- 1Bond Life Sciences Center, University of Missouri, Columbia, MO, United States
- 2Biomedical Sciences, University of Missouri, Columbia, MO, United States
- 3Thompson Center for Autism and Neurobehavioral Disorders, University of Missouri, Columbia, MO, United States
- 4Genetics Area Program, University of Missouri, Columbia, MO, United States
Brain sexual differentiation is orchestrated by precise coordination of sex steroid hormones. In some species, programming of select male brain regions is dependent upon aromatization of testosterone to estrogen. In mammals, these hormones surge during the organizational and activational periods that occur during perinatal development and adulthood, respectively. In various fish and reptiles, incubation temperature during a critical embryonic period results in male or female sexual differentiation, but this can be overridden in males by early exposure to estrogenic chemicals. Testes development in mammals requires a Y chromosome and testis determining gene SRY (in humans)/Sry (all other therian mammals), although there are notable exceptions. Two species of spiny rats: Amami spiny rat (Tokudaia osimensis) and Tokunoshima spiny rat (Tokudaia tokunoshimensis) and two species of mole voles (Ellobius lutescens and Ellobius tancrei), lack a Y chromosome/Sry and possess an XO chromosome system in both sexes. Such rodent species, prototherians (monotremes, who also lack Sry), and fish and reptile species that demonstrate temperature sex determination (TSD) seemingly call into question the requirement of Sry for brain sexual differentiation. This review will consider brain regions expressing SRY/Sry in humans and rodents, respectively, and potential roles of SRY/Sry in the brain will be discussed. The evidence from various taxa disputing the requirement of Sry for brain sexual differentiation in mammals (therians and prototherians) and certain fish and reptilian species will be examined. A comparative approach to address this question may elucidate other genes, pathways, and epigenetic modifications stimulating brain sexual differentiation in vertebrate species, including humans.
Introduction
In many sexually reproducing species, the undifferentiated gonad and brain are programmed to be either male or female. As discussed below, in mammalian species that rely on genetic sex determination (GSD), sexual differentiation of the gonad is dependent upon the Y chromosome and the SRY/Sry gene. However, brain sexual differentiation is generally orchestrated by a surge in gonadal steroid hormones during embryo development followed by a second surge later in adulthood. This paradigm is considered “organization-activational programming” and occurs in a sex-specific manner (Phoenix et al., 1959; Arnold and Breedlove, 1985; Morris et al., 2004). The organizational-activational concept was formulated based initially on findings in guinea pigs (Cavia porcellus). Female guinea pigs prenatally exposed to testosterone propionate during a specific developmental window had permanent rewiring of the neural circuitry and as adults, demonstrated male-pattern behaviors (Phoenix et al., 1959). Similar results were then obtained with other mammals (Phoenix et al., 1959; Arnold and Breedlove, 1985; Morris et al., 2004). The early brain programming is termed the organizational period and is characterized by spikes in androgens and/or estrogens such that the brain becomes permanently masculinized (Watson and Adkins-Regan, 1989a,b; Bowers et al., 2010; Konkle and McCarthy, 2011). In the developing mouse brain, sex chromosomes appear to interact with steroid hormones to increase the expression of aromatase expression, which converts androgens into estrogens, and this process appears to be through activation of neural estrogen receptor β (ESR2) (Cisternas et al., 2015, 2017). Masculinization of several brain regions, including the hippocampus, requires aromatization of testosterone to estrogen (Watson and Adkins-Regan, 1989a,b; Bowers et al., 2010; Konkle and McCarthy, 2011). Programming of the male brain though requires both defeminization and masculinization, which generally requires binding of estradiol to its cognate receptors, estrogen receptor-α (ESR1) and ESR2 (Naftolin et al., 1975; Lephart, 1996; McCarthy, 2010). MRI examination of the brain from women with androgen insensitivity syndrome (CAIS), who exhibit a male (46,XY) karyotype but lack functional androgen receptors, reveals that testosterone alone may, however, regulate development of certain brain regions (Savic et al., 2017). These include the somatosensory and visual cortices and their axonal connections to the frontal cortex, along with the connections from the amygdala.
For characteristic sex-dependent behaviors, especially those seen in males, a second spike in steroid hormones at adulthood (“activational period”) is required for full elaboration of sex-specific neurobehavioral pathways (Tsai et al., 2009; Menger et al., 2010; Nugent and McCarthy, 2011; Jasarevic et al., 2012; Matsuda et al., 2012; Chung and Auger, 2013). Steroid hormones at both periods may trigger epigenetic changes in various brain regions that underpin sex differences in brain responses (Tsai et al., 2009; Menger et al., 2010; Nugent and McCarthy, 2011; Jasarevic et al., 2012; Matsuda et al., 2012; Chung and Auger, 2013). However, the question remains as to potential other contributions of sex chromosomes and those genes residing on the X and Y in initiating brain sexual differentiation. One gene residing on the Y chromosome that that has received particular attention is SRY (in men)/Sry (in other therian mammals). For consistency, the gene annotation form Sry will be used throughout the manuscript, unless studies within humans or the human form of SRY are being described.
Sry is located on the Y chromosome in therian mammals, which includes placental mammals and marsupials (Delbridge and Graves, 1999; Graves, 2002, 2015; Marshall Graves, 2002; Cortez et al., 2014). SRY functions as a transcriptional activator modulating testis development, and thus it was named the testis determining factor (TDF) in therian mammals (Berta et al., 1990; Sinclair et al., 1990; Foster et al., 1992). The Sry gene presumably evolved from a degraded version of the SOX3 gene on the X chromosome that may itself have originated to guide brain development in both sexes (Graves, 2001). Sry is postulated to have evolved before the actual formation of the Y chromosome (Marshall Graves, 2002).
