- 1Department of Experimental Medical Science, Faculty of Medicine, Lund University, Lund, Sweden
- 2The Mina and Everard Goodman Faculty of Life Sciences, Bar-Ilan University, Ramat Gan, Israel
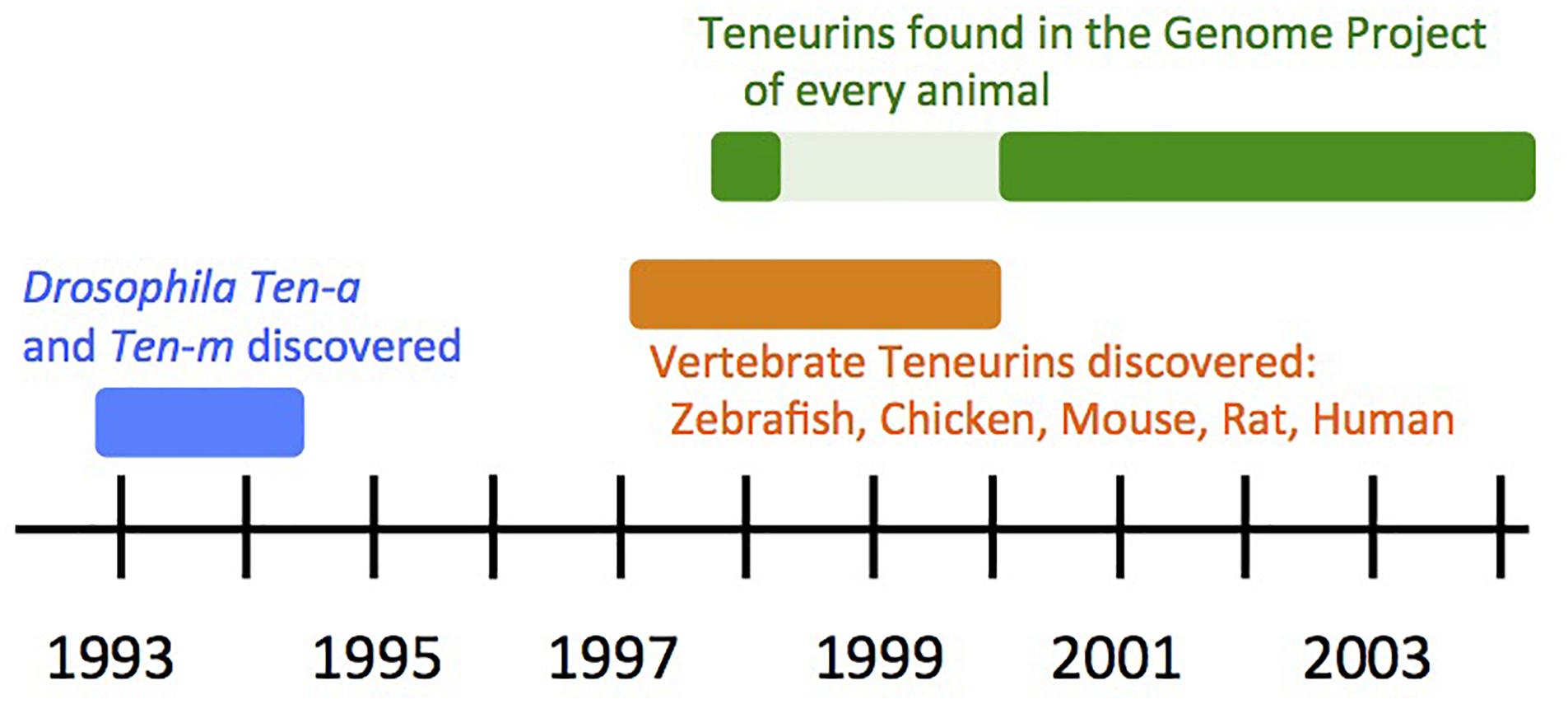
Teneurins were first discovered and published in 1993 and 1994, in Drosophila melanogaster as Ten-a and Ten-m. They were initially described as cell surface proteins, and as pair-rule genes. Later, they proved to be type II transmembrane proteins, and not to be pair-rule genes. Ten-m might nonetheless have had an ancestral function in clock-based segmentation as a Ten-m oscillator. The turn of the millennium saw a watershed of vertebrate Teneurin discovery, which was soon complemented by Teneurin protein annotations from whole genome sequence publications. Teneurins encode proteins with essentially invariant domain order and size. The first years of Teneurin studies in many experimental systems led to key insights, and a unified picture, of Teneurin proteins.
Fly Teneurins were First Described as Cell Surface Proteins, and as Pair-Rule Genes
Discovery of the Teneurins
The teneurins were discovered in the early 1990 when one of us (SB) tried to find the tenascin-C homologue in Drosophila. Tenascin-C is a six-armed extracellular matrix (ECM) molecule which displays many functions during development, morphogenesis and tissue homeostasis (Midwood et al., 2016). Since the Drosophila genome harbors a solid stock of basement membrane and other important ECM molecules (Broadie et al., 2011), it seemed conceivable to search for a Drosophila homologue of tenascin-C using PCR and degenerate primers. The tenascins are composed of several domains that appear in a repetitive manner such as the tenascin-type of EGF repeats or the fibronectin-type III (FN III) repeats. The carboxy terminus harbors a globular fibrinogen domain. Since all these above mentioned domains were found as parts of other Drosophila proteins, the question was which domain-specific primer pair would turn out to be fruitful. Of the many primers that were used in this approach, only the EGF-like domain proved successful, leading to the detection of the first Drosophila tenascin-type EGF-like repeats. These were then used to screen bacterial cDNA libraries that were optimized for long cDNAs (Brown and Kafatos, 1988; Brown et al., 1989) resulting in three overlapping cDNAs of 7.3 kb in length that altogether constituted a partial sequence of what had the potential to represent the Drosophila homologue of tenascin-C (Baumgartner and Chiquet-Ehrismann, 1993). The deduced amino acid (aa) sequence showed the presence of eight tenascin-type EGF repeats (Figure 1), as was the case in vertebrate tenascin-C (Midwood et al., 2016). At the amino terminus, a hydrophobic stretch of amino acids reminiscent of a signal or a transmembrane domain was found. C-terminally of the EGF-like repeat, an additional 100 aa were found that did not show any resemblance to FN III repeats, but soon the protein would run into a stop codon, leaving 4.3 kb of a putative 3′ untranslated region (UTR). Based on the deduced sequence information, the isolated composite cDNA was proposed to code for a 782 aa secreted protein and was subsequently called Ten-a (tenascin accessory) (Baumgartner and Chiquet-Ehrismann, 1993). In retrospect, the published Ten-a aa sequence from 1993 comprised only a partial sequence. This became also evident from comparing the transcript size on a Northern analysis which showed two large transcripts of 11 and 13 kb, respectively, which were developmentally regulated (Baumgartner and Chiquet-Ehrismann, 1993). The discrepancy of the length deduced from available cDNA and the actual transcript size by Northern analysis was attributed to an unusually long 5′ untranslated region (UTR) which later turned out not to be true. Indeed, it would take years to realize that the protein was indeed much larger (Fascetti and Baumgartner, 2002), because its coding part extended considerably in the carboxy terminal direction. This carboxy extension was also confirmed by the advent of the fully sequenced Drosophila genome (Adams et al., 2000).
