
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 26 November 2021
Sec. Neurogenomics
Volume 15 - 2021 | https://doi.org/10.3389/fnins.2021.705297
This article is part of the Research TopicGenetic Influence on the Onset, Progression and Treatment Response of Neuropsychiatric DiseasesView all 6 articles
Epilepsy is a common neurologic disorder characterized by intractable seizures, involving genetic factors. There is a need to develop reliable genetic markers to predict the risk of epilepsy and design effective therapies. Arsenite methyltransferase (AS3MT) catalyzes the biomethylation of arsenic and hence regulates arsenic metabolism. AS3MT variation has been linked to the progression of various diseases including schizophrenia and attention deficit or hyperactivity disorder. Whether genetic polymorphism of AS3MT contributes to epilepsy remains unclear. In this study, we investigated the association of AS3MT gene polymorphism with susceptibility to epilepsy in children from south China. We also explored the effect of AS3MT variation on the safety of antiepileptic drugs. Genotypic analysis for AS3MT rs7085104 was performed using samples from a Chinese cohort of 200 epileptic children and 244 healthy individuals. The results revealed a genetic association of AS3MT rs7085104 with susceptibility to pediatric epilepsy. Mutant homozygous GG genotype exhibited a lower susceptibility to childhood epilepsy than AA genotype. Carriers of AS3MT rs7085104 AA genotype exhibited a higher risk of digestive adverse drug reactions (dADRs) in children when treated with valproic acid (VPA) or oxcarbazepine (OXC). Additionally, bioinformatics analysis identified eight AS3MT target genes related to epilepsy and three AS3MT-associated genes in VPA-related dADRs. The effects of AS3MT on epilepsy might involve multiple targets including CNNM2, CACNB2, TRIM26, MTHFR, GSTM1, CYP17A1, NT5C2, and YBX3. This study reveals that AS3MT may be a new gene contributing to epileptogenesis. Hence, analysis of AS3MT polymorphisms will help to evaluate susceptibility to pediatric epilepsy and drug safety.
Epilepsy is a common chronic neurological disease among children. It is characterized by unpredictable lasting epileptic seizures and numerous serious neurobiological, cognitive, and psychosocial consequences and a high financial burden, therefore it is a major health concern worldwide (Beghi et al., 2005; Fisher et al., 2014). The overall risk of death in epileptic children is approximately ten times higher compared with the general population (Donner et al., 2017). Approximately 35 in every 100,000 Chinese children suffer epilepsy annually (Hu et al., 2015). Advances in neonatal and critical disease care during the last several decades have reduced premature mortality, thus resulting in increasing cases of pediatric epilepsy. Epilepsy etiology involves genetics, metabolism, infection, immunity, structure and unknown factors (Abramovici and Bagić, 2016; Scheffer et al., 2017). Antiepileptic medications are the main treatment strategies for epilepsy, and act by suppressing the seizures rather than modifying the disease process. Non-pharmacological approaches include curative surgery, palliative surgical procedures, and ketogenic diet. Epilepsy is regarded as a symptom condition with multiple risk factors and high genetic predisposition (Thijs et al., 2019). Since identification of CHRNA4 gene, the first epilepsy gene in 1995 (Steinlein et al., 1995), several studies have focused on developing reliable and potential markers of genetic polymorphism related to epilepsy. Currently, more than 500 epilepsy-associated genes have been identified (Wang et al., 2017). However, genetic basis of epilepsy has not been fully explored, therefore, further studies on epilepsy genomics are needed.
Arsenite methyltransferase (AS3MT) catalyzes transfer of a methyl group from S-adenosyl-L-methionine (SAMe) to trivalent arsenic resulting in biomethylation of arsenic in vivo and in vitro, thus it plays a critical role in arsenic metabolism (Lin et al., 2002; Liu et al., 2020). AS3MT (previously called CYT19) gene variation affects arsenic methylation activity in children and adults (Antonelli et al., 2014; De Loma et al., 2018). AS3MT gene is associated with various diseases including Borst-Jadassohn intraepidermal carcinoma of dermal system, liver injury, schizophrenia, and attention deficit or hyperactivity disorder (Li et al., 2016; Zhao et al., 2020). Previous studies indicated that the AS3MT gene was ubiquitously expressed in the mouse brain (cerebral cortex, hippocampus, and cerebellum) and adult human neurons and astrocytes, and upregulated during human stem cell differentiation toward neuronal fates (Sánchez-Peña et al., 2010; Li et al., 2016). A two-stage GWAS of schizophrenia with large sample sizes in the Han Chinese population showed a significant association for AS3MT 10q24.32 loci (Yu et al., 2017). AS3MT rs7085104 was a schizophrenia-associated risk implicated in alteration of striatal dopamine synthesis capacity (Duarte et al., 2016; D’Ambrosio et al., 2019). Schizophrenia is associated with epilepsy based on neuropathological, clinical, and epidemiological data and at genetic level (Vonberg and Bigdeli, 2016). However, studies have not explored the role of the gene mutation of AS3MT in epilepsy.
The aim of this study was to explore the role of AS3MT rs7085104 genetic polymorphism on the risk of pediatric epilepsy in southern Chinese children. Target pathway/function network of AS3MT in epilepsy was explored through bioinformatic analysis to predict underlying mechanisms. Furthermore, the role of AS3MT rs7085104 variation on adverse drug reactions (ADRs) associated with antiepileptic drugs (AEDs) was evaluated.
Pediatric patients with epilepsy (n = 200) and healthy individuals (n = 244) of Han Chinese descent from Shenzhen Baoan Women’s and Children’s Health Hospital were enrolled into this case-control study as described previously (Fan et al., 2020). All participants or their parents signed an informed consent approved by Baoan Women’s and Children’s Health Hospital ethics committee. Patients aged <16 years and >1 month were diagnosed with definitely characterized epilepsy or epileptic syndrome by two independent experienced clinicians following the latest updated version of International League Against Epilepsy (ILAE) commission’s classification (Scheffer et al., 2017). Detailed clinical data of all patients including basic demographic characteristics, epilepsy classification, age of seizure onset, family history, history of status epilepticus, abnormal delivery or delayed discharge from NICU, and therapeutic drugs were retrieved from the hospital information system (HIS) and through telephone follow-up and were analyzed retrospectively. Patients without a clear diagnosis of epilepsy and those with non-reliable seizure frequency, a history of pseudoseizures, or presence of any diseases that may aggravate epilepsy (n = 39) were excluded from this study. Participants in the control group were individuals with similar age with patients in the case group and were clinically free of epilepsy or had no family history of central nervous system disorders. Children missing the above-mentioned clinical data were excluded from this study.
Patients with pediatric epilepsy who had received one or more AEDs for at least 6 months were used for pharmacogenetic studies to explore the correlation between AS3MT and ADRs. Patients with incomplete medication records were excluded. Patients who developed AEDs-associated ADRs whose causative drugs could not be established were also excluded. Each ADR report was analyzed by two authors using the WHO-UMC causality assessment scale.
Peripheral venous blood samples were collected from 444 children during their regular outpatient visits to Baoan Women’s and Children’s Health Hospital. Genomic DNA was extracted from whole blood following the protocol described previously (Loparev et al., 1991). DNA samples were amplified using polymerase chain reaction (PCR). Genotyping primers of AS3MT rs7085104 used in this study were as follows: forward primer sequence: 5′-ACGTTGGATGTGGTCTCCGTTTTGGTGATG-3′; reverse primer sequence: 5′-ACGTTGGATGCACATTC CGTGAAATCCATC-3′ and extended primer sequence: 5′-TGTGCAGTTCTGTGAACTC-3′. The PCR reaction system comprised 10 ng template DNA and 0.5 μM of primers. PCR reaction was performed under the following conditions: initial denaturation set at 95°C for 2 min followed by 45 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 1 min followed by a final extension of 72°C for 5 min. PCR products were purified by shrimp alkaline phosphatase (SAP) treatment. The cycle sequencing reaction was then performed using Complete iPLEX® Gold Genotyping Reagents (Agena Bioscience™, San Diego, CA, United States). Genotyping of polymorphism was performed with Sequenom MassArray platform (Agena Bioscience, San Diego, CA, United States) and iPLEX Gold Assay. MassArray system comprises MassArray Analyzer mass spectrometer and integrated data analysis software.
