inverted mediolateral axis in the pallium and the position of ventricles. Because the lumen surface of the neural tube is critical to generating neural progenitor cells, changing the position of ventricles may cause alterations in the direction of the neuronal migration and orientation of neural fibers (Huesa et al., 2009; Wullimann, 2009; Yamamoto et al., 2017).
In fishes and mammals, considerable changes have been observed in the neuron number, pallium cytoarchitecture, and neural circuit complexity. Because components in Notch signaling are highly conserved, the activity of Notch signaling may be widely involved in multiple developmental events in the formation of the pallium such as the maintenance of neural and cortical progenitor cell pools, transition from aRGs to IPCs, and corticogenesis. Thus, we speculate that the evolutionary divergence in pallium formation may result from the dominant isoform switch, the distinct regulation mechanism, or the emergence of novel genes, which will be discussed in the following sections.
Dominant Isoform Switch: Comparison of Dll1/3 in Mammals and DeltaC/D in Zebrafish
In zebrafish, Notch ligands in the Delta family include DeltaA, DeltaB, DeltaD, and DeltaC, whereas Delta-like ligands include Dll1, Dll3, and Dll4 in mammals. DeltaA–D are expressed in the developing zebrafish pallium (Smithers et al., 2000; Mueller and Wullimann, 2003; Takano et al., 2011), whereas Dll1 and Dll3 (Nelson et al., 2013) but not Dll4 are expressed in the developing mammalian pallium (Herman et al., 2018). Comparing the DNA sequences of these delta genes with mouse Dll1 and Dll3 revealed that the sequences of DeltaD, DeltaA, and DeltaB are similar to that of mouse Dll1, whereas the sequence of DeltaC is similar to that of mouse Dll3 (Figure 2A). As fewer studies have examined the roles of Delta proteins in the development of the zebrafish pallium, we would like to briefly introduce the functions of Delta genes during somitogenesis and elaborate their possible implications in pallial development.
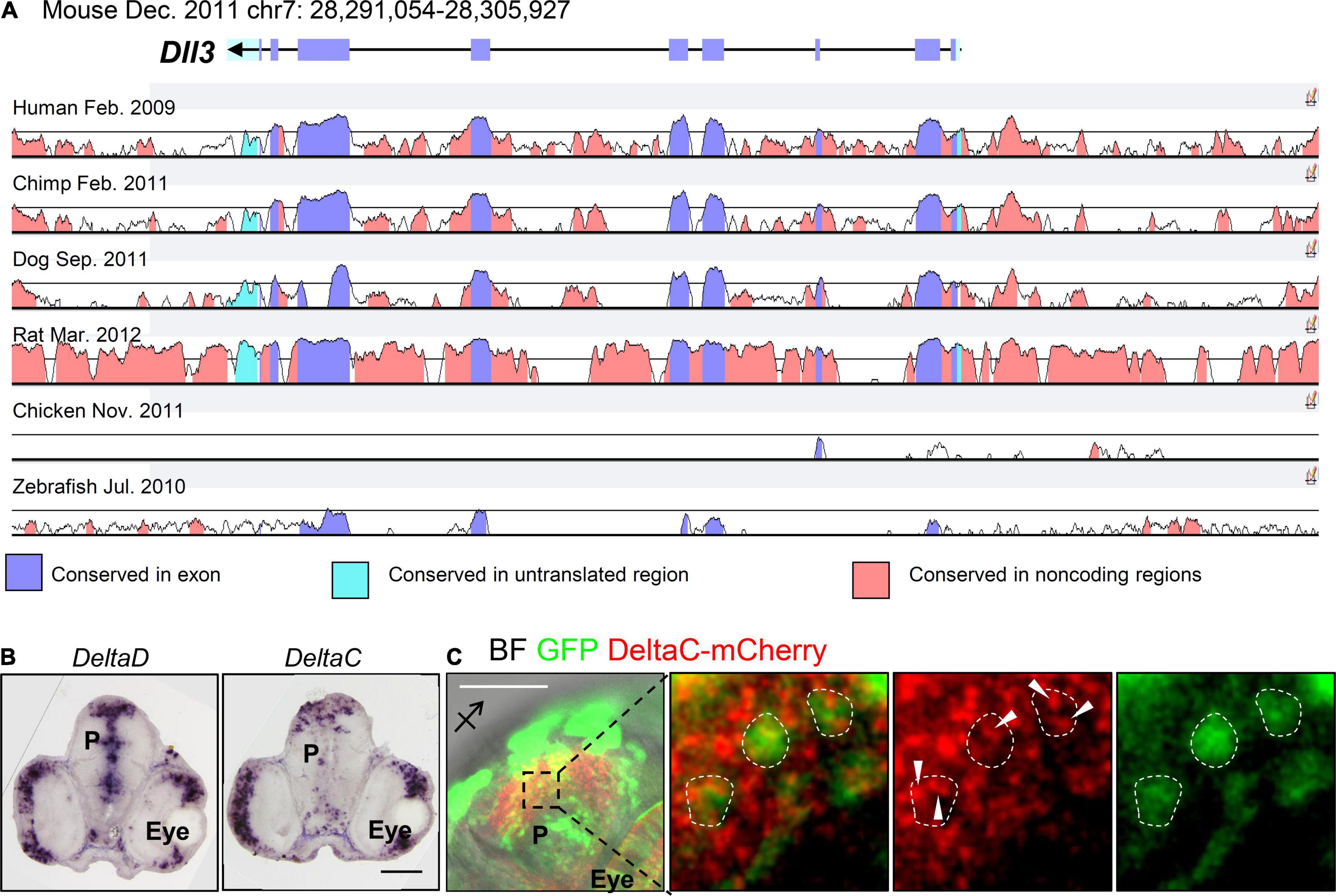
Figure 2. Conservation of Dll3 among species. (A) VISTA Browser display of a 14.9 kb fragment of the Dll3 region on Chr. 7 of the mouse genome (chr7: 28,291,054–28,305,927). VISTA plots are shown in the seven-way (Mouse-Human-Chimp-Dog-Rat-Chicken-Zebrafish) alignment. Based on the annotation, conserved regions above 50%/100 bp in VISTA are above the cutoff, and are colored under the curve with the indicated color. (B) Spatial expression of DeltaD and DeltaC in the 2 dpf zebrafish brain slice. (C) The expression pattern of DeltaC–mCherry in the zebrafish brain 2 days after introducing the plasmids carrying DeltaC–mCherry or GFP at 4-cell stage. BF: bright field. White dotted lines circle GFP-positive cells and arrows indicate puncta pattern of DeltaC-mCherry. Scale bar: 100 μm.
In zebrafish, somitogenesis is controlled by segmentation clock, which is coordinated by several components in Notch signaling including a Notch ligand (DeltaC) and the downstream target hairy/E(spl) genes (her1 and her7) with an oscillatory expression pattern. Both DeltaD and DeltaC are essential for somitogenesis, and DeltaC, as one of the oscillators, is critical for proper somite segmentation (Mara et al., 2007). Unexpectedly, a study using DeltaD mutant embryos suggested that DeltaD is required for the oscillation of her1, the downstream target gene of Notch signaling, whereas the expression of DeltaD is maintained at a constant level (Holley et al., 2000). These findings suggest that DeltaC and DeltaD expressed in different patterns have distinct functions during somitogenesis, and the deficiency of any protein causes defects in somite formation (Holley et al., 2000; Mara et al., 2007). Further examination by Wright et al. (2011) revealed the puncta expression pattern of DeltaC and DeltaD in the retina and hindbrain, and, notably, DeltaC and DeltaD were colocalized in the retina but not in the hindbrain. In cellular level, DeltaD may be either expressed in the cytoplasm or on the plasma membrane depending on the expression level of DeltaC in the presomitic mesoderm (PSM) during the formation of new somite (Wright et al., 2011). Mechanically, DeltaC is expressed as a soluble form to physically attract DeltaD away from the cell membrane to switch off DeltaD-mediated Notch signaling in the DeltaC-enriched region (Wright et al., 2011). In the developing pallium, DeltaD and DeltaC are both expressed, as demonstrated by our in situ hybridization data (DeltaD and DeltaC, Figure 2B) and previous studies (Smithers et al., 2000; Takano et al., 2011) and, in protein level, DeltaC was expressed in a puncta pattern in the cytoplasm, as shown by the DeltaC–mCherry strategy (Figure 2C). Considering the interplay between DeltaC and DeltaD in somitogenesis, DeltaC and DeltaD may play similar roles in regulating Notch signaling to control pallial formation.
