- Department of Developmental Neuroscience, United Centers for Advanced Research and Translational Medicine (ART), Tohoku University Graduate School of Medicine, Sendai, Japan
The Dmrt genes encode the transcription factor containing the DM (doublesex and mab-3) domain, an intertwined zinc finger-like DNA binding module. While Dmrt genes are mainly involved in the sexual development of various species, recent studies have revealed that Dmrt genes, which belong to the DmrtA subfamily, are differentially expressed in the embryonic brain and spinal cord and are essential for the development of the central nervous system. Herein, we summarize recent studies that reveal the multiple functions of the Dmrt genes in various aspects of vertebrate neural development, including brain patterning, neurogenesis, and the specification of neurons.
Introduction
The Dmrt (double-sex and mab-3 related transcription factor) genes encode a large family of transcription factors involved in sexual development (Zarkower, 2001; Kopp, 2012). First identified as Doublesex in Drosophila and MAB-3 in Caenorhabditis elegans (C. elegans), the Dmrt family proteins share a DM (doublesex and mab-3) domain that consists of a highly intertwined zinc finger DNA-binding motif (Erdman and Burtis, 1993; Raymond et al., 1998). Several Dmrt genes have been identified in vertebrates, including Dmrt1 to Dmrt8 in mice and humans (Bellefroid et al., 2013). Among these, the DmrtA subfamily members (DmrtAs, i.e., Dmrt3, Dmrta1, and Dmrta2) have a conserved DMA domain near the C-terminus in addition to the DM domain (Ottolenghi et al., 2002). It has been reported that the DMA domain is required for the activity of the Dmrt protein (Parlier et al., 2013). Moreover, the DMA domain in DMD-4 in C. elegans can bind to ubiquitin and stabilize the protein, thus playing a sex-specific role in synaptic connectivity (Bayer et al., 2020). Although each Dmrt acts as a transcriptional regulator, the DM domain proteins can form heterodimers on DNA, raising the possibility of combinatorial gene regulation by these proteins (Murphy et al., 2007).
In vertebrates, Dmrt-family genes are primarily involved in the development of sexual organs. As a representative function, Dmrt1 controls many aspects of testicular development, including the postnatal differentiation of germ cells and Sertoli cells (Raymond et al., 2000; Kim et al., 2007a). Dmrt7 localizes to spermatocytes, specifically the XY body, a domain where X and Y chromosomes are silenced and compartmentalized, which leads to the proper meiotic progression in the mouse testis (Kawamata and Nishimori, 2006; Kim et al., 2007b). In addition to the development of sexual organs, the Dmrt factors are involved in various events during embryonic development (Hong et al., 2007; Bellefroid et al., 2013). For example, Dmrt2 is necessary for the establishment of left-right asymmetry during somitogenesis (Saude et al., 2005; Liu et al., 2009; Lourenco et al., 2010). Based on recent findings, including ours, this review focuses on various functions of Dmrt genes, especially DmrtA subfamily members, in central nervous system (CNS) development.
Expression Patterns of Dmrt Genes in the Developing Central Nervous System
In various regions of the developing brain, DmrtAs are mainly expressed in neural stem/progenitor cells (NSPCs) in the ventricular zone (VZ), which lines the ventricular wall. In the spinal cord, the expression of DmrtAs are detected in specific neurons. We summarize the expression patterns of DmrtAs in each region of the CNS during development (Figure 1).
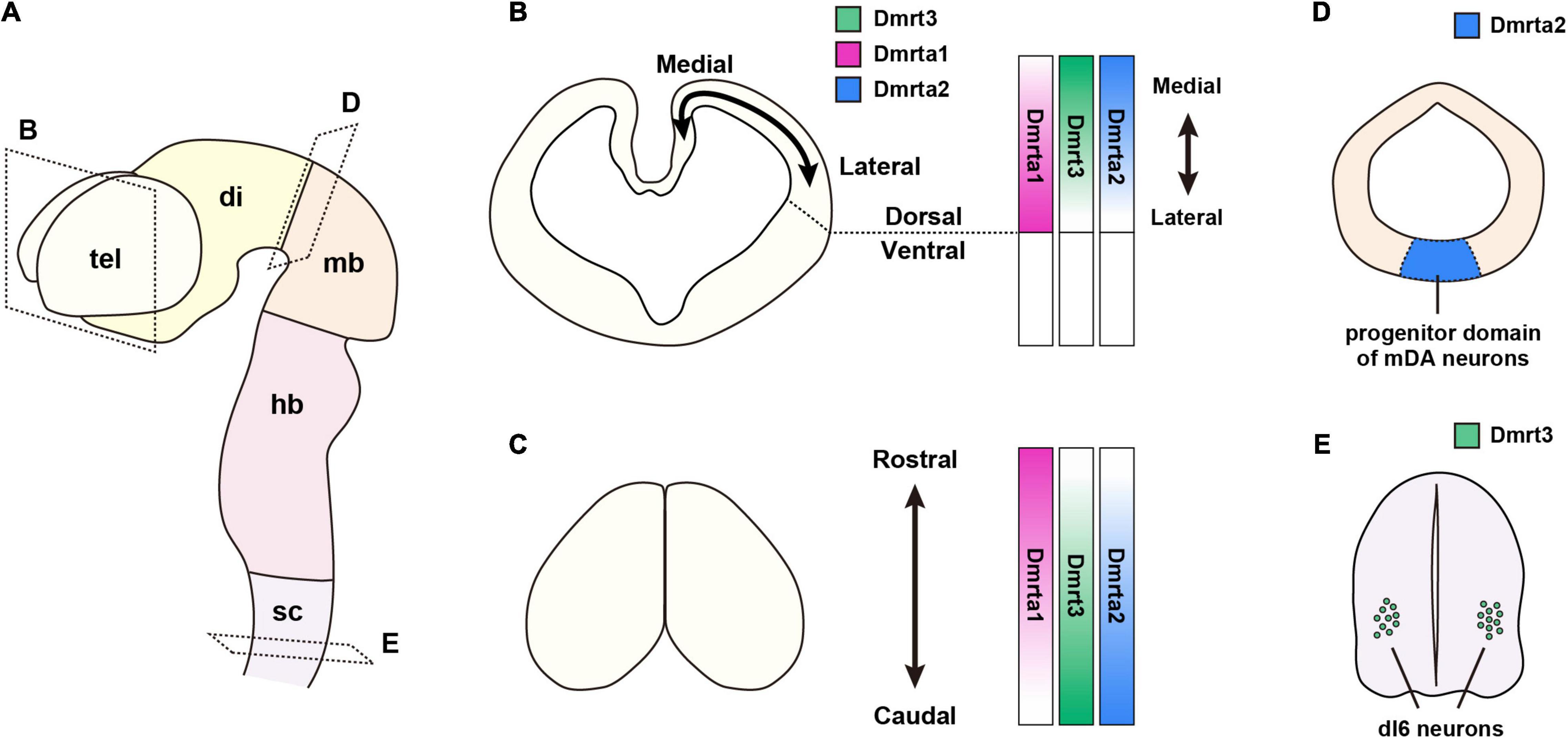
Figure 1. Expression patterns of DmrtA subfamily members in the embryonic mouse CNS. (A) Schematic representation of the mouse embryonic CNS at E11. (B–E) Expression pattern of DmrtAs. (B) Cross-sections of the telencephalon. Magenta indicates the expression of Dmrta1; green, Dmrt3; and blue, Dmrta2. DmrtAs are expressed in NSPCs in the dorsal telencephalon. Dmrta1 expression has a medial-low/lateral-high gradient, whereas Dmrt3 and Dmrta2 show a medial-high/lateral-low gradient. (C) Dorsal view of the telencephalon. Dmrta1 is expressed in a rostral-high/caudal-low gradient, whereas Dmrt3 and Dmrta2 show a rostral-low/caudal-high gradient. (D) Cross-sections of the midbrain. Dmrta2 is expressed in the progenitor domain of mDA neurons in the ventral midbrain. (E) Cross-sections of the spinal cord. Dmrt3 is specifically expressed in dI6 neurons in the spinal cord. CNS, central nervous system; di, diencephalon; dI, dorsal interneuron; DmrtAs, DmrtA subfamily members; hb, hindbrain; mb, midbrain; NSPCs, neural stem progenitor cells; sc, spinal cord; tel, telencephalon.
In the mouse telencephalon, mRNAs of Dmrt3, Dmrta1, and Dmrta2 are detected as early as embryonic day (E) 9.5, and their levels peak around E10.5–E12.5 (Konno et al., 2012). The expression of Dmrta1 in rats begins on E10.5 (corresponding to E8.5), an early stage of CNS development (Kikkawa et al., 2013). In non-rodent species, chick embryos begin to show a strong expression of Dmrt3 in the developing telencephalon by E2.5 (Smith et al., 2002). Xenopus Dmrta1 (XDmrt4) is initially expressed in the anterior neural ridge and is restricted to a part of the telencephalon by stage 35 (Huang et al., 2005). Medaka fish (Oryzias latipes) also show restricted expression in the dorsal telencephalon at stage 26, and the mRNA level gradually decreases thereafter (Winkler et al., 2004). Ciona, a member of the vertebrate sister group Urochordata, expresses Dmrt1 (related to Dmrta1 and Dmrta2 and with a DMA domain) in the anterior neural plate and is later restricted it to the anterior brain at the tailbud stage (Bellefroid et al., 2013). Thus, the expression of DmrtAs in the telencephalon appears to be conserved across species.