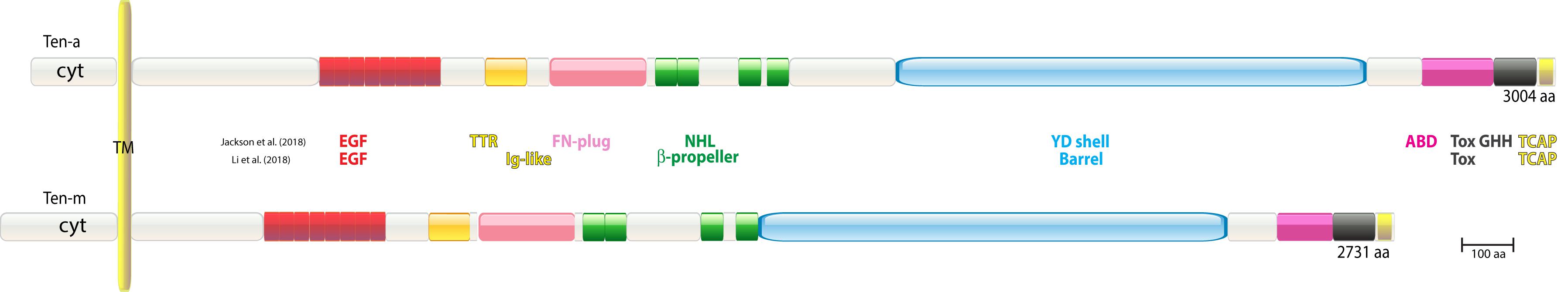
Figure 1. Domain structure of Drosophila teneurins. Only the major isoforms are shown. Domain structures are depicted according to the crystallization data of Jackson et al. (2018) and the cryo-EM data of Li et al. (2018), as they were identified and are drawn to scale. EGF, epidermal growth factor repeat; TTR, transthyretin; FN, fibronectin; NHL, NCL, HT2A Lin41; YD, YD-repeat motif; ABD, antibiotic binding domain; Tox GHH, Tox GHH fold (Zhang et al., 2012); TCAP, Teneurin C-terminal-associated Peptides; Ig, immunoglobulin.
(Baumgartner and Chiquet-Ehrismann, 1993) also showed a zoo blot equipped with DNA from Drosophila, leech, zebrafish, chicken, mouse, and human origin, as probed with chicken tenascin-C EGF sequences under low stringency. The blot revealed that the majority of genomes analyzed showed cross-hybridizing bands. These findings immediately opened the avenue for further quests/searches for tenascin-type EGF-like sequences not only in Drosophila, but later also in higher organisms (Mieda et al., 1999; Minet et al., 1999; Oohashi et al., 1999; Rubin et al., 1999; Feng et al., 2002).
The Drosophila lane in the zoo blot contained several cross-hybridizing bands, two of which could readily be ascribed to Ten-a. The Drosophila lane, however, revealed further unidentified bands, hence the hunt for further tenascin-EGF-like sequences was continued. To this end, one of us (S. B.) used a Ten-a EGF-like repeat probe and screened Drosophila genomic libraries under low-stringency conditions (McGinnis et al., 1984). Several cross-hybridizing phages were isolated that all mapped to a new locus (Baumgartner et al., 1994). Subsequently, overlapping cDNAs were isolated from this locus and were assembled. These cDNA clones covered two slightly smaller transcripts compared to Ten-a, 10.5 and 11.5 kb in size, respectively. Due to the fact that the protein encoded by the transcript of this new locus was apparently larger than that of Ten-a, this gene was termed Ten-m (tenascin major) (Baumgartner et al., 1994). At the time, it was proposed that the gene encoded a large secreted proteoglycan ECM molecule. Ten-a and Ten-m proteins’ structures and domains, as realized in 2018 terms (as described below), can be seen in Figure 1.
One of the two Teneurins was independently discovered in Drosophila melanogaster via an alternative approach: (Ten-m, as “odd Oz” by RW), in Levine et al. (1994). In 1990, a screen was carried out to uncover novel fly tyrosine kinase substrates of previously unknown classes. Drosophila proteins were highly immunopurified on an anti-phosphotyrosine antibody column, and the resulting phospho-protein collection was used to raise a bank of monoclonal antibodies. One of these specific monoclonals was directed against a greater than 300 kD protein that was later given the name Odd Oz (Odz, now Ten-m) (see Figure 1). That monoclonal was used for expression cloning of the Odz/Ten-m’s 11 kb transcript from an embryonic cDNA library (Zinn et al., 1988). Further mapping led to genomic cloning, chromosome mapping, mutant identification, and expression and phenotype characterizations (Levine et al., 1994). Two hydrophobic stretches in the predicted protein were interpreted as: (1), a signal peptide before a series of EGF-like repeats, followed by (2), a post-EGF transmembrane domain. The type-I transmembrane model was anchored by placement of the EGF-like repeats extracellularly. Yet this type-I model was also influenced by biases based on the phospho-tyrosine protein screen and consensus phosphorylation site motifs of the time. In fact, the second predicted “transmembrane” domain assignment was incorrect, and the assigned “signal peptide” sequence is the protein’s true transmembrane stretch. Odz/Ten-m is instead a type-II transmembrane protein, as all further Teneurins proved to be (see below).
Expression of the Founding Teneurins in Drosophila
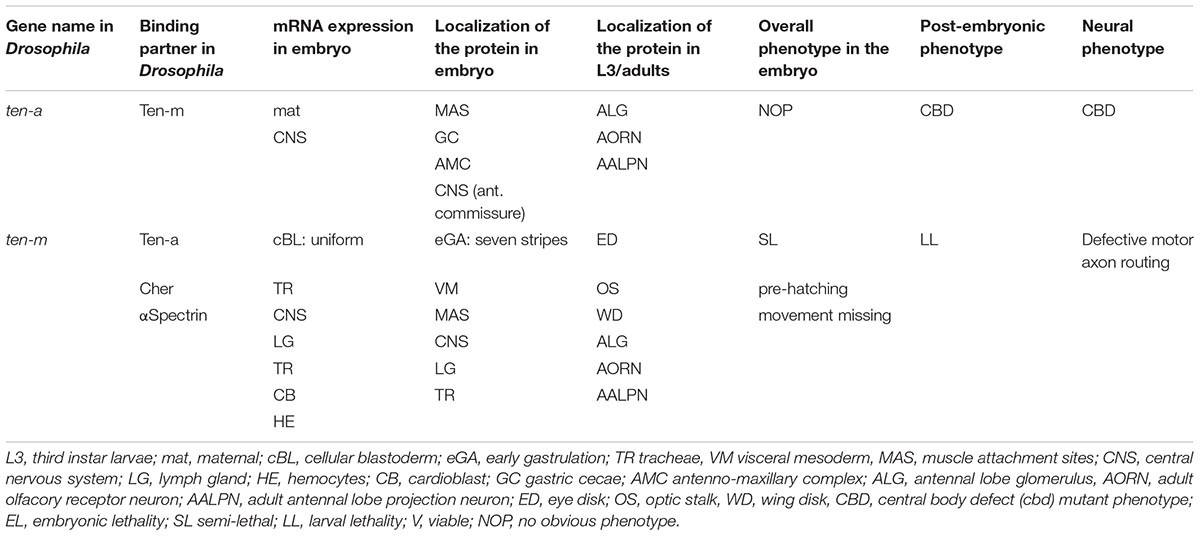
Both Ten-a and Ten-m genes were extensively analyzed with respect to their expression patterns during early Drosophila embryogenesis (Baumgartner and Chiquet-Ehrismann, 1993; Baumgartner et al., 1994; Levine et al., 1994; Fascetti and Baumgartner, 2002; Table 1). Predominant expression of both genes was in the central nervous system (CNS). In general, the Ten-m gene showed far more progenitor tissue labeled. Apart from its prominent CNS expression, Ten-m was found in the future tracheal cells, heart cells, lymph gland and hemocytes. Hence, the expression profile demanded further claims to call it “major.”