Protein function of AS3MT was explored through protein–protein interaction (PPI), which elucidates the role of regulation among proteins (Vella et al., 2017). STRING database v11.01, a public database containing interactions between known and predicted proteins (Szklarczyk et al., 2018), was used to construct PPI network of AS3MT. The minimum required interaction score was set as medium confidence (0.400) and the maximum number of interactors was less than 50 (1st shell not more than 50). The PPI network was further visualized using Cytoscape v3.6.0 to present AS3MT-related genes. Metascape was used for GO analysis and Reactome pathway analysis of potential targets of AS3MT in the present study. P-value < 0.01, minimum count of 3, and enrichment factor >1.5 were used for filtering terms. Membership similarity (Kappa score) > 0.3 was applied as the threshold to group which enriched terms into clusters. A subset of representative terms was then selected from the cluster and used to construct a network. Each term was presented as a circle node, and terms with a similarity score > 0.3 were linked edges. The network was visualized using Cytoscape (v3.6.0) software.
To further explore the identified disease-related genes, GeneCards2 and DisGeNET3 databases were used to retrieve epilepsy-associated genes and digestive diseases-associated genes. Genes obtained were further filtered based on the number of pubmed IDs (PMIDs) > 0 for DisGeNET and the relevance score ≥ 1 for GeneCards. To obtain potential targets of AS3MT in epilepsy or dADRs, genes related to AS3MT identified in PPI analysis were intersected with epilepsy-associated genes and digestive diseases-associated genes from DisGeNET and GeneCards databases.
All statistical analyses were performed using the Statistical Package for Social Sciences (SPSS) software (V25.0). Differences in age, gender, and AEDs therapy distribution between cases and controls were analyzed using Mann--Whitney U test and Chi-square test by preliminary cross-tabulations. SNPStats is a web-based tool designed for analyzing association studies. SNPStats Web Tool4 was used to calculate genotype distribution and minor allelic frequency using Exact test Hardy–Weinberg equilibrium calculations for AS3MT rs7085104 SNP in case and control groups. Data on SNPs’ allele contrast and five genetic inheritance models (codominant, dominant, recessive, overdominance, and additive genetic correlation) between the two groups were also analyzed. Odds ratio (OR) values and relative risk (95% CI) were calculated for multiple testing. Values with p < 0.05 were considered statistically significant. The power of this study for the minimum sample size calculation was computed for verification as described in our previous study (Fan et al., 2020). Power > 0.9 was obtained based on the frequency distribution of three AS3MT rs7085104 genotypes from participants in the control and case groups.
General clinical characteristics of all the participants including 200 cases and 244 controls in this study were analyzed. The case group comprised 85 (42.5%) females and 115 (57.5%) males with an average age of 2.99 ± 3.09 years at the first seizure onset and 3.4 ± 3.1 years at the time diagnosis was confirmed. The control group consisted of 112 (45.9%) females and 132 (54.1%) males with a mean age of 4.6 ± 2.7 years. Analysis showed that there were no significant differences in gender between the two groups. Twenty-three epileptic children at their perinatal stage experienced diseases like hypertensive disorders, preeclampsia and eclampsia, gestational diabetes and threatened abortion. Twenty-three children experienced abnormal delivery or delayed discharge from NICU with invasive mechanical ventilation owing to severe diseases. Forty-four children with epilepsy suffered febrile seizure and twenty patients had a history of status epilepticus. The family members of 31 children suffered from febrile seizures, epilepsy, inherited metabolic diseases, and chromosome abnormality, among whom ten children experienced febrile seizure.
AEDs used in our cases were VPA (n = 96, 52.2%), OXC (n = 49, 26.6%), and other AEDs (n = 39, 21.2%), which was consistent with ILAE guidelines (Glauser et al., 2013; Wilmshurst et al., 2015). Among 174 pediatric patients with epilepsy receiving AEDs, 123 children presented with ADRs including digestive adverse drug reactions (dADRs), skin reactions and neurological adverse reactions. Causality assessment using WHO UMC scale showed 3.7% certain, 85.9% probable, and 10.4% possible association of ADRs with AEDs. The percentage of patients presenting with ADRs was significantly higher compared with those without ADRs (64.55% vs. 35.45% for monotherapy and 81.25% vs. 18.75% for polytherapy). Polytherapy of AEDs resulted in more nADRs and cADRs compared with monotherapy (Supplementary Table S1). This finding implied that polytherapy highly predisposed children with epilepsy to ADRs compared with monotherapy (Table 1). The number of patients who presented with dADRs, nervous ADRs (nADRs), and cutaneous ADRs (cADRs) was 69 (39.7%), 58 (33.3%), and 30 (17.2%), respectively, mainly triggered by VPA and OXC (Figure 1A). Loss of appetite, dyspepsia, abdominal pain, diarrhea, and vomiting were the main manifestations of AEDs related-dADRs (Figure 1B). Number of pediatric patients with and without dADRs was significantly different in different groups based on age of diagnosis (OR = 2.11, 95%CI = 1.12–3.90, p = 0.0169) (Table 1). Age of diagnosis in patients with dADRs was 2.71 ± 2.55 years, which was significantly less than that of patients without dADRs (4.12 ± 3.21 years). The incidence rate of dADRs in patients with childhood epilepsy receiving VPA alone was higher than that of patients treated using VPA in combination with other AEDs.
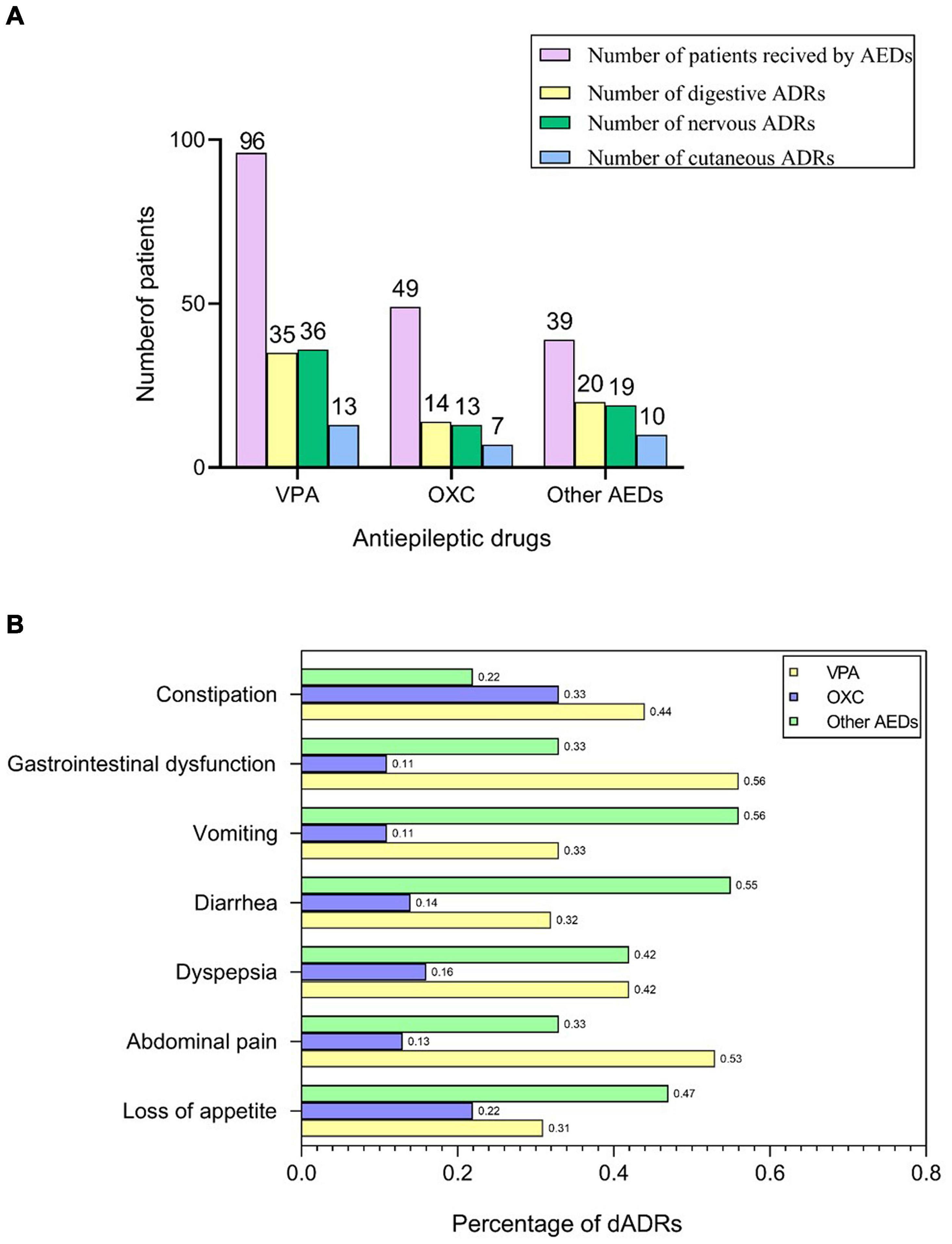
Figure 1. AEDs related-ADRs observed in our study. (A) Number of patients who received AEDs and presented with ADRs. (B) Frequency of dADRs in epileptic children.