Sry encodes a transcriptional factor that includes an HMG box as a DNA domain (Hacker et al., 1995) that activates Sry-related box 9 (Sox9) (Sekido and Lovell-Badge, 2008), cerebellin 4 precursor gene (Cbln4) (Bradford et al., 2009), Pod1 (Bhandari et al., 2011), and neurotrophin 3 (Nt3) (Clement et al., 2011). Other potential actions of Sry include repressing anti-testis forming genes (McElreavey et al., 1993), bending the DNA double helix (Pontiggia et al., 1994), acting as a cofactor within gene silencing complexes (Oh et al., 2005), a splicing factors for pre-mRNA (Ohe et al., 2002); also, the Sry transcript might function as a regulator for non-coding RNA (ncRNA) (Hansen et al., 2013). The last three mechanisms imply that Sry can stimulate epigenetic responses in the gonad and brain. Other epigenetic mechanisms regulated by Sry include DNA methylation, histone protein modifications, and in relation to regulation of ncRNA, Sry might act as a potential sponge for microRNAs (miRNAs) (Hansen et al., 2013; Sekido, 2014). This last epigenetic function is determined by which type of Sry is transcribed, as detailed below.
The Sry gene is intronless, the notable exception being in a single marsupial species, and besides the HMG box region, there is generally poor conservation of the Sry genomic sequence across taxa (Sekido, 2010). In mice, two types of Sry transcripts can originate depending on whether transcription is signaled through a proximal or distal promoter site (Jeske et al., 1995; Dolci et al., 1997). The first generates a linear Sry transcript, which will be translated into a functional protein. The second promoter site gives rise to a transcript that contains an inverted repeat on the 5′ and 3′ end, which results in formation of a stem-loop or circular transcript (Capel et al., 1993). Both transcripts are expressed in the testis and as detailed below, at various points in brain development (Capel et al., 1993; Mayer et al., 2000). Similar to other circular RNAs, the circular form of Sry (circSry) might act as a “miRNA sponge” that binds to ncRNAs and in the process mitigates their gene silencing ability (Hansen et al., 2013). In relation to the potential role of Sry in regulating brain sexual differentiation, circSry consists of 16 potential binding sites for miR-148 (Hansen et al., 2013), which is abundant in neuronal and other cell types (Obernosterer et al., 2006; Morton et al., 2008; Eskildsen et al., 2011). In hippocampal neurons, miR-138 inhibits the expression of acyl protein thioesterase 1, an enzyme that mediates depalmitylating α13 subunits of G proteins, which is required for enlargement of dendritic spines (Siegel et al., 2009).
Herein, I will review the evidence to date that Sry is involved in brain sexual differentiation in therian species that express this gene. In exploring the potential roles of Sry in the brain, we will consider the brain regions where it is expressed, the genes and pathways influenced by Sry, and neurobehavioral phenotypes of mice with alterations in Sry expression. While Sry might be essential for brain sexual differentiation in therian animals, there are mammalian, prototherian, and poikilothermic species that lack Sry and/or the Y chromosome. How sexual differentiation, including within the brain, occurs in such species lacking Sry will be discussed, along with potential alternative candidates for Sry. The primary goal in reviewing the results to date that span several taxa will be to determine whether Sry is required for brain sexual differentiation in most therian species and potential other genes that might compensate in Sry-deficient species.
Localization of SRY and Potential Roles of SRY in Brain Sexual Differentiation
Several studies have examined expression patterns and potential function of SRY/Sry in the brain in rodent models and humans. Ontogenetic examination of Sry expression in the brain of male mice reveals that the circular, presumably non-translated form, is expressed through embryonic days 11–19 (Mayer et al., 2000). After birth, linear, and translatable forms of Sry are identified in the diencephalon, midbrain, and cortex. The change from the circular to linear transcripts is presumably driven by a switch in promoter activation, as detailed above. A study with adult mice suggests that Sry is expressed in the hypothalamus and midbrain of males but not females (Lahr et al., 1995). Using autopsied samples, SRY protein has been identified in the hypothalamus and frontal and temporal cortex of adult men (Clepet et al., 1993; Mayer et al., 1998).
Another mouse study suggests that Sry is expressed in tyrosine hydroxylase (TH)-expressing neurons within the substantia nigra (SN) in adult males (Dewing et al., 2006). Inhibition of Sry with antisense oligodeoxynucleotides results in suppression of TH but without reducing neuronal numbers. The reduction in TH in turn leads to motor deficits in treated males. Similarly, SRY protein has been identified in TH-positive neurons within the SN pars compacta (SNc) in human males but not females (Czech et al., 2012). SRY also co-localizes with TH in the ventral tegmental area. When human retinoic-acid treated precursor neuronal NT2 cells differentiate into dopaminergic cells, TH, nuclear receptor related 1 protein (NURR1), dopamine receptor D2 (D2R), and SRY expression are upregulated. Suppression of SRY in human neuroblastoma cell line, M17, results in a depressed expression of TH, DOPA decarboxylase (DDC), dopamine β-hydroxylase (DBH), and monoamine oxidase A (MAO A) expression. These enzymes are primarily involved in dopamine synthesis and metabolism. In contrast, overexpression of SRY elevates the expression of these genes, which is associated with enhanced extracellular dopamine levels. When tested with a luciferase assay, SRY activates an upstream regulatory region within the human TH promoter/nigral enhancer. Dopamine neurons are greater in the SNc in males than females (Murray et al., 2003; Dewing et al., 2006; McArthur et al., 2007). Taken together, the collective results suggest that Sry/SRY in male mice and humans, respectively, acts as a positive regulator of catecholamine and dopaminergic synthesis and metabolism in the midbrain region. The findings might also help explain why men are more vulnerable to dopamine disorders, namely Parkinson's disease and schizophrenia.
To test further the role of SRY in regulating dopaminergic neurons, human male dopamine M17 cells were treated with a dopaminergic toxin, 6-hydroxydopamine (6-OHDA), which resulted in a significant upregulation of SRY (Czech et al., 2014). Corresponding to this increase, GADD45g, a known SRY regulator, was also elevated. Inhibition of SRY in treated M17 cells increased the levels of reactive oxygen species (ROS), upregulated the pro-apoptotic marker PUMA, and resulted in cell injury. In contrast, overexpression of SRY in 6-OHDA-treated female SH-SY5Y cells was protective and abolished the above cytotoxic effects. However, testing of even greater levels of 6-OHDA or chronic exposure to this toxin resulted in tapered upregulation of SRY, which may be due to gradual apoptosis of dopamine cells.
