
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurosci. , 01 August 2019
Sec. Neuropharmacology
Volume 13 - 2019 | https://doi.org/10.3389/fnins.2019.00771
Methamphetamine (METH) is a widespread illicit drug. If it is taken by pregnant women, it passes through the placenta and just as it affects the mother, it can impair the development of the offspring. The aim of our study was to identify candidates to investigate for changes in the gene expression in the specific regions of the brain associated with addiction to METH in rats. We examined the various areas of the central nervous system (striatum, hippocampus, prefrontal cortex) for signs of impairment in postnatal day 80 in experimental rats, whose mothers had been administered METH (5 mg/kg/day) during the entire gestation period. Changes in the gene expression at the mRNA level were determined by two techniques, microarray and real-time PCR. Results of two microarray trials were evaluated by LIMMA analysis. The first microarray trial detected either up-regulated or down-regulated expression of 2189 genes in the striatum; the second microarray trial detected either up-regulated or down-regulated expression of 1344 genes in the hippocampus of prenatally METH-exposed rats. We examined the expression of 10 genes using the real-time PCR technique. Differences in the gene expression were counted by the Mann–Whitney U-test. Significant changes were observed in the cocaine- and amphetamine-regulated transcript prepropeptide, tachykinin receptor 3, dopamine receptor D3 gene expression in the striatum regions, in the glucocorticoid nuclear receptor Nr3c1 gene expression in the prefrontal cortex and in the carboxylesterase 2 gene expression in the hippocampus of prenatally METH-exposed rats. The microarray technique also detected up-regulated expression of trace amine-associated receptor 7 h gene in the hippocampus of prenatally METH-exposed rats. We have identified susceptible genes; candidates for the study of an impairment related to methamphetamine addiction in the specific regions of the brain.
- The microarray technique detected either up-regulated or down-regulated expression of 2189 genes in the striatum of prenatally METH-exposed rats.
- The microarray technique detected either up-regulated or down-regulated expression of 1344 genes in the hippocampus of prenatally METH-exposed rats.
- We examined the expression of 10 genes using the real-time PCR technique.
- Prenatally administered METH significantly alters the expression of the glucocorticoid nuclear receptor Nr3c1 mRNA within prefrontal cortex of rats.
- Prenatally administered METH significantly alters the expression of the cocaine- and amphetamine-regulated transcript prepropeptide, the tachykinin receptor 3 and dopamine receptor D3 mRNA within striatum of rats.
- Prenatally administered METH significantly alters the expression of the carboxylesterase 2 mRNA within the hippocampus of rats.
- Prenatally administered METH alters the expression of the trace amine-associated receptor mRNA within hippocampus of rats
- Both techniques, microarray and real-time PCR, showed altered expression of the tachykinin receptor 3 and dopamine receptor D3 mRNA within striatum of rats.
Methamphetamine (METH) is one of the most common “hard” drugs taken by pregnant women (Marwick, 2000). It is also one of the most frequently used illicit drugs in the Czechia, Middle and Eastern Europe (Vavřinková et al., 2001; Šlamberová, 2012). Statistics show that only 17% of female drug abusers in the United States were primary METH users, but 38% had used it during pregnancy, because some drug-abused women replace other drugs for METH during pregnancy for its anorectic effect (Marwick, 2000). METH is a powerfully addictive stimulant that metabolizes slowly resulting in the drug having a long-lasting high (8 to 24 h) (Marwick, 2000). This might be the reason that makes it so popular. The National Institute of Drug Abuse (NIDA; part of NIH) in the United States decided to increase funding for studies testing the effects of METH administration during pregnancy. Research concerning the long-term effects of prenatal METH exposure is still in its infancy. Our laboratory specializes in investigating the effects of drugs (especially METH) on rat mothers and their progeny.
METH is an indirect agonist of monoamines, which works by disrupting multiple presynaptic and synaptic processes. The resultant increases in dopamine, norepinephrine, and serotonin contribute to its properties as well as inducing other central and peripheral effects (Thompson et al., 2009; Lloyd et al., 2013). In low doses, METH can elevate mood and increase alertness, concentration and energy. At higher doses, it can induce psychosis, breakdown of skeletal muscle, seizures and bleeding in the brain. Chronic, high-dose use can precipitate unpredictable and rapid mood swings, prominent delusions and violent behavior. Chronic abuse is associated with serious health complications including deficits in attention, memory, and executive functions in humans (Cadet and Krasnova, 2009). METH is known to have a high addiction liability (i.e., compulsive METH use) and dependence liability (i.e., withdrawal symptoms occur when METH use ceases) (ChEBI, 2018; Wikipedia contributors, 2019b). METH travels through the bloodstream to the brain, where it readily crosses the blood-brain barrier (Dattel, 1990; Burchfield et al., 1991). The similarity of the chemical structure of METH to monoamine neurotransmitters (dopamine and noradrenaline) determines its mechanism of action (Sulzer et al., 2005; Heal et al., 2013, Rambousek et al., 2014). METH affects the function and trafficking of DAT. It leads to increased release of dopamine from presynaptic terminals, as well as synaptic vesicles (Sulzer et al., 1995). Methamphetamine interacts with Taar1 receptors and this interaction results in inhibition of dopamine uptake, induction of dopamine efflux, and promotion of dopamine transporter internalization. TAAR1 mediates methamphetamine-induced regulation of dopamine transporter function and trafficking in brain striatal synaptosomes (Xie and Miller, 2009; Rutigliano et al., 2018). In Taar1-knock out mice sensitivity to amphetamine and methamphetamine was exacerbated (Lindemann et al., 2008; Achat-Mendes et al., 2012; Rutigliano et al., 2018). The neuropsychiatric complications might, in part, be related to drug-induced neurotoxic effects, which include damage to dopaminergic and serotonergic terminals, neuronal apoptosis, as well as activated astroglial and microglial cells in the brain (Cadet and Krasnova, 2009).
METH present in a drug-abusing women’s bloodstream, can pass through the placenta to a fetus and can also be secreted into breast milk (Winslow et al., 2007). Amphetamines have vasoactive effects, which result in the restrictions of the supply of nutrients to a developing fetus, causing the anorectic symptoms often reported (Winslow et al., 2007). Infants born from METH-abusing mothers were found to have significant growth reductions (Dixon and Bejar, 1989; Plessinger, 1998; Cui et al., 2006), cleft palate, cardiac defects (Plessinger, 1998; Cui et al., 2006), aggressive behavior (Eriksson et al., 1989; Cui et al., 2006), peer-related problems (Eriksson and Zetterstrom, 1994; Cui et al., 2006) and learning problems (Cernerud et al., 1996; Cui et al., 2006). Matera and colleagues reported severe brain malformations in a human infant exposed to amphetamine in utero, such as exencephaly, anomalies of gyria, and hemorrhage (Matera et al., 1968; Cui et al., 2006). Human infants exposed to methamphetamine in utero have tended to show a significant decrease in head circumference (Little et al., 1988; Dixon and Bejar, 1989; Cui et al., 2006; Winslow et al., 2007). Non-invasive studies of the brain using Echo or MRI have revealed intracranial hemorrhage (Dixon and Bejar, 1989; Cui et al., 2006) and volume decrease of subcortical area (Chang et al., 2004; Cui et al., 2006). The effects of METH-administered prenatally in clinical studies are summarized in Table 1.
Experimental studies demonstrated that administration of METH to pregnant rats leads to increased likelihood of abortion and maternal death (Acuff-Smith et al., 1992; Matějovská et al., 2014). Pregnant rats who receive METH have a shorter gestation period and have fewer litters of young than control mothers (Martin, 1975; Martin et al., 1976). METH-administered prenatally increases infant mortality, reduces weight gain, delays development and slows down reflexes (Martin et al., 1976; Cho et al., 1991; Acuff-Smith et al., 1996; Matějovská et al., 2014). Studies from our laboratory and others demonstrated that prenatal or neonatal exposure to METH seriously impairs the behavior of adult rats and their sensitivity to drugs (Šlamberová, 2012). Also, our studies demonstrated that maternal addiction during gestation and/or lactation periods impairs maternal behavior, which in turn has adverse effects on pup development (Šlamberová et al., 2005a,b, 2006). Such impairment caused by prenatal METH exposure can be partially suppressed by cross-fostering with control rats, demonstrating the positive effect of good postnatal foster care (Hrubá et al., 2009, 2010; Pometlová et al., 2009). On the other hand, maternal separation induces social stress and maternal physical stress, aggravating cognitive impairment and responses to stressors in the exposed offspring (Holubová et al., 2016, 2018). Thus, it seems that social factors, such as bad or good maternal care or maternal stress, can have significant influence on the development of rat offspring exposed to METH during gestation. In addition, one of our studies found that drug exposure during gestation may even influence second generation pups (Šlamberová et al., 2005d, 2007).
There are several brain structures that are associated with drug addiction. The most important are mesolimbic, and mesocortical pathways (Robinson and Becker, 1986) which includes the striatum, hippocampus and prefrontal cortex. Striatum coordinates multiple aspects of cognition, including both motor and action planning, decision-making, motivation, reinforcement, and reward perception (Yager et al., 2015). The hippocampus plays important roles in the consolidation of information from short-term memory to long-term memory, and in spatial memory that enables navigation (Anderson et al., 2007). The prefrontal cortex has been implicated in planning complex cognitive behavior, personality expression, decision making, and moderating social behavior (Yang and Raine, 2009). Various studies have found that prenatal exposure to psychostimulants results in enhanced responsiveness of mesostriatal monoaminergic projections (Bubeníková-Valešová et al., 2009; Fujáková-Lipski et al., 2017). Given that our previous studies demonstrated changes in locomotor activity, cognition as well as social behavior (Fialová et al., 2015; Šlamberová et al., 2015), types of behavior that are associated with the above mentioned brain structures, the aim of the present study was to identify candidates for changes in the gene expression in the striatum, hippocampus and prefrontal cortex in adult rats in PD 80, whose mothers had been administered METH (5 mg/kg/day) during the entire gestation period.
The procedures for animal experimentation utilized in this study were reviewed and approved by the Committee for Protection of Experimental Animals of Third faculty of Medicine and by Departmental Committee of Ministry of Education, Youth, and Sports and are in agreement with the Czech Government Requirements under the Policy of Humans Care of Laboratory Animals (no. 246/1992) and also comply with the regulations of the Ministry of Agriculture of the Czech Republic (no. 311/1997). Adult male (300–400 g) and female (250–300 g) Wistar rats from Charles River Laboratories International, Inc., were delivered by VELAZ (Prague, Czechia) and housed 4–5 per cage in a temperature-controlled (22–24.8°C) colony room with a standard 12 h light/dark cycle, lights on at 06.00 h. Animals were left undisturbed for 1 week with food and water ad libitum. After the acclimation period, the females were weighed and smeared using vaginal lavage to determine the phase of the estrous cycle. Females at the onset of the estrous phase of the estrous cycle were housed with sexually mature males for overnight (1 pair per cage) (Šlamberová et al., 2005a,b,c,d; Schutová et al., 2012). On the following day, the females were smeared and returned to their previous home cages. The fertilization was assigned as Day 1 of gestation based on the presence of sperm in the vaginal smear. On Day 21 of gestation, the females were removed from the group cages and placed into maternity cages (1 female per cage). The day of the delivery was counted as PD 0. The number of pups in each litter was adjusted to 12. Whenever possible, the same numbers of male and female pups were kept in each litter. To avoid litter bias, pups were cross-fostered on PD 1 so that one mother usually raised three male and three female pups of her own and three male and three female pups from another mother (see below). On PD 21, animals were weaned and housed in groups separated by sex. The light/dark cycle of the animals was reversed with lights on at 18.00 h. Animals were left undisturbed until adulthood (Schutová et al., 2012).
For prenatal exposure, the pregnant dams were divided into two groups: the METH group and the SAL group. The females from the METH group were administered D-methamphetamine HCl (Sigma Aldrich, Czechia) at a dose of 5 mg/kg/day and volume of 1 ml/kg/day s.c. for the duration of the gestation period (i.e., from the first to the last day of gestation) (Šlamberová et al., 2005a,b,d, 2006; Schutová et al., 2012). This METH dose was chosen because it results in similar fetal brain drug concentrations to those found in human infants of METH-abusing women (Acuff-Smith et al., 1996; Schutová et al., 2012). This dose is also the standard dose used in our experiments (Šlamberová et al., 2005a,b,d, 2006, 2007; Schutová et al., 2012). The females from the SAL group were administered 0.9% NaCl solution s.c. at the same time and in the same volume as METH. On PD 1, the offspring were marked according to the prenatal exposure using the intradermal application of black India ink; the METH group of prenatally affected offspring was marked in the left foot pad, and the SAL group of prenatally affected offspring was marked in the right foot pad.
Before we started the experiment in PD 80, the experimental animals (8 METH and 8 SAL rats) were weighed as were the parts of the brains from which the RNA was isolated. The isolation of total RNA (mixture of mRNA, tRNA, and rRNA) from the snap-frozen tissue was obtained in accordance with the published procedure by Zhu Shirley: Total RNA isolation and purification for microarray (Gilks et al., 2005). The cerebral parts of experimental rats, striatum, hippocampus and prefrontal cortex, were obtained and stored in TRI reagent (Sigma-Aldrich, United States). The tissue was homogenized, centrifuged, washed with chloroform, and precipitated in isopropanol. Total RNA from the striatum, hippocampus and prefrontal cortex was used in the microarray and real-time PCR techniques.