Other studies expanded the breadth of Ten-m expression profiles. Striking expression was documented in non-neuronal imaginal disk tissues, such as ring gland expression (Harvie et al., 1998), in the in sensory and motor neuron precursors in pupae, and in adult neuronal tissues (Levine et al., 1997). Expression in the eye, and influences of upsteam genes such as Glass on Ten-m expression, were observed (Treisman and Rubin, 1996). Hematopoietic cells showed Ten-m expression, such as plasmatocytes (Baumgartner et al., 1994; Braun et al., 1997).
Drosophila Ten-m/odz and Segmentation
Phenotypically for odz/Ten-m, multiple alleles from independent screens proved allelic, and displayed different severities of a pair-rule mutant phenotype (Baumgartner et al., 1994; Levine et al., 1994). This phenotype was very like that of odd paired (opa). Considerably later, the assigned pair-rule phenotype was instead attributed to mutant opa alleles in the genetic backgrounds of the odz/Ten-m strains (see below). On another note - in retrospect, previously created mutations in Ten-a existed that, appropriately, affect the fly brain and behavior. The gene central body defective (cbd), with several alleles known, had been isolated a decade before the gene was cloned and characterized (see Table 1; Heisenberg et al., 1985). The recognition that cbd mutations were Ten-a lesions occurred two decades later (Cheng et al., 2013).
In 2006, an indication that odz/Ten-m is not a pair-rule gene was published. Using new technologies that were developed, it was found that an entire 133 kb genomic clone covering Ten-m failed to rescue the attributed odz/Ten-m pair-rule phenotype (Venken et al., 2006). The concern arising from this finding led to a re-examination of all odz/Ten-m mutant lines displaying the pair-rule phenotype. Ultimately, the pair-rule phenotype proved to derive from odd paired (opa) mutations on the balancers in odz/Ten-m strains (Zheng et al., 2011).
The different odz/Ten-m mutations, and the balancers in their lines, came from separate mobilized-P-element screens (Cooley et al., 1988; Karpen and Spradling, 1992). The sources of the balancers for these screens were different. In addition, the many non-odz/Ten-m lines examined from these screens, with these balancers, displayed no pair-rule phenotype. The lines that were chosen to assess for odz/Ten-m lesions were based on genome position, and not pair-rule appearance, so phenotype was not a screening bias. Ten-m is deployed as seven stripes during late cellular blastoderm, but its mutants do not have pair-rule phenotypes. To this day, the reasons for the many co-incidences that led to the findings are still unclear. Unfortunately, a great deal of mis-directed work was subsequently carried out. A Ten-a maternal effect impact on segmentation was reported, then was later retracted (Rakovitsky et al., 2007; retracted 2012), despite the correct molecular data detailed there.
Ten-m Might Nonetheless Have a Segmentation Role: A Ten-m Oscillator?
One aspect of Ten-m expression was particularly interesting because it showed its transcripts relatively uniformly expressed during cellular blastoderm, while the Ten-m protein only minutes later was detected in seven stripes (Baumgartner et al., 1994; Levine et al., 1994; Figure 2C). This observation opened the avenue for proposing a function of Ten-m as an oscillator.
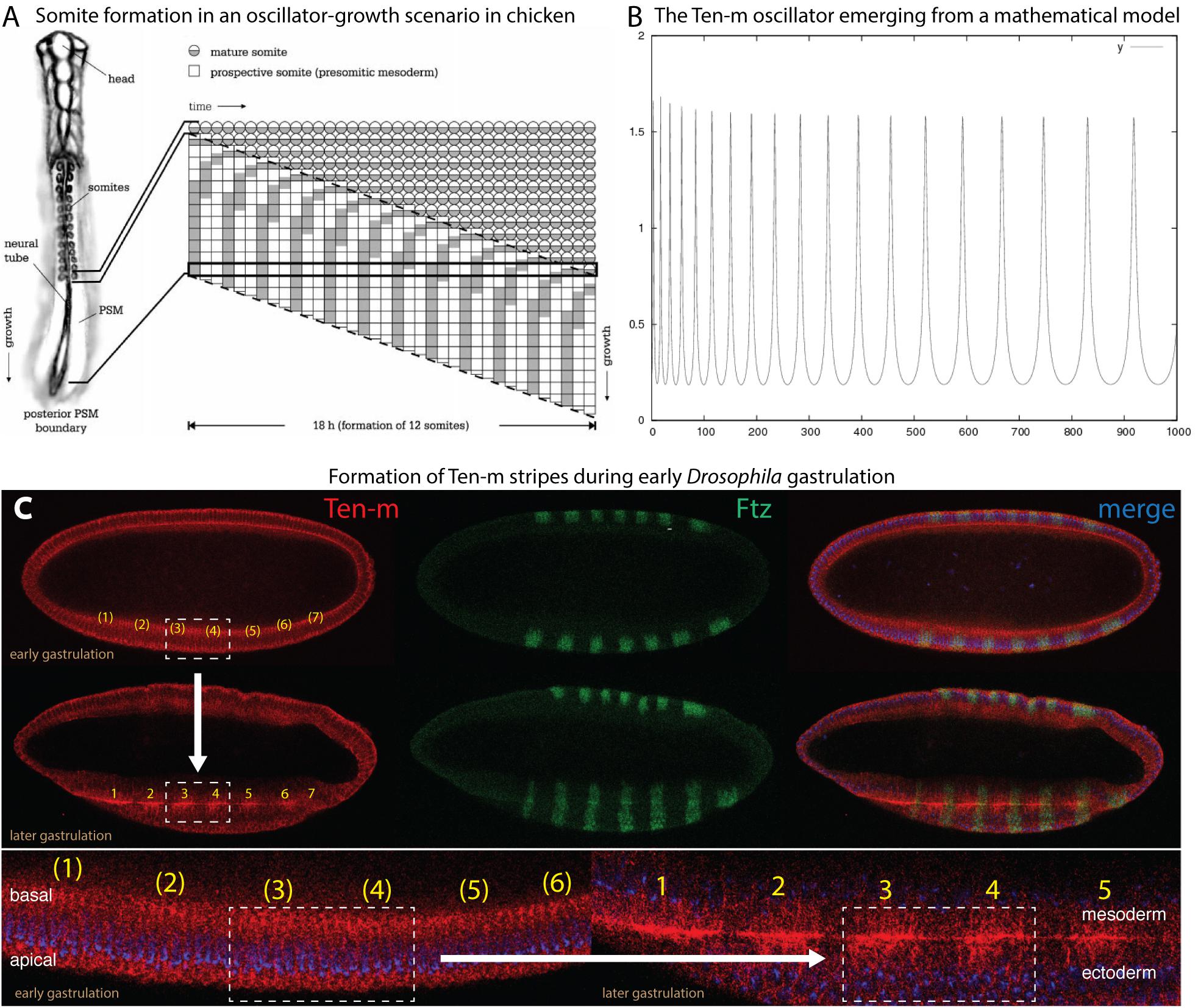
Figure 2. Proposed features of Ten-m as a biological oscillator. Data slightly modified from Hunding and Baumgartner (2017), see details therein. Reproduced with permission. (A) Somite formation in an oscillator-based system, as exemplified in chicken (Palmeirim et al., 1997). (B) The Ten-m oscillator as it emerges from a mathematical model. (C) Experimental evidence of emergence of Ten-m stripe formation during early Drosophila gastrulation, starting from ubiquitous Ten-m expression. Double-antibody staining reveals Ten-m in red and Fushi tarazu in green (for comparison). Top part shows the transition from ubiquitous Ten-m expression at early gastrulation to the formation of Ten-m stripes at somewhat later gastrulation (as exemplified of the boxed part comprising stripes 3 and 4 and indicated by an arrow). Bottom part shows enlargements of the formation of Ten-m stripes, again exemplified by stripe 3 and 4 formation and the boxed area. Note that Fushi tarazu (green) is already expressed in stripes from the very beginning, in contrast to Ten-m.
