- School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, United States
Conserved transcription factors termed “terminal selectors” regulate neuronal sub-type specification and differentiation through combinatorial transcriptional regulation of terminal differentiation genes. The unique combinations of terminal differentiation gene products in turn contribute to the functional identities of each neuron. One well-characterized terminal selector is COE (Collier/Olf/Ebf), which has been shown to activate cholinergic gene batteries in C. elegans motor neurons. However, its functions in other metazoans, particularly chordates, is less clear. Here we show that the sole COE ortholog in the non-vertebrate chordate Ciona robusta, Ebf, controls the expression of the cholinergic locus VAChT/ChAT in a single dorsal interneuron of the larval Motor Ganglion, which is presumed to be homologous to the vertebrate spinal cord. We propose that, while the function of Ebf as a regulator of cholinergic neuron identity conserved across bilaterians, its exact role may have diverged in different cholinergic neuron subtypes (e.g., interneurons vs. motor neurons) in chordate-specific motor circuits.
Introduction
The myriad functions of nervous systems are made possible by the rich functional diversity of neuronal types and subtypes that are generated and connected to one another. Each terminally differentiated neuron in a neural circuit differs in their morphological, biochemical, and electrical properties. These properties in turn are often defined by coordinated gene expression changes regulated by transcription factors termed “terminal selectors” (Etchberger et al., 2007; Allan and Thor, 2015; Hobert and Kratsios, 2019). Terminal selectors can act alone or in combination to regulate the transcription of genes encoding rate-limiting effectors of terminal differentiation features. However, transcription factors that act as terminal selectors in one context might not function as such in other contexts.
Many terminal selectors have been shown to be evolutionarily conserved throughout animals, like Pou4/Brn3-family homeodomain factors expressed in various bilaterian and cnidarian neuron types (Finney et al., 1988; Eng et al., 2007; Serrano-Saiz et al., 2013, 2018; Zhang et al., 2014; Tournière et al., 2020). Another example comes from the COE (Collier/Olf/Ebf) family of transcription factors. A COE ortholog, UNC-3 in C. elegans, was shown to initiate and maintain the transcription of cholinergic genes, encoding essential components required for synthesis, transport, and reuptake of the major neurotransmitter acetylcholine (Kratsios et al., 2012) in motor neurons. Similarly, UNC-3 was found to control cholinergic gene expression in premotor interneurons as well, suggesting a broader function in regulating the development of cholinergic neurotransmission (Pereira et al., 2015). The COE ortholog Ebf was also found to be important for cholinergic motor neuron development in the invertebrate chordate Ciona, suggesting its role as a cholinergic motor neuron terminal selector is conserved from nematodes to chordates (Kratsios et al., 2012). However, subsequent studies on COE orthologs in vertebrates found a more nuanced role in motor neuron differentiation: different EBF paralogs are expressed in distinct spinal cord motor neurons innervating different axial muscles in mouse (Catela et al., 2019). Furthermore, Ebf2 is required for differentiation of a subset of motor neurons innervating epaxial (back) muscles, but not for their expression of cholinergic genes like Vesicular acetylcholine transporter (VAChT) (Catela et al., 2019). Thus, the regulation of cholinergic gene expression in vertebrate motor neurons may have shifted away from Ebf to another transcription factor, Islet (Cho et al., 2014; Catela et al., 2019).
Given the emerging differences between cholinergic gene regulation in vertebrate and invertebrate motor neurons, we decided to further investigate the role of Ebf in the regulation of cholinergic neuron identity in Ciona. With recent advances in tissue-specific CRISPR/Cas9-mediated mutagenesis (Stolfi et al., 2014) and the mapping of the Ciona larval connectome (Ryan et al., 2016), we were able to expand on this work with greater resolution than before. Based on these new tools and insights, we show here that Ebf activates cholinergic gene expression in a single neuron in the dorsal motor ganglion (MG) identified by the connectome as the Ascending Motor Ganglion Neuron 5 (AMG5) (Ryan et al., 2018; Kourakis et al., 2019). Although AMG5 is a cholinergic neuron situated in the major motor control center of the larva, it does not synapse directly onto muscles but rather onto other neurons of the MG, including primary motor neurons. We discuss these findings in the context of different scenarios for the evolution of cholinergic gene regulation in chordate motor circuits.
Methods
Ciona robusta (intestinalis Type A) adults were collected in the San Diego, CA region (M-REP Consulting). Gametes were isolated and prepared for in vitro fertilization as previously described (Christiaen et al., 2009b). Dechorionated zygotes were transfected by electroporation as previously described (Christiaen et al., 2009a). All relevant sequences are described in Supplementary Sequence File 1. Embryos were raised at 20°C (unless otherwise stated) and fixed in MEMFA fixative (3.7% formaldehyde, 0.1 M MOPS pH7.4, 0.5 M NaCl, 1 mM EGTA, 2 mM MgSO4, 0.1% Triton-X100) for 15 min, rinsed once in PBS/0.4% Triton-X100/50 mM NH4Cl and once in PBS/0.1% Triton-X100, then finally mounted in 1X PBS/50% Glycerol/2% DABCO mounting solution. Images were acquired on inverted epifluorescence (Leica DMIL LED or DMi8) or scanning point confocal microscopes (Zeiss LSM 700). Confocal images were processed as maximum intensity Z projections using Zeiss LSM or ImageJ software. CRISPR/Cas9-mediated knockout of Ebf was performed using 30 μg FOG > Cas9 (Gandhi et al., 2017), 40 μg U6 > Ebf.C (Gandhi et al., 2017) or U6 > Control (Stolfi et al., 2014), 15 μg FOG > H2B:mCherry (Rothbächer et al., 2007; Gline et al., 2009), and 90 μg VAChT/ChAT -4315/-3886 + bpFOG > Unc-76:GFP. The Unc-76 tag has been previously used for efficient labeling of neurons especially their axons (Dynes and Ngai, 1998; Imai et al., 2009).
