
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurosci., 25 February 2022
Sec. Gut-Brain Axis
Volume 16 - 2022 | https://doi.org/10.3389/fnins.2022.836605
This article is part of the Research TopicRole of the Gut-Brain Axis in Neurodegenerative and Neurodevelopmental DisordersView all 5 articles
 Piyush Padhi1,2
Piyush Padhi1,2 Carter Worth1
Carter Worth1 Gary Zenitsky2
Gary Zenitsky2 Huajun Jin2
Huajun Jin2 Kumar Sambamurti3
Kumar Sambamurti3 Vellareddy Anantharam2
Vellareddy Anantharam2 Arthi Kanthasamy2
Arthi Kanthasamy2 Anumantha G. Kanthasamy1,2*
Anumantha G. Kanthasamy1,2*
The human gut microbiota is a complex, dynamic, and highly diverse community of microorganisms. Beginning as early as in utero fetal development and continuing through birth to late-stage adulthood, the crosstalk between the gut microbiome and brain is essential for modulating various metabolic, neurodevelopmental, and immune-related pathways. Conversely, microbial dysbiosis – defined as alterations in richness and relative abundances – of the gut is implicated in the pathogenesis of several chronic neurological and neurodegenerative disorders. Evidence from large-population cohort studies suggests that individuals with neurodegenerative conditions have an altered gut microbial composition as well as microbial and serum metabolomic profiles distinct from those in the healthy population. Dysbiosis is also linked to psychiatric and gastrointestinal complications – comorbidities often associated with the prodromal phase of Parkinson’s disease (PD) and Alzheimer’s disease (AD). Studies have identified potential mediators that link gut dysbiosis and neurological disorders. Recent findings have also elucidated the potential mechanisms of disease pathology in the enteric nervous system prior to the onset of neurodegeneration. This review highlights the functional pathways and mechanisms, particularly gut microbe-induced chronic inflammation, protein misfolding, propagation of disease-specific pathology, defective protein clearance, and autoimmune dysregulation, linking gut microbial dysbiosis and neurodegeneration. In addition, we also discuss how pathogenic transformation of microbial composition leads to increased endotoxin production and fewer beneficial metabolites, both of which could trigger immune cell activation and enteric neuronal dysfunction. These can further disrupt intestinal barrier permeability, aggravate the systemic pro-inflammatory state, impair blood–brain barrier permeability and recruit immune mediators leading to neuroinflammation and neurodegeneration. Continued biomedical advances in understanding the microbiota-gut-brain axis will extend the frontier of neurodegenerative disorders and enable the utilization of novel diagnostic and therapeutic strategies to mitigate the pathological burden of these diseases.
The harmonious symbiotic evolution of microbes is essential for normal neurodevelopment (Warner, 2019), immune maturation (Inlender et al., 2021), and protection against pathogens in humans and animals (Pickard et al., 2017). Microbes are detected in every exposed organ, including the skin, gastrointestinal (GI) tract, oral cavity, nares, and bronchial tracts. However, the greatest microbial species density and diversity are found in the GI tract. The number of GI microbes in humans is estimated at 1013–1014, consisting primarily of various bacteria and lower amounts of archaea, fungi, and viruses (Sender et al., 2016). Astonishingly, the number of bacteria present in our body shows a nearly equal ratio to human cells (1.3:1). It boasts immense metabolic capabilities as the gut-microbiota may contain up to 23 million genes – a number that dwarfs the human genome (Claesson et al., 2009; Sender et al., 2016; Tierney et al., 2019). Since gut-residing bacteria are vital for host survival, understanding their influence can broaden the horizon for diagnosis and therapy for complex multifactorial diseases, including neurodegenerative diseases.
The human gut microbiome is primarily composed of two dominant major bacterial phyla, Bacteroidetes and Firmicutes, composing 90% of the total gut bacteria with the remaining 10% represented by other phyla including Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia. Despite a predominant pattern that emerges among the higher taxonomic categories, the gut microbiota is highly dynamic and individualized based on a person’s early life composition, diet, exercise, lifestyle factors, and disease status (Matsumoto et al., 2008; Desai et al., 2016; Martin et al., 2016; Xu et al., 2019). Furthermore, the regional and temporal variations influence microbial diversity and the host’s response to disease states (Arumugam et al., 2011). The microbial composition diversifies from in utero to early and mid-stage adulthood, followed by a progressive collapse of healthy microbiota in late adulthood (Lahiri et al., 2009; Martinez et al., 2018; Stinson et al., 2019; Yang et al., 2019; Mitchell et al., 2020). Although the existence of a placental microbiome during the in utero stage of development is contentious, evidence suggests the presence of bacteria and short-chain fatty acids (SCFAs) in the meconium and amniotic fluid (Stinson et al., 2019). Furthermore, at birth, the infant’s microbiome is contingent on the mode of delivery (i.e., birth canal or Cesarean delivery). The mother’s skin or vaginal microbiome signature is imprinted, enabling early colonizers to shape the infant’s long-term microbial composition, diversity, and early immune development (Martinez et al., 2018; Yang et al., 2019; Mitchell et al., 2020). Aging, a major risk factor in the pathological progression of neurodegenerative disorders, is also associated with modulating the gut microbial ecosystem (Bosco and Noti, 2021). It contributes to a compositional shift in gut microbiota characterized by a dramatic decrease in the diversity and abundance of several beneficial bacteria (reviewed in Dinan and Cryan, 2017). Additionally, the host’s genetics and environmental factors such as diet or exposure to toxins can further augment disease states (reviewed in Cabrera et al., 2021). A classic example of diet-induced gut dysbiosis was recorded in the early 70’s, where individuals from southern India who migrated to the United Kingdom in the 1970s frequently suffered from severe vitamin B12 deficiency (Britt et al., 1970, 1971; Stewart et al., 1970; Roberts et al., 1973; Britt and Harper, 1976). The subjects who presented with normal Vitamin B12 levels were found to consume a diet similar to that of South India and a later investigation reported that the synthesis of vitamin B12 precursors was modulated by microbial flora in the small intestine (Mathan et al., 1974; Albert et al., 1980). Since then, it has become evidently clear from several pre-clinical animal and human cohort studies that the microbiome is crucial for shaping various metabolic, immunological, and neurodevelopmental homeostatic processes (De Vadder et al., 2018; Rothhammer et al., 2018; Wang et al., 2020), and that cumulative exposure to infections, poor diet, and antibiotics can trigger dysbiosis and exacerbate the progression of several chronic age-related disorders (reviewed in Soto et al., 2018; Fang et al., 2020; Bosco and Noti, 2021).
The symbiotic and competitive relationships between diverse bacterial communities and hosts are preserved by enteric networks spanning various gut environments, and the central nervous system (CNS). The microbiota establishes a bi-directional communication network with the brain via neural, endocrine, and metabolic signaling modalities, hence the popular reference to the microbiome-gut-brain axis (Figure 1). A dysfunctional gut environment and its implications in nervous system disorders have been known for over two decades. More recently, a growing number of associative and mechanistic studies in neurobehavior, inflammation, neurogenesis, and neurodegeneration have implicated gut-residing microbes as key modulators of several disease etiologies (Soto et al., 2018; Gershon, 2019; Ma et al., 2019; Fang et al., 2020). Specifically, in neurodegenerative disorders such as Parkinson’s disease (PD) and Alzheimer’s disease (AD), recent findings indicate that the detrimental alterations to microbial composition augment disease-specific pathology. This review aims to provide some mechanistic insights into how a shift in specific populations of microbes and their derived metabolites in the gut triggers and facilitates various disease processes, including chronic intestinal and systemic inflammation, protein misfolding and aggregation, dysfunctional protein clearance, and a heightened autoimmune response, thereby contributing to disease propagation in the brain. This review also discusses how the microbiota influences the enteric nervous system (ENS) and CNS and modulates neuro-immune synapses and enteric neuropathy.
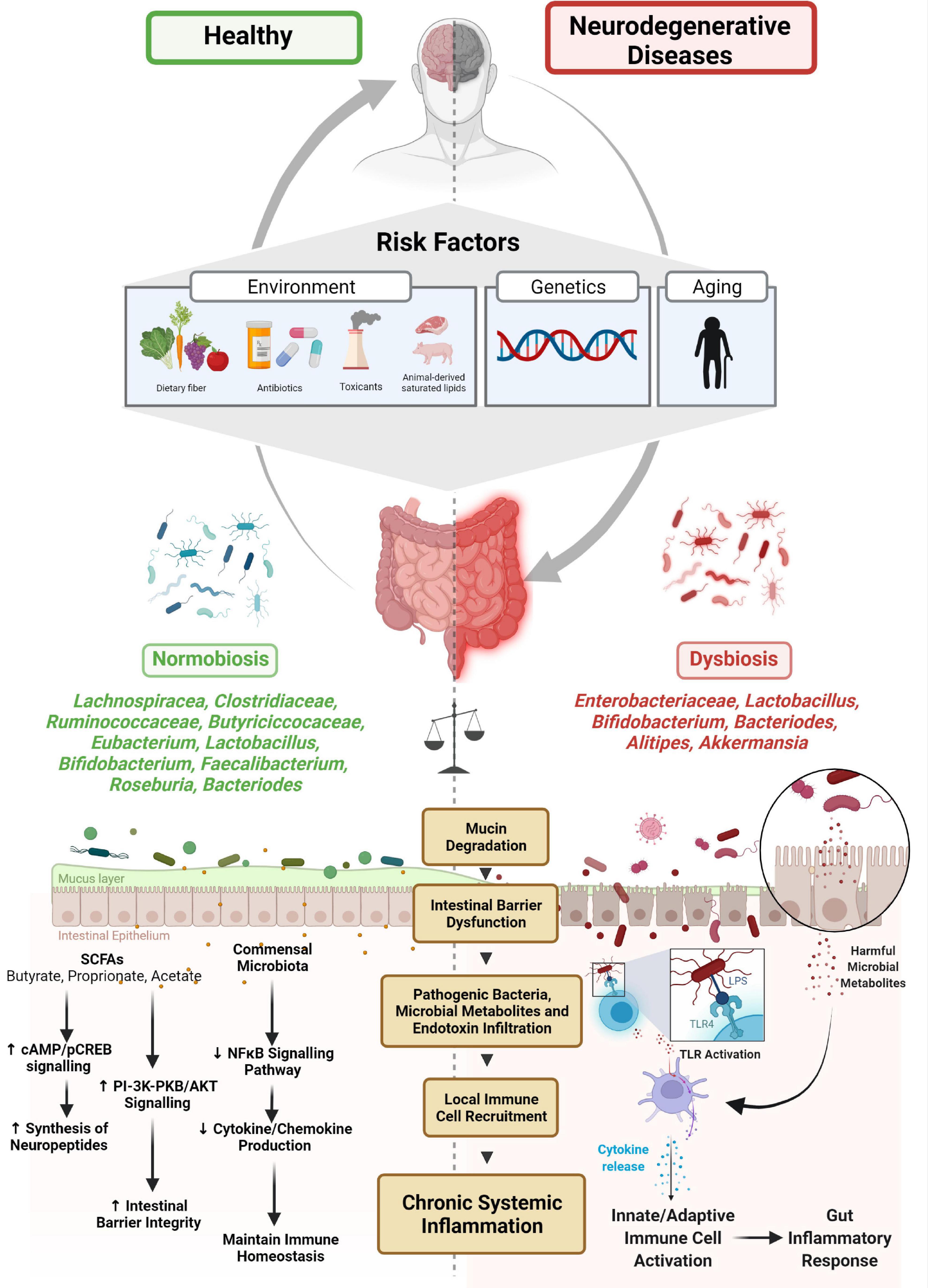
Figure 1. Microbiome-gut-brain axis and neurodegenerative disease. Several risk factors have been associated with the etiology of microbial dysbiosis and neurodegenerative disorders. The balance between normobiosis and dysbiosis within the gut microbiome is preserved by the presence or absence of high-abundant species (Lachnospiracea, Clostridiaceae, Ruminococcaceae, Eubacterium, Butyriciccocaceae, Lactobacillus, Bifidobacterium, Faecalibacterium, Roseburia, and Bacteroides) and low-abundant species (Enterobacteriaceae, Alistipes, and Akkermansia). In the presence of beneficial microbes, homeostatic mechanisms such as neuropeptide synthesis, promotion of intestinal barrier integrity and immune cell regulatory functions are maintained. Higher abundances of microbes are associated with dysbiosis, degrade mucin, dysregulate intestinal barrier, and enable pathogenic microbes and their metabolites and endotoxins to infiltrate, promoting local immune cell recruitment and triggering systemic inflammation.
Advances in genomic technologies allowed for massive cataloging of genomes from fecal microbes to be directly extracted and evaluated by gene ontology bioinformatics tools to understand the biological processes of the human-microbe hybrid (Backhed et al., 2005; Gill et al., 2006). Studies on the role of microbes in health were fostered by the setting up of National Institutes of Health (NIH)- and European-sponsored human microbiome projects that developed publicly available databases of healthy microbiota (see NIH-Human Microbiome Project and European MetaHIT consortium).
Microbial ecosystems are driven by the composition of distinct groups and subgroups of bacterial taxa cohabitating and interacting in a preferred community (Peterson et al., 2009; Arumugam et al., 2011). Using fecal metagenomic sequencing and 16S ribosomal RNA gene profiling from individuals from three continents, two landmark studies in 2011 and 2018 stratified the human gut microbiome into three distinct clusters, called Enterotypes 1 to 3 (Arumugam et al., 2011; Costea et al., 2018), each classified by a dominant genus: Bacteroides (Enterotype 1) and Prevotella (Enterotype 2) and a third Firmicutes dominant cluster with Ruminococcus (Enterotype 3) (Arumugam et al., 2011; Costea et al., 2018). More importantly, along with the dominant genera, a shared network of several co-occurring genera with similar functional properties was detected. For example, Bacteroides and its co-occurring enriched genera Parabacteroides share saccharolytic and proteolytic pathways, thereby deriving energy from diets enriched with animal protein fats and carbohydrate-rich foods. The functional attributes between the ‘microbial-cluster profiles’ were unique, varying in diversity, richness, and temporal stability, thereby supporting the notion that microbes exist in ecological niches within a particular gut environment, and cohabitating microbes share functionally similar metabolic environments (Costea et al., 2018; Jansma and El Aidy, 2021). Furthermore, cluster analyses revealed that the dominant genera with their co-occurring highly abundant microbes influence more macro-molecular functions (e.g., energy production pathways), and the less abundant microbial species detected in all three clusters supported the dominant genera through their involvement in more specialized functions (e.g., archaeon Methanobrevibacter, hydrogen producer and methanogen, and Desulfovibrio, a sulfate reducer) (Arumugam et al., 2011; Costea et al., 2018). Herein, high- and low-abundant species will be used to denote microbes with macro-molecular and specialized functions, respectively. The normobiosis of the gut ecosystem depends on a critical balance among the high- and low-abundant species and the degree to which selective pressures (e.g., environmental factors, genetic predisposition, and age of host) impact compositional features (Figure 1). It is evident from a growing number of studies that high-fat diets, drugs (antibiotics), and toxicants induce dysbiosis and improve the survivability of normally uncommon gut microbial species, leading to enhanced functional responses, pathogen infiltration, and further deterioration of gut health (Desai et al., 2016). Therefore, a detailed network mapping of high- and low-abundant, cohabitating microbes can generate ‘microbial-cluster profiles’ that provide a dynamic view of microbe-microbe and microbe-host interactions and help stratify the heterogeneous multifactorial disease types (Vascellari et al., 2021). Second, cluster analyses can aid in clinical subtyping while also identifying and diagnosing microbe-derived gene signatures as potential biomarkers for neurodegenerative disease (Qian et al., 2020). And finally, results obtained from meta-analyses from clinical data can be corroborated with a more ‘fine-tuned’ mechanistic evaluation in animal models to unravel microbe-mediated pathophysiological changes (Romano et al., 2021).
