
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Neurosci. , 25 February 2019
Sec. Sleep and Circadian Rhythms
Volume 13 - 2019 | https://doi.org/10.3389/fnins.2019.00139
This article is part of the Research Topic The Gating and Maintenance of sleep and Wake: New Circuits and Insights View all 24 articles
The suprachiasmatic nucleus (SCN) functions as the central circadian pacemaker in mammals and entrains to the environmental light/dark cycle. It is composed of multiple types of GABAergic neurons, and interneuronal communications among these neurons are essential for the circadian pacemaking of the SCN. However, the mechanisms underlying the SCN neuronal network remain unknown. This review will provide a brief overview of the current knowledge concerning the differential roles of multiple neuropeptides and neuropeptide-expressing neurons in the SCN, especially focusing on the emerging roles of arginine vasopressin-producing neurons uncovered by recent studies utilizing neuron type-specific genetic manipulations in mice.
The circadian oscillator of the hypothalamic suprachiasmatic nucleus (SCN) is the central pacemaker in mammals, orchestrating multiple circadian biological rhythms in the organism and being regulated according to the external light/dark conditions conveyed from the eye (Reppert and Weaver, 2002). The SCN contains ∼20,000 neurons, most of which are able to oscillate autonomously. Individual cellular oscillators (cellular clocks) are driven by autoregulatory transcriptional/translational feedback loops (TTFLs) of clock genes in concert with cytosolic signaling molecules, including cAMP and Ca2+ (Welsh et al., 2010; Herzog et al., 2017; Takahashi, 2017). Surprisingly, these intracellular molecular mechanisms are not unique to SCN cells but are shared with peripheral cells (Balsalobre et al., 1998). Rather, intercellular communications among SCN cells through the neuronal and diffusible network are the unique feature of the SCN that is responsible for the generation of highly robust and coherent oscillations as an ensemble (Welsh et al., 2010).
The SCN is a heterogeneous structure that consists of multiple types of GABAergic neurons (Antle and Silver, 2005). Many of them co-express neuropeptides, represented by. arginine vasopressin (AVP)-producing neurons located in the shell, the dorsomedial part, of the SCN, as well as by vasoactive intestinal peptide (VIP)-producing neurons and gastrin releasing peptide (GRP)-producing neurons in the core, the ventrolateral part, of the SCN (Figure 1A). Rhythmic Period (Per) expression in constant darkness (DD) is highest in the SCN shell (Hamada et al., 2004). In contrast, the SCN core contains retinorecipient neurons that respond immediately to the environmental light stimuli and communicate this information to the shell (Silver et al., 1996; Shigeyoshi et al., 1997). Two other input pathways from the median raphe and intergeniculate leaflet also terminate mainly in the SCN core, while afferents from the hypothalamus and limbic areas terminate mainly in the SCN shell (Moga and Moore, 