MAO A is encoded by the X chromosome, and it functions to catalyze the oxidative deamination of monoamine transmitters, including serotonin. SRY can activate the MAO A-promoter and catalytic activities when tested in a human male neuroblastoma BE(2)C cell line (Wu et al., 2009). Co-immunoprecipitation and ChIP assays reveal that SRY and specificity protein 1 (SP1) form a transcriptional complex and synergistically activate MAO A transcription.
Sry has been identified in catecholaminergic brain regions of male but not female rats (Milsted et al., 2004). TH is the rate-limiting step of catecholamines. PC12 cells co-transfected with Sry and a portion of the TH promoter connected to luciferase exhibit an increase in luciferase reporter activity. Transfection with a shorter construct lacking putative Sry sites was also induced by Sry. However, transfection with a construct lacking the AP1 site in the TH promoter was not responsive to Sry.
Transgenic Mouse Models for Sry and the Y Chromosome
To understand and uncouple the role of Sry in the brain from other genes on the Y chromosome, various transgenic mouse models have been created. When human SRY (hSRY) is inserted into single-cell mouse embryos (hSRYON), this early activation of SRY results in impaired brain neurogenesis and several other disorders, including inability of the resulting pups to suckle, development of fatty liver disease, arrested alveologenesis in the lung, sporadic myocardial fibrosis, and thymic hypoplasia (Kido et al., 2017). While the hSRYON pups are similar in size to wild-type (WT) mice at birth, they undergo retarded postnatal growth and development and die with multi-organ failure before 2 weeks of age. The findings suggest that while SRY might regulate various pathways in the brain and other organs, premature activation of this gene can be deleterious to normal development and physiological function of these organs.
Another approach that has been used to disentangle the role of Sry from other Y chromosome genes, has been to delete Sry from the Y chromosome and then insert it as a transgene on an autosomal chromosome in both XX and XY chromosome bearing mice (Wagner et al., 2004). XX and XY mice that harbor the Sry transgene showed enhanced expression of progesterone receptor immunoreactivity (PRir) in the anteroventral perventricular nucleus (AVPV), the medial preoptic area (MPOA), and the ventromedial nucleus compared to mice lacking the Sry transgene. The investigators ascribed the results to the fact that Sry transgene upregulated gonadal hormones, and that the results were independent of genetic sex for the brain cells. However, the differences in PRir could also be due to direct effects of Sry transgene expression in these brain regions, which was not tested in this study. Co-transfection studies, as detailed above, could be performed to determine if Sry directly upregulates the expression of PR, as has been identified with TH.
The breeding scheme with this transgenic mouse model above gives rise to the “four-core genotypes—FCG” mouse model (Figure 1; Arnold and Chen, 2009). Offspring from these mated pairs can be one of four different genotypes: XX (karotypically and gonadally female), XXSry+ (karoytypically female but gonadally male due to presence of autosomal Sry transgene), XYSry− (karyoptically male but gonadally female due to deletion of endogenous Sry gene), and XYSry+ (karyotypically and gonadally male). Behavioral testing with the FCG model that uncouples the potential effects of Sry from other Y chromosome-associated genes and mice gonadectomized at adulthood demonstrates that mice with two XX chromosomes consumed more food during daylight hours and had increased adiposity than XY mice (Chen et al., 2012). However, no Sry-related or sex chromosome dependent differences were detected in locomotor or anxiety-related measures (McPhie-Lalmansingh et al., 2008). In this study, however, differences in social behaviors between the FCG were noted. Female XYSry− mice spent more time following and sniffing an intruder compared to XX female mice. XX females were more likely to engage in asocial and digging behaviors compared to XXSry+ male mice.
Figure 1. Breeding of XXSry+ males with XX females results in offspring of four different genotype/phenotype combinations that in two cases demonstrate mismatches in terms of genetic vs. phenotypic sex of the gonad and likely the brain, as well. The resulting phenotypes could be due to a direct effect of the Sry gene or secondary alteration in gonadal sex steroid hormones.
Testing of intact mice with this same model revealed that XX and XY gonadally female (Sry−) mice were more active, consumed less food per body weight, and when tested in the elevated plus maze (EPM) and zero maze, these groups demonstrated enhanced anxiogenic behaviors compared to XX and XY gonadally male mice (Sry+) (Kopsida et al., 2013). Another study reported that gonadal males possess greater androgen receptor (Ar) expression in the suprachiasmatic nucleus (SCN) and suppressed photic phase-induced behavioral responses compared with gonadal female FCG mice (Kuljis et al., 2013). Adult gonadectomy to remove activational effects of steroid hormones results in XX individuals showing longer activity duration than those with XY chromosomes regardless of gonadal phenotype. Activational effects of gonadal hormones exerted a greater effect in regulating activity levels in gonadal male mice as opposed to gonadal female FCG mice.
The FCG mouse model has also revealed that latency to show aggression and pup retrieval are influenced by gonadal sex and sex chromosome status (Gatewood et al., 2006). Females but not males with XX sex chromosome differ from XY individuals. Vasopressin immunoreactivity in the lateral septum was elevated in gonadal males compared to gonadal females but also differed according to sex chromosome content. The findings suggest genes residing on the sex chromosomes, other than Sry, might modulate sex differences in neurobehavioral patterns. When subjected to chronic “stress” conditions, XY mice exhibit reduced expression in the frontal cortex for GABA/serotonin/dopamine-related genes relative to XX mice (Seney et al., 2013). Estradiol treatment of the FCG mice increased growth hormone (Gh) expression in the cerebellum and hippocampus independent of sex chromosome complement or gonadal sex (Quinnies et al., 2015). Conversely, in the hypothalamus, females had greater Gh expression than males, and XY females showed elevated Gh compared to XY males and XX females. In the arcuate nucleus, Gh was greater in XX compared to XY mice. Taken together, the findings suggest that sex chromosome complement regulates certain genes within discrete brain regions, but steroid hormones might also play a contributory role.