Frequency distribution of AS3MT rs7085104 genotype in cases and controls was shown in Table 2. Significant differences of frequency distribution between the two groups were observed. Allele analysis showed that allele A had a higher frequency among epileptic patients compared with that in healthy children (51.8% and 41.2%, respectively). Allele frequency of AS3MT rs7085104 in our study was consistent with that from the HapMap study. Distribution of AA, GA, and GG genotypes in the case group was 28.0%, 47.5%, and 24.5%, respectively, whereas the distribution of these genotypes in the control group was 16.8%, 48.8%, and 34.4% (p = 0.0068). Among three AS3MT rs7085104 genotypes, heterozygous GA genotype exhibited the highest frequency of distribution in both case group and control group. Carriers of two polymorphic AS3MT rs7085104 A alleles (mutant homozygous AA genotype) showed a higher epilepsy risk of childhood epilepsy (OR = 1.93, 95% CI = 1.22–3.04, p = 0.0046). However, mutant homozygous GG genotype exhibited lower susceptibility to childhood epilepsy compared with AA genotype (AA vs. GG: OR = 1.52, 95% CI = 1.16–1.98, p = 0.0019). These results indicated that children carrying AA diplotype of AS3MT rs7085104 had higher risk of epilepsy.
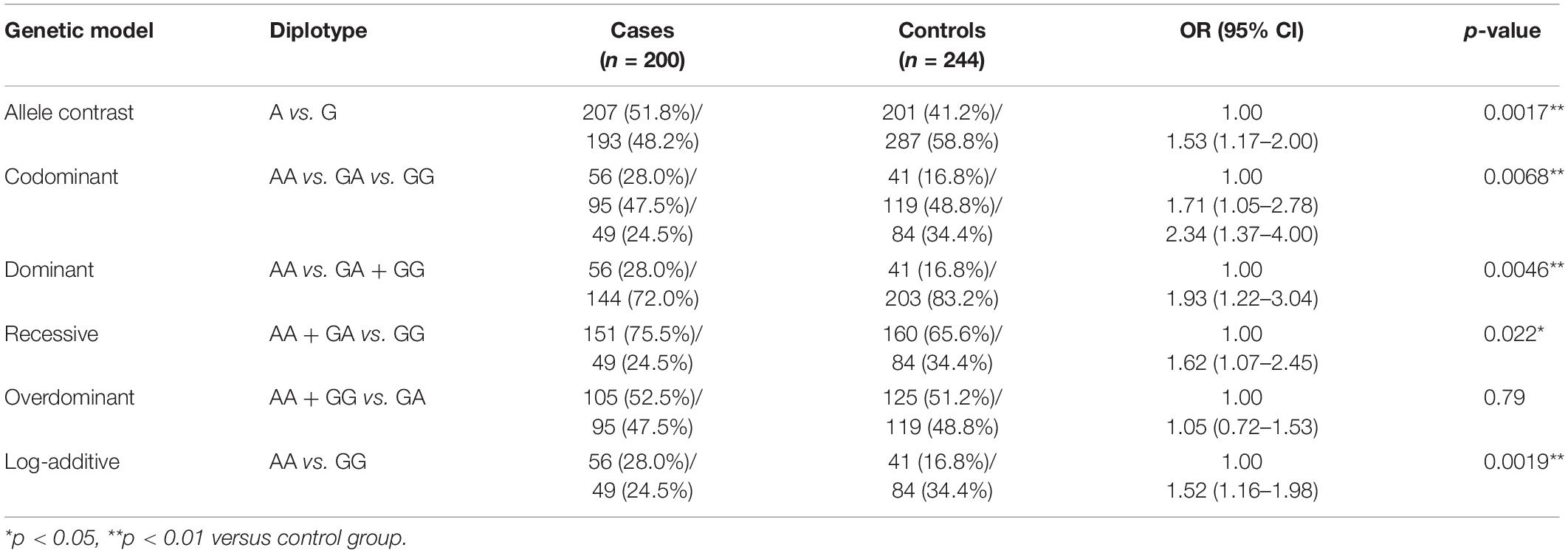
Table 2. Comparison of AS3MT rs7085104 diplotype distribution between epileptic children and healthy children.
Stratification analysis of AS3MT rs7085104 was further conducted using selected variables from these patients. Frequency distribution of AS3MT rs7085104 genotypes showed significant difference between cases and controls in the subgroup of female patients (p = 0.0021, Table 3). Epileptic patients with focal onset or generalized onset showed significant differences in genotype distribution of AS3MT rs7085104 compared with healthy individuals (p = 0.0005 and p = 0.0123, respectively). The proportions of AA genotype in patients with focal onset seizures and generalized onset seizures were significantly higher than those of the control group.
