
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 23 June 2021
Sec. Neurogenomics
Volume 15 - 2021 | https://doi.org/10.3389/fnins.2021.669224
This article is part of the Research Topic Epigenetic Regulation in Neurological Disorders and Diseases View all 9 articles
Spinal cord injury (SCI) is a devastating neurological disorder that affects thousands of individuals each year. Previously, our study in non-human primates with SCI demonstrated that methylprednisolone (MP) resulted in the dysfunction of neural stem cells (NSCs), which may help to explain the controversial roles of MP in SCI. However, the detailed mechanism is still unclear. In this manuscript, we investigated the LncRNA and mRNA expression profiles of NSCs treated with MP. A total of 63 differentially expressed LncRNAs and 174 differentially expressed mRNAs were identified. Gene ontology (GO) analysis showed that differentially expressed mRNAs were highly associated with terms related to regulation of external stimulation, secretion, and migration. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis results indicated that the PI3K–Akt signaling pathway contributed to the functions of MP treated NSCs. Besides, 3899 co-expression pairs were constructed among the differentially expressed LncRNA and mRNA, among which five predicted target mRNAs with the differentially expressed LncRNAs were identified. These results provide greater insight into the precise mechanisms of MP mediating NSC dysfunction in SCI.
Spinal cord injury (SCI) is a devastating neurological disorder that affects thousands of individuals each year. As about 150–180 hundred new cases increasing every year, the amount of the SCI patient population was estimated to be over 3 million cases around the world (David et al., 2019). However, not only the clinical treatment but also the functional rehabilitation is still the highly problematic issue in clinic, which greatly affects the physiological, psychological, and social activity of the SCI patients (Silva et al., 2014).
Methylprednisolone (MP), a kind of glucocorticoid, is used to be one of the most important medicine in SCI treatment at acute phase. Although several randomized controlled trial studies demonstrated the effectiveness of MP in SCI treatment (Bracken et al., 1984, 1990, 1997), more and more controversies arose recently because more complications but less effectiveness of MP were observed in patients with SCI (Evaniew et al., 2016). Previously, our study in non-human primates with SCI demonstrated that MP resulted in the dysfunction of neural stem cells (NSCs), which may help to explain the controversy of MP in SCI (Ye et al., 2018). NSCs are a kind of adult pluripotent stem cells located in nervous tissue in vivo (Ortiz-Alvarez et al., 2019). NSCs can differentiate into multiple cell lineages including neuron, astrocyte, and oligodendrocyte, which greatly contribute to nervous tissue repairment and regeneration (Tang et al., 2017). However, the molecular mechanism that MP affects the functions of NSCs is still unclear.
Long non-coding RNA (LncRNA), a novel member of non-coding RNA, is characterized by the length larger than 200 nt and without protein-coding potential (Ransohoff et al., 2018). Researches have demonstrated that LncRNA played an important role in regulation of cell function, which was greatly involved in physiological and pathological processes in vivo (Gil and Ulitsky, 2020). Moreover, it has been reported that LncRNA expression was up- or downregulated after glucocorticoid stimulation (Bunch, 2018). Whether LncRNAs contribute in the MP mediating NSC dysfunction needs to be addressed.
In this study, we investigate the expression profiles of LncRNA and mRNA of NSCs treated with MP using a microarray, followed by bioinformatics analysis. These results provide greater insight into the precise mechanisms of MP mediating NSC dysfunction in SCI.
This study was approved by the Committee for the Care and Use of Laboratory Animals of Sun Yat-sen University, Guangzhou, China. This study was performed in the Laboratory Animals Center of Sun Yat-sen University.
To isolate the NSCs from mice, neonatal C57/BL male mice were sacrificed through overdose of anesthesia. The spinal cord tissues of the neonatal C57/BL mice were separated and then thoroughly cut into pieces in a sterile environment. A total of 3 ml DMEM/F12 medium supplemented with 2% B27 supplement, 5 μg/ml heparin, 20 ng/ml bFGF, and 20 ng/ml EGF were added to resuspend the tissue. These suspensions were seeded in a 25-cm2 culture flask and cultured at 37°C humidified atmosphere with 5% CO2. Medium was changed every 3 days, and the adherent cells were removed. Suspended neural spheres containing NSCs were used for experiments.