Transcription activator-like effector nuclease (TALEN) technology is another method that has been used to generate transgenic knockout mice (Sung et al., 2013). Sry knockout (KO) mice have been created with TALEN via oocyte injection (Kato et al., 2013). Predictably, Sry KO mice possess female external and internal genitalia, low circulating testosterone concentrations, reduced fecundity to complete infertility with decreased number of oocytes but increased luteinized un-ruptured follicles, and feminized neurobehavioral programming. In the AVPV within the MPOA, WT males have greater number of calbindin immuoreactive cells than WT females. Sry KO mice exhibit similar number of calbindin positive cells as WT females but lower expression levels relative to that of WT males, particularly in the medial dorsal portion. Sry KO mice exhibit female typical reproductive (lordosis) behavior when paired with WT males. Generation of Sry-green fluorescent protein (GFP) mice confirmed that Sry is expressed in the gonad and brain in day 12 embryos (Wang et al., 2013).
Rodent Species Naturally Lacking a Y Chromosome and SRY
In therian mammals, sex determination is dependent upon the sex chromosome complement and in particular Sry on the Y chromosome. However, a handful of notable rodent exceptions have been identified. Two genera that include members lacking the Y chromosome and Sry are Ellobius and Tokudaia (Matthey, 1958; Fredga, 1988; Just et al., 1995; Soullier et al., 1998; Vogel et al., 1998; Sutou et al., 2001). Separate evolutionary events presumably resulted in select species within Ellobius and Tokudaia shedding the Y chromosome and Sry as these genera belong to distinct subfamilies (Arvecolinae and Murinae, respectively). Both of these genera possess an XO system in males and females, and the mechanisms underpinning gonadal and brain sex determination have remained elusive. Detailed below are genomic and gonadal gene expression studies with these two genera. To date, no research group has performed a global assessment of the brain transcriptomic profile in Y chromosome-deficient species within these two genera to determine which genes possibly compensate for the absence of Sry expression in the brain of these males.
Ellobius
The Transcaucasian mole vole (Ellobius lutescens) and Zaisan mole vole (Ellobius tancrei) lack the Y chromosome and Sry, whereas the closely related species, Southern mole vole (Ellobius fuscocapillus) and the three other mole voles within this genus have retained the XY system. This loss of the Y chromosome in the two Ellobius species likely occurred through two independent evolutionary events that occurred ~0.6 and 2–5 million years ago (MYA) for E. lutescens and Ellobius talpinus, respectively (Mulugeta et al., 2016). Within the evolutionary conserved region (ECV) of the testis-specific enhancer of SOX9 (TESCO), E. lutescens, Ellobius trancrei, and E. fuscocapillus display a 14 base pair (bp) deletion removing a highly conserved SOX/TCF site (Bagheri-Fam et al., 2012). This deletion may have resulted in upregulation of Sox9 in XX gonads and eventual destabilization of the XY/XX determining system in the Ellobius genus. E. lutescens and E. trancrei could have independently evolved Sry-independent mechanisms to stabilize sex determination, whereas, E. fuscocapillus continued to use Sry-dependent mechanisms. Zfy is also missing in E. lutescens and E. trancrei (Vogel et al., 1998). Other genes that have been eliminated as potential replacements for Sry in guiding sexual differentiation in E. lutescens and E. trancrei include nuclear receptor subfamily 5 group A member 1 (Nr5a1), dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 (Dax1), steroidogenic factor 1 (Sf1), forkhead box protein L2 (Foxl2/Pistr1), Sox9, Sox3, and doublesex and mab-3 related transcription factor 1 (Dmrt1) (Baumstark et al., 2001, 2005; Just et al., 2002, 2007).
Tokudaia
Ryuku (Amami and Tokunoshima) spiny rats (Tokudaia) are confined to three islands in the Nansei Shoto archipelago in Japan. The two species though vary in chromosome number with Tokudaia osmiensis (residing on the Amami-Oshima Island) having 2n = 25 chromosomes and Tokudaia tokunoshimensis (from the Tokunoshima Island) possessing 2n = 45 chromosomes. The Y chromosome and Sry are no longer extant in Amami spiny rat (Tokudaia osimensis) and Tokunoshima spiny rat (Tokudaia tokunoshimensis), and the loss in the two species likely occurred through separate evolutionary events (Soullier et al., 1998; Sutou et al., 2001). The sex-specific region that governs sex determination in XO Tokudaia species is likely very minute (Kobayashi et al., 2007). These species possess additional copies of Cbx2, which acts upstream of Sry (Kuroiwa et al., 2011). Another possible gene that might replace SRY-induced SOX9 expression in the testis of Amami spiny rats is Ets-related protein 71 (Er71). Er71 is also upregulated by SRY in species containing this gene (De Haro and Janknecht, 2005). In conjunction with SF1, SRY binds to TESCO (Sekido and Lovell-Badge, 2008). TESCO activity is lost in this spiny rat, but the activity of the Sox9 promoter is enhanced by ER71 in this species (Otake and Kuroiwa, 2016). Examination of other Y-linked genes in Amami spiny rats reveal that the original Rbmy1A1 has also been lost, but Eif2s3y and Kdm5d have been conserved due to translocation from the Y chromosome to the X chromosome and/or autosomes (Kuroiwa et al., 2010). The other Rbmy1A1 gene has been reduced to a pseudogene on an autosomal chromosome. Both Eif2s3y and Kdm5d are expressed in the gonads and brains of both sexes, thus arguing against their possible involvement in sexual differentiation. Testis-specific protein, Y encoded (Tspy) and Zfy are two other Y-associated genes that translocated to the distal part of the long arm of the X chromosome in the Amami spiny rat (Arakawa et al., 2002), suggesting that these genes are also not the initiators of sexual differentiation in this species.
