
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci., 16 March 2021
Sec. Neuroendocrine Science
Volume 15 - 2021 | https://doi.org/10.3389/fnins.2021.608102
Fetal alcohol exposure (FAE) causes various neurodevelopmental deficits in offspring, including reduced expression of the stress regulatory proopiomelanocortin (Pomc) gene and an elevated stress response for multiple generations via the male germline. Male germline-specific effects of FAE on the Pomc gene raises the question if the sex-determining region Y (SRY) may have a role in regulating Pomc gene expression. Using a transgenerational model of FAE in Fischer 344 rats, we determined the role of SRY in the regulation of the Pomc gene. FAEs, like on the Pomc gene, reduced Sry gene expression in sperm and the mediobasal hypothalamus (MBH) in male adult offspring. Fetal alcohol-induced inhibition of Sry gene expression was associated with increased Sry promoter DNA methylation. Additionally, fetal alcohol effects on the Sry gene persisted for three generations in the male germline but not in the female germline. Sry gene knockdown reduced the Pomc gene expression. Sry recruitment onto the Pomc promoter was found to be reduced in the hypothalamus of fetal alcohol-exposed rats compared to control rats. Pomc promoter luciferase activity was increased following Sry overexpression. A site-directed mutagenesis study revealed that SRY binding sites are required for POMC promoter transcription activity. Overall, these findings suggest that SRY plays a stimulatory role in the regulation of Pomc gene expression and may potentially contribute to the fetal alcohol-induced changes in the level of Pomc gene expression for multiple generations.
Fetal alcohol exposure (FAE) has been shown to elevate neuroendocrine response of the hypothalamic-pituitary-adrenal (HPA) axis to stress (Ogilvie and Rivier, 1997; Hellemans et al., 2010; Bakoyiannis et al., 2014; Sarkar et al., 2019), partly due to a lack of the inhibitory control of the proopiomelanocortin (POMC)-derived peptide in the hypothalamus (Sarkar et al., 2007; Wynne and Sarkar, 2013). Additionally, it has been shown that replenishment of POMC neurons by neuronal transplants in the hypothalamus prevents stress hyperresponse in fetal alcohol-exposed rats (Boyadjieva et al., 2009), suggesting that FAE suppresses the level of POMC to alter the function of the HPA axis. The FAE effect on POMC involves epigenetic modification of Pomc DNA methylation via recruitment of MeCP2 on Pomc promoter (Govorko et al., 2012; Gangisetty et al., 2014). Interestingly, FAE-induced changes in Pomc promoter DNA methylation bypasses epigenetic reprogramming during embryonic development and are transmitted for multiple generations only through the male—but not female germline (Govorko et al., 2012; Sarkar, 2016), suggesting the possibility of involvement of a male-specific chromosome/gene which overcomes developmental editing and regulates Pomc gene expression. Paternal transmission of the Y chromosome can trigger testicular differentiation from bipotential gonads in mammalian embryos. The non-pairing region of the Y chromosome (YNPAR) is transmitted from father to sons (Eyre-Walker, 1993) and has been proposed to bypass developmental molecular programming and therefore participates in the male-specific transgenerational transfer of the epigenetic mark (Li et al., 2002). The sex-determining region Y (SRY) is one of the functional genes involved in male reproduction that is mapped on YNPAR. It has been proposed that β-endorphin, a functional peptide of POMC, is regulated by YNPAR that comprises SRY (Botbol et al., 2011). SRY is a transcription factor involved in regulating several genes in male sex determination (Sekido and Lovell-Badge, 2008; Bhandari et al., 2011). SRY also regulates endocrine response to stress (Dickey et al., 2012). SRY binding sites were identified to be altered transgenerationally in the Sertoli cell of the testis in vinclozolin-treated male offspring (Skinner et al., 2015). We hypothesize that SRY may have a role in regulation of Pomc gene expression and may contribute to the transgenerational effects of FAE on Pomc. We determined FAE effects on Pomc and Sry expression in the mediobasal hypothalamus (MBH) and sperm. We also conducted Sry gene knockdown experiments in vitro and in vivo and determined its effect on Pomc expression. Additionally, SRY recruitment onto the Pomc promoter was assessed in MBH of FAE rats. Furthermore, we determined SRY binding motifs on Pomc promoter and evaluated SRY regulation through its binding sites using promoter luciferase activity. The results of these studies are presented here.
All rat studies were performed with approved protocols in compliance with the Association for the Assessment and Accreditation of Laboratory Animal Care and Rutgers University. Fisher 344 strain rats were obtained from Harlan Laboratories (Indianapolis, IN) and housed in a controlled condition at a constant temperature of 22°C and 12-h light/dark cycles throughout the study. These rats were bred in our animal facility and used for this study. On gestational day (GD) 7 through 21, rats were fed rat chow ad libitum (AD), a liquid diet containing ethanol (AF; 1.7–5.0% v/v from GD7-10 and 6.7% v/v from GD11-21; Bioserve, Inc., Frenchtown, NJ), or pair-fed (PF; Bioserve) an isocaloric liquid control diet (with alcohol calories replaced by maltose-dextrin). Previous studies have shown that the peak blood ethanol concentration is achieved in the range of 120–150 mg/dl in pregnant dams fed with this ethanol-containing liquid diet (Miller, 1992). AF and PF litters were cross-fostered, and the litter size was maintained at 8 pups/dam. Only one pup from each litter was used in an experimental measure. The male offspring animals were used for the study and were sacrificed at the age of 2–3 months. Transgenerational studies were conducted by breeding AF, PF, or AD rats with control animals of the opposite gender to produce two germlines. The male germline (AFM or PFM) was generated by breeding male (AF or PF) rats and their male offspring with control (AD) females, and the female germline (AFF or PFF) was generated by breeding female (AF or PF) rats and their female offspring with control (AD) males. All rats were sacrificed at 60–90 days after birth for further experimentation. MBH was excised from the whole brain for experiments. Caudal epididymis was collected from male rats in a small petri dish containing about 3 ml of RPMI medium (Sigma, St. Louis, MO). The sperm was released from epididymis in the medium, which was centrifuged at 2,000 × g for 15 min and the pellet was collected. The sperm pellet was washed one time with PBS and collected and stored until assayed for gene expression and methylation measurements.
Neural stem cells were prepared and differentiated to POMC/β-endorphin neurons using cAMP and pituitary adenylate cyclase-activating polypeptide (PACAP) and maintained in cultures as we described previously (Sarkar et al., 2008). The HEK293FT cell line was purchased from Life Technologies (Carlsbad, CA) and was grown in DMEM (high glucose) medium with 10% fetal bovine serum (FBS), 0.1 mM MEM non-essential amino acids, 6mM L-glutamine, 1 mM sodium pyruvate, and 1% Pen-Strep (Life Technologies) in a 37°C incubator with 7.5% CO2.