Suspended neural spheres were centrifuged at 1,500 rpm, and the supernatant was removed. Trypsin (0.25%) supplemented with 0.53 mM EDTA was used to digest the neural spheres into single cell NSCs, which were then seeded in a six-well plate at a density of 2 × 104 cells/cm2 using culture medium. MPs were added in the culture medium at a concentration of 10 μg/ml according to our previous study (Wang et al., 2014). Culture medium without MP was used as a control. After 24 h stimulation, NSCs were collected for experiments described below.
Three MP treated NSCs and control NSCs were collected as described above. Total RNA of NSCs was extracted using Trizol reagent and was then purified using NucleoSpin® RNA clean-up kits according to the instructions. RNA integrity was determined by formaldehyde denaturing gel electrophoresis. The purity and concentration of RNA were determined from OD260/280 readings using spectrophotometer. Total RNA was reverse transcribed into cDNA, which was successively labeled with fluorescent dye (Cy3-dCTP) and hybridized with LncRNA + mRNA Gene Expression Microarray V4.0 (4 × 180 K; CapitalBio Corp).
The microarrays prepared above were washed and scanned using G2565CA Microarray Scanner. Data were analyzed for data summarization, normalization, and quality control by using the GeneSpring software (version 13.0). Differentially expressed genes were identified according to the criterion of fold change > | 2.0| and P-values < 0.05.
Functional enrichment of the differentially expressed genes was performed using the Gene ontology (GO) analysis. The signaling pathways with statistic difference were identified using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database based on the differentially expressed genes. Coding–non-coding gene co-expression (CNC) networks were constructed based on the results of correlation analysis between mRNA and LncRNA expression, which meets the criterion of Pearson correlation coefficients > | 0.99| and P-values < 0.05. Based on the CNC network, the sequences of LncRNA and mRNA were compared and analyzed to identify the predicted target differentially expressed mRNA of the differentially expressed LncRNA.
RNAs of nine NSCs were isolated and reverse transcribed into cDNA. QRT-PCR was performed using SYBR Premix Ex Taq in the LightCycler 480 PCR System. All the data were normalized by GAPDH. The relative expression levels of LncRNA and mRNA were analyzed using the 2–△ △ Ct formula. The forward and reverse primers for each gene were present in Supplementary Table 1.