Detailed and differential expression patterns of DmrtAs in the telencephalon have been studied in mice. Focusing on the dorsoventral and mediolateral axes of the telencephalon, Dmrt3 and Dmrta2 are expressed with a medial-high/lateral-low gradient (Konno et al., 2012, 2019; Kikkawa et al., 2013), whereas Dmrta1 has a medial-low/lateral-high gradient in the dorsal telencephalon (Kikkawa et al., 2020 and our unpublished data; Figure 1B). In the rostral-caudal axis, Dmrt3 and Dmrta2 show a rostral-low/caudal-high gradient (Figure 1C). Conversely, Dmrta1 has a relatively rostral-high/caudal-low expression level (Kikkawa et al., 2020). Their functional differences will be discussed later in the sections on “brain patterning” and “corticogenesis.”
In the diencephalon, another structure subdivided from the forebrain, the expression pattern of the Dmrt gene is different from that in the neighboring telencephalon. In zebrafish, Dmrta2 is restricted to the ventral region of the diencephalon and hypothalamus (a part of the diencephalon) at the 6-somite stage (Guo et al., 2004; Yoshizawa et al., 2011). Xenopus also shows Dmrta2 expression in the ventral diencephalon as does the zebrafish (Parlier et al., 2013).
Dmrta2 expression in the mesencephalon (midbrain) is restricted ventrally, as in the diencephalon (Figure 1D). This expression pattern is found in platyfish, chicks, and mouse embryos (Veith et al., 2006; Gennet et al., 2011; Saulnier et al., 2013); however, it appears not to be the case in Xenopus (Parlier et al., 2013). In Dmrta1, our lacZ knock-in mice to the locus of Dmrta1 show β-galactosidase (β-gal) expression in the ventral midbrain (Kikkawa et al., 2020). The ventral midbrain is the primary origin of midbrain dopaminergic (mDA) neurons (Gale and Li, 2008). Interestingly, Dmrta2 is a regulator of the mouse ventral mesencephalic neural fate specification (Gennet et al., 2011) (see below).
Only Dmrt3 is expressed in the spinal cord. Unlike in brain regions, it is expressed in differentiated neurons but not in NSPCs. Dmrt3 expression in the embryonic spinal cord appears to be conserved among mice, chicks, and fish (Smith et al., 2002; Kim et al., 2003; Winkler et al., 2004; Li et al., 2008). Mouse embryonic data show that Dmrt3 is specifically expressed in the dorsal interneuron (dI) subtype, dI6 neurons, in the spinal cord and regulates their neuronal specification of the interneuron subpopulation (Andersson et al., 2012; Figure 1E) (see below).
As described above, DmrtAs are expressed during the early embryonic development of the CNS in various species. We discuss the function of DmrtAs in brain development in the following sections.
Functions of Dmrt Genes in Brain Patterning
The telencephalon is patterned by the combined action of different signaling centers, such as the rostral signaling center in the rostromedial forebrain secreting Fgfs, the dorsal cortical hem (CH) in the caudomedial telencephalon secreting Wnts and BMPs, and in the floor plate at the ventral midline of the embryonic forebrain secreting Sonic hedgehog (Shh) (Hebert and Fishell, 2008). These factors specifically regulate the transcription of target genes, thus defining the specific fate of cells and conferring positional information along the axis (Wilson and Houart, 2004; Rhinn et al., 2006; Monuki, 2007).
Since DmrtAs are strongly expressed in the telencephalon, many recent studies have reported their involvement in regionalization (Figure 2). Therefore, we would like to mention the roles of DmrtAs in telencephalic patterning, including the upstream and downstream molecular networks of DmrtAs.
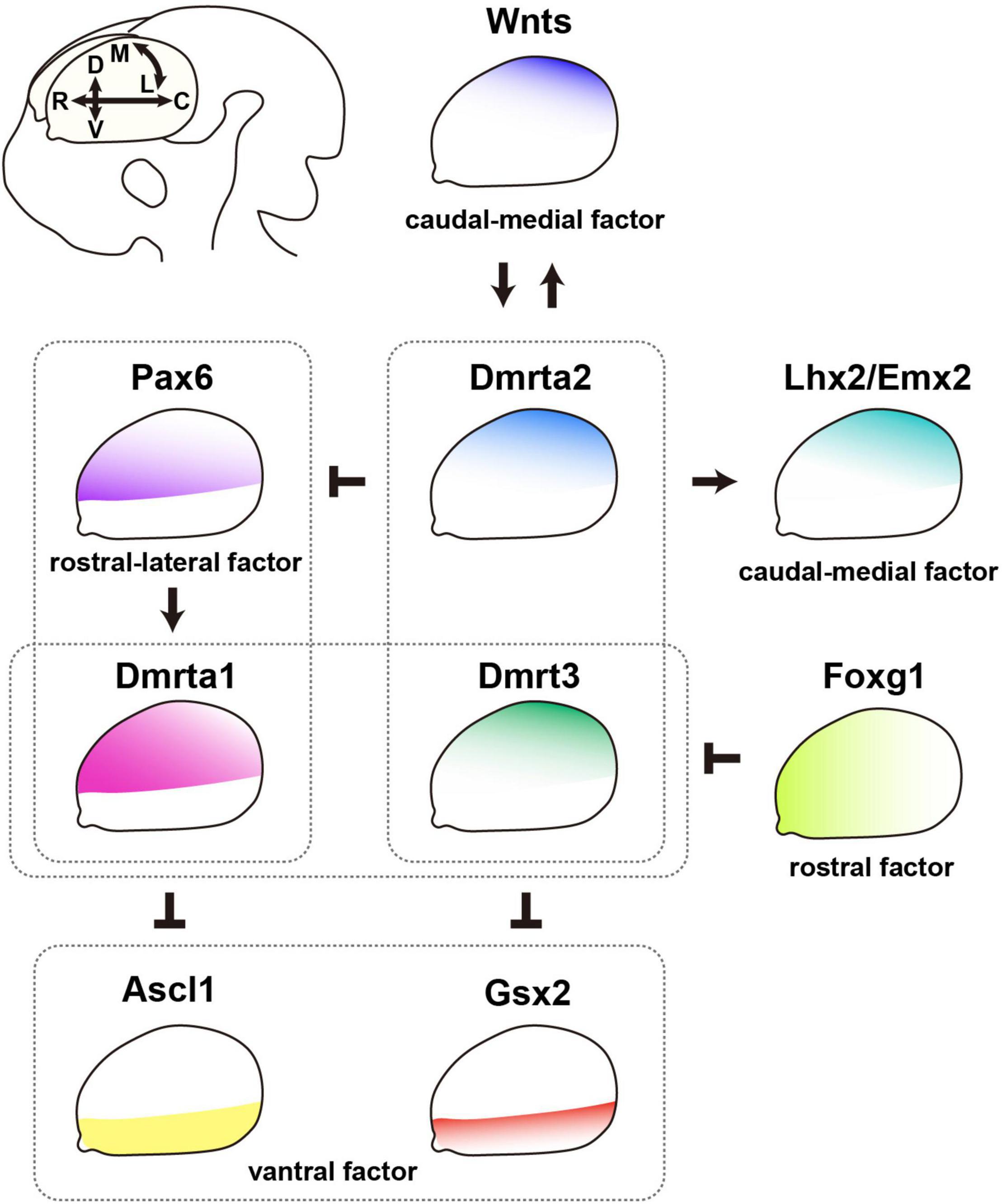
Figure 2. DmrtA subfamily members contribute to the patterning of the telencephalon. Regulatory networks of telencephalic patterning molecules related to the contribution of DmrtA subfamily members (DmrtAs). DmrtAs are involved in subdivision along the dorsoventral axis of the telencephalon. Dmrta1 is a direct target of the transcription factor Pax6, which exhibits a rostral-high/lateral-high gradient expression. Dmrta1 represses Ascl1 expression in the ventral telencephalon, while Dmrt3 and Dmrta2 directly repress the expression of Gsx2. Therefore, DmrtAs are essential for determining the dorsoventral identity of progenitor cells by repressing ventralization. DmrtAs play roles in mediolateral patterning of the dorsal telencephalon. The caudal-medial signaling center secretes Wnts that are required for the formation of the medial telencephalic structure. Wnts and DmrtAs mutually regulate each other. Dmrt3 and Dmrta2 mainly determine the proper patterning of the caudal-medial telencephalon. Dmrta1 supports medial telencephalic development. The rostral factor Foxg1 represses Dmrt3 and Dmrta1 expression, and this molecular pathway may be needed to acquire the caudal-medial fate specification. Furthermore, Dmrta2 modulates the expression of transcription factors such as Pax6, Emx2, and Lhx2, leading to the proper patterning of the neocortex. C, caudal region; D, dorsal region; L, lateral region; M, medial region; R, rostral region; V, ventral region.