The microarray technique was carried out using Genechip Rat Gene 2.0 ST Array (Affymetrix, United States) in the Genomics and Bioinformatics department, Institute of Molecular Genetics of the ASCR (Prague, Czechia). The procedure was carried out twice. Because of the limited resource only 6 properly isolated samples from striatum (from 3 METH and from 3 SAL rats) were examined to distinguish the mRNA expression level of 29489 genes by the microarray technique in the first trial. Also, because of the limited resource only 6 properly isolated samples from the hippocampus (from 3 METH and from 3 SAL rats) were examined to distinguish the mRNA expression level of 29489 genes by the microarray technique in the second trial. Results of both examinations were evaluated by LIMMA analysis in the Genomics and Bioinformatics department, Institute of Molecular Genetics of the ASCR (Prague, Czechia).
Total RNA (70 ng) from 8 METH and 8 SAL rats was processed using TaqMan Gene Expression Assays (ThermoFisher Scientific, United States) and TaqMan Gene Expression Master Mix (ThermoFisher Scientific, United States) by real-time PCR in the 7500 Fast Real-Time PCR System (ThermoFisher Scientific, United States). Together 6 genes, Gapdh, Pgk1, 18sRNA, B2m, Rps6, PPIA (Bernaudin et al., 2002; Hori et al., 2005; Shilling et al., 2006; Romanowski et al., 2007; Langnaese et al., 2008; Gubern et al., 2009) were assessed for their potential as being suitable reference genes in all three types of tissue. Reference genes, Rps6 and B2m were chosen according to the best reproducible results observed for gene expression in all three of tissues (striatum, hippocampus, prefrontal cortex), and taking into account the published results of another scientific group (Shilling et al., 2006). A quantification of gene expression was calculated by the Pfaffl method (Pfaffl, 2001; Šmehilová, 2011); exponential amplifications factors of Assays were equal to 1 (ThermoFisher Scientific, United States). Differences were counted by the Mann–Whitney U-test.
The first group of genes (Table 2), which was examined using real-time PCR, includes Bdnf, nuclear receptor subfamily 3 group C member 1 (Nr3c1), Esr1, insulin-like growth factor I receptor (Igfr1), and olfactory factor 522 (Olr522). The genes were chosen based on data published by various scientific groups (Middaugh, 1989; Cernerud et al., 1996; Wouldes et al., 2004; Šlamberová and Rokyta, 2005; Chang et al., 2007; Ago et al., 2009; Schutová et al., 2008, 2009, 2012; Lloyd et al., 2013; Macúchová et al., 2013, 2014; Leung et al., 2014; Šlamberová et al., 2014; Ren et al., 2016, 2017). The second group of genes (Table 3), which was examined using real-time PCR, includes Tacr3, dopamine receptor D3 (Drd3), Cartpt, Rps6ka a Foxp2 genes. The genes were chosen based on our results from the microarray technique and in accordance with data published by other scientific groups (Paris and Lorens, 1989; Gecz and Mulley, 2000; Couceyro et al., 2005; Le Foll et al., 2005; Krasnova et al., 2017). Together, the expression of 10 genes was examined using the real-time PCR technique.
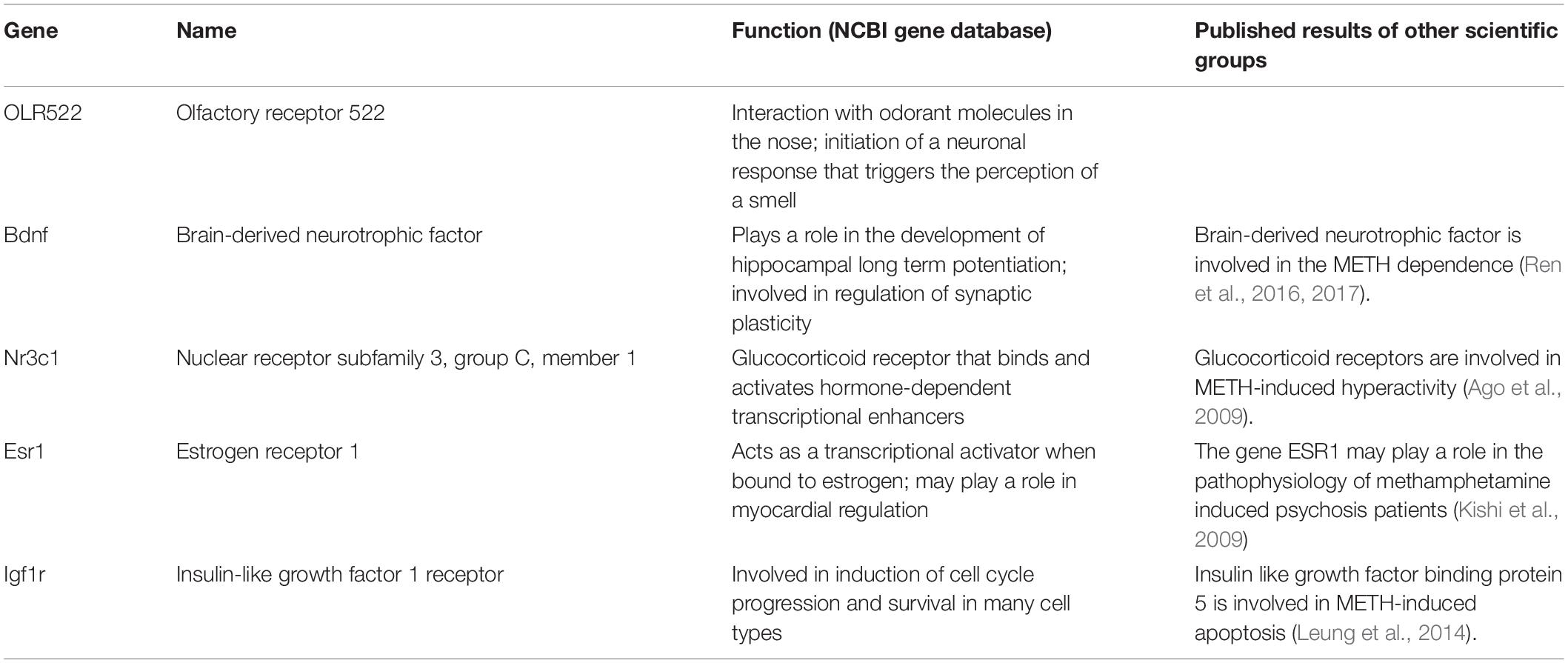
Table 2. The first group of examined genes with the description of their function according to the NCBI gene database.
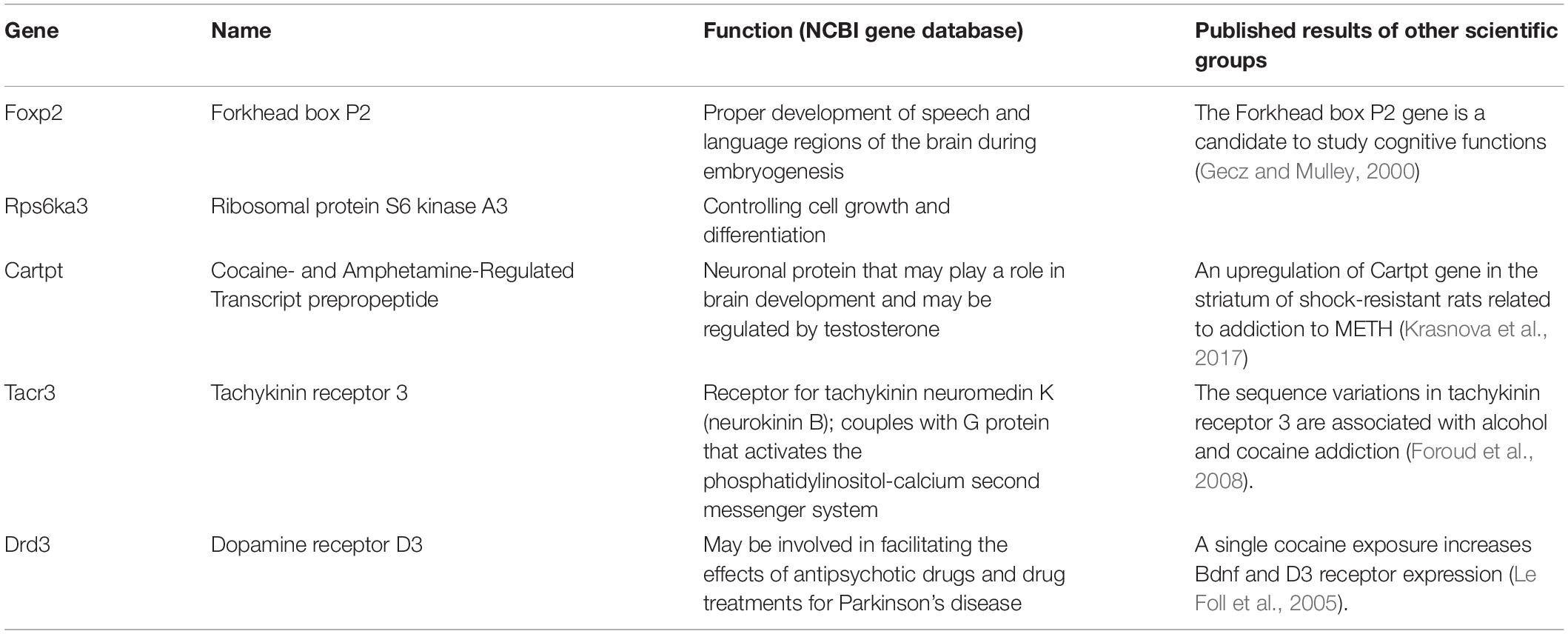
Table 3. The second group of examined genes with the description of their function according to the NCBI gene database.
The weights of SAL animals ranged from 285 to 360 g with a median of 310,5 g. The weights of METH animals ranged from 298 to 350 g with the median of 330 g. The animals were divided into boxes of four (1–4, 5–8).
The mRNA expression (cDNA) of 29489 genes was examined in 6 striatum samples: three samples from the prenatally METH-exposed rats and three samples from the prenatally SAL-exposed rats. The expression of approximately 2189 genes was either up-regulated or down-regulated (p < 0.05) in the striatum, however, the differences were very low. The borderline significant difference (p < 0.05 and logFC > 1) between METH and SAM samples was observed in the case of Drd3 and Tacr3 genes (Table 4) (Zoubková, 2019a). The significant difference (p < 0.05 and logFC > 1) between METH and SAM samples was observed in the case of Cartpt gene, olfactory receptor 522 and 1448 genes (Table 4) (Zoubková, 2019a). The non-significant difference between METH and SAM samples was observed in the case of Foxp2 and Rps6ka3 genes (Table 4) (Zoubková, 2019a).
The mRNA expression (cDNA) of 29489 genes was examined in 6 hippocampus samples: three samples from the prenatally METH-exposed rats and three samples from the prenatally SAL-exposed rats. The expression of approximately 1344 genes was either up-regulated or down-regulated (p < 0.05) in the hippocampus, however, the differences were very low. The borderline significant difference (p < 0.05 and logFC > 1) between METH and SAM samples was observed in the case of Taar7h gene. The significant difference (p < 0.05 and logFC > 1) between METH and SAM samples was observed in the case of carboxylesterase 2i (Ces2) and olfactory receptor 1726 genes (Table 5). The non-significant difference between METH and SAM samples was observed in the case of Tacr3 gene (Zoubková, 2019b).
For each of the tissue types (striatum, hippocampus and prefrontal cortex) the expression of chosen genes was examined using real-time PCR with TaqMan® Gene Expression Assays (ThermoFisher Scientific, United States). The measured values were normalized to the measured values of two reference genes, B2m and Rps6, and quantified by the Pfaffl method (calculation of ratio of target to reference gene). Significant differences between METH and SAM samples were evaluated by the Mann–Whitney U-test (Supplementary Table S1).
In the prefrontal cortex of prenatally METH-exposed rats, the mRNA expression of the Nr3c1 gene was significantly (p ≤ 0.01) increased compared to that in prenatally SAL-exposed rats (Figure 1). The measured values were normalized to the measured values of the reference B2m gene. However, the increase was not detected, when the measured values were normalized to the measured values of the reference Rps6 gene. The mRNA expression of the Foxp2 gene of prenatally METH-exposed rats was significantly (p ≤ 0.05) decreased compared to that in prenatally SAL-exposed rats in the prefrontal cortex, when the measured values were normalized to the measured values of the reference Rps6 gene. However, no difference was observed, when the measured values were normalized to the measured values of the reference B2m gene.