Statistical analysis was performed with SPSS software (version 24). Data were expressed as means ± SD, and P-values < 0.05 were considered as statistically significant difference.
Mouse NSCs with MP stimulation at a concentration of 10 μg/ml for 24 h as well as control NSCs were detected using a LncRNA–mRNA microarray. Results showed that a total of 63 differentially expressed LncRNAs were identified, which contained 39 upregulated LncRNAs and 24 downregulated LncRNAs in MP stimulated NSCs compared to control NSCs (Figure 1A). The top 10 LncRNAs with the largest significant difference are shown in Table 1. Moreover, there were 174 differentially expressed mRNAs in MP stimulated NSCs compared to the control group, including 89 upregulated mRNAs and 85 downregulated mRNAs (Figure 1B). The top 10 mRNAs with the largest significant difference are shown in Table 2. These results indicated that MP treatment altered the LncRNA and mRNA expression profiles of NSCs.
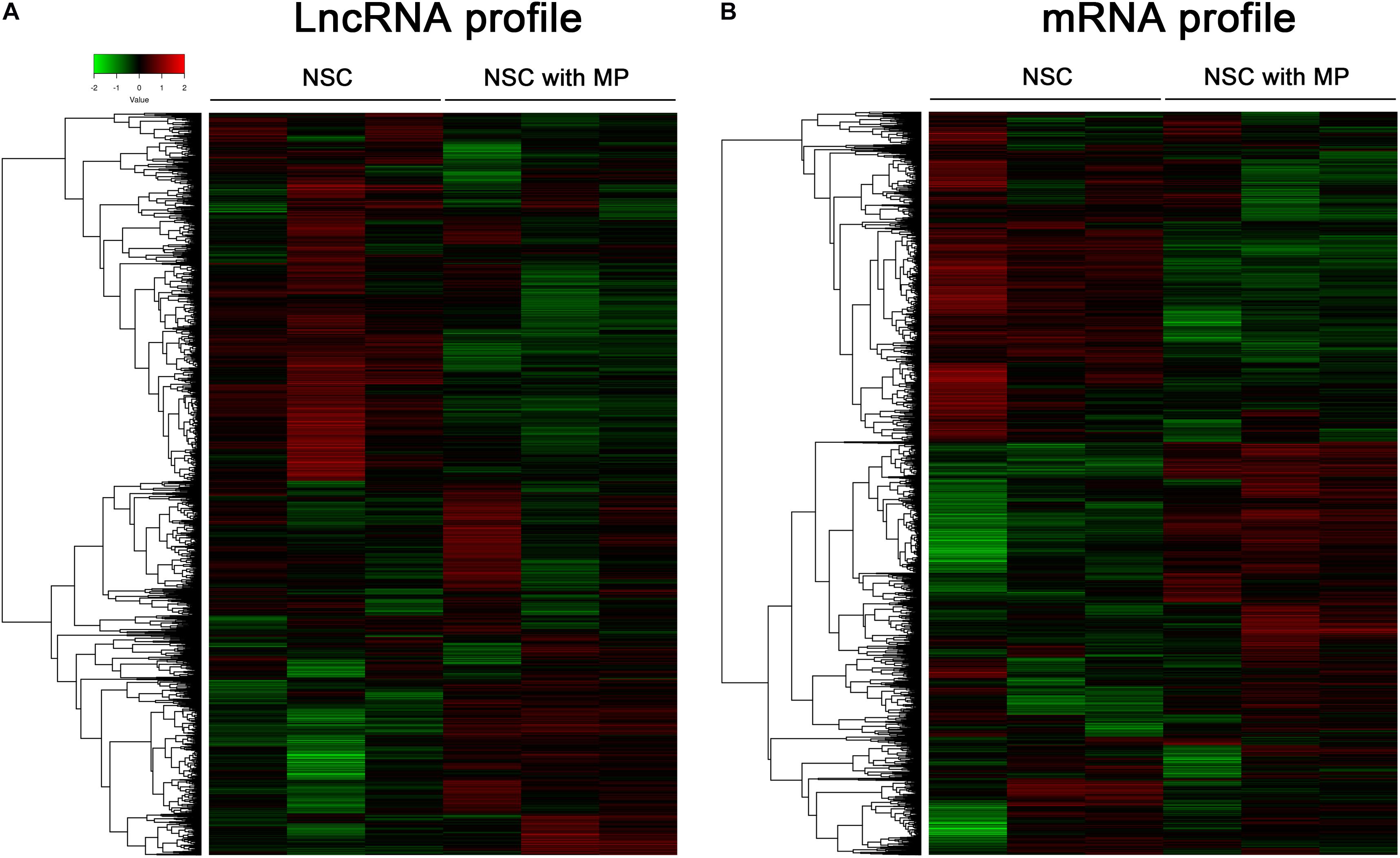
Figure 1. Clusters of differentially expressed LncRNA and mRNA of NSCs. (A) A total of 63 LncRNAs were differentially expressed in MP treated NSCs (n = 3) compared with control NSCs (n = 3). Hierarchical clustering showed a distinguishable LncRNA expression profile. (B) A total of 174 mRNAs were differentially expressed in MP treated NSCs (n = 3) compared with control NSCs (n = 3). Hierarchical clustering showed a distinguishable mRNA expression profile.
In order to speculate the possible function of NSCs affected by MP stimulation, GO analysis was performed. All differentially expressed mRNAs were classified into three domains, including biological process, molecular function, and cellular component. In the biological process domain, four GO terms of the top 10 GO terms for the differentially expressed mRNAs were about cell migration. Besides, response to external stimulus and secretion regulation were enriched in the top 10 GO terms of biological process domain (Figure 2A). In the molecular function domain, the top 5 GO terms were receptor ligand activity, receptor regulator activity, collagen binding, virus receptor activity, and hijacked molecular function (Figure 2B). Moreover, the top 5 GO terms of cellular component domain were extracellular matrix, collagen-containing extracellular matrix, endoplasmic reticulum lumen, platelet alpha granule, and specific granule (Figure 2C).
