
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neurosci., 30 April 2021
Sec. Neurodegeneration
Volume 15 - 2021 | https://doi.org/10.3389/fnins.2021.660379
 Yong Hui Nies1
Yong Hui Nies1 Nor Haliza Mohamad Najib1
Nor Haliza Mohamad Najib1 Wei Ling Lim2
Wei Ling Lim2 Mohd Amir Kamaruzzaman1
Mohd Amir Kamaruzzaman1 Mohamad Fairuz Yahaya1
Mohamad Fairuz Yahaya1 Seong Lin Teoh1*
Seong Lin Teoh1*Parkinson’s disease (PD) is a severely debilitating neurodegenerative disease, affecting the motor system, leading to resting tremor, cogwheel rigidity, bradykinesia, walking and gait difficulties, and postural instability. The severe loss of dopaminergic neurons in the substantia nigra pars compacta causes striatal dopamine deficiency and the presence of Lewy bodies indicates a pathological hallmark of PD. Although the current treatment of PD aims to preserve dopaminergic neurons or to replace dopamine depletion in the brain, it is notable that complete recovery from the disease is yet to be achieved. Given the complexity and multisystem effects of PD, the underlying mechanisms of PD pathogenesis are yet to be elucidated. The advancement of medical technologies has given some insights in understanding the mechanism and potential treatment of PD with a special interest in the role of microRNAs (miRNAs) to unravel the pathophysiology of PD. In PD patients, it was found that striatal brain tissue and dopaminergic neurons from the substantia nigra demonstrated dysregulated miRNAs expression profiles. Hence, dysregulation of miRNAs may contribute to the pathogenesis of PD through modulation of PD-associated gene and protein expression. This review will discuss recent findings on PD-associated miRNAs dysregulation, from the regulation of PD-associated genes, dopaminergic neuron survival, α-synuclein-induced inflammation and circulating miRNAs. The next section of this review also provides an update on the potential uses of miRNAs as diagnostic biomarkers and therapeutic tools for PD.
Parkinson’s disease (PD) is the second most common neurodegenerative disorder after Alzheimer’s disease. The clinical signs of PD involved dopamine (DA)-related motor symptoms such as bradykinesia, postural instability, resting tremor, walking and gait difficulties (Jellinger, 2015). Compelling evidences relating to the multisystem effects of PD reported that the primary feature defects in movement control are caused by the death of DA neurons within the substantia nigra pars compacta (SNpc) (Chen et al., 2019). In addition to the motor symptoms, PD patients also presented with other non-motor symptoms which may precede the development of motor symptoms such as olfactory dysfunction, autonomic dysfunction (respiratory, cardia, gastrointestinal, urogenital), sensory symptoms, sleep disorder, neuropsychiatric symptoms (cognitive, mood, dementia, hallucinations) and others (Azmin et al., 2014; Jellinger, 2015). Most PD patients exhibit impaired cognitive function, particularly the ability to concentrate, executive functioning, and experiencing varying degrees of dementia (Goedert et al., 2013; Irwin et al., 2013).
The incidence of PD is between 10 and 50/100,000 person/year, and the prevalence in the population is between 100 and 300/100,000 (Pringsheim et al., 2014). The prevalence and incidence rates of PD among the Eastern population are slightly lower compared to the Western population with a 51.3 – 176.9/100,000 prevalence rate. Genetic and environmental factors could be the reasons to explain a lower PD prevalence rate observed in Asia countries (Abbas et al., 2018). The number of individuals with PD continues to grow from 4.6 million in the year 2005 to an estimate of 9.3 million in 2030 in Western Europe (Dorsey et al., 2007). From the 6.1 million individuals worldwide diagnosed with PD in 2016, 2.9 million (47.5%) were women and 3.2 million (52.5%) were men with a ratio of 1: 1.5 (GBD 2016 Parkinson’s Disease Collaborators, 2018). The differences could be due to the neuroprotective role of estrogens in women, frequent occupational exposures in men and X-linked genetics factors. Aging is one of the known risk factors of PD. This is supported by a study in Olmsted County, Minnesota whereby an increased incidence rate of parkinsonism and PD was reported from 1976 to 2005, particularly in men of age 70 and above (Savica et al., 2016).
The major pathological features of PD are widespread α-synuclein aggregation and progressive degeneration of the nigrostriatal system, leading to the death of DA neurons in the SNpc. This eventually leads to the presentation of both motor and non-motor symptoms of PD (Fujita et al., 2014; Pang et al., 2019; Peball et al., 2020). Additionally, the non-motor symptoms of PD are also mediated by non-dopaminergic systems, and other structures outside the nigrostriatal system (Jellinger, 2017b). PD should be viewed in various aspects in terms of its etiological, molecular biological, anatomical, physiological and pathological aspects of neuropathogenesis. Although the exact etiology of PD is not well understood, different possible causes have been identified. Most PD cases are likely to involve an interplay of aging, genetic susceptibility and exposure to environmental factors (Figure 1; Pang et al., 2019).
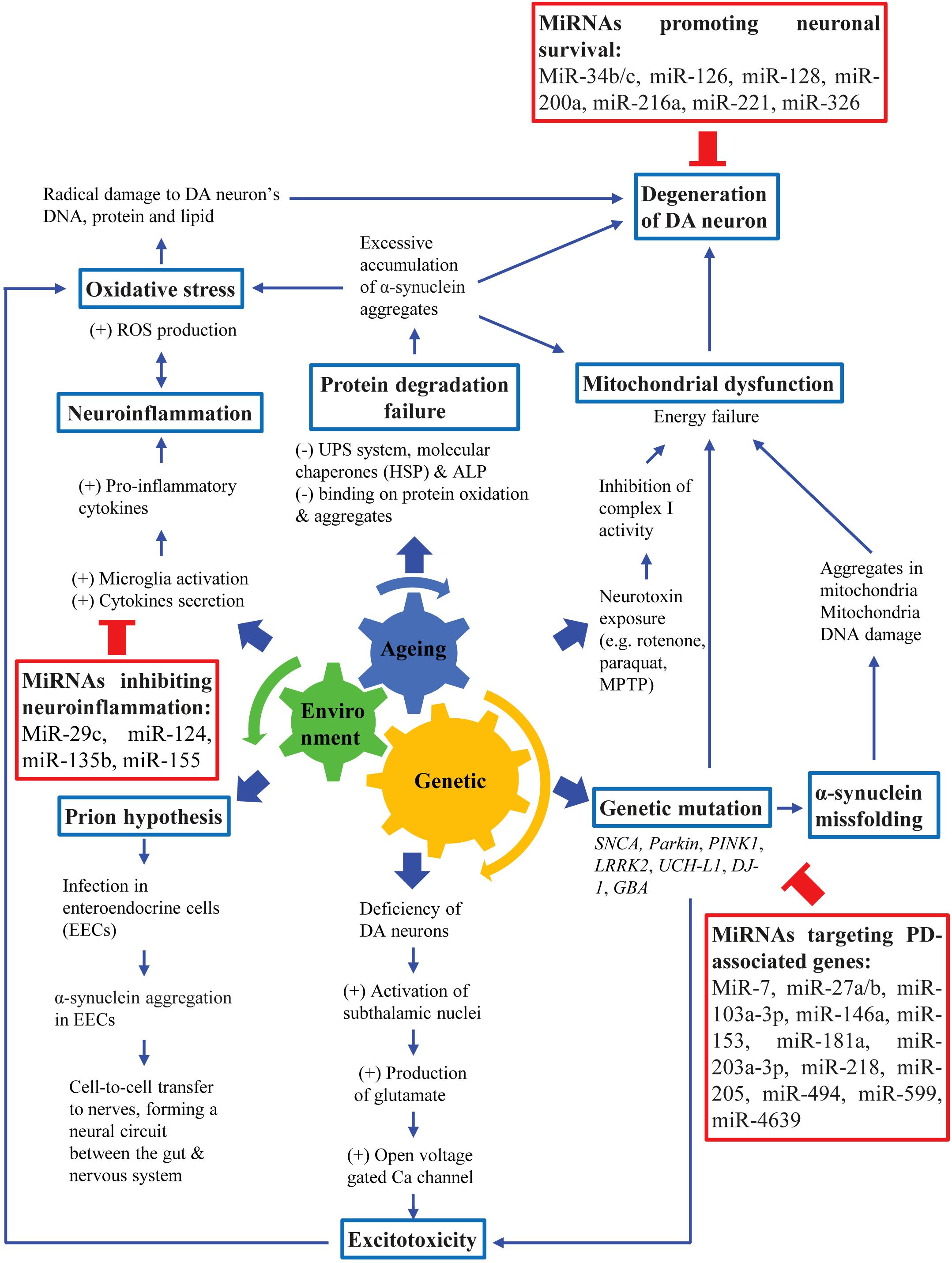
Figure 1. Pathogenesis of PD. PD pathogenesis involves an interplay among aging, genetic susceptibility and environmental factors, leading to loss of DA neurons which eventually lead to cardinal motor symptoms of PD.
Aging is a natural process that involves deregulation of numerous cellular functions and signaling pathways, such as arrested cell cycle, oxidative stress, mitochondrial dysfunction, autophagy and neuro-inflammation that are also implicated in the pathogenesis of neurodegenerative diseases (Dexter and Jenner, 2013; Jellinger, 2017a). Although age is linked to PD, aging itself is insufficient to cause PD. Therefore, it is likely that the association of both genetic and environmental factors are crucial to amplify the SNpc neuronal decline in normal aging to accelerate DA neuronal degeneration as observed in PD patients (Pang et al., 2019). Several PD-causing genes such as α-synuclein (SNCA), parkin (PARK2), Leucine-rich repeat kinase 2 (LRRK2), DJ-1 (PARK7) and PTEN-induced kinase 1 (PINK1) have been implicated in the pathogenesis of PD and have contributed to the creation of various genetic animal models (Lesage et al., 2020; Tolosa et al., 2020). Additionally, exposure to numerous environmental toxins such as pesticides and heavy metals has been known to increase the risk of PD. Neurotoxins such as 6-hydroxydopamine (6-OHDA), 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), paraquat and rotenone have been widely used to create PD animal models by selectively destroying DA neurons (Najib et al., 2020).