Gene expression levels of Pomc and Sry in rat MBH and sperm were measured by quantitative real-time polymerase chain reaction (RT-PCR) (SYBR Green assay). Total RNA from MBH and sperm were extracted using an RNeasy kit (Qiagen, Germantown, MD). Total RNA (1 μg) was converted to first-strand complementary DNA (cDNA) using a high-capacity cDNA reverse transcription kit (Life Technologies). The primer sequences used for Pomc, Sry, Gapdh, 18S, and Rpl19 are given in Table 1. Real-time quantitative PCR was performed at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 40 s in the Applied Biosystems 7,500 real-time PCR system (Foster City, CA). The quantity of target genes (Pomc, Sry) and the three reference genes (Gapdh, 18S, Rpl19) were measured using the standard curve method. Target-gene expression was normalized with reference gene expression levels.
Sense and antisense oligos (ODN2, ODN3) were used for Sry knockdown as it was described (Dewing et al., 2006). The sequences for the oligos were given in Table 1 and were designed using software from Exiqon. These oligos were purchased from Exiqon, and equal quantities were mixed in artificial cerebrospinal fluid at a final concentration of 1 μg/μl. For in vitro knockdown experiments, β-endorphin-producing neurons were seeded (25,000 cells/well) in a 12-well plate, and oligo 0.4 μg was added in 0.4 ml of medium in each well (final concentration of 1μg/ml medium) and incubated in a 37°C incubator with 7.5% CO2. The cells were harvested 48 h after transfection, and we pooled the cells from two wells in triplicate. Gene expression for Sry and Pomc was measured as it is described in section “Materials and Methods.” For in vivo knockdown experiments, oligos were injected into the third ventricle of the rats via intracerebroventricular infusion as we described earlier (Gangisetty et al., 2014). Male rats at the age of 90 days (n = 6) for each group were used for the experiment. Oligo mix prepared in artificial CSF (1 μg/μl) was injected 2 μl into each rat at the rate of 1 μl/min. Rats were allowed to recover, were sacrificed 72 h after oligo injection, and then brain tissue was collected. MBH was isolated from the brain and analyzed for Sry and Pomc gene expression as it was described in section “Materials and Methods.”
ChIP assays were performed using a ChIP assay kit (Millipore Sigma, Saint Louis, MO) following instructions from the manufacturer. MBH collected from the brain tissue was placed on a small petri dish on ice and was minced into tiny pieces. Proteins were crosslinked with DNA from a tissue sample in 1% formaldehyde in PBS with protease inhibitors at room temperature for 30 min. The crosslinking was quenched with 0.125M glycine, and the sample was collected by centrifugation and washed twice with ice-cold PBS. The sample was suspended in SDS lysis buffer (with protease inhibitor) and homogenized to get uniform cell suspension. The crosslinked DNA was sheared into fragments the size of about 400–500 bp with a setting of 30% amplitude, with 15 s pulse on, 30 s pulse off for 10–12 cycles using the Qsonica ultrasonic cell disruptor (Qsonica, Newton, CT). The sample was centrifuged at 13,000 × g for 10 min, and we collected sonicated cell supernatant. The sample was diluted 10-fold in ChIP dilution buffer, and we used an aliquot for input. The remaining diluted sample was precleared with salmon sperm DNA/protein-A agarose. A ChIP assay was performed with a precleared sonicated sample with rabbit polyclonal SRY antibody (Thermo Fisher Scientific, Waltham, MA) (10 μg/sample) using a ChIP assay kit (Millipore Sigma, Burlington, MA) as specified in the manufacturer’s instructions. Equal concentration of the normal rabbit IgG was employed as negative control. The eluted ChIP samples and input sonicated samples were reverse crosslinked with 5M NaCl and incubated at 65°C overnight. DNA was extracted from these samples using a DNeasy micro kit (Qiagen). Pomc promotor-specific ChIP PCR primers (sequences in Table 1) were used to amplify the SRY enriched Pomc gene promoter region. The quantitative RT-PCR using SYBR Green assay was performed on the Applied Biosystems 7500 real-time PCR system. PCR conditions were 5 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 40 s at 72°C. ChIP DNA sample’s quantity measures were normalized by their respective input sample’s DNA.
Pomc promoter region was PCR amplified from rat MBH genomic DNA using primers as listed in Table 1. The PCR product was cloned into basic PCR2.1 vector (Invitrogen, Carlsbad, CA), and the sequence was verified with M13R primer. The plasmid DNA was digested with Sac1 and Xho1 and cloned into PGL3 basic luciferase reporter vector (Promega, Madison, WI). The positive clones were sequence verified with RVP3, a sequencing primer for PGL3 vector. We constructed three different mutants with change in nucleotide sequences of SRY binding sites of the Pomc promoter. All three mutants were designed by altering SRY binding site 1 (mutant 1), binding site 2 (mutant 2), and 1 and 2 together (mutant 3, double mutant). We used PGL3 vector-containing wild-type Pomc promoter as a template to make these mutants using primers whose sequences are given in Table 1. A quick-change site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA) was used to make all mutants as per instructions from the manufacturer.
HEK293 cells were seeded 50,000 cells/well in a 96-well plate. The cells were transfected with 250 ng of reporter vector or vector-containing Pomc promoter wild-type or mutant plasmid DNA along with either Sry overexpression plasmid or empty vector (PCDNA3.0) using Lipofectamine 3000 reagent (Thermo Fisher Scientific). All cells were also transfected with a plasmid-carrying renilla luciferase for normalization purposes. After 24 h of transfection, we measured firefly luciferase and renilla luciferase activity using the Dual Glo luciferase assay system (Promega) in a BioTech model synergy HT plate reader (BioTech, Winooski, VT). Eight samples were used in each group in transfection studies. Luciferase activity was measured in duplicates.
Data were analyzed using Prism 5.0 (GraphPad Software). The data shown in the figures are mean ± standard error mean (SEM). The significant differences between different treatment groups were assessed with one-way analysis of variance (ANOVA) with post hoc analysis using the Newman-Keuls post hoc test. P < 0.05 was considered significant. The significant differences between two groups were analyzed using the unpaired t-test, and two-tailed p-value < 0.05 was considered significant.
We used a liquid diet model of alcohol feeding in pregnant rats between day 7 and 21 of pregnancy that is known to raise blood levels of alcohol in the range of 120–150 mg/dl (Miller, 1992) and produce offspring with endophenotypes similar to those found in human fetal alcohol spectrum disorders (e.g., high-anxiety behaviors, stress hyperresponsiveness, metabolic diseases; Ting and Lautt, 2006; Boyadjieva et al., 2009; Hellemans et al., 2010; Nash and Davies, 2017). We have also shown previously that FAE increases Pomc promoter DNA methylation and suppresses Pomc gene expression in the MBH and in sperm (Govorko et al., 2012). In this study, we confirmed that FAE increases Pomc DNA methylation and suppresses Pomc gene expression in the MBH and in sperm of Fischer 344 rats (Figures 1A,B,E,F). Furthermore, we show that FAE also increases Sry DNA methylation and suppresses Sry gene expression in the MBH and in sperm of Fischer 344 rats (Figures 1C,D,G,H). These data indicate that FAE results in a similar increase in DNA methylation and similar decrease in gene expression of both Pomc and Sry in sperm and the MBH of male offspring.