1997). SCN neurons project principally to areas within the diencephalon, especially to the subparaventricular zone, the area just dorsal to the SCN. The majority of efferent projections originates in the shell, but the core neurons also send efferent projections in a manner different from the shell neurons (Leak and Moore, 2001). Within the SCN, core neurons send projections densely to the shell, while fibers of shell neurons in the core are sparse (Leak et al., 1999).
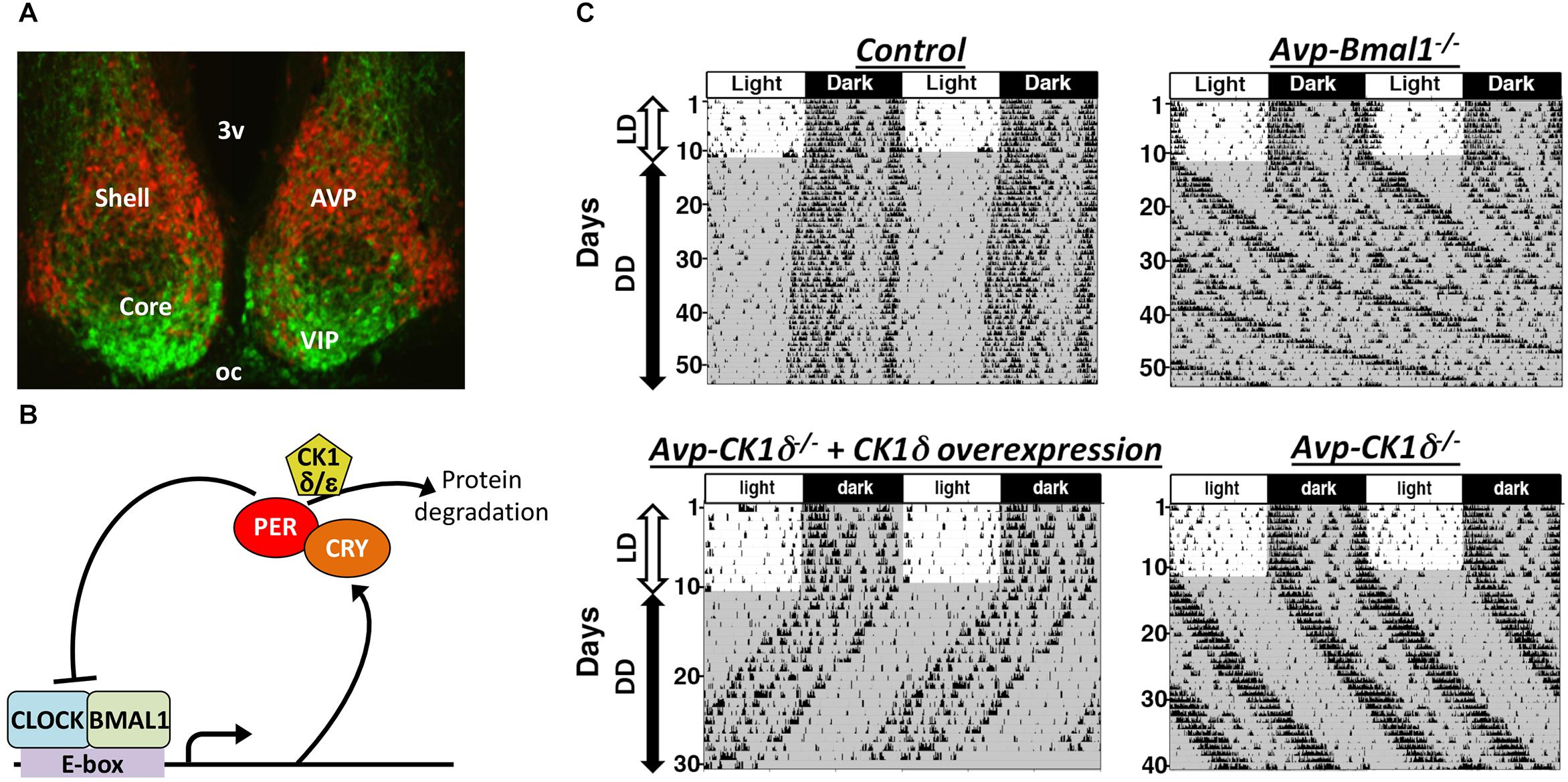
Figure 1. The SCN is composed of multiple types of clock neurons. (A) A coronal section of mouse SCN, showing the dorsal shell region delineated by the expression of tdTomato fluorescent protein in AVP neurons (red) and the ventral core region containing VIP neurons labeled immunofluorescently (green). 3v, third ventricle; oc, optic chiasm. (B) A simplified schematic of cellular clock, the molecular machinery of circadian clock composed of TTFLs of clock genes. Only the part relevant to this review is shown here. For the detailed mechanism, please see other reviews, such as Takahashi, 2017. Transcription factors CLOCK and BMAL1 bind to E-box sequences to drive expression of PER and CRY, which in turn suppress CLOCK-BMAL1 activity, closing a negative feedback loop. Phosphorylation of PER proteins by CK1δ enhances those proteins’ degradation and consequently accelerates the speed of cellular clocks. (C) Representative actograms of mice with AVP neuron-specific genetic manipulations. The free-running period is lengthened in both Avp-Bmal1-/- and Avp-CK1δ-/- mice, while the activity time was lengthened only in Avp-Bmal1-/- mice. Modified from Mieda et al. (2015, 2016).
Vasoactive intestinal peptide has been demonstrated to be especially important for the maintenance and synchronization of cellular clocks in the individual SCN neurons (Herzog et al., 2017). Thus, mice lacking Vip or VIP receptor Vipr2 gene demonstrate drastically weakened behavioral rhythms, often with multiple period components (Harmar et al., 2002; Colwell et al., 2003; Aton et al., 2005). At the cellular level, circadian oscillations of electrical firing and clock gene expression of individual SCN neurons (cellular clocks) are desynchronized in slices (Aton et al., 2005; Maywood et al., 2006; Brown et al., 2007). Furthermore, the numbers of rhythmic neurons are drastically reduced (Aton et al., 2005; Maywood et al., 2006; Brown et al., 2007). These observations suggest that VIP functions as a master synchronizer in the SCN.
Consistent with these observations, optogenetic activation of VIP neurons in the SCN phase-shifts the cellular clock (circadian PER2::LUC oscillation) in explants and entrains the behavior rhythm in vivo (Jones et al., 2015; Mazuski et al., 2018). In addition, chemogenetic inhibition of these neurons attenuates the light-induced phase-shift of circadian behavior rhythm (Jones et al., 2018). In explants, prolonged chemogenetic stimulation of these neurons further reprograms the global spatiotemporal dynamics of the SCN cellular clocks (Brancaccio et al., 2013).