Prototherian Mammals (Monotremes)
Mammals are comprised for two classification groups: therians, which includes placental mammals and marsupials) and prototherians (monotremes or egg-laying mammals). This divergence traces back to 190 million years ago (MYA). Animals within the monotreme order include the duck-billed platypus (Ornithorhynchus anatinus) and echidnas (Tachyglossus aculeatus, Zaglossus spp., and Meglibgwilia spp.). While both monotremes possess five sets of XY chromosomes, they lack Sry (Rens et al., 2007; Wallis et al., 2008). Platypuses contain five X and five Y chromosomes that appear to have originated due to translocation of an avian-like sex chromosome (Z) with orthologs of four other chicken chromosomes (Rens et al., 2007; Wallis et al., 2008). The echidna X and Y chromosomes are similar except for the fifth pair. The therian X chromosome resembles platypus autosome 6 and echidna autosome 16 chromosomes (Ferguson-Smith and Rens, 2010). Sry is absent in monotremes, and the related gene, Sox3 (which is present on the X chromosome in therians) resides on chromosome 6 of platypuses and chromosome 16 in echidnas (Wallis et al., 2007b). Sox9 mediates male sexual differentiation in therians. This gene and related Sox10 map to platypus autosomal chromosomes 15 and 10, respectively, which argues against both of them being the primary sex determining genes in prototherians (Wallis et al., 2007a). Other genes that have been eliminated as potential sex-determining genes in monotremes are Wilm's tumor 1 (Wt1), Sf1, LIM Homeobox 1 (Lhx1), LIM Homeobox 2 (Lhx2), Fibroblast Growth Factor 9 (Fgf9), Wnt Family Member 4 (Wnt4), R-Spondin 1 (Rspo1), and GATA Binding Protein 4 (Gata4) (Grafodatskaya et al., 2007). The most likely candidate regulating gonad and potentially brain sexual differentiation in these species is anti-Mullerian hormone (Amh) that resides on the shortest and oldest platypus Y5 chromosome (Cortez et al., 2014). Another potential candidate is complex protein gametologs on platypus Y5 (Crspy), which is exclusively expressed in the testis and kidneys of males (Tsend-Ayush et al., 2012). There is also a variant on the X1 chromosome (Crspx). While the gene(s) stimulating sexual differentiation in monotremes have remained elusive, these species are further evidence that gonad and brain sexual differentiation can occur in the absence of Sry.
Brain Sexual Differentiation in Example Fish and Reptilian Species Lacking Sex Chromosomes and/or Sry
This section provides a brief overview of some example fish and reptilian species that lack sex chromosomes and the equivalent to therian Sry. Many of these species rely on temperature sex determination (TSD) or other mechanisms to guide sexual differentiation of the gonad and possibly the brain. Several other review articles provide an in-depth discussion of the hormonal control of sexual differentiation in fish (Devlin and Nagahama, 2002; Le Page et al., 2010; Todd et al., 2016; Rosenfeld et al., 2017). The focus herein will be to provide a brief description of these processes in fish and reptilian species and those genes that might guide gonad and brain sexual differentiation in the absence of Sry.
Fish Species
Brain sexual differentiation in rodents and other vertebrate species is considered permanent (Arnold and Breedlove, 1985). However, sexualization in fish is strikingly plastic and likely an adaptive mechanism endowed in several fish species to deal with rapid environmental fluctuations occurring throughout an individual's lifespan (Todd et al., 2016). Common reproductive strategies in fish include sequential hermaphrodism where an individual begins life as one sex and then reemerges later on as the opposite sex. Within this category, there are those who exhibit protoandrous (male to female), protogynous (female to male), or serial (bidirectional) sex change. For instance, false clown anemonefish (Amphiprion ocellaris) demonstrate protandrous sex changes with a monogamous mating strategy such that a single fish can begin life as a male but then undergo sexual differentiation to become a female, which is likely modulated by transcriptomic changes at the brain and gonad level (Iwata et al., 2012). Sexual plasticity with ability to change gonadal and phenotypic sex is prominent in fish of the teleost lineage. Most teleost fish demonstrate environmental sex determination that is influenced by such factors as water temperature, oxygen levels, pH, fish density, social environment, and age (Nagahama, 2005; Le Page et al., 2010). The primary exception is medaka (Oryzias latipes), who demonstrate GSD. In medaka, the DM domain gene on the Y chromomosome/doublesex and mab-3 related transcription factor 1b on the Y chromosome (dmy/dmrt1bY) and dmrt1 appear to take the place of therian Sry in regulating sexual differentiation (Nanda et al., 2002; Matsuda, 2003; Kikuchi and Hamaguchi, 2013). In this species, dmy might stimulate male sexual differentiation by increasing gonadal soma derived growth factor (gsdf) and sox9a2 but suppressing the expression of rspo1 (Chakraborty et al., 2016). The gene wt1a appears to act as an upstream regulator of dmy.
Early exposure to exogenous steroid hormones during the critical period of sexual differentiation can alter the sex within select fish (Devlin and Nagahama, 2002). While brain aromatase activity decreases after birth in the mammalian brain, adult teleosts continue to demonstrate high expression of this enzyme due to the upregulation of two aromatase genes (aromatase A—cyp19a1a and aromatase B—cyp19a1b; Le Page et al., 2010). The latter form is only expressed in radial glial cells (RGC) in adult fish. This persistent expression pattern of neural aromatase in and associated high neurogenic activity suggest that throughout life the brain can be reprogrammed. In gonochoristic (an organism being one of at least two distinct sexes) and hermaphroditic fish species, suppression of cy19a1a in the gonad may be required for testicular differentiation (Guiguen et al., 2010). It is uncertain though whether there are SRY-like proteins that regulate the initial sexual differentiation and reprogramming of sex in those that demonstrate sexual plasticity. While fish do not express SRY, several studies have demonstrated that SRY-like proteins are expressed in the gonad and brain.
In Nile tilapia (Orechromis niloticus), 27 sox genes have been discovered (Wei et al., 2016). It is possible that this gene family has expanded greatly in tilapia and other teleost fishes due to genome duplication. Transcriptome analyses revealed that different sox genes show stage-specific and sex-dependent expression patterns during gonadal development. Six of the group B sox genes are exclusively expressed in the adult brain. In medaka, cDNA library screening revealed two forms of sox9 (sox9 and sox9lf) (Yokoi et al., 2002). The latter included an additional exon in the 5′ upstream region. The sox9 gene was abundant in the adult ovary, as determined by Northern blot and in situ hybridization analyses. In contrast, this gene was only detectable in the testis by qPCR.