In the past and first documented in the chicken hairy gene, it could be shown that periodically waves can arise from the posterior end of the elongating embryo. These waves move toward the anterior end where they come to a halt and add a segment during each period, as depicted in Figure 2A (Palmeirim et al., 1997). Later, a model emerged involving oscillation of the zebrafish hairy/Enhancer of split-related genes, her1 and her7 (Lewis, 2003). This model proposed that the Her protein would bind to its own promoter and inhibit its own transcription. It was then concluded that the delay would cause a biochemical oscillator because of the time difference between formation of the mRNA and the protein (Lewis, 2003). Posteriorly, cells are fed to the presomitic mesoderm (PSM) during this oscillation. When the embryo undergoes elongation, more oscillating cells are fed into the PSM that will reveal different phases. Subsequently, the cellular oscillation grows until the oscillation comes to a halt with the consequence that segmental borders will emerge. The mechanism is repeated when subsequent cells stop their oscillation. This mechanism enables that segments can be generated, starting from the anterior to the posterior.
The idea that Ten-m could be an oscillator originated from the observation that the Ten-m mRNA was uniformly expressed during nuclear cycle (nc) 14, but once the protein was synthesized, it started to emerge as seven stripes (Baumgartner et al., 1994; Levine et al., 1994; Hunding and Baumgartner, 2017). Since only the long nc 14 was long enough to synthesize the large primary transcript of Ten-m (115 kb, see Table 2) and to translate Ten-m (Prescott and Bender, 1962; Baumgartner et al., 1994), it appeared conceivable to assume that stripe formation was tightly linked to translation and to occur only during a limited time, i.e., during late nc 14. nc 14 is terminated once cellularization occurs, whereby the nuclei are wrapped by a membrane. Hence, once cellularization has taken place, signaling from the extracellular space is only possible with the help of a receptor residing at the surface of the cells.
As stated above, the Drosophila Ten-m gene encodes a large type II transmembrane protein (Figure 1) hence, it is located at the cell surface. Ten-m becomes localized to the membrane which grows from the apical side to the basal side thereby ensheathing the syncytial nuclei (Figure 2C). The large extracellular domain of Ten-m may be involved in forming homodimers, as was shown for Ten-a (Fascetti and Baumgartner, 2002) and mouse Teneurins (Feng et al., 2002; Berns et al., 2018). The dynamics of this process has properties proposed to have the potential to create a biochemical oscillator (Hunding and Baumgartner, 2017). Ten-m interaction at the membrane could lead to intracellular cleavage of Ten-m. This cytoplasmic fragment then translocates to the nucleus. As alluded to above, Ten-m is not transcribed in seven stripes, but rather appears fairly homogeneous along the A-P axis. The mechanism to solve this apparent discrepancy is so far not clear. However, it was proposed that the intracellular mechanism of the interplay between the protein and the membrane may lead to a spontaneous pattern-forming mechanism, as was reported from other biochemical oscillators (Hunding and Baumgartner, 2017). In fact, Ten-m fulfills most criteria of stripe formation based on a model originally described for prokaryotic cell division (Hunding and Engelhardt, 1995) and further developed by (Meinhardt and de Boer, 2001). This model has recently been recapitulated using in vitro data and expanded models (Loose et al., 2008).
Thus, the Ten-m oscillator is not caused by delayed translation as in the case for the zebrafish her1/her7 genes (Lewis, 2003), but could arise from cooperative membrane binding (Hunding and Baumgartner, 2017). To enable Ten-m to function as a signaling molecule, a mechanism was proposed that would involve regulated intramembrane proteolysis (RIP) (Brown et al., 2000; McCarthy et al., 2017). Indeed, reports could show that an intracellular tail part of vertebrate Teneurin 2 protein is proteolytically cleaved off, possibly via RIP. This short peptide is then translocated to the nucleus where it represses zic-1, a vertebrate counterpart of odd-paired (opa), a pair-rule gene in Drosophila (Bagutti et al., 2003). On the other hand, ubiquitously expressed Zic-1 leads to fast degradation of the short Teneurin 2 signaling peptide. The model of Hunding and Baumgartner (2017) included thus cooperative interaction of Ten-m with the membrane, intracellular cleavage and degradation (Figure 2B).
In summary, what these data would like to suggest is that, despite the fact that Ten-m mutants do not show a segmental phenotype, there might be an ancestral function of Ten-m in clock-based segmentation. The one established by the Notch signaling system was probably when the insects evolved, due to the fact that Notch receptor does not show an involvement in Drosophila segmentation. This is where Ten-m might come in and the field is eagerly waiting for data that support this hypothesis.
Watershed of Vertebrate Teneurin Discovery, 1998–2000, in the Pre-Vertebrate-Genome Era
The first publications of vertebrate Teneurin genes emerged from screens searching for other phenomena: studies of cancer rearrangements and gene changes; olfaction-related genes; and ER stress-related CHOP genes. A gene rearrangement encoding a fusion protein containing Teneurin 4 and Neuregulin 1 domains was identified in human breast tumor tissue (Schaefer et al., 1997). The resulting fusion protein contained only the pre-EGF amino-terminal portion of TENM4, but beyond ESTs, was the first harbinger of vertebrate Teneurins, as was later recognized (Wang et al., 1999). Soon thereafter, human Teneurin 1 was sequenced and named TNM (Figure 3), when it was found adjacent to the X-linked lymphoproliferative syndrome causative gene SH2D1A (Coffey et al., 1998). Teneurin 4 in mouse was uncovered in a screen for CHOP - dependent stress-induced genes, and was named DOC4 (Wang et al., 1998). Tenm4 came up twice in that screen (as DOC4 and DOC5), and was the first non-fly Teneurin to receive non-cursory treatment, with a number of pivotal observations made for the protein family as a whole. Some time later, first phenotypes for mouse Teneurin 4 were documented, when it was established that existing l7Rn3 mice were mutants (Lossie et al., 2005). In a search based on homology to E2 cysteine rich loops of odorant receptors, rat Neurestin (Teneurin 2) was found as a novel, non-odorant receptor, protein (Otaki and Firestein, 1999a). The characterization of Teneurin 2 in Neurestin papers also contributed key observations made for the protein family as a whole (Otaki and Firestein, 1999a,b).