On the other hand, VIP neurons may not play a significant role in the pacemaking of circadian rhythms by the SCN network, that is, determination of the circadian period, as discussed again later. Lee et al. demonstrated that overexpression of the ClockΔ19 transgene in VIP neurons had no effect on the behavioral free-running period, although such a ClockΔ19 overexpression lengthens the intrinsic period of cellular clocks in the manipulated cells (Table 1; Lee et al., 2015). These results contrasted clearly with the observations that the same genetic manipulations in SCN neurons expressing a neuropeptide neuromedin-S (NMS) lengthens the period of behavior rhythm: NMS neurons include VIP neurons, AVP neurons, and some other types of neurons. In conjunction with the observation that genetic ablation of cellar clocks specifically in NMS neurons disrupts circadian rhythms, Lee et al. (2015) concluded that NMS neurons act as essential pacemakers in the SCN.
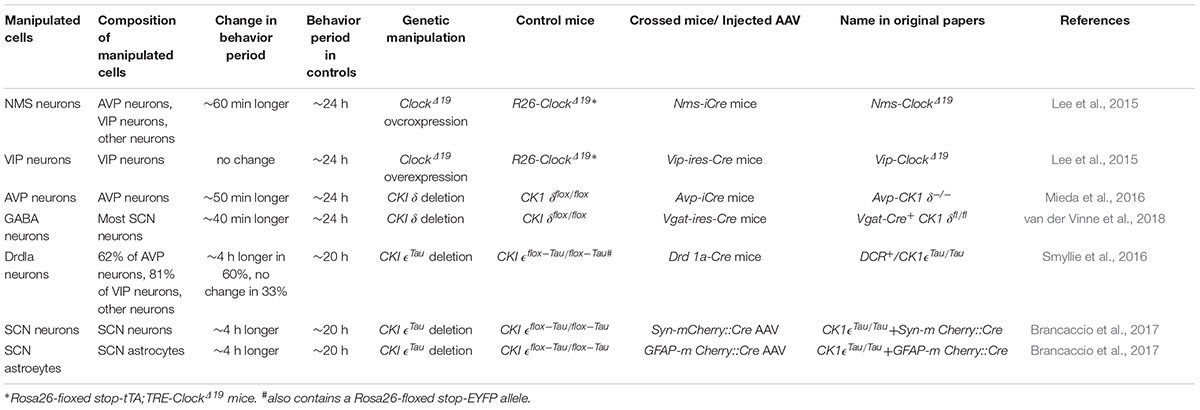
Table 1. Effects of neuron type-specific alterations of cellular circadian periods on the free-running periods of circadian behavior rhythms.
The concentration of AVP in the cerebrospinal fluid (CSF) daily fluctuates with a peak in the morning (Stark and Daniel, 1989; Kalsbeek et al., 2010). Such a circadian variation of CSF AVP level has been shown to originate from the AVP content in the SCN (Södersten et al., 1985). Indeed, the transcription of Avp gene in the SCN is under the control of cellular clocks, the molecular machinery of the circadian clock composed of TTFL of clock genes (Jin et al., 1999). In conjunction with the fact that AVP-deficient Brattleboro rats display attenuated circadian rhythms but little abnormality in circadian pacemaking, AVP has been considered to function as an SCN output (Groblewski et al., 1981; Brown and Nunez, 1989; Kalsbeek et al., 2010). A recent optogenetic study directly demonstrated clock-driven AVP neurotransmission that mediates anticipatory thirst prior to sleep (Gizowski et al., 2016).
Nevertheless, AVP may also play a minor but significant role in the coupling of SCN neurons. In coculture experiments of SCN explants, the requirement of AVP signaling for the synchronization of SCN neurons becomes manifest in the absence of VIP signaling (Maywood et al., 2011; Edwards et al., 2016; Ono et al., 2016). Deletion of V1a receptor, the principal AVP receptor of the SCN, lengthens the activity time in DD by approximately 100 min in mice, suggesting the attenuated coupling among SCN neurons (Li et al., 2009). A small number of these mice even show arrhythmicity. In another study, V1a-/-; V1b-/- mice were reported to immediately reentrain to phase-shifted LD cycles whereas their free-running rhythms are intact, indicating that interneuronal communication mediated by AVP make the SCN resistant to environmental perturbations such as jet lag (Yamaguchi et al., 2013). In contrast to VIP, Avp knockout mice are not available for the examination of circadian behavior rhythm, because they do not survive beyond postnatal day 7 (Yoshikawa et al., 2015). Therefore, development and study of SCN-specific knockout mice would further elucidate the physiological role of AVP signaling in the central circadian clock.