Results
We previously identified a predicted binding site for Ebf (CATTTGGG) approximately 4.1 kb upstream of the translation start codon of Ciona VAChT, based on the consensus sequence CCCNNGGG (Figure 1A; Kratsios et al., 2012). VAChT and Choline acetyltransferase (ChAT) form a cholinergic locus (henceforth referred to as VAChT/ChAT) in which alternative splicing of a shared transcript results in two distinct mRNAs coding for different effectors of cholinergic neurotransmission: VAChT (also known as Slc18a3) and ChAT. This peculiar arrangement is conserved from nematodes to chordates (Alfonso et al., 1993). A fluorescent reporter plasmid spanning -4315 bp upstream of the VAChT translation start that includes this predicted Ebf site was previously shown to drive expression in all cholinergic neurons of the larva (Figure 1B; Kratsios et al., 2012). This includes cholinergic neurons of the brain, MG, and in bipolar tail neurons.
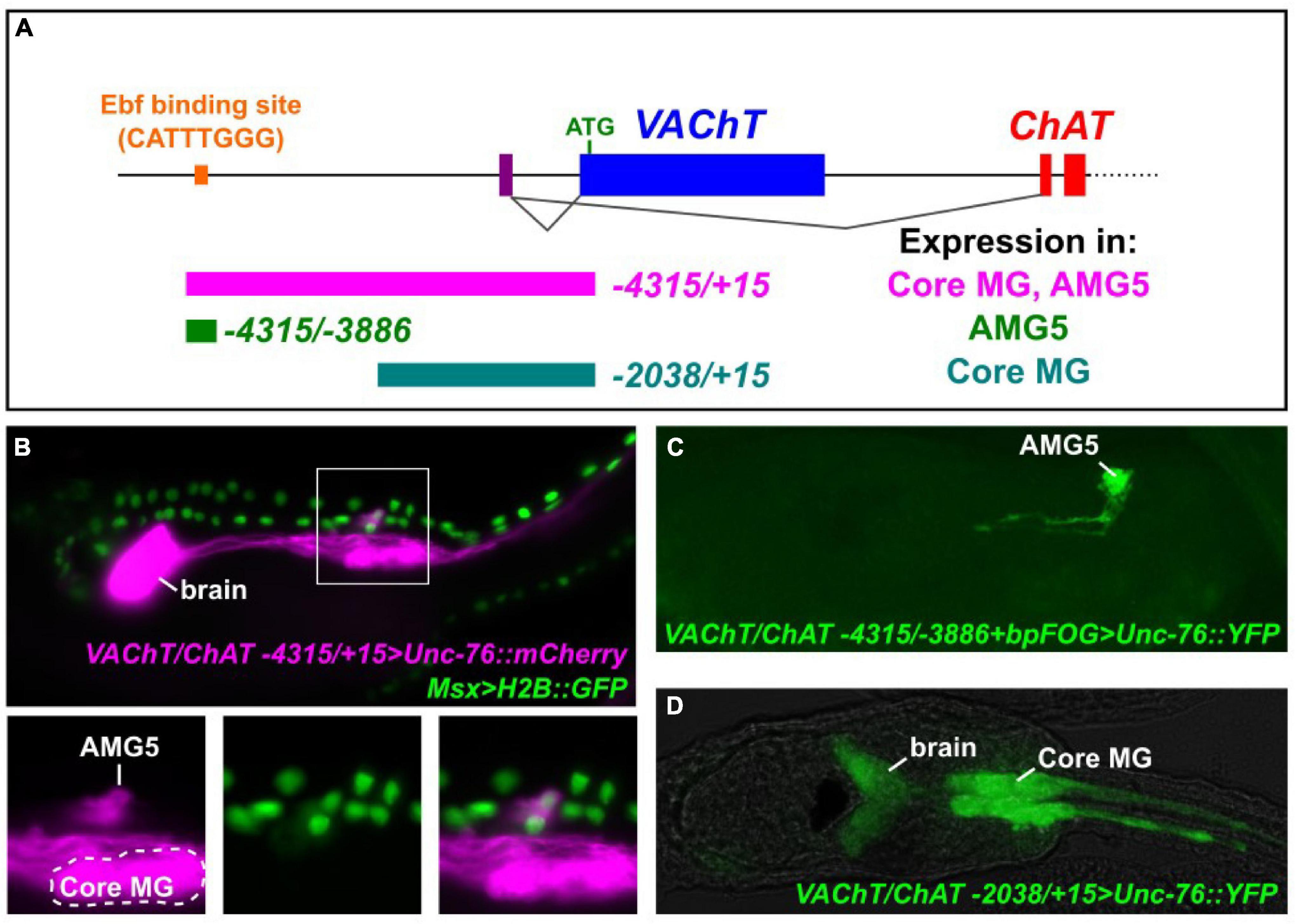
Figure 1. Expression of VAChT/ChAT cholinergic locus in the AMG5 neuron. (A) Cartoon schematic of the VAChT/ChAT locus encoding VAChT and ChAT from two mutually exclusive, alternatively spliced transcript variants sharing the same non-coding exon 1 (purple bar). Dotted line indicates additional exons encoding ChAT not shown. Colored bars underneath locus indicate cis-regulatory sequences tested, numbers reflect distance from the start codon (+ 1) of VAChT. Expression in “core” motor ganglion (MG) and Ascending Motor Ganglion neuron 5 (AMG5) was assessed qualitatively. An Ebf binding site previously reported (Kratsios et al., 2012) is situated -4111 bp upstream of VAChT. (B) Co-electroporation of VAChT/ChAT -4315/ + 15 > Unc-76:mCherry and Msx > H2B:GFP revealed co-expression in AMG5 at 19 h post-fertilization (hpf) at 20°C, a cholinergic interneuron of the dorsal MG, derived from the dorsal cells of the neural tube. (C) A minimal fragment surrounding the Ebf binding site is sufficient to drive AMG5-specific reporter expression at 18 hpf, 20°C. Image is a confocal Z stack projection. (D) A smaller proximal fragment from VAChT/ChAT (-2083/ + 15) lacking the Ebf binding site is sufficient to drive expression in other cholinergic neurons including those of the core MG, but not in AMG5, at 19 hpf, 18°C.