Screening of a microarray library and the lamprey (a primitive fishlike and jawless vertebrate) genome revealed genes within the sox B, D, E, and F subfamilies (Uy et al., 2012). Within these sub-families, several sox genes were expressed during embryogenesis in the neural crest. The rice field eel (Monopterus albus) possess at least five sox genes (Zhou et al., 2002). The sox1, sox4, and sox14 are intronless in the HMG box region, whereas, sox9 and sox17 contain an intron within this conserved region. The rice field eel sox17 transcript was identified in the gonads of male, female, and intersex individuals (those with an ovotestis), along with the brain and spleen (Wang et al., 2003). Three forms of sox17 have been identified in European sea bass (Dicentrarchus labrax) (Navarro-Martin et al., 2009). One encodes for a typical protein, the second results in a truncated protein, and third shows intron retention. The truncated form of sox17 was abundantly expressed in the brain and skin, whereas, the normal size transcript increased in the gonad at the onset of sexual differentiation (~150 days of age). From 250 days of age, this sox17 form was significantly greater in females, and the mRNA expression pattern correlated with gonadal aromatase activity. The promoter of cyp19a is hypermethylated in juvenile male sea bass but exposure to an elevated temperature increases cyp19a promoter methylation in females, which is associated with decreased expression of this gene (Navarro-Martin et al., 2011). These epigenetic differences are detected in the gonad but not the brain of this fish species. The sox19 gene is also abundant in the sea bass brain and differentiating ovary but not testis (Navarro-Martin et al., 2012).
The full length form of sox11a gene (Lc-sox11a) in large yellow croaker (Larimichthys crocea) also shows temporal—and sex-dependent expression differences in the gonad and brain (Jiang et al., 2015). In females, Lc-sox11a expression was greatest in the ovary followed by the brain, eye, and gill. Conversely, the expression pattern in males was highest in the brain followed by the testis and gill. This transcript was more abundant in the male compared to female brain. With age, Lc-sox11a expression increased in the ovary but decreased in the testes. The sox11a gene might also be important in neurogenesis in orange-spotted grouper (Epinephelus coioides), but it remains to be determine whether there are sex-dependent differences in this species (Zhang et al., 2010).
By screening a testis cDNA library from rainbow trout (Oncorhynchus mykiss) followed by Northern blot analyses, soxlz was identified, and the protein encoded by this gene bound strongly to an oligonucleotide sequence also recognized by mouse Sry and Sox5 (Takamatsu et al., 1995). Another SRY-related gene, soxp1, was identified in the pituitary gland of rainbow trout and showed higher levels of expression in immature fish (Ito et al., 1995).
While sox transcripts might stimulate sexual differentiation that results in an increase in testosterone and/or estrogen within the brain or gonad, their expression pattern might also in turn be influenced by endogenous and exogenous steroid hormones. For instance, exposure of adult male zebrafish (Danio rerio) for 2 weeks to 25 ng/L ethinyl estradiol (EE2, estrogen present in birth control pills) results in decreased expression of sox9a and nuclear receptor subfamily 5 group number 1b (nr5a2l) transcripts in the testis that presumably contributes to spawning failure in exposed males (Reyhanian Caspillo et al., 2014). Other expression changes observed in the testis of treated males included an increase in vitellogenin (vtg) but a decrease in estrogen receptor-β (esr2).
Other sex determining genes characterized in therian mammals have been discovered in different fish species, such as, anti-Müllerian hormone Y (amhy) in Patagonian perjerrey (Odontesthes hatcheri), gsdf in Oryzias luzonensis (related to medaka), anti-Müllerian hormone receptor type II (amhr2) in tiger pufferfish-fugu (Takifugu rubripes), and sexually dimorphic on the Y chromosome (sdY) in rainbow trout (Hattori et al., 2012; Kamiya et al., 2012; Myosho et al., 2012; Yano et al., 2012). Other potential sex determining genes in the brain and gonad of cichlid fish (so-called tribes: Eretmodini, Ectodini, Haplochromini, and Lamprologini) include members of the transforming growth factor-b (tgfb) superfamily, Wnt-pathway, sox, dm-domain, and high mobility group-box families (Bohne et al., 2014). A follow-up study by this group that tested East African cichlid fish (Astatotilapia burtoni) found that the earliest gonadal sexual differentiation markers appeared at 11–12 days post-fry (dpf) and included wnt4b and wt1a (Heule et al., 2014). Late testis genes in this species were cyp19a1a, gsdf, dmrt1, and gata4, and brain markers were catenin beta 1a (ctnnb1a), catenin beta 1b (ctnnb1b), dax1a, foxl2, foxl3, nanos1a, nanos1b, rspo1, steroidogenic factor (sf1), sox9a, and sox9b.
Examination of gene expression patterns in the female to male transition in the honeycomb grouper (Epinephelus merra), which demonstrates protogynous sex change, revealed that foxl2 was elevated in the brain, pituitary, ovary, and gill but was downregulated during late transitional stage to male (Alam et al., 2008). In contrast, dmrt1 expression increased with the initiation of spermatogenesis and complete formation of the testis. This gene might also be important in male sex determination in Chinese tongue sole (Cynoglossus semilaevis), a marine fish with ZW sex determination. TALEN injection of dmrt1 into ZZ mutant fish stimulates ovary-like testis development with disrupted spermatogenesis, an increase female-associated transcripts (foxl2 and cyp19a1a) in the gonad, but reduced expression of male-related genes (sox9a and amh) (Cui et al., 2017). The dmrt1 deficient ZZ fish outgrew ZZ male controls.
Reptilian Species
Various patterns of reproductive and sex-determining mechanisms are present within reptiles. Some reptile species exhibit gonochorism, whereas others are unisexual (parthenogenesis). In those with two sexes, sexual differentiation can occur via GSD with male (XX/XY) and female (ZZ/ZW) heterogamety or due to TSD (Ezaz et al., 2009; Wapstra and Warner, 2010). Two hundred out of 1,000 karyotyed reptilian species possess sex chromosomes. Those lacking sex chromosomes are predominantly featured in this section.
In the leopard gecko (Eublepharis macularius), incubation temperature of the egg during a critical period of embryonic development modulates gonadal sex and sex-differences in body growth, adult morphology, aggressiveness, reproductive physiology, and neurobehavioral organization and programming (Crews et al., 1998). By altering steroid hormone synthesis, especially for testosterone and estrogen, and increasing expression of either male or female temperature-sensitive transcripts, incubation temperature alone may program early brain development and later behavioral responses. Painted turtles (Chrysemys picta) also lack sex chromosomes and demonstrate TSD. When we exposed painted turtles eggs that were incubated at the male producing temperature (MPT) to the endocrine disrupting chemical, bisphenol A (BPA), it resulted in partial gonadal sex reversal to female, feminized their behavioral responses, and altered brain gene expression patterns, especially for transcripts associated with mitochondrial and ribosomal function relative to control males (Jandegian et al., 2015; Manshack et al., 2016, 2017).