Although the exact mechanism underlying the death of DA neurons in the SNpc and the presence of Lewy bodies and Lewy neurites remains unclear, mechanisms such as α-synuclein misfolding and their aggregation, mitochondrial dysfunction, oxidative stress, excitotoxicity, impairment of protein clearance pathways and “prion-like protein infection” have been suggested (Golpich et al., 2017; Maiti et al., 2017). The aggregation of α-synuclein has been commonly suggested as the neurodegenerative cause of Parkinsonism (Dickson, 2018). Although most of the α-synuclein immune-reactive pathology in PD is within SNpc and ventral tegmental area, both Lewy bodies and Lewy neurites are also found to be present in nearly all brainstem nuclei and fiber tracts (Seidel et al., 2015). The biological basis for selective vulnerability of DA neurons may be due to the pacemaker-like properties of these neurons that lead to frequent intracellular calcium transients (Guo et al., 2018). Calcium buffering may be relatively deficient in neurons involved in the nigrostriatal pathway compared to neurons in the mesolimbic pathway, which may cause dysregulated cellular homeostasis. DA neuronal death is associated with the loss of functional nuclear envelope integrity and release of pro-aggregant nuclear factors that may then initiate the aggregation of α-synuclein (Jiang et al., 2016). The initiation of aggregation may subsequently lead to spreading to other cells (Braak et al., 2004; Danzer et al., 2012; Dickson, 2018). Degeneration of DA neurons in the SNpc and diminished DA levels in PD patients resulted in less inhibition of striatal neuronal activity and the derangement of striatal neuronal firing. This then led to the loss of inhibitory effects on the globus pallidus and thalamus, which will then cause the excessive activation of the motor cortex. This led to the impairment of motor coordination and patients will exhibit PD-associated motor symptoms (Magrinelli et al., 2016).
MicroRNAs (miRNAs) are a family of small single-stranded non-coding RNA molecules (20 – 25 nucleotides in length) that bind to the 3′ untranslated regions (UTR) of the target mRNAs. The binding induces mRNA degradation and translational repression hence contribute to the post-transcriptional regulation of gene expression. MiRNAs also interact with 5′ UTR, coding sequence, gene promoters and transcription factors (Khee et al., 2014; O’Brien et al., 2018).
The first miRNA, lin-4 was discovered in 1993 as a developmental gene regulator of Caenorhabditis elegans. The lin-4 contained complementary sequences which enable its binding to the 3′ UTR sequence of lin-14 (Lee et al., 1993; Wightman et al., 1993). The binding of lin-4 to the lin-14 forms multiple RNA duplexes that lead to the down-regulation of lin-14 translation (Wightman et al., 1993). To date, around 48,860 mature miRNAs have been identified from 271 different species, including over 2,600 miRNAs from humans, and these data are registered in miRBase (the miRNA database1) (Kozomara et al., 2019).
The identification of target genes is important to understand the function of miRNAs. Currently, numerous bioinformatics and biological tools are used to identify miRNA target genes, which may then be validated with further experiments (Wang et al., 2017).
Dysregulation of miRNAs will result in development and progression of numerous disease, such as in cancers (Ibrahim et al., 2015). Studies have identified dysregulation of miRNAs expression in neurodegenerative diseases such as Alzheimer’s disease, amyotrophic lateral sclerosis and Huntington’s disease (Lee et al., 2011; Figueroa-Romero et al., 2016; Putteeraj et al., 2017). Similarly, in PD patients, the striatal brain tissue and DA neurons from the SNpc demonstrated dysregulated miRNAs expression profiles (Briggs et al., 2015; Nair and Ge, 2016). A total of 125 miRNAs was significantly altered in the post-mortem analysis of the prefrontal cortex from PD patients compared to the neuropathologically normal controls (Hoss et al., 2016). Therefore, it is believed that miRNA dysregulation may contribute to the pathogenesis of PD by regulating various genes and proteins related to PD. This review will discuss the recent findings of miRNAs dysregulation related to PD.
MiR-7, miR-153 and miR-203a-3p possess binding sites to the 3′ UTR of the SNCA gene. The expression of miR-7 is significantly reduced in the SNpc of PD patients (McMillan et al., 2017). It has been shown that over-expression of both miR-7 and miR-153 in the human embryonic kidney cell line, HEK293 cells and cortical neurons lead to significant reduction of SNCA mRNA and protein expression levels meanwhile miR-7 knockdown induces over-expression of α-synuclein protein level (Doxakis, 2010; McMillan et al., 2017). Both miRNAs regulate SNCA protein levels via a different pathway in which miR-7 inhibits its translation while mir-153 degrades the mRNA (Doxakis, 2010). Je and Kim (2017) demonstrated that exposure of 1-methyl-4-phenylpyridinium (MPP+, the active metabolite of MPTP) to the HEK293 cells relieved the miRNA-mediated translational suppression of SNCA, via MPP+-mediated mitochondrial reactive oxygen species (ROS) production. Interestingly, the over-expression of miR-7 and miR-153 in cortical neurons reduced MPP+-induced neurotoxicity by increasing neuronal viability, up-regulation of pro-survival BCL-2 protein and in-activation of pro-apoptotic caspase-3 (Fragkouli and Doxakis, 2014).
Another miRNA targeting SNCA, miR-203a-3p expression was down-regulated in MMP+-treated human dopaminergic neuroblastoma, SH-SY5Y cells (Jiang et al., 2020). MMP+ treatment led to reduce cell proliferation and induced apoptosis in the SH-SY5Y cells that then led to enhance expression of SNCA, p53 and cleaved Caspase-3 proteins of which were inhibited by the up-regulation of miR-203a-3p (Jiang et al., 2020). Other miRNAs that were identified as regulators of SNCA expression include the miR-30b, miR-34b/c, miR-214, and miR-433 (Wang et al., 2015; Consales et al., 2018; Tarale et al., 2018; Shen et al., 2020).
Mutations in the PRKN, encoding Parkin (an important E3 ubiquitin-protein ligase that mediates the elimination of damaged mitochondria via mitophagy), cause an autosomal recessive form of early onset PD (Kim et al., 2012; Zanellati et al., 2015). Mitochondrial dysfunction and the down-regulation of Parkin in the MPTP-treated mice and MPP+-treated SH-SY5Y cells (Zhou et al., 2020). Furthermore, Parkin protein expression was significantly down-regulated following miR-103a-3p over-expression while Parkin protein expression was up-regulated following miR-103a-3p knockout. This suggest that miR-103a-3p able to regulate Parkin expression, by binding to the 3′-UTR of Parkin mRNA (Zhou et al., 2020).
In a rotenone-induced PD rat model, miR-146a, a known inflammation regulatory miRNAs was identified as the most up-regulated miRNAs (Jauhari et al., 2020). The exposure of rotenone led to the activation of NF-κβ and induces the transcription of miR-146a that is responsible for the down-regulation of Parkin protein level (Jauhari et al., 2020). In contrast, down-regulation of miR-146a expression led to the inhibition of NF-κβ phosphorylation and increased Parkin level in the rotenone-treated SH-SY5Y cells (Jauhari et al., 2020).
PRKN was also identified as a target of other miRNAs such as miR-181a and miR-218. Over-expression of miR-181a significantly reduced Parkin mRNA and protein level, while inhibition of miR-181a increased Parkin expression (Cheng et al., 2016). In addition to PRKN, miR-181a also targets genes that control neurite growth namely SMAD1 and SMAD5 transcription factors (Hegarty et al., 2018). Inhibition of miR-181a expression in SH-SY5Y cells led to elevation of Smad1/5 proteins, as well as the neurite length and neurite branching (Hegarty et al., 2018). Similarly, down-regulation of miR-181a-5p confers neuroprotection to MPP+-treated cells by enhancing cell viability and suppression of cell apoptosis, LDH activity and inflammation (Guo and Hua, 2020). The data on miR-181a dysregulation reported in PD patients and axonal degeneration of DA neurons in early stages of PD suggest that inhibition of miR-181a may be a promising therapeutic approach for PD. MiR-218 also targets PRKN and negatively regulates PINK1/PRKN-mediated mitophagy. Over-expression of miR-218 down-regulated PRKN expression, leading to dysregulated mitophagy (Di Rita et al., 2020). However, miR-218 expression was found to be reduced in the patients and animal models (Xing et al., 2020; Ma et al., 2021).
Homozygous and compound heterozygous mutations in the PINK1 gene are associated with the early onset of PD (Krohn et al., 2020). The PINK1 gene encodes a PTEN-induced serine-threonine kinase 1 located in the mitochondria in which the mutations can lead to respiratory chain dysfunction and impairment in ATP production (Rango et al., 2020). MiR-27a and miR-27b suppressed the expression of PINK1 by direct binding to the 3′UTR of its mRNA (Kim J. et al., 2016). Thus, miR-27a/b inhibited accumulation of PINK1, which in turn prevented the translocation of Parkin to the mitochondria following mitochondrial damage hence leads to the inhibition of lysosomal degradation of damaged mitochondria (Kim J. et al., 2016). In addition to miR-27a/b, miR-140 has also been reported to target PINK1. Amyloid β-derived diffusible ligands-induced neurons transfected with miR-140 mimic exhibited up-regulated miR-140 and down-regulated PINK1 expressions. Reciprocal effects were seen following miR-140 inhibitor transfection (Liang et al., 2021).
The LRRK2 gene mutations are associated with familial and sporadic forms of PD (Gopalai et al., 2014). The LRRK2 protein level increased with no significant changes in its mRNA levels in the frontal cortex of sporadic PD patients hence suggesting the involvement of miRNAs in suppressing post-transcriptional regulation of LRRK2 (Cho et al., 2013). There were reduced miR-205 expression in PD patients, and the reduction showed inverse correlation between the expression of LRRK2 protein and miR-205 (Cho et al., 2013). Similarly, over-expression of miR-205 in HEK293T cells suppressed LRRK2 protein expression in vitro, while inhibition of miR-205 enhanced LRRK2 protein expression (Cho et al., 2013).
MiR-599 expression was down-regulated both in vivo [lipopolysaccharide (LPS) administered mice] and in vitro (MPP+-treated SH-SY5Y cells) PD models, which was associated with increased LRRK2 expression (Wu et al., 2019). Over-expression of miR-599 was shown to down-regulate LRRK2 expression while knockdown of miR-599 up-regulated LRRK2 expression in the MPP+-treated SH-SY5Y cells (Wu et al., 2019).