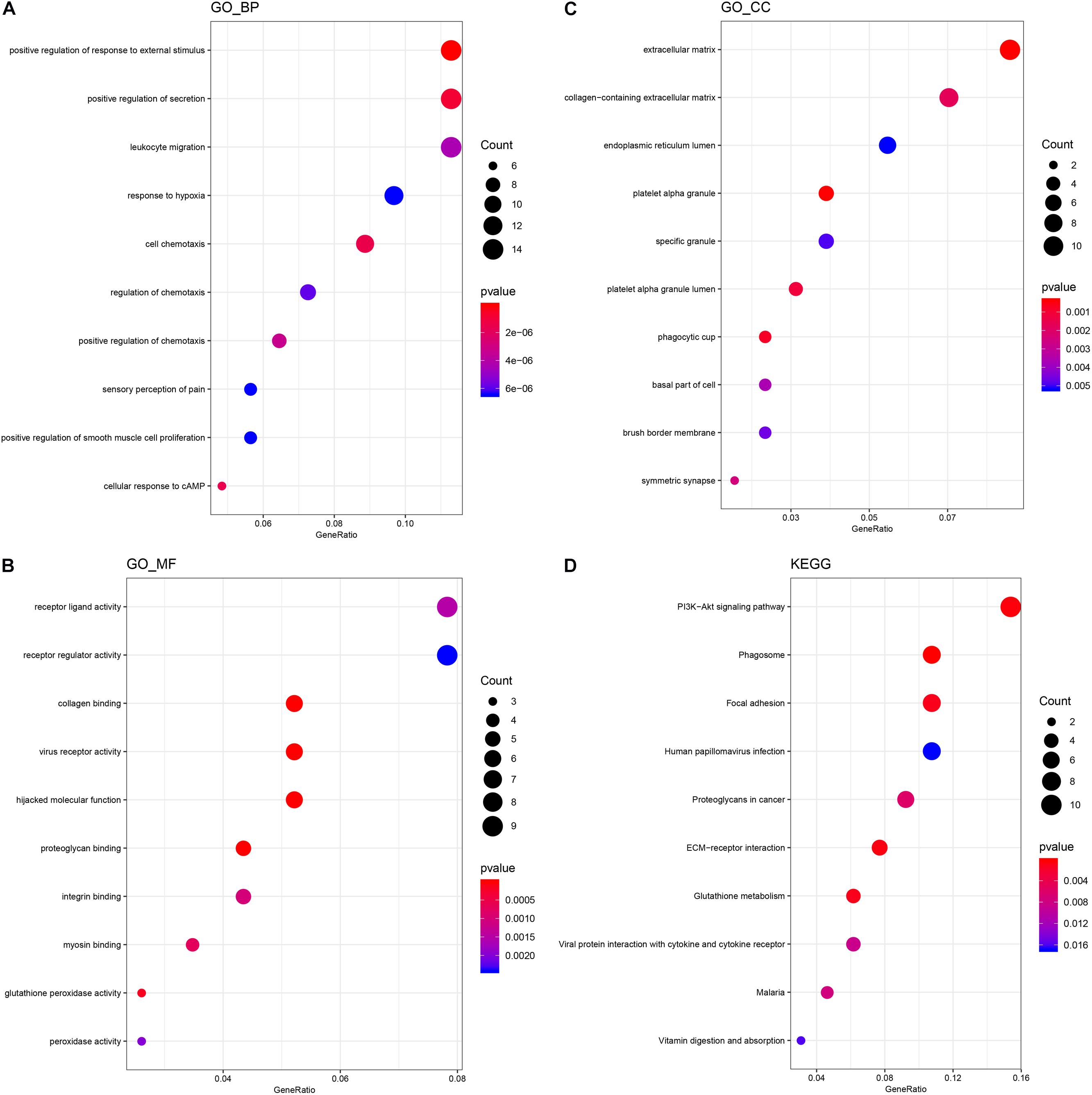
Figure 2. GO and KEGG analysis of differentially expressed mRNA. (A) The top 10 GO terms of the biological process domain. (B) The top 10 GO terms of the molecular function domain. (C) The top 10 GO terms of the cellular component domain. (D) The top 10 signaling pathways enriched by the KEGG database.
KEGG analysis was then performed to further speculate the functional signaling pathways of NSCs affected by MP stimulation. A total of 22 signaling pathways were enriched by KEGG analysis based on the differentially expressed mRNA. The top 5 signaling pathways in KEGG analysis are shown in Table 3. Among these signaling pathways, the PI3K–Akt signaling pathway was the largest significant difference pathway with the highest gene ratio (differentially expressed gene/total gene in pathway) (Figure 2D), indicating that the PI3K–Akt signaling pathway has an important role in MP stimulated NSCs.
A CNC network was constructed to clarify the relationship of the differentially expressed LncRNA and mRNA. There were 3899 co-expression pairs identified among the differentially expressed LncRNA and mRNA. Based on this CNC network, we investigated and identified five pairs of the differentially expressed LncRNA and their predicted target mRNA (Table 4). NONMMUT029251 and NONMMUT067949 positively regulated their target mRNA Galnt15 and Mmp12 separately, and NONMMUT028954 and NONMMUT028954 negatively regulated their target gene. The predicted regulatory mechanisms were cis-acting (four pairs) or as trans-acting (one pair). Moreover, the expression level of these LncRNA and mRNA was confirmed by qRT-PCR, which is consistent with the results of microarray (Figure 3).