Unlike other organ systems, the gut is under a constant state of inflammation, observed by a huge expanse of lymphocytes in the lamina propria and the intra-epithelial compartment of the small and large intestines (reviewed in Powell et al., 2017). It can defend from various pervasive pathogens while immunologically tolerating beneficial commensal microbes, densely distributed across a vast, convoluted surface area. Along with the presence of high- and low-abundant microbes, the maintenance of the intestinal epithelial barrier is further maintained by critical players, including immune cells (mononuclear macrophages, T-cells, and innate lymphoid cells), enteric glial cells (EGCs), and the intrinsic/extrinsic enteric-associated-neurons (iEANs/eEANs) of the ENS (Gabanyi et al., 2016; Gury-BenAri et al., 2016; Chen et al., 2018; Jarret et al., 2020; Muller et al., 2020b). The functions of such vital players have been reviewed by Powell et al. (2017) and Natale et al. (2021) and will not be addressed in this review. Given the knowledge gaps in understanding the etiology of neurodegenerative disease, emerging research in microbe-dependent modulation of the ENS and the microbe-ENS-brain axis provides clues into the emergence of enteric dysfunction and neurodegenerative diseases.
The ENS encompasses vast networks of extrinsic and intrinsic neuronal circuits that independently control endocrine functions, conduct transmucosal fluid movement, and regulate local blood flow and motility via reflex systems (reviewed in Rao and Gershon, 2016). Moreover, its intimate connection with gut microbiota is crucial for enteric neurogenesis and neuroprotection (De Vadder et al., 2018), while its colonization is essential for neuronal cell population density (Collins et al., 2014) and neuron-dependent motility reflexes (Delungahawatta et al., 2017). Depleting microbes by antibiotics reduces motility reflexes ex vivo (Delungahawatta et al., 2017), alters colonic motility and neurochemicals, and disrupts enteric neuronal networks in post-weaned animals (Hung et al., 2020). Similarly, a high-fat diet elicits microbial dysbiosis in juvenile rats, myenteric neuronal loss and decreased nitrogenic neurons (McMenamin et al., 2018). In addition to the enteric neurons, microbial colonization enables early postnatal EGC maturation (Inlender et al., 2021) and influences renewal and preservation for the enteric glial population (Kabouridis et al., 2015). Interestingly, veterans of the Persian Gulf War experience GI disorders similar to those of irritable bowl syndrome (IBS) and irritable bowel disease (IBD) (Koch and Emory, 2005). Studies in animal models of Gulf War Syndrome suggest gut-microbial dysbiosis likely increases immune activation mediated by the TLR4-S100β/RAGE-iNOS pathway of EGCs followed by redox instability and dysfunction of the GI barrier (Kimono et al., 2019), further implicating the microbiota as an important regulator of enteric neuronal and glial physiology, independent of CNS modulation.
Although many ENS functions are independent of CNS input, the ENS does function as an interface to detect intestinal luminal cues that trigger upstream CNS signaling pathways. In a landmark paper from Nature, Muller et al. (2020b) identified eEANs in contact with intestinal epithelial cells can detect microbes and their metabolites, specifically SCFAs, bile acids, and neuropeptides, via vagal afferents and subsequently sympathetically modulate gut activity via a CNS-directed circuit (Muller et al., 2020b). Upon loss of microbial signals, modeled by germ-free, gnotobiotic, and broad-spectrum antibiotic-treated rodents, eEANs sensed the changes in intestinal epithelial and mucosal areas in the lamina propria, relayed signals to sensory vagal afferents of the nodose ganglion and dorsal vagal complex, sympathetically activated glutaminergic neurons lateral to the paragigantocellular nucleus/rostral ventrolateral medulla (LPGi/RVLM) and downstream coeliac-superior mesenteric ganglia (CM-SMG), whereby efferent nerves reduced GI transit time. As this study comprehensively evaluated how microbes and their derived metabolites alter gut physiology by direct CNS-mediated gut sympathetic modulation, separate research implicates the role of the aryl hydrocarbon receptor (AHR) as a critical node in regulating gut physiology and neurogenesis (Obata et al., 2020; Wei et al., 2021). AHR, a transcriptional factor differentially expressed in various intestinal segments and dominant in colonic enteric neurons, integrates the signals from the luminal environment and regulates intestinal peristalsis upon microbial colonization (Obata et al., 2020). Furthermore, AHR signaling was also relevant in intestinal barrier function and the mucosal immune system (Metidji et al., 2018; Rothhammer and Quintana, 2019), further implicating its role as a regulatory node, central to maintaining gut homeostasis and a possible biomarker for GI disorders (Obata et al., 2020). Beyond the ENS, this transcriptional factor when selectively activated by the microbial metabolite indole produced by tryptophanase-expressing gut microbes interestingly induces neurogenic effects in the adult murine hippocampus as identified by elevated expression of β-catenin, Neurog2, and VEGF-α (Wei et al., 2021). While administration of antibiotics decreases adult neurogenesis (Mohle et al., 2016), the indole-AHR signaling pathway highlights microbiota as a direct modulator to hippocampal neurogenesis.
In addition to the sensory afferents and autonomic efferents of the extrinsic enteric system, the eEANs concurrently communicate and transmit information to the iEANs (Matheis et al., 2020). However, iEANs exhibit a more region-specific anatomical and gene expression profile than do eEANs, indicating their distinct functional role in maintaining GI functions. Recent evidence signifies the importance of microbes in regulating iEAN-mediated GI homeostasis (Matheis et al., 2020). As the diversity and abundance of microbes increase from the upper GI to distal sites, the number of iEANs likewise increases. The total cell count, morphology, and the gene expression profile of iEANs in the duodenum, ileum, and colonic myenteric plexus varies significantly, with a higher neuronal count in specific-pathogen-free mice compared to germ-free animals (Matheis et al., 2020). GI physiology necessitates the presence of commensal microbes. Compared to both germ-free and antibiotic-administered rodents, commensal microbiota lead to an increased synthesis of neuropeptides, measured indirectly by phosphorylated cAMP response element-binding protein (pCREB), neuropeptide transcripts and protein expression of somatostatin (SST) and cocaine- and amphetamine-regulated transcript (CART) in the ileum and colon. Additionally, the absence of microbiota, or the presence of broad-spectrum antibiotic-induced dysbiosis, triggered the NOD-like receptor family pyrin domain (PYD)-containing 6 (NLRP6)/Caspase 11 (Casp11) inflammasome-mediated pathway and subsequent loss of iEANs and enteric neuropathy (Muller et al., 2020a). Critically, this is especially relevant to GI-mediated complications commonly associated with neurodegenerative disorders, in which GI motility impairment, chronic low-grade inflammation, and intestinal nerve damage are widely observed (De Giorgio et al., 2004; Chalazonitis and Rao, 2018). Like neuron-glia interaction in the CNS, the ENS forms a ‘neuro-immune synapse’ with its local immune players to limit enteric neuropathy and maintain gut health. Recent evidence suggests that intestinal resident muscularis macrophages directly form synapses with iEANs and play an essential role in limiting iEAN neuropathy and mediating their pathogen-resistance mechanism (Matheis et al., 2020). Unlike the lamina propria-residing macrophages, which sense and respond via pathogen-induced phagocytotic mechanisms to clear dying or senescent intestinal epithelial cells, muscularis macrophages present a tissue-protective phenotype by catecholamine-mediated activation of the β2-adrenergic receptor signaling pathway, triggered upon enteric infection. This further highlights the essential role of microbial composition in altering gut-pathogen resistance mechanisms by sympathetic modulation at the neuro-immune synaptic junctions (De Schepper et al., 2018).
Parkinson’s disease is a heterogeneous disease with multiple subtypes (Chaudhuri et al., 2006; Lawton et al., 2018). However, before the onset of clinically diagnosable motor symptoms, most patients experience several non-motor symptoms, including depression, apathy, dementia, rapid eye movement (REM), sleep behavior disorder (RBD), and GI-associated disorders, including reduced gastric emptying and constipation (Chaudhuri et al., 2006; Seppi et al., 2011; Fasano et al., 2015). Although still controversial, several lines of evidence from metagenomic studies of PD patients and healthy age-matched individuals suggest dysbiosis within the gut microbiome of PD patients could modify the risk and progressively worsen disease status (Sampson et al., 2016; Qian et al., 2020; Romano et al., 2021). Despite methodological differences across studies, including inclusion criterion, sample collection, disease status, or additional confounders, contributing to divergent microbial profiles, the evidence of microbial dysbiosis in PD is consistent (Romano et al., 2021; Rosario et al., 2021). Individuals with PD have dramatically divergent microbial profiles compared to healthy controls (Romano et al., 2021). Dominant taxa (e.g., Lachnospiraceae, Ruminococcaceae, Faecalibacterium, Roseburia, and Butyriciccocaceae) that are part of the core microbial community specializing in carbohydrate and energy metabolism and involved in the production of butyrate and other SCFAs, decrease in PD patients, while the genera Akkermansia, Lactobacillus, and Bifidobacterium increase (Geirnaert et al., 2014; Vacca et al., 2020). Although Lactobacillus and Bifidobacterium spp. are considered probiotics and often associated with improving constipation in PD, it is unclear whether their increased abundance is due to a transient immune compensatory response in already immune-compromised patients (Suez et al., 2019; Tan et al., 2021).
As dietary fiber is vital for maintaining the colonic mucus barrier, fiber deficiency favors the proliferation of distinct microbial populations that degrade the colonic mucus layer and enable enhanced colonization and infiltration of opportunistic pathogens (Desai et al., 2016). Akkermansia spp., relatively sparse in healthy subjects, are consistently enriched in PD samples versus controls (Hill-Burns et al., 2017; Wallen et al., 2020; Rosario et al., 2021) and are increasingly abundant in the fecal microbiome of patients who experience constipation, a primary non-motor symptom of PD. Nishiwaki et al. (2020) implicated this genus in the neuropathological progression of PD by mechanisms of degrading mucin. Mucin degradation and altered intestinal O-glycans expression contribute to the eventual erosion of colonic mucosal layers, identified by decreased periodic acid-Schiff (PAS)-positive goblet cells, which could compromise the intestinal barrier (An et al., 2007; Cao et al., 2017) and increase susceptibility to pathogenic infiltration (Desai et al., 2016) (Figure 1). Indeed, an increased abundance of Akkermansia in PD individuals is correlated with an increased presence of co-occurring opportunistic pathogens (Wallen et al., 2020), thereby supporting the notion that higher levels of typically less common microbial species represent a risk factor in modifying PD pathophysiology (Wallen et al., 2020).
Like PD, the influence of microbial dysfunction is further implicated in the etiopathogenesis of AD. Although variability in compositional features exists in separate clinical cohort studies, AD patients demonstrate a differential abundance of several genera, mainly fewer Lachnospiraceae, Clostridiaceae and Ruminococcaceae, and Eubacterium, with more Bacteroides, Bifidobacterium, Enterobacteriaceae, Alistipes, and Akkermansia taxa (Vogt et al., 2017; Liu et al., 2019; Ling et al., 2020; Marizzoni et al., 2020; Lee et al., 2021b; Nara et al., 2021). Furthermore, selected genera such as Escherichia/Shigella isolated from participants’ stool samples have been shown to directly trigger a pro-inflammatory state and amyloid-β accumulation (Cattaneo et al., 2017). Amyloid deposition and peripheral inflammatory mediators are commonly linked to AD pathophysiology. The involvement of gut microbiota in triggering neuroinflammatory pathways was previously observed in antibiotic-treated rodents (Minter et al., 2016). In the APP (Swe)/[PS1(L166P)] transgenic (Tg) model of AD, germ-free status reduced cerebral Aβ load compared to conventionally raised APP rodents (Harach et al., 2017), implicating microbiota as a direct or indirect modifier of peripheral amyloidosis (discussed later in Section “Gut Microbiome-Mediated Fibril Formation”). Functional analyses of the gut microbiota in AD patients reveal broad metabolic changes, including alterations in bacterial cell motility, lipoic acid metabolism, and glycan degradation (Ling et al., 2020). How global functional changes are implicated in the pathophysiology of AD is yet to be uncovered. Specific bacterial taxa, such as Bacteriodetes spp., is associated with AD dementia. However, reports are conflicting as some suggest it could increase the risk of dementia, while others propose the opposite effect (Alkasir et al., 2017; Vogt et al., 2017; Saji et al., 2020). Bacteriodetes are common Gram-negative bacteria commensal to the human gut and are responsible for producing SCFAs in specific GI environments (Parada Venegas et al., 2019). However, in cases of chronic microbial dysbiosis induced by exposure to environmental toxicants, ingestion of a high-fat diet can cause changes in the GI environment that can enable distinct microbes to proliferate and infiltrate neighboring gut environments, and subsequently trigger deleterious inflammatory pathways (Figure 1) (Sun et al., 2018; Miao et al., 2021). Certain species within Bacteriodetes taxa, such as Bacteroides fragilis, contain lipopolysaccharide (LPS) in its outer membrane, and the toxin fragilysin, a pro-inflammatory zinc metalloprotease, can trigger systemic inflammation and amyloid fibrillogenesis (Lukiw, 2016; Zhao et al., 2017). Since aging is a primary risk factor in neurodegenerative diseases, it is likely that the age-dependent reduction in microbial species diversity, including alterations in relative abundances of region-specific microbes, further facilitates persistent degradation and damage of gut mucosal and gut epithelial layers and increases infiltrations of bacterial toxins (Qin et al., 2007; Peniche et al., 2018; Xu et al., 2019). Hyper-stimulation of low-grade inflammatory mediators triggers peripheral endotoxemia and a persistent inflammatory state, popularly called “inflammaging” (reviewed in Franceschi et al., 2018; Brown, 2019; Kowalski and Mulak, 2019; Jukic Peladic et al., 2021).
A clear pathophysiological link between immune and metabolic dysregulation and neurodegenerative disorders in the brain had been previously established (reviewed in Hammond et al., 2019; Yan et al., 2020). More recently, evaluation of microbiome-mediated gut dysregulation provides insight into the peripheral etiology of neurodegenerative disorders such as PD and AD.
Chronic GI inflammation increases the risk of PD (Lee et al., 2021a) and AD-dementia (Zhang et al., 2021). Emerging studies suggest that the enteric microbiome – neuroimmune system interaction could contribute to a novel etiopathogenesis of neurodegenerative disorders. Of the GI-associated conditions commonly associated with an increased risk of neurodegenerative disease, IBS (Liu et al., 2021) and IBD are the most relevant; however, this section will focus on IBD (Kim et al., 2021). The genetic and pathophysiological overlap between IBD and neurodegenerative diseases converges on shared mechanisms, representing early therapy targets.
Recent epidemiological evidence suggests that an increased incidence of IBD is associated with a 20–90% higher risk of PD (Lee et al., 2021a). Furthermore, younger IBD patients aged 40–65 years are at greater risk of developing PD (Kim et al., 2021); however, individuals already with PD are not at risk of developing IBD (Lee et al., 2021a). If chronic gut inflammation can trigger the onset of PD, perhaps early intervention strategies could prevent or slow the downstream pathology. Indeed, in a retrospective study, administration of anti-tumor necrosis factor (anti-TNF) to clinically diagnosed IBD patients led to a 78% reduction in the risk of developing PD, compared to untreated controls (Peter et al., 2018). A similar reduction was observed with the administration of other anti-inflammatory agents, including 5-aminosalicylic acid, azathioprine, and corticosteroids (Park et al., 2019; Pinel Rios et al., 2019). It is evident that, like in IBD, intestinal inflammation and intestinal barrier dysfunction are similarly apparent in PD. Increased inflammatory markers such as CD8B and NFκB p65 were identified in colon biopsies in PD patients, with reduced expression of the regulator protein G-protein signaling 10 (RSG10) in peripheral immune cells (Houser et al., 2021). RGS10 is expressed in myeloid-cell lines and is involved in the neuroprotective mechanism of inhibiting NFκB activity (Lee et al., 2011). RGS10 deficiency triggered intestinal inflammation in RGS10-knockout (KO) mice and augmented MPTP-induced nigrostriatal dopaminergic degeneration (Houser et al., 2021). The occurrence of morphological and functional alterations in the intestinal epithelial layer of PD patients is also evidenced by increased alpha-1-antitrypsin and zonulin in human fecal samples (van IJzendoorn and Derkinderen, 2019; Aho et al., 2021) and the reduction of zona occludens proteins (ZO-1) as revealed by immunofluorescence staining (Kuan et al., 2016). The gut microbiota plays a critical role in maintaining the mucosal intestinal barrier, and exposure to environmental toxicants can further exacerbate PD phenotype (Dodiya et al., 2020). Chronic administration of rotenone, a potent well-studied toxicant of PD, triggered tyrosine hydroxylase-positive neuronal loss in the substantia nigra in both conventionally raised animals (Kanthasamy et al., 2019; Bhattarai et al., 2021) and germ-free animals (Bhattarai et al., 2021). However, decreased motor control and motor coordination as well as disruption of the intestinal epithelial barrier were observed in rotenone-treated conventionally raised mice, but not in rotenone-administered germ-free mice, thereby linking the gut microbiota to PD etiology (Bhattarai et al., 2021). Intriguingly, an altered gut microbiota, a defective intestinal barrier, and gut inflammation were similarly observed in the MPTP mouse model of PD. Timed, subacute MPTP administration disrupted the intestinal barrier, immune status, and gut microbiota after the first dose. These effects recovered; however, the second MPTP dose further exacerbated gut dysbiosis and gut barrier dysfunction, possibly further implicating the microbiota as a modifier of developing PD (Xie et al., 2020).