DJ-1 (PARK7) is identified as a recessive familial PD gene in which loss of this gene resulted in the increased the susceptibility of cells to the damaging effect of oxidative stress, leading to the early onset of PD (Irrcher et al., 2010). MiR-494 binds to the 3′ UTR of DJ-1 and leads to the inhibition of DJ-1 transcription (Xiong et al., 2014). Over-expression of miR-494 in 3T3 cells resulted in decreased DJ-1 protein level and increased ROS production without changing the mRNA expression (Xiong et al., 2014). Similarly, over-expression of miR-494 in mice leads to a decrease in DJ-1 expression in the SNpc following MPTP administration, alongside the exacerbated degeneration of DA neurons in the SNpc and significant motor impairment (Xiong et al., 2014; Geng et al., 2018).
The miR-4639-5p level was up-regulated in the plasma of PD patients whereby miR-4639-5p negatively regulates DJ-1 in the post-transcriptional level (Chen et al., 2017). The up-regulation of miR-4639-5p led to the down-regulation of DJ-1 protein level hence resulted in severe oxidative stress and neuronal death in MPP+- and rotenone-treated SH-SY5Y cells (Chen et al., 2017). MiR-145-3p and miR-874 are among other miRNAs that were predicted to regulate DJ-1 expression, and both were shown to be highly expressed in the saliva of PD patients (Chen et al., 2020).
The expression of both miR-34b and miR-34c were shown to be down-regulated in various brain region of PD patients including the putamen, amygdala, substantia nigra, frontal cortex, and cerebellum (Minones-Moyano et al., 2011; Villar-Menendez et al., 2014). Inhibition of miR-34b/c expression in SH-SY5Y cells lead to reduced cell viability, mitochondrial dysfunction and increased ROS generation in the cells (Minones-Moyano et al., 2011). In addition, miR-34b/c inhibition could also lead to a significant reduction in DJ-1 and Parkin protein levels in which these proteins are associated with familial forms of PD (Minones-Moyano et al., 2011). In another study, miR-34b/c inhibition increased the α-synuclein levels and stimulate the aggregate formation, possibly contributing to PD pathogenesis (Kabaria et al., 2015).
MiR-126 was up-regulated in the SNpc DA neurons from PD patients (Kim W. et al., 2014). Over-expression of miR-126 compromised the survival of primary DA neurons and SH-SY5Y cells hence increasing their vulnerability to 6-OHDA toxicity (Kim W. et al., 2014). In addition to DA neurotoxins, miR-126 over-expression also increases neuronal vulnerability to staurosporine (a non-selective protein kinase inhibitor) and Alzheimer’s disease-associated amyloid β 1-42 peptides (Kim W. et al., 2016). The miR-126 over-expressing cells showed reduced expression of p85β, IRS-1 and SPRED1, reduced phosphorylated AKT and extracellular signal-regulated kinase (ERK) protein levels. These results suggest that miR-126 down-regulates insulin/IGF-1/phosphatidylinositol-3-kinase (PI3K)/AKT and ERK signaling cascades in promoting neuronal death in PD (Kim W. et al., 2014).
The expression of miR-128 decreased in the MPTP-induced PD mice (Zhou et al., 2018; Zhang G. et al., 2020). MiR-128 protects DA neurons from apoptosis and up-regulates the expression of Excitatory Amino Acid Transporter 4 (EAAT4) by binding to the axis inhibition protein 1 (AXIN1) (Zhou et al., 2018). Further study reported that the increased HIF-1α/miR-128-3p inhibited apoptosis of hippocampal neurons through Wnt/β-catenin signaling pathway activation via the suppression of AXIN1 (Zhang G. et al., 2020).
The expression of miR-200a was significantly up-regulated in the pheochromocytoma cell line PC12 cells and SH-SY5Y cells following exposure to MPP+ that leads to the induction of oxidative stress and cell apoptosis (Salimian et al., 2018; Talepoor Ardakani et al., 2019). Additionally, the transcript level of SIRT1 (a miR-200a target) showed significant down-regulation in the MPP+-treated cells, confirming that SIRT1 may induce DA neuronal apoptosis via P53 and FOXO signaling pathways (Salimian et al., 2018). In contrast, inhibition of miR-200a reduced the apoptosis and increased DA neuronal population in 6-OHDA-treated PD rats, via inhibition of the cAMP/PKA signaling pathway (Wu et al., 2018).
MiR-216a over-expression has shown to provide protection against MPP+-induced neurotoxicity in SH-SY5Y cells by significantly increase neuronal viability following MPP+ treatment (Yang et al., 2020). This was also associated with reduced neuronal apoptosis, ROS level and lipid peroxidation (Yang et al., 2020). MiR-216a promotes neuronal survival by down-regulating Bax that is involved as apoptotic regulators (Yang et al., 2020). The up-regulation of miR-216a also exerts its neuroprotective effects against ischemic brain injury by targeting JAK2/STAT3 pathways (Tian et al., 2018).
MiR-221 expression was significantly down-regulated in 6-OHDA-treated PC12 cells (Li et al., 2018). Over-expression of miR-221 cells treated with 6-OHDA and MPP+ significantly promotes cell viability and proliferation while inhibited cell apoptosis (Li et al., 2018; Oh et al., 2018). The brains of DJ-1–/– mouse and DJ-1 knock down SH-SY5Y cells demonstrated down-regulation of miR-221 (Oh et al., 2018). Additionally, miR-221 down-regulated the expression of pro-apoptotic proteins (bcl-2-like protein 11, bcl2 modifying factor, and bcl2 interacting protein 3-like) at basal conditions thus prevented the oxidative stress-induced increase of bcl-2-like protein 11 (Oh et al., 2018). Based on these findings, the modulation of miR-221 by DJ-1 could contribute to the pathogenesis of autosomal recessive inherited PD or sporadic PD, resulted from the loss-of-function mutations in DJ-1.
The expression of miR-326 was shown to be down-regulated in MPTP-induced PD mice (Zhao et al., 2019). MiR-326 over-expression ameliorates the motor dysfunction and the structural abnormality of the SNpc (increase content of DA, SOD, GSH-Px, and TH positive expression) in PD mice (Zhang et al., 2019). In addition, miR-326 overexpression also inhibits numerous immunomodulatory factors such as TNF-α, IL-1, IL-6, and INF-γ. Mice treated with miR-326 mimic inhibits expression of iNOS and neuronal apoptosis and promotes autophagy of DA neurons (Zhang et al., 2019; Zhao et al., 2019). Recent evidence also showed that miR-326 may play an important role as the key suppressor in the development of PD due to the suppression of pyroptotic cell death with the activation of NLRP3 inflammasome. It was shown that silencing the lncRNA homeobox transcript antisense RNA resulted in significant inhibition of neuronal damage through the suppression of NLRP3-mediated pyroptosis via the regulation of miR-326/ELAVL1 axis in PD model (Zhang Q. et al., 2020).
The serum miR-29 level was significantly down-regulated in PD patients, and were correlated with the disease severity (Bai et al., 2017). Over-expression of miR-29c attenuated DA neuronal death and α-synuclein aggregation in the SNpc of MPTP-treated mice. Furthermore, miR-29c exhibited anti-inflammatory properties by ameliorated the elevated pro-inflammatory cytokines (TNF-α, IL-1β, and IL-6) and apoptosis in the MPTP-treated mice (Wang et al., 2020b). Additionally, over-expression of miR-29c suppressed the LPS-induced microglial activation, via modulation of NOD-like receptor protein 3 (NLRP3) inflammasome by targeting nuclear factor of activated T-cells (NFAT) 5 (Wang et al., 2020a).
The expression of miR-124 decreased in the midbrain of the MPTP-induced PD mouse model (associated with reduced DA neurons, and decreased tyrosine hydroxylase and DA transporter expressions), MPP+-treated SH-SY5Y and MN9D cells (Kanagaraj et al., 2014; Liu et al., 2017). The reduced miR-124 expression increased the expression of calpain 1, leading to the activation of the p25/cdk5 pathway, which is known to be involved in the DA neuronal loss in PD (Kanagaraj et al., 2014). Additionally, miR-124 suppression increased cell autophagy-related protein expression (LC3 II and Beclin 1), increased phosphorylated-AMPK level while decreased p-mTOR expression suggest that miR-124 regulates DA neuronal apoptosis and autophagy by regulating the AMPK/mTOR pathway (Gong et al., 2016). In contrast, up-regulation of miR-124 through administration of miR-124 in the MPTP-treated mice attenuated the loss of DA neurons and striatal DA level caused by MPTP toxicity (Wang H. et al., 2016). Additionally, over-expression of miR-124 protects SH-SY5Y cells from cell death caused by microglial activation following LPS exposure suggesting the miR-124 role in the inhibition of neuroinflammation (Yao et al., 2018). In addition, over-expression of miR-124 effectively attenuated the expression of pro-inflammatory cytokines (iNOS, IL-6, and TNF-α) and promote the secretion of neuroprotective factors (TGF-β1 and IL-10) in LPS-induced immortalized murine microglial cell line BV2 cells (Yao et al., 2018). Further study suggested that miR-124 inhibits neuroinflammation by targeting sequestosome 1 (p62), phospho-p38 mitogen-activated protein kinases and autophagy (Yao et al., 2019).
Exposure of MPP+ to SH-SY5Y cells leads to a significant down-regulation of miR-135b (Zhang et al., 2017). Over-expression of miR-135b was able to attenuate neurotoxicity effects of MPP+: increased in cell viability, suppressed apoptosis, reduced level of cleaved caspase 3 (an apoptotic marker), and inhibited MPP+-induced TNF-α and IL-1β secretion (Zhang et al., 2017). Over-expression of miR-135b suppressed MPP+-induced increased quantity of pyroptotic cells and protein levels of various pyroptotic related genes (TXNIP, NLRP3, Caspase-1, ASC, GSDMDNterm and IL-1β) in the SH-SY5Y and PC-12 cells (Zeng et al., 2019).