While the “core” MG comprises bilateral pairs of neurons derived from the lateral rows of the neural tube which includes the two pairs of major primary, or lower motor neurons (Cole and Meinertzhagen, 2004; Ryan et al., 2016), a set of ascending motor ganglion (AMG) neurons is situated dorsal to the MG, relaying peripheral inputs to neurons of the core MG (Ryan et al., 2016, 2018). These have been shown to comprise a mix of cholinergic and GABAergic neurons and (previously called dvCNs and dvGNs, respectively) (Takamura et al., 2010). More recently, it was shown that AMG neurons 1–4, 6, and 7 are GABAergic, while AMG neuron 5 (AMG5) represents the sole cholinergic neuron in this cluster (Kourakis et al., 2019). Indeed, we found that the full-length (-4315/ + 15) VAChT/ChAT reporter labels a single cell just dorsal to the anterior half of the core MG, in the exact spot where we expect AMG5 to be (Figure 1B). Co-electroporation with the reporter Msx > H2B:GFP, which labels animal pole-derived lateral neural plate border cells that give rise to the dorsal row of the neural tube (Cole and Meinertzhagen, 2004; Russo et al., 2004; Stolfi et al., 2015; Figure 1B). This suggests the AMG5 neuron comes from one of the b8.19 blastomeres on either side of the embryo prior to neural tube closure (Cole and Meinertzhagen, 2004; Pasini et al., 2006). To our knowledge, this is the first evidence for the developmental origins of AMG5 from the Msx + dorsal row of the neural tube, although we could not ascertain whether this cell is invariantly derived from the left or right side of the embryo.
When we tested a smaller fragment surrounding the predicted Ebf site (-4315/-3886) in conjunction with a heterologous basal promoter (bpFOG) (Rothbächer et al., 2007), we found that this was sufficient to drive expression of fluorescent reporter solely in AMG5, but not any other cholinergic neuron (Figure 1C). This revealed the characteristic morphology of AMG5 as originally determined by the serial-section electron micrographs of the connectome study (Ryan et al., 2016, 2018; Ryan and Meinertzhagen, 2019), including unusual left and right ascending neurites (Figure 1C). Therefore we conclude that this region around the previously identified Ebf site corresponds to a cis-regulatory element that is sufficient to drive VAChT/ChAT in the cholinergic AMG5 neuron. In contrast, a shorter proximal fragment spanning -2083 bp upstream of VAChT (not encompassing the AMG5-specific element) was sufficient to drive expression in other cholinergic neurons, including the motor neurons and other “core” MG neurons (Figure 1D).
Because of the predicted Ebf binding site in this AMG5-specific VAChT/ChAT cis-regulatory element (-4315/-3886), we hypothesized that Ebf might be directly activating VAChT/ChAT expression (and thus cholinergic identity) specifically in AMG5. Consistent with this hypothesis, we found that fluorescent protein expression driven by Ebf cis-regulatory sequences (Stolfi and Levine, 2011; Razy-Krajka et al., 2014) also labeled AMG5 and other cholinergic neurons, but not other surrounding (GABAergic) AMG neurons (Figure 2A). Co-expression with VAChT/ChAT reporter was confirmed by co-electroporation of Ebf and AMG5-specific VAChT/ChAT reporter plasmids (Figure 2B). Further, mutating the predicted Ebf site from CATTTGGG to CATTGCC in the context of the -4315/-3886 cis-regulatory element completely abolished reporter activity in AMG5 (Figure 3A). To test if Ebf can activate this AMG5-specific element, we overexpressed it in the ventral sensory papilla of the larva using the Msx promoter (Russo et al., 2004; Figure 3B). This strategy had previously been used to test ectopic expression of the full-length VAChT/ChAT reporter plasmid in response to Ebf overexpression (Kratsios et al., 2012). As expected, Ebf overexpression activated the VAChT/ChAT -4315/-3886 reporter in the papilla 54% of larvae, compared to 0% of larvae upon overexpression of a negative control lacZ sequence instead (Figures 3B,C). Furthermore, Ebf overexpression did not activate the expression of the VAChT/ChAT -4315/-3886 reporter carrying the mutated Ebf site (CATTTGGG to CATTGCC, Figure 3C). Taken together, these data suggest that Ebf directly activates an AMG5-specific cis-regulatory element for VAChT/ChAT expression.