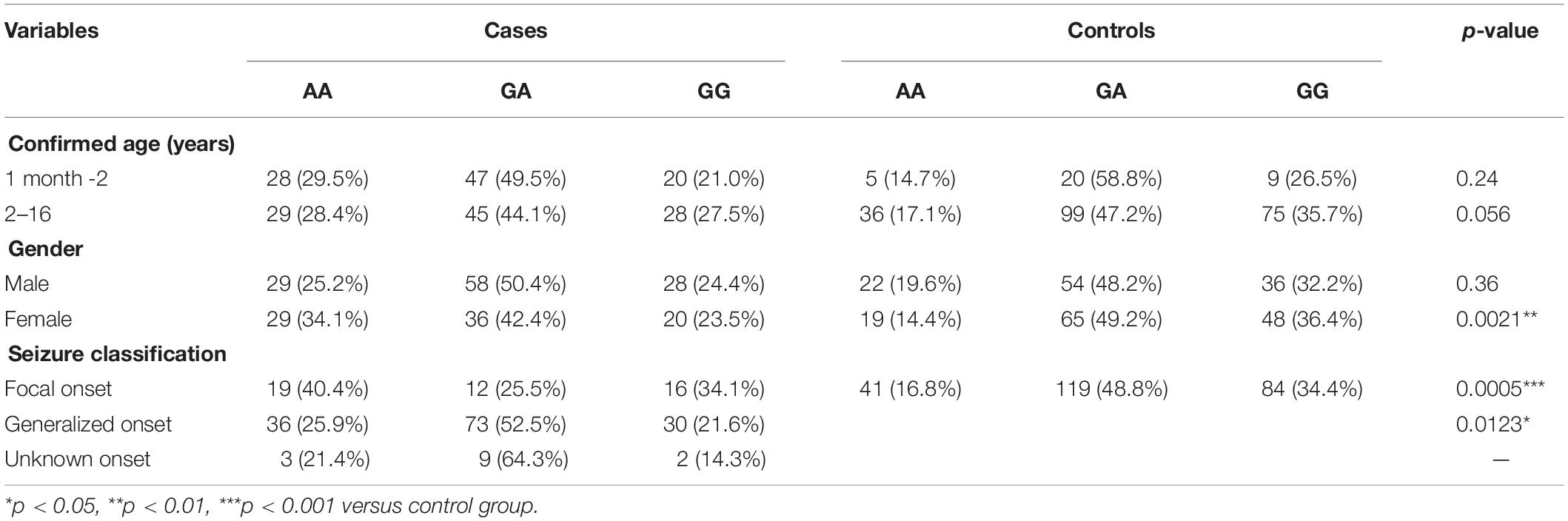
Table 3. Stratification analysis of AS3MT rs7085104 genotypes using selected variables in epileptic children and healthy children.
Frequency distribution of AS3MT rs7085104 genotypes in patients with and without ADRs in this study was further analyzed. Allele analysis showed that allele A was more frequent in epileptic children who suffered dADRs when receiving OXC than allele G (71.4% and 47.1%, respectively), and the frequency in patients without dADRs was on the contrary (Table 4). Carriers of AS3MT rs7085104 AA genotype showed a higher risk of dADRs after treatment with VPA or OXC (OR = 2.77, 95% CI = 1.12–6.86, p = 0.027 for VPA and OR = 4.50, 95% CI = 1.20–16.85, p = 0.023 for OXC) compared to those with GA and or GG genotypes. However, mutant homozygous GG genotype exhibited a lower predisposition to dADRs after administration with VPA or OXC drugs compared with AA genotype. These findings indicate that genetic polymorphism of AS3MT rs7085104 is correlated with the potential risk of dADRs caused by VPA and OXC. Furthermore, association of AS3MT rs7085104 polymorphism with other ADRs in central nervous system, skin and subcutaneous tissues was explored but no significant difference was observed (Supplementary Tables S2, S3).
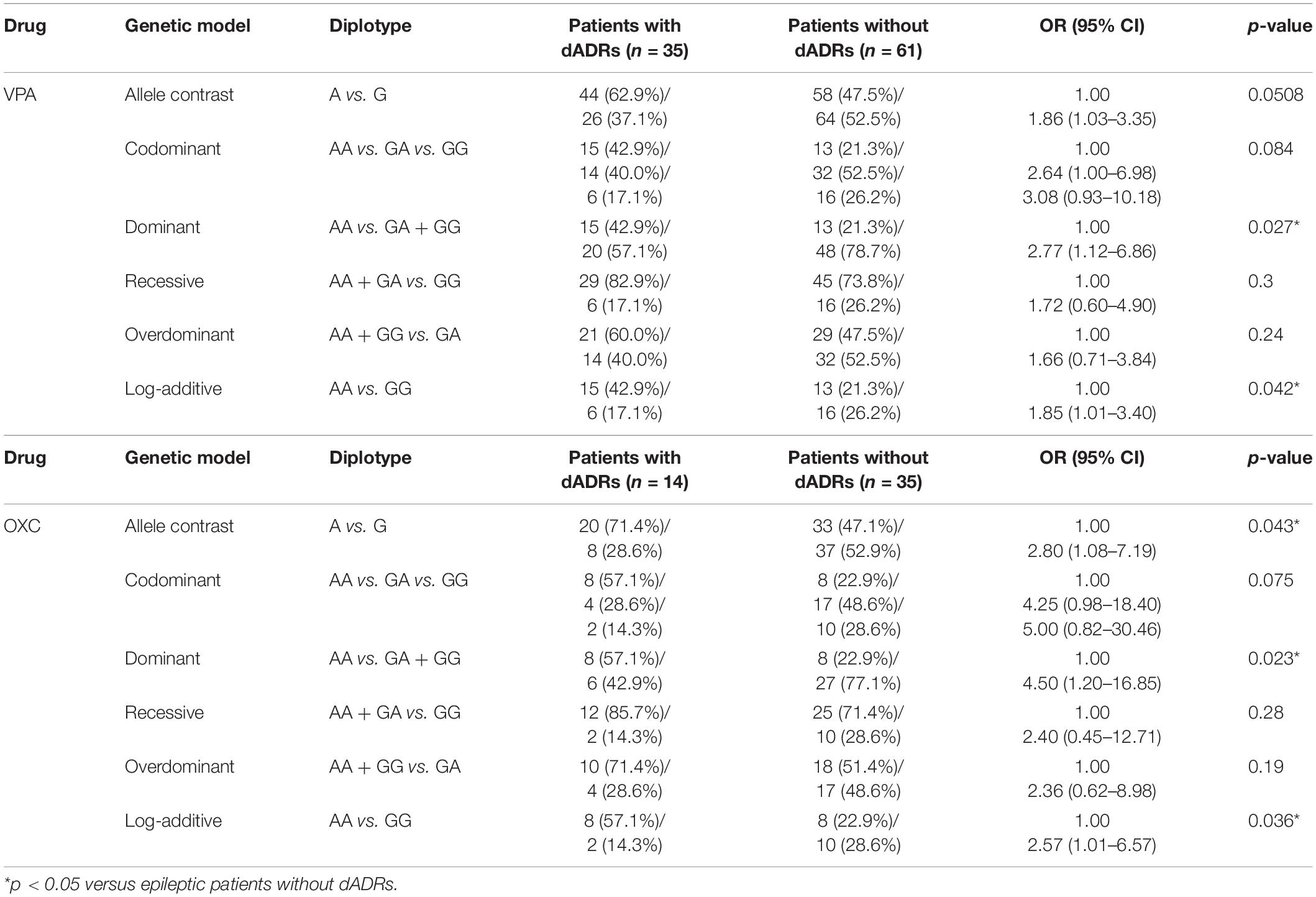
Table 4. Comparison of AS3MT rs7085104 diplotype distribution for epileptic children with or without dADRs after receiving AEDs.
STRING database was used to explore the functional relationships and interactions of AS3MT. Thirty-three genes were found to interact with AS3MT. A PPI network comprising 34 nodes and 86 edges was constructed (Figure 2A). These genes were implicated in a total of 154 pathways and functions and were mainly involved in “sulfur compound biosynthetic process,” “homocysteine metabolic process,” “cardiac muscle contraction,” “methylation,” and “cellular modified amino acid metabolic process”, which were the top five clusters based on p-values (Figures 2B–D). Furthermore, 1176 genes from DisGeNET and 5064 genes from GeneCards database were shown to be associated with epilepsy. After removing the duplicates from two databases, a total of 5166 epilepsy-associated genes were obtained. These genes were intersected with 33 genes related to AS3MT from PPI analysis. Eight common genes including MTHFR, GSTM1, CYP17A1, NT5C2, YBX3, CNNM2, CACNB2, and TRIM26 were identified as potential targets of AS3MT in epilepsy. CNNM2, NT5C2, and TRIM26 were the top three key genes related to AS3MT according to high-ranking interaction score. The PPI network between eight targets and AS3MT is shown in Figure 2E. In addition, 10282 digestive diseases-associated genes from GeneCards and DisGeNET, 1065 VPA-related genes, and 116 OXC-related genes from DrugBank and SwissTargetPrediction were obtained. Thirty-three interacting genes of AS3MT were further intersected with digestive diseases-associated genes and VPA or OXC-related genes. Three common genes including CYP17A1, GSTM1, and MTHFR were identified for AS3MT in VPA-related dADRs (Figure 2F), however, no common gene was identified in OXC-related dADRs. These results indicate that AS3MT plays a role in epilepsy and AEDs-related ADRs likely through multiple targets.