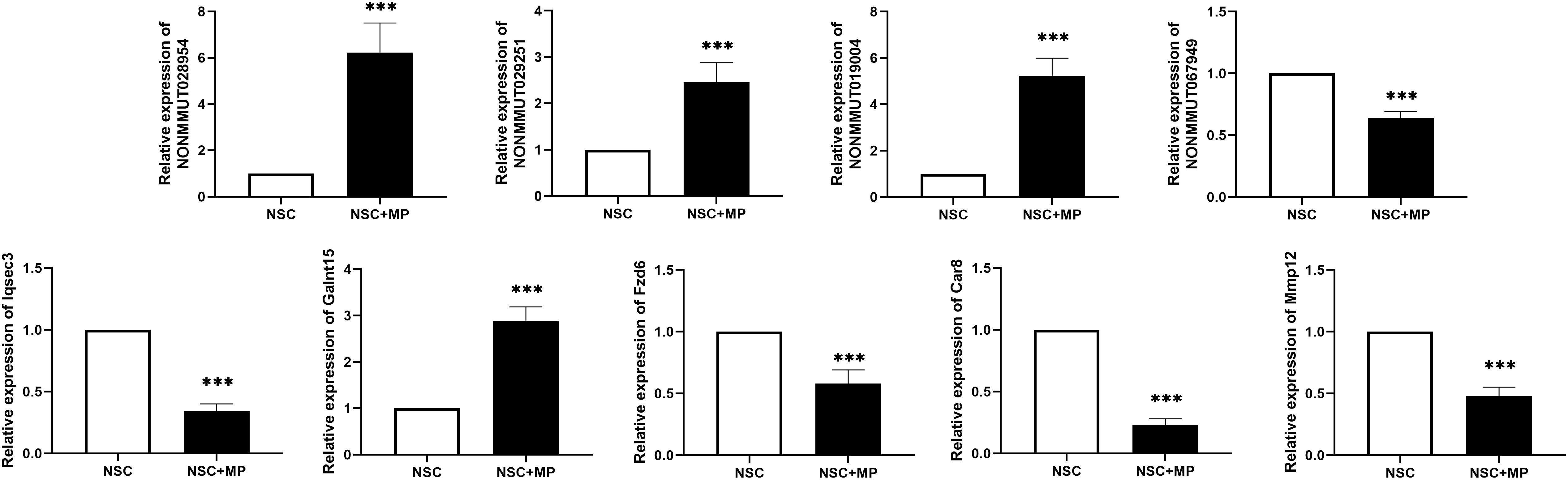
Figure 3. Differentially expressed LncRNA and mRNA confirmation by qRT-PCR. Five pairs of the differentially expressed LncRNAs and their predicted target mRNAs were confirmed by qRT-PCR. The mRNAs Iqsec3, Fzd6, Car8, and Mmp12 were downregulated, and Galnt15 was upregulated. NONMMUT067949 was downregulated, and NONMMUT029251, NONMMUT019004, and NONMMUT028954 were upregulated. N = 9, ***P < 0.001.
NSCs, a kind of adult multipotential stem cell, are one of the critical cells in nervous tissue repairment after SCI. Once the spine cord injury occurs, endogenous NSCs are firstly activated from quiescent status, followed by migration to the injured sites, and then exhibit their healing function through differentiation into neuron or secreting cytokines (Stenudd et al., 2015). Except for the endogenous NSCs, more and more studies have demonstrated that exogenous NSC transplantation improved the recovery of nervous tissue in SCI models, emphasizing the important role of NSCs in SCI treatment (Yousefifard et al., 2016). However, these reparative processes are widely and usually affected by kinds of factors in vivo and in vitro.
MP is a widely used glucocorticoid for many diseases in clinic, in which its effectiveness is nevertheless a controversial focus of SCI treatment (Ahuja et al., 2017). Recently, our research found that MP inhibited the proliferation of NSCs in a non-human primate with SCI. Besides, MP affected the differentiation potentials of NSCs in a rat model with SCI (Wang et al., 2014). Moreover, high dose MP after SCI significantly reduces the proliferation rate of NSCs in vitro (Schroter et al., 2009). These results remind us that the ineffectiveness of MP in SCI may partially come from its effect on NSCs, but the detailed mechanism is largely unknown.