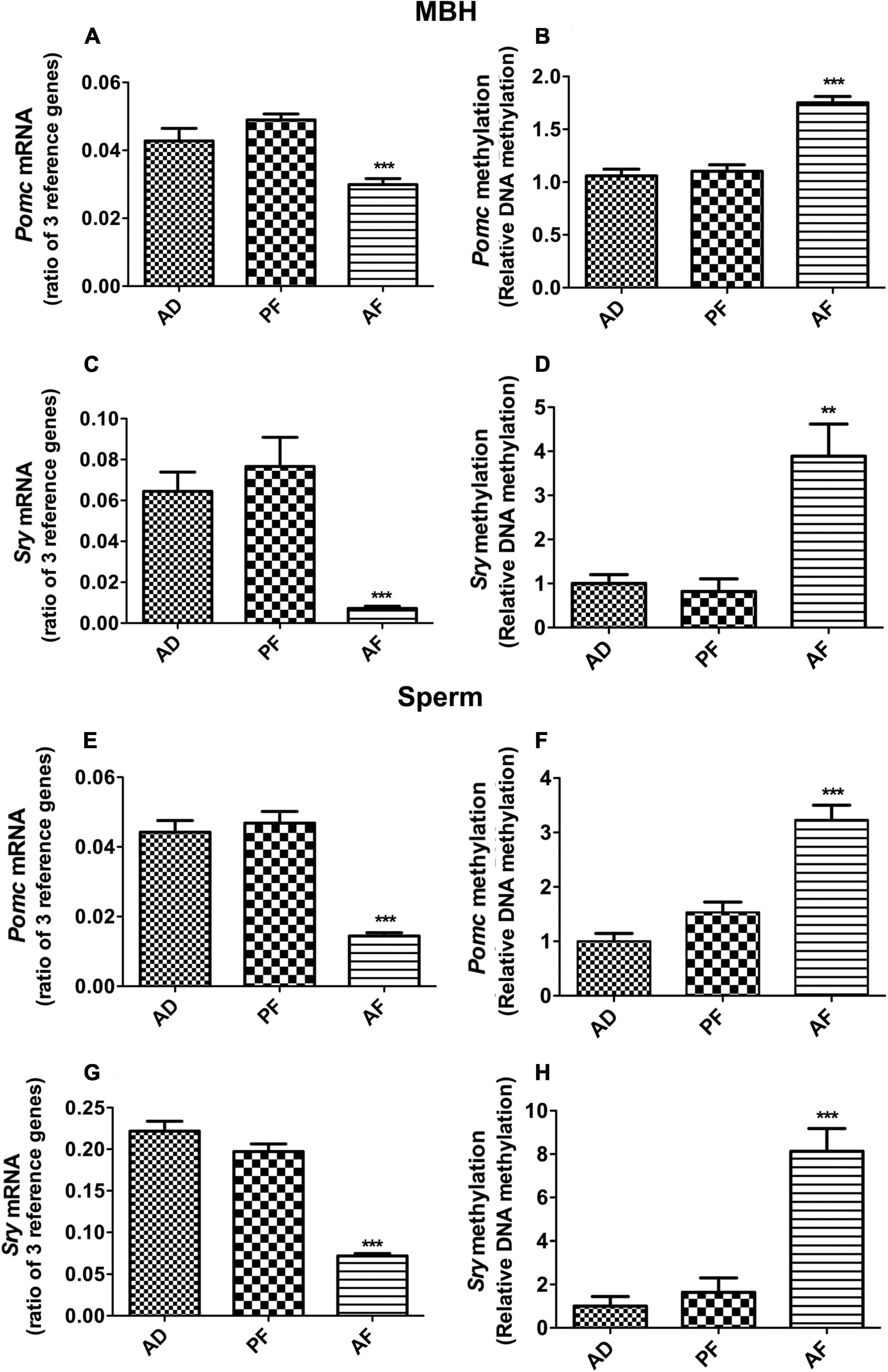
Figure 1. Effect of fetal alcohol exposure on the gene expression and promoter DNA methylation of Pomc and Sry genes in mediobasal hypothalamus (MBH) and sperm samples of F1 rat offspring. Pomc mRNA (A,E), Pomc promoter DNA methylation (B,F), Sry mRNA (C,G) and Sry promoter DNA methylation (D,H) levels in MBH and sperm samples of ad lib-fed (AD), pair-fed (PF), and alcohol-fed (AF) male rats from F1 generation offspring. Gene expression changes of Pomc and Sry genes were measured by quantitative RT PCR, and the quantities were normalized with ratios of three reference genes (Gapdh, 18S, Rpl19). Promoter DNA methylation of Pomc and Sry genes were measured by methylation-specific PCR, and methylation levels were measured as a ratio of methylated verses unmethylated DNA. Data are mean ± SEM (n = 6) and were analyzed using one-way ANOVA with the Newman-Keuls post hoc test; **p < 0.01, ***p < 0.001 between AF and controls (AD, PF).
Since FAE similarly affects the expression of Pomc and Sry genes in the F1 offspring, and FAE effects on Pomc gene expression have been shown to transmit for multiple generations (Govorko et al., 2012), we tested whether the FAE effect on Sry is also transmitted transgenerationally. We produced two different germlines, a male germline by breeding male fetal alcohol-exposed rats and their male offspring with normal females, and a female germline by breeding female fetal alcohol-exposed rats and their female offspring with normal males, as we have previously described (Govorko et al., 2012; Gangisetty et al., 2020). We also produced male (PFM) and female germlines (PFF) of control-fed rats or ad libitum-fed rats (AD). We measured the expression of Sry in F2 and F3 male offspring of male and female germlines to determine whether the fetal alcohol effect is transmitted through successive generations via germline. Like in the F1 generation, the F2 male progeny of male germlines (AFM) showed a significant reduction in Sry mRNA levels in the MBH (Figure 2A) and sperm (Figure 2C) as compared to the corresponding control groups. In the F3 generation, like in the F2 progeny, the F3 male progeny of the male germline (AFM) showed a reduction in Sry mRNA levels (Figures 2B,D). Other treatment groups did not differ from the control groups.