Arginine vasopressin neurons express neurotransmitters other than AVP, such as GABA and prokineticin 2 (Antle and Silver, 2005; Masumoto et al., 2006; Welsh et al., 2010). Multiple transmitters in one neuronal type may transmit differential information, as reported in orexigenic AgRP neurons in the hypothalamic arcuate nucleus and wake-stabilizing orexin neurons in the lateral and perifornical hypothalamus (Krashes et al., 2013; Muschamp et al., 2014; Schöne et al., 2014). Therefore, neurons producing AVP may play a more fundamental role in the circadian pacemaking of the SCN than the AVP molecule does. This hypothesis was testable by genetically manipulating AVP neurons using the Cre-loxP system. When Bmal1, an essential transcription factor of cellular clocks (Figure 1B; Bunger et al., 2000), was deleted specifically in AVP neurons (Avp-Bmal1-/- mice), mice demonstrated a significant impairment of the locomotor activity rhythm in DD (Figure 1C; Mieda et al., 2015). When released into DD, the interval between the activity onset and offset (activity time) gradually became expanded by approximately 5 h as compared with that in controls. Their free-running period was approximately 50 min longer on average than that of control mice. Furthermore, a small number of Avp-Bmal1-/- mice even demonstrated arrhythmicity. Importantly, Bmal1 restoration in AVP neurons of the SCN with the aid of a recombinant AAV vector reversed the circadian impairment of Avp-Bmal1-/- mice almost completely. These results indicate that the cellular circadian oscillation persists, but the mutual coupling between clock neurons regulating the onset and offset components of activity may be severely impaired in the SCN of Avp-Bmal1-/- mice.
In these mice, the circadian expression of factors involved in intercellular communications, including Avp, Prokineticin 2, and Rgs16, was considerably decreased in the SCN shell, where AVP neurons are located. In SCN explants, PER2::LUC oscillations in the shell cells were attenuated with highly variable and lengthened periods. Collectively, Bmal1-based cellular clocks of AVP neurons are likely to enhance the coupling of the SCN cells to generate robust circadian rhythms by regulating expression of multiple factors involved in interneuronal communications (Mieda et al., 2015).
By artificially manipulating the period of cellular clocks specifically in AVP neurons, the possibility that AVP neurons actively work as pacemaker cells to determine the period of circadian rhythm generated by the SCN network was examined (Mieda et al., 2016). It has been shown that the phosphorylation of PER proteins by casein kinase 1δ (CK1δ) regulates the speed of cellular clocks (Figure 1B; Herzog et al., 2017). Artificial lengthening of the cellular circadian period specifically in AVP neurons, achieved by deleting CK1δ in AVP neurons (Avp-CK1δ-/-), also lengthened the free-running period of behavior rhythm by approximately 50 min, while their activity time remained normal (Figure 1C and Table 1). Conversely, artificial shortening of the AVP neuronal circadian period, archived by overexpression of CK1δ1 in these neurons via focal injection of a Cre-dependent AAV expression vector, shortened the period of behavior rhythm (Figure 1C). Thus, the manipulation of CK1δ expression levels in AVP neurons of the SCN bidirectionally changed the free-running period of behavior rhythm, suggesting that AVP neurons do indeed regulate SCN pacemaking (Mieda et al., 2016).
How much do AVP neurons contribute to the period determination? Mice in which CK1δ was deleted in the entire SCN, using GABAergic neuron-specific Vgat-Cre driver mice, also showed a lengthened free-running period of behavior rhythm by approximately 40 min (Table 1; van der Vinne et al., 2018), which was comparable to that in Avp-CK1δ-/- mice. These data indicate that AVP neurons are the principal determinant of circadian period generated by the SCN network in vivo.
Because of the coherently lengthened free-running period of Avp-CK1δ-/- mice (Figure 1C), the cellular clocks (PER2::LUC oscillations) of the entire SCN were also expected to oscillate with a longer period in slices. Contrary to such an expectation, however, the SCN shell and core of Avp-CK1δ-/-; Per2::Luc mice transiently demonstrated different cellular periods in explants (Mieda et al., 2016). The period of the shell was longer, but this lengthening did not last into the subsequent cycles. These data suggest that the core modulated the shell in the prolonged SCN culture. Indeed, the lengthening of shell’s period in Avp-CK1δ-/-; Per2::Luc mice lasted for a longer duration when slices were surgically cut between the shell and core. A similar dissociation of behavior rhythm and PER2::LUC rhythm has also been observed in Avp-Bmal1-/- mice (Mieda et al., 2015). Thus, the intact structure of the SCN and/or its connections with other brain areas might be important for the coupling between SCN shell and core in vivo.