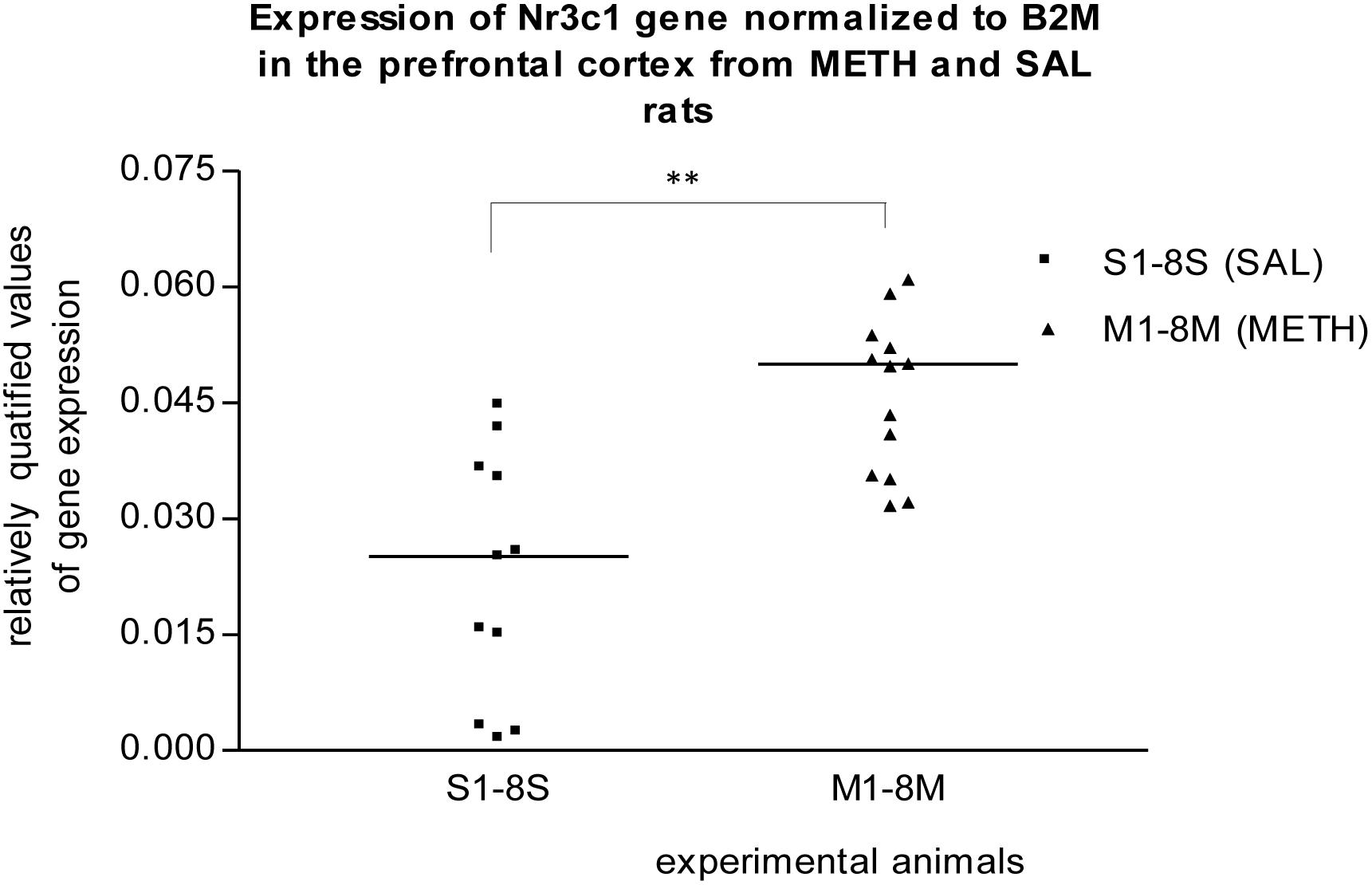
Figure 1. Significant difference (p ≤ 0.01) in the expression of Nr3c1 gene between METH and SAL samples in the prefrontal cortex. The results of the Nr3c1 gene expression were normalized to the results of the expression of the reference B2m gene. The values of METH rats are marked as M and the values of SAL rats are marked as S. The values are presented with a median. The number of values is higher than 8 M and 8 S, respectively 8 METH and 8 SAL, because we also used the remaining samples from the pilot study (S1–S3, M1–M5).
In the striatum of prenatally METH-exposed rats, the mRNA expression of Tacr3 gene (p < 0.01) and dopamine receptor D3 gene (p < 0.05) were significantly increased compared to that in prenatally SAL-exposed rats. The measured values were normalized to the measured values of the reference B2m gene (Figures 2, 3). No increase however was observed, when the measured values were normalized to the measured values of the reference Rps6 gene.
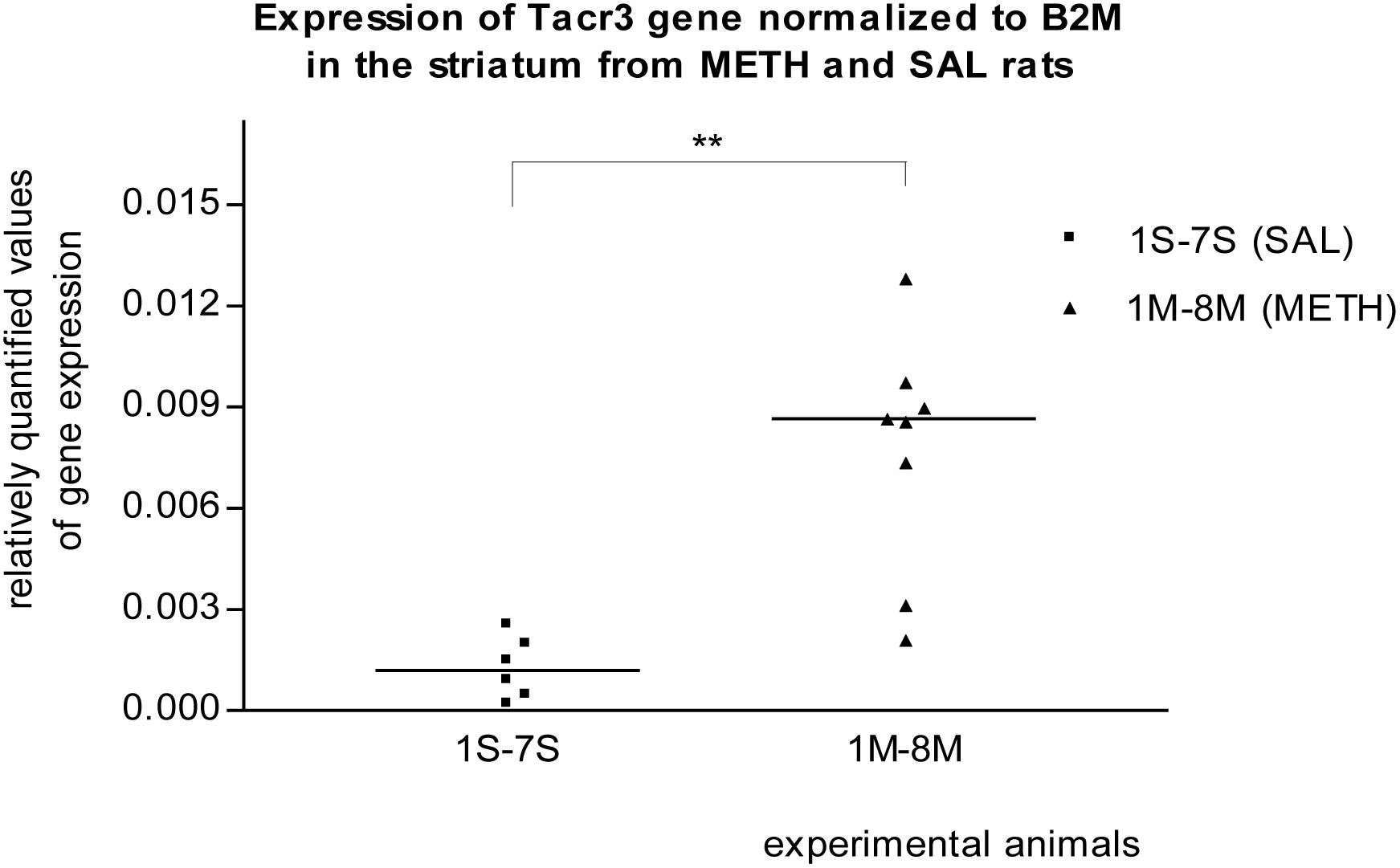
Figure 2. Significant difference (p ≤ 0.01) in the expression of the Tacr3 gene between METH and SAL samples in the striatum. The results of the Tacr3 gene expression were normalized to the results of the expression of the reference B2m gene. The values of METH rats are marked as M and the values of SAL rats are marked as S. The values are presented with a median.
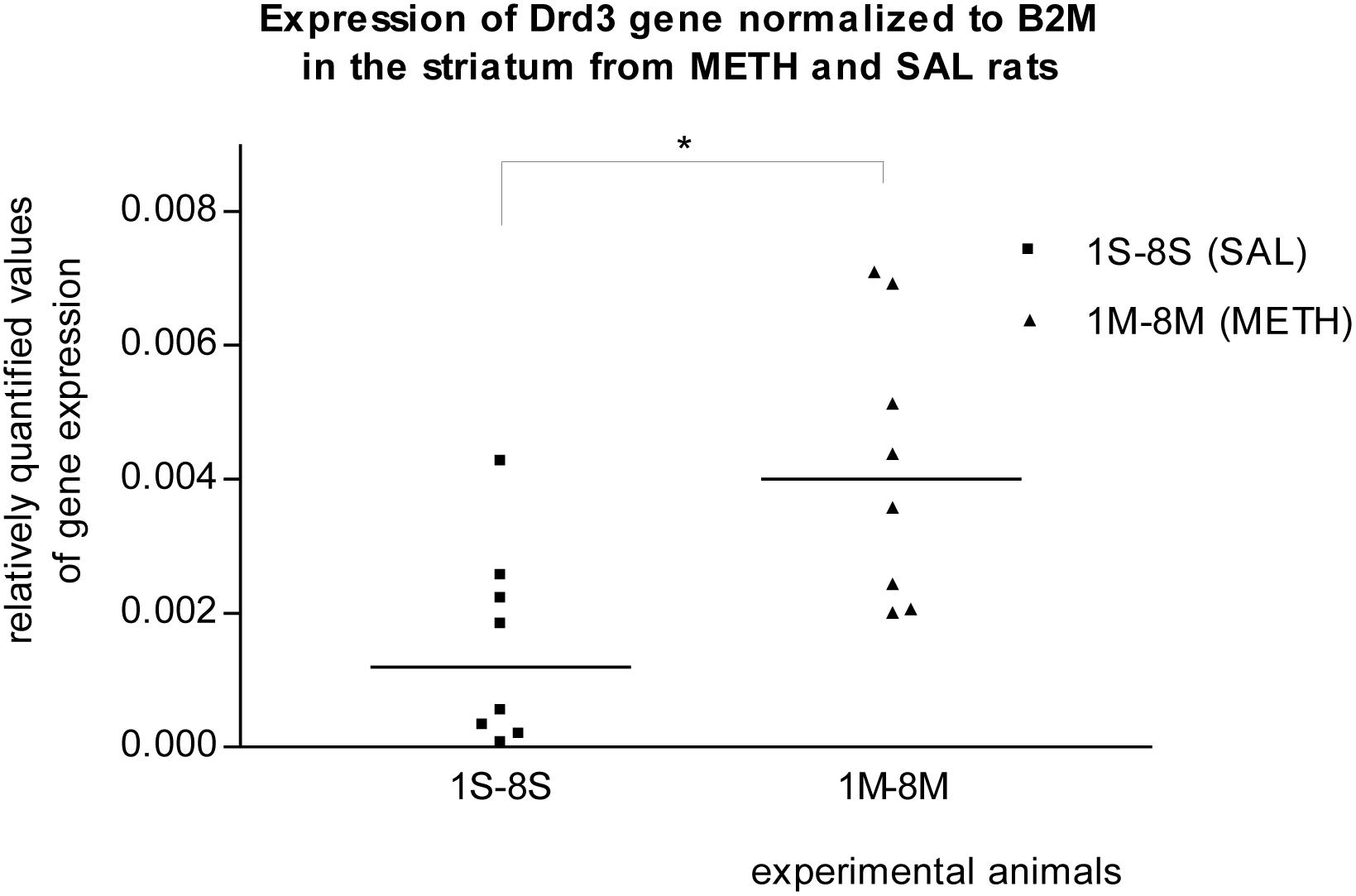
Figure 3. Significant difference (p ≤ 0.05) in the expression of the dopamine D3 receptor (Drd3) gene between METH and SAL samples in the striatum. The results of the Drd3 gene expression were normalized to the results of the expression of the reference B2m gene. The values of METH rats are marked as M and the values of SAL rats are marked as S. The values are presented with a median.
No differences in the mRNA expression were observed for any of the genes between prenatally METH-exposed rats and prenatally SAL-exposed rats in the hippocampus, even if the measured values were normalized to the measured values of either Rps6 or B2m reference genes.
Using a real-time PCR technique we examined changes in gene expression in the striatum, hippocampus, and prefrontal cortex. Using microarray we examined the changes in gene expression in the striatum and hippocampus of prenatally METH-exposed rats. Despite our expectations, we found only three genes in the striatum from approximately 2189 up-regulated or down-regulated genes, and found only two genes in the hippocampus from approximately 1344 up-regulated or down-regulated genes, whose mRNA expression were significantly altered (p < 0.05 and logFC > 1). In addition, according to our previous studies, particularly the striatum and hippocampus are structures that demonstrated the impairment associated with METH-induced behavioral changes (Bubeníková-Valešová et al., 2009; Fialová et al., 2015; Fujáková-Lipski et al., 2017). For that reason we did not perform the third microarray trial with the samples from the prefrontal cortex, where we did not expect many changes.