Subdivisions Along the Dorsoventral Axis of the Telencephalon
The telencephalon is patterned into two major subdivisions, the dorsal and ventral regions, called the pallium and subpallium, respectively (Puelles et al., 2000). NSPCs in the pallium produce excitatory neurons, whereas those in the subpallium produce interneurons (Anderson et al., 1997; Gorski et al., 2002). For proper patterning along the dorsoventral (DV) axis, it is essential that several transcription factors work in concert. For example, the loss of Pax6, a transcription factor expressed in the dorsal region, shows severe ventralization with a dorsal expansion of the expression of Gsx2, Ascl1, and Dlx1, which are involved in the production of interneurons from the ventral region; thus, Pax6 is crucial for patterning the dorsal and ventral telencephalon (Stoykova et al., 2000; Toresson et al., 2000). In particular, the positioning of the DV boundary region is defined by Pax6-Gsx2 mutual antagonism (Corbin et al., 2000; Toresson et al., 2000; Yun et al., 2001).
DmrtAs are also crucial for the determination of the DV axis. Recent Dmrt3 and Dmrta2 knock-out (KO) studies have clarified how they specify DV regional identity in progenitors as follows. Dmrt3;Dmrta2 double-KO (dKO) mice showed the most severe phenotype of DV regionalization of the telencephalon than Dmrta2 single-KO (sKO) mice, and Dmrt3 sKO mice showed the mildest phenotype (Desmaris et al., 2018; Konno et al., 2019). It is noteworthy that the expression of subpallium-specific markers (e.g., Gsx2, Dlx2, Ascl1) is shifted dorsally to the lateral part of the dorsal telencephalon in Dmrt3;Dmrta2 dKO mice (Desmaris et al., 2018). Furthermore, although Dmrta1 sKO mice did not have a phenotype of ventralization, the overexpression of Dmrta1 in the ventral telencephalon in rats induced the expression of the dorsal marker Neurog2 and repressed that of the ventral marker Ascl1 (Kikkawa et al., 2013). These results indicate that DmrtAs function as an essential determinant of progenitor cell DV identity by repressing ventralization.
Therefore, a question arises: what are the main target genes of DmrtA transcription factors that determine telencephalic identity? The enhancer activity of the Gsx2 locus, to which Dmrta2 binds, is found in the ventral telencephalon (Desmaris et al., 2018). A more comprehensive method, whole-genome chromatin immunoprecipitation-sequencing (ChIP-seq), revealed that Dmrt3 and Dmrta2 bind to the Gsx2 and Pax6 loci within ±100 kb from the transcription start site (Konno et al., 2019). The authors also generated transgenic mice to visualize the enhancer activity of Dmrt3/Dmrta2-binding sites at Gsx2 and Pax6. They found that the enhancer activities of the loci at Gsx2 and Pax6 bound by Dmrts are detected in the dorsal lateral ganglionic eminence (dLGE) and the dorsal telencephalon, where Gsx2 and Pax6 are expressed, respectively. Interestingly, a suppressive histone moiety, H3K27me3, is found in the Gsx2 enhancer bound by Dmrt3/Dmrta2 in their study, suggesting that DmrtAs may suppress the expression of Gsx2. Since the dLGE neighboring the DV boundary expresses a high level of Gsx2, its direct regulation may modulate the fate specification of the progenitors in the dorsal and ventral telencephalon.
Pax6 regulates the expression of various genes in the embryonic telencephalon (Sansom et al., 2009; Kikkawa et al., 2013, 2019; Walcher et al., 2013; Xie et al., 2013; Sun et al., 2015). We initially searched for downstream genes of Pax6 by comparing the transcriptomic profiles of telencephalic samples from wild-type and Pax6 null mutant rat embryos and found Dmrta1 as a novel target (Kikkawa et al., 2013) in addition to the various downstream molecules in the embryonic CNS (Shimoda et al., 2002; Arai et al., 2005; Numayama-Tsuruta et al., 2010; Shinohara et al., 2013; Inada et al., 2018). Pax6 ChIP-seq has later proved the direct regulation of the Dmrta1 gene by the Pax6 transcription factor in the E12.5 mouse forebrain (Sun et al., 2015). However, this Pax6-Dmrta1 pathway seems to work only in specific dorsal-lateral telencephalic regions. Dmrta1 expression was markedly decreased in the neocortex of Pax6 homozygous mutant rat embryos, although it remained in the CH (Kikkawa et al., 2013). Focusing on the functions in the formation of the CH, Dmrta1 supports the generation of the CH (Kikkawa et al., 2020), whereas Pax6 suppresses its fate specification of the CH (Godbole et al., 2017). Therefore, the Pax6-Dmrta1 pathway may work differently in the neocortex and extra-neocortical areas, such as the CH.
Patterning of the Dorsal Telencephalon Along the Mediolateral Axis
The dorsal telencephalon is divided into the neocortex, hippocampus, and dorsal midline, giving rise to the CH and choroid plexus. The dorsal midline of the telencephalon is known to express BMPs and Wnts (Furuta et al., 1997; Grove et al., 1998). BMP and Wnt signaling are required for the formation of the medial telencephalic structure that develops into the hippocampus and the dorsal midline region (Lee et al., 2000; Hebert et al., 2002), and both signaling pathways also regulate the expression of transcription factors such as Emx2 and Lhx2, which specify and expand the medial and dorsal parts of the telencephalon (Monuki et al., 2001; Theil et al., 2002). These molecular pathways are essential for patterning the dorsal telencephalon along the mediolateral axis.
In addition to these critical molecules that regulate the development of the medial telencephalon, it has become clear that DmrtAs contribute to the following events. Previous studies have demonstrated malformation of caudomedial telencephalic structures, that is, the CH that expresses Wnt3a and Bmp4, and the hippocampus, in Dmrta2 sKO mice (Konno et al., 2012; Saulnier et al., 2013; De Clercq et al., 2018). Dmrt3 sKO embryos also have shown defects in the CH formation (De Clercq et al., 2018; Kikkawa et al., 2020). These phenotypes are reasonable because Dmrt3 and Dmrta2 are expressed with medial-high/lateral-low and rostral-low/caudal-high gradients. Although the expression level of Dmrta1 was weak in the medial telencephalon, Dmrt3;Dmrta1 dKO mice exhibited more severe defects in medial structures compared to Dmrt3 sKO mice (Kikkawa et al., 2020). Therefore, Dmrta1 may have an additional support function for patterning medial telencephalic structures in cooperation with Dmrt3.
When do the defects in the medial telencephalon of Dmrt mutants begin? It has been reported that DmrtAs are expressed in the forebrain at early developmental stages, that is, E10.5–E12.5 (Konno et al., 2012; Kikkawa et al., 2013). Because Wnt3aCre-driven Dmrta2 conditional KO (cKO) mice showed normal morphology of the medial telencephalon (De Clercq et al., 2018), the apparent defects in the medial telencephalon by the ablation of DmrtAs may be due to the loss of their expression during earlier developmental stages before the initiation of recombination by Wnt3a-Cre at E10 in mice (Yoshida et al., 2006). These findings suggest that DmrtAs establish the dorsal midline structure, a signaling center that expresses Wnt and BMP in patterning the cerebral cortex.
Then, what molecules function upstream of DmrtAs to determine the dorsomedial patterning of the telencephalon? Dmrt gene expression was downregulated in Gli mutant mice, in which Wnt expression in the forebrain was severely affected (Hasenpusch-Theil et al., 2012). This study also clarifies that Dmrt3 is a direct Wnt target gene in the dorsomedial telencephalon by DNA binding and reporter gene assays. Furthermore, the expression of Dmrt3 and Dmrta2 were decreased by the overexpression of a dominant-negative form of Tcf3 and increased by the overexpression of a constitutively active form of β-catenin (Konno et al., 2012). Moreover, Dmrta2 expression was induced in organotypic slice cultures of mouse embryonic telencephalons treated with Chir that selectively inhibits GSK3β and activates Wnt signaling (Saulnier et al., 2013). This means that the secretory factor Wnts can directly regulate Dmrt expression and could contribute to adjusting the amount of Dmrts.
Arealization of the Neocortex
The neocortex has “primary” areas: the primary motor (M1, controls the voluntary movement of body parts), somatosensory (S1, processes the information received from the body), and visual (V1, processes the information received from eyes) areas. One of the hypotheses to determine the cortical arealization is the “protomap model.” The feature of progenitors in early development is predetermined by the combination of molecules, and then the progenitors differentiate into specific neurons, leading to the formation of different cortical regions (Rakic, 1988). Some transcription factors that contribute to the formation of the “protomap.” Rostralhigh-Pax6 and caudalhigh-Emx2 in progenitors preferentially impart the identities of the rostrolateral and caudomedial areas, respectively (Bishop et al., 2002). A study on the loss of Pax6/Emx2 indicates that they suppress each other’s expression (Muzio et al., 2002). Lhx2 is expressed in a graded manner (caudalhigh/medialhigh) in the telencephalon (Nakagawa et al., 1999; Monuki et al., 2001), acts as an essential determinant of cortical identity (Mangale et al., 2008), and is further required for the neocortical-paleocortical subdivision (Chou et al., 2009). Pax6 expression is decreased in the Lhx2 mutant in the dorsal telencephalon due to direct regulation by Lhx2 (Hou et al., 2013; Shetty et al., 2013). The combinatorial expression patterns of these transcription factors regulate the regionalization of the cerebral cortex.