In rodents, core neurons communicate with those in the shell, while there is less communication in the reverse direction (Leak et al., 1999). A recent study of mouse SCN connectome reported that, although the direct connection from AVP neurons to VIP neurons is extremely sparse, AVP neurons make plenty of contacts onto other types of neurons in the SCN core, such as GRP neurons, raising the possibility that AVP neurons communicate well with VIP neurons indirectly via those non-VIP core neurons (Varadarajan et al., 2018). Such asymmetric anatomical interactions between the SCN core and shell may make shell-to-core interaction more fragile in slices.
Lee et al. (2015) demonstrated that lengthening the cellular circadian period of NMS-producing SCN neurons by overexpression of ClockΔ19 lengthened the free-running period of behavior (Table 1). Nevertheless, NMS neurons are still a heterogeneous population that contains AVP neurons, VIP neurons, and other types of neurons (Lee et al., 2015), leaving the long-standing debate on the differential roles of the shell and core of the SCN on its pacemaking unresolved.
More recently, Smyllie et al. (2016) created chimeric mice by crossing Drd1a-Cre mice to floxed CK1𝜖Tau/Tau mice, whose SCN contained dopamine 1a receptor (Drd1a) cells (CK1𝜖-/- cells) with an intrinsic cellular period of 24 h alongside non-Drd1a cells (CK1𝜖Tau/Tau cells) with a period of 20 h (Table 1). Remarkably, 60% of these mice showed 24 h periods of behavior and SCN PER2::LUC rhythms, whereas 33% showed 20 h periods. Drd1a cells contain 63% of all SCN cells, including 62% of AVP neurons and 81% of VIP neurons (Smyllie et al., 2016). The fact that the behavioral period did not necessarily follow the cellular period of 80% of VIP neurons is consistent with the earlier finding by Lee et al. (2015) that lengthening the VIP neuronal cellular period had no effect on the behavioral free-running period. Collectively, these observations suggest that VIP neurons may not be directly involved in the pacemaking of the SCN, although VIP signaling plays a principal role in the synchronization of SCN neurons.
Taken in conjunction with data indicating that AVP neurons are involved in the SCN pacemaking (Mieda et al., 2016), as described earlier, the slight difference in the ratio of 24 h AVP neurons to 20 h AVP neurons in Smyllie et al. (2016) could exert a substantial impact on the period in which the chimeric SCN oscillates. In addition, the extent of lengthening in free-running period in mice lacking CK1δ in the entire SCN is comparable to that in mice with AVP neuron-specific CK1δ deletion (van der Vinne et al., 2018), as discussed earlier. These observations suggest that AVP neurons are the primary determinant of the period of circadian rhythm generated by the SCN network. VIP neurons may play a dominant role in the synchronization and phase regulation of SCN neurons, but their contribution in period determination may be little.
Recently, striking contributions of astrocytes of the SCN in the circadian pacemaking was reported (Barca-Mayo et al., 2017; Brancaccio et al., 2017; Tso et al., 2017). SCN astrocytes and neurons are likely to act as two arms of the central circadian pacemaker network, which shows oscillations anti-phasic to each other (Brancaccio et al., 2017). These neuronal and astrocytic oscillators are coupled via glutamate released from astrocytes, which increases presynaptic GABA release and subsequently suppresses neuronal activity of postsynaptic neurons during night. Floxed CK1𝜖Tau/Tau mice that originally had a free-running behavior period of 20 h changed the period to 24 h when CK1𝜖Tau alleles were deleted specifically in SCN astrocytes via viral Cre delivery, suggesting that SCN astrocytes can control the period of circadian behavior rhythms (Table 1). Interestingly, the same reversal of free-running period was observed when CK1𝜖Tau alleles were deleted specifically in SCN neurons of the same mice. Therefore, both SCN astrocytes and neurons are equally able to impart timekeeping information to the rest of the body (Brancaccio et al., 2017). However, these results may appear a little strange and difficult to interpret. Although astrocyte- and neuron-specific deletions of CK1𝜖Tau in floxed CK1𝜖Tau/Tau mice resulted in reversed temporal misalignments of the SCN – that is, the 20 h neuronal clock and the 24 h astrocytic clock, and vice versa – the chimeric mice always showed a free-running period of 24 h. One explanation for these observations may be that cellular clocks and the SCN network are optimized to work at 24 h and therefore would be advantaged in the chimeras over the 20 h cells, regardless of which cell type has been targeted (Brancaccio et al., 2017). It would be very interesting to examine whether artificial lengthening (by CK1δ deletion or ClockΔ19 overexpression) or shortening (by CK1δ1 overexpression) of the astrocytic cellular period from 24 h alters the free-running period of behavior rhythm as much as neuronal manipulations do. In any case, comprehensive understanding of the network principle of the SCN central circadian clock needs further study.