The microarray technique demonstrated significant decrease in the mRNA expression of the genes encoding the olfactory receptors 522 and 1448 in the striatum of prenatally METH-exposed rats. In the hippocampus of prenatally METH-exposed rats the microarray technique demonstrated significant decrease in the mRNA expression of the olfactory receptor 1726. Given that the olfactory receptors are only present in the Rattus norvegicus species (NCIB database) we did not further investigate the decreased mRNA of olfactory gene expression in the striatum and hippocampus, as such studies in rats would not contribute to our understanding of addiction to METH in humans.
Using real-time PCR 6 genes, Gapdh, Pgk1, 18sRNA, B2m, Rps6, and PPIA (Bernaudin et al., 2002; Hori et al., 2005; Shilling et al., 2006; Romanowski et al., 2007; Langnaese et al., 2008; Gubern et al., 2009) were assessed for their potential as being suitable reference genes in all three types of tissue. Reference genes, Rps6 and B2m, were chosen according to the best reproducible results observed for gene expression in all three types of tissues (striatum, hippocampus, prefrontal cortex).
Ribosomal Protein S6 is a major substrate of protein kinases in eukaryotic ribosomes (NCBI gene database). Recent studies concluded that cellular Ser/Thr kinase mTOR1 activation and rpS6 phosphorylation interrelated (Volovelsky et al., 2016; Huang et al., 2018). Activated mTORC1 complex initiates phosphorylation of rpS6 protein (Volovelsky et al., 2016; Huang et al., 2018), and METH decreases phosphorylation of mTORC1 and its downstream kinases (Liu et al., 2019). mTOR activation in the complexes with proteins, subsequent activation/phosphorylation of down-stream substrates (RSP6 protein) and METH-induced suppression of mTOR1 phosphorylation was also demonstrated in the nucleus accumbens (Huang et al., 2018). In summary, mTORC1 regulates cell proliferation through Rps6 phosphorylation and METH decreases phosphorylation of mTORC1. Given that RPS6 is related to METH metabolism, the results of analyses, where RPS6 is used as a reference gene, are questionable. Of note, the gene encoding Rps6ka3, which is Rps6 kinase, polypeptide 3, was one of 29489 up-regulated genes in the striatum in the first microarray trial.
Beta-2-Microglobulin is a serum protein found in association with the major histocompatibility complex class I heavy chain on the surface of nearly all nucleated cells (Gussow et al., 1987; NCBI gene database). No differences in the expression of the B2m gene were observed in the striatum or in the hippocampus during the microarray trials. The B2m gene was therefore considered to be a suitable reference gene for use in the real-time PCR studies, when examining all three types of tissues in the prenatally METH-exposed rats.
The previous studies demonstrated that prenatal METH exposure can impair the development of the neonatal central nervous system (Williams et al., 2003; Šlamberová et al., 2006; Smith et al., 2008; Rambousek et al., 2014) and the developing brain seems to be one of the primary targets of developmental toxicities of amphetamine and methamphetamine (Thompson et al., 2009; Lloyd et al., 2013). There was evidence of oxidative DNA damage in embryonic and fetal brains of mice caused by amphetamines (Jeng et al., 2005; Wong et al., 2008). METH can be activated to free radical intermediates that initiate ROS formation and cause oxidative DNA damage in the brain (Jeng et al., 2006; Wong et al., 2008; Jeng and Wells, 2010; Ramkissoon and Wells, 2011). The damage leads to long-term postnatal neurodevelopmental deficits via a mechanism different from that underlying the neurodegeneration observed in METH-exposed adults (Jeng et al., 2005).
Prenatal METH exposure induces abnormalities in the brain’s structure and chemistry, especially in the striatum (Chang et al., 2007; Derauf et al., 2012). Won et al. (2001) described regional dependent distribution of METH in pup brains on gestational day 14 with the highest concentration in the striatum (Won et al., 2001; Rambousek et al., 2014). Cart peptide was detected in the rat nucleus accumbens and due to its distribution in the brain and its modulation of dopamine systems, may be involved in aspects of reward and drug abuse (Hubert and Kuhar, 2005; Kuhar et al., 2005). In our project, the microarray technique demonstrated a significant increase in the mRNA expression level of the gene coding Cart prepropeptide in the striatum of prenatally METH-exposed rats. The differences in mRNA expression between prenatally METH-exposed and prenatally SAL-exposed rats using microarray were very low. It appears that the dose 5 mg/kg/day of METH administered to the mothers during the gestation period might be too low to cause strong significant impairment in the development of the striatum, hippocampus and prefrontal cortex of the offspring. This may be the reason why we did not detect significant differences in the Cartpt gene expression using real-time PCR technique. However, when comparing our results to those of older studies, it must be pointed out that also acute administration of methamphetamine increased Cartpt mRNA level in the nucleus accumbens (Ogden et al., 2004; Salinas et al., 2006; Jean et al., 2007; Rogge et al., 2008). Kuhar and colleagues demonstrated that Cart peptides have a role in drug abuse by virtue of the fact that they are modulators of mesolimbic function (Kuhar et al., 2005). We can speculate that the mechanism of METH regulation of addiction is via the CREB phosphorylation pathway. METH self-administration causes enrichment of phosphorylated CREB (pCREB) (Krasnova et al., 2013) and CREB may activate Cartpt expression (Rogge et al., 2008), and biologically active Cart peptide plays a role in reward and addiction (Entrez PubMed, 2019a; GeneCards, 2019a).
It was previously reported that METH self-administration was associated with transcriptional changes in genes that regulate transcription, synaptic transmission, and synaptic plasticity in the dorsal striatum (Krasnova et al., 2013). These molecular changes are associated with the transition from recreational drug use to addiction (Volkow et al., 2001; Krasnova et al., 2013). Dopamine Receptor D3 has the highest binding affinity for endogenous dopamine of all known dopamine receptors (Sokoloff et al., 1990; O’Brien and Gardner, 2005) and shows preferential neuroanatomical localization in the limbic area (Bouthenet et al., 1991; Le’vesque et al., 1992; Levant, 1998; Diaz et al., 2000; O’Brien and Gardner, 2005). In our real-time PCR studies, the expression of the Drd3 gene in the striatum of METH exposed rats appeared to be significantly (p ≤ 0.05) increased compared to that in SAL exposed rats. Borderline significant increased mRNA expression of dopamine receptor D3 gene in the striatum was also observed in the first microarray study. Le Foll et al. (2005) reported that a single administration of METH induces a transient increase in BDNF expression in the prefrontal cortex, associated with a long-lasting elevation in dopamine receptor D3 binding and mRNA levels in the nucleus accumbens (Le Foll et al., 2005; Sokoloff et al., 2006). From cocaine use receptors D3 appeared to be up-regulated in the nucleus accumbens (Staley and Mash, 1996; Segal et al., 1997; Neisewander et al., 2004; O’Brien and Gardner, 2005). Given that the effects of METH are believed to result primarily from the increased release of synthesized dopamine and inhibition of dopamine uptake (O’Brien and Gardner, 2005; Chen et al., 2014) and that elevated extracellular concentrations of dopamine lead to increased stimulation of dopaminergic receptors (Robinson and Becker, 1986; Pierce and Kumaresan, 2006; Rambousek et al., 2014) we could speculate that the basis of Drd3 contribution to the addiction to METH is in its sensitivity. A normalization of dopamine receptor D3 function may reduce vulnerability to relapse in psychostimulant abuse (Le Foll et al., 2005; Chen et al., 2014), and a blockade of the dopamine receptor D3 by an antagonist attenuated the incentive motivational effects of METH in rats and may have pharmacotherapeutic potential in the treatment of methamphetamine addiction (Heidbreder et al., 2005; Chen et al., 2014). Tachykinin Receptor 3 gene (NK-3 receptor) expression was significantly (p ≤ 0.01) increased in the striatum of METH exposed rats compared to that in SAL exposed rats in our real-time PCR studies. Borderline significant increased mRNA expression of the Tacr3 gene in striatum was also detected in the microarrays trial. Of further note, we observed that the expression of the Tacr3 gene was also non-significantly up-regulated in METH hippocampus samples in our second microarray study. This result provides new evidence of METH and Tacr3 relation or interaction. The previous findings described the relation between METH neurotoxicity and neurokinin-1 (NK-1) receptor. The striatal neurokinin-1 receptors modulated METH-induced production of nitric oxide (Wang and Angulo, 2011). METH induces pre- and post-synaptic damage in the striatum and can be prevented with pharmacological blockade of the NK-1 receptor (Yu et al., 2004; Tulloch et al., 2011). It was also previously reported that the NK-3 receptor (Tacr3) controls nucleus accumbens dopamine responses to cocaine in rats (Jocham et al., 2007; Barros et al., 2013) and to alcohol in humans (Foroud et al., 2008). In monkeys, repeated administration of cocaine induces a decrease in methylation of NK3 receptor gene (De Souza Silva et al., 2006; Silva et al., 2008) and the NK3 (Tacr3) antagonist blocks the behavioral effects of cocaine (Barros et al., 2013; Melamed et al., 2013). We can speculate that prenatal METH exposure, like cocaine, decreases the methylation status of the Tacr3 gene and increases the expression of the Tacr3 gene within striatum of rats. If the pharmacological blockage of tachykinin receptors 1 and 3 prevents the METH effects in the striatum, the Tacr3 and its gene is a suitable target for studying the addiction to METH.
Previous studies demonstrated that prenatal METH exposure may be neurotoxic to the developing brain; and prenatally METH-exposed children exhibited smaller hippocampus volumes compared with the control group correlated with poorer performance on sustained attention and delayed verbal memory (Chang et al., 2004). METH exposure early in life of mice caused sex- dependent impairments in object recognition, spatial learning, and memory in adulthood (Acevedo et al., 2007). In experimental rats, prenatal MA exposure caused an impairment of non-spatial memory (Šlamberová et al., 2014). Our microarray technique detected significant differences in the expression of the Ces2 gene in the hippocampus of prenatally METH- and SAL-exposed rats. Ces2 is a member of a large multigene family. The enzymes encoded by these genes are responsible for the hydrolysis of ester- and amide-bond-containing drugs such as cocaine and heroin. They also hydrolyze long-chain fatty acid esters and thioesters (Lian et al., 2018; Entrez PubMed, 2019b; GeneCards, 2019b; Wikipedia contributors, 2019a). The products of hydrolysis are generally more polar than the original ester resulting in an increase in water solubility, promoting renal elimination (Laizure et al., 2013). Mice and rats have plasma carboxylesterase activity, which scavenge many organophosphorus compounds, rendering animals more resistant to organophosphates (Duysen et al., 2011; Marrero-Rosado et al., 2018). The human carboxylesterases are located in the cytoplasm and endoplasmic reticulum of numerous tissues, but the greatest quantities are found in the liver and small intestine where they contribute significantly to the first-pass metabolic hydrolysis of substrate drugs (Imai et al., 2003, 2006; Ross and Crow, 2007; Taketani et al., 2007; Hatfield et al., 2011; Laizure et al., 2013). Our result provides evidence about prenatal METH-induced decrease in the expression of the Ces2 mRNA within the hippocampus of experimental rats. We suspect that the changes in the gene expression level in the hippocampus associated with addiction to METH would be more significant, if the prenatally METH-exposed experimental rats were subjected to stress-inducing conditions, such as social behavior, learning and memory tests. However, we can speculate that decreased expression of the Ces2 enzyme could contribute to such METH-induced impairment of hippocampus in rats. It is interesting to note that there is evidence of a clinically significant drug interaction of a carboxylesterase substrate and ethanol in humans. Ethanol-mediated inhibition of human Ces2 hydrolysis of cocaine was demonstrated in hepatic microsomes (Roberts et al., 1993; Laizure et al., 2013). The finding is an in vitro proof of an existence of metabolic process or signaling pathway, which could inhibit the function of Ces2 in mouse and human cells.
Our microarray technique detected up-regulated expression of Taar 7h gene in the hippocampus of prenatally METH-exposed rats. The specific function of Taar7h and other members of a family of Taars has not been determined yet (Rutigliano et al., 2018). Taars are widely distributed throughout peripheral and brain tissues. Some Taars appear to be olfactory receptors, at least in rodents (Liberles and Buck, 2006; Rutigliano et al., 2018). The most effects have been reported on the Taar1 receptor. The previous experiments revealed Taar1 expression in many distinct rodent CNS regions, namely, nucleus accumbens/olfactory tubercle, as well as limbic and monoaminergic areas, such as hippocampus (Borowsky et al., 2001; Bunzow et al., 2001; Rutigliano et al., 2018). Taar1 is a novel G protein-coupled receptor, which responds to a number of endogenous amines, as well as to number psychoactive drugs, i.e., amphetamines. Such sensitization effects of Taar1 to dopamine, serotonin and norepinephrine could account for its involvement in the rewarding effects of drugs of abuse. Accordingly, Taar1-knock out mice developed an earlier and longer-lasting methamphetamine induced conditioned place preference as compared to wild-type littermates (Achat-Mendes et al., 2012; Rutigliano et al., 2018). In addition, Taar1-knock out mice show increased locomotor response to amphetamine and, in the opposite case, the mice overexpressing Taar1 exhibited a lower response to the stimulating effects of amphetamine in terms of locomotor activity and monoamine release (Rutigliano et al., 2018). These findings are important in the understanding of our result of the increased expression of Taar7 gene and of the previous finding that adult female rats in diestrus and adult males postnatally exposed to METH via breast milk had decreased locomotion and exploratory behavior (Hrubá et al., 2012).