During mammalian neocortical development, Dll1 has been found to be expressed in neural progenitors with an oscillatory pattern (Shimojo et al., 2008). In situ hybridization in the E9.5 whole mount embryo indicated that Dll1 was expressed in the forebrain, whereas Dll3 was expressed only in the ventral region of the forebrain. During somite formation, Dll1 and Dll3 were differentially expressed in the posterior or anterior region of newly formed somite (Dunwoodie et al., 1997) and were both necessary for somitogenesis (Kusumi et al., 1998; Dunwoodie et al., 2002). Mutations in human DLL1 induce neurodevelopmental disorders with non-specific brain abnormalities (Fischer-Zirnsak et al., 2019), whereas mutations in DLL3 cause spondylocostal dysostosis with axial skeletal defects (Bulman et al., 2000). These pathological findings suggest the critical role of Dll1 in dorsal telencephalic development while Dll3 mainly functions in somitogenesis. The use of Dll3 cDNA to replace Dll1 gene resulted in embryonic lethality in transgenic mice, suggesting that at least some Dll1 functions cannot be replaced by Dll3 (Geffers et al., 2007). Besides, Dll1 was found on the plasma membrane, whereas Dll3 was observed in the cytosol with a puncta pattern in mouse PSM and cultured cell lines (Geffers et al., 2007; Chapman et al., 2011); this is similar to the distribution of DeltaC and DetlaD in zebrafish somitogenesis. Another reason to explain the interchangeable role of Dll1 by Dll3 is the absence of lysine within the intracellular domain of Dll3. As lysine in the intracellular domain would be ubiquitinated by the ubiquitin ligase, Mib1, to triggers the endocytosis to recycle the ligand on the signal sending cell and pulling Notch receptor on the signal sending cell to activate downstream signaling through exposing the S2 protease site (Ladi et al., 2005; Le Borgne et al., 2005; Sprinzak and Blacklow, 2021). Thus, Dll3 cannot be exhibited on the cell membrane to compensate the loss of Dll1. Besides the intracellular domain, Geffers et al. (2007) provided evidences showing N-terminal DSL domain and the first two EGF-like repeats of Dll1 were critical to activate Notch signaling and cannot be replaced by that of Dll3 using different forms of chimeric Dll1 and Dll3 fusion proteins. Komatsu et al. (2008) also reported that in addition to the DSL domain, the conserved DOS motif within the first two EGF-like repeats is vital for activating Notch signaling and suggested that the DOS motif may cooperate with the DSL domain in binding to the Notch receptor. Separate studies performing mutation and structural analysis have indicated the importance of the DOS motif in cell lines (Shimizu et al., 1999; Geffers et al., 2007; Cordle et al., 2008a) and C. elegans (Komatsu et al., 2008). However, mouse Dll3 and Dll4 and zebrafish DeltaC do not contain this DOS motif, which may explain why Dll3 is unable to activate Notch signaling in certain cell types (Ladi et al., 2005; Geffers et al., 2007). Although Komatsu et al. (2008) suggested that Notch ligands without the DOS motif, such as DeltaC and Dll3, may trigger non-canonical Notch signaling with non-canonical ligands with the DOS motif, the role of non-canonical Notch signal pathway in either neurogenesis or somitogenesis should be further confirmed.
Both zebrafish DeltaC and mouse Dll3 share some similar features such as the intracellular distribution and lack of a DOS motif. Mutation of either DeltaD or DeltaC in zebrafish would lead to defects in somite development (Holley et al., 2000; Mara et al., 2007), suggesting that DeltaD and DeltaC are both necessary for somitogenesis. Studies on human disorders have indicated that DLL1 is more crucial for the neocortical development (Fischer-Zirnsak et al., 2019), whereas DLL3 is more critical for somitogenesis (Bulman et al., 2000). Thus, during somite formation, the dominant isoform changes from DeltaD and DeltaC in zebrafish to DLL3 in humans. This may reflect an evolutionary change in dominant forms in distinct tissue development. As DeltaA–D are all expressed in the developing zebrafish pallium (Smithers et al., 2000; Mueller and Wullimann, 2003; Takano et al., 2011), the expression pattern of Delta-like genes in mice and clinical features of human diseases suggest that the compensation may occur in the developing zebrafish pallium but not in the developing human dorsal telencephalon. These findings imply that the dominant form regulating telencephalic development may switch during the course of evolution.
Distinct Regulatory Machinery Leads to Diverse Cortex Formation Among Species
Cortical development involves multiple neural and cortical progenitors to produce cortical neurons at the right place and correct time. After the onset of corticogenesis, aRGs derived from RGs produce neurons either through the direct or indirect pathway (Figure 3A, black arrows: direct pathway; green arrows: indirect pathway). In the indirect pathway, aRGs generate to IPCs before producing neurons. Promotion of the indirect neurogenic pathway may be an evolutionary event (Cárdenas et al., 2018; Cárdenas and Borrell, 2020). A comparative approach using multiple species such as snakes and the mammalian pallium demonstrated the dominance of direct neurogenesis, whereas indirect neurogenesis gradually replaces direct neurogenesis in higher animals such as mammals (Figure 3A). Borrell’s team identified this evolutionary trend and suggested its relation to the gradient expression of Robo/Dll1 in the pallium across different species (Borrell and Reillo, 2012; Borrell et al., 2012; Cárdenas and Borrell, 2020). During indirect neurogenesis, RGs generate IPCs before becoming neurons, and one IPC symmetrically divides again to generate two neurons (Miyata et al., 2004; Noctor et al., 2004). IPCs act as a source of Notch ligands (Mizutani et al., 2007) to maintain the RG cell pool in a feedback loop of Notch signaling (Kawaguchi et al., 2008; Yoon et al., 2008; Lui et al., 2011; Nelson et al., 2013). Kawaguchi demonstrated that Dll1-positive cells in the VZ/SVZ of the E13.5 mouse neocortex were separated from those with active Notch signaling, and conditional Dll1 knockout mice driven by Nestin-Cre showed neuronal prematuration, suggesting that Dll1 can maintain neural progenitors in an undifferentiated state (Kawaguchi et al., 2008). Yoon et al. (2008) used Mib1 knockout mice in their study. Mib1 is a RING-type E3 ubiquitin ligase that promotes the endocytosis of canonical Notch ligands. They demonstrated that Mib1-positive cells may provide the Dll1 ligand to activate Notch signaling in adjacent cells in vitro. In addition, most Mib1-positive cells including IPCs and neurons can serve as Dll1 sources to activate Notch signaling in surrounding RGs (Yoon et al., 2008) which was supported by the asymmetric distribution of Dll1 and Mib1 during the asymmetric division of a neural progenitor to produce a progenitor and a neuron (Tozer et al., 2017).