Beneficial bacteria such as the genera Ruminococcaceae and Lachnospiraceae play an essential role in strengthening the intestinal barrier by producing SCFAs (Baxter et al., 2019). PD patients consistently display lower abundances of these microbes, thereby significantly lowering concentrations of SCFAs, including acetate, propionate, and butyrate (Aho et al., 2021; Baert et al., 2021). As these critical dietary metabolites were previously discussed to regulate the mucosal immune system and protect gut epithelial layers in IBD (Parada Venegas et al., 2019) and NDs (Pellegrini et al., 2018; Wang et al., 2020), a recent report suggests that propionate increases the expression of ZO-1 and occludin, thereby improving intestinal epithelial integrity by a serine-threonine kinase (AKT) signaling pathway (Huang et al., 2021). Additionally, certain conventionally dominant commensal bacteria like Faecalibacterium prausnitzii, which plays a critical role in maintaining immune homeostasis by inhibiting the NFκB signaling pathway and cytokine production and increasing colonic epithelial tight junction proteins, is significantly reduced in both IBD and PD (Laval et al., 2015; Lopez-Siles et al., 2016; Pellegrini et al., 2018). Therefore, a shift in the community of beneficial microbes decreases anti-inflammatory microbial mediators, thereby contributing to a persistent loss in gut barrier integrity and a constipation phenotype often noted in PD patients. Grun et al. (2020) reported that commonly prescribed PD drugs such as COMT inhibitors can further lower the abundance of F. prausnitzii and SCFAs concentrations, thereby enhancing the drug’s bioactivity and toxicity and further damaging the gut epithelial barrier leading to infiltration of other opportunistic pathogens (reviewed in Weersma et al., 2020).
In addition to exposure to environmental toxicants, several genetic risk factors are shared among IBD and PD patients (ROH3P, HLA, CCNY, LRRK2, MAPT, SYMPK, RSPH6A, GUCY1A3, HLA, BTNL2, and TRIM10). The most significant is the Leucine-rich-repeat kinase 2 (LRRK2), a multimeric protein with both kinase and GTPase activity associated with autosomal dominant PD. A missense mutation (G2019S) within LRRK2 modifies α-synuclein pathology in a mouse PD model and increases kinase GTPase activity to inhibit autophagy and augment the pro-inflammatory response in mouse models of colitis (Takagawa et al., 2018; Bieri et al., 2019). Similarly, an autosomal recessive gene associated with early Parkinsonism, DJ-1 (PARK 7), is dysfunctional in human colonic tissue samples as well as in in vitro and in vivo experimental models of colitis (Zhang et al., 2020). As both IBD and PD have compromised intestinal barrier functions, DJ-1 deficiency promotes cytokine-mediated inflammation (IL-1β, IL-6, TNFα, and TGFB1) and apoptosis of intestinal epithelial cells via a p53-dependent mechanism in vitro, in ex vivo colonic sacs, and in vivo (van IJzendoorn and Derkinderen, 2019; Zhang et al., 2020; Lippai et al., 2021). This further supports the hypothesis that the onset, progression and severity of IBD and PD are aggravated by gene-environment interactions occurring at both the host and gut microbiome levels.
Commonalities also exist between IBD incidence and increased risk of AD. A recent Korean cohort study identified that IBD patients ≥65 years suffer an increased risk of AD compared to controls (Kim et al., 2021). These findings further supported a Taiwanese cohort study suggesting the overall incidence of AD dementia is significantly elevated among patients with IBD (Zhang et al., 2021). However, unlike PD, the average age of patients with IBD with dementia was ≥65 years, thereby enabling the possibility of future research investigating the functional consequence of an age-dependent reduction in commensal microbial diversity and richness on AD-dementia. In a recent in vivo study utilizing an aging mouse model of IBD, chronic colitis triggered spatial and memory deficits (He et al., 2021). Dextran sulfate sodium (DSS)-induced colitis caused amyloid plaque accumulation, compromised glymphatic clearance and increased infiltration of gut-derived T-cells, and triggered cortical and hippocampal degeneration mediated by NACHT-LRR and PYD-containing protein 3 (NLRP3) inflammasome expression (He et al., 2021). The absence of NLRP3 protected against neuroinflammatory and neurodegenerative processes in DSS-induced colitis in aged animals. Although a defined mechanistic evaluation of microbe-dependent pathology of AD-dementia is yet to be uncovered, previous studies indicate that AD pathogenesis is perhaps associated with age-dependent depletion of healthy microbes with concurrently increased burden of infectious opportune, enterotoxigenic bacteria and viruses, i.e., Bacteroides fragilis and herpes simplex virus-1 (HSV-1) (Desai et al., 2016; Lukiw et al., 2021). Both B. fragilis and HSV-1 are commensal to the human host and play a critical role in stimulating the innate immune response by the NFκB – microRNA-146a signaling pathway (Li et al., 2020). HSV-1 is noted to trigger peripheral infection and is relevant in the progression of AD (reviewed in McManus and Heneka, 2017; Zhao and Lukiw, 2018). As mentioned earlier, an enterotoxigenic form of B. fragilis can release a unique pro-inflammatory LPS-subtype and fragilysin, leading to an impaired intestinal paracellular and transcellular epithelial barrier, thereby allowing an already ‘leaky’ barrier a gateway for the enhanced entry of microbiome-derived neuro- and entero-toxins into the circulation (Lukiw et al., 2021). Enterotoxigenic B. fragilis triggered GI inflammation in colonic epithelial cells via G-protein coupled receptor-35 (GPCR35) binding. Colitis triggered in mouse models by enterotoxigenic B. fragilis signals bound to GPCR35 was reversed by a GPCR35 antagonist (Boleij et al., 2021).
MicroRNAs (miRNA) are single-stranded, short RNAs that post-transcriptionally regulate gene expression by binding to the 3′ untranslated region (UTR) and silencing the target genes (Chopra et al., 2020; Li et al., 2020). They can be detected in the brain, biofluids [urine, cerebrospinal fluid (CSF), and blood], and, more recently, in stool samples (Tarallo et al., 2021). With a growing number of studies elucidating the role of miRNAs in various disease processes, the gut-residing microbes can further modulate miRNA expression and regulate host pathophysiology. Exciting work by Cao et al. (2021) demonstrated that enterotoxigenic B. fragilis downregulated METTL14-mediated N6-methyladenosine (METTL14/m6A) methylation, thereby blocking the processing of miRNA-149-3p (miR-149-3p). The downregulation of exosome-derived miR-149-3p contributed to T-helper type 17 (Th17) cell differentiation, promoting intestinal inflammation (Cao et al., 2021). Plasma exosomal miR-149-3p was similarly reduced in IBD patients, implying miRNA is a potential biomarker for systemic immune dysregulation. As a growing number of studies implicate distinct microbial species in triggering intestinal inflammation, further mechanistic exploration is necessary to prove causality in neurodegenerative diseases.
In PD, α-synuclein inclusion bodies evidenced in the brainstem, locus coeruleus, and dorsal motor nucleus of the vagus (DMV), were also present in the stomach, duodenum, colon (Shannon et al., 2012; Sanchez-Ferro et al., 2015), and enteric submucosal Meissner’s plexus (Kupsky et al., 1987; Wakabayashi et al., 1988; Braak et al., 2006) in both autopsied PD patients and biopsy samples of the prodromal stage of PD (Shannon et al., 2012; Hilton et al., 2014). Although the interaction between genetic risk factors and environmental toxins likely initiates the oligomerization, aggregation, and propagation of α-synuclein, gut microbiota-derived products can potentiate the pathologic process and further modify disease biology. When the fecal microbiota from PD patients was transplanted to an α-synuclein overexpression (ASO) mouse model, the mice displayed exacerbated α-synuclein inclusion bodies and PD-motor and GI deficits compared to mice transplanted with the microbiome from healthy donors (Sampson et al., 2016, 2020; Hill-Burns et al., 2017). Since a complex microbiota was found to be necessary for worsening α-synuclein pathology, it is likely that a combination of the increase in harmful bacteria, decrease in beneficial bacteria, and similar changes in their derived products trigger and facilitate α-synuclein fibril formation, propagation, and disease pathology (Figure 2).
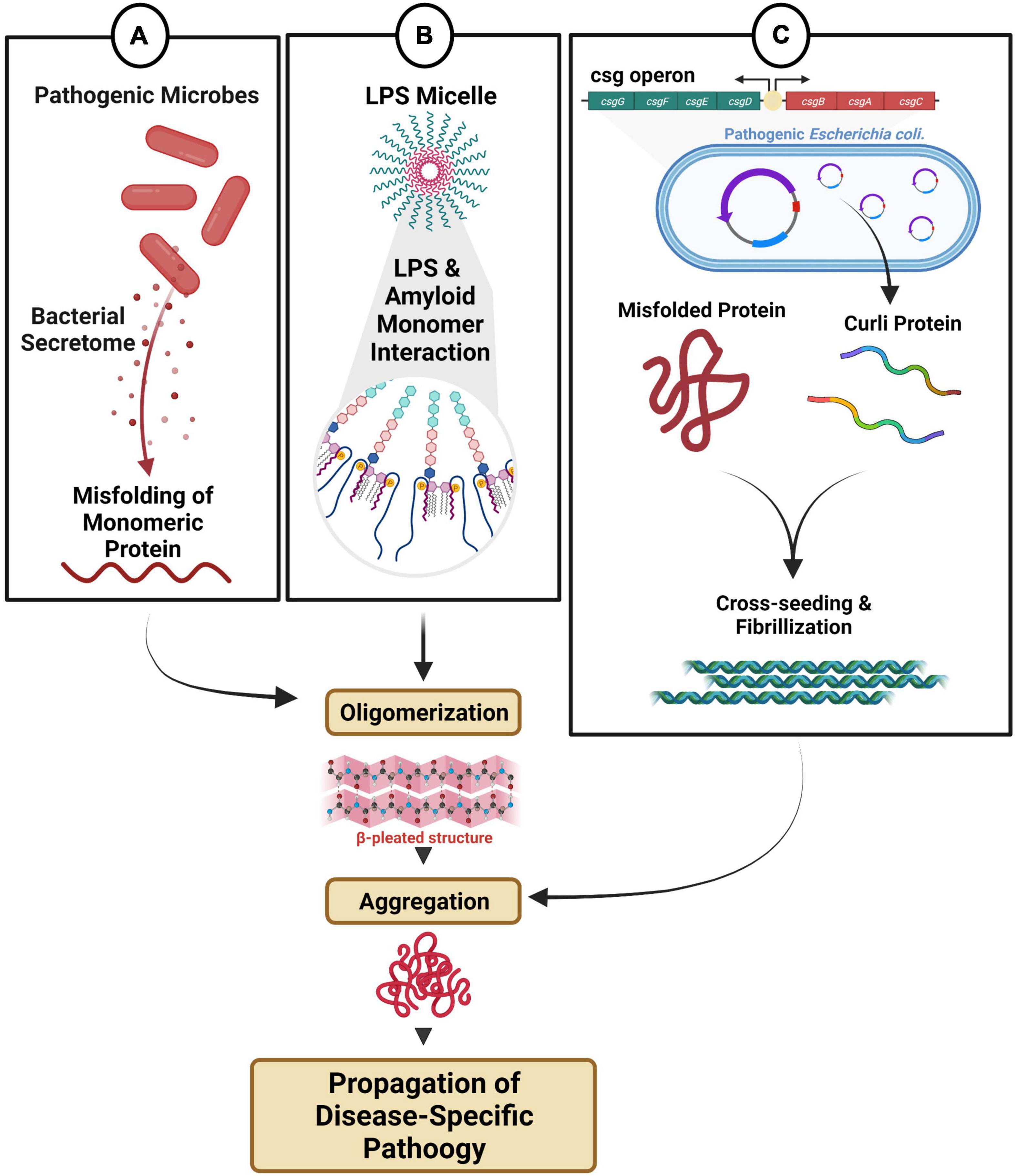
Figure 2. Microbial dysbiosis-mediated fibril formation. (A) Distinct microbes secrete metabolites that can directly and/or indirectly modulate fibril formation. (B) Endotoxin and LPS micelles can interact with amyloid monomeric protein triggering oligomerization and aggregation. (C) Pathogenic bacteria with csgA/B operon produce amyloid curli proteins, which can participate in cross-seeding, fibrillization and aggregation.
The genus Akkermansia is consistently enriched in human PD patients versus healthy controls, and the data corroborate recent findings in an A53T α-synuclein Tg non-human primate model. Investigators noted a higher diversity of microbial flora with significant increases in abundance of Akkermansia, Sybergistetes, and Eggerthella lenta (Nishiwaki et al., 2020; Yan et al., 2021). As a mucin-degrading bacteria, Akkermansia spp. are utilized as a microbial biomarker for determining failure in bacteriotherapy for treating colitis and are implicated in the progression of PD and AD pathology by promoting a pro-inflammatory state (Ihekweazu et al., 2019; Dodiya et al., 2020). While a definitive characterization of Akkermansia spp. in neurodegenerative disorders is yet to be elucidated, recent studies in the chronically stressed rotenone-induced PD model indicate elevated abundances of Akkermansia spp. could modulate fibril formation and exacerbate α-synuclein expression in the colon (Dodiya et al., 2020). Furthermore, an unpublished report suggests that extracellular mucin modulates the release of the Akkermansia protein secretome, which dysregulates calcium homeostasis, leading to the promotion of calcium uptake in the mitochondria of entero-endocrine cells, triggering the formation of reactive oxygen species and subsequent α-synuclein phosphorylation, aggregation, and deposition in vitro (Amorim Neto et al., 2021). Although these results are limited to in vitro evaluation, it highlights previously unknown functions of distinct microbes in influencing the formation of pathogenic fibrils (Figure 2A). Furthermore, it is conceivable that an already altered microbial community structure in PD, causing an overabundance of pathogenic microbes and a reduction in resident protective microbes, could facilitate α-synuclein aggregation and a PD disease phenotype. A recent strain-level meta analysis of gut microbiota of PD patients supports this notion as a reduced abundance of Bacteroides ovatus was noted. B. ovatus is a resident protective microbe that converts dietary flavanols to phenolic acids. Phenolic acids such as 3-hydroxybenzoic acid (3-HBA), 3,4-dihydroxybenzoic acid (3,4-diHBA), and 3-(3-hydroxyphenyl)propionic acid (3-HPPA), produced as a consequence of microbial fermentation, interfere with the assembly of monomeric forms of α-synuclein into its protofibrils in vitro, and upon administration, improved locomotor activity in an A53T Drosophila model of synucleinopathy (Ho et al., 2019). Additionally, Bacillus subtilis, a commensal microbe, can inhibit the aggregation of α-synuclein in both young and aged Caenorhabditis elegans models of synucleinopathy in varied pathways (Goya et al., 2020). B. subtilis induced biofilm formation (mediated from matrix protein TasA), produced nitric oxide, and upregulated numerous protective pathways, including the sphingolipid metabolic pathway responsible for inhibiting and reversing α-synuclein aggregation (Goya et al., 2020). Adult roundworms that fed on vegetative B. subtilis activate downstream transcriptional factors DAF-16/FOXO in the insulin-like receptor DAF-2 pathway to reduce α-synuclein aggregation. Familial PD caused by a mutation in glucocerebrosidase (GBA1) leads to the metabolic dysregulation of the important bioactive lipid ceramide and imbalance of its intermediate glucosylceramide (Plotegher et al., 2019). B. subtilis was found to further exhibit anti-aggregation properties by regulating the sphingolipid metabolic pathway involving upregulation of LAGR-1/CERS1 (ceramide synthase), ASM-3/SMPD1 (acid sphingomyelinase), and downregulation of SPTL-3/SPTLC2 (serine palmitoyltransferase), therefore, reducing glucosylceramide (Goya et al., 2020). Overall, these studies highlight the dynamic abundances of pathogenic and protective microbes and their metabolic products in propagating or limiting α-synuclein aggregation.