Previous studies reported the prenatal METH-induced impairment of prefrontal cortex in experimental rats (Fujáková-Lipski et al., 2017; Hrebíčková et al., 2017) and in humans (Thompson et al., 2009). Mizoguchi and Yamada (2019) reported that patients addicted to methamphetamine exhibit impaired cognitive functions such as executive function, attention, social cognition, flexibility, and working memory (Mizoguchi and Yamada, 2019). From our studies we found that the expression of the Foxp2 gene was significantly (p ≤ 0.05) changed using real-time PCR and non-significantly changed using microarray technique in the prefrontal cortex of prenatally METH-exposed rats compared to the gene expression in prefrontal cortex of prenatally SAL-exposed rats. The data from real-time technique were normalized to the expression of the reference Rps6 gene. Given the fact that RPS6 phosphorylation is related to the metabolism of methamphetamine, the result from real-time PCR is questionable. However, the function of the neural learning system is modulated in the case of Foxp2 mutation (Chandrasekaran et al., 2015) and some cases of speech impairment were observed in prenatally METH-exposed children in some clinical trials (Cernerud et al., 1996; Wouldes et al., 2004).
In response to various stimuli, including stress, glucocorticoids coordinate metabolic, endocrine, immune, and nervous system responses (Liu et al., 1997; Tronche et al., 1999). In the brain, the glucocorticoid receptor has been thought to modulate emotional behavior, cognitive functions, and addictive states (Tronche et al., 1999). Liu et al. (1997) studied neuroendocrine responses to stress in rats and observed that adult rats, the offspring of mothers that exhibited more licking and grooming of pups during the first 10 days of life, exhibited increased glucocorticoid receptor mRNA expression in the hippocampus (Liu et al., 1997). From our studies, no apparent significant increases in gene expression were observed in the hippocampus of prenatally METH-exposed rats. Our results from the real-time PCR trials demonstrated that the expression of the Nr3c1 gene was significantly (p ≤ 0.01) increased in the prefrontal cortex of prenatally METH-exposed rats compared to that in prenatally SAL-exposed rats. Champagne et al. (2003) and Weaver et al. (2004) studied neuroendocrine responses to stress and Kosten et al. (2014) studied neuroendocrine responses to childhood abuse and both confirmed that variations in the maternal behavior of pup-licking are associated with the methylation status of the Nr3c1 gene (Champagne et al., 2003; Weaver et al., 2004; Kosten and Nielsen, 2014; Kosten et al., 2014). Future research should examine if prenatal METH exposure is able to alter the methylation status of the Nr3c1 gene. However, our finding is in accordance with findings reported by other scientific groups, that an increased motor activity and declined adaptability (Middaugh, 1989), change of behavior in an unknown environment, and an improvement of cognitive functions and anxiety (Schutová et al., 2008, 2009, 2012; Šlamberová et al., 2015) are associated to METH in gestation. The glucocorticoid nuclear receptor Nr3c1 and its gene are suitable targets for studying the changes associated with addiction to METH.
The differences in the mRNA expression between prenatally METH-exposed and prenatally SAL-exposed rats using both techniques were very low. It appears that the dose 5 mg/kg/day of METH administered to the mothers during the gestation period might be too low to cause strong significant impairment in the development of the striatum, hippocampus and prefrontal cortex of the offspring. We have not demonstrated robust changes in the gene expression levels of prenatally METH-exposed rats. We isolated total RNA from adult rats on PD 80. It is possible that the changes in the gene expression just after birth could have been more robust since postnatally induced permanent behavioral changes have an impact on gene expression in the brain and can dramatically change prenatally adjusted a gene expression scenario that can disappear in adulthood.
The aims of our project were fulfilled. We have identified susceptible genes, candidates to the study of an impairment related to addiction to METH. Cartpt, Drd3, Tacr3 in the striatum and the Ces2 in the hippocampus have been found to be associated with addiction to METH. Future research should continue to confirm or refute our finding about an association between prenatal exposure to METH and the changes in the expression of Nr3c1 and Foxp2 genes in the prefrontal cortex. Further research is needed to confirm and investigate the novel finding of prenatally METH-induced decrease of Ces2 mRNA and prenatally METH-induced increase of Taar7h mRNA in the hippocampus. Our plan is to study the changes in the gene expression level in the striatum, hippocampus and prefrontal cortex of prenatally METH-exposed rats during the different stages of gestation. And in addition, we intend to study what genetic and epigenetic changes occur, when prenatally METH-exposed rats are subjected to varying stressful conditions.
The datasets generated for this study can be found in Figshare, https://figshare.com/articles/Results_of_second_microarray_trial/8081120; https://figshare.com/articles/Results_of_first_microarray_trial/8081072.
The study, the protocols and procedures with experimental animals were reviewed and approved by the Committee for Protection of Experimental Animals of Third faculty of Medicine and by Departmental Committee of Ministry of Education, Youth, and Sports. They are also in agreement with the Czech Government Requirements under the Policy of Humans Care of Laboratory Animals (no. 246/1992) and also comply with the regulations of the Ministry of Agriculture of the Czech Republic (no. 311/1997).
HZ isolated the RNA, chose the second group of the genes, carried out the real-time PCR experiments, analyzed data, and wrote the manuscript. AT participated in the real-time PCR experiments and in the data analysis. KN carried out the experiments with animals. MÈ participated in study design and coordination, chose the first group of the genes, and revised the manuscript. RŠ conceived the study, carried out the experiments with animals, and revised the manuscript. All authors approved the final version of the manuscript for submission.
This work was supported by the grant project 18-03806S from Grant Agency of the Czech Republic, by program PROGRES Q35 and 260388/SVV/2019 from Charles University, and project PharmaBrain CZ.02.1.01/0.0/0.0/16_025/0007444 funded from OP VVV.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Morna Mc Nish for checking the spelling and grammar in the manuscript and Marie Kvasnicová for great laboratory assistance.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnins.2019.00771/full#supplementary-material
ASCR, Academy of Sciences of the Czech Republic; B2m, Beta-2-microglobulin; Bdnf, Brain-derived neurotrophic factor; Cartpt, Cocaine- And Amphetamine-Regulated Transcript prepropeptide; Ces2, Carboxylesterase 2; DAT, dopamine transporters; Drd3, Dopamine receptor D3; Esr1, Estrogen receptor 1; Foxp2, Forkhead box P2; Gapdh, Glyceraldehyde-3-phosphate dehydrogenase; Igfr1, Insulin-like growth factor 1; METH, Methamphetamine; mTORC1, mammalian target of rapamycin complex 1; NK-1, neurokinin 1; NK-3, neurokinin 3; Nr3c1, Nuclear receptor subfamily 3group c member 1; Olr1448, Olfactory receptor 1448; Olr1726, Olfactory receptor 1726; Olr522, Olfactory receptor 522; PD, Postnatal day; Pgk1, Phosphoglycerate kinase 1; PPIA, Peptidylprolyl isomerase A; Rps6, Ribosomal protein S6; Rps6ka3, Ribosomal protein S6 kinase A3; s.c., Subcutaneously; SAL, Saline; Ser/Thr, Serine/threonine; Taar, trace amine-associated receptor; Taar7h, trace amine-associated receptor 7h; Tacr3, Tachykinin receptor 3.
Acevedo, S. F., de Esch, I. J. P., and Raber, J. (2007). Sex- and histamine-dependent long-term cognitive effects of methamphetamine exposure. Neuropsychopharmacology 32, 665–672. doi: 10.1038/sj.npp.1301091
Achat-Mendes, C., Lynch, L. J., Sullivan, K. A., Vallender, E. J., and Miller, G. M. (2012). Augmentation of methamphetamine-induced behaviors in transgenic mice lacking the trace amine-associated receptor 1. Pharmacol. Biochem. Behav. 101, 201–207. doi: 10.1016/j.pbb.2011.10.025
Acuff-Smith, K. D., George, M., Lorens, S. A., and Vorhees, C. V. (1992). Preliminary evidence for methamphetamine-induced behavioral and ocular effects in rat offspring following exposure during early organogenesis. Psychopharmacology 109, 255–263. doi: 10.1007/bf02245871
Acuff-Smith, K. D., Schilling, M. A., Fisher, J. E., and Voorhees, C. V. (1996). Stage-specific effects of prenatal d-methamphetamine exposure on behavioral and eye development in rats. Neurotoxicol. Teratol. 18, 199–215. doi: 10.1016/0892-0362(95)02015-2
Ago, Y., Arikawa, S., Yata, M., Yano, K., Abe, M., Takuma, K., et al. (2009). Role of prefrontal dopaminergic neurotransmission in glucocorticoid receptor-mediated modulation of methamphetamine-induced hyperactivity. Synapse 63, 7–14. doi: 10.1002/syn.20575
Anderson, P., Morris, R., Amaral, D., Bliss, T., and O’Keefe, J. (2007). “The Hippocampal Formation,” in The Hippocampus Book, First Edn, eds P. Anderson, R. Morris, D. Amaral, T. Bliss, and J. O’Keefe (New York, NY: Oxford University Press).
Barros, M., Dempster, E. L., Illott, N., Chabrawi, S., Maior, R. S., Tomaz, C., et al. (2013). Decreased methylation of the NK3 receptor coding gene (TACR3) after cocaine-induced place preference in marmoset monkeys. Addict. Biol. 18, 452–454. doi: 10.1111/j.1369-1600.2011.00409.x
Bernaudin, M., Tang, Y., Reilly, M., Petit, E., and Sharp, F. R. (2002). Brain genomic response following hypoxia and re-oxygenation in the neonatal rat. Identification of genes that might contribute to hypoxia-induced ischemic tolerance. J. Biol. Chem. 277, 39728–39738. doi: 10.1074/jbc.M204619200
Billing, L., Eriksson, M., Jonsson, B., Steneroth, G., and Zetterström, R. (1994). The influence of environmental factors on behavioral problems in 8-year-old children exposed to amphetamine during fetal life. Child Abuse Negl. 18, 3–9. doi: 10.1016/0145-2134(94)90091-4
Borowsky, B., Adham, N., Jones, K. A., Raddatz, R., Artymyshyn, R., Ogozalek, K. L., et al. (2001). Trace amines: identification of a family of mammalian G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 8966–8971. doi: 10.1073/pnas.151105198
Bouthenet, M. L., Souil, E., Martres, M. P., Sokoloff, P., Giros, B., and Schwartz, J. C. (1991). Localization of dopamine D3 receptor mRNA in the rat brain using in situ hybridization histochemistry: comparison with dopamine D2 receptor mRNA. Brain Res. 564, 203–219. doi: 10.1016/0006-8993(91)91456-b
Bubeníková-Valešová, V., Kačer, P., Syslová, K., Rambousek, L., Janovský, M., Schutová, B., et al. (2009). Prenatal methamphetamine exposure affects the mesolimbic dopaminergic system and behavior in adult offspring. Int. J. Dev. Neurosci. 27, 525–530. doi: 10.1016/j.ijdevneu.2009.06.012
Bunzow, J. R., Sonders, M. S., Arttamangkul, S., Harrison, L. M., Zhang, G., Quigley, D. I., et al. (2001). Amphetamine, 3,4-methylenedioxymethamphetamine, lysergic acid diethylamide, and metabolites of the catecholamine neurotransmitters are agonists of a rat trace amine receptor. Mol. Pharmacol. 60, 1181–1188. doi: 10.1124/mol.60.6.1181
Burchfield, D. J., Lucas, V. W., Abrams, R. M., Miller, R. L., and DeVane, C. L. (1991). Disposition and pharmacodynamics of methamphetamine in pregnant sheep. JAMA 265, 1968–1973. doi: 10.1001/jama.265.15.1968
Cadet, J. L., and Krasnova, I. N. (2009). Molecular bases of methamphetamine-induced neurodegeneration. Int. Rev. Neurobiol. 88, 101–119. doi: 10.1016/S0074-7742(09)88005-7
Cernerud, L., Eriksson, M., Jonsson, B., Steneroth, G., and Zetterström, R. (1996). Amphetamine addiction during pregnancy: 14- year follow-up of growth and school performance. Acta Paediatr. 85, 204–208. doi: 10.1111/j.1651-2227.1996.tb13993.x
Champagne, F. A., Francis, D. D., Mar, A., and Meaney, M. J. (2003). Variations in maternal care in the rat as a mediating influence for the effects of environment on development. Physiol. Behav. 79, 359–371. doi: 10.1016/s0031-9384(03)00149-5
Chandrasekaran, B., Yi, H. G., Blanco, N. J., McGeary, J. E., and Maddox, W. T. (2015). Enhanced procedural learning of speech sound categories in a genetic variant of FOXP2. J. Neurosci. 35, 7808–7812. doi: 10.1523/JNEUROSCI.4706-14.2015
Chang, L., Alicata, D., Ernst, T., and Volkow, N. (2007). Structural and metabolic brain changes in the striatum associated with methamphetamine abuse. Addiction 102, 16–32. doi: 10.1111/j.1360-0443.2006.01782.x
Chang, L., Smith, L. M., LoPresti, C., Yonekura, M. L., Kuo, J., Walot, I., et al. (2004). Smaller subcortical volumes and cognitive deficits in children with prenatal methamphetamine exposure. Psychiatry Res. 132, 95–106. doi: 10.1016/j.pscychresns.2004.06.004
ChEBI (2018). The European Bioinformatics Institute (EMBL-EBI), CHEBI:6809 – Methamphetamine. Available at: https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:6809 (accessed June 23, 2018).