NSC functions were under the control of kinds of intracellular regulatory mechanisms at gene transcription and protein translation levels, among which LncRNAs were one of the most important mechanisms (Kim, 2016; Ramos et al., 2016). LncRNAs widely exist in cells and regulate the cells’ function through trans/cis-acting or endogenous competitive RNA (ceRNA) mechanisms (Guil and Esteller, 2015). Previously, many studies have shown the large involvement of LncRNA in regulating functions of NSCs. A LncRNA named Pnky was determined to be a neural-specific LncRNA to inhibit the neuronal differentiation of NSCs (Ramos et al., 2015). Moreover, another LncRNA, LncRNA-158, was proven to promote the NSC differentiation into oligodendrocytes through the NFκB signaling pathway (Li et al., 2018). Nevertheless, the LncRNA expression profiles are easily affected by external stimulation, which in turn results in function alternation of NSCs (Lu et al., 2018). In this study, in order to figure out the role of LncRNA in MP mediating NSC dysfunction, a LncRNA–mRNA microarray was performed to scan their expression in NSCs after MP stimulation. There were 63 differentially expressed LncRNAs and 174 differentially expressed mRNAs identified in MP treated NSCs compared to those control NSCs without stimulation. These results indicated that not only the mRNA expression profile but also the LncRNA expression profile was altered by MP treatment. Moreover, the possibility that MP affected the NSC function through regulation of their LncRNA expression existed.
A cell’s mRNA expression profile reflects its status and function. To investigate the MP affected functions of NSCs, GO was performed to enrich the differentially expressed mRNA. In the biological process domain of GO terms, functions about cell migration, secretion, and stimulation reaction were enriched, suggesting that MP stimulation at a concentration of 10 μg/ml for 24 h mainly affects these functions of NSCs. It is worth noting that NSCs migrating to the sites of injury and secreting large amount of neurotrophic factors are two key steps in neural restoration except for differentiation directly (Stenudd et al., 2015). Therefore, we suggest that MP may also affect the NSC migration and secretion potential in addition to the previously reported ability in proliferation and differentiation. To explore the enriched signaling pathway, we then performed the KEGG analysis based on the differentially expressed mRNA profile. Although 22 signaling pathways were enriched, the PI3K–Akt signaling pathway was determined to be the largest significant difference pathway with the highest gene ratio. It has been widely proven that the PI3K–Akt signaling pathway contributed greatly to the functional regulation of NSCs (Lee et al., 2016; Zhang et al., 2017; Wang et al., 2018). To our knowledge, for the first time, we demonstrated that MP may affect the NSCs by modulating the PI3K–Akt signaling pathway. Song et al. (2017) found that LncRNA named IGF2AS could inactivate the Akt signaling pathway and then protected the NSC derived neurons from apoptosis. We speculated that differentially expressed LncRNA may activate or inactivate the PI3K–Akt signaling pathway, which ultimately caused the dysfunction of NSCs. Functional experiments in vivo and in vitro should be performed to confirm this hypothesis.
LncRNAs always show a positive or negative correlation in expression level to their target gene (Chen, 2016). To further clarify the relationship and regulatory mechanism of differentially expressed mRNA and LncRNA, we identified the predicted target mRNA and their related LncRNA using bioinformatics methods based on the CNC network of those differentially expressed mRNA and LncRNA in this study. Among the five identified pair, NONMMUT067949-MMP12 was the most remarkable combination. Previous study by Shan et al. demonstrated that MMP12 was a critical molecular in the activation and development of NSCs (Shan et al., 2018). As our results, NONMMUT067949 may be positively related to MMP12 expression. After MP stimulation, NONMMUT067949 expression decreased, which downregulated the expression of MMP12 through cis-acting mechanism, and finally resulted in the dysfunction of NSCs. These relationships and the regulatory mechanisms are also need to be addressed in the future study.
In summary, we investigated the LncRNA and mRNA expression profiles of NSCs treated with MP. This study may help to understand the mechanism of MP to regulate NSCs’ function and the role of LncRNA in this process. However, limitations still exist in this study. The detailed regulatory mechanisms of these identified LncRNA are still unknown in cells and mice models. Besides, the LncRNA expression profiles of NSCs treated by MP at different concentrations or stimulation times have not been clarified. Further studies should be addressed to elucidate these questions.