Since the loss of CH affects cortical size and patterning (Caronia-Brown et al., 2014), the reduction in cortical size in Dmrt mutants may be due to the developmental impairment of CH formation. However, deleting Dmrta2 after CH formation still decreases the cortical size and changes the area map; the V1 area, which is the caudomedial neocortical region, was reduced in Emx1Cre- and NestinCre-driven Dmrta2 cKO mice on postnatal day 7 (De Clercq et al., 2018). Conversely, Dmrta2Tg/+; Emx1Cre mice with excess Dmrta2 in the cortical primordium showed enlarged V1 and reduced size of the S1 and M1 areas. They also revealed a decrease in Lhx2 and Emx2 and the expansion of Pax6 to the caudal cortex in Dmrta2 cKO embryos. From these results, the level of Dmrta2 seems to regulate the expression of these transcription factors that determine the rostral-caudal patterning during cortical development and construct the proper neocortical area map.
Functions of Dmrt Genes in Corticogenesis
Maintenance of NSPCs and Their Differentiation Into Neurons
NSPCs maintain their population by self-renewal and produce neurons by differentiation during embryogenesis. At an early stage of development, that is, E9–E11 in the mouse telencephalon, NSPCs undergo symmetric division in the VZ, producing daughter cells with similar fates to give rise to more progenitors. As development proceeds, NSPCs start to divide asymmetrically, producing one apical progenitor (AP) positive for Pax6 with self-renewing capability and one differentiated neuron, or one intermediate progenitor (IP) positive for Tbr2, which divides symmetrically in the subventricular zone (SVZ) and generates a pair of IPs or neurons (Gotz and Huttner, 2005; Huttner and Kosodo, 2005). Subsequently, newly born neurons migrate radially from the VZ/SVZ to the upper area of the telencephalon. In this section, we introduce the multiple roles of DmrtAs expressed in NSPCs in the telencephalon in cell proliferation and differentiation regulation.
Dmrta2 is expressed in NSPCs derived from mouse embryonic stem cells (ESCs) (Young et al., 2017). It maintains their proliferation by positively regulating Hes1 expression via Dmrta2 bound to the locus of Hes1 (Young et al., 2017), a significant Notch target gene that inhibits neuronal differentiation through negative regulation of proneural genes (Kageyama et al., 2020). Consistently, the neurogenic gene Neurog2 was upregulated in the medial region of the dorsal telencephalon of Dmrta2 sKO mice (Saulnier et al., 2013). Furthermore, in Dmrt3, transgenic mice overexpressing Dmrt3 in NSPCs under the control of the Nestin enhancer showed a planar expansion of the ventricular surface, indicating an increased NSPC pool and decreased Tbr2-positive IP cells (Konno et al., 2019). These results suggest that Dmrt3 and Dmrta2 are involved in the maintenance of NSPCs in the dorsal telencephalon.
In contrast, DmrtAs seem to contribute to neuronal differentiation. A zebrafish mutant of the ha2 locus, encoding Dmrta2, reduced the expression of neurog1 and impaired telencephalic neurogenesis (Yoshizawa et al., 2011). In addition to the brain, Xenopus Dmrta1 and Dmrta2 promoted neurogenesis in the olfactory placode (Huang et al., 2005; Parlier et al., 2013). Although Dmrt3 and Dmrta2 are involved in the maintenance of NSPCs, as mentioned above, the ratio of Tbr2+ to the total number of progenitors was lower, while that of Pax6+ cells was higher, in Dmrta2 sKO mice than in wild type mice in the lateral cortex (Ratie et al., 2020). This suggests that the APs could generate fewer IPs and/or that the timing of their differentiation could be delayed. Our data showed that the knockdown of Dmrta1 or Dmrt3 in the rat dorsal telencephalon reduces Neurog2, while the overexpression of Dmrta1 in the ventral telencephalon induces ectopic Neurog2 expression in the restricted lateral cortex and near the DV boundary region (Kikkawa et al., 2013). Based on the above reports, the function of DmrtAs in neurogenesis is not simple, and its function may be altered in a region-dependent manner within the telencephalon.
Fate Determination of NSPCs Into Either Neurons or Glial Cells
The sequential production of neurons and glia from NSPCs is a critical event during CNS development. In the mammalian neocortex, NSPCs at the early developmental stage generate neuronal cells, whereas those at the late stage generate mainly glial cells, including astrocytes and oligodendrocytes (Qian et al., 2000). The appropriate neurogenic-to-gliogenic switch in NSPCs is essential for the production of proper numbers of neurons and glia. Various molecular mechanisms have been clarified; for example, BMP, Fgf, and Notch signaling induce the cell fate switch from neuronal to glial cells (Miller and Gauthier, 2007). The transcription factor Lhx2 suppresses astrogliogenesis and promotes neurogenesis in the developing hippocampus, but not in the neocortex, indicating the spatial-specific regulation of NSPCs to neuron/glia fate specification (Subramanian et al., 2011).
There is an exciting finding that Dmrta2 is involved in the cell fate switch from neurons to astrocytes. The loss of Dmrta2 increased the population of GFAP-positive cultured astrocytes taken from the embryonic hippocampus (Muralidharan et al., 2017). They also showed that the loss of Lhx2 also induces astrogliogenesis, and the phenotype is rescued by the overexpression of Dmrta2. These results suggest that Dmrta2 and Lhx2 reciprocally regulate each other and that this pathway is involved in the neuron-glia cell-fate switch, emphasizing the novel role of Dmrta2 as a neurogenic factor. It is reasonable to assume that this phenotype is limited to the hippocampus, but not the cortex, because both Lhx2 and Dmrta2 are strongly expressed in the medial region and have already been reported to be involved in hippocampal development (Bulchand et al., 2001; Konno et al., 2012; Saulnier et al., 2013; De Clercq et al., 2018).
In the Lhx2-Dmrta2 pathway, ChIP-quantitative PCR using the embryonic hippocampus showed that Lhx2 binds to the Dmrta2 locus, indicating that Lhx2 could directly regulate the expression of Dmrta2 (Muralidharan et al., 2017). Interestingly, the locus bound by Dmrta2 is evolutionarily highly conserved among species. If this mechanism of the neuron-glia cell fate switch by the Lhx-Dmrta2 pathway could be conserved among different organisms, it may provide a deeper insight into the poorly understood neuron-glia switch in other species.
Production of Early Born Neurons
It is unclear which neuronal subtypes are regulated by DmrtAs. The expression of DmrtAs is abundant in developmental stages when early born neurons called Cajal-Retzius (CR) cells are produced (Konno et al., 2012; Kikkawa et al., 2013). CR cells are among the first neurons to be generated (between E9.5 and E13.5) in mice (Hevner et al., 2003; Takiguchi-Hayashi et al., 2004). CR cells are generated from specific extra-neocortical regions, that is, (1) the pallial septum, which is located adjacent to a rostral signaling center in the rostromedial telencephalon; (2) the boundary between the pallium and subpallium (pallial-subpallial boundary; PSB); and (3) the CH in the caudomedial telencephalon, and later migrate tangentially over long distances from their original production sites, gradually covering the surface of the cortex (Meyer et al., 2002; Takiguchi-Hayashi et al., 2004; Bielle et al., 2005; Yoshida et al., 2006; Garcia-Moreno et al., 2007; Imayoshi et al., 2008; Gu et al., 2009; Tissir et al., 2009). CR cells appear to play critical roles in the radial migration of subsequently born cortical neurons and eventually in the laminar organization of the cortex (D’Arcangelo et al., 1995; Ogawa et al., 1995; Super et al., 2000).
We revealed the reduced production of CH-derived CR cells in Dmrt3 sKO mice, especially in Dmrta1 and Dmrt3 dKO mice (Kikkawa et al., 2020). The reduction in CR cell production is consistent with the developmental impairment of the CH from which CR cells are produced. Dmrta2 sKO mice also have defects in the CR cells (Saulnier et al., 2013). These results suggest that DmrtAs cooperatively maintain the appropriate number of CR cells derived from specific source regions by defining the patterning of the medial telencephalon. For CR cell production, one of the candidate molecules working upstream of Dmrt3 and Dmrta1 may be forkhead box G1 (Foxg1). Foxg1 cKO mice (Foxg1tetOFoxg1 line, repressing Foxg1 transgene expression in the presence of doxycycline) showed an upregulated expression of Dmrt3 and Dmrta1 in the dorsal telencephalon (Kumamoto et al., 2013). ChIP-seq for Foxg1 revealed direct binding to the intronic sequences of Dmrt3 upstream and Dmrta1 downstream. Thus, the pathway by which Foxg1 represses Dmrt3 and Dmrta1 expression may critically affect telencephalic regionalization. This hypothesis is consistent with the converse phenotypes in the formation of CH in Dmrt mutants versus Foxg1 KO mice, showing region expansion (Dou et al., 1999; Martynoga et al., 2005; Hanashima et al., 2007).
Another early born neurons are subplate (SP) neurons generated before the excitatory neurons in the cortex; that is, between E10.5 and E12.5, in the mouse (Price et al., 1997). Glutamatergic SP neurons are derived from the cortex and rostral medial telencephalic wall (Yoshida et al., 1997; Shinozaki et al., 2002; Garcia-Moreno et al., 2008; Pedraza et al., 2014). SP neurons contribute to the establishment of the initial neural circuits between the cortex and thalamus during neocortical development (McConnell et al., 1989; Ghosh et al., 1990). SP neurons are also critical for inducing the multipolar-to-bipolar transition, which leads to a faster migration mode (Ohtaka-Maruyama et al., 2018).