Chen, Y., Song, R., Yang, R. F., Wu, N., and Li, J. (2014). A novel dopamine D3 receptor antagonist YQA14 inhibits methamphetamine self-administration and relapse to drug-seeking behavior in rats. Eur. J. Pharmacol. 743, 126–132. doi: 10.1016/j.ejphar.2014.09.026
Cho, D. H., Lyu, H. M., Lee, H. B., Kim, P. Y., and Chin, K. (1991). Behavioral teratogenicity of methamphetamine. J. Toxicol. Sci. 16, 37–49. doi: 10.2131/jts.16.supplementi_37
Couceyro, P. R., Evans, C., McKinzie, A., Mitchell, D., Dube, M., Hagshenas, L., et al. (2005). Cocaine- and amphetamine-regulated transcript (CART) peptides modulate the locomotor and motivational properties of psychostimulants. J. Pharmacol. Exp. Ther. 315, 1091–1100. doi: 10.1124/jpet.105.091678
Cui, C., Sakata-Haga, H., Ohta, K., Nishida, M., Yashiki, M., Sawada, K., et al. (2006). Histological brain alterations following prenatal methamphetamine exposure in rats. Cong.. Anom. Kyoto 46, 180–187. doi: 10.1111/j.1741-4520.2006.00126.x
De Souza Silva, M. A., Mello, E. L. Jr., Müller, C. P., Jocham, G., Maior, R. S., Huston, J. P., et al. (2006). The tachykinin NK3 receptor antagonist SR142801 blocks the behavioral effects of cocaine in marmoset monkeys. Eur. J. Pharmacol. 536, 269–278. doi: 10.1016/j.ejphar.2006.03.010
Derauf, C., Lester, B. M., Neyzi, N., Kekatpure, M., Gracia, L., Davis, J., et al. (2012). Subcortical and cortical structural central nervous system changes and attention processing deficits in preschool-aged children with prenatal methamphetamine and tobacco exposure. Dev. Neurosci. 34, 327–341. doi: 10.1159/000341119
Diaz, J., Pilon, C., Le Foll, B., Gros, C., Triller, A., Schwartz, J. C., et al. (2000). Dopamine D3 receptors expressed by all mesencephalic dopamine neurons. J. Neurosci. 20, 8677–8684. doi: 10.1523/jneurosci.20-23-08677.2000
Diaz, S. D., Smith, L. M., LaGasse, L. L., Derauf, C., Newman, E., Shah, R., et al. (2014). Effects of prenatal methamphetamine exposure on behavioral and cognitive findings at 7.5 years. J. Pediatr. 164, 1333–1338. doi: 10.1016/j.jpeds.2014.01.053
Dixon, S. D., and Bejar, R. (1989). Echoencephalographic findings in neonates associated with maternal cocaine and methamphetamine use: incidence and clinical correlates. J. Pediatr. 115, 770–778. doi: 10.1016/s0022-3476(89)80661-4
Duysen, E. G., Koentgen, F., Williams, G. R., Timperley, C. M., Schopfer, L. M., Cerasoli, D. M., et al. (2011). Production of ES1 plasma carboxylesterase knockout mice for toxicity studies. Chem. Res. Toxicol. 24, 1891–1898. doi: 10.1021/tx200237a
Entrez PubMed (2019a). Gene Database, “CARTPT CART Prepropeptide”, Gene ID: 29131. Available at: https://www.ncbi.nlm.nih.gov/gene/29131 (accessed June 8, 2019).
Entrez PubMed (2019b). Gene Database, “CES2 carboxylesterase 2“, Gene ID: 8824. Available at: https://www.ncbi.nlm.nih.gov/gene/8824 (accessed June 8, 2019).
Eriksson, M., Billing, L., Steneroth, G., and Zetterstrom, R. (1989). Health and development of 8-year-old children whose mothers abused amphetamine during pregnancy. Acta Paediatr. Scand. 78, 944–949. doi: 10.1111/j.1651-2227.1989.tb11179.x
Eriksson, M., Larsson, G., Winbladh, B., and Zetterstrom, R. (1978). The influence of amphetamine addiction on pregnancy and the newborn infant. Acta Paediatr. Scand. 67, 95–99. doi: 10.1111/j.1651-2227.1978.tb16283.x
Eriksson, M., Larsson, G., and Zetterstrom, R. (1981). Amphetamine addiction and pregnancy. II. Pregnancy, delivery and the neonatal period. Socio-medical aspects. Acta Obstet. Gynecol. Scand. 60, 253–259. doi: 10.3109/00016348109158127
Eriksson, M., and Zetterstrom, R. (1994). Amphetamine addiction during pregnancy: 10-year follow-up. Acta Paediatr. Suppl. 404, 27–31. doi: 10.1111/j.1651-2227.1994.tb13380.x
Fialová, M., Šírová, J., Bubeníková-Valešová, V., and Šlamerová, R. (2015). The effect of prenatal methamphetamine exposure on recognition memory in adult rats. Prague Med. Rep. 116, 31–39. doi: 10.14712/23362936.2015.43
Foroud, T., Wetherill, L. F., Kramer, J., Tischfield, J. A., Nurnberger, J. I. Jr., Schuckit, M. A., et al. (2008). The tachykinin receptor 3 is associated with alcohol and cocaine dependence. Alcohol. Clin. Exp. Res. 32, 1023–1030. doi: 10.1111/j.1530-0277.2008.00663.x
Fujáková-Lipski, M., Kaping, D., Šírová, J., Horáček, J., Páleníček, T., Zach, P., et al. (2017). Trans-generational neurochemical modulation of methamphetamine in the adult brain of the Wistar rat. Arch. Toxicol. 91, 3373–3384. doi: 10.1007/s00204-017-1969-y
Gecz, J., and Mulley, J. (2000). Genes for cognitive function: developments on the X. Genome Res. 10, 157–163. doi: 10.1101/gr.10.2.157
GeneCards (2019a). Human Gene Database “Cartpt gene”. Available at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CARTPT (accessed June 8, 2019).
GeneCards (2019b). Human Gene Database “CES2 gene”. Available at: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CES2 (accessed June 8, 2019).
Gilks, C. B., Vanderhyden, B. C., Zhu, S., van de Rijn, M., and Longacre, T. A. (2005). Distinction between serous tumors of low malignant potential and serous carcinomas based on global mRNA expression profiling. Gynecol. Oncol. 96, 684–694. doi: 10.1016/j.ygyno.2004.11.039
Gubern, C., Hurtado, O., Rodríguez, R., Morales, J. R., Romera, V. G., Moro, M. A., et al. (2009). Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol. Biol. 10:57. doi: 10.1186/1471-2199-10-57
Gussow, D., Rein, R., Ginjaar, I., Hochstenbach, F., Seemann, G., Kottman, A., et al. (1987). The human beta-2-microglobulin gene. Primary structure and definition of the transcriptional unit. J. Immunol. 139, 3132–3138.
Hatfield, M. J., Tsurkan, L., Garrett, M., Shaver, T. M., Hyatt, J. L., Edwards, C. C., et al. (2011). Organ-specific carboxylesterase profiling identifies the small intestine and kidney as major contributors of activation of the anticancer prodrug CPT-11. Biochem. Pharmacol. 81, 24–31. doi: 10.1016/j.bcp.2010.09.001
Heal, D. J., Smith, S. L., Gosden, J., and Nutt, D. J. (2013). Amphetamine, past and present – a pharmacological and clinical perspective. J. Psychopharmacol. 27, 479–496. doi: 10.1177/0269881113482532
Heidbreder, C. A., Gardner, E. L., Xi, Z. X., Thanos, P. K., Mugnaini, M., Hagan, J. J., et al. (2005). The role of central dopamine D3 receptors in drug addiction: a review of pharmacological evidence. Brain Res. Brain Res. Rev. 49, 77–105. doi: 10.1016/j.brainresrev.2004.12.033
Holubová, A., Lukášková, I., Tomášová, N., Šuhajdová, M., and Šlamberová, R. (2018). Early postnatal stress impairs cognitive functions of male rats persisting until adulthood. Front. Behav. Neurosci. 12:176. doi: 10.3389/fnbeh.2018.00176
Holubová, A., Štofková, A., Jurčovičová, J., and Šlamberová, R. (2016). The effect of neonatal maternal stress on plasma levels of adrenocorticotropic hormone, corticosterone, leptin, and ghrelin in adult male rats exposed to acute heterotypic stressor. Physiol. Res. 65, S557–S566.
Hori, S., Ohtsuki, S., Ichinowatari, M., Yokota, T., Kanda, T., and Terasaki, T. (2005). Selective gene silencing of rat ATP-binding cassette G2 transporter in an in vitro blood-brain barrier model by short interfering RNA. J. Neurochem. 93, 63–71. doi: 10.1111/j.1471-4159.2004.02994.x
Hrebíčková, I., Ševčíková, M., Macúchová, E., and Šlamberová, R. (2017). How methamphetamine exposure during different neurodevelopmental stages affects social behavior of adult rats? Physiol. Behav. 179, 391–400. doi: 10.1016/j.physbeh.2017.07.009
Hrubá, L., Schutová, B., Pometlová, M., Rokyta, R., and Šlamberová, R. (2010). Effect of methamphetamine exposure and cross-fostering on cognitive function in adult male rats. Behav. Brain Res. 208, 63–71. doi: 10.1016/j.bbr.2009.11.001
Hrubá, L., Schutova, B., and Šlamberová, R. (2012). Sex differences in anxiety-like behavior and locomotor activity following prenatal and postnatal methamphetamine exposure in adult rats. Physiol. Behav. 105, 364–370. doi: 10.1016/j.physbeh.2011.08.016
Hrubá, L., Schutová, B., Šlamberová, R., Pometlová, M., and Rokyta, R. (2009). Effect of methamphetamine exposure and cross-fostering on sensorimotor development of male and female rat pups. Dev. Psychobiol. 51, 73–83. doi: 10.1002/dev.20346
Huang, S. H., Wu, W. R., Lee, L. M., Huang, P. R., and Chen, J. C. (2018). mTOR signaling in the nucleus accumbens mediates behavioral sensitization to methamphetamine. Prog. Neuropsychopharmacol. Biol. Psychiatry 86, 331–339. doi: 10.1016/j.pnpbp.2018.03.017
Hubert, G. W., and Kuhar, M. J. (2005). Colocalization of CART with substance P but not enkephalin in the rat nucleus accumbens. Brain Res. 1050, 8–14. doi: 10.1016/j.brainres.2005.05.025
Imai, T., Taketani, M., Shii, M., Hosokawa, M., and Chiba, K. (2006). Substrate specificity of carboxylesterase isozymes and their contribution to hydrolase activity in human liver and small intestine. Drug Metab. Dispos. 34, 1734–1741. doi: 10.1124/dmd.106.009381
Imai, T., Yoshigae, Y., Hosokawa, M., Chiba, K., and Otagiri, M. (2003). Evidence for the involvement of a pulmonary first-pass effect via carboxylesterase in the disposition of a propranolol ester derivative after intravenous administration. J. Pharmacol. Exp. Ther. 307, 1234–1242. doi: 10.1124/jpet.103.056499
Jean, A., Conductier, G., Manrique, C., Bouras, C., Berta, P., Gen, R., et al. (2007). Anorexia induced by activation of serotonin 5-HT4 receptors is mediated by increases in CART in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 104, 16335–16340. doi: 10.1073/pnas.0701471104
Jeng, W., Ramkissoon, A., Parman, T., and Wells, P. G. (2006). Prostaglandin H synthase-catalyzed bioactivation of amphetamines to free radical intermediates that cause CNS regional DNA oxidation and nerve terminal neurodegeneration. FASEB J. 20, 638–650. doi: 10.1096/fj.05-5271com
Jeng, W., and Wells, P. G. (2010). Reduced 3,4-methylenedioxymethamphetamine (Ecstasy)-initiated oxidative DNA damage and neurodegeneration in prostaglandin H synthase-1 knockout mice. ACS Chem. Neurosci. 1, 366–380. doi: 10.1021/cn900022w
Jeng, W., Wong, A. W., Ting-A-Kee, R., and Wells, P. G. (2005). Methamphetamine-enhanced embryonic oxidative DNA damage and neurodevelopmental deficits. Free Radical Biol. Med. 39, 317–326. doi: 10.1016/j.freeradbiomed.2005.03.015
Jocham, G., Lauber, A. C., Müller, C. P., Huston, J. P., and de Souza Silva, M. A. (2007). Neurokinin 3 receptor activation potentiates the psychomotor and nucleus accumbens dopamine response to cocaine, but not its place conditioning effects. Eur. J. Neurosci. 25, 2457–2472. doi: 10.1111/j.1460-9568.2007.05491.x
Kishi, T., Ikeda, M., Kitajima, T., Yamanouchi, Y., Kinoshita, Y., Kawashima, K., et al. (2009). A functional polymorphism in estrogen receptor alpha gene is associated with Japanese methamphetamine induced psychosis. Prog. Neuropsychopharmacol. Biol. Psychiatry 33, 895–898. doi: 10.1016/j.pnpbp.2009.04.008
Kosten, T. A., Huang, W., and Nielsen, D. A. (2014). Sex and litter effects on anxiety and DNA methylation levels of stress and neurotrophin genes in adolescent rats. Dev. Psychobiol. 56, 392–406. doi: 10.1002/dev.21106