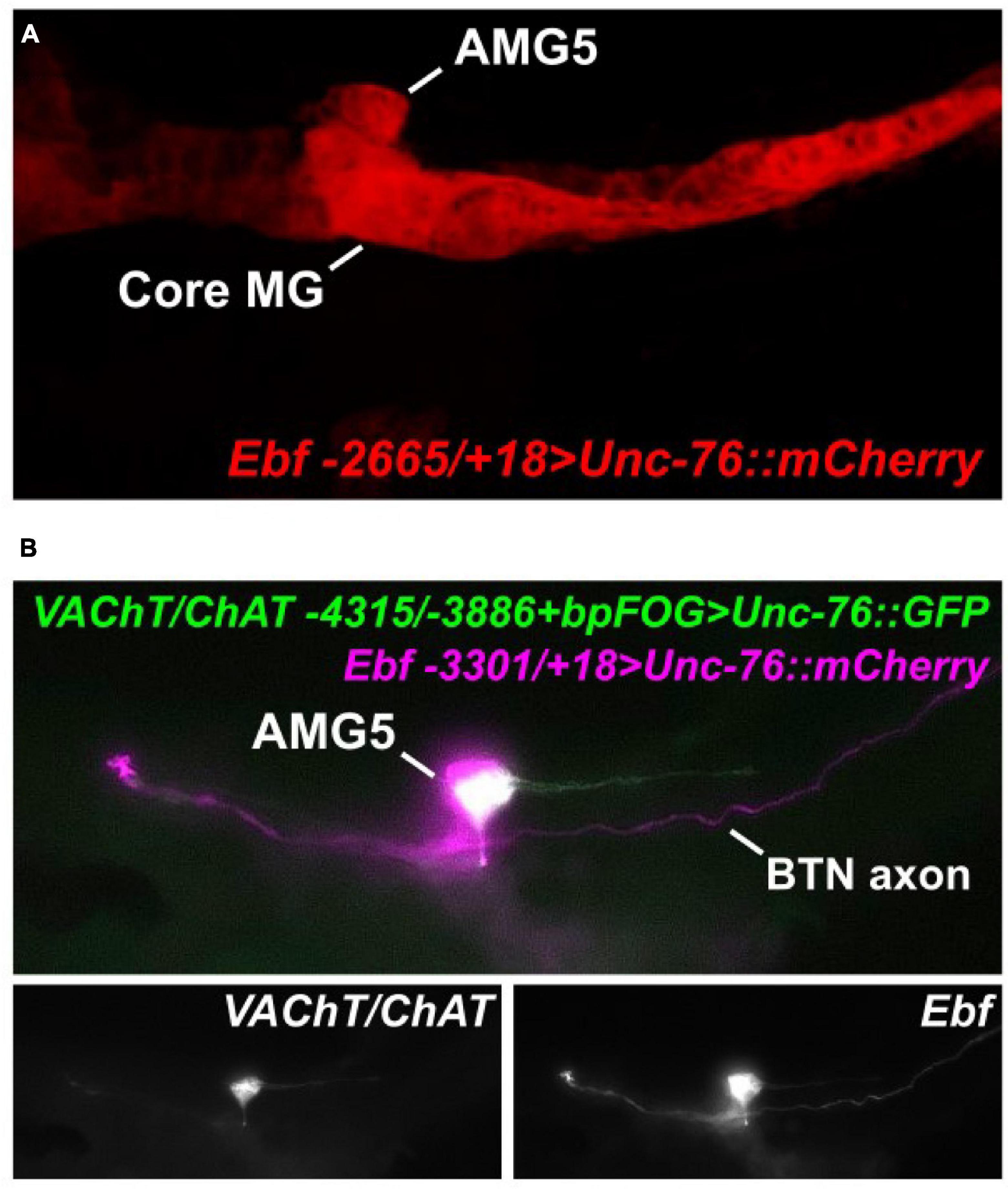
Figure 2. AMG5 co-expresses VAChT/ChAT and Ebf. (A) An Ebf reporter plasmid labels various CNS neurons including AMG5 at 18 h post-fertilization (hpf) at 20°C. Image is a confocal Z stack projection. (B) Co-electroporation of the Ebf reporter and the minimal AMG5-specific VAChT/ChAT reporter revealed co-expression in AMG5 at 17 hpf, 20°C. In this example, the larva shows mosaic incorporation of the plasmids only in the animal pole-derived lineages, which give rise to AMG5 and the Bipolar Tail Neuron (BTN) that also expresses Ebf (Stolfi et al., 2015), but not the core MG.
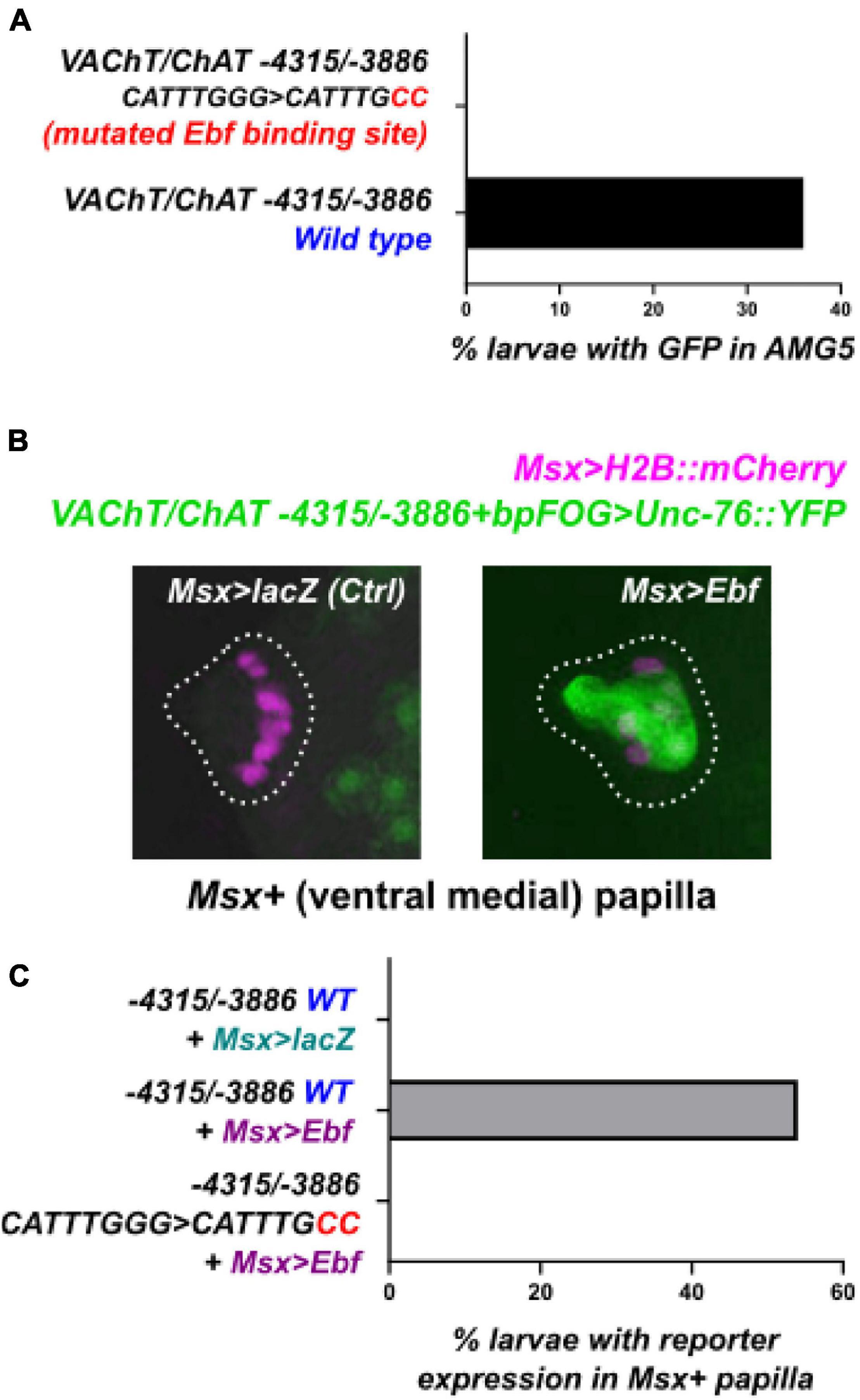
Figure 3. An Ebf binding site is necessary for VAChT/ChAT reporter activity. (A) Mutating the Ebf binding site in the minimal AMG5-specific VAChT/ChAT cis-regulatory element was sufficient to abolish its activity in AMG5. Mutated reporter n = 50, wild-type reporter n = 44. (B) The AMG5-specific VAChT/ChAT reporter can be ectopically activated in ventral papilla cells at 15.5 h post-fertilization (hpf) at 20°C, by overexpressing Ebf there (using Msx > Ebf). No expression is seen in the negative control electroporated with the neutral construct Msx > lacZ instead. Images are confocal Z stack projections. (C) Yet Ebf overexpression in the ventral papilla does not cause ectopic expression of the mutated VAChT/ChAT reporter, suggesting the Ebf binding site is indispensable for Ebf-mediated activation of this element. For each of the three conditions, n = 50.
To verify that Ebf acts in trans to activate this minimal AMG5-specific VAChT/ChAT cis-regulatory element, we performed tissue-specific CRISPR/Cas9-mediated knockout of Ebf in Ciona embryos. We co-electroporated a highly efficient, previously validated single-chain guide RNA (sgRNA) vector targeting Ebf (Gandhi et al., 2017) together with FOG > Cas9, which drives Cas9 specifically in the animal pole-derived blastomeres at the 8-cell stage (Rothbächer et al., 2007; Gandhi et al., 2017). This combination was sufficient to abolish expression of VAChT/ChAT -4315/-3886 reporter in AMG5 (Figure 4). Taking these manipulations in cis and trans together, we conclude that Ebf binds to and activates a distal cis-regulatory element of VAChT/ChAT that is activated only in AMG5, the sole cholinergic neuron in this region immediately dorsal to the “core” MG.
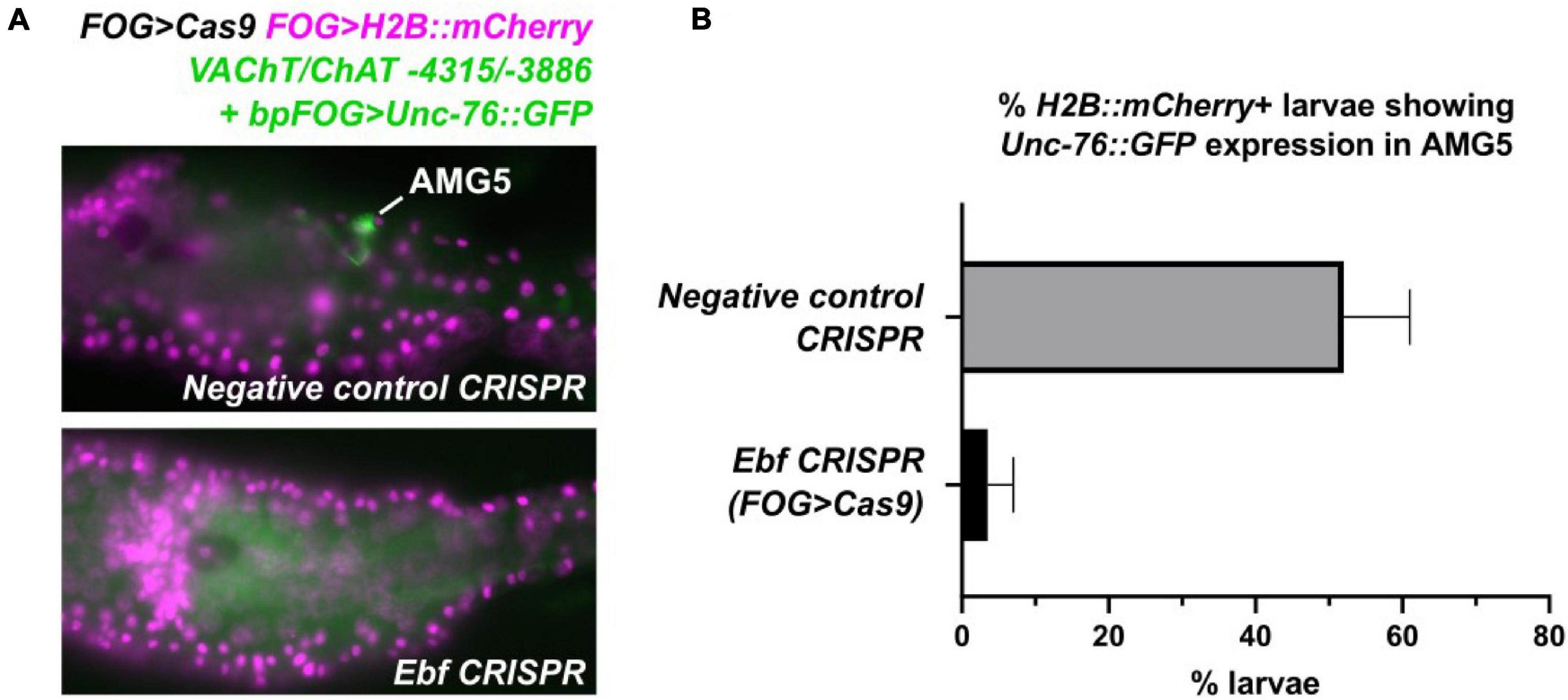