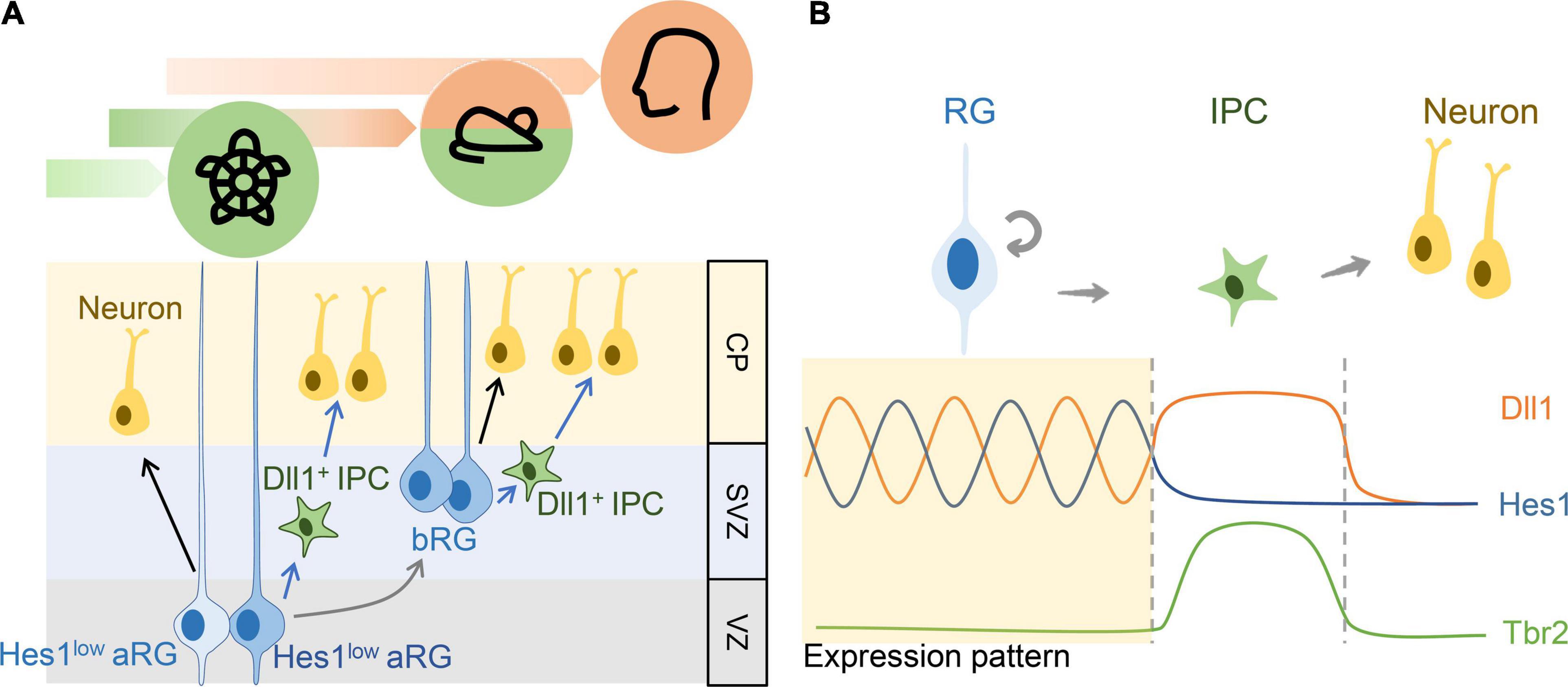
Figure 3. The role of Notch signaling in the balance between self-renewal and differentiation in the neural progenitors. (A) The schematic diagram showing the direct (black arrows) or indirect modes (blue arrows) of neurogenesis and the expression of Notch signaling components in the developing turtle, mouse, and human cortex. (B) The schematic diagram showing the dynamic expression pattern of key factors, Hes1 and Dll1, in Notch signaling and IPC gene Tbr2. The upregulation of Tbr2 follows the sustained Dll1 expression and the downregulation of Hes1 expression.
Nelson et al. (2013) categorized the major expression of Dll1 into two clusters in the SVZ and VZ at the E14.5 mouse neocortex similar to that reported in the Allen Brain Atlas1 (Figure 3A). The in situ hybridization results from the Allen Brain Atlas showed that Dll1 was expressed in the VZ and SVZ but not in the cortical plate in the cortices at various developmental stages. In addition, the number of Dll1-expressing cells gradually decreased over the course of development. Through multiphoton microscopy, Nelson discovered that Dll1-positive IPCs could contact Hes1/5-positive RGs through dynamic and transient elongate processes to maintain the RG cell pool. In addition, they suggested that other Notch ligands may participate in Notch signaling such as Dll3 in IPCs and Jag1 in RGs (Nelson et al., 2013). On the basis of the findings of these studies, we summarized that the oscillatory expression of Notch ligands such as Dll1 or Jag1 (Nelson et al., 2013) and receptors (Notch1/2) (Mase et al., 2021) can maintain RGs in the cell cycle (Figure 3). After differentiation into IPCs, Notch ligands are expressed at a constant level because Notch signaling cannot be activated to produce downstream Hes1, which represses Dll1 expression in a negative feedback loop (Figure 3B; Mizutani et al., 2007). With a constant expression of Dll1, IPCs can act as a Dll1 source to activate Notch signaling in RGs through ligand–receptor interaction. Oscillated Hes1 and Dll1 may occur only in RGs but not in IPCs. Although the detailed mechanism remains unclear, Notch signaling is believed to be inactivated in IPCs due to the lack of Notch receptors or the presence of molecules that inhibit the activation of Notch signaling. Finally, the expression of Dll1 is the lowest in neurons (Figure 3B). In the brains, such as reptiles and birds, which lack IPCs because they primarily rely on direct neurogenesis during pallium development, Notch ligands (Dll1 or Jag1) are produced solely by RGs. However, Notch ligands in RGs are oscillatory in response to the negative regulator Hes protein, which may explain the smaller RG pool in reptiles than in mammals due to the lack of an alternative source of Notch ligands (Figure 3A). During the development of the neocortex in humans, a significant increase in the number and types of neural/cortical progenitors contribute to a larger size of the dorsal telencephalon than in other vertebrates. The enriched bRG is the most prominent feature for massive cortical expansion. bRG is derived from aRGs similar to the derivation of IPCs from aRGs. Similar to direct and indirect neurogenic pathways, bRG may produce neurons directly or generate basal IPC before producing neurons. Notch signaling is activated in bRGs based on the expression of Hes1. In addition, according to Nelson’s study with Dll1d2YFP reporter, many neural/cortical progenitors, including RGs, bRGs and IPs, express Dll1, and basal IPCs can maintain bRG proliferation through physical contact with bRGs in the SVZ (Figure 3B; Nelson et al., 2013; Govindan and Jabaudon, 2017).
Nomura found that neural progenitors in the pallium of geckos required a long time to differentiate into neurons compared with other species of amniotes (Nomura et al., 2013). They applied pulse labeling to monitor the period from neural progenitors to neurons in the pallium of mouse (M. musculus), gecko (Poekilocerus pictus), turtle (Pelodiscus sinensis), and chick (Gallus gallus). Compared with the neural progenitors of other species, neural progenitors in geckos required a longer period to differentiate, twice as those required by mice and chicks. Although cortical progenitors stay in the progenitor stage for a longer period in geckos than in mice, the number of mitotic cells was lower in geckos, suggesting that the size of the neural progenitor pool may not be associated with the duration in the progenitor stage. Furthermore, they used a CBF1-driven reporter to monitor Notch activity in neural progenitors. Neural progenitors of geckos exhibited higher Notch activity than those of other species. Notably, the distribution of neural progenitors with active Notch signaling differed among species: mosaic in mice, turtles, and chicks but homogenous in geckos (Nomura et al., 2013). Two possibilities can explain the mosaic distribution of neural/cortical progenitors. First, this distribution may result from the oscillated expression pattern of Notch signaling components (Shimojo et al., 2008; Ohtsuka and Kageyama, 2021b). Second, this pattern may be due to scattered IPCs in the neural progenitor pool to deliver the Notch ligands (Mizutani et al., 2007). In either possibility, the cross-species study suggested that differences in the duration of neural progenitor differentiation and the number of mitotic cells within amniotes may be linked to the spatial distribution of neural/cortical progenitor cells, which may be uniform or mosaic. Thus, the emergence of the mosaic distribution of neural/cortical progenitor may be an evolutionary key to the expansion of the telencephalon.