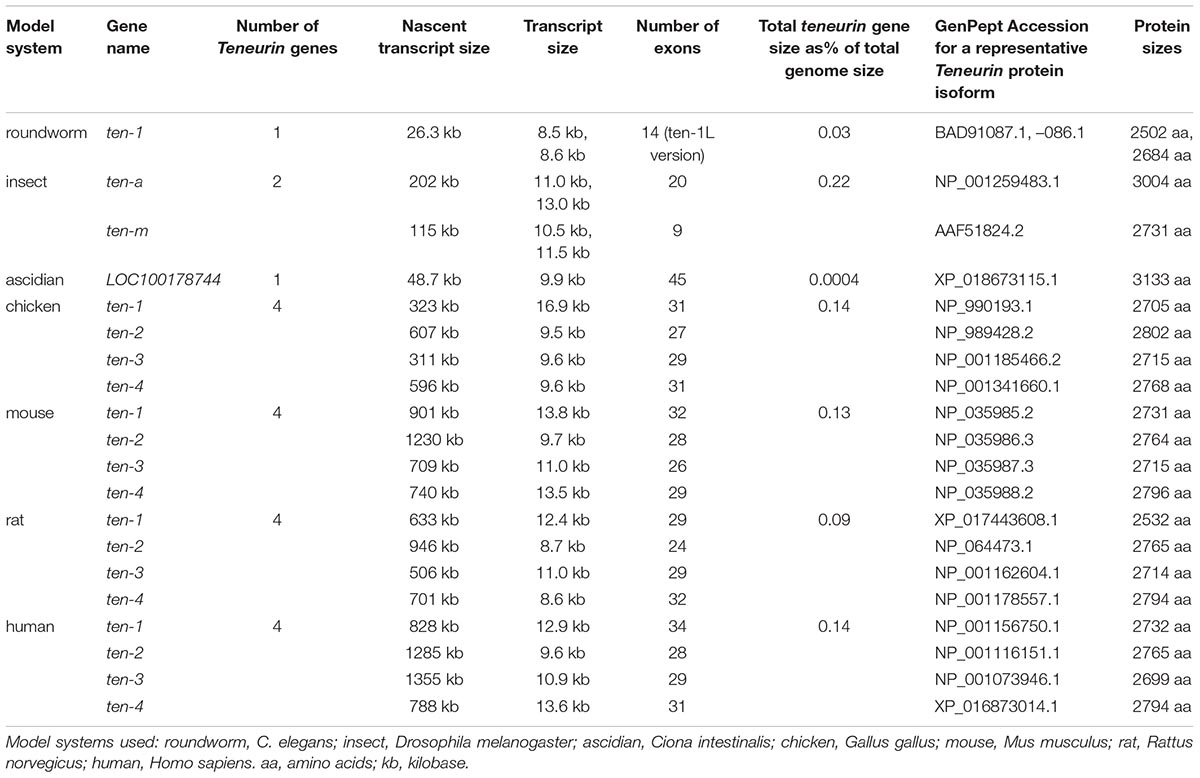
Meanwhile, efforts directed specifically at identifying and cloning vertebrate Teneurin by homology to the fly genes were underway in three species, and were reported in 1999 and 2000. The four paralog types in chicken were identified, Tenm1–Tenm4 (Minet et al., 1999; Rubin et al., 1999), and this work continued with many wide-ranging discoveries and publications. Four corresponding mouse paralogs were found and well characterized (Oohashi et al., 1999), and were also independently sequenced and mapped (Ben-Zur and Wides, 1999; Ben-Zur et al., 2000). Two of the four of these paralog types were also uncovered in zebrafish (Mieda et al., 1999). The rat and human Teneurin genes mentioned above were retrospectively assigned to their paralog-type number. Thus, at the end of the “pre-vertebrate genome sequence” era, five vertebrate species had been proven to bear Teneurins, with a four-copy content apparent as the common, and likely conserved, paralog complement (Figure 3).
Analysis From the First Complete Genomes: Teneurins Form a Distinct, Animal, Family
With the completion of the Caenorhabditis elegans genome, a single full length Teneurin, Ten-1, was evident (Figure 3; C. elegans Sequencing Consortium, 1998). Its protein and function were characterized and described (Drabikowski et al., 2005). The Drosophila melanogaster genome encoded a Ten-a the length of the original Ten-m (Adams et al., 2000), as was later described (Fascetti and Baumgartner, 2002). A nematode singleton Teneurin, in contrast to a pair of paralogs, Ten-a and Ten-m, in insects, held true in the nematode C. briggsae (Stein et al., 2003), the mosquito Anopheles gambiae (Holt et al., 2002), and the silkworm Bombyx mori (Mita et al., 2004), genomes. Vertebrate genomes, including human (Lander et al., 2001; Venter et al., 2001), mouse (Mouse Genome Sequencing et al., 2002; Mural et al., 2002), rat (Gibbs et al., 2004), and chicken (International Chicken Genome Sequencing Consortium, 2004) validated that the four paralogs Tenm1, 2, 3 and 4 were a fixed vertebrate feature. Contemporaneously, non-vertebrate chordates, such as the ascidian Ciona intestinalis, proved to have a single Teneurin gene (Dehal et al., 2002). This indicated that Teneurins were quadruplicated sometime during chordate or early vertebrate evolution. A representative list of these Teneurin genes appears as Table 2. The proteins, which all maintain the same domain order, are of roughly the same size. Their protein lengths are reflected in their mature transcript sizes. Their nascent transcripts, however, are consistently of unusually large size, as is often seen for highly developmentally regulated genes. As a consequence, the Teneurin genes’ lengths occupy an “over-sized” fraction of total genome sizes (Table 2).
In contrast, Teneurins were not found in the kingdoms of plants or fungi. The earliest sequenced genomes of: the yeasts Saccharomyces cerevisiae (Goffeau et al., 1996) and Saccharomyces pombe (Wood et al., 2002); plants Arabidopsis (The Arabidopsis Genome Initiative, 2000) and rice (Goff et al., 2002; Yu et al., 2002), and the first other protists and fungi revealed no Teneurins. No eukaryotic homologous sequences could be found at all, outside of those to the Teneurin’s EGF-like domains. The only other Teneurin domains with homology to any proteins were rhs (recombination hot spot)-like elements otherwise found only in a small number of bacteria (Minet et al., 1999; Minet and Chiquet-Ehrismann, 2000).