A recent study has demonstrated that the production of SP neurons is significantly decreased by the loss of Dmrt3 and Dmrta2 (Ratie et al., 2020). To clarify the time window of the effect of Dmrta2 in SP neuron formation, they disrupted Dmrta2 in cortical progenitors from E10.5 in Dmrta2Lox/Lox;Emx1Cre mice and from E11.5 in Dmrta2Lox/Lox;NestinCre mice. The SP neurogenesis was only developmentally delayed in Dmrta2Lox/Lox;Emx1Cre mice and no phenotypic change was observed in Dmrta2Lox/Lox;NestinCre mice. Thus, the continuous expression of Dmrta2 from an earlier stage is likely necessary for the production and specification of SP neurons. Interestingly, the Dmrta2 protein is continuously detected in SP neurons, where Dmrta2 may regulate cortical neuron migration (Ratie et al., 2020).
Functions of Dmrt Genes in the Specification of Neural Stem/Progenitor Cells and Neurons Outside the Developing Telencephalon
So far, we have focused mainly on neurogenesis in the telencephalon. As mentioned earlier, Dmrta2 is also localized to the ventral-medial mesencephalic NSPCs, giving rise to mDA neurons that secrete the neurotransmitter dopamine. mDA neurons are generated from E10.5 to E14.5, with neuronal birth peaking between E11 and E12 (Bayer et al., 1995). Many factors function during mDA neurogenesis both in the expansion of the NSPC pool and in the proper specification of mDA neuronal fate (Gale and Li, 2008). Intriguingly, Dmrta2 is strongly expressed from E10.5 to E12.5 in the progenitor domains and promoted the expression of Foxa2, Lmx1a, and Msx1, (Gennet et al., 2011) which are transcription factors required for the specification of ventral-medial cell identities. Although the loss of Dmrta2 did not affect neurogenesis (Gennet et al., 2011), Dmrta2 repressed other genes in the ventral-lateral region, independent of Shh, which mainly contributes to the production of mDA neurons. In the telencephalon, a double knockdown of Dmrt3 and Dmrta2 did not affect the expression of Gli1 and Ptch1, which are involved in the Shh signaling pathway (Konno et al., 2019). From these results, the fate specification of the progenitors by Dmrts in both the telencephalon and midbrain seems to be regulated independently of the Shh signaling pathway. Since the β-gal signal was detected in the ventral midbrain of lacZ knock-in mice to the locus of Dmrta1 (Kikkawa et al., 2020), it would be interesting to know if there could be some functional overlap between Dmrta1 and Dmrta2.
A combinatorial expression of transcription factors specifies the dorsal spinal cord lineages where NSPCs differentiate into specific interneurons in the spinal cord. Dmrt3 is not expressed in NSPCs but in dI6 neurons, originating from dI6 progenitors at around E11.5 (Andersson et al., 2012). These dI6 neurons have two populations: Dmrt3- and Wt1-expressing interneurons. Their Dmrt3 KO mice showed an increased number of Wt1+ neurons, suggesting a fate change in the Dmrt3+ population within a specific subset of dI6 neurons. Consistent with the phenotype of the altered subpopulation of interneurons in the spinal cord, the Dmrt3 KO mice exhibited defects in the coordinated locomotor network controlling limb movements. A premature stop codon by DMRT3 mutation in horses changes locomotion patterns, which is favorable for harnessing racing performances (Andersson et al., 2012; Perry et al., 2019). Zebrafish studies have also reported that Dmrt3-expressing neurons contribute to locomotor activity (Del Pozo et al., 2020; Satou et al., 2020). These results suggest a conserved function of Dmrt3 in the spinal cord of vertebrates.
Perspectives on the Role of Dmrt in Human Pathogenesis
Although little is known about the function of DmrtAs in human diseases, several studies have recently shown that DmrtAs are involved in neurological diseases. One case report examined a consanguineous family with three confirmed siblings affected by a severe prenatal neurodevelopmental disorder characterized by severe progressive microcephaly (Urquhart et al., 2016). Their exome sequencing identified a homozygous single base pair deletion in DMRTA2 that lead to a frameshift variant. This phenotype in microcephaly appears to be consistent with cortical malformations in Dmrta2 KO mice as mentioned above. The second report identified the DMRT3 enhancer as a candidate involved in the pathogenesis of spastic cerebral palsy (Kubota et al., 2018), a disease that affects the movement and posture and is caused by a genetic abnormality in 30% of cerebral palsy cases (Fahey et al., 2017). The deletion of the enhancer, including the retinoic acid (RA) receptor/retinoid X receptor (RAR/RXR) complex-binding motif, has been identified in patients with spastic cerebral palsy (Lerer et al., 2005; Vanzo et al., 2013; Kubota et al., 2018). High-throughput chromosome conformation capture (Hi-C) data revealed that this enhancer region contacts the DMRT3 promoter (Kubota et al., 2018). Furthermore, RA stimulation upregulated the expression of Dmrt3 in embryonal carcinoma cells (Chatagnon et al., 2015). These results suggest that the transcription of Dmrt3 is decreased in the absence of the enhancer, leading to cerebral palsy. Many patients with spastic cerebral palsy find it difficult to move their arms and legs smoothly, possibly with impaired limb movement due to DMRT3 dysfunction in the interneurons of the spinal cord.
Conclusion
It is gradually becoming clear that members of DmrtA transcription factors contribute to multiple aspects of CNS development; however, their targets are still poorly understood despite their common structure containing the DM domain, which is a zinc finger DNA-binding motif. Since Dmrts form heterodimers with each other, it is possible that the transcriptional regulation is not straightforward and may be a source of complexity. As the molecular structure and expression patterns of DmrtAs are evolutionarily conserved, further analyses of these transcription factors will lead to an understanding of the shared mechanism of CNS development among various species. Recently, the involvement of DMRT in the malformation of the human brain has been identified (Urquhart et al., 2016); further findings of the DmrtA subfamily in human pathogenesis may lead to the elucidation of new insights for the understanding of diseases caused by developmental abnormalities in the CNS.
Author Contributions
TK drafted the manuscript and created the Figures. TK and NO discussed and revised the manuscript. Both authors contributed to the study and have approved the final manuscript.
Funding
This work was supported by JSPS KAKENHI for TK (#20K07237) and NO (#19H03318).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Anderson, S. A., Eisenstat, D. D., Shi, L., and Rubenstein, J. L. (1997). Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278, 474–476. doi: 10.1126/science.278.5337.474
Andersson, L. S., Larhammar, M., Memic, F., Wootz, H., Schwochow, D., Rubin, C. J., et al. (2012). Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646. doi: 10.1038/nature11399
Arai, Y., Funatsu, N., Numayama-Tsuruta, K., Nomura, T., Nakamura, S., and Osumi, N. (2005). Role of Fabp7, a downstream gene of Pax6, in the maintenance of neuroepithelial cells during early embryonic development of the rat cortex. J. Neurosci. 25, 9752–9761. doi: 10.1523/JNEUROSCI.2512-05.2005
Bayer, E. A., Stecky, R. C., Neal, L., Katsamba, P. S., Ahlsen, G., Balaji, V., et al. (2020). Ubiquitin-dependent regulation of a conserved DMRT protein controls sexually dimorphic synaptic connectivity and behavior. Elife 9:59614. doi: 10.7554/eLife.59614
Bayer, S. A., Wills, K. V., Triarhou, L. C., and Ghetti, B. (1995). Time of neuron origin and gradients of neurogenesis in midbrain dopaminergic neurons in the mouse. Exp. Brain Res. 105, 191–199. doi: 10.1007/BF00240955
Bellefroid, E. J., Leclere, L., Saulnier, A., Keruzore, M., Sirakov, M., Vervoort, M., et al. (2013). Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell Mol. Life Sci. 70, 3829–3845. doi: 10.1007/s00018-013-1288-2
Bielle, F., Griveau, A., Narboux-Neme, N., Vigneau, S., Sigrist, M., Arber, S., et al. (2005). Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat. Neurosci. 8, 1002–1012. doi: 10.1038/nn1511
Bishop, K. M., Rubenstein, J. L., and O’Leary, D. D. (2002). Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. J. Neurosci. 22, 7627–7638.
Bulchand, S., Grove, E. A., Porter, F. D., and Tole, S. (2001). LIM-homeodomain gene Lhx2 regulates the formation of the cortical hem. Mech. Dev. 100, 165–175. doi: 10.1016/s0925-4773(00)00515-3
Caronia-Brown, G., Yoshida, M., Gulden, F., Assimacopoulos, S., and Grove, E. A. (2014). The cortical hem regulates the size and patterning of neocortex. Development 141, 2855–2865. doi: 10.1242/dev.106914
Chatagnon, A., Veber, P., Morin, V., Bedo, J., Triqueneaux, G., Semon, M., et al. (2015). RAR/RXR binding dynamics distinguish pluripotency from differentiation associated cis-regulatory elements. Nucleic Acids Res. 43, 4833–4854. doi: 10.1093/nar/gkv370
Chou, S. J., Perez-Garcia, C. G., Kroll, T. T., and O’Leary, D. D. (2009). Lhx2 specifies regional fate in Emx1 lineage of telencephalic progenitors generating cerebral cortex. Nat. Neurosci. 12, 1381–1389. doi: 10.1038/nn.2427
Corbin, J. G., Gaiano, N., Machold, R. P., Langston, A., and Fishell, G. (2000). The Gsh2 homeodomain gene controls multiple aspects of telencephalic development. Development 127, 5007–5020.