Novel Human-Specific Genes
Notch signaling is essential for self-renewal in RGs to maintain the progenitor cell pool during cortical development. Expansion of the neural progenitor pool and a prolonged neural progenitor self-renewal period are believed to be critical events in cerebral cortex evolution (Hansen et al., 2010; Lui et al., 2011; Borrell and Reillo, 2012; Geschwind and Rakic, 2013). Recently, a human-specific NOTCH2 partial duplicated paralog, NOTCH2NL gene, was found to be expressed in both human aRG and bRGs and to improve the expansion of cortical progenitors by activating NOTCH signaling through interrupting the cis-inhibition of DLL1 (Suzuki et al., 2018). Overexpression of NOTCH2NL in embryonic mice or human organoids prolonged the self-renewal stage and delayed neuronal differentiation, resulting in clonal expansion in neural progenitors (Fiddes et al., 2018; Suzuki et al., 2018). By contrast, NOTCH2NL knockout accelerated neuronal differentiation and reduced neurogenesis (Fiddes et al., 2018). Investigations on underlying mechanisms revealed that NOTCH2NLs would interact with NOTCH receptors and inhibit cell autonomous DLL1 (NOTCH ligand) function to enhance NOTCH activity during corticogenesis (Fiddes et al., 2018; Suzuki et al., 2018). In addition, the deletion or duplication of NOTCH2NL genes in humans induced microcephaly and megacephaly, respectively, suggesting the crucial role of NOTCH2NL in human neocortical development (Fiddes et al., 2018). Hence, enhancing NOTCH signaling at a proper level may contribute to cortical evolution.
Conclusion and Future Perspectives
Notch signaling is highly conserved among species and regulates a wide range of developmental processes. It had been demonstrated that the activity of the canonical Notch signaling pathway determines the size of the neural progenitor pool and the initiation of neural differentiation during the telencephalon development in amniotes (Nomura et al., 2013; Cárdenas et al., 2018). However, it remains unclear how Notch signaling contributes to the formation of the telencephalon in anamniotes and how the conserved Notch signaling contributes to the establishment of distinct telencephalic cytoarchitecture in different species. To facilitate the multiple roles of Notch signaling, it may utilize different combinations of ligands (Nelson et al., 2013) and receptors (Mase et al., 2021), interact with other signaling amniotes (Cárdenas et al., 2018; Cárdenas and Borrell, 2020; Ohtsuka and Kageyama, 2021a) or novel genes (Fiddes et al., 2018; Suzuki et al., 2018). Further, the involvement of non-canonical Notch signaling would improve the complex regulations by Notch signaling in orchestrated multiple developmental processes. These evidences demonstrate the delicate regulation of Notch signaling is capable of activating distinct downstream machinery in either the developmental processes or evolution. Thus, to delineate the whole pictures of Notch signaling is believed to decode the mystery underlying the brain evolution to acquire higher cognitive functions in mammals.
Author Contributions
F-SN and P-SH collected the relevant research for the review and wrote the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Ministry of Science and Technology (MOST 108-2320-B-010-046-MY3), National Health Research Institute (NHRI-EX111-11007NC), Yen Tjing Ling Medical Foundation (CI-110-4), and Brain Research Center, National Yang Ming Chiao Tung University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Aaku-Saraste, E., Hellwig, A., and Huttner, W. B. (1996). Loss of occludin and functional tight junctions, but not ZO-1, during neural tube closure–remodeling of the neuroepithelium prior to neurogenesis. Dev. Biol. 180, 664–679. doi: 10.1006/dbio.1996.0336
Andersen, P., Uosaki, H., Shenje, L. T., and Kwon, C. (2012). Non-canonical notch signaling: emerging role and mechanism. Trends Cell Biol. 22, 257–265. doi: 10.1016/j.tcb.2012.02.003
Andrews, W., Barber, M., Hernadez-Miranda, L. R., Xian, J., Rakic, S., Sundaresan, V., et al. (2008). The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev. Biol. 313, 648–658. doi: 10.1016/j.ydbio.2007.10.052
Apelqvist, Å, Li, H., Sommer, L., Beatus, P., Anderson, D. J., Honjo, T., et al. (1999). notch signalling controls pancreatic cell differentiation. Nature 400, 877–881. doi: 10.1038/23716
Artavanis-Tsakonas, S., Rand, M. D., and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770–776. doi: 10.1126/science.284.5415.770
Baek, J. H., Hatakeyama, J., Sakamoto, S., Ohtsuka, T., and Kageyama, R. (2006). Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 133, 2467–2476. doi: 10.1242/dev.02403
Belloni, E., Muenke, M., Roessler, E., Traverse, G., Siegel-Bartelt, J., Frumkin, A., et al. (1996). Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat. Genet. 14, 353–356. doi: 10.1038/ng1196-353
Bertrand, N., and Dahmane, N. (2006). Sonic hedgehog signaling in forebrain development and its interactions with pathways that modify its effects. Trends Cell Biol. 16, 597–605. doi: 10.1016/j.tcb.2006.09.007
Borrell, V., Cárdenas, A., Ciceri, G., Galcerán, J., Flames, N., Pla, R., et al. (2012). Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron 76, 338–352. doi: 10.1016/j.neuron.2012.08.003
Borrell, V., and Reillo, I. (2012). Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev. Neurobiol. 72, 955–971. doi: 10.1002/dneu.22013
Bray, S. J. (2006). Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689. doi: 10.1038/nrm2009
Briscoe, S. D., and Ragsdale, C. W. (2018a). Homology, neocortex, and the evolution of developmental mechanisms. Science 362, 190–193. doi: 10.1126/science.aau3711
Briscoe, S. D., and Ragsdale, C. W. (2018b). Molecular anatomy of the alligator dorsal telencephalon. J. Comp. Neurol. 526, 1613–1646. doi: 10.1002/cne.24427
Brose, K., Bland, K. S., Wang, K. H., Arnott, D., Henzel, W., Goodman, C. S., et al. (1999). Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806.