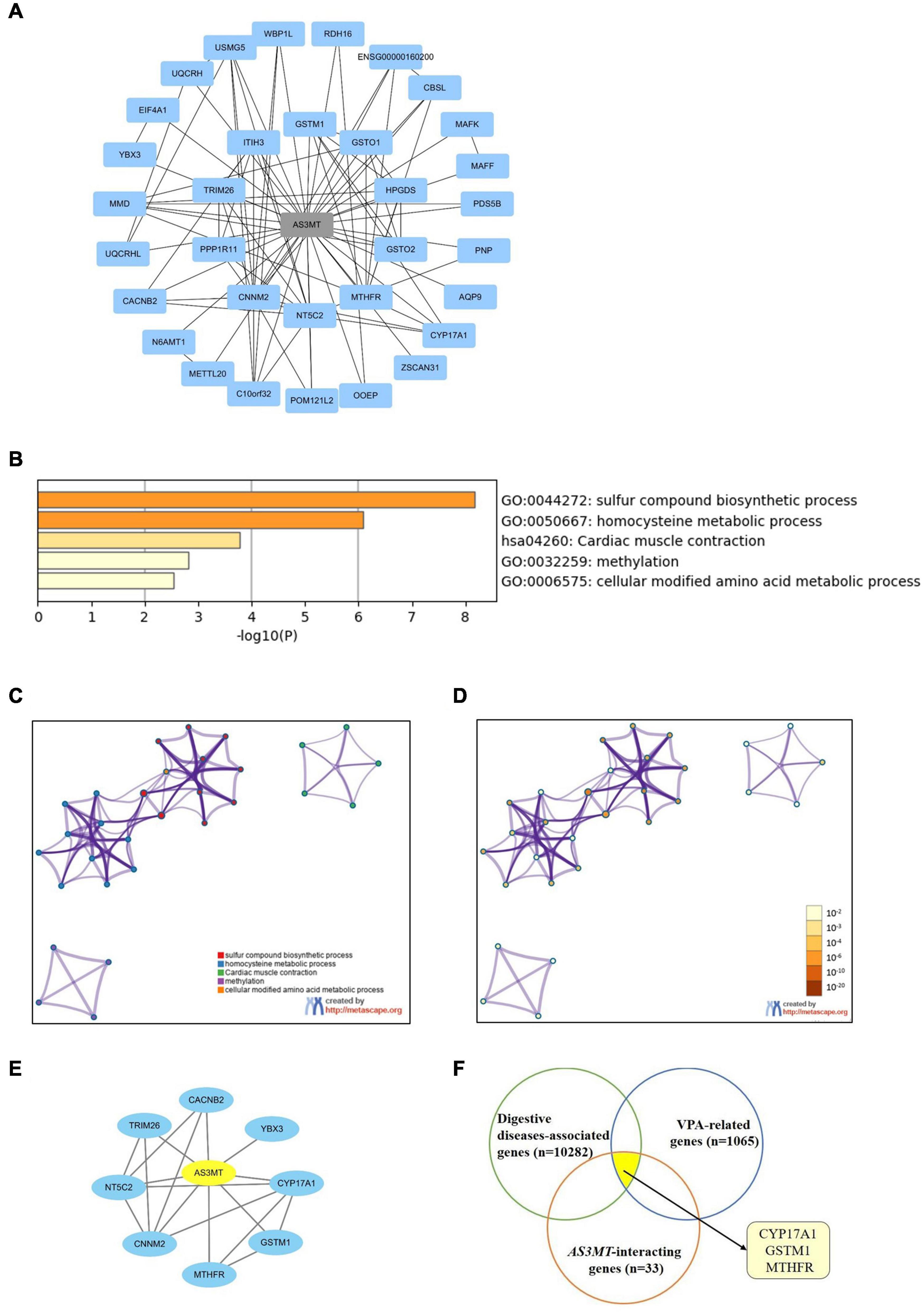
Figure 2. Network of AS3MT related genes implicated in epilepsy and VPA-related dADRs using bioinformatic analysis. (A) PPI network of AS3MT comprising 34 nodes and 86 edges. Each target protein was represented by a node. (B) Functional enrichment analysis of 33 target genes related with AS3MT. The top non-redundant enrichment clusters identified using Metascape and statistical significance of one per cluster represented by a discrete color scale. (C,D) Metascape visualization of the interactome network. Nodes are colored based on their identities (C) and p-values (D). Each circle node represents a term, where its size is proportional to the number of input genes clustering into that term. The color represents cluster identity of the node. A darker color represents high statistical significance of the node. (E) PPI network between AS3MT and 8 common genes associated with epilepsy comprising 9 nodes and 18 edges. (F) Three common genes identified for AS3MT in VPA-related dADRs.
Epilepsy, a common neurologic disease, has a genetic component contributed by multiple genes (Wang et al., 2017; Thijs et al., 2019). More than half of all epilepsy cases have a genetic basis. A major challenge on elimination of epilepsy is the inherent complexity and heterogeneity of known epileptic syndromes and differential genetic susceptibilities exhibited by patients at risk. Genetic testing as a routine diagnostic strategy of epilepsy can be effective for diagnosis and for guiding treatment (Pal et al., 2010). Mutations of several genes have been reported to exert a significant effect on susceptibility to idiopathic epilepsies and associated comorbidities (Canavati et al., 2019; Fan et al., 2020; Song et al., 2020). The largest genome-wide association study (GWAS) in epilepsy published by ILAE Consortium revealed sixteen genome-wide significant loci with eleven novel loci (Abou-Khalil et al., 2018; Perucca et al., 2020).
AS3MT (previously called CYT19) gene located in 10q24.32, encodes a 43 kDa cytosolic protein which is a cysteine rich enzyme that transfers a methyl group from S-adenosyl-L-methionine to trivalent arsenical (Lin et al., 2002; Liu et al., 2020). AS3MT plays an important role in catalysis of biomethylation of arsenic in vivo and in vitro. Previous studies reported that arsenic exposure had a toxic effect on the nervous system, causing nervous system dysfunction and psychosis such as polyneuropathy, EEG abnormalities and, in extreme cases, hallucinations, disorientation and agitation (Rodríguez et al., 2003). Genetic factors such as gene polymorphisms related to arsenic metabolism are used to determine arsenic concentrations in the human body (Chernoff et al., 2020). Arsenic methylation efficiency in children and adults is affected by AS3MT variation (Antonelli et al., 2014; De Loma et al., 2018). AS3MT is mainly expressed in human adrenal glands, liver, heart, kidney, and brain (Su et al., 2002). Moreover, AS3MT expression is highly expressed in adult human neurons and astrocytes during human stem cell differentiation toward neuronal fates and in brains of patients with schizophrenia compared with controls (Li et al., 2016). AS3MT gene was associated with schizophrenia and attention deficit or hyperactivity disorder (Li et al., 2016; Zhao et al., 2020). Notably, AS3MT rs7085104 as a schizophrenia-associated risk SNP altered striatal dopamine synthesis capacity (Duarte et al., 2016; D’Ambrosio et al., 2019; Cai et al., 2021). Dopamine pathway was regarded as a new regulating mechanism for epilepsy. There was a positive genetic correlation between schizophrenia and epilepsy (Vonberg and Bigdeli, 2016). Whether the SNP of AS3MT rs7085104 played a role in pediatric epilepsy remained unknown and thus we aimed to explore it. The findings of our study showed that frequency distribution of AS3MT rs7085104 genotypes was associated with pediatric epilepsy in a Han Chinese population from Southern China. Allele A showed higher frequency in children with epilepsy compared with healthy controls. Carries of AS3MT rs7085104 AA genotype showed a higher epilepsy risk, whereas children with GG genotype were less predisposed to epilepsy. The human-specific variable number of tandem repeat (VNTR) and AS3MT rs7085104 were significantly associated with AS3MTd2d3 mRNA expression in brains of Han Chinese donors with the A allele at rs7085104 predicting higher mRNA levels of this isoform (Cai et al., 2021). Additionally, another SNP rs10883795 was found to be associated with AS3MT expression levels and the mRNA levels of nearby genes, including CNNM2 and NT5C2 (Yu et al., 2017).