MiR-155 demonstrated enhanced expression in the SNpc of PD mice model produced by adeno-associated-virus-mediated expression of α-synuclein (Thome et al., 2016). The deletion of miR-155 in mice prevents α-synuclein-associated neurodegeneration of DA neurons, which corresponds to the reduced microgliosis (Thome et al., 2016). Prolong exposure of TNF-α (PD patients demonstrated increased TNF-α level in the serum and CSF) to SH-SY5Y cells induced apoptosis, dysregulation of complex I and mitochondrial oxidative stress (Prajapati et al., 2015). TNF-α exposure also resulted in the up-regulation of numerous miRNAs, including miR-155, which may target nuclear encoded subunits of mitochondrial complex-I (NADH:Ubiquinone Oxidoreductase Core Subunit V1) (Prajapati et al., 2015). In addition, DJ-1 deficiency increased the expression of miR-155 in microglia and astrocytes, which down-regulates the expression of suppressor of cytokine signaling 1 (SOCS1) (Kim J. H. et al., 2014). Taken together, miR-155 modulates microglia-mediated inflammation by down-regulating SOCS1 expression and increasing the production of cytokine and nitric oxide. This leads to inefficient termination of STAT1-induced inflammatory response, which may cause neuronal death (Cardoso et al., 2012; Kim J. H. et al., 2014).
Disease modeling using induced pluripotent stem cell (iPSC) has shown to be useful by providing cellular insight into disease pathology in various neurodegenerative diseases, including PD (Doss and Sachinidis, 2019). For example, iPSC-derived DA neurons from sporadic PD and monogenic LRRK2-associated PD patients exhibited global DNA hyper-methylation changes (Fernandez-Santiago et al., 2019). An explorative genome-wide study of miRNAs expression levels in iPSC-derived DA neurons from PD patients identified an up-regulation of 5 miRNAs (miR-9-5p, miR-135a-5p, miR-135b-5p, miR-449a, and miR-449b-5p) and down-regulation of 5 miRNAs (miR-141-3p, miR-199a-5p, miR-299-5p, miR-518e-3p, and miR-519a-3p) (Tolosa et al., 2018). The genes targeted by the identified miRNAs were involved in regulating cytoskeleton, axonal transport, cell adhesion and cell survival in PD.
Circulating miRNAs are secreted from cells and have been found in various biological fluids, such as plasma, serum, saliva, milk, urine, seminal plasma, amniotic fluid, cerebrospinal fluid (CSF), peritoneal fluid, and pleural fluid (Turchinovich et al., 2012; Faruq and Vecchione, 2015). Extracellular miRNAs can either be contained within membranous vesicles, or are associated with Argonaute proteins (Turchinovich et al., 2012). The circulating miRNAs are stable in different conditions as they are resistant to RNases activity, high or low pH, multiple freeze-thaw cycles and long-term storage at room temperature thus making them suitable to be used as disease biomarkers (Turchinovich et al., 2012; Makarova et al., 2015). Various studies have described the use of circulating miRNAs as non-invasive biomarkers for the diagnosis of different diseases, including cancers, liver diseases, respiratory diseases, kidney diseases, and myocardial infarction (Arrese et al., 2015; Tiberio et al., 2015; Coskunpinar et al., 2016; Huttenhofer and Mayer, 2017; Teoh and Das, 2017; Pattarayan et al., 2018).
Circulating miRNAs profile has been analyzed from CSF, serum, plasma, peripheral blood mononuclear cells (PBMCs), and saliva collected from PD patients (summarized in Table 2; Gui et al., 2015; Ding et al., 2016; Ma et al., 2016; Cao et al., 2017; Li et al., 2017; Marques et al., 2017; Schwienbacher et al., 2017; Caggiu et al., 2018; Chen et al., 2018; Yang et al., 2019b; Cressatti et al., 2020; Ozdilek and Demircan, 2020; Xie et al., 2020). These studies have identified numerous miRNAs as potential PD biomarkers. In an example, the expression of miR-144-5p, miR-200a-3p, and miR-542-3p that were dysregulated in A53T mutant α-synuclein transgenic mice, were also shown to be up-regulated in the CSF of PD patients (Mo et al., 2017). Additionally, the reduced CSF miR-626 and elevated plasma miR-105-5p levels may be able to be used as a biomarker to differentiate PD from other neurodegenerative diseases, for example, Alzheimer’s disease (Yang et al., 2019a; Qin et al., 2021). Furthermore, apart from identifying PD, circulating miRNAs such as miR-29a, miR-29c, miR-30c-5p, miR-132-3p, miR-146a-5p, and miR-373 were found to be correlated with the severity of PD (Bai et al., 2017; Shu et al., 2020; Zhang L. et al., 2020).
The accurate diagnosis of PD is difficult, especially in its early stages. PD is normally diagnosed with the presence of motor symptoms in which loss of more than 50% DA neurons were reported in PD patients (Kordower et al., 2013). Early detection and monitoring of PD would ensure that the available treatment could potentially slow down the progression of the disease. Dos Santos et al. (2018) demonstrated that CSF obtained from patients in the early stage of PD express increased Let-7f-5p level and reduced miR-27a-3p and miR-423-5p levels. The healthy controls, on the other hand, express high levels of miR-125a-5p and low levels of miR-151a-3p in the CSF. Alternatively, serum expression of miR-141, miR-214, miR-146b-5p, and miR-193a-3p should be reduced in the early stage of PD patients compared to the controls (Dong et al., 2016). Another recent study showed that a compound down-regulation of SRRM2 and miR-27a-3p with an up-regulation of miR-27b-3p in PBMCs of PD patients is associated with the early stage onset of the disease (Fazeli et al., 2020). Additionally, circulating miRNAs also differ between early-onset and late-onset PDs. Sulaiman et al. (2020) reported a total of 5 miRNAs were found in early-onset PD patients in which 1 miRNA was up-regulated (miR-29b-3p) and 4 miRNAs were down-regulated (miR-297, 4462, 1909-5p, and 346) and the predicted targets of these miRNAs are involved in DA synapse regulation.
Despite the promising results from previous studies showing the potential of miRNAs as PD biomarkers, there are limited overlapping miRNAs identified across different studies, even from the same biological fluid. This was observed in 7 studies analyzing serum obtained from PD patients as shown in Table 1, miR-195 was found to be up-regulated in only 2 studies, while miR-146-5p, miR-214, and miR-221 were down-regulated as demonstrated in another 2 studies, suggesting the low reproducibility of these miRNAs as a disease biomarker. To increase the specificity and reproducibility of miRNAs as potential PD biomarkers, methods of miRNAs detection in samples and adequately quality-controlled data processing need to be standardized in future studies (Wang J. et al., 2016). Additionally, to increase the accuracy of miRNAs as a biomarker, Patil et al. (2019) demonstrated the use of a combination of serum miRNAs such as miR-335-5p/miR-3613-3p, miR-335-5p/miR-6865-3p, and miR-335-5p/miR-3613-3p/miR-6865-3p that showed a high degree of discriminatory accuracy in a large cohort.
Since miRNAs can regulate multiple genes, they may serve as potential therapeutic tools for PD. MiRNAs can be used to silence up-regulated genes in PD, while anti-miRNAs can be used to restore a down-regulated target gene. To overcome the issue of instability due to enzyme degradation, consideration of miRNA-based therapeutics would require a safe and stable delivery system to protect the miRNAs from degradation and facilitate their ability to cross the blood-brain-barrier (Wen, 2016).
Previous studies have demonstrated decreased miR-124 expression in various in vivo and in vitro PD models, whereas administration of miR-124 was able to reverse the loss of DA neurons and inhibit neuroinflammation in the MPP+-treated SH-SY5Y cells, MPP+-treated MN9D cells and MPTP-treated mice (Wang H. et al., 2016; Yao et al., 2018, 2019). Polymeric nanoparticles which are stable in systemic circulation can be used to deliver miR-124 to a specific region of the brain. Rabies virus glycoprotein (RVG29)-linked miR-124-loaded polymeric nanoparticles were able to cross through the in vitro blood-brain-barrier model (Gan et al., 2019). Similarly to the in vitro study, administration of the miR-124-loaded polymeric nanoparticles into the lateral ventricle of the MPTP-administered mice was able to inhibit Mitogen activated protein kinase kinase kinase-3 (MEKK3 which plays a critical role in mediating NF-κB activation) signaling and the activation of microglia (Gan et al., 2019).
As PD is also characterized by loss of DA neurons in the SNpc, enhancing adult neuroregeneration (generation of functional neurons from adult neural precursors) could be one therapeutic approach to treat PD. In mammals, adult neurogenesis occurs throughout life in restricted brain regions, which include the dentate gyrus of the hippocampus and from the subventricular zone of the lateral ventricle (Ming and Song, 2011). miR-124 loaded nanoparticles administered to the subventricular zone cells primary culture did not affect overall cell proliferation. However, this treatment increased neuroblast proliferation while decreasing the proliferation of astrocytic-like cells, by repressing the expression of non-neuronal genes (Sox9 and Jagged1) (Saraiva et al., 2016). Intraventricular administration of miR-124 loaded nanoparticles in vivo increased the number of subventricular zone-derived neuroblasts in the striatum of 6-OHDA-treated mice. This data support the potential of miRNAs as therapeutic strategies to induce neuroregeneration and to promote brain repair (Saraiva et al., 2016).
This review reported that the dysregulation of several miRNAs has a significant role in the pathogenesis of PD. These miRNAs can be classified into those which are involved in the regulation of PD-associated genes, DA neuronal health and α-synuclein-induced neurodegeneration. Understanding the roles of these miRNAs not only shed some light on the pathogenesis of PD, but could also serve as potential therapeutic targets since regulating specific microRNAs was found to have beneficial effects in the in vitro and in vivo models of PD. Further studies are therefore essential to explore further functions of these miRNAs as well as others that are yet to be discovered in the pathogenesis and therapeutic options of PD.