Bulman, M. P., Kusumi, K., Frayling, T. M., Mckeown, C., Garrett, C., Lander, E. S., et al. (2000). Mutations in the human delta homologue, DLL3, cause axial skeletal defects in spondylocostal dysostosis. Nat. Genet. 24, 438–441. doi: 10.1038/74307
Bush, G., Miyamoto, A., Denault, J.-B., Leduc, R., and Weinmaster, G. (2001). Ligand-induced signaling in the absence of furin processing of notch1. Dev. Biol. 229, 494–502. doi: 10.1006/dbio.2000.9992
Cárdenas, A., and Borrell, V. (2020). Molecular and cellular evolution of corticogenesis in amniotes. Cell. Mol. Life Sci. 77, 1435–1460. doi: 10.1007/s00018-019-03315-x
Cárdenas, A., Villalba, A., De Juan Romero, C., Picó, E., Kyrousi, C., Tzika, A. C., et al. (2018). Evolution of cortical neurogenesis in amniotes controlled by robo signaling levels. Cell 174, 590.e21–606.e21. doi: 10.1016/j.cell.2018.06.007
Castro, D. S., Skowronska-Krawczyk, D., Armant, O., Donaldson, I. J., Parras, C., Hunt, C., et al. (2006). Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell 11, 831–844. doi: 10.1016/j.devcel.2006.10.006
Chapman, G., Sparrow, D. B., Kremmer, E., and Dunwoodie, S. L. (2011). Notch inhibition by the ligand Delta-Like 3 defines the mechanism of abnormal vertebral segmentation in spondylocostal dysostosis. Hum. Mol. Genet. 20, 905–916. doi: 10.1093/hmg/ddq529
Chen, C. C., Winkler, C. M., Pfenning, A. R., and Jarvis, E. D. (2013). Molecular profiling of the developing avian telencephalon: regional timing and brain subdivision continuities. J. Comp. Neurol. 521, 3666–3701. doi: 10.1002/cne.23406
Chen, H., Thiagalingam, A., Chopra, H., Borges, M. W., Feder, J. N., Nelkin, B. D., et al. (1997). Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc. Natl. Acad. Sci. 94, 5355–5360. doi: 10.1073/pnas.94.10.5355
Chen, N., and Greenwald, I. (2004). The lateral signal for LIN-12/notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6, 183–192. doi: 10.1016/s1534-5807(04)00021-8
Cinquin, O. (2007). Understanding the somitogenesis clock: what’s missing? Mech. Dev. 124, 501–517. doi: 10.1016/j.mod.2007.06.004
Cordle, J., Johnson, S., Tay, J. Z. Y., Roversi, P., Wilkin, M. B., De Madrid, B. H., et al. (2008a). A conserved face of the jagged/serrate DSL domain is involved in notch trans-activation and cis-inhibition. Nat. Struct. Mol. Biol. 15, 849–857. doi: 10.1038/nsmb.1457
Cordle, J., Redfieldz, C., Stacey, M., Van Der Merwe, P. A., Willis, A. C., Champion, B. R., et al. (2008b). Localization of the delta-like-1-binding site in human notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283, 11785–11793. doi: 10.1074/jbc.M708424200
del Álamo, D., Rouault, H., and Schweisguth, F. (2011). Mechanism and significance of cis-inhibition in notch signalling. Curr. Biol. 21, R40–R47. doi: 10.1016/j.cub.2010.10.034
Dexter, J. S. (1914). The analysis of a case of continuous variation in Drosophila by a study of its linkage relations. Am. Nat. 48, 712–758.
Dickson, B. J., and Gilestro, G. F. (2006). Regulation of commissural axon pathfinding by slit and its Robo receptors. Annu. Rev. Cell Dev. Biol. 22, 651–675. doi: 10.1146/annurev.cellbio.21.090704.151234
D’souza, B., Meloty-Kapella, L., and Weinmaster, G. (2010). Canonical and non-canonical notch ligands. Curr. Top. Dev. Biol. 92, 73–129. doi: 10.1016/S0070-2153(10)92003-6
D’souza, B., Miyamoto, A., and Weinmaster, G. (2008). The many facets of notch ligands. Oncogene 27, 5148–5167. doi: 10.1038/onc.2008.229
Dugas-Ford, J., Rowell, J. J., and Ragsdale, C. W. (2012). Cell-type homologies and the origins of the neocortex. Proc. Natl. Acad. Sci. 109, 16974–16979. doi: 10.1073/pnas.1204773109
Dunwoodie, S. L., Clements, M., Sparrow, D. B., Sa, X., Conlon, R. A., and Beddington, R. S. (2002). Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129, 1795–1806. doi: 10.1242/dev.129.7.1795
Dunwoodie, S. L., Henrique, D., Harrison, S. M., and Beddington, R. S. (1997). Mouse Dll3: a novel divergent delta gene which may complement the function of other delta homologues during early pattern formation in the mouse embryo. Development 124, 3065–3076. doi: 10.1242/dev.124.16.3065
Echelard, Y., Epstein, D. J., St-Jacques, B., Shen, L., Mohler, J., Mcmahon, J. A., et al. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417–1430. doi: 10.1016/0092-8674(93)90627-3
Eldadah, Z. A., Hamosh, A., Biery, N. J., Montgomery, R. A., Duke, M., Elkins, R., et al. (2001). Familial tetralogy of fallot caused by mutation in the jagged1 gene. Hum. Mol. Genet. 10, 163–169. doi: 10.1093/hmg/10.2.163
Englund, C., Fink, A., Lau, C., Pham, D., Daza, R. A., Bulfone, A., et al. (2005). Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 25, 247–251. doi: 10.1523/JNEUROSCI.2899-04.2005
Feng, L., Hatten, M. E., and Heintz, N. (1994). Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron 12, 895–908. doi: 10.1016/0896-6273(94)90341-7
Fiddes, I. T., Lodewijk, G. A., Mooring, M., Bosworth, C. M., Ewing, A. D., Mantalas, G. L., et al. (2018). Human-specific NOTCH2NL genes affect notch signaling and cortical neurogenesis. Cell 173, 1356.e–1369.e. doi: 10.1016/j.cell.2018.03.051
Fietz, S. A., Kelava, I., Vogt, J., Wilsch-Brauninger, M., Stenzel, D., Fish, J. L., et al. (2010). OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat. Neurosci. 13, 690–699. doi: 10.1038/nn.2553
Fischer-Zirnsak, B., Segebrecht, L., Schubach, M., Charles, P., Alderman, E., Brown, K., et al. (2019). Haploinsufficiency of the notch ligand DLL1 causes variable neurodevelopmental disorders. Am. J. Hum. Genet. 105, 631–639. doi: 10.1016/j.ajhg.2019.07.002
Fleming, R. J. (1998). Structural conservation of notch receptors and ligands. Semin. Cell Dev. Biol. 9, 599–607. doi: 10.1006/scdb.1998.0260
Gaiano, N., Nye, J. S., and Fishell, G. (2000). Radial glial identity is promoted by notch1 signaling in the murine forebrain. Neuron 26, 395–404. doi: 10.1016/s0896-6273(00)81172-1
Geffers, I., Serth, K., Chapman, G., Jaekel, R., Schuster-Gossler, K., Cordes, R., et al. (2007). Divergent functions and distinct localization of the notch ligands DLL1 and DLL3 in vivo. J. Cell Biol. 178, 465–476. doi: 10.1083/jcb.200702009
Gertz, C. C., and Kriegstein, A. R. (2015). Neuronal migration dynamics in the developing ferret cortex. J. Neurosci. 35, 14307–14315. doi: 10.1523/JNEUROSCI.2198-15.2015
Geschwind, D. H., and Rakic, P. (2013). Cortical evolution: judge the brain by its cover. Neuron 80, 633–647. doi: 10.1016/j.neuron.2013.10.045
Go, M. J., Eastman, D. S., and Artavanis-Tsakonas, S. (1998). Cell proliferation control by notch signaling in Drosophila development. Development 125, 2031–2040. doi: 10.1242/dev.125.11.2031
Goffinet, A. M., Daumerie, C., Langerwerf, B., and Pieau, C. (1986). Neurogenesis in reptilian cortical structures: 3H-thymidine autoradiographic analysis. J. Comp. Neurol. 243, 106–116. doi: 10.1002/cne.902430109
Gordon, W. R., Vardar-Ulu, D., Histen, G., Sanchez-Irizarry, C., Aster, J. C., and Blacklow, S. C. (2007). Structural basis for autoinhibition of notch. Nat. Struct. Mol. Biol. 14, 295–300. doi: 10.1038/nsmb1227
Govindan, S., and Jabaudon, D. (2017). Coupling progenitor and neuronal diversity in the developing neocortex. FEBS Lett. 591, 3960–3977. doi: 10.1002/1873-3468.12846
Guillemot, F. (2005). Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr. Opin. Cell Biol. 17, 639–647. doi: 10.1016/j.ceb.2005.09.006
Hansen, D. V., Lui, J. H., Parker, P. R., and Kriegstein, A. R. (2010). Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464, 554–561. doi: 10.1038/nature08845
Hansson, E. M., Lanner, F., Das, D., Mutvei, A., Marklund, U., Ericson, J., et al. (2010). Control of notch-ligand endocytosis by ligand-receptor interaction. J. Cell Sci. 123, 2931–2942. doi: 10.1242/jcs.073239
Hatakeyama, J., Bessho, Y., Katoh, K., Ookawara, S., Fujioka, M., Guillemot, F., et al. (2004). Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 131, 5539–5550. doi: 10.1242/dev.01436
Henderson, S. T., Gao, D., Christensen, S., and Kimble, J. (1997). Functional domains of LAG-2, a putative signaling ligand for LIN-12 and GLP-1 receptors in Caenorhabditis elegans. Mol. Biol. Cell 8, 1751–1762. doi: 10.1091/mbc.8.9.1751
Henderson, S. T., Gao, D., Lambie, E. J., and Kimble, J. (1994). lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 120, 2913–2924. doi: 10.1242/dev.120.10.2913
Herman, A., Rhyner, A., Devine, P., Marrelli, S., Bruneau, B., and Wythe, J. (2018). A novel reporter allele for monitoring Dll4 expression within the embryonic and adult mouse. Biol. Open 7:bio026799. doi: 10.1242/bio.026799
Holley, S. A., Geisler, R., and Nüsslein-Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678–1690.