Use of antiepileptic drugs is the main treatment strategy for pediatric epilepsy. VPA, OXC, levetiracetam (LEV), and lamotrigine (LTG) are the main AEDs used for treatment of epilepsy in children. Children are generally at higher risk of toxic effects compared with adults. In our study, high incidences of ADRs associated with AEDs including dADRs, cADRs, and nADRs were observed. Notably, VPA was associated with most ADRs observed in this study, which was consistent with previous reports (Buchanan, 1992; Kaushik et al., 2019). Polytherapy significantly increased risk of ADRs in children. AEDs therapy in pediatric patients requires attention on safety and tolerability from clinicians. Knowledge of different mechanisms of AEDs-related adverse effects is essential to ensure safe treatment of pediatric epilepsy. Pharmacogenomics is a valuable tool for exploring association of inter-individual genetic variations with treatment response in children. AS3MT gene polymorphism was implicated in pediatric epilepsy, therefore, the role of gene mutation in responsiveness and safety of AEDs was further explored in this study. AS3MT rs7085104 polymorphism was correlated with potential risk of AEDs-induced dADRs. In addition, AA genotype predisposed pediatric children to dADRs after treatment with VPA or OXC.
Potential mechanisms of AS3MT in epilepsy were further explored through bioinformatics analysis. Eight common genes between AS3MT and epilepsy were identified. Out of the eight genes, CNNM2, NT5C2, and TRIM26 were the top three key genes associated with AS3MT. NT5C2 and CNNM2 are involved in etiology and pathogenesis of schizophrenia and have been confirmed as schizophrenia susceptibility genes (Duarte et al., 2016; Guan et al., 2016). NT5C2 encodes a phosphatase involved in cellular purine metabolism, which is associated with disorders characterized by psychiatric and psychomotor disturbances (Duarte et al., 2019). NT5C2 has a significantly high affinity for adenosine monophosphate and is involved in the extensive transcriptional programming which regulates cell maintenance, proliferation, migration, and differentiation during neurodevelopment (Itoh, 2013). Cyclin M2 (CNNM2) encodes a member of the protein family containing a cyclin box motif. It plays a role in brain development, neurological functioning and Mg2+ homeostasis. Mutations in CNNM2 gene are implicated in mental retardation and seizures in patients with hypomagnesemia (Arjona et al., 2014; Accogli et al., 2019). The tripartite motif-containing 26 (TRIM26), localized in cytoplasmic bodies, comprises three zinc-binding domains, including RING, B-box type 1 and B-box type 2 domains, and a coiled-coil region, which have DNA-binding activity. TRIM26-associated diseases include neural tube defects and its related pathways are implicated in interferon gamma signaling and innate immune system. TRIM26 is associated with pathogenesis of schizophrenia (de Jong et al., 2012). Other common genes including MTHFR, GSTM1, CYP17A1, and CACNB2 are implicated in neurological disorders such as epilepsy (Scher et al., 2011; Glister et al., 2012; Chbili et al., 2014; Kim and State, 2014). Furthermore, due to lack of direct evidence for AS3MT on dADRs, bioinformatics analysis was performed to explore the relationship between AS3MT and dADRs induced by VPA or OXC drugs. CYP17A1, GSTM1, and MTHFR were identified as the common genes of AS3MT in VPA-related dADRs whereas no common genes were identified in OXC-related dADRs. It was found that the CYP17A1 rs743572 was in strong linkage disequilibrium (LD) with the AS3MT SNPs and had similar effects on the metabolite profiles (Schläwicke Engström et al., 2009; Gomez-Rubio et al., 2010). Previous studies reported that GSTM1 and MTHFR were associated with adverse reactions of VPA (Roy et al., 2008; Fricke-Galindo et al., 2018). Therefore, we deduce that AS3MT may exert its effect on epilepsy and VPA-induced dADRs through these genes.
Our study population included epileptic children with different types of and etiologies. Due to the relatively small sample size, the association between AS3MT and the specific epilepsy subtypes was not explored and no significant difference was observed for ADRs from other systems in our study. In further study, we will increase the sample size and further explore the role of AS3MT in different epilepsy subtypes and other AEDs related ADRs. Our study only investigated single SNP, which underestimate moderate and large effect sizes for epilepsy with numerous risk SNPs due to non-collapsibility of the odds ratio. The polygenic model of epilepsy and the LD between AS3MT with other nearby genes will be further investigated in genomic association studies. Furthermore, findings from bioinformatics analysis lack experimental validation. Therefore, in vitro and in vivo studies should be performed to elucidate the underlying mechanisms of AS3MT in epilepsy and dADRs. In addition, gene polymorphism may possibly alter gene transcription, therefore, AS3MT mRNA and protein expression in different genotypes of AS3MT rs7085104 should be determined to provide insights into the molecular mechanisms of AS3MT in pediatric epilepsy and drug safety.
Epilepsy, one of the most common serious brain conditions, is a disease with complex symptoms, multiple risk factors, and a strong genetic component. Our study revealed that genetic polymorphism of AS3MT rs7085104 was associated with the susceptibility to pediatric epilepsy and risk of AEDs-related dADRs. Mutant homozygous GG genotype exhibited a lower susceptibility to childhood epilepsy than AA genotype. Carriers of AS3MT rs7085104 AA genotype tended to have a higher risk of dADRs for children after treatment with VPA or OXC. Bioinformatics analysis revealed that the effects of AS3MT on epilepsy likely involved multiple targets including MTHFR, GSTM1, CYP17A1, NT5C2, YBX3, CNNM2, CACNB2, and TRIM26. AS3MT may be a novel potential gene implicated in pathogenesis of epilepsies. Therefore, exploring AS3MT gene polymorphism will provide information on understanding susceptibility to pediatric epilepsy and drug safety.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by Baoan Women’s and Children’s Health Hospital Ethics Committee. Written informed consent was obtained from the individual(s), and minor(s)’ legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
XF and HX: conceptualization. YC, JL, and HG: data curation. YC, JL, and WL: formal analysis. XF: funding acquisition. XF and XL: methodology. HX: supervision. YC, JL, and QC: project administration. XF and YC: writing—original draft. XF and HX: writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (Grant 81603185), the Natural Science Foundation of Guangdong Province (Grant 2020A1515011452), and the basic medical and health research project of Baoan district (2019JD356).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2021.705297/full#supplementary-material
ADRs, adverse drug reactions; AEDs, antiepileptic drugs; AS3MT, arsenite methyltransferase; cADRs, cutaneous adverse drug reactions; CNNM2, cyclin and CBS domain divalent metal cation transport mediator 2; dADRs, digestive adverse drug reactions; LD, linkage disequilibrium; nADRs, nervous adverse drug reactions; OXC, oxcarbazepine; PCR, polymerase chain reaction; PPI, protein-protein interaction; SAMe, S-adenosyl -L-methionine; SNP, single nucleotide polymorphism; TRIM26, tripartite motif containing 26; VPA, valproic acid.