The data of microarrays can be obtained from the NCBI SRA database at this link: https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA727403&o=acc_s%3Aa.
JY, YT, and ZX designed research studies, conducted the experiments, analyzed the data, and wrote the manuscript. MM and YL conducted the experiments, analyzed data, and wrote the manuscript. KD conducted the experiments. All authors contributed to the article and approved the submitted version.
This study was financially supported by the National Natural Science Foundation of China (81801224) and the Science and Technology Development Funding Project of Guangdong Province (2017A030310219 and 2017A030310554).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2021.669224/full#supplementary-material
Ahuja, C. S., Schroeder, G. D., Vaccaro, A. R., and Fehlings, M. G. (2017). Spinal cord injury-what are the controversies? J. Orthop. Trauma 31(Suppl. 4), S7–S13.
Bracken, M. B., Collins, W. F., Freeman, D. F., Shepard, M. J., Wagner, F. W., Silten, R. M., et al. (1984). Efficacy of methylprednisolone in acute spinal cord injury. JAMA 251, 45–52. doi: 10.1001/jama.1984.03340250025015
Bracken, M. B., Shepard, M. J., Collins, W. F., Holford, T. R., Young, W., Baskin, D. S., et al. (1990). A randomized, controlled trial of methylprednisolone or naloxone in the treatment of acute spinal-cord injury. results of the second national acute spinal cord injury study. N. Engl. J. Med. 322, 1405–1411.
Bracken, M. B., Shepard, M. J., Holford, T. R., Leo-Summers, L., Aldrich, E. F., Fazl, M., et al. (1997). Administration of methylprednisolone for 24 or 48 hours or tirilazad mesylate for 48 hours in the treatment of acute spinal cord injury. results of the third national acute spinal cord injury randomized controlled trial. national acute spinal cord injury study. JAMA 277, 1597–1604. doi: 10.1001/jama.277.20.1597
Bunch, H. (2018). Gene regulation of mammalian long non-coding RNA. Mol. Genet. Genomics 293, 1–15. doi: 10.1007/s00438-017-1370-9
Chen, L. L. (2016). Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41, 761–772. doi: 10.1016/j.tibs.2016.07.003
David, G., Mohammadi, S., Martin, A. R., Cohen-Adad, J., Weiskopf, N., Thompson, A., et al. (2019). Traumatic and nontraumatic spinal cord injury: pathological insights from neuroimaging. Nat. Rev. Neurol. 15, 718–731. doi: 10.1038/s41582-019-0270-5
Evaniew, N., Belley-Cote, E. P., Fallah, N., Noonan, V. K., Rivers, C. S., and Dvorak, M. F. (2016). Methylprednisolone for the treatment of patients with acute spinal cord injuries: a systematic review and meta-analysis. J. Neurotrauma 33, 468–481. doi: 10.1089/neu.2015.4192
Gil, N., and Ulitsky, I. (2020). Regulation of gene expression by cis-acting long non-coding RNAs. Nat. Rev. Genet. 21, 102–117. doi: 10.1038/s41576-019-0184-5
Guil, S., and Esteller, M. (2015). RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem. Sci. 40, 248–256. doi: 10.1016/j.tibs.2015.03.001
Kim, D. Y. (2016). Post-transcriptional regulation of gene expression in neural stem cells. Cell Biochem. Funct. 34, 197–208. doi: 10.1002/cbf.3181
Lee, J. E., Lim, M. S., Park, J. H., Park, C. H., and Koh, H. C. (2016). S6K promotes dopaminergic neuronal differentiation through PI3K/Akt/mTOR-dependent signaling pathways in human neural stem cells. Mol. Neurobiol. 53, 3771–3782. doi: 10.1007/s12035-015-9325-9
Li, Y., Guo, B., Yang, R., Xiao, Z., Gao, X., Yu, J., et al. (2018). A novel long noncoding RNA lnc158 promotes the differentiation of mouse neural precursor cells into oligodendrocytes by targeting nuclear factor-IB. Neuroreport 29, 1121–1128. doi: 10.1097/wnr.0000000000001083
Lu, G., Xu, H., Zhao, W., Zhang, J., Rao, D., and Xu, S. (2018). Upregulation of long noncoding RNA Gadd45a is associated with sevoflurane-induced neurotoxicity in rat neural stem cells. Neuroreport 29, 605–614. doi: 10.1097/wnr.0000000000000980
Ortiz-Alvarez, G., Daclin, M., Shihavuddin, A., Lansade, P., Fortoul, A., Faucourt, M., et al. (2019). Adult neural stem cells and multiciliated ependymal cells share a common lineage regulated by the geminin family members. Neuron 102, 159–172.e7.