In PD and AD individuals, an increased abundance of Enterobacteriaceae taxa, particularly Escherichia coli, is associated with increased concentrations of LPS and the bacterial amyloid protein curli (Scheperjans et al., 2015; Li et al., 2019; Sampson et al., 2020). LPS is a major component of the cell wall of Gram-negative bacteria and is the most well-characterized and understood endotoxin that triggers chronic inflammation and neurodegeneration (Qin et al., 2007), which is found in high concentrations in the blood, gut, skin, and gums during bacterial infections. Several studies show that LPS promotes the aggregation of peripheral amyloid, tau, and α-synuclein, thereby accelerating fibrillogenesis (Lee et al., 2008; Brown, 2019). Notably, NMR studies reveal that α-synuclein aggregation kinetics depends on direct heteromolecular interaction with the LPS-structural motif, leading to downstream nucleation events and stable fibril forms (Figure 2B) (Bhattacharyya et al., 2019). Furthermore, the bacterial curli protein is endogenously produced from microbial species (E. coli/Salmonella spp.) as they contain the csgA/B operon for expression of CsgA/B for biofilm formation. Biofilm formation enables commensal E. coli to persist in the colon’s outer mucus layer. However, other strains of E. coli that are more invasive, aggregative, pathogenic, hemorrhagic and toxigenic can adhere to the intestinal epithelium and trigger an immune response (Rossi et al., 2018). As curli protein is one of the major components of the bacterial extracellular matrix, it was shown to accelerate fibrilization by cross-seeding and aggregation of α-synuclein and β-amyloid (Ivanova et al., 2021). Although the trigger that contributes to fibrillization and pathological cross-seeding curli protein is unclear, a genome-wide screening analysis identified that bacterial curli amyloid was the distinguishing factor for prompting cross-seed α-synuclein aggregation and dysfunction of mitochondrial cellular respiration in both an in vitro neuroblastoma cell line and in vivo A53T α-synuclein over-expressing C. elegans model (Sampson et al., 2020; Wang et al., 2021). These findings were further confirmed in a Thy1-ASO mouse model, where curli-producing bacteria (E. coli strain MC4100) amplified α-synuclein neuropathology in the midbrain and exacerbated motor and GI deficits (Sampson et al., 2020). Bacterial native amyloid monomeric CsgA protein augmented aggregation of α-synuclein proto-fibrils but did not trigger the acceleration of α-synuclein monomer to its oligomeric form, as no interaction occurred between CsgA and α-synuclein monomer in vitro (Sampson et al., 2020). Possibly, other transient mechanisms, e.g., CsgA and α-synuclein interaction with Toll-like receptor-2 (Tukel et al., 2009) are involved in triggering either CsgA or α-synuclein oligomerization (Sampson et al., 2020). Although additional studies are warranted, the curli protein is also known to promote aggregation and neurodegeneration of amyloid-β, SOD1-G85R, and huntingtin in AD, ALS, and Huntington disease models, respectively (Wang et al., 2021). Thus, the pro-fibrillogenic status of amyloid curli protein is a functional consequence of altered gut microbial and metabolic composition (Figure 2C).
Aggregated misfolded protein and autophagolysosomal-proteasomal pathways participate in a vicious cycle that causes cytotoxicity and exacerbates hallmark pathologies of neurodegenerative disorders (Monaco and Fraldi, 2020). Previous studies in CNS point to inflammatory mediators as triggers of the dysfunctional autophagy-lysosomal pathway. Recent studies report similar mechanistic pathways contributing to early seeding and formation of peripheral inclusion bodies in the gut. Upon direct intramuscular injection of pre-formed α-synuclein fibrils in the duodenum of ASO mice, investigators report a heightened inflammatory response (IL-6) in the duodenum 7 days post-inoculation (dpi), which likely suggests an early protective response to maintain enteric health (Schafer et al., 1999; Challis et al., 2020). The preliminary inflammatory response was followed by increased fractalkine levels (a marker for macrophage recruitment), heightened levels of macrophage colony-stimulating factor (MCSF, a marker for macrophage recruitment and differentiation), and recruitment of Iba1+ macrophages in a time-dependent manner. Additionally, no changes to myenteric neuronal cell count were observed; however, the neuronal cell volume had significantly decreased seven dpi, which recovered by 21 dpi. On the contrary, the GFAP+ myenteric EGC count and volume showed a sustained increase over time, reflective of reactive gliosis (Challis et al., 2020). Accumulation of pathological α-synuclein aggregates and a heightened state of gliosis indicate impairment of relevant protein clearance pathways (Fellner et al., 2011). Of the several lysosomal proteins, variants of the major lysosomal enzyme GBA1 are implicated with increased PD risk (Atashrazm et al., 2018). α-Synuclein aggregates can directly inhibit GCase activity and set up a futile feed-forward loop to increase both α-synuclein levels and neurotoxicity (Mazzulli et al., 2011). In the gut, α-synuclein fibrils inhibited the GBA1 function, leading to loss of ENS connectivity and GI functional deficits in aged ASO mice, not young mice (Challis et al., 2020). Thus, age-dependent loss of GBA1 function and heightened susceptibility to peripheral α-synuclein pathology could depend on the loss of ‘healthy’ microbes. As discussed earlier, microbes such as B. subtilis can reduce α-synuclein load directly by activating the DAF-2 signaling pathway and indirectly by regulating the sphingolipid metabolic pathway (Goya et al., 2020). Since B. subtilis abundance is reduced in an age-dependent manner, supporting GBA1 function by microbial supplementation could limit α-synuclein pathology.
Failure of the autophagosomal-proteasomal functions accounts for a substantial percentage of sporadic PD cases (reviewed in Maloney and Lahiri, 2016; Zheng et al., 2016). Interestingly, recent reports suggest that appendectomies lowered PD risk, while individuals with an intact vermiform appendix showed increased levels of truncated soluble α-synuclein, capable of oligomerization and propagation (Killinger et al., 2018). As a vestigial organ, the appendix is indeed a reservoir of beneficial microbes, and it is suggested to be involved in repopulating the large intestine with commensal flora after diarrhea and, in general, maintains intestinal health (Randal Bollinger et al., 2007; Im et al., 2011; Guinane et al., 2013; Sahami et al., 2016). Furthermore, the appendix contains mucosa and submucosa rich with macrophages, follicular dendritic cells, and lymphocytes, which aid in detecting and tolerizing the body to host and foreign antigens (Kooij et al., 2016). But a growing number of studies have identified that the vermiform appendix can also contribute to the development of a chronic intestinal inflammatory state (Sahami et al., 2016). The presence of the appendix significantly correlates with the development of ulcerative colitis, while an inverse association holds for individuals with appendectomies (Sahami et al., 2016). Similar findings were observed in murine colitis models (Mizoguchi et al., 1996; Farkas et al., 2005). Additionally, in analyzing the role of the cecal patch in GF-mice, Masahata et al. (2014) revealed that removal of cecal lymphoid tissue in animals decreased colonic IgA+ cell accumulation in the large intestine and altered fecal microbiota composition. Although a clear mechanistic perspective is lacking, it can be conceptualized that gut microbial dysbiosis (and its metabolites) modulates innate and adaptive immune players by recruiting immune secreting cells from the appendix, thereby priming the immune system for an enhanced inflammatory response and a possible source of α-synuclein misfolding (Masahata et al., 2014; Morrison and Preston, 2016; Zheng et al., 2020b). Although it is well characterized that pathological α-synuclein propagates in a prion-like fashion from the stomach and duodenum to the brain via the vagus (Kim et al., 2019; Van Den Berge et al., 2019), how the appendix facilitates the pathological spread is unanswered since the evidence shows increased levels of α-synuclein with significant enrichment within the axonal varicosities of the mucosal plexus (Gray et al., 2014). Furthermore, the pathological forms of truncated α-synuclein were also identified within a healthy human appendix (Killinger et al., 2018). Interestingly, accumulation of α-synuclein was found in intralysosomal sites within appendiceal mucosal CD68+ macrophages (Gordevicius et al., 2021). As mucosal immune cells contain endolysosomal proteins responsible for the breakdown of α-synuclein, deeper investigations into the appendix of PD patients identified epigenetic silencing via DNA methylation of genes related to the production of autophagic protein and lysosomal degradation pathways (Gordevicius et al., 2021). Although the factors that promote this dysregulation are poorly understood in PD, recent data suggest that increased α-synuclein levels can reduce autophagic flux by decreasing the autophagosomal membrane-associated protein LC3, resulting in further accumulation of α-synuclein and cytotoxicity (Figure 3A) (Lei et al., 2019). Furthermore, the shift in microbial species triggers gut inflammatory pathways and weakens homeostatic autophagosomal-proteasomal functions (necessary for suppressing inflammation). Deteriorated clearance mechanism leads to the accumulation and sensitization of α-synuclein in vivo, thereby increasing the risk of developing PD (Lei et al., 2019). Similarly, the gut microbiota modulate β-amyloid and tau AD pathology. In a study in the 5XFAD mouse model of AD, microbial dysbiosis was noted to be associated with activation of the endolysosomal CCAAT/enhancer-binding protein-β (C/EBP-β) – asparagine endopeptidase (AEP) pathway (Chen et al., 2020). AEP is present in the endolysosome and is implicated in causing aberrant cleaving of amyloid precursor protein (APP N585) and tau N368 leading to plaques and neurofibrillary tangles (NFTs), while C/EBP-β is involved in inducing expression of proinflammatory signaling molecules in microglia and astrocytes and participating in a vicious feedback loop that worsens in an age-dependent manner (Basurto-Islas et al., 2018; Wang et al., 2018). Implanting gut microbiota from aged 3xTg AD mice (APP Swedish, MAPT P301L, and PSEN1 M146V) to young 3xTg mice increased intestinal permeability and temporally augmented C/EBPβ/AEP activation (Chen et al., 2020). These findings support temporal enteric neuronal loss, increased inflammation in the gut, and increased expression of AEP and C/EBPβ in the brain. Interestingly, antibiotic administration diminished the C/EBP-β/AEP signaling pathway in the brain, reduced proinflammatory Iba+ microglia signals, abolished the aggregation of Aβ fibrils measured by thioflavin S assay, and restored cognitive functions (Chen et al., 2020). As these findings do not directly implicate a specific microbial population, age-dependent reductions in diversity and richness of microbial species, with increased gut permeability and infiltration of toxic microbial metabolites such as LPS and amyloids, likely stimulate peripheral C/EBP-β/AEP signaling, and thereby progressive Aβ and NFT pathologies (Chen et al., 2020).
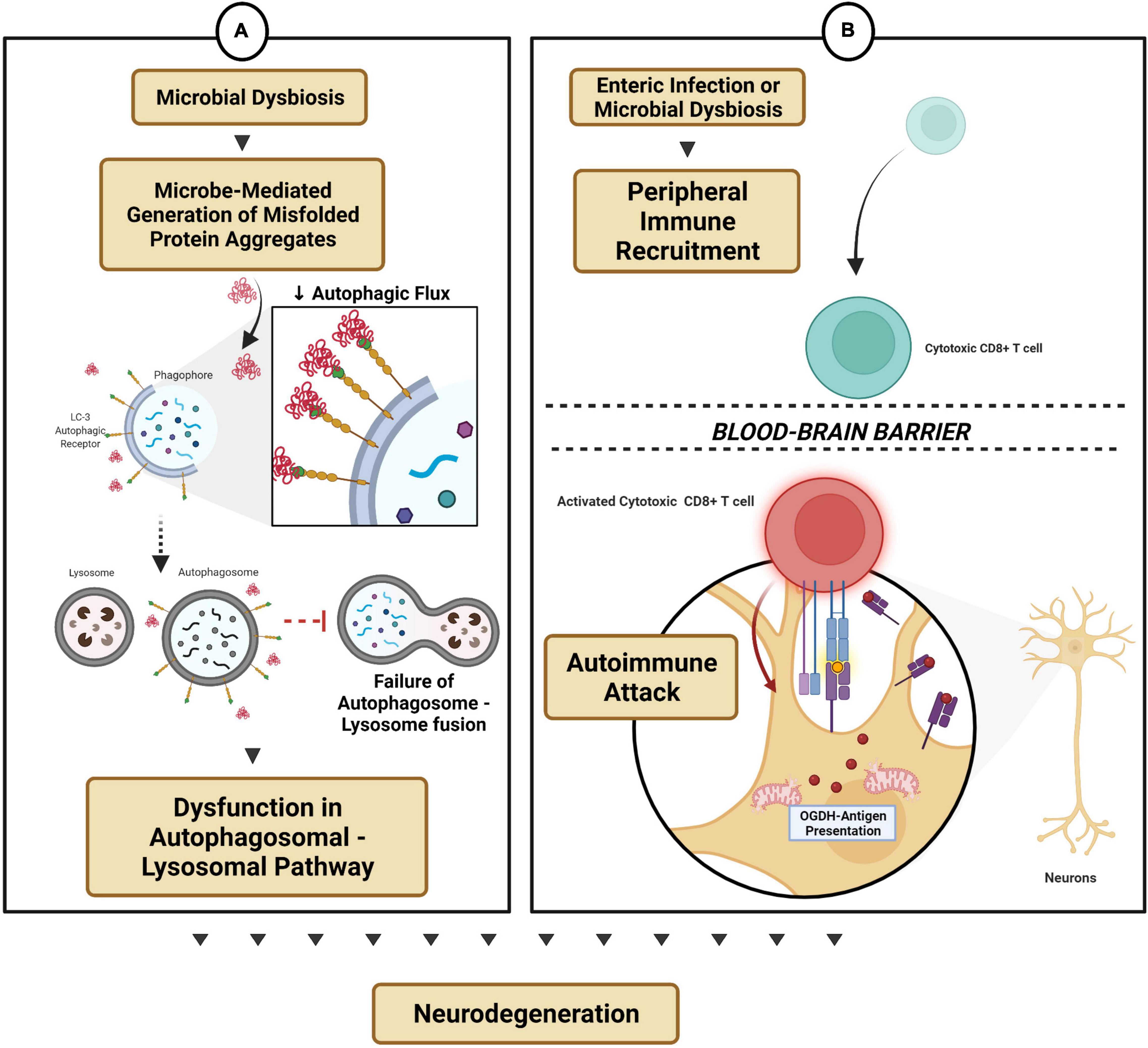
Figure 3. Microbial dysbiosis-modulated protein mechanisms and autoimmune functions. (A) Microbe-mediated generation of protein aggregates reduces autophagic flux and thereby dysregulates the autophagosomal-lysosomal pathway. (B) Enteric infection triggers peripheral immune recruitment into the CNS by gaining access across the blood–brain barrier, thereby triggering an autoimmune attack on subsets of neurons (e.g., dopaminergic neurons).