A previous pioneering study utilized chimera mice of wild type and long-period ClockΔ19/Δ19 mutant cells to address the network mechanism of the circadian period determination by the SCN (Low-Zeddies and Takahashi, 2001). In these mice, random subsets of wild type SCN cells were replaced with ClockΔ19/Δ19 cells. The proportion of ClockΔ19/Δ19 versus wild type cells largely determined circadian behavior in chimeric individuals. However, the intermediate periods were observed in some but not evident in all balanced chimeras. This fact indicates that the emergence of intermediate periods is dependent on not only the proportion but also the distribution of wild type and ClockΔ19/Δ19 cells (Low-Zeddies and Takahashi, 2001), suggesting unequal contributions among SCN cells to the period determination. Cell type-specific manipulations of the cellular circadian period described earlier in this review further support such an idea that there exist cells that function as the dominant pacemaking elements in the SCN network, a likely candidate of which may be AVP neurons.
Thus, as the cellular clocks have molecular mechanisms to determine their period, amplitude, and phase within the individual cells, the SCN may have multicellular and network mechanisms to determine the period, amplitude, and phase of the circadian rhythm it generates, which is not a simple summation of multiple cellular clocks. In other words, there exists functional localization within the SCN. The characterization of Avp-Bmal1-/- mice and Avp-CK1δ-/- mice definitively demonstrated that cellular clocks of SCN AVP neurons play a critical role in the generation of robust circadian behavior rhythm through the regulation of the coupling of SCN neurons, as well as in the determination of the circadian period. Additional manipulations of cellular clocks and neuronal properties in various combinations of neuron types and genetic-engineering techniques would provide further information to comprehensively understand the principle of the SCN neural network as the central circadian pacemaker.
The author confirms being the sole contributor of this work and has approved it for publication.
This work was supported by MEXT/JSPS KAKENHI Grant Numbers JP16H05120, JP18H04941, JP18K19421, and JP18H04972.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Antle, M. C., and Silver, R. (2005). Orchestrating time: arrangements of the brain circadian clock. Trends Neurosci. 28, 145–151. doi: 10.1016/j.tins.2005.01.003
Aton, S. J., Colwell, C. S., Harmar, A. J., Waschek, J., and Herzog, E. D. (2005). Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat. Neurosci. 8, 476–483. doi: 10.1038/nn1419
Balsalobre, A., Damiola, F., and Schibler, U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. doi: 10.1016/S0092-8674(00)81199-X
Barca-Mayo, O., Pons-Espinal, M., Follert, P., Armirotti, A., Berdondini, L., and De Pietri Tonelli, D. (2017). Astrocyte deletion of Bmal1 alters daily locomotor activity and cognitive functions via GABA signalling. Nat. Commun. 8, 1–14. doi: 10.1038/ncomms14336
Brancaccio, M., Maywood, E. S., Chesham, J. E., Loudon, A. S. I., and Hastings, M. H. (2013). A Gq-Ca2+Axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron 78, 714–728. doi: 10.1016/j.neuron.2013.03.011
Brancaccio, M., Patton, A. P., Chesham, J. E., Maywood, E. S., and Hastings, M. H. (2017). Astrocytes control circadian timekeeping in the suprachiasmatic nucleus via glutamatergic signaling. Neuron 93, 1420.e5–1435.e5. doi: 10.1016/j.neuron.2017.02.030
Brown, M. H., and Nunez, A. A. (1989). Vasopressin-deficient rats show a reduced amplitude of the circadian sleep rhythm. Physiol. Behav. 46, 759–762. doi: 10.1016/0031-9384(89)90364-8
Brown, T. M., Colwell, C. S., Waschek, J. A., and Piggins, H. D. (2007). Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J. Neurophysiol. 97, 2553–2558. doi: 10.1152/jn.01206.2006
Bunger, M. K., Wilsbacher, L. D., Moran, S. M., Clendenin, C., Radcliffe, L. A., Hogenesch, J. B., et al. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009–1017. doi: 10.1016/S0092-8674(00)00205-1
Colwell, C. S., Michel, S., Itri, J., Rodriguez, W., Tam, J., Lelievre, V., et al. (2003). Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am. J. Physiol. Integr. Comp. Physiol. 285, R939–R949. doi: 10.1152/ajpregu.00200.2003