D’Arcangelo, G., Miao, G. G., Chen, S. C., Soares, H. D., Morgan, J. I., and Curran, T. (1995). A protein related to extracellular matrix proteins deleted in the mouse mutant reeler. Nature 374, 719–723. doi: 10.1038/374719a0
De Clercq, S., Keruzore, M., Desmaris, E., Pollart, C., Assimacopoulos, S., Preillon, J., et al. (2018). DMRT5 Together with DMRT3 Directly Controls Hippocampus Development and Neocortical Area Map Formation. Cereb. Cortex 28, 493–509. doi: 10.1093/cercor/bhw384
Del Pozo, A., Manuel, R., Iglesias Gonzalez, A. B., Koning, H. K., Habicher, J., Zhang, H., et al. (2020). Behavioral Characterization of dmrt3a Mutant Zebrafish Reveals Crucial Aspects of Vertebrate Locomotion through Phenotypes Related to Acceleration. eNeuro 7:2020. doi: 10.1523/ENEURO.0047-20.2020
Desmaris, E., Keruzore, M., Saulnier, A., Ratie, L., Assimacopoulos, S., De Clercq, S., et al. (2018). DMRT5, DMRT3, and EMX2 Cooperatively Repress Gsx2 at the Pallium-Subpallium Boundary to Maintain Cortical Identity in Dorsal Telencephalic Progenitors. J. Neurosci. 38, 9105–9121. doi: 10.1523/JNEUROSCI.0375-18.2018
Dou, C. L., Li, S., and Lai, E. (1999). Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb. Cortex 9, 543–550. doi: 10.1093/cercor/9.6.543
Erdman, S. E., and Burtis, K. C. (1993). The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12, 527–535.
Fahey, M. C., Maclennan, A. H., Kretzschmar, D., Gecz, J., and Kruer, M. C. (2017). The genetic basis of cerebral palsy. Dev. Med. Child Neurol. 59, 462–469. doi: 10.1111/dmcn.13363
Furuta, Y., Piston, D. W., and Hogan, B. L. (1997). Bone morphogenetic proteins (BMPs) as regulators of dorsal forebrain development. Development 124, 2203–2212.
Gale, E., and Li, M. (2008). Midbrain dopaminergic neuron fate specification: Of mice and embryonic stem cells. Mol. Brain 1:8. doi: 10.1186/1756-6606-1-8
Garcia-Moreno, F., Lopez-Mascaraque, L., and De Carlos, J. A. (2007). Origins and migratory routes of murine Cajal-Retzius cells. J. Comp. Neurol. 500, 419–432. doi: 10.1002/cne.21128
Garcia-Moreno, F., Lopez-Mascaraque, L., and de Carlos, J. A. (2008). Early telencephalic migration topographically converging in the olfactory cortex. Cereb. Cortex 18, 1239–1252. doi: 10.1093/cercor/bhm154
Gennet, N., Gale, E., Nan, X., Farley, E., Takacs, K., Oberwallner, B., et al. (2011). Doublesex and mab-3-related transcription factor 5 promotes midbrain dopaminergic identity in pluripotent stem cells by enforcing a ventral-medial progenitor fate. Proc. Natl. Acad. Sci. U S A. 108, 9131–9136. doi: 10.1073/pnas.1016679108
Ghosh, A., Antonini, A., McConnell, S. K., and Shatz, C. J. (1990). Requirement for subplate neurons in the formation of thalamocortical connections. Nature 347, 179–181. doi: 10.1038/347179a0
Godbole, G., Roy, A., Shetty, A. S., and Tole, S. (2017). Novel functions of LHX2 and PAX6 in the developing telencephalon revealed upon combined loss of both genes. Neural Dev. 12:19. doi: 10.1186/s13064-017-0097-y
Gorski, J. A., Talley, T., Qiu, M., Puelles, L., Rubenstein, J. L., and Jones, K. R. (2002). Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 22, 6309–6314.
Gotz, M., and Huttner, W. B. (2005). The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 6, 777–788. doi: 10.1038/nrm1739
Grove, E. A., Tole, S., Limon, J., Yip, L. W., and Ragsdale, C. W. (1998). The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development 125, 2315–2325.
Gu, X., Yan, Y., Li, H., He, D., Pleasure, S. J., and Zhao, C. (2009). Characterization of the Frizzled10-CreER transgenic mouse: an inducible Cre line for the study of Cajal-Retzius cell development. Genesis 47, 210–216. doi: 10.1002/dvg.20472
Guo, Y., Li, Q., Gao, S., Zhou, X., He, Y., Shang, X., et al. (2004). Molecular cloning, characterization, and expression in brain and gonad of Dmrt5 of zebrafish. Biochem. Biophys. Res. Commun. 324, 569–575. doi: 10.1016/j.bbrc.2004.09.085
Hanashima, C., Fernandes, M., Hebert, J. M., and Fishell, G. (2007). The role of Foxg1 and dorsal midline signaling in the generation of Cajal-Retzius subtypes. J. Neurosci. 27, 11103–11111. doi: 10.1523/JNEUROSCI.1066-07.2007
Hasenpusch-Theil, K., Magnani, D., Amaniti, E. M., Han, L., Armstrong, D., and Theil, T. (2012). Transcriptional analysis of Gli3 mutants identifies Wnt target genes in the developing hippocampus. Cereb. Cortex 22, 2878–2893. doi: 10.1093/cercor/bhr365
Hebert, J. M., and Fishell, G. (2008). The genetics of early telencephalon patterning: some assembly required. Nat. Rev. Neurosci. 9, 678–685. doi: 10.1038/nrn2463
Hebert, J. M., Mishina, Y., and McConnell, S. K. (2002). BMP signaling is required locally to pattern the dorsal telencephalic midline. Neuron 35, 1029–1041. doi: 10.1016/s0896-6273(02)00900-5
Hevner, R. F., Neogi, T., Englund, C., Daza, R. A., and Fink, A. (2003). Cajal-Retzius cells in the mouse: transcription factors, neurotransmitters, and birthdays suggest a pallial origin. Brain Res. Dev. Brain Res. 141, 39–53.
Hong, C. S., Park, B. Y., and Saint-Jeannet, J. P. (2007). The function of Dmrt genes in vertebrate development: it is not just about sex. Dev. Biol. 310, 1–9. doi: 10.1016/j.ydbio.2007.07.035
Hou, P. S., Chuang, C. Y., Kao, C. F., Chou, S. J., Stone, L., Ho, H. N., et al. (2013). LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 41, 7753–7770. doi: 10.1093/nar/gkt567
Huang, X., Hong, C. S., O’Donnell, M., and Saint-Jeannet, J. P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. U S A. 102, 11349–11354. doi: 10.1073/pnas.0505106102
Huttner, W. B., and Kosodo, Y. (2005). Symmetric versus asymmetric cell division during neurogenesis in the developing vertebrate central nervous system. Curr. Opin. Cell Biol. 17, 648–657. doi: 10.1016/j.ceb.2005.10.005
Imayoshi, I., Shimogori, T., Ohtsuka, T., and Kageyama, R. (2008). Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development 135, 2531–2541. doi: 10.1242/dev.021535
Inada, H., Numayama-Tsuruta, K., Mochizuki, K., Sasaki-Hoshino, M., and Osumi, N. (2018). Pax6-dependent regulation of the rat Fabp7 promoter activity. Genes Cells 2018:12623. doi: 10.1111/gtc.12623
Kageyama, R., Ochi, S., Sueda, R., and Shimojo, H. (2020). The significance of gene expression dynamics in neural stem cell regulation. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 96, 351–363. doi: 10.2183/pjab.96.026
Kawamata, M., and Nishimori, K. (2006). Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 580, 6442–6446. doi: 10.1016/j.febslet.2006.10.066
Kikkawa, T., Casingal, C. R., Chun, S. H., Shinohara, H., Hiraoka, K., and Osumi, N. (2019). The role of Pax6 in brain development and its impact on pathogenesis of autism spectrum disorder. Brain Res. 1705, 95–103. doi: 10.1016/j.brainres.2018.02.041
Kikkawa, T., Obayashi, T., Takahashi, M., Fukuzaki-Dohi, U., Numayama-Tsuruta, K., and Osumi, N. (2013). Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes Cells 18, 636–649. doi: 10.1111/gtc.12061
Kikkawa, T., Sakayori, N., Yuuki, H., Katsuyama, Y., Matsuzaki, F., Konno, D., et al. (2020). Dmrt genes participate in the development of Cajal-Retzius cells derived from the cortical hem in the telencephalon. Dev. Dyn. 249, 698–710. doi: 10.1002/dvdy.156
Kim, S., Bardwell, V. J., and Zarkower, D. (2007a). Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev. Biol. 307, 314–327. doi: 10.1016/j.ydbio.2007.04.046
Kim, S., Kettlewell, J. R., Anderson, R. C., Bardwell, V. J., and Zarkower, D. (2003). Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 3, 77–82.