Kosten, T. A., and Nielsen, D. A. (2014). Litter and sex effects on maternal behavior and DNA methylation of the Nr3c1 exon 17 promoter gene in hippocampus and cerebellum. Int. J. Dev. Neurosci. 36, 5–12. doi: 10.1016/j.ijdevneu.2014.03.010
Krasnova, I. N., Chiflikyan, M., Justinova, Z., McCoy, M. T., Ladenheim, B., Jayanthi, S., et al. (2013). CREB phosphorylation regulates striatal transcriptional responses in the self-administration model of methamphetamine addiction in the rat. Neurobiol. Dis 58, 132–143. doi: 10.1016/j.nbd.2013.05.009
Krasnova, I. N., Gerra, M. C., Walther, D., Jayanthi, S., Ladenheim, B., Mccoy, M. T., et al. (2017). Compulsive methamphetamine taking in the presence of punishment is associated with increased oxytocin expression in the nucleus accumbens of rats. Sci. Rep. 7:8331. doi: 10.1038/s41598-017-08898-8
Kuhar, M. J., Jaworski, J. N., Hubert, G. W., Philpot, K. B., and Dominguez, G. (2005). Cocaine- and amphetamine-regulated transcript peptides play a role in drug abuse and are potential therapeutic targets. AAPS J. 7, E259–E265. doi: 10.1208/aapsj070125
LaGasse, L. L., Derauf, C., Smith, L. M., Newman, E., Shah, R., Neal, C., et al. (2012). Prenatal methamphetamine exposure and childhood behavior problems at 3 and 5 years of age. Pediatrics 129, 681–688. doi: 10.1542/peds.2011-2209
Laizure, S. C., Herring, V., Hu, Z., Witbrodt, K., and Parker, R. B. (2013). The role of human carboxylesterases in drug metabolism: have we overlooked their importance? Pharmacotherapy. 33, 210–222. doi: 10.1002/phar.1194
Langnaese, K., John, R., Schweizer, H., Ebmeyer, U., and Keilhoff, G. (2008). Selection of reference genes for quantitative real-time PCR in a rat asphyxial cardiac arrest model. BMC Mol. Biol. 9:53. doi: 10.1186/1471-2199-9-53
Le Foll, B., Diaz, J., and Sokoloff, P. (2005). A single cocaine exposure increases BDNF and D-3 receptor expression: implications for drug-conditioning. Neuroreport 16, 175–178. doi: 10.1097/00001756-200502080-00022
Leung, K. P., Qu, Y. H., Qiao, D. F., Xie, W. B., Li, D. R., Xu, J. T., et al. (2014). Critical role of insulin-like growth factor binding protein-5 in methamphetamine-induced apoptosis in cardiomyocytes. Mol. Med. Rep. 10, 2306–2312. doi: 10.3892/mmr.2014.2572
Levant, B. (1998). Differential distribution of D3 dopamine receptors in the brains of several mammalian species. Brain Res. 800, 269–274. doi: 10.1016/S0006-8993(98)00529-0
Le’vesque, D., Diaz, J., Pilon, C., Martres, M. P., Giros, B., Souil, E., et al. (1992). Identification, characterization, and localization of the dopamine D3 receptor in rat brain using 7-[3H]hydroxy-N,N-di-n-propyl-2-aminotetralin. Proc. Natl. Acad. Sci. U.S.A. 89, 8155–8159. doi: 10.1073/pnas.89.17.8155
Lian, J., Nelson, R., and Lehner, R. (2018). Carboxylesterases in lipid metabolism: from mouse to human. Protein Cell. 9, 178–195. doi: 10.1007/s13238-017-0437-z
Liberles, S. D., and Buck, L. B. (2006). A second class of chemosensory receptors in the olfactory epithelium. Nature 442, 645–650. doi: 10.1038/nature05066
Lindemann, L., Meyer, C. A., Jeanneau, K., Bradaia, A., Ozmen, L., Bluethmann, H., et al. (2008). Trace amine-associated receptor 1 modulates dopaminergic activity. J. Pharmacol. Exp. Ther. 324, 948–956. doi: 10.1124/jpet.107.132647
Little, B. B., Snell, L. M., and Gilstrap, L. C. III (1988). Methamphetamine use during pregnancy: outcome and fetal effects. Obstet. Gynecol. 72, 541–544.
Liu, D., Diorio, J., Tannenbaum, B., Caldji, C., Francis, D., Freedman, A., et al. (1997). Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science 277, 1659–1662. doi: 10.1126/science.277.5332.1659
Liu, H. Q., An, Y. W., Hu, A. Z., Li, M. H., Wu, J. L., Liu, L., et al. (2019). Critical roles of the PI3K-Akt-mTOR signaling pathway in apoptosis and autophagy of astrocytes induced by methamphetamine. Open Chem. 17, 96–104. doi: 10.1515/chem-2019-0015
Lloyd, S. A., Oltean, C., Pass, H., Phillips, B., Staton, K., Robertson, C. L., et al. (2013). Prenatal exposure to psychostimulants increases impulsivity, compulsivity, and motivation for rewards in adult mice. Physiol. Behav. 119, 43–51. doi: 10.1016/j.physbeh.2013.05.038
Macúchová, E., Nohejlová, K., and Šlamberová, R. (2014). Gender differences in the effect of adult amphetamine on cognitive functions of rats prenatally exposed to methamphetamine. Behav. Brain Res. 270, 8–17. doi: 10.1016/j.bbr.2014.04.040
Macúchová, E., Nohejlová-Deykun, K., and Šlamberová, R. (2013). Effect of methamphetamine on cognitive functions of adult female rats prenatally exposed to the same drug. Physiol. Res. 62, S89–S98. doi: 10.1016/j.bbr.2014.04.040
Marrero-Rosado, B., de Araujo Furtado, M., Schultz, V., Stone, M., Kundrick, E., Walker, E., et al. (2018). Soman-induced status epilepticus, epileptogenesis, and neuropathology in carboxylesterase knockout mice treated with midazolam. Epilepsia 59, 2206–2218. doi: 10.1111/epi.14582
Martin, J. C. (1975). Effects on offspring of chronic maternal methamphetamine exposure. Dev. Psychobiol. 8, 397–404. doi: 10.1002/dev.420080504
Martin, J. C., Martin, D. C., Radow, B., and Sigman, G. (1976). Growth, development and activity in rat offspring following maternal drug exposure. Exp. Aging Res. 2, 235–251. doi: 10.1080/03610737608257179
Marwick, C. (2000). NIDA seeking data on effect of fetal exposure to methamphetamine. JAMA 283, 2225–2226. doi: 10.1001/jama.283.17.2225
Matějovská, I., Bernášková, K., and Šlamberová, R. (2014). Effect of prenatal methamphetamine exposure and challenge dose of the same drug in adulthood on epileptiform activity induced by electrical stimulation in female rats. Neuroscience 257, 130–138. doi: 10.1016/j.neuroscience.2013.10.069
Matera, R. F., Zabala, H., and Jimenez, A. P. (1968). Bifid exencephalia. Teratogen action of amphetamine. Int. Surg. 50, 79–85.
Melamed, J. L., de Souza Silva, M. A., Tomaz, C., Müller, C. P., Huston, J. P., and Barros, M. (2013). Sensitization of hypervigilance effects of cocaine can be induced by NK3 receptor activation in marmoset monkeys. Drug Alcohol Depend 128, 155–160. doi: 10.1016/j.drugalcdep.2012.08.020
Middaugh, L. D. (1989). Prenatal amphetamine effects on behavior: possible mediation by brain monoamines. Ann. N. Y. Acad. Sci. 562, 308–318. doi: 10.1111/j.1749-6632.1989.tb21028.x
Mizoguchi, H., and Yamada, K. (2019). Methamphetamine use causes cognitive impairment and altered decision-making. Neurochem. Int. 124, 106–113. doi: 10.1016/j.neuint.2018.12.019
Neisewander, J. L., Fuchs, R. A., Tran-Nguyen, L. T., Weber, S. M., Coffey, G. P., and Joyce, J. N. (2004). Increases in dopamine D3 receptor binding in rats receiving a cocaine challenge at various time points after cocaine self-administration: implications for cocaine-seeking behavior. Neuropsychopharmacology 29, 1479–1487. doi: 10.1038/sj.npp.1300456
O’Brien, C. P., and Gardner, E. L. (2005). Critical assessment of how to study addiction and its treatment: human and non-human animal models. Pharmacol. Ther. 108, 18–58. doi: 10.1016/j.pharmthera.2005.06.018
Ogden, C. A., Rich, M. E., Schork, N. J., Paulus, M. P., Geyer, M. A., Lohr, J. B., et al. (2004). Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol. Psychiatry 9, 1007–1029. doi: 10.1038/sj.mp.4001547
Oro, A. S., and Dixon, S. D. (1987). Perinatal cocaine and methamphetamine exposure: maternal and neonatal correlates. J. Pediatr. 111, 571–578. doi: 10.1016/s0022-3476(87)80125-7
Paris, J. M., and Lorens, S. A. (1989). A dose-response analysis of intra-raphe tachykinin-induced hyperactivity. J. Pharmacol. Exp. Ther. 251, 388–393.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29:e45.
Pierce, R. C., and Kumaresan, V. (2006). The mesolimbic dopamine system: the final com- mon pathway for the reinforcing effect of drugs of abuse? Neurosci. Biobehav. Rev. 30, 215–238. doi: 10.1016/j.neubiorev.2005.04.016
Piper, B. J., Acevedo, S. F., Kolchugina, G. K., Butler, R. W., Corbett, S. M., Honeycutt, E. B., et al. (2011). Abnormalities in parentally rated executive function in methamphetamine/polysubstance exposed children. Pharmacol. Biochem. Behav. 98, 432–439. doi: 10.1016/j.pbb.2011.02.013
Plessinger, M. A. (1998). Prenatal exposure to amphetamines. Risks and adverse outcomes in pregnancy. Obstet. Gynecol. Clin. North Am. 25, 119–138.
Pometlová, M., Hrubá, L., Slamberová, R., and Rokyta, R. (2009). Cross-fostering effect on postnatal development of rat pups exposed to methamphetamine during gestation and preweaning periods. Int. J. Dev. Neurosci. 27, 149–155. doi: 10.1016/j.ijdevneu.2008.11.006
Rambousek, L., Kacer, P., Syslova, K., Bumba, J., Bubenikova-Valesova, V., and Slamberova, R. (2014). Sex differences in methamphetamine pharmacokinetics in adult rats and its transfer to pups through the placental membrane and breast milk. Drug Alcohol Depend 139, 138–144. doi: 10.1016/j.drugalcdep.2014.03.023
Ramkissoon, A., and Wells, P. G. (2011). Human prostaglandin H synthase (hPHS)21- and hPHS-2-dependent bioactivation, oxidative macromolecular damage and cytotoxicity of dopamine, its precursor and metabolites. Free Radical Biol. Med. 50, 295–304. doi: 10.1016/j.freeradbiomed.2010.11.010
Ren, W., Luan, X., Zhang, J., Gutteea, P., Cai, Y., Zhao, J., et al. (2017). Brain-derived neurotrophic factor levels and depression during methamphetamine withdrawal. J. Affect. Disord. 221, 165–171. doi: 10.1016/j.jad.2017.06.017
Ren, W., Tao, J., Wei, Y., Su, H., Zhang, J., Xie, Y., et al. (2016). Time-dependent serum brain-derived neurotrophic factor decline during methamphetamine withdrawal. Medicine 95:e2604. doi: 10.1097/MD.0000000000002604
Roberts, S. M., Harbison, R. D., and James, R. C. (1993). Inhibition by ethanol of the metabolism of cocaine to benzoylecgonine and ecgonine methyl ester in mouse and human liver. Drug Metab. Dispos. 21, 537–541.
Robinson, T. E., and Becker, J. B. (1986). Enduring changes in brain and behavior produced by chronic amphetamine administration: a review and evaluation of animal models of amphetamine psychosis. Brain Res. 11, 157–198. doi: 10.1016/s0006-8993(86)80193-7
Rogge, G., Jones, D., Hubert, G. W., Lin, Y., and Kuhar, M. J. (2008). CART peptides: regulators of body weight, reward and other functions. Nat. Rev. Neurosci. 9, 747–758. doi: 10.1038/nrn2493
Romanowski, T., Markiewicz, A., Bednarz, N., and Bielawski, K. P. (2007). Housekeeping genes as a reference in quantitative real-time RT-PCR. Postepy. Hig. Med. Dosw. 61, 500–510.
Ross, M. K., and Crow, J. A. (2007). Human carboxylesterases and their role in xenobiotic and endobiotic metabolism. J. Biochem. Mol. Toxicol. 21, 187–196. doi: 10.1002/jbt.20178
Rutigliano, G., Accorroni, A., and Zucchi, R. (2018). The case for TAAR1 as a modulator of central nervous system function. Front. Pharmacol. 8:987. doi: 10.3389/fphar.2017.00987
Salinas, A., Wilde, J. D., and Maldve, R. E. (2006). Ethanol enhancement of cocaine- and amphetamine-regulated transcript mRNA and peptide expression in the nucleus accumbens. J. Neurochem. 2006, 408–415. doi: 10.1111/j.1471-4159.2006.03745.x
Schutová, B., Hrubá, L., Pometlová, M., Deykun, K., and Šlamberová, R. (2008). Impact of methamphetamine administered prenatally and in adulthood on cognitive functions of male rats tested in Morris water maze. Prague Med. Rep. 109, 62–70.