The peripheral etiology of neurodegenerative disorders links the role of gut microbiota in influencing T-cell differentiation and autoimmune regulation. CD4+ T-cell subsets, T-helper 17 (Th17), and regulatory T cells (Treg) play a vital role by limiting α-synuclein-mediated microglial activation and reducing the activity of CD8+ cytotoxic T-cell and natural killer (NK) cells during the neuroinflammation processes in PD (Chen et al., 2018). Interestingly, CD8+ T-cell infiltration in the substantia nigra pars compacta was observed in the brain in early pre-symptomatic PD, which correlated with α-synuclein aggregate accumulation and eventual dopaminergic neuronal cell loss (Galiano-Landeira et al., 2020). Microbial flora plays a crucial role in regulating and differentiating CD4+ T-cell subsets (Geuking and Burkhard, 2020). Critical Th17/Treg imbalances have been linked to PD pathogenesis (Chen et al., 2015; Zheng et al., 2020b). Understanding microbiome-mediated immunomodulation arises from studies in B. fragilis, an important gut-residing bacterium that expresses a capsular protein, polysaccharide A (PSA). PSA can bind to B-cells, triggering Treg cells to release IL-10 and thereby protect against herpes simplex encephalitis (Sittipo et al., 2018; Ramakrishna et al., 2019). Additionally, PSA protected against pathogenic infections, intestinal inflammatory diseases and triggered anti-inflammatory signaling pathways by plasmacytoid dendritic cells and the Toll-like receptor-2-mediated CD9 signaling mechanism (Erturk-Hasdemir et al., 2019). A longitudinal Japanese PD cohort study reported a gradual reduction in the absolute abundances of B. fragilis quantified by targeted rRNA qRT-PCR (Minato et al., 2017; Zheng et al., 2020a). Although limited by sample size, the findings suggest that intestinal dysbiosis can trigger an autoimmune response and participate in the pathogenesis of PD. Indeed, Pink1-KO mice, a model that is typically asymptomatic of PD and void of dopaminergic degeneration, exhibited a pronounced reduction in the density of tyrosine hydroxylase-positive neurons in the dorsal and ventral striatum and dopamine transporters following infection by a pathogenic Gram-negative bacterium, Citrobacter rodentium (Matheoud et al., 2019). Mutation within the genes of PINK1 and PRKN ubiquitin ligase impairs the process of mitophagy and is associated with the early onset of PD (Ge et al., 2020). Infection by the mouse intestinal pathogen C. rodentium exclusively led to the increased presentation of mitochondrial antigen protein 2-oxoglutarate dehydrogenase (OGDH) on its matrix, expression of the MHC class I molecule on the surface of dopaminergic neurons, infiltration of mitochondrial specific CD8+ T-cells into the brain, dopaminergic neuronal dysfunction and motor impairment (Figure 3B) (Matheoud et al., 2019; Quinn et al., 2020). A follow-up investigation reported that microbial diversity post-infection by C. rodentium was similar in both Pink1-KO mice and their littermate controls, however, the immune activation response was varied (Cannon et al., 2020). At the peak of infection, the level of butyric acid, an SCFA, was disproportionally increased in Pink1-KO mice, possibly implying a compensatory response in early-stage PD pathology (Cannon et al., 2020). Nevertheless, a more detailed mechanistic evaluation is necessary to understand the genetic and environmental roles of microbial immunomodulation and adaptive immunity in the etiology of PD.
As mentioned, a pro-inflammatory state is required to recruit peripheral immune cells into the brain parenchyma, and dysregulation within this process can promote neurodegeneration (Scheld et al., 2016). Since the gut is a gateway for environmental pathogens to enter, and the CNS and ENS participate in pathogen resistance mechanisms, the microbial flora and their metabolites are essential for promoting tight-junction expression and maintaining blood–brain barrier (BBB) function (Braniste et al., 2014). Indeed, microbial dysbiosis, induced by enterobacterial infection, enabled peripheral immune hemocyte recruitment by triggering brain-reactive oxidative species and aggravating neurodegeneration in a Drosophila model of AD (Wu et al., 2017). Furthermore, since T-cells are actively recruited into the brain during a pro-inflammatory response, astrocytes modulate the anti-inflammatory response by activating the TNF-related apoptosis-inducing ligand (TRAIL) (Sanmarco et al., 2021). As TRAIL-expressing astrocytes have been previously demonstrated to induce T-cell apoptosis and downstream CNS inflammation, notably, the expression of this receptor was dependent on meningeal IFNγ expression from NK cells, and the microbiota was known to induce IFNγ expression from NK cells (Burgaletto et al., 2020; Sanmarco et al., 2021). Enteric infection decreases TRAIL expression, thereby promoting subsets of astrocytes to become pathogenic in experimental autoimmune encephalomyelitis, a model for multiple sclerosis (Sanmarco et al., 2021). NK cells are typically prevented from crossing the BBB; however, T-cells can alter their phenotype for increased expression of cell adhesion molecules, chemokine and cytokine receptors, and matrix-degrading enzymes for BBB attachment-gaining entrance (Flugel et al., 2001). Thus, normobiosis within the microbiota enables a basal homeostatic physiological immune response by modulating immune effector sites in the CNS. Bacterial infection or microbial dysbiosis can limit immunoregulatory activity in neuroglial cells, thereby exacerbating neuroinflammation and neurodegeneration. Beyond peripheral immune recruitment, microbial-derived SCFAs and tryptophan metabolites readily gain access through the BBB and into the CNS and act directly on astrocytes and microglia via AHR, SCFAs, and the FFAR2 receptor, respectively, for modulating maturation, function, and inflammation (Braniste et al., 2014; Erny et al., 2015; Rothhammer et al., 2016, 2018; Powell et al., 2017; Wekerle, 2017).
To summarize, the microbiota is a vital modulator for several disease etiologies, including neurodegenerative diseases. As correlative studies in large-cohort populations provide a global view, mechanistic elucidation permits the identification of well-defined molecular pathways that have immense implications for early biomarker discovery and the development of novel therapies. This review attempts to provide some mechanistic insight (Figures 1–3) into how the bi-directional network between the gut microbiota and the brain can not only protect but also trigger and aid chronic gut inflammation, promote a global pro-inflammatory state, modify and contribute to protein misfolding processes, dysregulate the autophagic-lysosomal protein clearance mechanism, differentiate T-cell subsets to facilitate a deleterious autoimmune response, and finally modulate immune recruitment and BBB status. We also discussed how the gut microbiota and the ENS maintain gut physiology and limit enteric neuropathy by CNS-mediated circuits and neuro-immune synapses. As this field is rapidly evolving, and a growing number of studies support the notion that alterations in the microbiome and disease-specific pathologies occur several years before the onset of neurodegeneration, a more refined diagnostic procedure and development of novel intervention strategies (e.g., microbiome-based therapeutics) targeting the microbiome will greatly benefit translational discovery efforts in neurodegeneration.
PP conceived and wrote the manuscript with the help of CW and AGK. KS, GZ, HJ, VA, and AK edited and provided critical feedback for approval of the final manuscript. AGK supervised, conceived the topics with PP, and extended crucial input to support the final manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by NIH R61NS112441, R01 NS100090, R01 ES026892, R21 AG062378, and R01 EB026533.
AGK has an equity interest in PK Biosciences Corporation and Probiome Therapeutics located in Ames, IA, United States. The terms of this arrangement have been reviewed and approved by Iowa State University and University of Georgia in accordance with its conflict-of-interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank the members of AK’s lab for providing intellectual insights into the manuscript. We thank both Iowa State University and the University of Georgia for providing the infrastructure and resources required to complete our research. We also acknowledge Johnny Isakson Endowed Chair, Llyod Endowed Chair, Armbrust Endowment, and Talbot Fellowship award to our research. All figures were created with BioRender.com.
Aho, V. T. E., Houser, M. C., Pereira, P. A. B., Chang, J., Rudi, K., Paulin, L., et al. (2021). Relationships of gut microbiota, short-chain fatty acids, inflammation, and the gut barrier in Parkinson’s disease. Mol. Neurodegener. 16:6. doi: 10.1186/s13024-021-00427-6
Albert, M. J., Mathan, V. I., and Baker, S. J. (1980). Vitamin B12 synthesis by human small intestinal bacteria. Nature 283, 781–782. doi: 10.1038/283781a0
Alkasir, R., Li, J., Li, X., Jin, M., and Zhu, B. (2017). Human gut microbiota: the links with dementia development. Protein Cell 8, 90–102. doi: 10.1007/s13238-016-0338-6
Amorim Neto, D. P., Bosque, B. P., Pereira de Godoy, J. V., Rodrigues, P. V., Meneses, D. D., Tostes, K., et al. (2021). Akkermansia muciniphila secretome promotes α-synuclein aggregation in enteroendocrine cells. bioRxiv doi: 10.1101/2021.02.12.430931
An, G., Wei, B., Xia, B., McDaniel, J. M., Ju, T., Cummings, R. D., et al. (2007). Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 204, 1417–1429. doi: 10.1084/jem.20061929
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Atashrazm, F., Hammond, D., Perera, G., Dobson-Stone, C., Mueller, N., Pickford, R., et al. (2018). Reduced glucocerebrosidase activity in monocytes from patients with Parkinson’s disease. Sci. Rep. 8:15446. doi: 10.1038/s41598-018-33921-x
Backhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Baert, F., Matthys, C., Maselyne, J., Van Poucke, C., Van Coillie, E., Bergmans, B., et al. (2021). Parkinson’s disease patients’ short chain fatty acids production capacity after in vitro fecal fiber fermentation. NPJ Parkinsons Dis. 7:72. doi: 10.1038/s41531-021-00215-5
Basurto-Islas, G., Gu, J. H., Tung, Y. C., Liu, F., and Iqbal, K. (2018). Mechanism of Tau hyperphosphorylation involving lysosomal enzyme asparagine endopeptidase in a mouse model of brain ischemia. J. Alzheimers Dis. 63, 821–833. doi: 10.3233/JAD-170715
Baxter, N. T., Schmidt, A. W., Venkataraman, A., Kim, K. S., Waldron, C., and Schmidt, T. M. (2019). Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. mBio 10, 1–13. doi: 10.1128/mBio.02566-18
Bhattacharyya, D., Mohite, G. M., Krishnamoorthy, J., Gayen, N., Mehra, S., Navalkar, A., et al. (2019). Lipopolysaccharide from gut microbiota modulates alpha-synuclein aggregation and alters its biological function. ACS Chem. Neurosci. 10, 2229–2236. doi: 10.1021/acschemneuro.8b00733
Bhattarai, Y., Si, J., Pu, M., Ross, O. A., McLean, P. J., Till, L., et al. (2021). Role of gut microbiota in regulating gastrointestinal dysfunction and motor symptoms in a mouse model of Parkinson’s disease. Gut Microbes 13:1866974. doi: 10.1080/19490976.2020.1866974
Bieri, G., Brahic, M., Bousset, L., Couthouis, J., Kramer, N. J., Ma, R., et al. (2019). LRRK2 modifies alpha-syn pathology and spread in mouse models and human neurons. Acta Neuropathol. 137, 961–980. doi: 10.1007/s00401-019-01995-0
Boleij, A., Fathi, P., Dalton, W., Park, B., Wu, X., Huso, D., et al. (2021). G-protein coupled receptor 35 (GPR35) regulates the colonic epithelial cell response to enterotoxigenic Bacteroides fragilis. Commun. Biol. 4:585. doi: 10.1038/s42003-021-02014-3
Bosco, N., and Noti, M. (2021). The aging gut microbiome and its impact on host immunity. Genes Immun. 22, 289–303. doi: 10.1038/s41435-021-00126-8
Braak, H., de Vos, R. A., Bohl, J., and Del Tredici, K. (2006). Gastric alpha-synuclein immunoreactive inclusions in Meissner’s and Auerbach’s plexuses in cases staged for Parkinson’s disease-related brain pathology. Neurosci. Lett. 396, 67–72. doi: 10.1016/j.neulet.2005.11.012
Braniste, V., Al-Asmakh, M., Kowal, C., Anuar, F., Abbaspour, A., Toth, M., et al. (2014). The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6:263ra158. doi: 10.1126/scitranslmed.3009759
Britt, R. P., Harper, C., and Spray, G. H. (1971). Megaloblastic anaemia among Indians in Britain. Q. J. Med. 40, 499–520.