SLT planned the conceptual ideas and proof outline. YHN, NHMN, MAK, and SLT wrote the manuscript. WLL, MFY, and SLT critically revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the Ministry of Higher Education Malaysia (FRGS-1-2018-SKK08-UKM-03-5) and Universiti Kebangsaan Malaysia (GGPM-2017-082).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbas, M. M., Xu, Z., and Tan, L. C. S. (2018). Epidemiology of Parkinson’s disease - East versus west. Mov. Disord. Clin. Pract. 5, 14–28. doi: 10.1002/mdc3.12568
Arrese, M., Eguchi, A., and Feldstein, A. E. (2015). Circulating microRNAs: emerging biomarkers of liver disease. Semin. Liver Dis. 35, 43–54. doi: 10.1055/s-0034-1397348
Azmin, S., Khairul Anuar, A. M., Tan, H. J., Nafisah, W. Y., Raymond, A. A., Hanita, O., et al. (2014). Nonmotor symptoms in a malaysian Parkinson’s disease population. Parkinsons Dis. 2014:472157. doi: 10.1155/2014/472157
Bai, X., Tang, Y., Yu, M., Wu, L., Liu, F., Ni, J., et al. (2017). Downregulation of blood serum microRNA 29 family in patients with Parkinson’s disease. Sci. Rep. 7:5411. doi: 10.1038/s41598-017-03887-3
Braak, H., Ghebremedhin, E., Rub, U., Bratzke, H., and Del Tredici, K. (2004). Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 318, 121–134. doi: 10.1007/s00441-004-0956-9
Briggs, C. E., Wang, Y., Kong, B., Woo, T. U., Iyer, L. K., and Sonntag, K. C. (2015). Midbrain dopamine neurons in Parkinson’s disease exhibit a dysregulated miRNA and target-gene network. Brain Res. 1618, 111–121. doi: 10.1016/j.brainres.2015.05.021
Caggiu, E., Paulus, K., Mameli, G., Arru, G., Sechi, G. P., and Sechi, L. A. (2018). Differential expression of miRNA 155 and miRNA 146a in Parkinson’s disease patients. eNeurologicalSci 13, 1–4. doi: 10.1016/j.ensci.2018.09.002
Cao, X. Y., Lu, J. M., Zhao, Z. Q., Li, M. C., Lu, T., An, X. S., et al. (2017). MicroRNA biomarkers of Parkinson’s disease in serum exosome-like microvesicles. Neurosci. Lett. 644, 94–99. doi: 10.1016/j.neulet.2017.02.045
Cardoso, A. L., Guedes, J. R., Pereira de Almeida, L., and Pedroso de Lima, M. C. (2012). miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 135, 73–88. doi: 10.1111/j.1365-2567.2011.03514.x
Chen, B., Wen, X., Jiang, H., Wang, J., Song, N., and Xie, J. (2019). Interactions between iron and α-synuclein pathology in Parkinson’s disease. Free Radic. Biol. Med. 141, 253–260. doi: 10.1016/j.freeradbiomed.2019.06.024
Chen, L., Yang, J., Lu, J., Cao, S., Zhao, Q., and Yu, Z. (2018). Identification of aberrant circulating miRNAs in Parkinson’s disease plasma samples. Brain Behav. 8:e00941. doi: 10.1002/brb3.941
Chen, Y., Gao, C., Sun, Q., Pan, H., Huang, P., Ding, J., et al. (2017). MicroRNA-4639 is a regulator of DJ-1 expression and a potential early diagnostic marker for Parkinson’s disease. Front. Aging Neurosci. 9:232. doi: 10.3389/fnagi.2017.00232
Chen, Y., Zheng, J., Su, L., Chen, F., Zhu, R., Chen, X., et al. (2020). Increased salivary microRNAs that regulate DJ-1 gene expression as potential markers for Parkinson’s disease. Front. Aging Neurosci. 12:210. doi: 10.3389/fnagi.2020.00210
Cheng, M., Liu, L., Lao, Y., Liao, W., Liao, M., Luo, X., et al. (2016). MicroRNA-181a suppresses parkin-mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler-induced apoptosis. Oncotarget 7, 42274–42287. doi: 10.18632/oncotarget.9786
Cho, H. J., Liu, G., Jin, S. M., Parisiadou, L., Xie, C., Yu, J., et al. (2013). MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 22, 608–620. doi: 10.1093/hmg/dds470
Consales, C., Cirotti, C., Filomeni, G., Panatta, M., Butera, A., Merla, C., et al. (2018). Fifty-hertz magnetic field affects the epigenetic modulation of the miR-34b/c in neuronal cells. Mol. Neurobiol. 55, 5698–5714. doi: 10.1007/s12035-017-0791-0
Coskunpinar, E., Cakmak, H. A., Kalkan, A. K., Tiryakioglu, N. O., Erturk, M., and Ongen, Z. (2016). Circulating miR-221-3p as a novel marker for early prediction of acute myocardial infarction. Gene 591, 90–96. doi: 10.1016/j.gene.2016.06.059
Cressatti, M., Juwara, L., Galindez, J. M., Velly, A. M., Nkurunziza, E. S., Marier, S., et al. (2020). Salivary microR-153 and microR-223 levels as potential diagnostic biomarkers of idiopathic Parkinson’s disease. Mov. Disord. 35, 468–477. doi: 10.1002/mds.27935
Danzer, K. M., Kranich, L. R., Ruf, W. P., Cagsal-Getkin, O., Winslow, A. R., Zhu, L., et al. (2012). Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol. Neurodegener. 7:42. doi: 10.1186/1750-1326-7-42
Dexter, D. T., and Jenner, P. (2013). Parkinson disease: from pathology to molecular disease mechanisms. Free Radic. Biol. Med. 62, 132–144. doi: 10.1016/j.freeradbiomed.2013.01.018
Di Rita, A., Maiorino, T., Bruqi, K., Volpicelli, F., Bellenchi, G. C., and Strappazzon, F. (2020). miR-218 inhibits mitochondrial clearance by targeting PRKN E3 ubiquitin ligase. Int. J. Mol. Sci. 21:355. doi: 10.3390/ijms21010355
Dickson, D. W. (2018). Neuropathology of Parkinson disease. Parkinsonism Relat. Disord. 46(Suppl. 1), S30–S33. doi: 10.1016/j.parkreldis.2017.07.033
Ding, H., Huang, Z., Chen, M., Wang, C., Chen, X., Chen, J., et al. (2016). Identification of a panel of five serum miRNAs as a biomarker for Parkinson’s disease. Parkinsonism Relat. Disord. 22, 68–73. doi: 10.1016/j.parkreldis.2015.11.014
Dong, H., Wang, C., Lu, S., Yu, C., Huang, L., Feng, W., et al. (2016). A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson’s disease. Biomarkers 21, 129–137. doi: 10.3109/1354750X.2015.1118544
Dorsey, E. R., Constantinescu, R., Thompson, J. P., Biglan, K. M., Holloway, R. G., Kieburtz, K., et al. (2007). Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 68, 384–386. doi: 10.1212/01.wnl.0000247740.47667.03
Dos Santos, M. C. T., Barreto-Sanz, M. A., Correia, B. R. S., Bell, R., Widnall, C., Perez, L. T., et al. (2018). miRNA-based signatures in cerebrospinal fluid as potential diagnostic tools for early stage Parkinson’s disease. Oncotarget 9, 17455–17465. doi: 10.18632/oncotarget.24736
Doss, M. X., and Sachinidis, A. (2019). Current challenges of iPSC-based disease modeling and therapeutic implications. Cells 8:403. doi: 10.3390/cells8050403
Doxakis, E. (2010). Post-transcriptional regulation of α-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 285, 12726–12734. doi: 10.1074/jbc.M109.086827
Faruq, O., and Vecchione, A. (2015). microRNA: diagnostic perspective. Front. Med. 2:51. doi: 10.3389/fmed.2015.00051
Fazeli, S., Motovali-Bashi, M., Peymani, M., Hashemi, M. S., Etemadifar, M., Nasr-Esfahani, M. H., et al. (2020). A compound downregulation of SRRM2 and miR-27a-3p with upregulation of miR-27b-3p in PBMCs of Parkinson’s patients is associated with the early stage onset of disease. PLoS One 15:e0240855. doi: 10.1371/journal.pone.0240855
Fernandez-Santiago, R., Merkel, A., Castellano, G., Heath, S., Raya, A., Tolosa, E., et al. (2019). Whole-genome DNA hyper-methylation in iPSC-derived dopaminergic neurons from Parkinson’s disease patients. Clin Epigenetics 11:108. doi: 10.1186/s13148-019-0701-6
Figueroa-Romero, C., Hur, J., Lunn, J. S., Paez-Colasante, X., Bender, D. E., Yung, R., et al. (2016). Expression of microRNAs in human post-mortem amyotrophic lateral sclerosis spinal cords provides insight into disease mechanisms. Mol. Cell. Neurosci. 71, 34–45. doi: 10.1016/j.mcn.2015.12.008
Fragkouli, A., and Doxakis, E. (2014). miR-7 and miR-153 protect neurons against MPP+-induced cell death via upregulation of mTOR pathway. Front. Cell. Neurosci. 8:182. doi: 10.3389/fncel.2014.00182
Fujita, K. A., Ostaszewski, M., Matsuoka, Y., Ghosh, S., Glaab, E., Trefois, C., et al. (2014). Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol. Neurobiol. 49, 88–102. doi: 10.1007/s12035-013-8489-4
Gan, L., Li, Z., Lv, Q., and Huang, W. (2019). Rabies virus glycoprotein (RVG29)-linked microRNA-124-loaded polymeric nanoparticles inhibit neuroinflammation in a Parkinson’s disease model. Int. J. Pharm. 567:118449. doi: 10.1016/j.ijpharm.2019.118449
GBD 2016 Parkinson’s Disease Collaborators (2018). Global, regional, and national burden of Parkinson’s disease, 1990-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 17, 939–953. doi: 10.1016/S1474-4422(18)30295-3
Geng, L., Zhang, T., Liu, W., and Chen, Y. (2018). miR-494-3p modulates the progression of in vitro and in vivo Parkinson’s disease models by targeting SIRT3. Neurosci. Lett. 675, 23–30. doi: 10.1016/j.neulet.2018.03.037
Goedert, M., Spillantini, M. G., Del Tredici, K., and Braak, H. (2013). 100 years of Lewy pathology. Nat. Rev. Neurol. 9, 13–24. doi: 10.1038/nrneurol.2012.242
Golpich, M., Amini, E., Mohamed, Z., Azman Ali, R., Mohamed Ibrahim, N., and Ahmadiani, A. (2017). Mitochondrial dysfunction and biogenesis in neurodegenerative diseases: pathogenesis and treatment. CNS Neurosci. Ther. 23, 5–22. doi: 10.1111/cns.12655
Gong, X., Wang, H., Ye, Y., Shu, Y., Deng, Y., He, X., et al. (2016). miR-124 regulates cell apoptosis and autophagy in dopaminergic neurons and protects them by regulating AMPK/mTOR pathway in Parkinson’s disease. Am. J. Transl. Res. 8, 2127–2137.