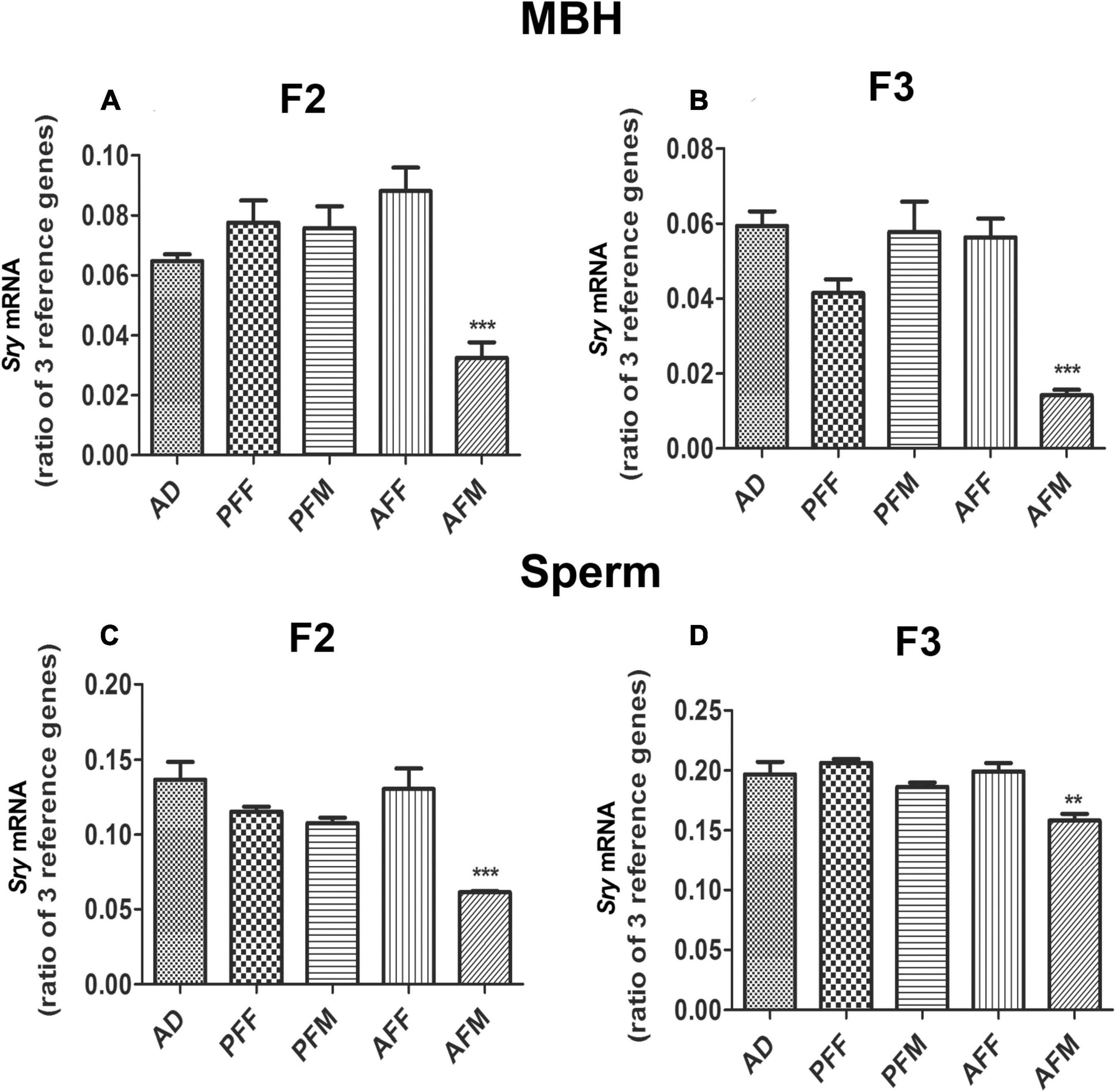
Figure 2. Transgenerational Effect of Fetal Alcohol Exposure on Sry Expression in MBH and Sperm Samples of Rat Offspring. Sry mRNA levels in MBH of F2 (A) and F3 (B) and in sperm of F2 (C) and of F3 (D) male rat offspring of ad lib-fed (AD), pair-fed male (PFM), pair-fed female (PFF), alcohol-fed male (AFM), or alcohol-fed female (AFF) rats. Sry mRNA levels were measured by quantitative RT PCR, and quantities were normalized using three reference genes (Gapdh, 18S, Rpl19). Data are mean ± SEM (n = 8) and were analyzed by one-way ANOVA with the Newman-Keuls post hoc test; **p < 0.01, ***p < 0.001 between AFM and controls (AD, PFF, PFM, AFF) in F2 and F3 generations.
SRY has been shown to regulate the endocrine response to stress (Dickey et al., 2012). Hence the possibility is raised that the Sry gene might have a regulatory role in the Pomc gene control mechanism. To test this hypothesis, we first determined the effect of Sry gene knockdown, using antisense oligo specific to Sry, on Pomc gene expression in POMC/(β-endorphin neurons in cultures. The results show that antisense Sry oligo significantly reduced the levels of Sry mRNA as compared to those in the sense oligo-treated control. Additionally, the antisense Sry oligo treatment reduced the level of Pomc mRNA in these neurons as compared to those treated with control sense oligo (Figures 3A,B). We also tested the effect of Sry gene knockdown by antisense oligo in vivo, and results are shown in Figures 3C,D. We found that Sry antisense oligo effectively reduced Sry mRNA expression in the MBH (Figure 3C). Furthermore, Sry antisense oligo treatment reduced the Pomc mRNA level in the MBH as compared to those in sense oligo-treated controls (Figure 3D). These results indicate that SRY has a regulatory role in Pomc expression.
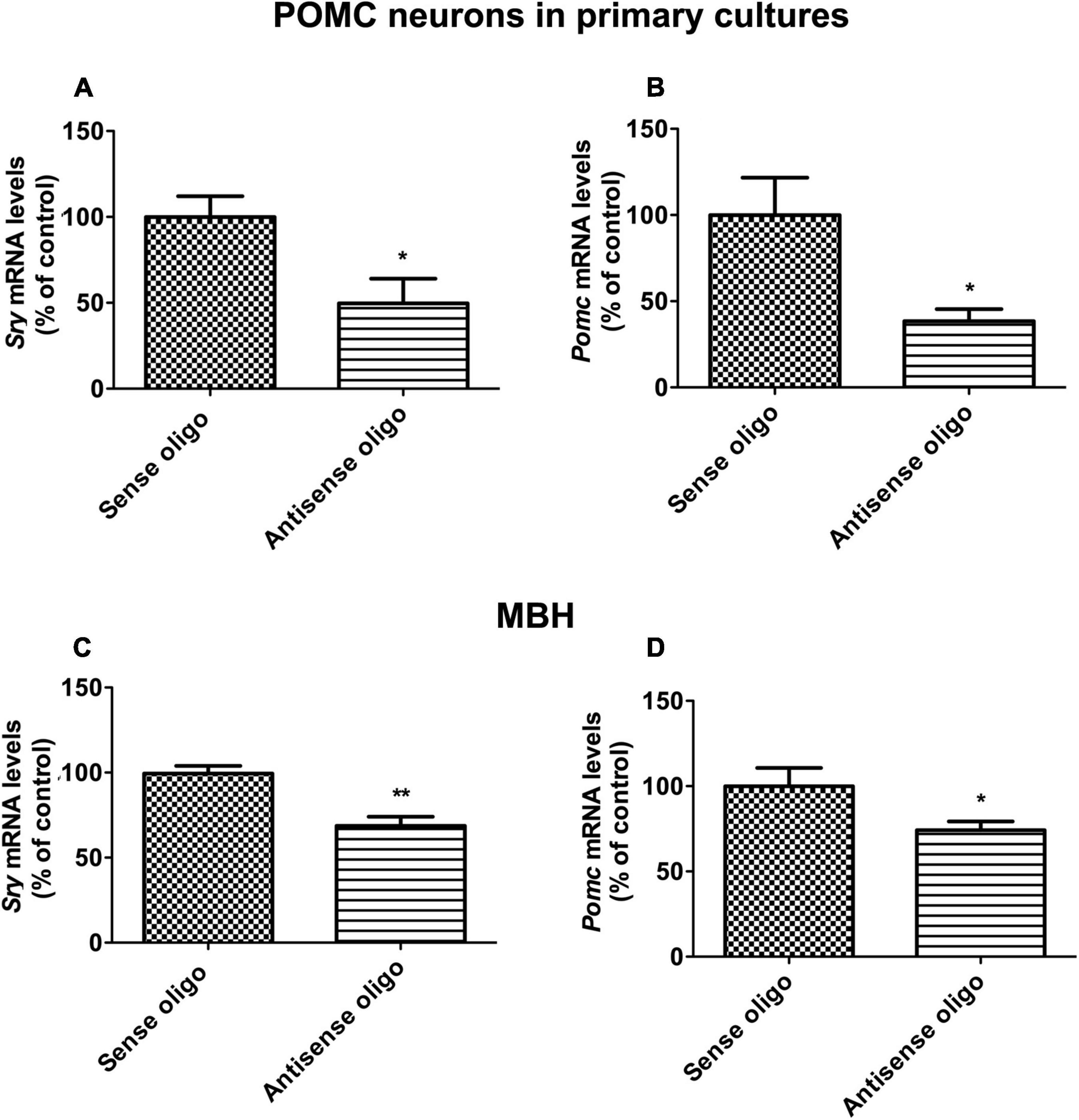
Figure 3. Sry gene knockdown effects on POMC expression in vitro and in vivo. In situ differentiated POMC neuronal cells from neuronal stem cells were transfected with antisense Sry oligo and sense control oligo. After 24 h of transfection, Sry mRNA (A) and Pomc mRNA (B) levels were measured by quantitative real-time PCR. Sry knockdown was performed in vivo through intracerebroventricular (icv) infusion of antisense Sry oligo and sense control oligo in male rats. After 72 h of infusion, Sry mRNA (C) and Pomc mRNA (D) levels were measured in MBH. Gene expression data was presented as percentage of sense control. Data are mean ± SEM (n = 6) and were analyzed using the unpaired t-test. *p < 0.05, **p < 0.01, between antisense oligo and sense oligo.