Britt, R. P., and Harper, C. M. (1976). Vitamin-B12 deficiency in Asian immigrants. Lancet 2:799. doi: 10.1016/s0140-6736(76)90629-2
Britt, R. P., Stranc, W., and Harper, C. (1970). Pernicious anaemia in Indian immigrants in the London area. Br. J. Haematol. 18, 637–642. doi: 10.1111/j.1365-2141.1970.tb01588.x
Brown, G. C. (2019). The endotoxin hypothesis of neurodegeneration. J. Neuroinflammation 16:180. doi: 10.1186/s12974-019-1564-7
Burgaletto, C., Munafo, A., Di Benedetto, G., De Francisci, C., Caraci, F., Di Mauro, R., et al. (2020). The immune system on the TRAIL of Alzheimer’s disease. J. Neuroinflammation 17:298. doi: 10.1186/s12974-020-01968-1
Cabrera, C., Vicens, P., and Torrente, M. (2021). Modifiable risk factors for dementia: the role of gut microbiota. Curr. Alzheimer Res. 18, 993–1009. doi: 10.2174/1567205018666211215152411
Cannon, T., Sinha, A., Trudeau, L. E., Maurice, C. F., and Gruenheid, S. (2020). Characterization of the intestinal microbiota during Citrobacter rodentium infection in a mouse model of infection-triggered Parkinson’s disease. Gut Microbes 12, 1–11. doi: 10.1080/19490976.2020.1830694
Cao, H., Liu, X., An, Y., Zhou, G., Liu, Y., Xu, M., et al. (2017). Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci. Rep. 7:10322. doi: 10.1038/s41598-017-10835-8
Cao, Y., Wang, Z., Yan, Y., Ji, L., He, J., Xuan, B., et al. (2021). Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology 161, 1552.e12–1566.e12. doi: 10.1053/j.gastro.2021.08.003
Cattaneo, A., Cattane, N., Galluzzi, S., Provasi, S., Lopizzo, N., Festari, C., et al. (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 49, 60–68. doi: 10.1016/j.neurobiolaging.2016.08.019
Chalazonitis, A., and Rao, M. (2018). Enteric nervous system manifestations of neurodegenerative disease. Brain Res. 1693(Pt B), 207–213. doi: 10.1016/j.brainres.2018.01.011
Challis, C., Hori, A., Sampson, T. R., Yoo, B. B., Challis, R. C., Hamilton, A. M., et al. (2020). Gut-seeded alpha-synuclein fibrils promote gut dysfunction and brain pathology specifically in aged mice. Nat. Neurosci. 23, 327–336. doi: 10.1038/s41593-020-0589-7
Chaudhuri, K. R., Healy, D. G., Schapira, A. H., and National Institute for Clinical, E. (2006). Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol. 5, 235–245. doi: 10.1016/S1474-4422(06)70373-8
Chen, C., Ahn, E. H., Kang, S. S., Liu, X., Alam, A., and Ye, K. (2020). Gut dysbiosis contributes to amyloid pathology, associated with C/EBPbeta/AEP signaling activation in Alzheimer’s disease mouse model. Sci. Adv. 6:eaba0466. doi: 10.1126/sciadv.aba0466
Chen, Y., Qi, B., Xu, W., Ma, B., Li, L., Chen, Q., et al. (2015). Clinical correlation of peripheral CD4+cell subsets, their imbalance and Parkinson’s disease. Mol. Med. Rep. 12, 6105–6111. doi: 10.3892/mmr.2015.4136
Chen, Z., Chen, S., and Liu, J. (2018). The role of T cells in the pathogenesis of Parkinson’s disease. Prog. Neurobiol. 169, 1–23. doi: 10.1016/j.pneurobio.2018.08.002
Chopra, N., Wang, R., Maloney, B., Nho, K., Beck, J. S., Pourshafie, N., et al. (2020). MicroRNA-298 reduces levels of human amyloid-beta precursor protein (APP), beta-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 26, 5636–5657. doi: 10.1038/s41380-019-0610-2
Claesson, M. J., O’Sullivan, O., Wang, Q., Nikkila, J., Marchesi, J. R., Smidt, H., et al. (2009). Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS One 4:e6669. doi: 10.1371/journal.pone.0006669
Collins, J., Borojevic, R., Verdu, E. F., Huizinga, J. D., and Ratcliffe, E. M. (2014). Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 26, 98–107. doi: 10.1111/nmo.12236
Costea, P. I., Hildebrand, F., Arumugam, M., Backhed, F., Blaser, M. J., Bushman, F. D., et al. (2018). Enterotypes in the landscape of gut microbial community composition. Nat. Microbiol. 3, 8–16. doi: 10.1038/s41564-017-0072-8
De Giorgio, R., Guerrini, S., Barbara, G., Stanghellini, V., De Ponti, F., Corinaldesi, R., et al. (2004). Inflammatory neuropathies of the enteric nervous system. Gastroenterology 126, 1872–1883. doi: 10.1053/j.gastro.2004.02.024
De Schepper, S., Verheijden, S., Aguilera-Lizarraga, J., Viola, M. F., Boesmans, W., Stakenborg, N., et al. (2018). Self-maintaining gut macrophages are essential for intestinal homeostasis. Cell 175, 400.e13–415.e13. doi: 10.1016/j.cell.2018.07.048
De Vadder, F., Grasset, E., Manneras Holm, L., Karsenty, G., Macpherson, A. J., Olofsson, L. E., et al. (2018). Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc. Natl. Acad. Sci. U.S.A. 115, 6458–6463. doi: 10.1073/pnas.1720017115
Delungahawatta, T., Amin, J. Y., Stanisz, A. M., Bienenstock, J., Forsythe, P., and Kunze, W. A. (2017). Antibiotic driven changes in gut motility suggest direct modulation of enteric nervous system. Front. Neurosci. 11:588. doi: 10.3389/fnins.2017.00588
Desai, M. S., Seekatz, A. M., Koropatkin, N. M., Kamada, N., Hickey, C. A., Wolter, M., et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339.e21–1353.e21. doi: 10.1016/j.cell.2016.10.043
Dinan, T. G., and Cryan, J. F. (2017). Gut instincts: microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 595, 489–503. doi: 10.1113/JP273106
Dodiya, H. B., Forsyth, C. B., Voigt, R. M., Engen, P. A., Patel, J., Shaikh, M., et al. (2020). Chronic stress-induced gut dysfunction exacerbates Parkinson’s disease phenotype and pathology in a rotenone-induced mouse model of Parkinson’s disease. Neurobiol. Dis. 135:104352. doi: 10.1016/j.nbd.2018.12.012
Erny, D., Hrabe de Angelis, A. L., Jaitin, D., Wieghofer, P., Staszewski, O., David, E., et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18, 965–977. doi: 10.1038/nn.4030
Erturk-Hasdemir, D., Oh, S. F., Okan, N. A., Stefanetti, G., Gazzaniga, F. S., Seeberger, P. H., et al. (2019). Symbionts exploit complex signaling to educate the immune system. Proc. Natl. Acad. Sci. U.S.A. 116, 26157–26166. doi: 10.1073/pnas.1915978116
Fang, P., Kazmi, S. A., Jameson, K. G., and Hsiao, E. Y. (2020). The microbiome as a modifier of neurodegenerative disease risk. Cell Host Microbe 28, 201–222. doi: 10.1016/j.chom.2020.06.008
Farkas, S. A., Hornung, M., Sattler, C., Steinbauer, M., Anthuber, M., Obermeier, F., et al. (2005). Preferential migration of CD62L cells into the appendix in mice with experimental chronic colitis. Eur. Surg. Res. 37, 115–122. doi: 10.1159/000084543
Fasano, A., Visanji, N. P., Liu, L. W., Lang, A. E., and Pfeiffer, R. F. (2015). Gastrointestinal dysfunction in Parkinson’s disease. Lancet Neurol. 14, 625–639. doi: 10.1016/S1474-4422(15)00007-1
Fellner, L., Jellinger, K. A., Wenning, G. K., and Stefanova, N. (2011). Glial dysfunction in the pathogenesis of alpha-synucleinopathies: emerging concepts. Acta Neuropathol. 121, 675–693. doi: 10.1007/s00401-011-0833-z
Flugel, A., Berkowicz, T., Ritter, T., Labeur, M., Jenne, D. E., Li, Z., et al. (2001). Migratory activity and functional changes of green fluorescent effector cells before and during experimental autoimmune encephalomyelitis. Immunity 14, 547–560. doi: 10.1016/s1074-7613(01)00143-1
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C., and Santoro, A. (2018). Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14, 576–590. doi: 10.1038/s41574-018-0059-4
Gabanyi, I., Muller, P. A., Feighery, L., Oliveira, T. Y., Costa-Pinto, F. A., and Mucida, D. (2016). Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell 164, 378–391. doi: 10.1016/j.cell.2015.12.023
Galiano-Landeira, J., Torra, A., Vila, M., and Bove, J. (2020). CD8 T cell nigral infiltration precedes synucleinopathy in early stages of Parkinson’s disease. Brain 143, 3717–3733. doi: 10.1093/brain/awaa269
Ge, P., Dawson, V. L., and Dawson, T. M. (2020). PINK1 and Parkin mitochondrial quality control: a source of regional vulnerability in Parkinson’s disease. Mol. Neurodegener. 15:20. doi: 10.1186/s13024-020-00367-7
Geirnaert, A., Steyaert, A., Eeckhaut, V., Debruyne, B., Arends, J. B., Van Immerseel, F., et al. (2014). Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe 30, 70–74. doi: 10.1016/j.anaerobe.2014.08.010
Gershon, M. D. (2019). The Second Brain: A Groundbreaking New Understanding of Nervous Disorders of the Stomach and Intestine. New York, NY: HarperCollins.
Geuking, M. B., and Burkhard, R. (2020). Microbial modulation of intestinal T helper cell responses and implications for disease and therapy. Mucosal Immunol. 13, 855–866. doi: 10.1038/s41385-020-00335-w
Gill, S. R., Pop, M., Deboy, R. T., Eckburg, P. B., Turnbaugh, P. J., Samuel, B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359. doi: 10.1126/science.1124234
Gordevicius, J., Li, P., Marshall, L. L., Killinger, B. A., Lang, S., Ensink, E., et al. (2021). Epigenetic inactivation of the autophagy-lysosomal system in appendix in Parkinson’s disease. Nat. Commun. 12:5134. doi: 10.1038/s41467-021-25474-x
Goya, M. E., Xue, F., Sampedro-Torres-Quevedo, C., Arnaouteli, S., Riquelme-Dominguez, L., Romanowski, A., et al. (2020). Probiotic Bacillus subtilis protects against alpha-synuclein aggregation in C. elegans. Cell Rep. 30, 367.e7–380.e7. doi: 10.1016/j.celrep.2019.12.078
Gray, M. T., Munoz, D. G., Gray, D. A., Schlossmacher, M. G., and Woulfe, J. M. (2014). Alpha-synuclein in the appendiceal mucosa of neurologically intact subjects. Mov. Disord. 29, 991–998. doi: 10.1002/mds.25779
Grun, D., Zimmer, V. C., Kauffmann, J., Spiegel, J., Dillmann, U., Schwiertz, A., et al. (2020). Impact of oral COMT-inhibitors on gut microbiota and short chain fatty acids in Parkinson’s disease. Parkinsonism Relat. Disord. 70, 20–22. doi: 10.1016/j.parkreldis.2019.11.020
Guinane, C. M., Tadrous, A., Fouhy, F., Ryan, C. A., Dempsey, E. M., Murphy, B., et al. (2013). Microbial composition of human appendices from patients following appendectomy. mBio 4:e00366-12. doi: 10.1128/mBio.00366-12
Gury-BenAri, M., Thaiss, C. A., Serafini, N., Winter, D. R., Giladi, A., Lara-Astiaso, D., et al. (2016). The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231.e13–1246.e13. doi: 10.1016/j.cell.2016.07.043
Hammond, T. R., Marsh, S. E., and Stevens, B. (2019). Immune signaling in neurodegeneration. Immunity 50, 955–974. doi: 10.1016/j.immuni.2019.03.016
Harach, T., Marungruang, N., Duthilleul, N., Cheatham, V., Mc Coy, K. D., Frisoni, G., et al. (2017). Erratum: reduction of abeta amyloid pathology in appps1 transgenic mice in the absence of gut microbiota. Sci. Rep. 7:46856. doi: 10.1038/srep46856
He, X. F., Li, L. L., Xian, W. B., Li, M. Y., Zhang, L. Y., Xu, J. H., et al. (2021). Chronic colitis exacerbates NLRP3-dependent neuroinflammation and cognitive impairment in middle-aged brain. J. Neuroinflammation 18:153. doi: 10.1186/s12974-021-02199-8
Hill-Burns, E. M., Debelius, J. W., Morton, J. T., Wissemann, W. T., Lewis, M. R., Wallen, Z. D., et al. (2017). Parkinson’s disease and Parkinson’s disease medications have distinct signatures of the gut microbiome. Mov. Disord. 32, 739–749. doi: 10.1002/mds.26942
Hilton, D., Stephens, M., Kirk, L., Edwards, P., Potter, R., Zajicek, J., et al. (2014). Accumulation of alpha-synuclein in the bowel of patients in the pre-clinical phase of Parkinson’s disease. Acta Neuropathol. 127, 235–241. doi: 10.1007/s00401-013-1214-6
Ho, L., Zhao, D., Ono, K., Ruan, K., Mogno, I., Tsuji, M., et al. (2019). Heterogeneity in gut microbiota drive polyphenol metabolism that influences alpha-synuclein misfolding and toxicity. J. Nutr. Biochem. 64, 170–181. doi: 10.1016/j.jnutbio.2018.10.019
Houser, M. C., Caudle, W. M., Chang, J., Kannarkat, G. T., Yang, Y., Kelly, S. D., et al. (2021). Experimental colitis promotes sustained, sex-dependent, T-cell-associated neuroinflammation and parkinsonian neuropathology. Acta Neuropathol. Commun. 9:139. doi: 10.1186/s40478-021-01240-4
Huang, T., Shi, H., Xu, Y., and Ji, L. (2021). The gut microbiota metabolite propionate ameliorates intestinal epithelial barrier dysfunction-mediated Parkinson’s disease via the AKT signaling pathway. Neuroreport 32, 244–251. doi: 10.1097/WNR.0000000000001585
Hung, L. Y., Parathan, P., Boonma, P., Wu, Q., Wang, Y., Haag, A., et al. (2020). Antibiotic exposure postweaning disrupts the neurochemistry and function of enteric neurons mediating colonic motor activity. Am. J. Physiol. Gastrointest. Liver Physiol. 318, G1042–G1053. doi: 10.1152/ajpgi.00088.2020
Ihekweazu, F. D., Fofanova, T. Y., Queliza, K., Nagy-Szakal, D., Stewart, C. J., Engevik, M. A., et al. (2019). Bacteroides ovatus ATCC 8483 monotherapy is superior to traditional fecal transplant and multi-strain bacteriotherapy in a murine colitis model. Gut Microbes 10, 504–520. doi: 10.1080/19490976.2018.1560753
Im, G. Y., Modayil, R. J., Lin, C. T., Geier, S. J., Katz, D. S., Feuerman, M., et al. (2011). The appendix may protect against Clostridium difficile recurrence. Clin. Gastroenterol. Hepatol. 9, 1072–1077. doi: 10.1016/j.cgh.2011.06.006
Inlender, T., Nissim-Eliraz, E., Stavely, R., Hotta, R., Goldstein, A. M., Yagel, S., et al. (2021). Homeostasis of mucosal glial cells in human gut is independent of microbiota. Sci. Rep. 11:12796. doi: 10.1038/s41598-021-92384-9
Ivanova, M. I., Lin, Y., Lee, Y. H., Zheng, J., and Ramamoorthy, A. (2021). Biophysical processes underlying cross-seeding in amyloid aggregation and implications in amyloid pathology. Biophys. Chem. 269:106507. doi: 10.1016/j.bpc.2020.106507
Jansma, J., and El Aidy, S. (2021). Understanding the host-microbe interactions using metabolic modeling. Microbiome 9:16. doi: 10.1186/s40168-020-00955-1
Jarret, A., Jackson, R., Duizer, C., Healy, M. E., Zhao, J., Rone, J. M., et al. (2020). Enteric nervous system-derived IL-18 orchestrates mucosal barrier immunity. Cell 180, 50–63. doi: 10.1016/j.cell.2019.12.016
Jukic Peladic, N., Dell’Aquila, G., Carrieri, B., Maggio, M., Cherubini, A., and Orlandoni, P. (2021). Potential role of probiotics for inflammaging: a narrative review. Nutrients 13:2919. doi: 10.3390/nu13092919
Kabouridis, P. S., Lasrado, R., McCallum, S., Chng, S. H., Snippert, H. J., Clevers, H., et al. (2015). Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 85, 289–295. doi: 10.1016/j.neuron.2014.12.037
Kanthasamy, A., Jin, H., Charli, A., Vellareddy, A., and Kanthasamy, A. (2019). Environmental neurotoxicant-induced dopaminergic neurodegeneration: a potential link to impaired neuroinflammatory mechanisms. Pharmacol. Ther. 197, 61–82. doi: 10.1016/j.pharmthera.2019.01.001
Killinger, B. A., Madaj, Z., Sikora, J. W., Rey, N., Haas, A. J., Vepa, Y., et al. (2018). The vermiform appendix impacts the risk of developing Parkinson’s disease. Sci. Transl. Med. 10:eaar5280. doi: 10.1126/scitranslmed.aar5280
Kim, G. H., Lee, Y. C., Kim, T. J., Kim, E. R., Hong, S. N., Chang, D. K., et al. (2021). Risk of neurodegenerative diseases in patients with inflammatory bowel disease: a nationwide population-based cohort study. J. Crohns Colitis doi: 10.1093/ecco-jcc/jjab162, Online ahead of print,
Kim, S., Kwon, S. H., Kam, T. I., Panicker, N., Karuppagounder, S. S., Lee, S., et al. (2019). Transneuronal propagation of pathologic alpha-synuclein from the gut to the brain models Parkinson’s disease. Neuron 103, 627.e7–641.e7. doi: 10.1016/j.neuron.2019.05.035
Kimono, D., Sarkar, S., Albadrani, M., Seth, R., Bose, D., Mondal, A., et al. (2019). Dysbiosis-associated enteric glial cell immune-activation and redox imbalance modulate tight junction protein expression in gulf war illness pathology. Front. Physiol. 10:1229. doi: 10.3389/fphys.2019.01229
Koch, T. R., and Emory, T. S. (2005). Evaluation of chronic gastrointestinal symptoms following persian gulf war exposure. Mil. Med. 170, 696–700. doi: 10.7205/milmed.170.8.696
Kooij, I. A., Sahami, S., Meijer, S. L., Buskens, C. J., and Te Velde, A. A. (2016). The immunology of the vermiform appendix: a review of the literature. Clin. Exp. Immunol. 186, 1–9. doi: 10.1111/cei.12821
Kowalski, K., and Mulak, A. (2019). Brain-gut-microbiota axis in Alzheimer’s disease. J. Neurogastroenterol. Motil. 25, 48–60. doi: 10.5056/jnm18087
Kuan, W. L., Bennett, N., He, X., Skepper, J. N., Martynyuk, N., Wijeyekoon, R., et al. (2016). Alpha-synuclein pre-formed fibrils impair tight junction protein expression without affecting cerebral endothelial cell function. Exp. Neurol. 285(Pt A), 72–81. doi: 10.1016/j.expneurol.2016.09.003
Kupsky, W. J., Grimes, M. M., Sweeting, J., Bertsch, R., and Cote, L. J. (1987). Parkinson’s disease and megacolon: concentric hyaline inclusions (Lewy bodies) in enteric ganglion cells. Neurology 37, 1253–1255. doi: 10.1212/wnl.37.7.1253
Lahiri, D. K., Maloney, B., and Zawia, N. H. (2009). The LEARn model: an epigenetic explanation for idiopathic neurobiological diseases. Mol. Psychiatry 14, 992–1003. doi: 10.1038/mp.2009.82
Laval, L., Martin, R., Natividad, J. N., Chain, F., Miquel, S., Desclee de Maredsous, C., et al. (2015). Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes 6, 1–9. doi: 10.4161/19490976.2014.990784
Lawton, M., Ben-Shlomo, Y., May, M. T., Baig, F., Barber, T. R., Klein, J. C., et al. (2018). Developing and validating Parkinson’s disease subtypes and their motor and cognitive progression. J. Neurol. Neurosurg. Psychiatry 89, 1279–1287. doi: 10.1136/jnnp-2018-318337
Lee, H. S., Lobbestael, E., Vermeire, S., Sabino, J., and Cleynen, I. (2021a). Inflammatory bowel disease and Parkinson’s disease: common pathophysiological links. Gut 70, 408–417. doi: 10.1136/gutjnl-2020-322429
Lee, J., Choi, M. K., Kim, J., Chun, S., Kim, H. G., Lee, H., et al. (2021b). Development and optimization of a rapid colorimetric membrane immunoassay for Porphyromonas gingivalis. J. Microbiol. Biotechnol. 31, 705–709. doi: 10.4014/jmb.2103.03029
Lee, J. K., Chung, J., McAlpine, F. E., and Tansey, M. G. (2011). Regulator of G-protein signaling-10 negatively regulates NF-kappaB in microglia and neuroprotects dopaminergic neurons in hemiparkinsonian rats. J. Neurosci. 31, 11879–11888. doi: 10.1523/JNEUROSCI.1002-11.2011
Lee, J. W., Lee, Y. K., Yuk, D. Y., Choi, D. Y., Ban, S. B., Oh, K. W., et al. (2008). Neuro-inflammation induced by lipopolysaccharide causes cognitive impairment through enhancement of beta-amyloid generation. J. Neuroinflammation 5:37. doi: 10.1186/1742-2094-5-37
Lei, Z., Cao, G., and Wei, G. (2019). A30P mutant alpha-synuclein impairs autophagic flux by inactivating JNK signaling to enhance ZKSCAN3 activity in midbrain dopaminergic neurons. Cell Death Dis. 10:133. doi: 10.1038/s41419-019-1364-0
Li, B., He, Y., Ma, J., Huang, P., Du, J., Cao, L., et al. (2019). Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 15, 1357–1366. doi: 10.1016/j.jalz.2019.07.002
Li, M., Chen, W. D., and Wang, Y. D. (2020). The roles of the gut microbiota-miRNA interaction in the host pathophysiology. Mol. Med. 26:101. doi: 10.1186/s10020-020-00234-7
Ling, Z., Zhu, M., Yan, X., Cheng, Y., Shao, L., Liu, X., et al. (2020). Structural and functional dysbiosis of fecal microbiota in chinese patients with Alzheimer’s disease. Front. Cell Dev. Biol. 8:634069. doi: 10.3389/fcell.2020.634069
Lippai, R., Veres-Szekely, A., Sziksz, E., Iwakura, Y., Pap, D., Rokonay, R., et al. (2021). Immunomodulatory role of Parkinson’s disease 7 in inflammatory bowel disease. Sci. Rep. 11:14582. doi: 10.1038/s41598-021-93671-1
Liu, B., Sjolander, A., Pedersen, N. L., Ludvigsson, J. F., Chen, H., Fang, F., et al. (2021). Irritable bowel syndrome and Parkinson’s disease risk: register-based studies. NPJ Parkinsons Dis. 7:5. doi: 10.1038/s41531-020-00145-8
Liu, P., Wu, L., Peng, G., Han, Y., Tang, R., Ge, J., et al. (2019). Altered microbiomes distinguish Alzheimer’s disease from amnestic mild cognitive impairment and health in a Chinese cohort. Brain Behav. Immun. 80, 633–643. doi: 10.1016/j.bbi.2019.05.008
Lopez-Siles, M., Martinez-Medina, M., Suris-Valls, R., Aldeguer, X., Sabat-Mir, M., Duncan, S. H., et al. (2016). Changes in the abundance of Faecalibacterium prausnitzii phylogroups I and II in the intestinal mucosa of inflammatory bowel disease and patients with colorectal cancer. Inflamm. Bowel Dis. 22, 28–41. doi: 10.1097/MIB.0000000000000590
Lukiw, W. J. (2016). Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer’s disease. Front. Microbiol. 7:1544. doi: 10.3389/fmicb.2016.01544
Lukiw, W. J., Arceneaux, L., Li, W., Bond, T., and Zhao, Y. (2021). Gastrointestinal (GI)-tract microbiome derived neurotoxins and their potential contribution to inflammatory neurodegeneration in Alzheimer’s disease (AD). J. Alzheimers Dis. Parkinsonism 11:525.