There is interest in identifying transcripts in the gonad and/or brain that are altered due to TSD in select reptilian species. Several genes were identified to be elevated in the gonad of American alligators (Alligator mississippiensis) after incubation at the MPT (33.5°C), including wnt11, histone demethylase kdm6b, and transcription factor cebpa (Yatsu et al., 2016). The alligator ortholog of the transient receptor potential cation channel subfamily V member 4 (trpv4) is another gene associated with the MPT and male sexual differentiation (Yatsu et al., 2015). The genes sf1 and wt1 are activated in the gonad of painted turtles during the thermosensitive period and dosage-sensitive sex reversal, and dax1 may also contribute to male development (Valenzuela et al., 2013).
Australian central bearded dragons (Pogona vitticeps) typically demonstrate GSD. However, this can be overridden when eggs are incubated at higher temperatures, which gives rise to sex-reversed female offspring. In females generated through high temperature but not normal chromosomal females, an intron is retained in mature transcripts from two Jumonji family genes, jarid2 and jmjd (Deveson et al., 2017). JARID2 is part of the master chromatin modifier Polycomb Repressive Complex 2, and mammalian Sry is regulated by an independent but closely related JUMONJI family member. The same introns are also retained in jarid2/jmjd3 transcripts in embryonic gonads from TSD alligators and turtles. The intron retention in these transcripts might induce epigenetic changes that override chromosomal sex-determining mechanisms, leading to sex reversal at elevated incubation temperatures or a switch from GSD to TSD. These genes might also compensate for the absence of Sry and stimulate organizational changes within the brain of these male reptile species.
Investigation of the genome of the lizard Calotes versicolor, which demonstrates GSD, with a human SRY probe showed hybridization in all males but also in select females (Ganesh et al., 1997). Thus, it is not clear whether sry might be involved in sex determination of this reptilian species possessing sex chromosomes. Studies have also been conducted to determine whether sry-related genes might compensate for the absence of sry in reptiles with TSD. The sox8 gene is expressed in the gonad of both male and female red-eared slider turtles (Trachemys scripta), and thus it is unlikely to be responsible for sex determination in turtles with TSD (Takada et al., 2004). While dax1 is unlikely to be involved in sexual differentiation in sea turtles (Lepidochelys olivacea), upregulation of dmrt1 and downregulation of sox9 might exert a role in male and female sexual differentiation, respectively (Torres Maldonado et al., 2002). The fact that sox9 expression occurs after amh suggests that in American alligators (A. mississippiensis), it is not the primary initiator of male sex determination (Western et al., 1999a,b). Instead, gonad and brain sex determination in reptilian species with TSD may be due to gene(s) unrelated to Sry, such as, those listed previously.
Discussion
The focus of this review has been to examine the evidence to date that SRY is required for brain sexual differentiation. The evidence to date in laboratory rodents and humans suggest that Sry/SRY is expressed in certain brain regions in males, especially the cortex, midbrain, hypothalamus, and SN (Clepet et al., 1993; Lahr et al., 1995; Mayer et al., 1998, 2000; Dewing et al., 2006; Czech et al., 2012). The primary enzymes and pathways in the brain altered by SRY include TH, MAO A, and dopaminergic pathways (Milsted et al., 2004; Dewing et al., 2006; Wu et al., 2009; Czech et al., 2012, 2014; Figure 2). The latter effects of SRY might account for men being more susceptible for dopaminergic-based neurological disorders, e.g., Parkinson's disease and schizophrenia.
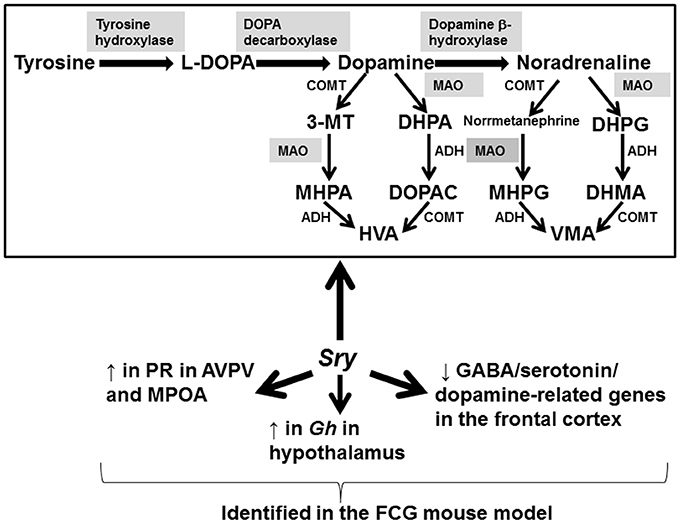
Figure 2. Genes, proteins, and pathways affected by SRY. Studies examining wild-type male mice, the FCG mouse model, and human brain samples/neuronal cells reveal these genes and pathways are primarily affected by the expression of Sry/SRY, respectively. Shaded enzymes are altered by Sry/SRY expression. 3-MT, 3-methoxytyramine; DHPA, 3,4 dihydrophenylacetaldehyde; MHPA, 3-methoxy-4-hydroxyphenylacetaldehyde; HVA, homovanillic acid; DHPA, 3,4-dihydroxyphenylacetic acid; DOPAC, 3,4-dihydroxyphenylacetic acid; MHPG, 3-methoxy-4-hydroxyphenylglycol; DHPG, dihydroxyphenylglycol; DHMA, 3,4-dihydroxymandelic acid; VMA, vanillylmandelic acid; ADH, alcohol dehydrogenase; MAO, monoamine oxidase; COMT, catechol-O-methyltransferase.