Hubaud, A., and Pourquié, O. (2014). Signalling dynamics in vertebrate segmentation. Nat. Rev. Mol. Cell Biol. 15, 709–721. doi: 10.1038/nrm3891
Huesa, G., Anadón, R., Folgueira, M., and Yáñez, J. (2009). “Evolution of the Pallium in Fishes,” in Encyclopedia of Neuroscience, eds M. D. Binder, N. Hirokawa, and U. Windhorst (Berlin: Springer), 1400–1404. doi: 10.1007/978-3-540-29678-2_3166
Imayoshi, I., Isomura, A., Harima, Y., Kawaguchi, K., Kori, H., Miyachi, H., et al. (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–1208. doi: 10.1126/science.1242366
Jarriault, S., Bail, O. L., Hirsinger, E., Pourquié, O., Logeat, F., Strong, C. F., et al. (1998). Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423–7431. doi: 10.1128/MCB.18.12.7423
Jarvis, E. D. (2009). “Evolution of the Pallium in Birds and Reptiles,” in Encyclopedia of Neuroscience, eds M. D. Binder, N. Hirokawa, and U. Windhorst (Berlin: Springer), 1390–1400. doi: 10.1007/978-3-540-29678-2_3165
Kageyama, R., Niwa, Y., Isomura, A., González, A., and Harima, Y. (2012). Oscillatory gene expression and somitogenesis. Wiley Interdiscip. Rev. Dev. Biol. 1, 629–641. doi: 10.1002/wdev.46
Karten, H. J. (1991). Homology and evolutionary origins of the ‘neocortex’. Brain Behav. Evol. 38, 264–272. doi: 10.1159/000114393
Karten, H. J., and Shimizu, T. (1989). The origins of neocortex: connections and lamination as distinct events in evolution. J. Cogn. Neurosci. 1, 291–301. doi: 10.1162/jocn.1989.1.4.291
Kawaguchi, D., Yoshimatsu, T., Hozumi, K., and Gotoh, Y. (2008). Selection of differentiating cells by different levels of delta-like 1 among neural precursor cells in the developing mouse telencephalon. Development 135, 3849–3858. doi: 10.1242/dev.024570
Kobayashi, T., and Kageyama, R. (2011). Hes1 oscillations contribute to heterogeneous differentiation responses in embryonic stem cells. Genes 2, 219–228. doi: 10.3390/genes2010219
Kobayashi, T., Mizuno, H., Imayoshi, I., Furusawa, C., Shirahige, K., and Kageyama, R. (2009). The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875. doi: 10.1101/gad.1823109
Komada, M. (2012). Sonic hedgehog signaling coordinates the proliferation and differentiation of neural stem/progenitor cells by regulating cell cycle kinetics during development of the neocortex. Congenit. Anom. 52, 72–77. doi: 10.1111/j.1741-4520.2012.00368.x
Komada, M., Saitsu, H., Kinboshi, M., Miura, T., Shiota, K., and Ishibashi, M. (2008). Hedgehog signaling is involved in development of the neocortex. Development 135, 2717–2727. doi: 10.1242/dev.015891
Komatsu, H., Chao, M. Y., Larkins-Ford, J., Corkins, M. E., Somers, G. A., Tucey, T., et al. (2008). OSM-11 facilitates LIN-12 notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 6:e196. doi: 10.1371/journal.pbio.0060196
Koo, B.-K., Lim, H.-S., Song, R., Yoon, M.-J., Yoon, K.-J., Moon, J.-S., et al. (2005). Mind bomb 1 is essential for generating functional notch ligands to activate notch. Development 132, 3459–3470. doi: 10.1242/dev.01922
Kopan, R., and Ilagan, M. X. G. (2009). The canonical notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233. doi: 10.1016/j.cell.2009.03.045
Kusumi, K., Sun, E. S., Kerrebrock, A. W., Bronson, R. T., Chi, D. C., Bulotsky, M. S., et al. (1998). The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat. Genet. 19, 274–278. doi: 10.1038/961
Ladi, E., Nichols, J. T., Ge, W., Miyamoto, A., Yao, C., Yang, L. T., et al. (2005). The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol. 170, 983–992. doi: 10.1083/jcb.200503113
Le Borgne, R., Bardin, A., and Schweisguth, F. (2005). The roles of receptor and ligand endocytosis in regulating notch signaling. Development 132, 1751–1762. doi: 10.1242/dev.01789
LeBon, L., Lee, T. V., Sprinzak, D., Jafar-Nejad, H., and Elowitz, M. B. (2014). Fringe proteins modulate notch-ligand cis and trans interactions to specify signaling states. eLife 3:e02950. doi: 10.7554/eLife.02950
Levitt, P., and Rakic, P. (1980). Immunoperoxidase localization of glial fibrillary acidic protein in radial glial cells and astrocytes of the developing rhesus monkey brain. J. Comp. Neurol. 193, 815–840. doi: 10.1002/cne.901930316
Liao, B.-K., Jörg, D. J., and Oates, A. C. (2016). Faster embryonic segmentation through elevated delta-notch signalling. Nat. Commun. 7, 1–12. doi: 10.1038/ncomms11861
Llinares-Benadero, C., and Borrell, V. (2019). Deconstructing cortical folding: genetic, cellular and mechanical determinants. Nat. Rev. Neurosci. 20, 161–176. doi: 10.1038/s41583-018-0112-2
Lubman, O. Y., Ilagan, M. X. G., Kopan, R., and Barrick, D. (2007). Quantitative dissection of the notch: CSL interaction: insights into the notch-mediated transcriptional switch. J. Mol. Biol. 365, 577–589. doi: 10.1016/j.jmb.2006.09.071
Lui, J. H., Hansen, D. V., and Kriegstein, A. R. (2011). Development and evolution of the human neocortex. Cell 146, 18–36.