Figure 4. Tissue-specific CRISPR/Cas9-mediated knockout of Ebf abolishes VAChT/ChAT expression in AMG5. (A) F0 larvae co-electroporated with FOG > Cas9, FOG > H2B:mCherry, and the AMG5-specific VAChT/ChAT reporter (-4315/-3886 + bpFOG > Unc-76:GFP), and either a negative control guide RNA or an Ebf-targeting guide RNA (Gandhi et al., 2017) were scored for GFP signal in AMG5 at 17 h post-fertilization at 20°C. FOG > Cas9 restricts CRISPR activity to the animal pole-derived cells from which AMG5 is born. (B) Scoring results of negative control and Ebf CRISPR larvae, averaged over two replicates. Negative control: replicate 1 n = 54, replicate 2 n = 49. Ebf CRISPR: replicate 1 n = 58, replicate 2 n = 43. Error bars indicate range between replicates. These results indicate that knocking out Ebf in the animal pole mostly abolishes VAChT/ChAT reporter expression in AMG5.
As for a possible earlier role for Ebf that could explain the loss of VAChT/ChAT reporter expression in AMG5, Ebf is not expressed zygotically in the lineage that gives rise to AMG5, before that cell is born. This is known thanks to detailed in situ hybridization screens (Imai et al., 2004, 2006, 2009). Ebf expression in the early embryo is non-existent until the late gastrula stage, when it comes up in the A9.32 pair of blastomeres and a pair of tail tip cells (none of which give rise to the AMG neurons). Therefore, we find it highly unlikely that CRISPR knockout of Ebf in the animal pole using FOG > Cas9 is abrogating AMG5 specification prior to its birth.
Discussion
We show here that VAChT/ChAT reporter gene expression in the dorsal motor ganglion region of Ciona requires the transcription factor Ebf. Since Ebf and VAChT/ChAT are co-expressed in a single neuron (recently identified as AMG5) in this region, we suspect that it may act as a classically defined terminal selector (Etchberger et al., 2007; Allan and Thor, 2015; Hobert and Kratsios, 2019) of cholinergic identity in this cell. The AMG neurons are a synaptically interconnected cluster of 7 ascending interneurons that receive synaptic connections from a variety of neurons processing diverse sensory inputs including light, gravity, touch, and possibly chemosensation. Their synaptic targets include neurons of the core MG and neurons of the brain (Ryan et al., 2016). It was recently shown that VAChT/ChAT is expressed in a single AMG neuron (AMG5), while the remaining 6 AMG neurons express Vesicular GABA transporter (VGAT) instead, indicating a GABAergic identity (Kourakis et al., 2019). AMG5 is a single multipolar neuron situated right on the midline surrounded by