Kim, S., Namekawa, S. H., Niswander, L. M., Ward, J. O., Lee, J. T., Bardwell, V. J., et al. (2007b). A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 3:e62. doi: 10.1371/journal.pgen.0030062
Konno, D., Iwashita, M., Satoh, Y., Momiyama, A., Abe, T., Kiyonari, H., et al. (2012). The mammalian DM domain transcription factor Dmrta2 is required for early embryonic development of the cerebral cortex. PLoS One 7:e46577. doi: 10.1371/journal.pone.0046577
Konno, D., Kishida, C., Maehara, K., Ohkawa, Y., Kiyonari, H., Okada, S., et al. (2019). Dmrt factors determine the positional information of cerebral cortical progenitors via differential suppression of homeobox genes. Development 146:174243. doi: 10.1242/dev.174243
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175–184. doi: 10.1016/j.tig.2012.02.002
Kubota, N., Yokoyama, T., Hoshi, N., and Suyama, M. (2018). Identification of a candidate enhancer for DMRT3 involved in spastic cerebral palsy pathogenesis. Biochem. Biophys. Res. Commun. 496, 133–139. doi: 10.1016/j.bbrc.2018.01.011
Kumamoto, T., Toma, K., Gunadi, McKenna, W. L., Kasukawa, T., Katzman, S., et al. (2013). Foxg1 coordinates the switch from nonradially to radially migrating glutamatergic subtypes in the neocortex through spatiotemporal repression. Cell Rep. 3, 931–945. doi: 10.1016/j.celrep.2013.02.023
Lee, S. M., Tole, S., Grove, E., and McMahon, A. P. (2000). A local Wnt-3a signal is required for development of the mammalian hippocampus. Development 127, 457–467.
Lerer, I., Sagi, M., Meiner, V., Cohen, T., Zlotogora, J., and Abeliovich, D. (2005). Deletion of the ANKRD15 gene at 9p24.3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum. Mol. Genet. 14, 3911–3920. doi: 10.1093/hmg/ddi415
Li, Q., Zhou, X., Guo, Y., Shang, X., Chen, H., Lu, H., et al. (2008). Nuclear localization, DNA binding and restricted expression in neural and germ cells of zebrafish Dmrt3. Biol. Cell 100, 453–463. doi: 10.1042/BC20070114
Liu, S., Li, Z., and Gui, J. F. (2009). Fish-specific duplicated dmrt2b contributes to a divergent function through Hedgehog pathway and maintains left-right asymmetry establishment function. PLoS One 4:e7261. doi: 10.1371/journal.pone.0007261
Lourenco, R., Lopes, S. S., and Saude, L. (2010). Left-right function of dmrt2 genes is not conserved between zebrafish and mouse. PLoS One 5:e14438. doi: 10.1371/journal.pone.0014438
Mangale, V. S., Hirokawa, K. E., Satyaki, P. R., Gokulchandran, N., Chikbire, S., Subramanian, L., et al. (2008). Lhx2 selector activity specifies cortical identity and suppresses hippocampal organizer fate. Science 319, 304–309. doi: 10.1126/science.1151695
Martynoga, B., Morrison, H., Price, D. J., and Mason, J. O. (2005). Foxg1 is required for specification of ventral telencephalon and region-specific regulation of dorsal telencephalic precursor proliferation and apoptosis. Dev. Biol. 283, 113–127. doi: 10.1016/j.ydbio.2005.04.005
McConnell, S. K., Ghosh, A., and Shatz, C. J. (1989). Subplate neurons pioneer the first axon pathway from the cerebral cortex. Science 245, 978–982. doi: 10.1126/science.2475909
Meyer, G., Perez-Garcia, C. G., Abraham, H., and Caput, D. (2002). Expression of p73 and Reelin in the developing human cortex. J. Neurosci. 22, 4973–4986.
Miller, F. D., and Gauthier, A. S. (2007). Timing is everything: making neurons versus glia in the developing cortex. Neuron 54, 357–369. doi: 10.1016/j.neuron.2007.04.019
Monuki, E. S. (2007). The morphogen signaling network in forebrain development and holoprosencephaly. J. Neuropathol. Exp. Neurol. 66, 566–575. doi: 10.1097/nen.0b013e3180986e1b
Monuki, E. S., Porter, F. D., and Walsh, C. A. (2001). Patterning of the dorsal telencephalon and cerebral cortex by a roof plate-Lhx2 pathway. Neuron 32, 591–604. doi: 10.1016/s0896-6273(01)00504-9
Muralidharan, B., Keruzore, M., Pradhan, S. J., Roy, B., Shetty, A. S., Kinare, V., et al. (2017). Dmrt5, a Novel Neurogenic Factor, Reciprocally Regulates Lhx2 to Control the Neuron-Glia Cell-Fate Switch in the Developing Hippocampus. J. Neurosci. 37, 11245–11254. doi: 10.1523/JNEUROSCI.1535-17.2017
Murphy, M. W., Zarkower, D., and Bardwell, V. J. (2007). Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol. Biol. 8:58. doi: 10.1186/1471-2199-8-58
Muzio, L., Di Benedetto, B., Stoykova, A., Boncinelli, E., Gruss, P., and Mallamaci, A. (2002). Emx2 and Pax6 control regionalization of the pre-neuronogenic cortical primordium. Cereb. Cortex 12, 129–139. doi: 10.1093/cercor/12.2.129
Nakagawa, Y., Johnson, J. E., and O’Leary, D. D. (1999). Graded and areal expression patterns of regulatory genes and cadherins in embryonic neocortex independent of thalamocortical input. J. Neurosci. 19, 10877–10885.
Numayama-Tsuruta, K., Arai, Y., Takahashi, M., Sasaki-Hoshino, M., Funatsu, N., Nakamura, S., et al. (2010). Downstream genes of Pax6 revealed by comprehensive transcriptome profiling in the developing rat hindbrain. BMC Dev. Biol. 10:6. doi: 10.1186/1471-213X-10-6
Ogawa, M., Miyata, T., Nakajima, K., Yagyu, K., Seike, M., Ikenaka, K., et al. (1995). The reeler gene-associated antigen on Cajal-Retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron 14, 899–912.
Ohtaka-Maruyama, C., Okamoto, M., Endo, K., Oshima, M., Kaneko, N., Yura, K., et al. (2018). Synaptic transmission from subplate neurons controls radial migration of neocortical neurons. Science 360, 313–317. doi: 10.1126/science.aar2866
Ottolenghi, C., Fellous, M., Barbieri, M., and McElreavey, K. (2002). Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex-related genes and the evolution of sex determination. Genomics 79, 333–343. doi: 10.1006/geno.2002.6711
Parlier, D., Moers, V., Van Campenhout, C., Preillon, J., Leclere, L., Saulnier, A., et al. (2013). The Xenopus doublesex-related gene Dmrt5 is required for olfactory placode neurogenesis. Dev. Biol. 373, 39–52. doi: 10.1016/j.ydbio.2012.10.003
Pedraza, M., Hoerder-Suabedissen, A., Albert-Maestro, M. A., Molnar, Z., and De Carlos, J. A. (2014). Extracortical origin of some murine subplate cell populations. Proc. Natl. Acad. Sci. U S A. 111, 8613–8618. doi: 10.1073/pnas.1323816111
Perry, S., Larhammar, M., Vieillard, J., Nagaraja, C., Hilscher, M. M., Tafreshiha, A., et al. (2019). Characterization of Dmrt3-Derived Neurons Suggest a Role within Locomotor Circuits. J. Neurosci. 39, 1771–1782. doi: 10.1523/JNEUROSCI.0326-18.2018
Price, D. J., Aslam, S., Tasker, L., and Gillies, K. (1997). Fates of the earliest generated cells in the developing murine neocortex. J. Comp. Neurol. 377, 414–422.
Puelles, L., Kuwana, E., Puelles, E., Bulfone, A., Shimamura, K., Keleher, J., et al. (2000). Pallial and subpallial derivatives in the embryonic chick and mouse telencephalon, traced by the expression of the genes Dlx-2, Emx-1, Nkx-2.1, Pax-6, and Tbr-1. J. Comp. Neurol. 424, 409–438.
Qian, X., Shen, Q., Goderie, S. K., He, W., Capela, A., Davis, A. A., et al. (2000). Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron 28, 69–80. doi: 10.1016/s0896-6273(00)00086-6
Rakic, P. (1988). Specification of cerebral cortical areas. Science 241, 170–176. doi: 10.1126/science.3291116
Ratie, L., Desmaris, E., Garcia-Moreno, F., Hoerder-Suabedissen, A., Kelman, A., Theil, T., et al. (2020). Loss of Dmrt5 Affects the Formation of the Subplate and Early Corticogenesis. Cereb. Cortex 30, 3296–3312. doi: 10.1093/cercor/bhz310
Raymond, C. S., Murphy, M. W., O’Sullivan, M. G., Bardwell, V. J., and Zarkower, D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587–2595.