Lukaszewicz, A., Savatier, P., Cortay, V., Giroud, P., Huissoud, C., Berland, M., et al. (2005). G1 phase regulation, area-specific cell cycle control, and cytoarchitectonics in the primate cortex. Neuron 47, 353–364. doi: 10.1016/j.neuron.2005.06.032
Luzzati, F. (2015). A hypothesis for the evolution of the upper layers of the neocortex through co-option of the olfactory cortex developmental program. Front. Neurosci. 9:162. doi: 10.3389/fnins.2015.00162
Malatesta, P., Hartfuss, E., and Gotz, M. (2000). Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development 127, 5253–5263. doi: 10.1242/dev.127.24.5253
Mara, A., Schroeder, J., Chalouni, C., and Holley, S. A. (2007). Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat. Cell Biol. 9, 523–530. doi: 10.1038/ncb1578
Mase, S., Shitamukai, A., Wu, Q., Morimoto, M., Gridley, T., and Matsuzaki, F. (2021). notch1 and notch2 collaboratively maintain radial glial cells in mouse neurogenesis. Neurosci. Res. 170, 122–132. doi: 10.1016/j.neures.2020.11.007
Matsuda, M., Yamanaka, Y., Uemura, M., Osawa, M., Saito, M. K., Nagahashi, A., et al. (2020). Recapitulating the human segmentation clock with pluripotent stem cells. Nature 580, 124–129. doi: 10.1038/s41586-020-2144-9
Mehta, B., and Bhat, K. M. (2001). Slit signaling promotes the terminal asymmetric division of neural precursor cells in the Drosophila CNS. Development 128, 3161–3168. doi: 10.1242/dev.128.16.3161
Miyata, T., Kawaguchi, A., Saito, K., Kawano, M., Muto, T., and Ogawa, M. (2004). Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development 131, 3133–3145. doi: 10.1242/dev.01173
Miyoshi, G., and Fishell, G. (2012). Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron 74, 1045–1058. doi: 10.1016/j.neuron.2012.04.025
Mizutani, K.-I., Yoon, K., Dang, L., Tokunaga, A., and Gaiano, N. (2007). Differential notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449, 351–355. doi: 10.1038/nature06090
Mohler, J. (1988). Requirements for hedgehog, a segmental polarity gene, in patterning larval and adult cuticle of Drosophila. Genetics 120, 1061–1072. doi: 10.1093/genetics/120.4.1061
Morgan, T. H. (1911). The origin of nine wing mutations in drosophila. Science 33, 496–499. doi: 10.1126/science.33.848.496
Mueller, T., and Wullimann, M. F. (2003). Anatomy of neurogenesis in the early zebrafish brain. Dev. Brain Res. 140, 137–155. doi: 10.1016/s0165-3806(02)00583-7
Murone, M., Rosenthal, A., and De Sauvage, F. J. (1999). Sonic hedgehog signaling by the patched–smoothened receptor complex. Curr. Biol. 9, 76–84. doi: 10.1016/s0960-9822(99)80018-9
Nandagopal, N., Santat, L. A., and Elowitz, M. B. (2019). Cis-activation in the notch signaling pathway. eLife 8:e37880. doi: 10.7554/eLife.37880
Nelson, B. R., Hodge, R. D., Bedogni, F., and Hevner, R. F. (2013). Dynamic interactions between intermediate neurogenic progenitors and radial glia in embryonic mouse neocortex: potential role in Dll1-Notch signaling. J. Neurosci. 33, 9122–9139. doi: 10.1523/JNEUROSCI.0791-13.2013
Noctor, S. C., Flint, A. C., Weissman, T. A., Dammerman, R. S., and Kriegstein, A. R. (2001). Neurons derived from radial glial cells establish radial units in neocortex. Nature 409, 714–720. doi: 10.1038/35055553
Noctor, S. C., Flint, A. C., Weissman, T. A., Wong, W. S., Clinton, B. K., and Kriegstein, A. R. (2002). Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J. Neurosci. 22, 3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002
Noctor, S. C., Martínez-Cerdeño, V., Ivic, L., and Kriegstein, A. R. (2004). Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 7, 136–144. doi: 10.1038/nn1172
Nofziger, D., Miyamoto, A., Lyons, K. M., and Weinmaster, G. (1999). Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development 126, 1689–1702. doi: 10.1242/dev.126.8.1689
Nomura, T., Gotoh, H., and Ono, K. (2013). Changes in the regulation of cortical neurogenesis contribute to encephalization during amniote brain evolution. Nat. Commun. 4:2206. doi: 10.1038/ncomms3206
Nomura, T., Ohtaka-Maruyama, C., Kiyonari, H., Gotoh, H., and Ono, K. (2020). Changes in wnt-dependent neuronal morphology underlie the anatomical diversification of neocortical homologs in amniotes. Cell Rep. 31:107592. doi: 10.1016/j.celrep.2020.107592
Nüsslein-Volhard, C., and Wieschaus, E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795–801. doi: 10.1038/287795a0
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H., and Kageyama, R. (2020). Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147:dev182204. doi: 10.1242/dev.182204
Ohtsuka, T., and Kageyama, R. (2021a). Dual activation of Shh and Notch signaling induces dramatic enlargement of neocortical surface area. Neurosci. Res. Online ahead of print., doi: 10.1016/j.neures.2021.09.006,
Ohtsuka, T., and Kageyama, R. (2021b). Hes1 overexpression leads to expansion of embryonic neural stem cell pool and stem cell reservoir in the postnatal brain. Development 148:dev189191. doi: 10.1242/dev.189191
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi: 10.1016/s0092-8674(00)80451-1
Purow, B. W., Haque, R. M., Noel, M. W., Su, Q., Burdick, M. J., Lee, J., et al. (2005). Expression of notch-1 and its ligands, delta-like-1 and jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 65, 2353–2363. doi: 10.1158/0008-5472.CAN-04-1890
Rakic, P. (1972). Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 145, 61–83. doi: 10.1002/cne.901450105
Rakic, P. (1974). Neurons in rhesus monkey visual cortex: systematic relation between time of origin and eventual disposition. Science 183, 425–427. doi: 10.1126/science.183.4123.425
Ramon y Cajal, S, and Azoulay, L. (1955). Histologie du Systeme Nerveux de L’homme et Des Vertebres. Paris: Maloine.
Rao, Z., Handford, P., Mayhew, M., Knott, V., Brownlee, G. G., and Stuartz, D. (1995). The structure of a Ca2+-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 82, 131–141. doi: 10.1016/0092-8674(95)90059-4
Raya, Á, Kawakami, Y., Rodríguez-Esteban, C., Ibañes, M., Rasskin-Gutman, D., Rodríguez-León, J., et al. (2004). Notch activity acts as a sensor for extracellular calcium during vertebrate left–right determination. Nature 427, 121–128. doi: 10.1038/nature02190
Reillo, I., De Juan Romero, C., García-Cabezas, M., and Borrell, V. (2011). A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb. Cortex 21, 1674–1694. doi: 10.1093/cercor/bhq238
Roelink, H., Porter, J., Chiang, C., Tanabe, Y., Chang, D., Beachy, P., et al. (1995). Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell 81, 445–455. doi: 10.1016/0092-8674(95)90397-6
Roessler, E., Belloni, E., Gaudenz, K., Jay, P., Berta, P., Scherer, S. W., et al. (1996). Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 14, 357–360. doi: 10.1038/ng1196-357
Ronchini, C., and Capobianco, A. J. (2001). Induction of cyclin D1 transcription and CDK2 activity by Notchic: implication for cell cycle disruption in transformation by Notchic. Mol. Cell. Biol. 21, 5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001
Saade, M., Gutiérrez-Vallejo, I., Le Dréau, G., Rabadán, M. A., Miguez, D. G., Buceta, J., et al. (2013). Sonic hedgehog signaling switches the mode of division in the developing nervous system. Cell Rep. 4, 492–503. doi: 10.1016/j.celrep.2013.06.038
Sang, L., Coller, H. A., and Roberts, J. M. (2008). Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100. doi: 10.1126/science.1155998