the remaining AMG neurons, with left and right axon branches projecting anteriorly. It receives heavy inputs from glutamatergic posterior apical trunk epidermal neurons (pATENs) and is presynaptic to prominent cholinergic neurons of the core MG including Motor Neuron 1 (MN1) and the Descending Decussating Neuron (ddN) (Ryan et al., 2016). Given its cholinergic identity and unique position and connectivity in the connectome, its function may be to trigger swimming behavior though cholinergic excitation of primary motor neurons, relaying an as-of-yet unidentified sensory stimulus transduced by the pATENs.
In C. elegans, the Ebf ortholog UNC-3 is a terminal selector for cholinergic neuron fate (Kratsios et al., 2012), suggesting a deep evolutionary history of this transcription factor as a determinant of cholinergic neuron fate. While the most abundant and prominent cholinergic neuron type in C. elegans is the motor neuron, UNC-3 does regulate cholinergic gene expression in cholinergic interneurons as well (Pereira et al., 2015). Similarly, expression of Ebf2 and other Ebf paralogs is seen in various neuronal precursors of the developing mammalian spinal cord, including those in the dorsal horn (Catela et al., 2019), where sparse expression of cholinergic genes has been observed (Mesnage et al., 2011). Given the role of dorsal horn interneurons in sensory integration, and the developmental origin of AMG neurons from the dorsal row of cells of the Ciona neural tube, AMG5 might be a homolog of these rare cholinergic interneurons of the mammalian spinal cord dorsal horn.
Although we previously used a dominant-repressor form of Ebf to suggest its role in specifying Ciona cholinergic motor neurons (Kratsios et al., 2012), we show here that a shorter VAChT/ChAT cis-regulatory fragment lacking a key Ebf binding site is sufficient to drive expression in motor neurons, but not AMG5. Therefore, it is likely that Ebf directly activates VAChT/ChAT in AMG5 but not necessarily in the neurons of the “core” MG that includes the primary motor neurons of the larva. This does not rule out a role for Ebf in regulating the activation and/or maintenance of other cholinergic effectors or more generic terminal differentiation genes in primary motor neurons. Additionally, there are cholinergic neurons, including primary motor neurons, in the post-metamorphic adults (Hozumi et al., 2015; Jokura et al., 2020), and it remains entirely unknown if these depend on Ebf for their specification and/or cholinergic fate.