In order to understand how SRY regulates Pomc gene expression, we first evaluated SRY binding sites on the Pomc promoter using the TRANSFAC bioinformatics web tool which uses TRANSFAC version 8.31 and then performed a ChIP assay using primers spanning the SRY binding site to identify its enrichment on the Pomc promoter (Figure 4A). The web tool predicted SRY binding sites within 1 kb of the promoter region of the Pomc gene. ChIP assay data revealed that SRY is enriched on the Pomc promoter in the MBH, and its enrichment is significantly reduced in AF offspring compared to AD and PF offspring (Figure 4B). These results suggest that FAE reduces SRY binding on the Pomc promoter in the MBH.
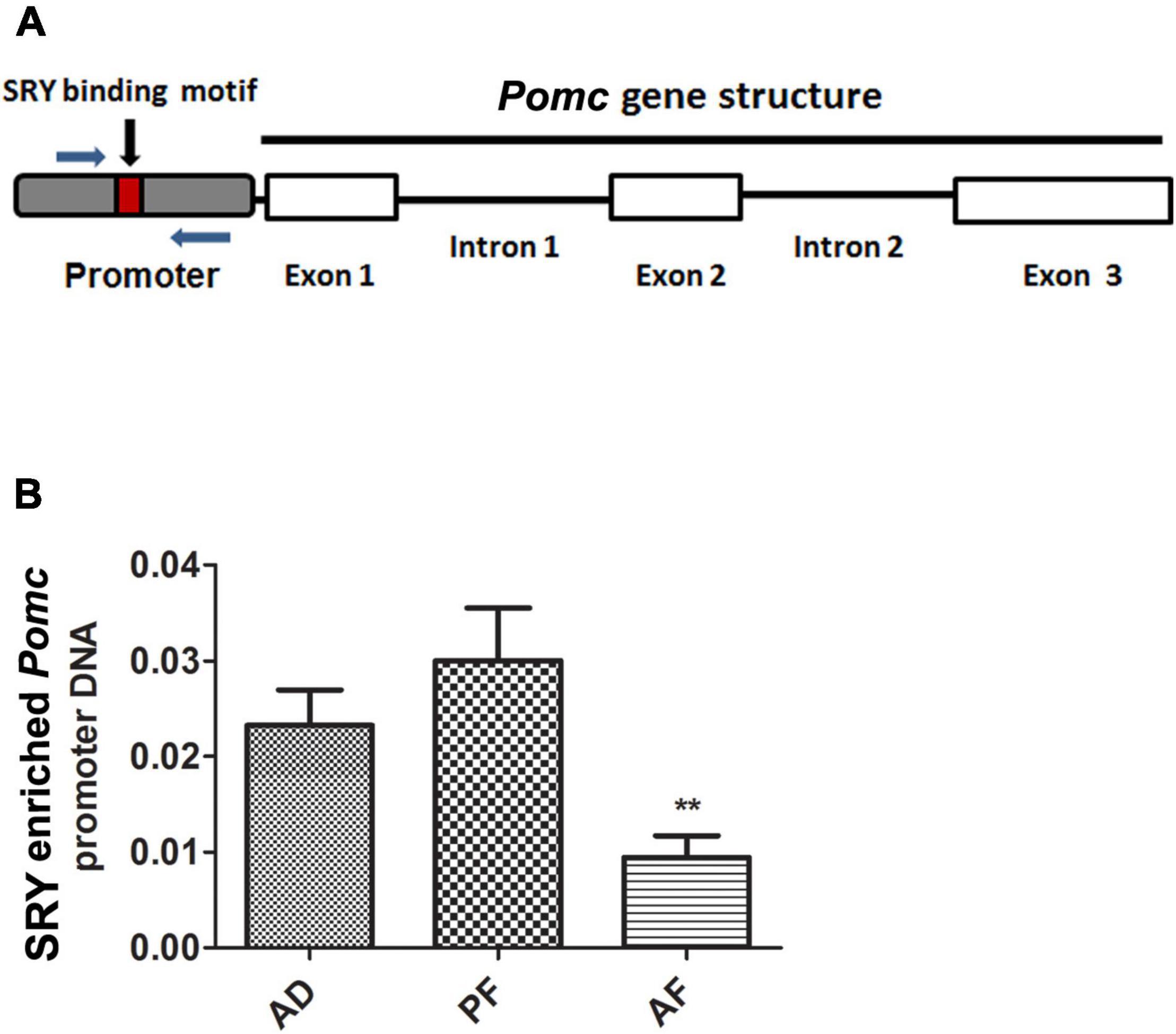
Figure 4. SRY recruitment onto the Pomc promoter in MBH of FAE rat offspring. A schematic diagram of Pomc gene structure comprising three exons and promoter. A small box within the promoter sequence represents the SRY binding motif. Arrows represent the ChIP primers spanning the SRY binding motif in Pomc promoter (A). SRY recruitment onto the Pomc promoter in MBH of AD, PF, and AF rat offspring were measured by ChIP assay. SRY-enriched Pomc promoter DNA was amplified by quantitative RT PCR using primers specific for Pomc promoter. SRY-enriched Pomc promoter DNA was normalized with input DNA (B). Data are mean ± SEM (n = 6) and were analyzed using one-way ANOVA with the Newman-Keuls post hoc test; **p < 0.01 between AF and controls (AD, PF).