Schutová, B., Hrubá, L., Pometlová, M., Deykun, K., and Šlamberová, R. (2009). Cognitive functions and drug sensitivity in adult male rats prenatally exposed to methamphetamine. Physiol. Res. 58, 741–750.
Schutová, B., Hrubá, L., Rokyta, R., and Šlamberová, R. (2012). Gender differences in behavioral changes elicited by prenatal methamphetamine exposure and application of the same drug in adulthood. Dev. Psychobiol. 55, 232–242. doi: 10.1002/dev.21016
Segal, D. M., Moraes, C. T., and Mash, D. C. (1997). Up-regulation of D-3 dopamine receptor mRNA in the nucleus accumbens of human cocaine fatalities. Mol. Brain Res.. 45, 335–339. doi: 10.1016/s0169-328x(97)00025-9
Shilling, P. D., Kuczenski, R., Segal, D. S., Barrett, T. B., and Kelsoe, J. R. (2006). Differential regulation of immediate-early gene expression in the prefrontal cortex of rats with a high vs low behavioral response to methamphetamine. Neuropsychopharmacology 31, 2359–2367. doi: 10.1038/sj.npp.1301162
Silva, M. A., Jocham, G., Barros, M., Tomaz, C., and Müller, C. P. (2008). Neurokinin 3 receptor modulation of the behavioral and neurochemical effects of cocaine in rats and monkeys. Rev. Neurosci. 19, 101–111.
Šlamberová, R. (2012). Drugs in pregnancy: the effects on mother and her progeny. Physiol. Res. 61, S123–S135.
Šlamberová, R., Charousová, P., and Pometlová, M. (2005a). Maternal behavior is impaired by methamphetamine administered during pre-mating, gestation and lactation. Reprod Toxicol. 20, 103–110. doi: 10.1016/j.reprotox.2004.11.010
Šlamberová, R., Charousová, P., and Pometlová, M. (2005b). Methamphetamine administration during gestation impairs maternal behavior. Dev. Psychobiol. 46, 57–65. doi: 10.1002/dev.20042
Šlamberová, R., Pometlová, M., Syllabová, L., and Mancusková, M. (2005c). Learning in the Place navigation task, not the New learning task, is altered by prenatal methamphetamine exposure. Brain Res. Dev. Brain Res. 157, 217–219. doi: 10.1016/j.devbrainres.2005.04.005
Šlamberová, R., Riley, M. A., and Vathy, I. (2005d). Cross-generational effect of prenatal morphine exposure on neurobehavioral development of rat pups. Physiol. Res. 54, 655–660.
Šlamberová, R., Pometlová, M., and Charousová, P. (2006). Postnatal development of rat pups is altered by prenatal methamphetamine exposure. Prog. Neuropsychopharmacol. Biol. Psychiatry 30, 82–88. doi: 10.1016/j.pnpbp.2005.06.006
Šlamberová, R., Pometlová, M., Macúchová, E., Nohejlová, K., Sruchlik, A., and Valeš, K. (2015). Do the effects of prenatal exposure and acute treatment of methamphetamine on anxiety vary depending on the animal model used? Behav. Brain Res. 292, 361–369. doi: 10.1016/j.bbr.2015.07.001
Šlamberová, R., Pometlová, M., and Rokyta, R. (2007). Effect of methamphetamine exposure during prenatal and preweaning periods lasts for generations in rats. Dev. Psychobiol. 49, 312–322. doi: 10.1002/dev.20203
Šlamberová, R., and Rokyta, R. (2005). Seizure susceptibility in prenatally methamphetamine-exposed adult female rats. Brain Res. 1060, 193–197. doi: 10.1016/j.brainres.2005.08.034
Šlamberová, R., Vrajová, M., Schutová, B., Mertlová, M., Macúchová, E., Nohejlová, K., et al. (2014). Prenatal methamphetamine exposure induces long-lasting alterations in memory and development of NMDA receptors in the hippocampus. Physiol. Res. 63, S547–S558.
Šmehilová, M. (2011). Principy a využití qPCR, Pokroèilé biochemické a biotechnologické metody. Available at: https://docplayer.cz/35816720-Principy-a-vyuzit-iti-qpcr-kbc-bam-pokrocil-rni-biologie.html (accessed June 12, 2019).
Smith, L. M., Lagasse, L. L., Derauf, C., Grant, P., Shah, R., Arria, A., et al. (2008). Prenatal methamphetamine use and neonatal neurobehavioral outcome. Neurotoxicol. Teratol. 30, 20–28. doi: 10.1016/j.ntt.2007.09.005
Sokoloff, P., Diaz, J., Le Foll, B., Guillin, O., Leriche, L., Bezard, E., et al. (2006). The dopamine D3 receptor: a therapeutic target for the treatment of neuropsychiatric disorders. CNS Neurol. Disord. Drug Targets 5, 25–43. doi: 10.2174/187152706784111551
Sokoloff, P., Giros, B., Martres, M. P., Bouthenet, M. L., and Schwartz, J. C. (1990). Molecular cloning and characterization of a novel dopamine receptor (D3) as a target for neuroleptics. Nature 347, 146–151. doi: 10.1038/347146a0
Staley, J. K., and Mash, D. C. (1996). Adaptive increase in D3 dopamine receptors in the brain reward circuits of human cocaine fatalities. J. Neurosci. 16, 6100–6106. doi: 10.1523/jneurosci.16-19-06100.1996
Stewart, J. L., and Meeker, J. E. (1997). Fetal and infant deaths associated with maternal methamphetamine abuse. J. Anal. Toxicol. 21, 515–517. doi: 10.1093/jat/21.6.515
Sulzer, D., Chen, T. K., Lau, Y. Y., Kristensen, H., Rayport, S., and Ewing, A. (1995). Amphetamine redistributes dopamine from synaptic vesicles to the cytosol and promotes reverse transport. J. Neurosci. 15, 4102–4108. doi: 10.1523/jneurosci.15-05-04102.1995
Sulzer, D., Sonders, M. S., Poulsen, N. W., and Galli, A. (2005). Mechanisms of neurotransmitter release by amphetamines: a review. Prog. Neurobiol. 75, 406–433. doi: 10.1016/j.pneurobio.2005.04.003
Taketani, M., Shii, M., Ohura, K., Ninomiya, S., and Imai, T. (2007). Carboxylesterase in the liver and small intestine of experimental animals and human. Life Sci. 81, 924–932. doi: 10.1016/j.lfs.2007.07.026
Thompson, B. L., Levitt, P., and Stanwood, G. D. (2009). Prenatal exposure to drugs: effects on brain development and implications for policy and education. Nat. Rev. Neurosci. 10, 303–312. doi: 10.1038/nrn2598
Tronche, F., Kellendonk, C., Kretz, O., Gass, P., Anlag, K., Orban, P. C., et al. (1999). Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 1, 99–103. doi: 10.1038/12703
Tulloch, I., Ghazaryan, N., Mexhitaj, I., Ordonez, D., and Angulo, J. A. (2011). Role of neurokinin-1 and dopamine receptors on the striatal methamphetamine-induced proliferation of new cells in mice. Brain Res. 1399, 33–39. doi: 10.1016/j.brainres.2011.05.017
van Baar, A. L., Fleury, P., Soepatmi, S., Ultee, C. A., and Wesselman, P. J. (1989). Neonatal behavior after drug dependent pregnancy. Arch. Dis. Child. 64, 235–240. doi: 10.1136/adc.64.2.235
Vavřinková, B., Binder, T., and Živný, J. (2001). Characteristics of a population of drug dependent pregnant women in the Czech Republic. Ceska. Gynekol. 66, 285–291.
Volkow, N. D., Chang, L., Wang, G. J., Fowler, J. S., Leonido-Yee, M., Franceschi, D., et al. (2001). Association of dopamine transporter reduction with psychomotor impairment in methamphetamine abusers. Am. J. Psychiatry 158, 377–382. doi: 10.1176/appi.ajp.158.3.377
Volovelsky, O., Cohen, G., Kenig, A., Wasserman, G., Dreazen, A., Meyuhas, O., et al. (2016). Phosphorylation of ribosomal protein S6 mediates mammalian target of rapamycin complex 1–induced parathyroid cell proliferation in secondary hyperparathyroidism. J. Am. Soc. Nephrol. 27, 1091–1101. doi: 10.1681/ASN.2015040339
Wang, J., and Angulo, J. A. (2011). Synergism between methamphetamine and the neuropeptide substance P on the production of nitric oxide in the striatum of mice. Brain Res. 1369, 131–139. doi: 10.1016/j.brainres.2010.11.017
Weaver, I. C., Cervoni, N., Champagne, F. A., D’Alessio, A. C., Sharma, S., Seckl, J. R., et al. (2004). Epigenetic programming by maternal behavior. Nat. Neurosci. 7, 847–854. doi: 10.1038/nn1276
Wikipedia contributors (2019a). “Carboxylesterase 2,” Wikipedia, The Free Encyclopedia. Available at: https://en.wikipedia.org/w/index.php?title=Carboxylesterase_2&oldid=889466082 (accessed June 9, 2019).
Wikipedia contributors (2019b). “Methamphetamine,” Wikipedia, The Free Encyclopedia. Available at: https://en.wikipedia.org/w/index.php?title=Methamphetamine&oldid=898948968 (accessed June 3, 2019).
Williams, M. T., Morford, L. L., Wood, S. L., Wallace, T. L., Fukumura, M., Broening, H. W., et al. (2003). Developmental d-methamphetamine treatment selectively induces spatial navigation impairments inreference memory in the Morris water maze while sparing working memory. Synapse 48, 138–148. doi: 10.1002/syn.10159
Winslow, B. T., Voorhees, K. I., and Pehl, K. A. (2007). Methamphetamine abuse. Am. Fam. Phys. 76, 1169–1174.
Won, L., Bubula, N., McCoy, H., and Heller, A. (2001). Methamphetamine concentrations in fetal and maternal brain following prenatal exposure. Neurotoxicol. Teratol. 23, 349–354. doi: 10.1016/S0892-0362(01)00151-9
Wong, A. W., McCallum, G. P., Jeng, W., and Wells, P. G. (2008). Oxoguanine glycosylase 1 protects against methamphetamine-enhanced fetal brain oxidative DNA damage and neurodevelopmental deficits. J. Neurosci. 28, 9047–9054. doi: 10.1523/JNEUROSCI.2557-08.2008
Wouldes, T., LaGasse, L., Sheridan, J., and Lester, B. (2004). Maternal methamphetamine use during pregnancy and child outcome: what do we know? N. Z. Med. J. 117:U1180.
Xie, Z., and Miller, G. M. (2009). A receptor mechanism for methamphetamine action in dopamine transporter regulation in brain. J. Pharmacol. Exp. Ther. 330, 316–325. doi: 10.1124/jpet.109.153775
Yager, L. M., Garcia, A. F., Wunsch, A. M., and Ferguson, S. M. (2015). The ins and outs of the striatum: role in drug addiction. Neuroscience 301, 529–541. doi: 10.1016/j.neuroscience.2015.06.033
Yang, Y., and Raine, A. (2009). Prefrontal structural and functional brain imaging findings in antisocial, violent, and psychopathic individuals: a meta-analysis. Psychiatry Res. 174, 81–88. doi: 10.1016/j.pscychresns.2009.03.012
Yu, J., Wang, J., Cadet, J. L., and Angulo, J. A. (2004). Histological evidence supporting a role for the striatal neurokinin-1 receptor in methamphetamine-induced neurotoxicity in the mouse brain. Brain Res. 1007, 124–131. doi: 10.1016/j.brainres.2004.01.077
Zoubková, H. (2019a). Data from: Results of First Microarray Trial of METH and SAL Exposed Rats. Available at: https://figshare.com/articles/Results_of_first_microarray_trial/8081072 (accessed June 18, 2019).
Zoubková, H. (2019b). Data from: Results of Second Microarray Trial METH and SAL Exposed Rats. Available at: https://figshare.com/articles/Results_of_second_microarray_trial/8081120 (accessed June 18, 2019).
Keywords: prenatal, methamphetamine, striatum, hippocampus, prefrontal cortex, receptor, microarray, real-time PCR
Citation: Zoubková H, Tomášková A, Nohejlová K, Černá M and Šlamberová R (2019) Prenatal Exposure to Methamphetamine: Up-Regulation of Brain Receptor Genes. Front. Neurosci. 13:771. doi: 10.3389/fnins.2019.00771
Received: 25 April 2019; Accepted: 09 July 2019;
Published: 01 August 2019.
Edited by:
Juan J. Canales, University of Tasmania, AustraliaReviewed by:
Karen K. Szumlinski, University of California, Santa Barbara, United StatesCopyright © 2019 Zoubková, Tomášková, Nohejlová, Černá and Šlamberová. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hana Zoubková, aGFuYS56b3Via292YUBsZjMuY3VuaS5jeg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.