Overall, in the first 10 years that Teneurins were studied, they were recognized as animal specific genes (Figure 3), with two paralogs in insects, and four paralogs in vertebrates. These were reviewed with an eye toward an evident ancient duplication, and an evident ancient quadruplication, in insects and vertebrates, respectively, (Tucker and Chiquet-Ehrismann, 2006; Tucker et al., 2007). These reviews also recognized that Teneurin proteins are largely invariant throughout evolution, with no domain content or order variation. For a more recent evolutionary history of the family, see the Wides article in this volume. For more recent views of Teneurins and their structure, see the Tucker (2018), and DePew et al. (2019) article, in this volume.
From Teneurin’s First Decade: Key Insights Into Its Protein
The Drosophila Teneurin homologs were initially described as ECM molecules (Baumgartner et al., 1994), or as type I transmembrane proteins (Levine et al., 1994). The first recognition that Teneurins were in fact type II single pass transmembrane proteins came for mouse Teneurin4, when it was discovered as DOC4 (Wang et al., 1998). This was validated rigorously when all four mouse genes were sequenced, and when their extracellular portions were imaged by electron microscopy (Oohashi et al., 1999). Several studies established that Teneurins are deployed to the cell membrane as protein dimers, but their full homo- and hetero-dimerization combinatorial repertoire was first methodically shown in mouse (Feng et al., 2002). Among protein – protein interactions proven for Teneurins, perhaps the first was the fly Ten-m RGD motif interaction with integrins (Graner et al., 1998). Broader still, and iconic for Teneurin function, was the discovery of homophilic interactions in chicken (Rubin et al., 2002). While a great deal still needs to be done to nest Teneurins within a complete pathway, their homophilic, and cross-paralog-homophilic, extracellular contacts are at the heart of their signaling role. Proteolytic cleavages at many sites by many proteases are also central to varied aspects of Teneurin protein function, and have been documented since the first works published. They are too numerous to be related here, but two perhaps suggest the most important functional implications. The cleavage and release of intracellular domains, and their freedom to then enter the nucleus to impact transcription was first described in chicken (Bagutti et al., 2003; Nunes et al., 2005). The cleavage of their extreme carboxy-terminal amino acids to yield independent, biologically active TCAPs (Teneurin C-terminal-associated Peptides) occurs in many important systems (Qian et al., 2004).
Interestingly, the studies on the Teneurin domain structure were recently complemented by two reports showing data based on crystallization and cryo-EM analyses, respectively (Jackson et al., 2018; Li et al., 2018). These in principle confirmed the sequence-based data, however, they revealed that most central domains merged into a large and centrally located 200 kD superfold. They also disclosed new findings, e.g., by highlighting the NHL domain (Figure 1) as a particularly well exposed domain where homophilic interactions between teneurins were ascribed (Berns et al., 2018). Moreover, alternative splicing within the NHL domain would allow modulation of this homophilic interaction. Based on sequence comparisons, both Ten-a and Ten-m follow the domain structure that the most-recent crystallization and cryo-EM data defined. Hence, the domain structure as drawn in Figure 1 likely holds true. Evolutionarily, the 200 kD superfold was adopted as a whole structure from bacteria. This was recognized in these two papers Jackson et al. (2018), Li et al. (2018), and in Ferralli et al. (2018). Teneurin’s Latrophilin binding, and its implications, was discovered well after the first decade, and is extensively treated in other articles in this volume.
Author Contributions
SB and RW collaborated and contributed equally to this paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
SB thanks the Swedish Research Council, the Ekhaga, Nilsson-Ehle and Erik Philip-Sörensen Foundations for support.
References
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
The Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. doi: 10.1038/35048692
Bagutti, C., Forro, G., Ferralli, J., Rubin, B., and Chiquet-Ehrismann, R. (2003). The intracellular domain of teneurin-2 has a nuclear function and represses zic-1-mediated transcription. J. Cell Sci. 116(Pt 14), 2957–2966. doi: 10.1242/jcs.00603
Baumgartner, S., and Chiquet-Ehrismann, R. (1993). Tena, a Drosophila gene related to tenascin, shows selective transcript localization. Mech. Dev. 40, 165–176. doi: 10.1016/0925-4773(93)90074-8
Baumgartner, S., Martin, D., Hagios, C., and Chiquet-Ehrismann, R. (1994). Tenm, a Drosophila gene related to tenascin, is a new pair-rule gene. EMBO J. 13, 3728–3740. doi: 10.1002/j.1460-2075.1994.tb06682.x
Ben-Zur, T., Feige, E., Motro, B., and Wides, R. (2000). The mammalian Odz gene family: homologs of a Drosophila pair-rule gene with expression implying distinct yet overlapping developmental roles. Dev. Biol. 217, 107–120. doi: 10.1006/dbio.1999.9532
Ben-Zur, T., and Wides, R. (1999). Mapping homologs of Drosophila odd Oz (odz): Doc4/Odz4 to mouse chromosome 7, Odz1 to mouse chromosome 11; and ODZ3 to human chromosome Xq25. Genomics 58, 102–103. doi: 10.1006/geno.1999.5798
Berns, D. S., DeNardo, L. A., Pederick, D. T., and Luo, L. (2018). Teneurin-3 controls topographic circuit assembly in the hippocampus. Nature 554, 328–333. doi: 10.1038/nature25463
Braun, A., Lemaitre, B., Lanot, R., Zachary, D., and Meister, M. (1997). Drosophila immunity: analysis of larval hemocytes by P-element-mediated enhancer trap. Genetics 147, 623–634.
Broadie, K., Baumgartner, S., and Prokop, A. (2011). Extracellular matrix and its receptors in Drosophila neural development. Dev. Neurobiol. 71, 1102–1130. doi: 10.1002/dneu.20935
Brown, M. S., Ye, J., Rawson, R. B., and Goldstein, J. L. (2000). Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398. doi: 10.1016/S0092-8674(00)80675-3
Brown, N. H., and Kafatos, F. C. (1988). Functional cDNA libraries from Drosophila embryos. J. Mol. Biol. 203, 425–437. doi: 10.1016/0022-2836(88)90010-1
Brown, N. H., King, D. L., Wilcox, M., and Kafatos, F. C. (1989). Developmentally regulated alternative splicing of Drosophila integrin PS2 alpha transcripts. Cell 59, 185–195. doi: 10.1016/0092-8674(89)90880-5
Cheng, X., Jiang, H., Li, W., Lv, H., Gong, Z., and Liu, L. (2013). Ten-a affects the fusion of central complex primordia in Drosophila. PLoS One 8:e57129. doi: 10.1371/journal.pone.0057129
Coffey, A. J., Brooksbank, R. A., Brandau, O., Oohashi, T., Howell, G. R., Bentley, D. R., et al. (1998). Host response to EBV infection in X-linked lymphoproliferative disease results from mutations in an SH2-domain encoding gene. Nat. Genet. 20, 129–135. doi: 10.1038/2424
Cooley, L., Berg, C., and Spradling, A. (1988). Controlling P element insertional mutagenesis. Trends Genet. 4, 254–258. doi: 10.1016/0168-9525(88)90032-7