In order to test that SRY may directly target Pomc promoter to increase its expression, we transfected a reporter plasmid carrying Pomc promoter along with a Sry overexpression plasmid or empty vector in HEK293 cells and measured luciferase activity. We analyzed the Pomc promoter region around 1 kb upstream of exon1 to detect SRY binding motifs using a bioinformatics web tool. We used a stringent threshold with > 95% sequence similarity and < 5% dissimilarity in the sequence to predict SRY binding sites on the Pomc promoter. It predicted three different SRY binding sites relative to exon 1 (−97, −577, and −747) in the Pomc promoter. We cloned the Pomc promoter upstream of firefly luciferase in a basic reporter luciferase vector (Figure 5A). Furthermore, we determined firefly luciferase activity as a measure of Pomc promoter and renilla luciferase activity for normalization purposes. We found that SRY significantly increased Pomc promoter luciferase activity compared to the vector control (Figure 5B). These results suggest that SRY regulate Pomc expression by acting on its binding motifs of POMC promoter.
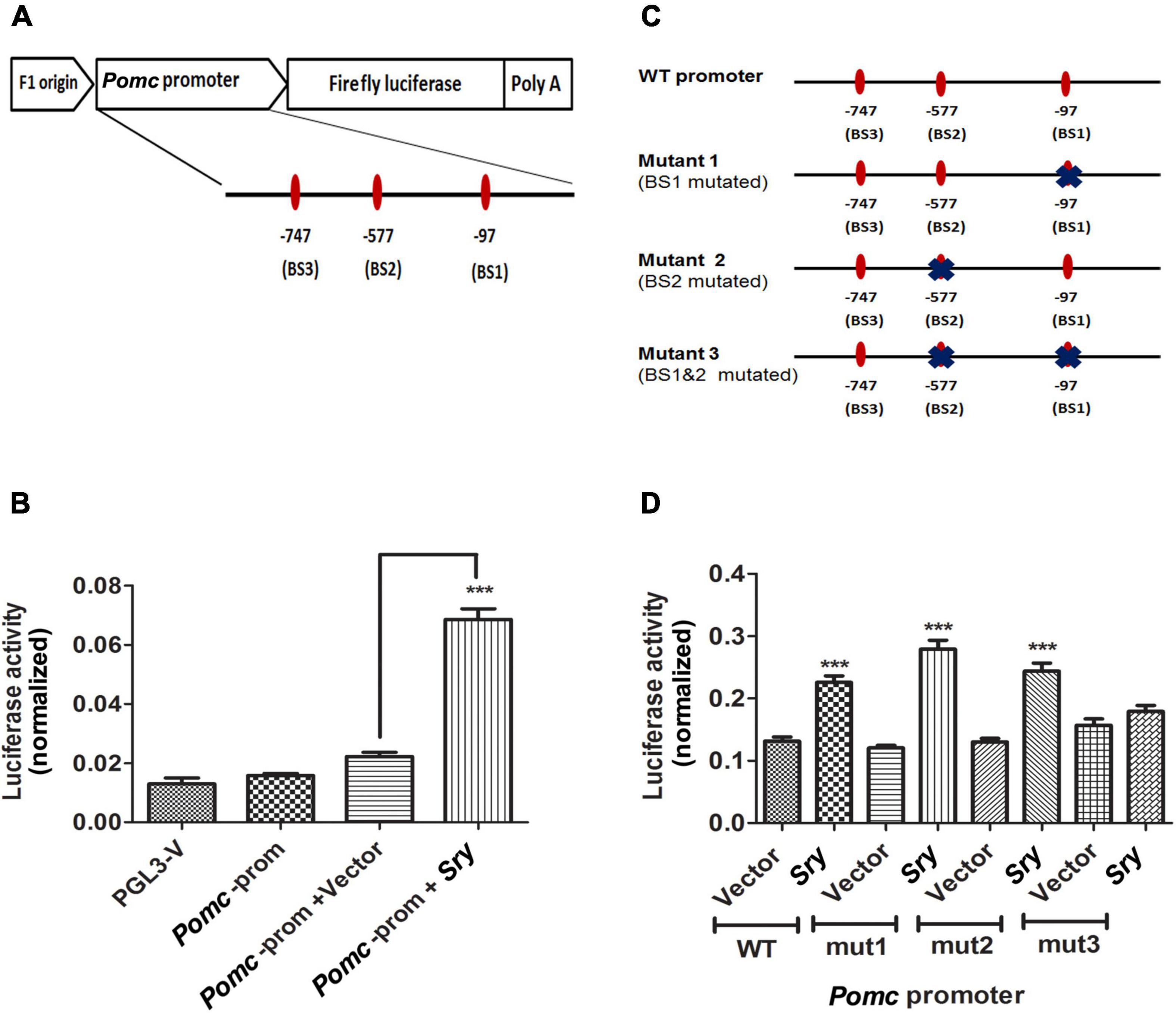
Figure 5. SRY directly targets its binding sites on Pomc promoter to regulate Pomc expression. A schematic representation of rat Pomc promoter was cloned upstream of firefly luciferase in a reporter luciferase vector. The three SRY binding sites (BS1, BS2, and BS3) are represented as ovals along with the horizontal line represented as the POMC promoter region. Each SRY binding site is indicated with its location relative to the exon 1 (A). Pomc promoter luciferase activity is measured in HEK293 cells transfected with a reporter plasmid containing Pomc promoter along with Sry overexpression plasmid or vector alone. Pomc promoter activity is measured as firefly luciferase activity normalized with renilla luciferase (B). Pomc promoter containing wild-type SRY binding sites and mutants with altered SRY binding sites represented in a schematic diagram were cloned in a reporter vector upstream of firefly luciferase, thereby generating 3 different mutants. Each mutation on the SRY binding site was marked with X on an oval shape representing the binding site (C). Pomc promoter activity of HEK293 cells transfected with a reporter plasmid carrying Pomc promoter with SRY binding sites, wild-type or mutant1 (mut1) or mutant2 (mut2) or mutant3 (mut3), as represented in Figure 5C along with SRY overexpression plasmid or vector. Pomc promoter activity is measured as firefly luciferase activity normalized with renilla luciferase (D). Data are mean ± SEM (n = 8) per group and were analyzed using one-way ANOVA with the Newman-Keuls post hoc test. ***p < 0.001 between SRY vs. vector.
We further confirmed the specificity of SRY binding onto its binding sites on Pomc promoter by mutating SRY binding sites using site-directed mutagenesis. We made three different mutant constructs by altering two of the SRY binding sites either singly or together, as shown in Figure 5C. We found that SRY increased Pomc promoter luciferase activity using the wild-type (WT) promoter sequence compared to vector. We also found that in mutant 1 (mut1: binding site1 mutated) and in mutant 2 (mut2: binding site2 mutated), SRY increased Pomc promoter luciferase activity similarly to that of the wild-type plasmid (Figure 5D). However, in mutant 3 (mut3: dual mutation of binding site1 and 2), SRY did not increase Pomc promoter activity compared to vector control, and it is in contrast to the wild-type promoter sequence (Figure 5D). Our results indicate that the mutation of a single binding site did not produce any effects, whereas the mutation of two binding sites significantly affected the SRY-induced Pomc promoter activity. Our results revealed that SRY may regulate Pomc gene expression by binding to two of its binding sites on the Pomc promoter.