Taken together, our results may help bridge the seemingly divergent roles of Ebf/UNC-3 in regulating motor neuron differentiation in C. elegans and mammals. While in C. elegans UNC-3 regulates cholinergic gene expression in primary motor neurons (Kratsios et al., 2012), Ebf factors in mouse regulate other aspects of motor neuron differentiation independently of their cholinergic identity (Catela et al., 2019). In the Ciona larva, we see a possible evolutionary intermediate between these two extremes. While Ebf directly regulates VAChT/ChAT expression in a cholinergic neuron of the motor ganglion that is immediately presynaptic to the primary motor neurons of the larva, its role in the primary motor neurons may reflect a more vertebrate-like function. Alternatively, there may be greater genetic redundancy in regulation of cholinergic gene expression in chordate motor neurons. A broader phylogenetic sampling may answer whether any of these (nematode, tunicate, vertebrate) closely resemble the ancestral condition in the last bilaterian common ancestor.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SP and AS designed, performed, and interpreted the experiments. AS wrote the manuscript, with edits and suggestions by SP. Both authors contributed to the article and approved the submitted version.
Funding
This work was funded by the NSF grant 1940743 (IOS) and NIH grant GM143326 to AS, and an ARCS Foundation fellowship to SP.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Susanne Gibboney, Florian Razy-Krajka, and all members of the laboratory for advice and technical assistance. We thank Paschalis Kratsios, Oliver Hobert, and Mike Levine for their advice and support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2021.784649/full#supplementary-material
References
Alfonso, A., Grundahl, K., Duerr, J. S., Han, H.-P., and Rand, J. B. (1993). The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science 261, 617–619. doi: 10.1126/science.8342028
Allan, D. W., and Thor, S. (2015). Transcriptional selectors, masters, and combinatorial codes: regulatory principles of neural subtype specification. Wiley Interdiscip. Rev. Dev. Biol. 4, 505–528. doi: 10.1002/wdev.191
Catela, C., Correa, E., Wen, K., Aburas, J., Croci, L., Consalez, G. G., et al. (2019). An ancient role for collier/Olf/Ebf (COE)-type transcription factors in axial motor neuron development. Neural Dev. 14:2. doi: 10.1186/s13064-018-0125-6
Cho, H.-H., Cargnin, F., Kim, Y., Lee, B., Kwon, R.-J., Nam, H., et al. (2014). Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 10:e1004280. doi: 10.1371/journal.pgen.1004280
Christiaen, L., Wagner, E., Shi, W., and Levine, M. (2009b). Isolation of sea squirt (Ciona) gametes, fertilization, dechorionation, and development. Cold Spring Harb. Protoc. 2009:pdb.prot5344. doi: 10.1101/pdb.prot5344
Christiaen, L., Wagner, E., Shi, W., and Levine, M. (2009a). Electroporation of transgenic DNAs in the sea squirt Ciona. Cold Spring Harb. Protoc. 2009:pdb.prot5345. doi: 10.1101/pdb.prot5345
Cole, A. G., and Meinertzhagen, I. A. (2004). The central nervous system of the ascidian larva: mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev. Biol. 271, 239–262. doi: 10.1016/j.ydbio.2004.04.001
Dynes, J. L., and Ngai, J. (1998). Pathfinding of olfactory neuron axons to stereotyped glomerular targets revealed by dynamic imaging in living zebrafish embryos. Neuron 20, 1081–1091. doi: 10.1016/s0896-6273(00)80490-0
Eng, S. R., Dykes, I. M., Lanier, J., Fedtsova, N., and Turner, E. E. (2007). POU-domain factor Brn3a regulates both distinct and common programs of gene expression in the spinal and trigeminal sensory ganglia. Neural Dev. 2, 1–17. doi: 10.1186/1749-8104-2-3
Etchberger, J. F., Lorch, A., Sleumer, M. C., Zapf, R., Jones, S. J., Marra, M. A., et al. (2007). The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev. 21, 1653–1674. doi: 10.1101/gad.1560107
Finney, M., Ruvkun, G., and Horvitz, H. R. (1988). The C. elegans cell lineage and differentiation gene unc-86 encodes a protein with a homeodomain and extended similarity to transcription factors. Cell 55, 757–769. doi: 10.1016/0092-8674(88)90132-8
Gandhi, S., Haeussler, M., Razy-Krajka, F., Christiaen, L., and Stolfi, A. (2017). Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev. Biol. 425, 8–20. doi: 10.1016/j.ydbio.2017.03.003
Gline, S. E., Kuo, D. H., Stolfi, A., and Weisblat, D. A. (2009). High resolution cell lineage tracing reveals developmental variability in leech. Dev. Dyn. 238, 3139–3151. doi: 10.1002/dvdy.22158
Hobert, O., and Kratsios, P. (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr. Opin. Neurobiol. 56, 97–105. doi: 10.1016/j.conb.2018.12.006
Hozumi, A., Horie, T., and Sasakura, Y. (2015). Neuronal map reveals the highly regionalized pattern of the juvenile central nervous system of the ascidian Ciona intestinalis. Dev. Dyn. 244, 1375–1393. doi: 10.1002/dvdy.24317
Imai, K. S., Hino, K., Yagi, K., Satoh, N., and Satou, Y. (2004). Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047–4058. doi: 10.1242/dev.01270
Imai, K. S., Levine, M., Satoh, N., and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183–1187. doi: 10.1126/science.1123404
Imai, K. S., Stolfi, A., Levine, M., and Satou, Y. (2009). Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development 136, 285–293. doi: 10.1242/dev.026419
Jokura, K., Nishino, J. M., Ogasawara, M., and Nishino, A. (2020). An α7-related nicotinic acetylcholine receptor mediates the ciliary arrest response in pharyngeal gill slits of Ciona. J. Exp. Biol. 223:jeb209320.