Ramos, A. D., Andersen, R. E., Liu, S. J., Nowakowski, T. J., Hong, S. J., Gertz, C., et al. (2015). The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16, 439–447. doi: 10.1016/j.stem.2015.02.007
Ramos, A. D., Attenello, F. J., and Lim, D. A. (2016). Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci. Lett. 625, 70–79. doi: 10.1016/j.neulet.2015.12.025
Ransohoff, J. D., Wei, Y., and Khavari, P. A. (2018). The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 19, 143–157. doi: 10.1038/nrm.2017.104
Schroter, A., Lustenberger, R. M., Obermair, F. J., and Thallmair, M. (2009). High-dose corticosteroids after spinal cord injury reduce neural progenitor cell proliferation. Neuroscience 161, 753–763. doi: 10.1016/j.neuroscience.2009.04.016
Shan, X., Tomlinson, L., Yang, Q., and Colognato, H. (2018). Distinct requirements for extracellular and intracellular MMP12 in the development of the adult V-SVZ neural stem cell niche. Stem Cell Rep. 10, 984–999. doi: 10.1016/j.stemcr.2018.01.038
Silva, N. A., Sousa, N., Reis, R. L., and Salgado, A. J. (2014). From basics to clinical: a comprehensive review on spinal cord injury. Prog. Neurobiol. 114, 25–57. doi: 10.1016/j.pneurobio.2013.11.002
Song, C., Song, C., Chen, K., and Zhang, X. (2017). Inhibition of long non-coding RNA IGF2AS protects apoptosis and neuronal loss in anesthetic-damaged mouse neural stem cell derived neurons. Biomed. Pharmacother 85, 218–224. doi: 10.1016/j.biopha.2016.10.094
Stenudd, M., Sabelstrom, H., and Frisen, J. (2015). Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 72, 235–237. doi: 10.1001/jamaneurol.2014.2927
Tang, Y., Yu, P., and Cheng, L. (2017). Current progress in the derivation and therapeutic application of neural stem cells. Cell Death Dis. 8:e3108. doi: 10.1038/cddis.2017.504
Wang, S., Huang, L., Zhang, Y., Peng, Y., Wang, X., and Peng, Y. (2018). Protective effects of L-3-n-Butylphthalide against H2O2-Induced injury in neural stem cells by activation of PI3K/Akt and Mash1 pathway. Neuroscience 393, 164–174. doi: 10.1016/j.neuroscience.2018.10.003
Wang, W., Wang, P., Li, S., Yang, J., Liang, X., Tang, Y., et al. (2014). Methylprednisolone inhibits the proliferation and affects the differentiation of rat spinal cord-derived neural progenitor cells cultured in low oxygen conditions by inhibiting HIF-1alpha and Hes1 in vitro. Int. J. Mol. Med. 34, 788–795. doi: 10.3892/ijmm.2014.1835
Ye, J., Qin, Y., Tang, Y., Ma, M., Wang, P., Huang, L., et al. (2018). Methylprednisolone inhibits the proliferation of endogenous neural stem cells in nonhuman primates with spinal cord injury. J. Neurosurg. Spine 29, 199–207. doi: 10.3171/2017.12.spine17669
Yousefifard, M., Rahimi-Movaghar, V., Nasirinezhad, F., Baikpour, M., Safari, S., Saadat, S., et al. (2016). Neural stem/progenitor cell transplantation for spinal cord injury treatment; a systematic review and meta-analysis. Neuroscience 322, 377–397.
Keywords: neural stem cells, methylprednisolone, long non-coding RNA, mRNA, spine injury
Citation: Tang Y, Xie Z, Ma M, Duan K, Li Y and Ye J (2021) LncRNA and mRNA Expression Profiles in Methylprednisolone Stimulated Neural Stem Cells. Front. Neurosci. 15:669224. doi: 10.3389/fnins.2021.669224
Received: 18 February 2021; Accepted: 18 May 2021;
Published: 23 June 2021.
Edited by:
Jee-Yeon Hwang, Creighton University, United StatesReviewed by:
Salva Mena-Mollá, University of Valencia, SpainCopyright © 2021 Tang, Xie, Ma, Duan, Li and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jichao Ye, eWVqaWNoQG1haWwuc3lzdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.