Abou-Khalil, B., Auce, P., Avbersek, A., Bahlo, M., Balding, D. J., Bast, T., et al. (2018). Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat. Commun. 9:5269. doi: 10.1038/s41467-018-07524-z
Abramovici, S., and Bagić, A. (2016). “Chapter 10 - Epidemiology of epilepsy,” in Handbook of Clinical Neurology, eds M. J. Aminoff, F. Boller, and D. F. Swaab (Amsterdam: Elsevier), 159–171.
Accogli, A., Scala, M., Calcagno, A., Napoli, F., Di Iorgi, N., Arrigo, S., et al. (2019). CNNM2 homozygous mutations cause severe refractory hypomagnesemia, epileptic encephalopathy and brain malformations. Eur. J. Med. Genet. 62, 198–203. doi: 10.1016/j.ejmg.2018.07.014
Antonelli, R., Shao, K., Thomas, D. J., Sams, R. II, and Cowden, J. (2014). AS3MT, GSTO, and PNP polymorphisms: impact on arsenic methylation and implications for disease susceptibility. Environ. Res. 132, 156–167. doi: 10.1016/j.envres.2014.03.012
Arjona, F. J., de Baaij, J. H., Schlingmann, K. P., Lameris, A. L., van Wijk, E., Flik, G., et al. (2014). CNNM2 mutations cause impaired brain development and seizures in patients with hypomagnesemia. PLoS Genet. 10:e1004267. doi: 10.1371/journal.pgen.1004267
Beghi, E., Frigeni, B., Beghi, M., De Compadri, P., and Garattini, L. (2005). A review of the costs of managing childhood epilepsy. Pharmacoeconomics 23, 27–45. doi: 10.2165/00019053-200523010-00003
Buchanan, N. (1992). The occurrence, management and outcome of antiepileptic drug side effects in 767 patients. Seizure 1, 89–98.
Cai, X., Yang, Z. H., Li, H. J., Xiao, X., Li, M., and Chang, H. (2021). A Human-Specific Schizophrenia Risk Tandem Repeat Affects Alternative Splicing of a Human-Unique Isoform AS3MTd2d3 and Mushroom Dendritic Spine Density. Schizophr. Bull. 47, 219–227. doi: 10.1093/schbul/sbaa098
Canavati, C., Klein, K. M., Afawi, Z., Pendziwiat, M., Abu Rayyan, A., Kamal, L., et al. (2019). Inclusion of hemimegalencephaly into the phenotypic spectrum of NPRL3 pathogenic variants in familial focal epilepsy with variable foci. Epilepsia 60, e67–e73. doi: 10.1111/epi.15665
Chbili, C., B’Chir, F., Ben Fredj, M., Saguem, B. N., Ben Amor, S., Ben Ammou, S., et al. (2014). Effects of glutathione S-transferase M1 and T1 deletions on epilepsy risk among a Tunisian population. Epilepsy Res. 108, 1168–1173. doi: 10.1016/j.eplepsyres.2014.05.009
Chernoff, M., Tong, L., Demanelis, K., Vander Griend, D., Ahsan, H., and Pierce, B. L. (2020). Genetic Determinants of Reduced Arsenic Metabolism Efficiency in the 10q24.32 Region Are Associated With Reduced AS3MT Expression in Multiple Human Tissue Types. Toxicol. Sci. 176, 382–395. doi: 10.1093/toxsci/kfaa075
D’Ambrosio, E., Dahoun, T., Pardiñas, A. F., Veronese, M., Bloomfield, M. A. P., Jauhar, S., et al. (2019). The effect of a genetic variant at the schizophrenia associated AS3MT/BORCS7 locus on striatal dopamine function: a PET imaging study. Psychiatry Res. Neuroimaging 291, 34–41. doi: 10.1016/j.pscychresns.2019.07.005
de Jong, S., van Eijk, K. R., Zeegers, D. W., Strengman, E., Janson, E., Veldink, J. H., et al. (2012). Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur. J. Hum. Genet. 20, 1004–1008. doi: 10.1038/ejhg.2012.38
De Loma, J., Skröder, H., Raqib, R., Vahter, M., and Broberg, K. (2018). Arsenite methyltransferase (AS3MT) polymorphisms and arsenic methylation in children in rural Bangladesh. Toxicol. Appl. Pharmacol. 357, 80–87. doi: 10.1016/j.taap.2018.08.020
Donner, E. J., Camfield, P., Brooks, L., Buchhalter, J., Camfield, C., Loddenkemper, T., et al. (2017). Understanding Death in Children With Epilepsy. Pediatr. Neurol. 70, 7–15. doi: 10.1016/j.pediatrneurol.2017.01.011
Duarte, R. R. R., Bachtel, N. D., Côtel, M.-C., Lee, S. H., Selvackadunco, S., Watson, I. A., et al. (2019). The Psychiatric Risk Gene NT5C2 Regulates Adenosine Monophosphate-Activated Protein Kinase Signaling and Protein Translation in Human Neural Progenitor Cells. Biol. Psychiatry 86, 120–130. doi: 10.1016/j.biopsych.2019.03.977
Duarte, R. R. R., Troakes, C., Nolan, M., Srivastava, D. P., Murray, R. M., and Bray, N. J. (2016). Genome-wide significant schizophrenia risk variation on chromosome 10q24 is associated with altered cis-regulation of BORCS7, AS3MT, and NT5C2 in the human brain. Am. J. Med. Genet. B Neuropsychiatr. Genet. 171, 806–814. doi: 10.1002/ajmg.b.32445
Fan, X., Chen, Y., Li, W., Xia, H., Liu, B., Guo, H., et al. (2020). Genetic Polymorphism of ADORA2A Is Associated With the Risk of Epilepsy and Predisposition to Neurologic Comorbidity in Chinese Southern Children. Front. Neurosci. 14:590605. doi: 10.3389/fnins.2020.590605
Fisher, R. S., Acevedo, C., Arzimanoglou, A., Bogacz, A., Cross, J. H., Elger, C. E., et al. (2014). ILAE official report: a practical clinical definition of epilepsy. Epilepsia 55, 475–482. doi: 10.1111/epi.12550
Fricke-Galindo, I., Jung-Cook, H., LLerena, A., and López-López, M. (2018). Pharmacogenetics of adverse reactions to antiepileptic drugs. Neurologia 33, 165–176. doi: 10.1016/j.nrl.2015.03.005
Glauser, T., Ben-Menachem, E., Bourgeois, B., Cnaan, A., Guerreiro, C., Kälviäinen, R., et al. (2013). Updated ILAE evidence review of antiepileptic drug efficacy and effectiveness as initial monotherapy for epileptic seizures and syndromes. Epilepsia 54, 551–563. doi: 10.1111/epi.12074
Glister, C., Satchell, L., Michael, A. E., Bicknell, A. B., and Knight, P. G. (2012). The anti-epileptic drug valproic acid (VPA) inhibits steroidogenesis in bovine theca and granulosa cells in vitro. PLoS One 7:e49553. doi: 10.1371/journal.pone.0049553
Gomez-Rubio, P., Meza-Montenegro, M. M., Cantu-Soto, E., and Klimecki, W. T. (2010). Genetic association between intronic variants in AS3MT and arsenic methylation efficiency is focused on a large linkage disequilibrium cluster in chromosome 10. J. Appl. Toxicol. 30, 260–270. doi: 10.1002/jat.1492
Guan, F., Zhang, T., Li, L., Fu, D., Lin, H., Chen, G., et al. (2016). Two-stage replication of previous genome-wide association studies of AS3MT-CNNM2-NT5C2 gene cluster region in a large schizophrenia case-control sample from Han Chinese population. Schizophr. Res. 176, 125–130. doi: 10.1016/j.schres.2016.07.004
Hu, Y. M., Jiang, Z. F., Sheng, K. L., and Shen, Y. (2015). Zhufutang Practical Pediatrics. Beijing: people’s Medical publishing house.