C. elegans Sequencing Consortium (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012–2018.
Dehal, P., Satou, Y., Campbell, R. K., Chapman, J., Degnan, B., De Tomasoand, A., et al. (2002). The draft genome of ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2167. doi: 10.1126/science.1080049
DePew, A. T., Aimino, M. A., and Mosca, T. J. (2019). Tenets of teneurin: conserved mechanisms regulate diverse developmental processes in the Drosophila nervous system. Front. Neurosci. 13:27. doi: 10.3389/fnins.2019.00027
Drabikowski, K., Trzebiatowska, A., and Chiquet-Ehrismann, R. (2005). ten-1, an essential gene for germ cell development, epidermal morphogenesis, gonad migration, and neuronal pathfinding in Caenorhabditis elegans. Dev. Biol. 282, 27–38. doi: 10.1016/j.ydbio.2005.02.017
Fascetti, N., and Baumgartner, S. (2002). Expression of Drosophila Ten-a, a dimeric receptor during embryonic development. Mech. Dev. 114, 197–200. doi: 10.1016/S0925-4773(02)00055-2
Feng, K., Zhou, X. H., Oohashi, T., Morgelin, M., Lustig, A., Hirakawa, S., et al. (2002). All four members of the Ten-m/Odz family of transmembrane proteins form dimers. J. Biol. Chem. 277, 26128–26135. doi: 10.1074/jbc.M203722200
Ferralli, J., Tucker, R. P., and Chiquet-Ehrismann, R. (2018). The teneurin C-terminal domain possesses nuclease activity and is apoptogenic. Biol Open 7:bio031765. doi: 10.1242/bio.031765
Gibbs, R. A., Weinstock, G. M., Metzker, M. L., Muzny, D. M., Sodergren, E. J., Scherer, S., et al. (2004). Genome Sequencing Project Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521. doi: 10.1038/nature02426
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa ssp, L., japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Goffeau, A., Barrell, B. G., Bussey, H., Davis, R. W., Dujon, B., Feldmann, H., et al. (1996). Life with 6000 genes. Science 546, 563–567. doi: 10.1126/science.274.5287.546
Graner, M. W., Bunch, T. A., Baumgartner, S., Kerschen, A., and Brower, D. L. (1998). Splice variants of the Drosophila PS2 integrins differentially interact with RGD-containing fragments of the extracellular proteins tiggrin, ten-m, and D-laminin 2. J. Biol. Chem. 273, 18235–18241. doi: 10.1074/jbc.273.29.18235
Harvie, P. D., Filippova, M., and Bryant, P. J. (1998). Genes expressed in the ring gland, the major endocrine organ of Drosophila melanogaster. Genetics 149, 217–231.
Heisenberg, M., Borst, A., Wagner, S., and Byers, D. (1985). Drosophila mushroom body mutants are deficient in olfactory learning. J. Neurogenet. 2, 1–30. doi: 10.3109/01677068509100140
Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., et al. (2002). The genome sequence of the malaria mosquito Anopheles gambiae. Science 298, 129–149. doi: 10.1126/science.1076181
Hunding, A., and Baumgartner, S. (2017). Ancient role of ten-m/odz in segmentation and the transition from sequential to syncytial segmentation. Hereditas 154:8. doi: 10.1186/s41065-017-0029-1
Hunding, A., and Engelhardt, R. (1995). Early biological morphogenesis and nonlinear dynamics. J. Theor. Biol. 173, 401–413. doi: 10.1006/jtbi.1995.0072
International Chicken Genome Sequencing Consortium (2004). Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432, 695–716.
Jackson, V. A., Meijer, D. H., Carrasquero, M., van Bezouwen, L. S., Lowe, E. D., Kleanthous, C., et al. (2018). Structures of teneurin adhesion receptors reveal an ancient fold for cell-cell interaction. Nat. Commun. 9:1079. doi: 10.1038/s41467-018-03460-0
Karpen, G. H., and Spradling, A. C. (1992). Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132, 737–753.
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., et al. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. doi: 10.1038/35057062
Levine, A., Bashan-Ahrend, A., Budai-Hadrian, O., Gartenberg, D., Menasherow, S., and Wides, R. (1994). Odd Oz: a novel Drosophila pair rule gene. Cell 77, 587–598. doi: 10.1016/0092-8674(94)90220-8
Levine, A., Weiss, C., and Wides, R. (1997). Expression of the pair-rule gene odd Oz (odz) in imaginal tissues. Dev. Dyn. 209, 1–14. doi: 10.1002/(SICI)1097-0177(199705)209:1<1::AID-AJA1>3.0.CO;2-M
Lewis, J. (2003). Autoinhibition with Transcriptional Delay. Curr. Biol. 13, 1398–1408. doi: 10.1016/S0960-9822(03)00534-7
Li, J., Shalev-Benami, M., Sando, R., Jiang, X., Kibrom, A., Wang, J., et al. (2018). Structural basis for teneurin function in circuit-wiring: a toxin motif at the synapse. Cell 173:735-748.e15. doi: 10.1016/j.cell.2018.03.036
Loose, M., Fischer-Friedrich, E., Ries, J., Kruse, K., and Schwille, P. (2008). Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792. doi: 10.1126/science.1154413
Lossie, A. C., Nakamura, H., Thomas, S. E., and Justice, M. J. (2005). Mutation of l7Rn3 shows that Odz4 is required for mouse gastrulation. Genetics 169, 285–299. doi: 10.1534/genetics.104.034967
McCarthy, A. J., Coleman-Vaughan, C., and McCarthy, J. V. (2017). Regulated intramembrane proteolysis: emergent role in cell signalling pathways. Biochem. Soc. Trans. 45, 1185–1202. doi: 10.1042/BST20170002
McGinnis, W., Levine, M. S., Hafen, E., Kuroiwa, A., and Gehring, W. J. (1984). A conserved DNA sequence in homoeotic genes of the Drosophila antennapedia and bithorax complexes. Nature 308, 428–433. doi: 10.1038/308428a0
Meinhardt, H., and de Boer, P. A. J. (2001). Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc. Natl. Acad. Sci. U.S.A. 98, 14202–14207. doi: 10.1073/pnas.251216598
Midwood, K. S., Chiquet, M., Tucker, R. P., and Orend, G. (2016). Tenascin-C at a glance. J. Cell Sci. 129, 4321–4327. doi: 10.1242/jcs.190546
Mieda, M., Kikuchi, Y., Hirate, Y., Aoki, M., and Okamoto, H. (1999). Compartmentalized expression of zebrafish ten-m3 and ten-m4, homologues of the Drosophila ten(m)/odd Oz gene, in the central nervous system. Mech. Dev. 87, 223–227. doi: 10.1016/S0925-4773(99)00155-0
Minet, A. D., and Chiquet-Ehrismann, R. (2000). Phylogenetic analysis of teneurin genes and comparison to the rearrangement hot spot elements of E. coli. Gene 257, 87–97. doi: 10.1016/S0378-1119(00)00388-7
Minet, A. D., Rubin, B. P., Tucker, R. P., Baumgartner, S., and Chiquet-Ehrismann, R. (1999). Teneurin-1, a vertebrate homologue of the Drosophila pair-rule gene ten-m, is a neuronal protein with a novel type of heparin-binding domain. J. Cell Sci. 112(Pt 12), 2019–2032.