In this study, we showed that FAE reduced Sry gene expression both in sperm and the MBH, and this effect on Sry gene expression persisted in F2 and F3 generations of male offspring that are derived from the male germline but not from the female germline. This transgenerational pattern of male germline transmission of fetal alcohol effects on Sry gene expression is similar to those seen in the fetal alcohol effect on Pomc gene expression (Govorko et al., 2012), suggesting that an interaction between these two genes might exist. The Sry gene on the Y chromosome is a master male sex-determining factor known to induce testis differentiation from bipotential gonads (Sinclair et al., 1990; Koopman et al., 1991). It is also responsible for generating male-specific properties of neuronal cells. SRY expression is localized in different regions of the brain, including the hypothalamus and substantia nigra of the midbrain regions that are involved in sex differentiation (Dewing et al., 2006). Studies in animal models have shown a possible effect of the specific part of the Y chromosome (YNPAR) that comprises SRY on β-endorphin, a functional peptide of POMC (Botbol et al., 2011). This raises the possibility that SRY may participate in mediation of fetal alcohol effect on the Pomc gene in males.
We demonstrated in this study that Sry gene knockdown effectively reduced Sry mRNA expression, as well as reduced the Pomc mRNA level, in both β-endorphin neurons in cultures and in the rat MBH (where β-endorphin neurons are accumulated) in vivo. Additionally, ChIP assay data showed that SRY is enriched on the Pomc promoter in the MBH, and its enrichment is significantly reduced in fetal alcohol-exposed male offspring. Furthermore, we confirmed the specificity of SRY binding on Pomc promoter by mutating SRY binding sites using site-directed mutagenesis and showing that SRY binding on Pomc promoter is required for its action. These data suggest for the first time that SRY regulates Pomc gene expression by binding on the Pomc promoter.
The present data also identify the sites where SRY binds to Pomc promoter to enhance its activity. A bioinformatics web tool used with a stringent threshold of more than 95% sequence similarity predicted three SRY binding sites (1, 2, and 3) located (−97, −577, −747) upstream of exon1. We found that SRY-increased Pomc promoter activity was unchanged when binding sites 1 or 2 were mutated individually, but it was lost when both of these two sites were mutated simultaneously, indicating a requirement of SRY binding to sites 1 and 2 to promote Pomc gene expression. A similar mechanism has been reported in a study where SRY binding to two sites on the Tcf21 promoter are shown to be required for Tcf21 gene transcription during male gonadal sex determination (Bhandari et al., 2011).
Our data highlights the novel finding that FAE-induced alteration of Sry expression and promoter methylation correlate with Pomc in the hypothalamus and sperm. Other evidence from our study, such as Sry knockdown altering Pomc expression, SRY binding on Pomc promoter, and SRY-induced Pomc promoter activity, clearly suggests that SRY plays a role in the regulation of Pomc. However, our study did not conclude whether SRY mediates the FAE-induced male germline transmission of POMC expression changes in the hypothalamus for multiple generations. Further studies determining the role of SRY on FAE-modified Pomc transcription in F2 and F3 offspring are needed to establish the role of this transcription factor in the transgenerational effects of FAE.
The germline transmission of phenotypic and epigenetic traits was reported mostly in animal studies. Some human studies provided evidence for intergenerational transmission of risk variants with the paternal lineage (Liu et al., 2005; Zuo et al., 2013). Studies also showed a grandparent with a major depressive disorder with alcohol abuse greatly increased the risk of an individual having a major depressive disorder (Olino et al., 2008). Other studies also showed that grandmothers of children with fetal alcohol syndrome were found to have a much greater rate of history of alcohol abuse, suggesting a generational phenomenon in FASDs (Kvigne et al., 2008). These human studies support the concept of multigenerational and transgenerational inheritance of psychiatric disorders with alcohol drinking. However, more clinical studies are needed to examine FASD manifestations with paternal lineage and the mechanism behind this phenomenon.
In conclusion, the current study provide some insights to our understanding of the mechanism of fetal alcohol-induced male germline transmission. More studies are required to understand the exact mechanism by which SRY plays a critical role in transmitting fetal alcohol-induced changes of Pomc gene expression through the male germline.
All relevant data are included in this manuscript. Any requests for materials will be fulfilled upon request through appropriate means.
All rat studies were performed with approved protocols in compliance with the Association for the Assessment and Accreditation of Laboratory Animal Care and Institutional Animal Care and Use Program, Rutgers, The State University of New Jersey Rutgers University.
DS designed the project and wrote the manuscript. OG helped in research design, participated in breeding and feeding the animals, contributed to sample collections, conducted biochemical assays, and helped in writing the manuscript. EM participated in conducting biochemical assays. All authors contributed to the article and approved the submitted version.
This work was supported by a National Institutes of Health Grant R01 AA025359.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ajay Palagani for conducting animal breeding and Stacey Pontoriero for editorial assistance.
AD, fed ad libitum with rat chow; AF, fed with a liquid diet containing 6.7% alcohol; AFF, AF female; AFM, AF male; ANOVA, one-way analysis of variance; ChIP, chromatin immunoprecipitation; FAE, fetal alcohol exposure; FBS, fetal bovine serum; GD, gestational day; HPA, hypothalamic-pituitary-adrenal; MBH, mediobasal hypothalamus; PACAP, pituitary adenylate cyclase-activating polypeptide; PF, pair-fed with isocaloric liquid diet; PFF, PF female; PFM, PF male; POMC, proopiomelanocortin; RT-PCR:, real-time polymerase chain reaction; SEM, standard error mean; SRY, sex-determining region Y.