Itoh, R. (2013). Enzymatic properties and physiological roles of cytosolic 5’-nucleotidase II. Curr. Med. Chem. 20, 4260–4284. doi: 10.2174/0929867311320340006
Kaushik, S., Chopra, D., Sharma, S., and Aneja, S. (2019). Adverse Drug Reactions of Anti-Epileptic Drugs in Children with Epilepsy: a Cross-Sectional Study. Curr. Drug Saf. 14, 217–224. doi: 10.2174/1574886314666190311112710
Kim, Y. S., and State, M. W. (2014). Recent challenges to the psychiatric diagnostic nosology: a focus on the genetics and genomics of neurodevelopmental disorders. Int. J. Epidemiol. 43, 465–475. doi: 10.1093/ije/dyu037
Li, M., Jaffe, A. E., Straub, R. E., Tao, R., Shin, J. H., Wang, Y., et al. (2016). A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat. Med. 22, 649–656. doi: 10.1038/nm.4096
Lin, S., Shi, Q., Nix, F. B., Styblo, M., Beck, M. A., Herbin-Davis, K. M., et al. (2002). A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 277, 10795–10803. doi: 10.1074/jbc.M110246200
Liu, W. S., Wang, X. Y., Lu, J., Zhang, Y. M., Ye, X. M., Li, J. M., et al. (2020). Polymorphisms in arsenic (+3 oxidation state) methyltransferase (AS3MT) predict the occurrence of hyperleukocytosis and arsenic metabolism in APL patients treated with As(2)O(3). Arch. Toxicol. 94, 1203–1213. doi: 10.1007/s00204-020-02686-6
Loparev, V. N., Cartas, M. A., Monken, C. E., Velpandi, A., and Srinivasan, A. (1991). An efficient and simple method of DNA extraction from whole blood and cell lines to identify infectious agents. J. Virol. Methods 34, 105–112. doi: 10.1016/0166-0934(91)90126-k
Pal, D. K., Pong, A. W., and Chung, W. K. (2010). Genetic evaluation and counseling for epilepsy. Nat. Rev. Neurol. 6, 445–453. doi: 10.1038/nrneurol.2010.92
Perucca, P., Bahlo, M., and Berkovic, S. F. (2020). The Genetics of Epilepsy. Annu. Rev. Genomics Hum. Genet. 21, 205–230. doi: 10.1146/annurev-genom-120219-074937
Rodríguez, V. M., Jiménez-Capdeville, M. E., and Giordano, M. (2003). The effects of arsenic exposure on the nervous system. Toxicol. Lett. 145, 1–18. doi: 10.1016/s0378-4274(03)00262-5
Roy, M., Leclerc, D., Wu, Q., Gupta, S., Kruger, W. D., and Rozen, R. (2008). Valproic acid increases expression of methylenetetrahydrofolate reductase (MTHFR) and induces lower teratogenicity in MTHFR deficiency. J. Cell Biochem. 105, 467–476. doi: 10.1002/jcb.21847
Sánchez-Peña, L. C., Petrosyan, P., Morales, M., González, N. B., Gutiérrez-Ospina, G., Del Razo, L. M., et al. (2010). Arsenic species, AS3MT amount, and AS3MT gene expression in different brain regions of mouse exposed to arsenite. Environ. Res. 110, 428–434. doi: 10.1016/j.envres.2010.01.007
Scheffer, I. E., Berkovic, S., Capovilla, G., Connolly, M. B., French, J., Guilhoto, L., et al. (2017). ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia 58, 512–521. doi: 10.1111/epi.13709
Scher, A. I., Wu, H., Tsao, J. W., Blom, H. J., Feit, P., Nevin, R. L., et al. (2011). MTHFR C677T genotype as a risk factor for epilepsy including post-traumatic epilepsy in a representative military cohort. J. Neurotrauma 28, 1739–1745. doi: 10.1089/neu.2011.1982
Schläwicke Engström, K., Nermell, B., Concha, G., Strömberg, U., Vahter, M., and Broberg, K. (2009). Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutat. Res. 667, 4–14. doi: 10.1016/j.mrfmmm.2008.07.003
Song, L. L., Wang, Y. M., Wu, X. X., Zhu, L. X., Pan, F., and Chen, Y. M. (2020). Correlation between AT1R gene polymorphism and epilepsy secondary to cerebral infarction. Eur. Rev. Med. Pharmacol. Sci. 24, 6873–6880. doi: 10.26355/eurrev_202006_21677
Steinlein, O. K., Mulley, J. C., Propping, P., Wallace, R. H., Phillips, H. A., Sutherland, G. R., et al. (1995). A missense mutation in the neuronal nicotinic acetylcholine receptor alpha 4 subunit is associated with autosomal dominant nocturnal frontal lobe epilepsy. Nat. Genet. 11, 201–203. doi: 10.1038/ng1095-201
Su, A. I., Cooke, M. P., Ching, K. A., Hakak, Y., Walker, J. R., Wiltshire, T., et al. (2002). Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. U. S. A. 99, 4465–4470. doi: 10.1073/pnas.012025199
Szklarczyk, D., Gable, A. L., Lyon, D., Junge, A., Wyder, S., Huerta-Cepas, J., et al. (2018). STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. doi: 10.1093/nar/gky1131
Thijs, R. D., Surges, R., O’Brien, T. J., and Sander, J. W. (2019). Epilepsy in adults. Lancet 393, 689–701. doi: 10.1016/s0140-6736(18)32596-0
Vella, D., Zoppis, I., Mauri, G., Mauri, P., and Di Silvestre, D. (2017). From protein-protein interactions to protein co-expression networks: a new perspective to evaluate large-scale proteomic data. EURASIP J. Bioinform. Syst. Biol. 2017:6. doi: 10.1186/s13637-017-0059-z
Vonberg, F. W., and Bigdeli, T. B. (2016). Genetic Correlation Between Schizophrenia and Epilepsy. JAMA Neurol. 73, 125–126.
Wang, J., Lin, Z. J., Liu, L., Xu, H. Q., Shi, Y. W., Yi, Y. H., et al. (2017). Epilepsy-associated genes. Seizure 44, 11–20. doi: 10.1016/j.seizure.2016.11.030
Wilmshurst, J. M., Gaillard, W. D., Vinayan, K. P., Tsuchida, T. N., Plouin, P., Van Bogaert, P., et al. (2015). Summary of recommendations for the management of infantile seizures: task Force Report for the ILAE Commission of Pediatrics. Epilepsia 56, 1185–1197. doi: 10.1111/epi.13057
Yu, H., Yan, H., Li, J., Li, Z., Zhang, X., Ma, Y., et al. (2017). Common variants on 2p16.1, 6p22.1 and 10q24.32 are associated with schizophrenia in Han Chinese population. Mol. Psychiatry 22, 954–960. doi: 10.1038/mp.2016.212
Keywords: pediatric epilepsy, AS3MT, gene polymorphism, susceptibility, adverse drug reaction, valproic acid, oxcarbazepine
Citation: Fan X, Chen Y, Lu J, Li W, Li X, Guo H, Chen Q, Yang Y and Xia H (2021) AS3MT Polymorphism: A Risk Factor for Epilepsy Susceptibility and Adverse Drug Reactions to Valproic Acid and Oxcarbazepine Treatment in Children From South China. Front. Neurosci. 15:705297. doi: 10.3389/fnins.2021.705297
Received: 05 May 2021; Accepted: 14 October 2021;
Published: 26 November 2021.
Edited by:
Hakon Grydeland, University of Oslo, NorwayReviewed by:
Yanan Chen, Nanjing Medical University, ChinaCopyright © 2021 Fan, Chen, Lu, Li, Li, Guo, Chen, Yang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hanbing Xia, eGhiMjU4OEAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.