Mita, K., Kasahara, M., Sasaki, S., Nagayasu, Y., Yamada, T., Kanamori, H., et al. (2004). The genome sequence of silkworm, Bombyx mori. DNA Res. 11, 27–35. doi: 10.1093/dnares/11.1.27
Mouse Genome Sequencing, C., Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi: 10.1038/nature01262
Mural, R. J., Adams, M. D., Myers, E. W., Smith, H. O., Miklos, G. L., Wides, R., et al. (2002). A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296, 1661–1671.
Nunes, S. M., Ferralli, J., Choi, K., Brown-Luedi, M., Minet, A. D., and Chiquet-Ehrismann, R. (2005). The intracellular domain of teneurin-1 interacts with MBD1 and CAP/ponsin resulting in subcellular codistribution and translocation to the nuclear matrix. Exp. Cell Res. 305, 122–132. doi: 10.1016/j.yexcr.2004.12.020
Oohashi, T., Zhou, X. H., Feng, K., Richter, B., Morgelin, M., Perez, M. T., et al. (1999). Mouse ten-m/Odz is a new family of dimeric type II transmembrane proteins expressed in many tissues. J. Cell Biol. 145, 563–577. doi: 10.1083/jcb.145.3.563
Otaki, J. M., and Firestein, S. (1999a). Neurestin: putative transmembrane molecule implicated in neuronal development. Dev. Biol. 212, 165–181.
Otaki, J. M., and Firestein, S. (1999b). Segregated expression of neurestin in the developing olfactory bulb. Neuroreport 10, 2677–2680.
Palmeirim, I., Henrique, D., Ish-Horowicz, D., and Pourquié, O. (1997). Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell 91, 639–648. doi: 10.1016/S0092-8674(00)80451-1
Prescott, D. M., and Bender, M. A. (1962). Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp. Cell Res. 26, 260–268. doi: 10.1016/0014-4827(62)90176-3
Qian, X., Barsyte-Lovejoy, D., Wang, L., Chewpoy, B., Gautam, N., Al Chawaf, A., et al. (2004). Cloning and characterization of teneurin C-terminus associated peptide (TCAP)-3 from the hypothalamus of an adult rainbow trout (Oncorhynchus mykiss). Gen. Comp. Endocrinol. 137, 205–216. doi: 10.1016/j.ygcen.2004.02.007
Rakovitsky, N., Buganim, Y., Swissa, T., Kinel-Tahan, Y., Brenner, S., Cohen, M. A., et al. (2007). Drosophila Ten-a is a maternal pair-rule and patterning gene. Mech. Dev. 124, 911–924. doi: 10.1016/j.mod.2007.08.003
Rubin, B. P., Tucker, R. P., Brown-Luedi, M., Martin, D., and Chiquet-Ehrismann, R. (2002). Teneurin 2 is expressed by the neurons of the thalamofugal visual system in situ and promotes homophilic cell-cell adhesion in vitro. Development 129, 4697–4705.
Rubin, B. P., Tucker, R. P., Martin, D., and Chiquet-Ehrismann, R. (1999). Teneurins: a novel family of neuronal cell surface proteins in vertebrates, homologous to the Drosophila pair-rule gene product Ten-m. Dev. Biol. 216, 195–209. doi: 10.1006/dbio.1999.9503
Schaefer, G., Fitzpatrick, V. D., and Sliwkowski, M. X. (1997). Gamma-heregulin: a novel heregulin isoform that is an autocrine growth factor for the human breast cancer cell line, MDA-MB-175. Oncogene 15, 1385–1394. doi: 10.1038/sj.onc.1201317
Stein, L. D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M. R., Chen, N., et al. (2003). The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1:E45. doi: 10.1371/journal.pbio.0000045
Treisman, J. E., and Rubin, G. M. (1996). Targets of glass regulation in the Drosophila eye disc. Mech. Dev. 56, 17–24. doi: 10.1016/0925-4773(96)00508-4
Tucker, R. P. (2018). Teneurins: domain architecture, evolutionary origins, and patterns of expression. Front. Neurosci. 12:938. doi: 10.3389/fnins.2018.00938
Tucker, R. P., and Chiquet-Ehrismann, R. (2006). Teneurins: a conserved family of transmembrane proteins involved in intercellular signaling during development. Dev. Biol. 290, 237–245. doi: 10.1016/j.ydbio.2005.11.038
Tucker, R. P., Kenzelmann, D., Trzebiatowska, A., and Chiquet-Ehrismann, R. (2007). Teneurins: transmembrane proteins with fundamental roles in development. Int. J. Biochem. Cell Biol. 39, 292–297. doi: 10.1016/j.biocel.2006.09.012
Venken, K. J., He, Y., Hoskins, R. A., and Bellen, H. J. (2006). P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in melanogaster, D. Science 314, 1747–1751. doi: 10.1126/science.1134426
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., et al. (2001). The sequence of the human genome. Science 291, 1304–1351. doi: 10.1126/science.1058040
Wang, X. Z., Jolicoeur, E. M., Conte, N., Chaffanet, M., Zhang, Y., Mozziconacci, M. J., et al. (1999). gamma-heregulin is the product of a chromosomal translocation fusing the DOC4 and HGL/NRG1 genes in the MDA-MB-175 breast cancer cell line. Oncogene 18, 5718–5721. doi: 10.1038/sj.onc.1202950
Wang, X. Z., Kuroda, M., Sok, J., Batchvarova, N., Kimmel, R., Chung, P., et al. (1998). Identification of novel stress-induced genes downstream of chop. EMBO J. 17, 3619–3630. doi: 10.1093/emboj/17.13.3619
Wood, V., Gwilliam, R., Rajandream, M. A., Lyne, M., Lyne, R., Stewart, A., et al. (2002). The genome sequence of schizosaccharomyces pombe. Nature 415, 871–880. doi: 10.1038/nature724
Yu, J., Hu, S., Wang, J., Wong, G. K., Li, S., Liu, B., et al. (2002). A draft sequence of the rice genome (Oryza sativa ssp, L. indica). Science 296, 79–92. doi: 10.1126/science.1068037
Zhang, D. R., de Souza, F., Anantharaman, V. L., Iyer, M., and Aravind, L. (2012). Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7:18. doi: 10.1186/1745-6150-7-18
Zheng, L., Michelson, Y., Freger, V., Avraham, Z., Venken, K. J., and Bellen, H. J. (2011). Drosophila Ten-m and filamin affect motor neuron growth cone guidance. PLoS One 6:e22956. doi: 10.1371/journal.pone.0022956
Keywords: teneurin, ten-m, ten-a, TENM, ODZ, latrophilin, type II transmembrane protein, Drosophila
Citation: Baumgartner S and Wides R (2019) Discovery of Teneurins. Front. Neurosci. 13:230. doi: 10.3389/fnins.2019.00230
Received: 11 October 2018; Accepted: 27 February 2019;
Published: 19 March 2019.
Edited by:
Richard P. Tucker, University of California, Davis, United StatesReviewed by:
Timothy Mosca, Thomas Jefferson University, United StatesKrzysztof Drabikowski, Institute of Biochemistry and Biophysics (PAN), Poland
Copyright © 2019 Baumgartner and Wides. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ron Wides, cm9ud2lkZXNAeWFob28uY29t
 Stefan Baumgartner
Stefan Baumgartner Ron Wides
Ron Wides