Gopalai, A. A., Lim, S. Y., Chua, J. Y., Tey, S., Lim, T. T., Mohamed Ibrahim, N., et al. (2014). LRRK2 G2385R and R1628P mutations are associated with an increased risk of Parkinson’s disease in the Malaysian population. Biomed. Res. Int. 2014:867321. doi: 10.1155/2014/867321
Gui, Y., Liu, H., Zhang, L., Lv, W., and Hu, X. (2015). Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 6, 37043–37053. doi: 10.18632/oncotarget.6158
Guo, J. D., Zhao, X., Li, Y., Li, G. R., and Liu, X. L. (2018). Damage to dopaminergic neurons by oxidative stress in Parkinson’s disease (Review). Int. J. Mol. Med. 41, 1817–1825. doi: 10.3892/ijmm.2018.3406
Guo, Y., and Hua, X. (2020). MicroRNA-181a-5p down-regulation presents neuroprotective effect in 1-methyl-4-phenylpyridinium-induced Parkinson’s disease: an in vitro study. J. Biomat. Tissue Eng. 10, 63–70. doi: 10.1166/jbt.2020.2219
Hegarty, S. V., Sullivan, A. M., and O’Keeffe, G. W. (2018). Inhibition of miR-181a promotes midbrain neuronal growth through a Smad1/5-dependent mechanism: implications for Parkinson’s disease. Neuronal Signal. 2:NS20170181. doi: 10.1042/NS20170181
Hoss, A. G., Labadorf, A., Beach, T. G., Latourelle, J. C., and Myers, R. H. (2016). microRNA profiles in Parkinson’s disease prefrontal cortex. Front. Aging Neurosci. 8:36. doi: 10.3389/fnagi.2016.00036
Huttenhofer, A., and Mayer, G. (2017). Circulating miRNAs as biomarkers of kidney disease. Clin. Kidney J. 10, 27–29. doi: 10.1093/ckj/sfw075
Ibrahim, F. F., Jamal, R., Syafruddin, S. E., Ab Mutalib, N. S., Saidin, S., MdZin, R. R., et al. (2015). MicroRNA-200c and microRNA-31 regulate proliferation, colony formation, migration and invasion in serous ovarian cancer. J. Ovarian Res. 8:56. doi: 10.1186/s13048-015-0186-7
Irrcher, I., Aleyasin, H., Seifert, E. L., Hewitt, S. J., Chhabra, S., Phillips, M., et al. (2010). Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 19, 3734–3746. doi: 10.1093/hmg/ddq288
Irwin, D. J., Lee, V. M., and Trojanowski, J. Q. (2013). Parkinson’s disease dementia: convergence of α-synuclein, tau and amyloid-β pathologies. Nat. Rev. Neurosci. 14, 626–636. doi: 10.1038/nrn3549
Jauhari, A., Singh, T., Mishra, S., Shankar, J., and Yadav, S. (2020). Coordinated action of miR-146a and Parkin gene regulate rotenone-induced neurodegeneration. Toxicol. Sci. 176, 433–445. doi: 10.1093/toxsci/kfaa066
Je, G., and Kim, Y. S. (2017). Mitochondrial ROS-mediated post-transcriptional regulation of α-synuclein through miR-7 and miR-153. Neurosci. Lett. 661, 132–136. doi: 10.1016/j.neulet.2017.09.065
Jellinger, K. A. (2015). Neuropathobiology of non-motor symptoms in Parkinson disease. J. Neural Transm. 122, 1429–1440. doi: 10.1007/s00702-015-1405-5
Jellinger, K. A. (2017a). “Neuropathology of movement disorders,” in Movement Disorders Curricula, eds C. Falup-Pecurariu, J. Ferreira, and P. K. C. Martinez-Martin (Vienna: Springer), 43–48. doi: 10.1007/978-3-7091-1628-9_5
Jellinger, K. A. (2017b). Neuropathology of nonmotor symptoms of Parkinson’s disease. Int. Rev. Neurobiol. 133, 13–62. doi: 10.1016/bs.irn.2017.05.005
Jiang, J., Xiang, R., Lu, J., Wang, X., and Gu, X. (2020). MicroRNA-203a-3p regulates the apoptosis of MPP+ Induced SH-SY5Y cells by targeting A-synuclein. J. Biomat. Tissue Eng. 10, 838–844. doi: 10.1166/jbt.2020.2320
Jiang, P., Gan, M., Yen, S. H., Moussaud, S., McLean, P. J., and Dickson, D. W. (2016). Proaggregant nuclear factor(s) trigger rapid formation of α-synuclein aggregates in apoptotic neurons. Acta Neuropathol. 132, 77–91. doi: 10.1007/s00401-016-1542-4
Kabaria, S., Choi, D. C., Chaudhuri, A. D., Mouradian, M. M., and Junn, E. (2015). Inhibition of miR-34b and miR-34c enhances α-synuclein expression in Parkinson’s disease. FEBS Lett. 589, 319–325. doi: 10.1016/j.febslet.2014.12.014
Kanagaraj, N., Beiping, H., Dheen, S. T., and Tay, S. S. (2014). Downregulation of miR-124 in MPTP-treated mouse model of Parkinson’s disease and MPP iodide-treated MN9D cells modulates the expression of the calpain/cdk5 pathway proteins. Neuroscience 272, 167–179. doi: 10.1016/j.neuroscience.2014.04.039
Khee, S. G., Yusof, Y. A., and Makpol, S. (2014). Expression of senescence-associated microRNAs and target genes in cellular aging and modulation by tocotrienol-rich fraction. Oxid. Med. Cell. Longev. 2014:725929. doi: 10.1155/2014/725929
Kim, J., Fiesel, F. C., Belmonte, K. C., Hudec, R., Wang, W. X., Kim, C., et al. (2016). miR-27a and miR-27b regulate autophagic clearance of damaged mitochondria by targeting PTEN-induced putative kinase 1 (PINK1). Mol. Neurodegener. 11:55. doi: 10.1186/s13024-016-0121-4
Kim, J. H., Jou, I., and Joe, E. H. (2014). Suppression of miR-155 expression in IFN-γ-treated astrocytes and microglia by DJ-1: a possible mechanism for maintaining SOCS1 expression. Exp. Neurobiol. 23, 148–154. doi: 10.5607/en.2014.23.2.148
Kim, S. Y., Seong, M. W., Jeon, B. S., Kim, S. Y., Ko, H. S., Kim, J. Y., et al. (2012). Phase analysis identifies compound heterozygous deletions of the PARK2 gene in patients with early-onset Parkinson disease. Clin. Genet. 82, 77–82. doi: 10.1111/j.1399-0004.2011.01693.x
Kim, W., Lee, Y., McKenna, N. D., Yi, M., Simunovic, F., Wang, Y., et al. (2014). miR-126 contributes to Parkinson’s disease by dysregulating the insulin-like growth factor/phosphoinositide 3-kinase signaling. Neurobiol. Aging 35, 1712–1721. doi: 10.1016/j.neurobiolaging.2014.01.021
Kim, W., Noh, H., Lee, Y., Jeon, J., Shanmugavadivu, A., McPhie, D. L., et al. (2016). MiR-126 regulates growth factor activities and vulnerability to toxic insult in neurons. Mol. Neurobiol. 53, 95–108. doi: 10.1007/s12035-014-8989-x
Kordower, J. H., Olanow, C. W., Dodiya, H. B., Chu, Y., Beach, T. G., Adler, C. H., et al. (2013). Disease duration and the integrity of the nigrostriatal system in Parkinson’s disease. Brain 136(Pt 8), 2419–2431. doi: 10.1093/brain/awt192
Kozomara, A., Birgaoanu, M., and Griffiths-Jones, S. (2019). miRBase: from microRNA sequences to function. Nucleic Acids Res. 47, D155–D162. doi: 10.1093/nar/gky1141
Krohn, L., Grenn, F. P., Makarious, M. B., Kim, J. J., Bandres-Ciga, S., Roosen, D. A., et al. (2020). Comprehensive assessment of PINK1 variants in Parkinson’s disease. Neurobiol. Aging 91, 168.e1–168.e5. doi: 10.1016/j.neurobiolaging.2020.03.003
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854. doi: 10.1016/0092-8674(93)90529-Y
Lee, S. T., Chu, K., Im, W. S., Yoon, H. J., Im, J. Y., Park, J. E., et al. (2011). Altered microRNA regulation in Huntington’s disease models. Exp. Neurol. 227, 172–179. doi: 10.1016/j.expneurol.2010.10.012
Lesage, S., Lunati, A., Houot, M., Romdhan, S. B., Clot, F., Tesson, C., et al. (2020). Characterization of recessive Parkinson disease in a large multicenter study. Ann. Neurol. 88, 843–850. doi: 10.1002/ana.25787
Li, L., Xu, J., Wu, M., and Hu, J. M. (2018). Protective role of microRNA-221 in Parkinson’s disease. Bratislava Med. J. 119, 22–27. doi: 10.4149/BLL_2018_005
Li, N., Pan, X., Zhang, J., Ma, A., Yang, S., Ma, J., et al. (2017). Plasma levels of miR-137 and miR-124 are associated with Parkinson’s disease but not with Parkinson’s disease with depression. Neurol. Sci. 38, 761–767. doi: 10.1007/s10072-017-2841-9
Liang, C., Mu, Y., Tian, H., Wang, D., Zhang, S., Wang, H., et al. (2021). MicroRNA-140 silencing represses the incidence of Alzheimer’s disease. Neurosci. Lett. 135674. doi: 10.1016/j.neulet.2021.135674 [Epub ahead of print].