Edwards, M. D., Brancaccio, M., Chesham, J. E., Maywood, E. S., and Hastings, M. H. (2016). Rhythmic expression of cryptochrome induces the circadian clock of arrhythmic suprachiasmatic nuclei through arginine vasopressin signaling. Proc. Natl. Acad. Sci. U.S.A. 113, 2732–2737. doi: 10.1073/pnas.1519044113
Gizowski, C., Zaelzer, C., and Bourque, C. W. (2016). Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature 537, 685–688. doi: 10.1038/nature19756
Groblewski, T. A., Nunez, A. A., and Gold, R. M. (1981). Circadian rhythms in vasopressin deficient rats. Brain Res. Bull. 6, 125–130. doi: 10.1016/S0361-9230(81)80036-6
Hamada, T., Antle, M. C., and Silver, R. (2004). Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur. J. Neurosci. 19, 1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x
Harmar, A. J., Marston, H. M., Shen, S., Spratt, C., West, K. M., Sheward, W. J., et al. (2002). The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell 109, 497–508. doi: 10.1016/S0092-8674(02)00736-5
Herzog, E. D., Hermanstyne, T., Smyllie, N. J., and Hastings, M. H. (2017). Regulating the suprachiasmatic nucleus (SCN) circadian clockwork: interplay between cell-autonomous and circuit-level mechanisms. Cold Spring Harb. Perspect. Biol. 9:a027706. doi: 10.1101/cshperspect.a027706
Jin, X., Shearman, L. P., Weaver, D. R., Zylka, M. J., de Vries, G. J., and Reppert, S. M. (1999). A Molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell 96, 57–68. doi: 10.1016/S0092-8674(00)80959-9
Jones, J. R., Simon, T., Lones, L., and Herzog, E. D. (2018). SCN VIP neurons are essential for normal light-mediated resetting of the circadian system. J. Neurosci. 38, 7986–7995. doi: 10.1523/JNEUROSCI.1322-18.2018
Jones, J. R., Tackenberg, M. C., and Mcmahon, D. G. (2015). Manipulating circadian clock neuron firing rate resets molecular circadian rhythms and behavior. Nat. Neurosci. 18, 373–377. doi: 10.1038/nn.3937
Kalsbeek, A., Fliers, E., Hofman, M. A., Swaab, D. F., and Buijs, R. M. (2010). Vasopressin and the output of the hypothalamic biological clock. J. Neuroendocrinol. 22, 362–372. doi: 10.1111/j.1365-2826.2010.01956.x
Krashes, M. J., Shah, B. P., Koda, S., and Lowell, B. B. (2013). Rapid versus delayed stimulation of feeding by the endogenously released agRP neuron mediators GABA, NPY, and AgRP. Cell Metab. 18, 588–595. doi: 10.1016/j.cmet.2013.09.009
Leak, R. K., Card, J. P., and Moore, R. Y. (1999). Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 819, 23–32. doi: 10.1016/S0006-8993(98)01317-1
Leak, R. K., and Moore, R. Y. (2001). Topographic organization of suprachiasmatic nucleus projection neurons. J. Comp. Neurol. 433, 312–334. doi: 10.1002/cne.1142
Lee, I. T., Chang, A. S., Manandhar, M., Shan, Y., Fan, J., Izumo, M., et al. (2015). Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron 85, 1086–1102. doi: 10.1016/j.neuron.2015.02.006
Li, J.-D., Burton, K. J., Zhang, C., Hu, S.-B., and Zhou, Q.-Y. (2009). Vasopressin receptor V1a regulates circadian rhythms of locomotor activity and expression of clock-controlled genes in the suprachiasmatic nuclei. Am. J. Physiol. Integr. Comp. Physiol. 296, R824–R830. doi: 10.1152/ajpregu.90463.2008
Low-Zeddies, S. S., and Takahashi, J. S. (2001). Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell 105, 25–42. doi: 10.1016/S0092-8674(01)00294-X
Masumoto, K. H., Nagano, M., Takashima, N., Hayasaka, N., Hiyama, H., Matsumoto, S. I., et al. (2006). Distinct localization of prokineticin 2 and prokineticin receptor 2 mRNAs in the rat suprachiasmatic nucleus. Eur. J. Neurosci. 23, 2959–2970. doi: 10.1111/j.1460-9568.2006.04834.x
Maywood, E. S., Chesham, J. E., O’Brien, J. A., and Hastings, M. H. (2011). A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc. Natl. Acad. Sci. 108, 14306–14311. doi: 10.1073/pnas.1101767108
Maywood, E. S., Reddy, A. B., Wong, G. K. Y., O’Neill, J. S., O’Brien, J. A., McMahon, D. G., et al. (2006). Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr. Biol. 16, 599–605. doi: 10.1016/j.cub.2006.02.023
Mazuski, C., Abel, J. H., Chen, S. P., Hermanstyne, T. O., Jones, J. R., Simon, T., et al. (2018). Entrainment of circadian rhythms depends on firing rates and neuropeptide release of VIP SCN neurons. Neuron 99, 555.e5–563.e5. doi: 10.1016/j.neuron.2018.06.029