Ma, Q., Xing, C., Long, W., Wang, H. Y., Liu, Q., and Wang, R. F. (2019). Impact of microbiota on central nervous system and neurological diseases: the gut-brain axis. J. Neuroinflammation 16:53. doi: 10.1186/s12974-019-1434-3
Maloney, B., and Lahiri, D. K. (2016). Epigenetics of dementia: understanding the disease as a transformation rather than a state. Lancet Neurol. 15, 760–774. doi: 10.1016/S1474-4422(16)00065-X
Marizzoni, M., Cattaneo, A., Mirabelli, P., Festari, C., Lopizzo, N., Nicolosi, V., et al. (2020). Short-chain fatty acids and lipopolysaccharide as mediators between gut dysbiosis and amyloid pathology in Alzheimer’s disease. J. Alzheimers Dis. 78, 683–697. doi: 10.3233/JAD-200306
Martin, R., Makino, H., Cetinyurek Yavuz, A., Ben-Amor, K., Roelofs, M., Ishikawa, E., et al. (2016). Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498
Martinez, I., Maldonado-Gomez, M. X., Gomes-Neto, J. C., Kittana, H., Ding, H., Schmaltz, R., et al. (2018). Experimental evaluation of the importance of colonization history in early-life gut microbiota assembly. Elife 7:e36521. doi: 10.7554/eLife.36521
Masahata, K., Umemoto, E., Kayama, H., Kotani, M., Nakamura, S., Kurakawa, T., et al. (2014). Generation of colonic IgA-secreting cells in the caecal patch. Nat. Commun. 5:3704. doi: 10.1038/ncomms4704
Mathan, V. I., Babior, B. M., and Donaldson, R. M. Jr. (1974). Kinetics of the attachment of intrinsic factor-bound cobamides to ileal receptors. J. Clin. Invest. 54, 598–608. doi: 10.1172/jci107797
Matheis, F., Muller, P. A., Graves, C. L., Gabanyi, I., Kerner, Z. J., Costa-Borges, D., et al. (2020). Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell 180, 64.e16–78.e16. doi: 10.1016/j.cell.2019.12.002
Matheoud, D., Cannon, T., Voisin, A., Penttinen, A. M., Ramet, L., Fahmy, A. M., et al. (2019). Intestinal infection triggers Parkinson’s disease-like symptoms in Pink1(-/-) mice. Nature 571, 565–569. doi: 10.1038/s41586-019-1405-y
Matsumoto, M., Inoue, R., Tsukahara, T., Ushida, K., Chiji, H., Matsubara, N., et al. (2008). Voluntary running exercise alters microbiota composition and increases n-butyrate concentration in the rat cecum. Biosci. Biotechnol. Biochem. 72, 572–576. doi: 10.1271/bbb.70474
Mazzulli, J. R., Xu, Y. H., Sun, Y., Knight, A. L., McLean, P. J., Caldwell, G. A., et al. (2011). Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell 146, 37–52. doi: 10.1016/j.cell.2011.06.001
McManus, R. M., and Heneka, M. T. (2017). Role of neuroinflammation in neurodegeneration: new insights. Alzheimers Res. Ther. 9:14. doi: 10.1186/s13195-017-0241-2
McMenamin, C. A., Clyburn, C., and Browning, K. N. (2018). High-fat diet during the perinatal period induces loss of myenteric nitrergic neurons and increases enteric glial density, prior to the development of obesity. Neuroscience 393, 369–380. doi: 10.1016/j.neuroscience.2018.09.033
Metidji, A., Omenetti, S., Crotta, S., Li, Y., Nye, E., Ross, E., et al. (2018). The environmental sensor AHR protects from inflammatory damage by maintaining intestinal stem cell homeostasis and barrier integrity. Immunity 49, 353.e5–362.e5. doi: 10.1016/j.immuni.2018.07.010
Miao, Z. H., Zhou, W. X., Cheng, R. Y., Liang, H. J., Jiang, F. L., Shen, X., et al. (2021). Dysbiosis of intestinal microbiota in early life aggravates high-fat diet induced dysmetabolism in adult mice. BMC Microbiol. 21:209. doi: 10.1186/s12866-021-02263-6
Minato, T., Maeda, T., Fujisawa, Y., Tsuji, H., Nomoto, K., Ohno, K., et al. (2017). Progression of Parkinson’s disease is associated with gut dysbiosis: two-year follow-up study. PLoS One 12:e0187307. doi: 10.1371/journal.pone.0187307
Minter, M. R., Zhang, C., Leone, V., Ringus, D. L., Zhang, X., Oyler-Castrillo, P., et al. (2016). Antibiotic-induced perturbations in gut microbial diversity influences neuro-inflammation and amyloidosis in a murine model of Alzheimer’s disease. Sci. Rep. 6:30028. doi: 10.1038/srep30028
Mitchell, C. M., Mazzoni, C., Hogstrom, L., Bryant, A., Bergerat, A., Cher, A., et al. (2020). Delivery mode affects stability of early infant gut microbiota. Cell Rep. Med. 1:100156. doi: 10.1016/j.xcrm.2020.100156
Mizoguchi, A., Mizoguchi, E., Chiba, C., and Bhan, A. K. (1996). Role of appendix in the development of inflammatory bowel disease in TCR-alpha mutant mice. J. Exp. Med. 184, 707–715. doi: 10.1084/jem.184.2.707
Mohle, L., Mattei, D., Heimesaat, M. M., Bereswill, S., Fischer, A., Alutis, M., et al. (2016). Ly6C(hi) monocytes provide a link between antibiotic-induced changes in gut microbiota and adult hippocampal neurogenesis. Cell Rep. 15, 1945–1956. doi: 10.1016/j.celrep.2016.04.074
Monaco, A., and Fraldi, A. (2020). Protein aggregation and dysfunction of autophagy-lysosomal pathway: a vicious cycle in lysosomal storage diseases. Front. Mol. Neurosci. 13:37. doi: 10.3389/fnmol.2020.00037
Morrison, D. J., and Preston, T. (2016). Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 7, 189–200. doi: 10.1080/19490976.2015.1134082
Muller, P. A., Matheis, F., Schneeberger, M., Kerner, Z., Jove, V., and Mucida, D. (2020a). Microbiota-modulated CART(+) enteric neurons autonomously regulate blood glucose. Science 370, 314–321. doi: 10.1126/science.abd6176
Muller, P. A., Schneeberger, M., Matheis, F., Wang, P., Kerner, Z., Ilanges, A., et al. (2020b). Microbiota modulate sympathetic neurons via a gut-brain circuit. Nature 583, 441–446. doi: 10.1038/s41586-020-2474-7
Nara, P. L., Sindelar, D., Penn, M. S., Potempa, J., and Griffin, W. S. T. (2021). Porphyromonas gingivalis outer membrane vesicles as the major driver of and explanation for neuropathogenesis, the cholinergic hypothesis, iron dyshomeostasis, and salivary lactoferrin in Alzheimer’s disease. J. Alzheimers Dis. 82, 1417–1450. doi: 10.3233/JAD-210448
Natale, G., Ryskalin, L., Morucci, G., Lazzeri, G., Frati, A., and Fornai, F. (2021). The baseline structure of the enteric nervous system and its role in Parkinson’s disease. Life 11:732. doi: 10.3390/life11080732
Nishiwaki, H., Ito, M., Ishida, T., Hamaguchi, T., Maeda, T., Kashihara, K., et al. (2020). Meta-analysis of gut dysbiosis in Parkinson’s disease. Mov. Disord. 35, 1626–1635. doi: 10.1002/mds.28119
Obata, Y., Castano, A., Boeing, S., Bon-Frauches, A. C., Fung, C., Fallesen, T., et al. (2020). Neuronal programming by microbiota regulates intestinal physiology. Nature 578, 284–289. doi: 10.1038/s41586-020-1975-8
Parada Venegas, D., De la Fuente, M. K., Landskron, G., Gonzalez, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Park, S., Kim, J., Chun, J., Han, K., Soh, H., Kang, E. A., et al. (2019). Patients with inflammatory bowel disease are at an increased risk of Parkinson’s Disease: a south Korean nationwide population-based study. J. Clin. Med. 8:1191. doi: 10.3390/jcm8081191
Pellegrini, C., Antonioli, L., Colucci, R., Blandizzi, C., and Fornai, M. (2018). Interplay among gut microbiota, intestinal mucosal barrier and enteric neuro-immune system: a common path to neurodegenerative diseases? Acta Neuropathol. 136, 345–361. doi: 10.1007/s00401-018-1856-5
Peniche, A. G., Spinler, J. K., Boonma, P., Savidge, T. C., and Dann, S. M. (2018). Aging impairs protective host defenses against Clostridioides (Clostridium) difficile infection in mice by suppressing neutrophil and IL-22 mediated immunity. Anaerobe 54, 83–91. doi: 10.1016/j.anaerobe.2018.07.011
Peter, I., Dubinsky, M., Bressman, S., Park, A., Lu, C., Chen, N., et al. (2018). Anti-tumor necrosis factor therapy and incidence of parkinson disease among patients with inflammatory bowel disease. JAMA Neurol. 75, 939–946. doi: 10.1001/jamaneurol.2018.0605
Peterson, J., Garges, S., Giovanni, M., McInnes, P., Wang, L., Schloss, J. A., et al. (2009). The NIH human microbiome project. Genome Res. 19, 2317–2323. doi: 10.1101/gr.096651.109
Pickard, J. M., Zeng, M. Y., Caruso, R., and Nunez, G. (2017). Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567
Pinel Rios, J., Madrid Navarro, C. J., Perez Navarro, M. J., Cabello Tapia, M. J., Pina Vera, M. J., Campos Arillo, V., et al. (2019). Association of Parkinson’s disease and treatment with aminosalicylates in inflammatory bowel disease: a cross-sectional study in a Spain drug dispensation records. BMJ Open 9:e025574. doi: 10.1136/bmjopen-2018-025574
Plotegher, N., Bubacco, L., Greggio, E., and Civiero, L. (2019). Ceramides in Parkinson’s disease: from recent evidence to new hypotheses. Front. Neurosci. 13:330. doi: 10.3389/fnins.2019.00330
Powell, N., Walker, M. M., and Talley, N. J. (2017). The mucosal immune system: master regulator of bidirectional gut-brain communications. Nat. Rev. Gastroenterol. Hepatol. 14, 143–159. doi: 10.1038/nrgastro.2016.191
Qian, Y., Yang, X., Xu, S., Huang, P., Li, B., Du, J., et al. (2020). Gut metagenomics-derived genes as potential biomarkers of Parkinson’s disease. Brain 143, 2474–2489. doi: 10.1093/brain/awaa201
Qin, L., Wu, X., Block, M. L., Liu, Y., Breese, G. R., Hong, J. S., et al. (2007). Systemic LPS causes chronic neuroinflammation and progressive neurodegeneration. Glia 55, 453–462. doi: 10.1002/glia.20467
Quinn, P. M. J., Moreira, P. I., Ambrosio, A. F., and Alves, C. H. (2020). PINK1/PARKIN signalling in neurodegeneration and neuroinflammation. Acta Neuropathol. Commun. 8:189. doi: 10.1186/s40478-020-01062-w
Ramakrishna, C., Kujawski, M., Chu, H., Li, L., Mazmanian, S. K., and Cantin, E. M. (2019). Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat. Commun. 10:2153. doi: 10.1038/s41467-019-09884-6
Randal Bollinger, R., Barbas, A. S., Bush, E. L., Lin, S. S., and Parker, W. (2007). Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. J. Theor. Biol. 249, 826–831. doi: 10.1016/j.jtbi.2007.08.032
Rao, M., and Gershon, M. D. (2016). The bowel and beyond: the enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 13, 517–528. doi: 10.1038/nrgastro.2016.107
Roberts, P. D., James, H., Petrie, A., Morgan, J. O., and Hoffbrand, A. V. (1973). Vitamin B 12 status in pregnancy among immigrants to Britain. Br. Med. J. 3, 67–72. doi: 10.1136/bmj.3.5871.67
Romano, S., Savva, G. M., Bedarf, J. R., Charles, I. G., Hildebrand, F., and Narbad, A. (2021). Meta-analysis of the Parkinson’s disease gut microbiome suggests alterations linked to intestinal inflammation. NPJ Parkinsons Dis. 7:27. doi: 10.1038/s41531-021-00156-z
Rosario, D., Bidkhori, G., Lee, S., Bedarf, J., Hildebrand, F., Le Chatelier, E., et al. (2021). Systematic analysis of gut microbiome reveals the role of bacterial folate and homocysteine metabolism in Parkinson’s disease. Cell Rep. 34:108807. doi: 10.1016/j.celrep.2021.108807
Rossi, E., Cimdins, A., Luthje, P., Brauner, A., Sjoling, A., Landini, P., et al. (2018). “It’s a gut feeling” - Escherichia coli biofilm formation in the gastrointestinal tract environment. Crit. Rev. Microbiol. 44, 1–30. doi: 10.1080/1040841X.2017.1303660
Rothhammer, V., Borucki, D. M., Tjon, E. C., Takenaka, M. C., Chao, C. C., Ardura-Fabregat, A., et al. (2018). Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728. doi: 10.1038/s41586-018-0119-x
Rothhammer, V., Mascanfroni, I. D., Bunse, L., Takenaka, M. C., Kenison, J. E., Mayo, L., et al. (2016). Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597. doi: 10.1038/nm.4106
Rothhammer, V., and Quintana, F. J. (2019). The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 19, 184–197. doi: 10.1038/s41577-019-0125-8
Sahami, S., Kooij, I. A., Meijer, S. L., Van den Brink, G. R., Buskens, C. J., and Te Velde, A. A. (2016). The link between the appendix and ulcerative colitis: clinical relevance and potential immunological mechanisms. Am. J. Gastroenterol. 111, 163–169. doi: 10.1038/ajg.2015.301