Liu, W., Zhang, Q., Zhang, J., Pan, W., Zhao, J., and Xu, Y. (2017). Long non-coding RNA MALAT1 contributes to cell apoptosis by sponging miR-124 in Parkinson disease. Cell Biosci. 7:19. doi: 10.1186/s13578-017-0147-5
Ma, W., Li, Y., Wang, C., Xu, F., Wang, M., and Liu, Y. (2016). Serum miR-221 serves as a biomarker for Parkinson’s disease. Cell Biochem. Funct. 34, 511–515. doi: 10.1002/cbf.3224
Ma, X., Zhang, H., Yin, H., Geng, S., Liu, Y., Liu, C., et al. (2021). Up-regulated microRNA-218-5p ameliorates the damage of dopaminergic neurons in rats with Parkinson’s disease via suppression of LASP1. Brain Res. Bull. 166, 92–101. doi: 10.1016/j.brainresbull.2020.10.019
Magrinelli, F., Picelli, A., Tocco, P., Federico, A., Roncari, L., Smania, N., et al. (2016). Pathophysiology of motor dysfunction in Parkinson’s disease as the rationale for drug treatment and rehabilitation. Parkinsons Dis. 2016:9832839. doi: 10.1155/2016/9832839
Maiti, P., Manna, J., and Dunbar, G. L. (2017). Current understanding of the molecular mechanisms in Parkinson’s disease: targets for potential treatments. Transl. Neurodegen. 6:28. doi: 10.1186/s40035-017-0099-z
Makarova, J. A., Shkurnikov, M. U., Turchinovich, A. A., Tonevitsky, A. G., and Grigoriev, A. I. (2015). Circulating microRNAs. Biochemistry 80, 1117–1126. doi: 10.1134/S0006297915090035
Marques, T. M., Kuiperij, H. B., Bruinsma, I. B., van Rumund, A., Aerts, M. B., Esselink, R. A. J., et al. (2017). MicroRNAs in cerebrospinal fluid as potential biomarkers for Parkinson’s disease and multiple system atrophy. Mol. Neurobiol. 54, 7736–7745. doi: 10.1007/s12035-016-0253-0
McMillan, K. J., Murray, T. K., Bengoa-Vergniory, N., Cordero-Llana, O., Cooper, J., Buckley, A., et al. (2017). Loss of microRNA-7 regulation leads to α-synuclein accumulation and dopaminergic neuronal loss in vivo. Mol. Ther. 25, 2404–2414. doi: 10.1016/j.ymthe.2017.08.017
Ming, G. L., and Song, H. (2011). Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron 70, 687–702. doi: 10.1016/j.neuron.2011.05.001
Minones-Moyano, E., Porta, S., Escaramis, G., Rabionet, R., Iraola, S., Kagerbauer, B., et al. (2011). MicroRNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum. Mol. Genet. 20, 3067–3078. doi: 10.1093/hmg/ddr210
Mo, M., Xiao, Y., Huang, S., Cen, L., Chen, X., Zhang, L., et al. (2017). MicroRNA expressing profiles in A53T mutant alpha-synuclein transgenic mice and Parkinsonian. Oncotarget 8, 15–28. doi: 10.18632/oncotarget.13905
Nair, V. D., and Ge, Y. (2016). Alterations of miRNAs reveal a dysregulated molecular regulatory network in Parkinson’s disease striatum. Neurosci. Lett. 629, 99–104. doi: 10.1016/j.neulet.2016.06.061
Najib, N. H. M., Nies, Y. H., Abd Halim, S. A. S., Yahaya, M. F., Das, S., Lim, W. L., et al. (2020). Modeling Parkinson’s disease in zebrafish. CNS Neurol. Disord. Drug Targets 19, 386–399. doi: 10.2174/1871527319666200708124117
O’Brien, J., Hayder, H., Zayed, Y., and Peng, C. (2018). Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 9:402. doi: 10.3389/fendo.2018.00402
Oh, S. E., Park, H. J., He, L., Skibiel, C., Junn, E., and Mouradian, M. M. (2018). The Parkinson’s disease gene product DJ-1 modulates miR-221 to promote neuronal survival against oxidative stress. Redox Biol. 19, 62–73. doi: 10.1016/j.redox.2018.07.021
Ozdilek, B., and Demircan, B. (2020). Serum microRNA expression levels in Turkish patients with Parkinson’s disease. Int. J. Neurosci. 1–9. doi: 10.1080/00207454.2020.1784165
Pang, S. Y., Ho, P. W., Liu, H. F., Leung, C. T., Li, L., Chang, E. E. S., et al. (2019). The interplay of aging, genetics and environmental factors in the pathogenesis of Parkinson’s disease. Transl. Neurodegen. 8:23. doi: 10.1186/s40035-019-0165-9
Patil, K. S., Basak, I., Dalen, I., Hoedt, E., Lange, J., Lunde, K. A., et al. (2019). Combinatory microRNA serum signatures as classifiers of Parkinson’s disease. Parkinsonism Relat. Disord. 64, 202–210. doi: 10.1016/j.parkreldis.2019.04.010
Pattarayan, D., Thimmulappa, R. K., Ravikumar, V., and Rajasekaran, S. (2018). Diagnostic potential of extracellular microRNA in respiratory diseases. Clin. Rev. Allergy Immunol. 54, 480–492. doi: 10.1007/s12016-016-8589-9
Peball, M., Krismer, F., Knaus, H. G., Djamshidian, A., Werkmann, M., Carbone, F., et al. (2020). Non-motor symptoms in Parkinson’s disease are reduced by nabilone. Ann. Neurol. 88, 712–722. doi: 10.1002/ana.25864
Prajapati, P., Sripada, L., Singh, K., Bhatelia, K., Singh, R., and Singh, R. (2015). TNF-α regulates miRNA targeting mitochondrial complex-I and induces cell death in dopaminergic cells. Biochim. Biophys. Acta 1852, 451–461. doi: 10.1016/j.bbadis.2014.11.019
Pringsheim, T., Jette, N., Frolkis, A., and Steeves, T. D. (2014). The prevalence of Parkinson’s disease: a systematic review and meta-analysis. Mov. Disord. 29, 1583–1590. doi: 10.1002/mds.25945
Putteeraj, M., Fairuz, Y. M., and Teoh, S. L. (2017). MicroRNA dysregulation in Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 16, 1000–1009. doi: 10.2174/1871527316666170807142311
Qin, L. X., Tan, J. Q., Zhang, H. N., Tang, J. G., Jiang, B., Shen, X. M., et al. (2021). Preliminary study of hsa-mir-626 change in the cerebrospinal fluid in Parkinson’s disease. Neurol India 69, 115–118. doi: 10.4103/0028-3886.310102
Rango, M., Dossi, G., Squarcina, L., and Bonifati, C. (2020). Brain mitochondrial impairment in early-onset Parkinson’s disease with or without PINK1 mutation. Mov. Disord. 35, 504–507. doi: 10.1002/mds.27946
Salimian, N., Peymani, M., Ghaedi, K., and Nasr Esfahani, M. H. (2018). Modulation in miR-200a/SIRT1axis is associated with apoptosis in MPP+-induced SH-SY5Y cells. Gene 674, 25–30. doi: 10.1016/j.gene.2018.06.061
Saraiva, C., Paiva, J., Santos, T., Ferreira, L., and Bernardino, L. (2016). MicroRNA-124 loaded nanoparticles enhance brain repair in Parkinson’s disease. J. Control. Release 235, 291–305. doi: 10.1016/j.jconrel.2016.06.005
Savica, R., Grossardt, B. R., Bower, J. H., Ahlskog, J. E., and Rocca, W. A. (2016). Time trends in the incidence of Parkinson disease. JAMA Neurol. 73, 981–989. doi: 10.1001/jamaneurol.2016.0947
Schwienbacher, C., Foco, L., Picard, A., Corradi, E., Serafin, A., Panzer, J., et al. (2017). Plasma and white blood cells show different miRNA expression profiles in Parkinson’s disease. J. Mol. Neurosci. 62, 244–254. doi: 10.1007/s12031-017-0926-9
Seidel, K., Mahlke, J., Siswanto, S., Kruger, R., Heinsen, H., Auburger, G., et al. (2015). The brainstem pathologies of Parkinson’s disease and dementia with Lewy bodies. Brain Pathol. 25, 121–135. doi: 10.1111/bpa.12168
Shen, Y. F., Zhu, Z. Y., Qian, S. X., Xu, C. Y., and Wang, Y. P. (2020). miR-30b protects nigrostriatal dopaminergic neurons from MPP(+)-induced neurotoxicity via SNCA. Brain Behav. 10:e01567. doi: 10.1002/brb3.1567
Shu, Y., Qian, J., and Wang, C. (2020). Aberrant expression of microRNA-132-3p and microRNA-146a-5p in Parkinson’s disease patients. Open Life Sci. 15, 647–653. doi: 10.1515/biol-2020-0060
Sulaiman, S. A., Ainaa Muhsin, N. I., Rasyadan Arshad, A., Wan Mohamad Nazarie, W. F., Jamal, R., Mohamed Ibrahim, N., et al. (2020). Differential expression of circulating miRNAs in Parkinson’s disease patients: potential early biomarker? Neurol. Asia 25, 319–329.