Bakoyiannis, I., Gkioka, E., Pergialiotis, V., Mastroleon, I., Prodromidou, A., Vlachos, G. D., et al. (2014). Fetal alcohol spectrum disorders and cognitive functions of young children. Rev. Neurosci. 25, 631–639. doi: 10.1515/revneuro-2014-0029
Bhandari, R. K., Sadler-Riggleman, I., Clement, T. M., and Skinner, M. K. (2011). Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS One 6:e19935. doi: 10.1371/journal.pone.0019935
Botbol, M., Roubertoux, P. L., Carlier, M., Trabado, S., Brailly-Tabard, S., Perez-Diaz, F., et al. (2011). Modulation of brain beta-endorphin concentration by the specific part of the Y chromosome in mice. PLoS One 6:e16704. doi: 10.1371/journal.pone.0016704
Boyadjieva, N. I., Ortiguela, M., Arjona, A., Cheng, X., and Sarkar, D. K. (2009). Beta-endorphin neuronal cell transplant reduces corticotropin releasing hormone hyperresponse to lipopolysaccharide and eliminates natural killer cell functional deficiencies in fetal alcohol exposed rats. Alcohol. Clin. Exp. Res. 33, 931–937. doi: 10.1111/j.1530-0277.2009.00911.x
Dewing, P., Chiang, C. W., Sinchak, K., Sim, H., Fernagut, P.-O., Kelly, S., et al. (2006). Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 16, 415–420. doi: 10.1016/j.cub.2006.01.017
Dickey, C., Toot, J., Terwilliger, M., Payne, R., Turner, M., and Ely, D. (2012). The SHR Y chromosome increases cardiovascular, endocrine, and behavioral responses to stress compared to the WKY Y chromosome. Physiol. Behav. 106, 101–108. doi: 10.1016/j.physbeh.2012.01.009
Eyre-Walker, A. (1993). Recombination and mammalian genome evolution. Proc. Biol. Sci. 252, 237–243. doi: 10.1098/rspb.1993.0071
Gangisetty, O., Bekdash, R., Maglakelidze, G., and Sarkar, D. K. (2014). Fetal alcohol exposure alters proopiomelanocortin gene expression and hypothalamic-pituitary-adrenal axis function via increasing MeCP2 expression in the hypothalamus. PLoS One 9:e113228. doi: 10.1371/journal.pone.0113228
Gangisetty, O., Palagani, A., and Sarkar, D. K. (2020). Transgenerational inheritance of fetal alcohol exposure adverse effects on immune gene interferon - Y. Clin. Epigenetics 12:70. doi: 10.1186/s13148-020-00859-9
Govorko, D., Bekdash, R. A., Zhang, C., and Sarkar, D. K. (2012). Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol. Psychiatry 72, 378–388. doi: 10.1016/j.biopsych.2012.04.006
Hellemans, K. G., Sliwowska, J. H., Verma, P., and Weinberg, J. (2010). Prenatal alcohol exposure: fetal programming and later life vulnerability to stress, depression and anxiety disorders. Neurosci. Biobehav. Rev. 34, 791–807. doi: 10.1016/j.neubiorev.2009.06.004
Koopman, P., Gubbay, J., Vivian, N., Goodfellow, P., and Lovell-Badge, R. (1991). Male development of chromosomally female mice transgenic for Sry. Nature 351, 117–121. doi: 10.1038/351117a0
Kvigne, V. L., Leonardson, G. R., Borzelleca, J., and Welty, T. K. (2008). Characteristics of grandmothers who have grandchildren with fetal alcohol syndrome or incomplete fetal alcohol syndrome. Matern. Child Health J. 12, 760–765. doi: 10.1007/s10995-007-0308-y
Li, Y., Kirschmann, D. A., and Wallrath, L. L. (2002). Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. U.S.A. 99(Suppl. 4), 16462–16469. doi: 10.1073/pnas.162371699
Liu, X. Q., Greenwood, C. M., Wang, K. S., and Paterson, A. D. (2005). A genome scan for parent-of-origin linkage effects in alcoholism. BMC Genet. 6(Suppl. 1):S160. doi: 10.1186/1471-2156-6-S1-S160
Miller, M. W. (1992). Circadian rhythm of cell proliferation in the telencephalic ventricular zone: effect of in utero exposure to ethanol. Brain Res. 595, 17–24. doi: 10.1016/0006-8993(92)91447-m
Nash, A., and Davies, L. (2017). Fetal alcohol spectrum disorders: what pediatric providers need to know. J. Pediatr. Health Care 31, 594–606. doi: 10.1016/j.pedhc.2017.04.002
Ogilvie, K. M., and Rivier, C. (1997). Prenatal alcohol exposure results in hyperactivity of the hypothalamic-pituitary-adrenal axis of the offspring: modulation by fostering at birth and postnatal handling. Alcohol. Clin. Exp. Res. 21, 424–429. doi: 10.1111/j.1530-0277.1997.tb03786.x
Olino, T. M., Pettit, J. W., Klein, D. N., Allen, N. B., Seeley, J. R., and Lewinsohn, P. M. (2008). Influence of parental and grandparental major depressive disorder on behavior problems in early childhood: a three-generation study. J. Am. Acad. Child Adolesc. Psychiatry 47, 53–60. doi: 10.1097/chi.0b013e31815a6ae6
Sarkar, D. K. (2016). Male germline transmits fetal alcohol epigenetic marks for multiple generations: a review. Addict. Biol. 21, 23–34. doi: 10.1111/adb.12186
Sarkar, D. K., Boyadjieva, N. I., Chen, C. P., Ortiguela, M., Reuhl, K., Clement, E. M., et al. (2008). Cyclic adenosine monophosphate differentiated beta-endorphin neurons promote immune function and prevent prostate cancer growth. Proc. Natl. Acad. Sci. U.S.A. 105, 9105–9110. doi: 10.1073/pnas.0800289105
Sarkar, D. K., Gangisetty, O., Wozniak, J. R., Eckerle, J. K., Georgieff, M. K., Foroud, T. M., et al. (2019). Persistent changes in stress-regulatory genes in pregnant women or children exposed prenatally to alcohol. Alcohol. Clin. Exp. Res. 43, 1887–1897. doi: 10.1111/acer.14148
Sarkar, D. K., Kuhn, P., Marano, J., Chen, C., and Boyadjieva, N. (2007). Alcohol exposure during the developmental period induces beta-endorphin neuronal death and causes alteration in the opioid control of stress axis function. Endocrinology 148, 2828–2834. doi: 10.1210/en.2006-1606
Sekido, R., and Lovell-Badge, R. (2008). Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930–934. doi: 10.1038/nature06944
Sinclair, A. H., Berta, P., Palmer, M. S., Hawkins, J. R., Griffiths, B. L., Smith, M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. doi: 10.1038/346240a0
Skinner, M. K., Bhandari, R. K., Haque, M. M., and Nilsson, E. E. (2015). Environmentally induced epigenetic transgenerational inheritance of altered SRY genomic binding during gonadal sex determination. Environ. Epigenet. 1:dvv004. doi: 10.1093/eep/dvv004
Ting, J. W., and Lautt, W. W. (2006). The effect of acute, chronic, and prenatal ethanol exposure on insulin sensitivity. Pharmacol. Ther. 111, 346–373. doi: 10.1016/j.pharmthera.2005.10.004
Wynne, O., and Sarkar, D. K. (2013). “Stress and neuroendocrine-immune interaction: a therapeutic role of beta endorphin,” in The Wiley-Blackwell Handbook of Psychoneuroimmunology, eds A. Kusnecov and H. Anisman (Oxford: John Wiley & Sons), 198–211. doi: 10.1002/9781118314814
Keywords: fetal alcohol exposure, SRY, POMC, epigenetic, transcriptional regulation
Citation: Gangisetty O, Mead EA and Sarkar DK (2021) Sex-Determining Region Y Controls the Effects of Fetal Alcohol Exposure on Proopiomelanocortin Gene Expression. Front. Neurosci. 15:608102. doi: 10.3389/fnins.2021.608102
Received: 19 September 2020; Accepted: 17 February 2021;
Published: 16 March 2021.
Edited by:
Vance L. Trudeau, University of Ottawa, CanadaReviewed by:
Ben Nephew, Worcester Polytechnic Institute, United StatesCopyright © 2021 Gangisetty, Mead and Sarkar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipak K. Sarkar, ZGlwYWsuc2Fya2FyQHJ1dGdlcnMuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.