Raymond, C. S., Shamu, C. E., Shen, M. M., Seifert, K. J., Hirsch, B., Hodgkin, J., et al. (1998). Evidence for evolutionary conservation of sex-determining genes. Nature 391, 691–695. doi: 10.1038/35618
Rhinn, M., Picker, A., and Brand, M. (2006). Global and local mechanisms of forebrain and midbrain patterning. Curr. Opin. Neurobiol. 16, 5–12. doi: 10.1016/j.conb.2006.01.005
Sansom, S. N., Griffiths, D. S., Faedo, A., Kleinjan, D. J., Ruan, Y., Smith, J., et al. (2009). The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 5:e1000511. doi: 10.1371/journal.pgen.1000511
Satou, C., Sugioka, T., Uemura, Y., Shimazaki, T., Zmarz, P., Kimura, Y., et al. (2020). Functional Diversity of Glycinergic Commissural Inhibitory Neurons in Larval Zebrafish. Cell Rep. 30, 3036–3050e3034. doi: 10.1016/j.celrep.2020.02.015
Saude, L., Lourenco, R., Goncalves, A., and Palmeirim, I. (2005). terra is a left-right asymmetry gene required for left-right synchronization of the segmentation clock. Nat. Cell Biol. 7, 918–920. doi: 10.1038/ncb1294
Saulnier, A., Keruzore, M., De Clercq, S., Bar, I., Moers, V., Magnani, D., et al. (2013). The doublesex homolog Dmrt5 is required for the development of the caudomedial cerebral cortex in mammals. Cereb. Cortex 23, 2552–2567. doi: 10.1093/cercor/bhs234
Shetty, A. S., Godbole, G., Maheshwari, U., Padmanabhan, H., Chaudhary, R., Muralidharan, B., et al. (2013). Lhx2 regulates a cortex-specific mechanism for barrel formation. Proc. Natl. Acad. Sci. U S A. 110, E4913–E4921. doi: 10.1073/pnas.1311158110
Shimoda, Y., Tajima, Y., Osanai, T., Katsume, A., Kohara, M., Kudo, T., et al. (2002). Pax6 controls the expression of Lewis x epitope in the embryonic forebrain by regulating alpha 1,3-fucosyltransferase IX expression. J. Biol. Chem. 277, 2033–2039. doi: 10.1074/jbc.M108495200
Shinohara, H., Sakayori, N., Takahashi, M., and Osumi, N. (2013). Ninein is essential for the maintenance of the cortical progenitor character by anchoring the centrosome to microtubules. Biol. Open 2, 739–749. doi: 10.1242/bio.20135231
Shinozaki, K., Miyagi, T., Yoshida, M., Miyata, T., Ogawa, M., Aizawa, S., et al. (2002). Absence of Cajal-Retzius cells and subplate neurons associated with defects of tangential cell migration from ganglionic eminence in Emx1/2 double mutant cerebral cortex. Development 129, 3479–3492.
Smith, C. A., Hurley, T. M., McClive, P. J., and Sinclair, A. H. (2002). Restricted expression of DMRT3 in chicken and mouse embryos. Mech. Dev. 119(Suppl. 1), S73–S76.
Stoykova, A., Treichel, D., Hallonet, M., and Gruss, P. (2000). Pax6 modulates the dorsoventral patterning of the mammalian telencephalon. J. Neurosci. 20, 8042–8050.
Subramanian, L., Sarkar, A., Shetty, A. S., Muralidharan, B., Padmanabhan, H., Piper, M., et al. (2011). Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl. Acad. Sci. U S A. 108, E265–E274. doi: 10.1073/pnas.1101109108
Sun, J., Rockowitz, S., Xie, Q., Ashery-Padan, R., Zheng, D., and Cvekl, A. (2015). Identification of in vivo DNA-binding mechanisms of Pax6 and reconstruction of Pax6-dependent gene regulatory networks during forebrain and lens development. Nucleic Acids Res. 2015:gkv589. doi: 10.1093/nar/gkv589
Super, H., Del Rio, J. A., Martinez, A., Perez-Sust, P., and Soriano, E. (2000). Disruption of neuronal migration and radial glia in the developing cerebral cortex following ablation of Cajal-Retzius cells. Cereb. Cortex 10, 602–613.
Takiguchi-Hayashi, K., Sekiguchi, M., Ashigaki, S., Takamatsu, M., Hasegawa, H., Suzuki-Migishima, R., et al. (2004). Generation of reelin-positive marginal zone cells from the caudomedial wall of telencephalic vesicles. J. Neurosci. 24, 2286–2295. doi: 10.1523/JNEUROSCI.4671-03.2004
Theil, T., Aydin, S., Koch, S., Grotewold, L., and Ruther, U. (2002). Wnt and Bmp signalling cooperatively regulate graded Emx2 expression in the dorsal telencephalon. Development 129, 3045–3054.
Tissir, F., Ravni, A., Achouri, Y., Riethmacher, D., Meyer, G., and Goffinet, A. M. (2009). DeltaNp73 regulates neuronal survival in vivo. Proc. Natl. Acad. Sci. U S A. 106, 16871–16876. doi: 10.1073/pnas.0903191106
Toresson, H., Potter, S. S., and Campbell, K. (2000). Genetic control of dorsal-ventral identity in the telencephalon: opposing roles for Pax6 and Gsh2. Development 127, 4361–4371.
Urquhart, J. E., Beaman, G., Byers, H., Roberts, N. A., Chervinsky, E., O’Sullivan, J., et al. (2016). DMRTA2 (DMRT5) is mutated in a novel cortical brain malformation. Clin. Genet. 89, 724–727. doi: 10.1111/cge.12734
Vanzo, R. J., Martin, M. M., Sdano, M. R., and South, S. T. (2013). Familial KANK1 deletion that does not follow expected imprinting pattern. Eur. J. Med. Genet. 56, 256–259. doi: 10.1016/j.ejmg.2013.02.006
Veith, A. M., Schafer, M., Kluver, N., Schmidt, C., Schultheis, C., Schartl, M., et al. (2006). Tissue-specific expression of dmrt genes in embryos and adults of the platyfish Xiphophorus maculatus. Zebrafish 3, 325–337. doi: 10.1089/zeb.2006.3.325
Walcher, T., Xie, Q., Sun, J., Irmler, M., Beckers, J., Ozturk, T., et al. (2013). Functional dissection of the paired domain of Pax6 reveals molecular mechanisms of coordinating neurogenesis and proliferation. Development 140, 1123–1136. doi: 10.1242/dev.082875
Wilson, S. W., and Houart, C. (2004). Early steps in the development of the forebrain. Dev. Cell 6, 167–181. doi: 10.1016/s1534-5807(04)00027-9
Winkler, C., Hornung, U., Kondo, M., Neuner, C., Duschl, J., Shima, A., et al. (2004). Developmentally regulated and non-sex-specific expression of autosomal dmrt genes in embryos of the Medaka fish (Oryzias latipes). Mech. Dev. 121, 997–1005. doi: 10.1016/j.mod.2004.03.018
Xie, Q., Yang, Y., Huang, J., Ninkovic, J., Walcher, T., Wolf, L., et al. (2013). Pax6 interactions with chromatin and identification of its novel direct target genes in lens and forebrain. PLoS One 8:e54507. doi: 10.1371/journal.pone.0054507
Yoshida, M., Assimacopoulos, S., Jones, K. R., and Grove, E. A. (2006). Massive loss of Cajal-Retzius cells does not disrupt neocortical layer order. Development 133, 537–545. doi: 10.1242/dev.02209
Yoshida, M., Suda, Y., Matsuo, I., Miyamoto, N., Takeda, N., Kuratani, S., et al. (1997). Emx1 and Emx2 functions in development of dorsal telencephalon. Development 124, 101–111.
Yoshizawa, A., Nakahara, Y., Izawa, T., Ishitani, T., Tsutsumi, M., Kuroiwa, A., et al. (2011). Zebrafish Dmrta2 regulates neurogenesis in the telencephalon. Genes Cells 16, 1097–1109. doi: 10.1111/j.1365-2443.2011.01555.x
Young, F. I., Keruzore, M., Nan, X., Gennet, N., Bellefroid, E. J., and Li, M. (2017). The doublesex-related Dmrta2 safeguards neural progenitor maintenance involving transcriptional regulation of Hes1. Proc. Natl. Acad. Sci. U S A. 114, E5599–E5607. doi: 10.1073/pnas.1705186114
Yun, K., Potter, S., and Rubenstein, J. L. (2001). Gsh2 and Pax6 play complementary roles in dorsoventral patterning of the mammalian telencephalon. Development 128, 193–205.
Keywords: DmrtA subfamily, patterning, neurogenesis, neuronal specification, corticogenesis
Citation: Kikkawa T and Osumi N (2021) Multiple Functions of the Dmrt Genes in the Development of the Central Nervous System. Front. Neurosci. 15:789583. doi: 10.3389/fnins.2021.789583
Received: 05 October 2021; Accepted: 22 November 2021;
Published: 09 December 2021.
Edited by:
Takuma Kumamoto, Tokyo Metropolitan Institute of Medical Science, JapanReviewed by:
Masakazu Namihira, National Institute of Advanced Industrial Science and Technology (AIST), JapanEric Joseph Bellefroid, Université Libre de Bruxelles, Belgium
Copyright © 2021 Kikkawa and Osumi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Takako Kikkawa, a2lra2F3YUBtZWQudG9ob2t1LmFjLmpw
 Takako Kikkawa
Takako Kikkawa Noriko Osumi
Noriko Osumi