Talepoor Ardakani, M., Rostamian Delavar, M., Baghi, M., Nasr-Esfahani, M. H., Kiani-Esfahani, A., and Ghaedi, K. (2019). Upregulation of miR-200a and miR-204 in MPP+ -treated differentiated PC12 cells as a model of Parkinson’s disease. Mol. Genet. Genomic Med. 7:e548. doi: 10.1002/mgg3.548
Tarale, P., Daiwile, A. P., Sivanesan, S., Stoger, R., Bafana, A., Naoghare, P. K., et al. (2018). Manganese exposure: linking down-regulation of miRNA-7 and miRNA-433 with α-synuclein overexpression and risk of idiopathic Parkinson’s disease. Toxicol. In Vitro 46, 94–101. doi: 10.1016/j.tiv.2017.10.003
Teoh, S. L., and Das, S. (2017). The role of microRNAs in diagnosis, prognosis, metastasis and resistant cases in breast cancer. Curr. Pharm. Des. 23, 1845–1859. doi: 10.2174/1381612822666161027120043
Thome, A. D., Harms, A. S., Volpicelli-Daley, L. A., and Standaert, D. G. (2016). microRNA-155 regulates alpha-synuclein-induced inflammatory responses in models of Parkinson disease. J. Neurosci. 36, 2383–2390. doi: 10.1523/JNEUROSCI.3900-15.2016
Tian, Y. S., Zhong, D., Liu, Q. Q., Zhao, X. L., Sun, H. X., Jin, J., et al. (2018). Upregulation of miR-216a exerts neuroprotective effects against ischemic injury through negatively regulating JAK2/STAT3-involved apoptosis and inflammatory pathways. J. Neurosurg. 130, 977–988. doi: 10.3171/2017.5.JNS163165
Tiberio, P., Callari, M., Angeloni, V., Daidone, M. G., and Appierto, V. (2015). Challenges in using circulating miRNAs as cancer biomarkers. Biomed. Res. Int. 2015:731479. doi: 10.1155/2015/731479
Tolosa, E., Botta-Orfila, T., Morato, X., Calatayud, C., Ferrer-Lorente, R., Marti, M. J., et al. (2018). MicroRNA alterations in iPSC-derived dopaminergic neurons from Parkinson disease patients. Neurobiol. Aging 69, 283–291. doi: 10.1016/j.neurobiolaging.2018.05.032
Tolosa, E., Vila, M., Klein, C., and Rascol, O. (2020). LRRK2 in Parkinson disease: challenges of clinical trials. Nat. Rev. Neurol. 16, 97–107. doi: 10.1038/s41582-019-0301-2
Turchinovich, A., Weiz, L., and Burwinkel, B. (2012). Extracellular miRNAs: the mystery of their origin and function. Trends Biochem. Sci. 37, 460–465. doi: 10.1016/j.tibs.2012.08.003
Villar-Menendez, I., Porta, S., Buira, S. P., Pereira-Veiga, T., Diaz-Sanchez, S., Albasanz, J. L., et al. (2014). Increased striatal adenosine A2A receptor levels is an early event in Parkinson’s disease-related pathology and it is potentially regulated by miR-34b. Neurobiol. Dis. 69, 206–214. doi: 10.1016/j.nbd.2014.05.030
Wang, H., Ye, Y., Zhu, Z., Mo, L., Lin, C., Wang, Q., et al. (2016). MiR-124 regulates apoptosis and autophagy process in MPTP model of Parkinson’s disease by targeting to Bim. Brain Pathol. 26, 167–176. doi: 10.1111/bpa.12267
Wang, J., Chen, J., and Sen, S. (2016). MicroRNA as biomarkers and diagnostics. J. Cell. Physiol. 231, 25–30. doi: 10.1002/jcp.25056
Wang, R., Li, Q., He, Y., Yang, Y., Ma, Q., and Li, C. (2020a). miR-29c-3p inhibits microglial NLRP3 inflammasome activation by targeting NFAT5 in Parkinson’s disease. Genes Cells 25, 364–374. doi: 10.1111/gtc.12764
Wang, R., Yang, Y., Wang, H., He, Y., and Li, C. (2020b). MiR-29c protects against inflammation and apoptosis in Parkinson’s disease model in vivo and in vitro by targeting SP1. Clin. Exp. Pharmacol. Physiol. 47, 372–382. doi: 10.1111/1440-1681.13212
Wang, Y., Yang, Z., and Le, W. (2017). Tiny but mighty: promising roles of microRNAs in the diagnosis and treatment of Parkinson’s disease. Neurosci. Bull. 33, 543–551. doi: 10.1007/s12264-017-0160-z
Wang, Z. H., Zhang, J. L., Duan, Y. L., Zhang, Q. S., Li, G. F., and Zheng, D. L. (2015). MicroRNA-214 participates in the neuroprotective effect of Resveratrol via inhibiting α-synuclein expression in MPTP-induced Parkinson’s disease mouse. Biomed. Pharmacother. 74, 252–256. doi: 10.1016/j.biopha.2015.08.025
Wen, M. M. (2016). Getting miRNA therapeutics into the target cells for neurodegenerative diseases: a mini-review. Front. Mol. Neurosci. 9:129. doi: 10.3389/fnmol.2016.00129
Wightman, B., Ha, I., and Ruvkun, G. (1993). Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862. doi: 10.1016/0092-8674(93)90530-4
Wu, D. M., Wang, S., Wen, X., Han, X. R., Wang, Y. J., Shen, M., et al. (2018). Inhibition of microRNA-200a upregulates the expression of striatal dopamine receptor D2 to repress apoptosis of striatum via the cAMP/PKA signaling pathway in rats with Parkinson’s disease. Cell. Physiol. Biochem. 51, 1600–1615. doi: 10.1159/000495649
Wu, Q., Xi, D. Z., and Wang, Y. H. (2019). MicroRNA-599 regulates the development of Parkinson’s disease through mediating LRRK2 expression. Eur. Rev. Med. Pharmacol. Sci. 23, 724–731. doi: 10.26355/eurrev_201901_16886
Xie, S., Niu, W., Xu, F., Wang, Y., Hu, S., and Niu, C. (2020). Differential expression and significance of miRNAs in plasma extracellular vesicles of patients with Parkinson’s disease. Int. J. Neurosci 1–16. doi: 10.1080/00207454.2020.1835899 [Epub ahead of print].
Xing, R. X., Li, L. G., Liu, X. W., Tian, B. X., and Cheng, Y. (2020). Down regulation of miR-218, miR-124, and miR-144 relates to Parkinson’s disease via activating NF-κB signaling. Kaohsiung J. Med. Sci. 36, 786–792. doi: 10.1002/kjm2.12241
Xiong, R., Wang, Z., Zhao, Z., Li, H., Chen, W., Zhang, B., et al. (2014). MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration. Neurobiol. Aging 35, 705–714. doi: 10.1016/j.neurobiolaging.2013.09.027
Yang, X., Zhang, M., Wei, M., Wang, A., Deng, Y., and Cao, H. (2020). MicroRNA-216a inhibits neuronal apoptosis in a cellular Parkinson’s disease model by targeting Bax. Metab. Brain Dis. 35, 627–635. doi: 10.1007/s11011-020-00546-x
Yang, Z., Li, T., Cui, Y., Li, S., Cheng, C., Shen, B., et al. (2019a). Elevated plasma microRNA-105-5p level in patients with idiopathic Parkinson’s disease: a potential disease biomarker. Front. Neurosci. 13:218. doi: 10.3389/fnins.2019.00218
Yang, Z., Li, T., Li, S., Wei, M., Qi, H., Shen, B., et al. (2019b). Altered expression levels of microRNA-132 and Nurr1 in peripheral blood of Parkinson’s disease: potential disease biomarkers. ACS Chem. Neurosci. 10, 2243–2249. doi: 10.1021/acschemneuro.8b00460
Yao, L., Ye, Y., Mao, H., Lu, F., He, X., Lu, G., et al. (2018). MicroRNA-124 regulates the expression of MEKK3 in the inflammatory pathogenesis of Parkinson’s disease. J. Neuroinflammation 15:13. doi: 10.1186/s12974-018-1053-4
Yao, L., Zhu, Z., Wu, J., Zhang, Y., Zhang, H., Sun, X., et al. (2019). MicroRNA-124 regulates the expression of p62/p38 and promotes autophagy in the inflammatory pathogenesis of Parkinson’s disease. FASEB J. 33, 8648–8665. doi: 10.1096/fj.201900363R
Zanellati, M. C., Monti, V., Barzaghi, C., Reale, C., Nardocci, N., Albanese, A., et al. (2015). Mitochondrial dysfunction in Parkinson disease: evidence in mutant PARK2 fibroblasts. Front. Genet. 6:78. doi: 10.3389/fgene.2015.00078
Zeng, R., Luo, D. X., Li, H. P., Zhang, Q. S., Lei, S. S., and Chen, J. H. (2019). MicroRNA-135b alleviates MPP+-mediated Parkinson’s disease in in vitro model through suppressing FoxO1-induced NLRP3 inflammasome and pyroptosis. J. Clin. Neurosci. 65, 125–133. doi: 10.1016/j.jocn.2019.04.004
Zhang, G., Chen, L., Liu, J., Jin, Y., Lin, Z., Du, S., et al. (2020). HIF-1α/microRNA-128-3p axis protects hippocampal neurons from apoptosis via the Axin1-mediated Wnt/β-catenin signaling pathway in Parkinson’s disease models. Aging 12, 4067–4081. doi: 10.18632/aging.102636
Zhang, J., Liu, W., Wang, Y., Zhao, S., and Chang, N. (2017). miR-135b plays a neuroprotective role by targeting GSK3β in MPP+-intoxicated SH-SY5Y cells. Dis. Markers 2017:5806146. doi: 10.1155/2017/5806146
Zhang, L., Zhang, J., Wang, K., and Wang, R. (2020). Serum microRNA-30c-5p and microRNA-373 expressions as potential biomarkers for Parkinson’s disease. All Life 13, 194–200. doi: 10.1080/26895293.2020.1741453
Zhang, Q., Huang, X. M., Liao, J. X., Dong, Y. K., Zhu, J. L., He, C. C., et al. (2020). LncRNA HOTAIR promotes neuronal damage through facilitating NLRP3 mediated-pyroptosis activation in Parkinson’s disease via regulation of miR-326/ELAVL1 Axis. Cell. Mol. Neurobiol. doi: 10.1007/s10571-020-00946-8 [Epub ahead of print].
Zhang, Y., Xu, W., Nan, S., Chang, M., and Fan, J. (2019). MicroRNA-326 inhibits apoptosis and promotes proliferation of dopaminergic neurons in Parkinson’s disease through suppression of KLK7-mediated MAPK signaling pathway. J. Mol. Neurosci. 69, 197–214. doi: 10.1007/s12031-019-01349-1
Zhao, X. H., Wang, Y. B., Yang, J., Liu, H. Q., and Wang, L. L. (2019). MicroRNA-326 suppresses iNOS expression and promotes autophagy of dopaminergic neurons through the JNK signaling by targeting XBP1 in a mouse model of Parkinson’s disease. J. Cell. Biochem. 120, 14995–15006. doi: 10.1002/jcb.28761
Zhou, J., Zhao, Y., Li, Z., Zhu, M., Wang, Z., Li, Y., et al. (2020). miR-103a-3p regulates mitophagy in Parkinson’s disease through Parkin/Ambra1 signaling. Pharmacol. Res. 160:105197. doi: 10.1016/j.phrs.2020.105197
Zhou, L., Yang, L., Li, Y. J., Mei, R., Yu, H. L., Gong, Y., et al. (2018). MicroRNA-128 protects dopamine neurons from apoptosis and upregulates the expression of excitatory amino acid transporter 4 in Parkinson’s disease by binding to AXIN1. Cell. Physiol. Biochem. 51, 2275–2289. doi: 10.1159/000495872
Keywords: microRNA, non-coding RNA, Parkinson’s disease, neurodegenerative disease, biomarker
Citation: Nies YH, Mohamad Najib NH, Lim WL, Kamaruzzaman MA, Yahaya MF and Teoh SL (2021) MicroRNA Dysregulation in Parkinson’s Disease: A Narrative Review. Front. Neurosci. 15:660379. doi: 10.3389/fnins.2021.660379
Received: 29 January 2021; Accepted: 09 April 2021;
Published: 30 April 2021.
Edited by:
Victor Tapias, Weill Cornell Medicine, United StatesReviewed by:
Gerard O’Keeffe, University College Cork, IrelandCopyright © 2021 Nies, Mohamad Najib, Lim, Kamaruzzaman, Yahaya and Teoh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seong Lin Teoh, dGVvaHNlb25nbGluQHlhaG9vLmNvbS5teQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.