Kourakis, M. J., Borba, C., Zhang, A., Newman-Smith, E., Salas, P., Manjunath, B., et al. (2019). Parallel visual circuitry in a basal chordate. eLife 8:e44753. doi: 10.7554/eLife.44753
Kratsios, P., Stolfi, A., Levine, M., and Hobert, O. (2012). Coordinated regulation of cholinergic motor neuron traits through a conserved terminal selector gene. Nat. Neurosci. 15, 205–214. doi: 10.1038/nn.2989
Mesnage, B., Gaillard, S., Godin, A. G., Rodeau, J.-L., Hammer, M., Von Engelhardt, J., et al. (2011). Morphological and functional characterization of cholinergic interneurons in the dorsal horn of the mouse spinal cord. J. Comp. Neurol. 519, 3139–3158. doi: 10.1002/cne.22668
Pasini, A., Amiel, A., Rothbächer, U., Roure, A., Lemaire, P., and Darras, S. (2006). Formation of the ascidian epidermal sensory neurons: insights into the origin of the chordate peripheral nervous system. PLoS Biol. 4:e225. doi: 10.1371/journal.pbio.0040225
Pereira, L., Kratsios, P., Serrano-Saiz, E., Sheftel, H., Mayo, A. E., Hall, D. H., et al. (2015). A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife 4:e12432. doi: 10.7554/eLife.12432
Razy-Krajka, F., Lam, K., Wang, W., Stolfi, A., Joly, M., Bonneau, R., et al. (2014). Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev. Cell 29, 263–276. doi: 10.1016/j.devcel.2014.04.001
Rothbächer, U., Bertrand, V., Lamy, C., and Lemaire, P. (2007). A combinatorial code of maternal GATA, Ets and β-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development 134, 4023–4032. doi: 10.1242/dev.010850
Russo, M. T., Donizetti, A., Locascio, A., D’Aniello, S., Amoroso, A., Aniello, F., et al. (2004). Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 267, 517–528. doi: 10.1016/j.ydbio.2003.11.005
Ryan, K., Lu, Z., and Meinertzhagen, I. A. (2016). The CNS connectome of a tadpole larva of Ciona intestinalis (L.) highlights sidedness in the brain of a chordate sibling. Elife 5:e16962. doi: 10.7554/eLife.16962
Ryan, K., Lu, Z., and Meinertzhagen, I. A. (2018). The peripheral nervous system of the ascidian tadpole larva: types of neurons and their synaptic networks. J. Comp. Neurol. 526, 583–608. doi: 10.1002/cne.24353
Ryan, K., and Meinertzhagen, I. A. (2019). Neuronal identity: the neuron types of a simple chordate sibling, the tadpole larva of Ciona intestinalis. Curr. Opin. Neurobiol. 56, 47–60. doi: 10.1016/j.conb.2018.10.015
Serrano-Saiz, E., Leyva-Díaz, E., De La Cruz, E., and Hobert, O. (2018). BRN3-type POU Homeobox Genes Maintain the Identity of Mature Postmitotic Neurons in Nematodes and Mice. Curr. Biol. 28, 2813–2823.e2.
Serrano-Saiz, E., Poole, R. J., Felton, T., Zhang, F., De La Cruz, E. D., and Hobert, O. (2013). Modular control of glutamatergic neuronal identity in C. elegans by distinct homeodomain proteins. Cell 155, 659–673. doi: 10.1016/j.cell.2013.09.052
Stolfi, A., Gandhi, S., Salek, F., and Christiaen, L. (2014). Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120. doi: 10.1242/dev.114488
Stolfi, A., and Levine, M. (2011). Neuronal subtype specification in the spinal cord of a protovertebrate. Development 138, 995–1004. doi: 10.1242/dev.061507
Stolfi, A., Ryan, K., Meinertzhagen, I. A., and Christiaen, L. (2015). Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 527, 371–374. doi: 10.1038/nature15758
Takamura, K., Minamida, N., and Okabe, S. (2010). Neural map of the larval central nervous system in the ascidian Ciona intestinalis. Zoolog. Sci. 27, 191–203.
Tournière, O., Dolan, D., Richards, G. S., Sunagar, K., Columbus-Shenkar, Y. Y., Moran, Y., et al. (2020). NvPOU4/Brain3 Functions as a Terminal Selector Gene in the Nervous System of the Cnidarian Nematostella vectensis. Cell Rep. 30, 4473–4489.e5.
Keywords: Ebf, COE, cholinergic, Ciona, tunicates, motor ganglion, acetylcholine
Citation: Popsuj S and Stolfi A (2021) Ebf Activates Expression of a Cholinergic Locus in a Multipolar Motor Ganglion Interneuron Subtype in Ciona. Front. Neurosci. 15:784649. doi: 10.3389/fnins.2021.784649
Received: 28 September 2021; Accepted: 30 November 2021;
Published: 17 December 2021.
Edited by:
Luisa Cochella, Research Institute of Molecular Pathology (IMP), AustriaReviewed by:
Takehiro Kusakabe, Konan University, JapanPaschalis Kratsios, University of Chicago, United States
Copyright © 2021 Popsuj and Stolfi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto Stolfi, YWxiZXJ0by5zdG9sZmlAYmlvc2NpLmdhdGVjaC5lZHU=
 Sydney Popsuj
Sydney Popsuj Alberto Stolfi
Alberto Stolfi