Shawber, C., Nofziger, D., Hsieh, J., Lindsell, C., Bogler, O., Hayward, D., et al. (1996). Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development 122, 3765–3773. doi: 10.1242/dev.122.12.3765
Sherwood, D. R., and McClay, D. R. (1997). Identification and localization of a sea urchin Notch homologue: insights into vegetal plate regionalization and Notch receptor regulation. Development 124, 3363–3374. doi: 10.1242/dev.124.17.3363
Shibata, T., Yamada, K., Watanabe, M., Ikenaka, K., Wada, K., Tanaka, K., et al. (1997). Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J. Neurosci. 17, 9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997
Shikata, Y., Okada, T., Hashimoto, M., Ellis, T., Matsumaru, D., Shiroishi, T., et al. (2011). Ptch1-mediated dosage-dependent action of Shh signaling regulates neural progenitor development at late gestational stages. Dev. Biol. 349, 147–159. doi: 10.1016/j.ydbio.2010.10.014
Shimizu, K., Chiba, S., Kumano, K., Hosoya, N., Takahashi, T., Kanda, Y., et al. (1999). Mouse jagged1 physically interacts with notch2 and other notch receptors: assessment by quantitative methods. J. Biol. Chem. 274, 32961–32969. doi: 10.1074/jbc.274.46.32961
Shimojo, H., Isomura, A., Ohtsuka, T., Kori, H., Miyachi, H., and Kageyama, R. (2016). Oscillatory control of Delta-like1 in cell interactions regulates dynamic gene expression and tissue morphogenesis. Genes Dev. 30, 102–116. doi: 10.1101/gad.270785.115
Shimojo, H., Ohtsuka, T., and Kageyama, R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. doi: 10.1016/j.neuron.2008.02.014
Smart, I. H., Dehay, C., Giroud, P., Berland, M., and Kennedy, H. (2002). Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 12, 37–53. doi: 10.1093/cercor/12.1.37
Smithers, L., Haddon, C., Jiang, Y.-J., and Lewis, J. (2000). Sequence and embryonic expression of deltaC in the zebrafish. Mech. Dev. 90, 119–123. doi: 10.1016/s0925-4773(99)00231-2
Sprinzak, D., and Blacklow, S. C. (2021). Biophysics of notch signaling. Annu. Rev. Biophys. 50, 157–189. doi: 10.1146/annurev-biophys-101920-082204
Sprinzak, D., Lakhanpal, A., Lebon, L., Santat, L. A., Fontes, M. E., Anderson, G. A., et al. (2010). Cis-interactions between notch and delta generate mutually exclusive signalling states. Nature 465, 86–90. doi: 10.1038/nature08959
Sueda, R., Imayoshi, I., Harima, Y., and Kageyama, R. (2019). High Hes1 expression and resultant Ascl1 suppression regulate quiescent vs. active neural stem cells in the adult mouse brain. Genes Dev. 33, 511–523. doi: 10.1101/gad.323196.118
Suzuki, I., and Hirata, T. (2014). A common developmental plan for neocortical gene-expressing neurons in the pallium of the domestic chicken Gallus gallus domesticus and the Chinese softshell turtle Pelodiscus sinensis. Front. Neuroanat. 8:20. doi: 10.3389/fnana.2014.00020
Suzuki, I. K., Gacquer, D., Van Heurck, R., Kumar, D., Wojno, M., Bilheu, A., et al. (2018). Human-specific NOTCH2NL genes expand cortical neurogenesis through delta/notch regulation. Cell 173, 1370.e16–1384.e16. doi: 10.1016/j.cell.2018.03.067
Suzuki, I. K., Kawasaki, T., Gojobori, T., and Hirata, T. (2012). The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev. Cell 22, 863–870. doi: 10.1016/j.devcel.2012.01.004
Takano, A., Zochi, R., Hibi, M., Terashima, T., and Katsuyama, Y. (2011). Function of strawberry notch family genes in the zebrafish brain development. Kobe J. Med. Sci. 56, E220–E230.
Tax, F. E., Yeargers, J. J., and Thomas, J. H. (1994). Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with delta and serrate of Drosophila. Nature 368, 150–154. doi: 10.1038/368150a0
Tekendo-Ngongang, C., Muenke, M., and Kruszka, P. (1993). “Holoprosencephaly overview,” in GeneReviews(§), eds M. P. Adam, H. H. Ardinger, R. A. Pagon, S. E. Wallace, L. J. H. Bean, G. Mirzaa, et al. (Seattle, WA: University of Washington, Seattle).
Tomita, K., Hattori, M., Nakamura, E., Nakanishi, S., Minato, N., and Kageyama, R. (1999). The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 13, 1203–1210. doi: 10.1101/gad.13.9.1203
Tosches, M. A., Yamawaki, T. M., Naumann, R. K., Jacobi, A. A., Tushev, G., and Laurent, G. (2018). Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888. doi: 10.1126/science.aar4237
Tozer, S., Baek, C., Fischer, E., Goiame, R., and Morin, X. (2017). Differential routing of mindbomb1 via centriolar satellites regulates asymmetric divisions of neural progenitors. Neuron 93, 542–551.e4. doi: 10.1016/j.neuron.2016.12.042
Tsai, H., Hardisty, R. E., Rhodes, C., Kiernan, A. E., Roby, P., Tymowska-Lalanne, Z., et al. (2001). The mouse slalom mutant demonstrates a role for Jagged1 in neuroepithelial patterning in the organ of Corti. Hum. Mol. Genet. 10, 507–512. doi: 10.1093/hmg/10.5.507
Turnpenny, P. D., Alman, B., Cornier, A. S., Giampietro, P. F., Offiah, A., Tassy, O., et al. (2007). Abnormal vertebral segmentation and the notch signaling pathway in man. Dev. Dyn. 236, 1456–1474. doi: 10.1002/dvdy.21182
Warthen, D., Moore, E., Kamath, B., Morrissette, J., Sanchez, P., Piccoli, D., et al. (2006). Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum. Mutat. 27, 436–443. doi: 10.1002/humu.20310
Weng, A. P., Millholland, J. M., Yashiro-Ohtani, Y., Arcangeli, M. L., Lau, A., Wai, C., et al. (2006). c-Myc is an important direct target of notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 20, 2096–2109. doi: 10.1101/gad.1450406
Wickström, M., Dyberg, C., Shimokawa, T., Milosevic, J., Baryawno, N., Fuskevåg, O. M., et al. (2013). Targeting the hedgehog signal transduction pathway at the level of GLI inhibits neuroblastoma cell growth in vitro and in vivo. Int. J. Cancer 132, 1516–1524. doi: 10.1002/ijc.27820
Wright, G. J., Giudicelli, F., Soza-Ried, C., Hanisch, A., Ariza-Mcnaughton, L., and Lewis, J. (2011). DeltaC and DeltaD interact as notch ligands in the zebrafish segmentation clock. Development 138, 2947–2956. doi: 10.1242/dev.066654
Wullimann, M. F. (2009). Secondary neurogenesis and telencephalic organization in zebrafish and mice: a brief review. Integr. Zool. 4, 123–133. doi: 10.1111/j.1749-4877.2008.00140.x
Wullimann, M. F., and Mueller, T. (2004). Teleostean and mammalian forebrains contrasted: evidence from genes to behavior. J. Comp. Neurol. 475, 143–162. doi: 10.1002/cne.20183
Yamamoto, K., Bloch, S., and Vernier, P. (2017). New perspective on the regionalization of the anterior forebrain in Osteichthyes. Dev. Growth Differ. 59, 175–187. doi: 10.1111/dgd.12348
Yoon, K.-J., Koo, B.-K., Im, S.-K., Jeong, H.-W., Ghim, J., Kwon, M.-C., et al. (2008). Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron 58, 519–531. doi: 10.1016/j.neuron.2008.03.018
Zeier, H., and Karten, H. J. (1971). The archistriatum of the pigeon: organization of afferent and efferent connections. Brain Res. 31, 313–326. doi: 10.1016/0006-8993(71)90185-5
Keywords: Notch, cortical development, cortical evolution, neurogenesis, HES, DLL
Citation: Nian F-S and Hou P-S (2022) Evolving Roles of Notch Signaling in Cortical Development. Front. Neurosci. 16:844410. doi: 10.3389/fnins.2022.844410
Received: 28 December 2021; Accepted: 15 February 2022;
Published: 29 March 2022.
Edited by:
Takuma Kumamoto, Tokyo Metropolitan Institute of Medical Science, JapanCopyright © 2022 Nian and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pei-Shan Hou, cHNob3VAbnljdS5lZHUudHc=
 Fang-Shin Nian
Fang-Shin Nian Pei-Shan Hou
Pei-Shan Hou