Saji, N., Murotani, K., Hisada, T., Kunihiro, T., Tsuduki, T., Sugimoto, T., et al. (2020). Relationship between dementia and gut microbiome-associated metabolites: a cross-sectional study in Japan. Sci. Rep. 10:8088. doi: 10.1038/s41598-020-65196-6
Sampson, T. R., Challis, C., Jain, N., Moiseyenko, A., Ladinsky, M. S., Shastri, G. G., et al. (2020). A gut bacterial amyloid promotes alpha-synuclein aggregation and motor impairment in mice. Elife 9:1191. doi: 10.7554/eLife.53111
Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., et al. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469.e12–1480.e12. doi: 10.1016/j.cell.2016.11.018
Sanchez-Ferro, A., Rabano, A., Catalan, M. J., Rodriguez-Valcarcel, F. C., Fernandez Diez, S., Herreros-Rodriguez, J., et al. (2015). In vivo gastric detection of alpha-synuclein inclusions in Parkinson’s disease. Mov. Disord. 30, 517–524. doi: 10.1002/mds.25988
Sanmarco, L. M., Wheeler, M. A., Gutierrez-Vazquez, C., Polonio, C. M., Linnerbauer, M., Pinho-Ribeiro, F. A., et al. (2021). Gut-licensed IFNgamma(+) NK cells drive LAMP1(+)TRAIL(+) anti-inflammatory astrocytes. Nature 590, 473–479. doi: 10.1038/s41586-020-03116-4
Schafer, K. H., Mestres, P., Marz, P., and Rose-John, S. (1999). The IL-6/sIL-6R fusion protein hyper-IL-6 promotes neurite outgrowth and neuron survival in cultured enteric neurons. J. Interferon Cytokine Res. 19, 527–532. doi: 10.1089/107999099313974
Scheld, M., Ruther, B. J., Grosse-Veldmann, R., Ohl, K., Tenbrock, K., Dreymuller, D., et al. (2016). Neurodegeneration triggers peripheral immune cell recruitment into the forebrain. J. Neurosci. 36, 1410–1415. doi: 10.1523/JNEUROSCI.2456-15.2016
Scheperjans, F., Aho, V., Pereira, P. A., Koskinen, K., Paulin, L., Pekkonen, E., et al. (2015). Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov. Disord. 30, 350–358. doi: 10.1002/mds.26069
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Seppi, K., Weintraub, D., Coelho, M., Perez-Lloret, S., Fox, S. H., Katzenschlager, R., et al. (2011). The movement disorder society evidence-based medicine review update: treatments for the non-motor symptoms of Parkinson’s disease. Mov. Disord. 26, (Suppl. 3), S42–S80. doi: 10.1002/mds.23884
Shannon, K. M., Keshavarzian, A., Mutlu, E., Dodiya, H. B., Daian, D., Jaglin, J. A., et al. (2012). Alpha-synuclein in colonic submucosa in early untreated Parkinson’s disease. Mov. Disord. 27, 709–715. doi: 10.1002/mds.23838
Sittipo, P., Lobionda, S., Choi, K., Sari, I. N., Kwon, H. Y., and Lee, Y. K. (2018). Toll-like Receptor 2-mediated suppression of colorectal cancer pathogenesis by polysaccharide a from Bacteroides fragilis. Front. Microbiol. 9:1588. doi: 10.3389/fmicb.2018.01588
Soto, M., Herzog, C., Pacheco, J. A., Fujisaka, S., Bullock, K., Clish, C. B., et al. (2018). Gut microbiota modulate neurobehavior through changes in brain insulin sensitivity and metabolism. Mol. Psychiatry 23, 2287–2301. doi: 10.1038/s41380-018-0086-5
Stewart, J. S., Roberts, P. D., and Hoffbrand, A. V. (1970). Response of dietary vitamin-B12 deficiency to physiological oral doses of cyanocobalamin. Lancet 2, 542–545. doi: 10.1016/s0140-6736(70)91347-4
Stinson, L. F., Boyce, M. C., Payne, M. S., and Keelan, J. A. (2019). The not-so-sterile womb: evidence that the human fetus is exposed to bacteria prior to birth. Front. Microbiol. 10:1124. doi: 10.3389/fmicb.2019.01124
Suez, J., Zmora, N., Segal, E., and Elinav, E. (2019). The pros, cons, and many unknowns of probiotics. Nat. Med. 25, 716–729. doi: 10.1038/s41591-019-0439-x
Sun, M. F., Zhu, Y. L., Zhou, Z. L., Jia, X. B., Xu, Y. D., Yang, Q., et al. (2018). Neuroprotective effects of fecal microbiota transplantation on MPTP-induced Parkinson’s disease mice: gut microbiota, glial reaction and TLR4/TNF-alpha signaling pathway. Brain Behav. Immun. 70, 48–60. doi: 10.1016/j.bbi.2018.02.005
Takagawa, T., Kitani, A., Fuss, I., Levine, B., Brant, S. R., Peter, I., et al. (2018). An increase in LRRK2 suppresses autophagy and enhances Dectin-1-induced immunity in a mouse model of colitis. Sci. Transl. Med. 10:eaan8162. doi: 10.1126/scitranslmed.aan8162
Tan, A. H., Hor, J. W., Chong, C. W., and Lim, S. Y. (2021). Probiotics for Parkinson’s disease: current evidence and future directions. JGH Open 5, 414–419. doi: 10.1002/jgh3.12450
Tarallo, S., Ferrero, G., De Filippis, F., Francavilla, A., Pasolli, E., Panero, V., et al. (2021). Stool microRNA profiles reflect different dietary and gut microbiome patterns in healthy individuals. Gut doi: 10.1136/gutjnl-2021-325168 Online ahead of print,
Tierney, B. T., Yang, Z., Luber, J. M., Beaudin, M., Wibowo, M. C., Baek, C., et al. (2019). The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe 26, 283.e8–295.e8. doi: 10.1016/j.chom.2019.07.008
Tukel, C., Wilson, R. P., Nishimori, J. H., Pezeshki, M., Chromy, B. A., and Baumler, A. J. (2009). Responses to amyloids of microbial and host origin are mediated through toll-like receptor 2. Cell Host Microbe 6, 45–53. doi: 10.1016/j.chom.2009.05.020
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut Lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573
Van Den Berge, N., Ferreira, N., Gram, H., Mikkelsen, T. W., Alstrup, A. K. O., Casadei, N., et al. (2019). Evidence for bidirectional and trans-synaptic parasympathetic and sympathetic propagation of alpha-synuclein in rats. Acta Neuropathol. 138, 535–550. doi: 10.1007/s00401-019-02040-w
van IJzendoorn, S. C. D., and Derkinderen, P. (2019). The intestinal barrier in Parkinson’s disease: current state of knowledge. J. Parkinsons Dis. 9, S323–S329. doi: 10.3233/JPD-191707
Vascellari, S., Melis, M., Palmas, V., Pisanu, S., Serra, A., Perra, D., et al. (2021). Clinical phenotypes of Parkinson’s disease associate with distinct gut microbiota and metabolome enterotypes. Biomolecules 11:144. doi: 10.3390/biom11020144
Vogt, N. M., Kerby, R. L., Dill-McFarland, K. A., Harding, S. J., Merluzzi, A. P., Johnson, S. C., et al. (2017). Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7:13537. doi: 10.1038/s41598-017-13601-y
Wakabayashi, K., Takahashi, H., Takeda, S., Ohama, E., and Ikuta, F. (1988). Parkinson’s disease: the presence of Lewy bodies in Auerbach’s and Meissner’s plexuses. Acta Neuropathol. 76, 217–221. doi: 10.1007/BF00687767
Wallen, Z. D., Appah, M., Dean, M. N., Sesler, C. L., Factor, S. A., Molho, E., et al. (2020). Characterizing dysbiosis of gut microbiome in PD: evidence for overabundance of opportunistic pathogens. NPJ Parkinsons Dis. 6:11. doi: 10.1038/s41531-020-0112-6
Wang, C., Lau, C. Y., Ma, F., and Zheng, C. (2021). Genome-wide screen identifies curli amyloid fibril as a bacterial component promoting host neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 118:e2106504118. doi: 10.1073/pnas.2106504118
Wang, H., Liu, X., Chen, S., and Ye, K. (2018). Spatiotemporal activation of the C/EBPbeta/delta-secretase axis regulates the pathogenesis of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 115, E12427–E12434. doi: 10.1073/pnas.1815915115
Wang, R. X., Lee, J. S., Campbell, E. L., and Colgan, S. P. (2020). Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. U.S.A. 117, 11648–11657. doi: 10.1073/pnas.1917597117
Warner, B. B. (2019). The contribution of the gut microbiome to neurodevelopment and neuropsychiatric disorders. Pediatr. Res. 85, 216–224. doi: 10.1038/s41390-018-0191-9
Weersma, R. K., Zhernakova, A., and Fu, J. (2020). Interaction between drugs and the gut microbiome. Gut 69, 1510–1519. doi: 10.1136/gutjnl-2019-320204
Wei, G. Z., Martin, K. A., Xing, P. Y., Agrawal, R., Whiley, L., Wood, T. K., et al. (2021). Tryptophan-metabolizing gut microbes regulate adult neurogenesis via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. U.S.A. 118:e2021091118. doi: 10.1073/pnas.2021091118
Wekerle, H. (2017). Brain autoimmunity and intestinal microbiota: 100 trillion game changers. Trends Immunol. 38, 483–497. doi: 10.1016/j.it.2017.03.008
Wu, S. C., Cao, Z. S., Chang, K. M., and Juang, J. L. (2017). Intestinal microbial dysbiosis aggravates the progression of Alzheimer’s disease in Drosophila. Nat. Commun. 8:24. doi: 10.1038/s41467-017-00040-6
Xie, W., Gao, J., Jiang, R., Liu, X., Lai, F., Tang, Y., et al. (2020). Twice subacute MPTP administrations induced time-dependent dopaminergic neurodegeneration and inflammation in midbrain and ileum, as well as gut microbiota disorders in PD mice. Neurotoxicology 76, 200–212. doi: 10.1016/j.neuro.2019.11.009
Xu, C., Zhu, H., and Qiu, P. (2019). Aging progression of human gut microbiota. BMC Microbiol. 19:236. doi: 10.1186/s12866-019-1616-2
Yan, X., Hu, Y., Wang, B., Wang, S., and Zhang, X. (2020). Metabolic dysregulation contributes to the progression of Alzheimer’s disease. Front. Neurosci. 14:530219. doi: 10.3389/fnins.2020.530219
Yan, Y., Ren, S., Duan, Y., Lu, C., Niu, Y., Wang, Z., et al. (2021). Gut microbiota and metabolites of alpha-synuclein transgenic monkey models with early stage of Parkinson’s disease. NPJ Biofilms Microbiomes 7:69. doi: 10.1038/s41522-021-00242-3
Yang, R., Gao, R., Cui, S., Zhong, H., Zhang, X., Chen, Y., et al. (2019). Dynamic signatures of gut microbiota and influences of delivery and feeding modes during the first 6 months of life. Physiol. Genomics 51, 368–378. doi: 10.1152/physiolgenomics.00026.2019
Zhang, B., Wang, H. E., Bai, Y. M., Tsai, S. J., Su, T. P., Chen, T. J., et al. (2021). Inflammatory bowel disease is associated with higher dementia risk: a nationwide longitudinal study. Gut 70, 85–91. doi: 10.1136/gutjnl-2020-320789
Zhang, J., Xu, M., Zhou, W., Li, D., Zhang, H., Chen, Y., et al. (2020). Deficiency in the anti-apoptotic protein DJ-1 promotes intestinal epithelial cell apoptosis and aggravates inflammatory bowel disease via p53. J. Biol. Chem. 295, 4237–4251. doi: 10.1074/jbc.RA119.010143
Zhao, Y., Jaber, V., and Lukiw, W. J. (2017). Secretory products of the human gi tract microbiome and their potential impact on Alzheimer’s disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front. Cell Infect. Microbiol. 7:318. doi: 10.3389/fcimb.2017.00318
Zhao, Y., and Lukiw, W. J. (2018). Microbiome-mediated upregulation of MicroRNA-146a in sporadic Alzheimer’s disease. Front. Neurol. 9:145. doi: 10.3389/fneur.2018.00145
Zheng, D., Liwinski, T., and Elinav, E. (2020a). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Zheng, Q., Huang, T., Zhang, L., Zhou, Y., Luo, H., Xu, H., et al. (2016). Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 8:303. doi: 10.3389/fnagi.2016.00303
Keywords: neurodegeneration, microbiome, neuroinflammation, protein aggregation, gut microbiota, gut metabolome, gut inflammation, Alzheimer’s and Parkinson’s diseases
Citation: Padhi P, Worth C, Zenitsky G, Jin H, Sambamurti K, Anantharam V, Kanthasamy A and Kanthasamy AG (2022) Mechanistic Insights Into Gut Microbiome Dysbiosis-Mediated Neuroimmune Dysregulation and Protein Misfolding and Clearance in the Pathogenesis of Chronic Neurodegenerative Disorders. Front. Neurosci. 16:836605. doi: 10.3389/fnins.2022.836605
Received: 15 December 2021; Accepted: 04 February 2022;
Published: 25 February 2022.
Edited by:
Dilshan Shanaka Harischandra, Covance, United StatesReviewed by:
Amy Shepherd, Boston Children’s Hospital and Harvard Medical School, United StatesCopyright © 2022 Padhi, Worth, Zenitsky, Jin, Sambamurti, Anantharam, Kanthasamy and Kanthasamy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anumantha G. Kanthasamy, YW51bWFudGhhLmthbnRoYXNhbXlAdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.