Mieda, M., Okamoto, H., and Sakurai, T. (2016). Manipulating the cellular circadian period of arginine vasopressin neurons alters the behavioral circadian period. Curr. Biol. 26, 2535–2542. doi: 10.1016/j.cub.2016.07.022
Mieda, M., Ono, D., Hasegawa, E., Okamoto, H., Honma, K., Honma, S., et al. (2015). Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116. doi: 10.1016/j.neuron.2015.02.005
Moga, M. M., and Moore, R. Y. (1997). Organization of neural inputs to the suprachiasmatic nucleus in the rat. J. Comp. Neurol. 389, 508–534. doi: 10.1002/(SICI)1096-9861(19971222)389:3<508::AID-CNE11>3.0.CO;2-H
Muschamp, J. W., Hollander, J. A., Thompson, J. L., Voren, G., Hassinger, L. C., Onvani, S., et al. (2014). Hypocretin (orexin) facilitates reward by attenuating the antireward effects of its cotransmitter dynorphin in ventral tegmental area. Proc. Natl. Acad. Sci. 111, E1648–E1655. doi: 10.1073/pnas.1315542111
Ono, D., Honma, S., and Honma, K. (2016). Differential roles of AVP and VIP signaling in the postnatal changes of neural networks for coherent circadian rhythms in the SCN. Sci. Adv. 2, 1–13. doi: 10.1126/sciadv.1600960
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Schöne, C., Apergis-Schoute, J., Sakurai, T., Adamantidis, A., and Burdakov, D. (2014). Coreleased orexin and glutamate evoke nonredundant spike outputs and computations in histamine neurons. Cell Rep. 7, 697–704. doi: 10.1016/j.celrep.2014.03.055
Shigeyoshi, Y., Taguchi, K., Yamamoto, S., Takekida, S., Yan, L., Tei, H., et al. (1997). Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell 91, 1043–1053. doi: 10.1016/S0092-8674(00)80494-8
Silver, R., Romero, M. T., Besmer, H. R., Leak, R., Nunez, J. M., and LeSauter, J. (1996). Calbindin-D28K cells in the hamster SCN express light-induced Fos. Neuroreport 7, 1224–1228. doi: 10.1097/00001756-199604260-00026
Smyllie, N. J., Chesham, J. E., Hamnett, R., Maywood, E. S., and Hastings, M. H. (2016). Temporally chimeric mice reveal flexibility of circadian period-setting in the suprachiasmatic nucleus. Proc. Natl. Acad. Sci. 113, 3657–3662. doi: 10.1073/pnas.1511351113
Södersten, P., De Vries, G. J., Buijs, R. M., and Melin, P. (1985). A daily rhythm in behavioral vasopressin sensitivity and brain vasopressin concentrations. Neurosci. Lett. 58, 37–41. doi: 10.1016/0304-3940(85)90325-8
Stark, R. I., and Daniel, S. S. (1989). Circadian rhythm of vasopressin levels in cerebrospinal fluid of the fetus: effect of continuous light. Endocrinology 124, 3095–3101. doi: 10.1210/endo-124-6-3095
Takahashi, J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179. doi: 10.1038/nrg.2016.150
Tso, C. F., Simon, T., Greenlaw, A. C., Puri, T., Mieda, M., and Herzog, E. D. (2017). Astrocytes regulate daily rhythms in the suprachiasmatic nucleus and behavior. Curr. Biol. 27, 1055–1061. doi: 10.1016/j.cub.2017.02.037
van der Vinne, V., Swoap, S. J., Vajtay, T. J., and Weaver, D. R. (2018). Desynchrony between brain and peripheral clocks caused by CK1δ/𝜀 disruption in GABA neurons does not lead to adverse metabolic outcomes. Proc. Natl. Acad. Sci. 115, E2437–E2446. doi: 10.1073/pnas.1712324115
Varadarajan, S., Tajiri, M., Jain, R., Holt, R., Ahmed, Q., LeSauter, J., et al. (2018). Connectome of the suprachiasmatic nucleus: new evidence of the core-shell relationship. eNeuro 5:ENEURO.205-18.2018. doi: 10.1523/ENEURO.0205-18.2018
Welsh, D. K., Takahashi, J. S., and Kay, S. A. (2010). Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577. doi: 10.1146/annurev-physiol-021909-135919
Yamaguchi, Y., Suzuki, T., Mizoro, Y., Kori, H., Okada, K., Chen, Y., et al. (2013). Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science 342, 85–90. doi: 10.1126/science.1238599
Keywords: circadian rhythm, suprachiasmatic nucleus, vasopressin, vasoactive intestinal peptide, neural network
Citation: Mieda M (2019) The Network Mechanism of the Central Circadian Pacemaker of the SCN: Do AVP Neurons Play a More Critical Role Than Expected? Front. Neurosci. 13:139. doi: 10.3389/fnins.2019.00139
Received: 08 December 2018; Accepted: 07 February 2019;
Published: 25 February 2019.
Edited by:
Michael Lazarus, University of Tsukuba, JapanCopyright © 2019 Mieda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michihiro Mieda, bWllZGFAbWVkLmthbmF6YXdhLXUuYWMuanA=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.