While SRY may be required for normal male brain function in many therian males, early induction of this gene can also impair neurogenesis as revealed by the hSRYON mice (Kido et al., 2017). The FCG mouse model has been useful in uncoupling the effects of Sry from other Y chromosome-associated genes (Wagner et al., 2004; Gatewood et al., 2006; Arnold and Chen, 2009; Chen et al., 2012; Kopsida et al., 2013; Kuljis et al., 2013; Seney et al., 2013; Quinnies et al., 2015). The collective results from this model indicate that Sry and other sex chromosome associated genes interact with steroid hormones to affect neurobehavioral programming as evidenced by the fact that the phenotypes observed depend upon whether the animals were intact or gonadectomized. For instance, gonadectomized XX females consumed more food and showed increased adiposity compared to XY mice (Chen et al., 2012). Conversely, intact XX and XYSry− mice demonstrated increased activity levels, consumed less food, and showed more anxiety-like behaviors (Kopsida et al., 2013). Social and parenting behaviors are also influenced by Sry and other Y chromosome associated genes (Gatewood et al., 2006; McPhie-Lalmansingh et al., 2008). The FCG mouse model reveals that Sry mediates neural expression of PR in the AVPV, MPOA, and ventromedial nucleus, GABA/serotonin/dopamine-related genes within the frontal cortex, and Gh expression in the hypothalamus (Figure 2; Wagner et al., 2004; Seney et al., 2013; Quinnies et al., 2015). Future transcriptomic and proteomic analyses of several brain regions within the FCG mouse model will likely elucidate other genes and pathways regulated by Sry. While this mouse model, along with the Sry KO mice generated via TALEN (Kato et al., 2013), have been useful in dissecting potential roles of Sry, future transgenic technology should specifically ablate Sry in brain neuronal cells. Such a deletion will allow further elucidation of the role of Sry in brain sexual differentiation and presumably distinguish secondary effects due to alterations in gonadal steroid hormones.
Surprisingly, there are rodent species within Ellobius and Tokudaia that have lost the Y chromosome and Sry. Such species might be useful in understanding how brain sexual differentiation can occur in the absence of Sry. While it is still not fully clear what presumably autosomally-associated genes might direct gonadal and brain sexual differentiation, several genes have been eliminated as possible candidates in both genus. These include Zfy, Nrd5a1, Dax1, Df1, Foxl2/Pistr1, Sox9, Sox3, Dmrt1, Tspy, Eif2s3y, and Kdm5d (Baumstark et al., 2001, 2005; Arakawa et al., 2002; Just et al., 2002, 2007; Kuroiwa et al., 2010). Two genes that might stimulate male gonad sexual differentiation in Amami spiny rats are Cbx2 and Er71 (Kuroiwa et al., 2011; Otake and Kuroiwa, 2016). However, it is not clear if these genes or possibly other transcripts are involved in brain sexual differentiation in this species and Ellobius spp. that lack Sry. To address this critical gap in our understanding, global transcriptomic profiles of the brain for these genuses and species would be useful. It would be of interest to determine whether similar transcripts are involved in programming the brain for both genuses and species devoid of the Y chromosome. On the other hand, it might be that brain sexual differentiation in the varying genuses and species is directed by unique signature pattern of transcript(s). Such findings would suggest that multiple pathways exist to sculpt brain masculinization even within therian mammals.
The prototherian mammals or monotremes further demonstrate that the presence of Sry is not a requisite for gonad and brain sexual differentiation as these species lack this gene. Both male and female duck-billed platypuses and echidnas contain five sets of X and Y chromosomes that resemble avian sex chromosomes (Rens et al., 2007; Wallis et al., 2008). As above, more genes have been discarded in searching for those that may regulate sexual differentiation in monotremes. The rejected gene pool includes Sox3, Sox9, Sox10, Wt1, Sf1, Lhx1, Lhx2, Fgf9, Wnt4, Rspo1, and Gata4 (Grafodatskaya et al., 2007; Wallis et al., 2007a,b). Amh and Crspy remain as viable contenders in duck-billed platypuses, and possibly echidnas (Tsend-Ayush et al., 2012; Cortez et al., 2014). CRISPR/Cas9 deletion of Amh and/or Crspy might reveal one or both of these genes to be essential in initiating male sexual differentiation in monotremes.
While sexual differentiation in the above species is mediated through genetic (albeit not necessarily involving Sry or Y chromosome associated genes) sex determination, fish, and reptilian species can demonstrate GSD and/or TSD. Unlike most other taxa, sexualization in fish is exceptionally plastic, which may confer an evolutionary advantage in rapidly changing environmental conditions (Todd et al., 2016). The sex determining genes in fish appear to vary across species, and several sox forms have been implicated in governing sexual differentiation in diverse fish species (Ito et al., 1995; Takamatsu et al., 1995; Yokoi et al., 2002; Zhou et al., 2002; Wang et al., 2003; Navarro-Martin et al., 2009, 2012; Zhang et al., 2010; Uy et al., 2012; Reyhanian Caspillo et al., 2014; Jiang et al., 2015). Other gene networks regulating sexual differentiation have been well characterized in medaka. In males of this species, evidence suggests that wt1a upregulates dmy, which increases gsdf and sox9a2 but suppresses the expression of rspo1 (Nanda et al., 2002; Matsuda, 2003; Kikuchi and Hamaguchi, 2013; Chakraborty et al., 2016). The genes dmrt1bY and dmrt1 also seemingly compensate for the absence of therian Sry. Other example genes that might regulate sexual differentiation of the gonad and brain in other fish species include amhy, amhr2, sdY, wnt4b, cyp19a1a, and foxl2 (Alam et al., 2008; Guiguen et al., 2010; Le Page et al., 2010; Hattori et al., 2012; Kamiya et al., 2012; Myosho et al., 2012; Yano et al., 2012; Heule et al., 2014; Cui et al., 2017).
In those reptilian species that possess TSD, such American alligators, painted turtles and sea turtles, wnt11, cebpa, trpv4, sf1, wt1, dax1, dmrt1, and sox9 may contribute to male gonad sexual differentiation (Torres Maldonado et al., 2002; Valenzuela et al., 2013; Yatsu et al., 2015, 2016). However, the same genes might not regulate gonad sexual differentiation across all those with TSD. While dax1 might be involved in painted turtles, it is not a regulatory factor in sea turtles (Torres Maldonado et al., 2002; Valenzuela et al., 2013). The gene sox9 might be important in sea turtles, but it is presumably not essential in alligators (Western et al., 1999a,b; Torres Maldonado et al., 2002). It is also not clear though if the same genes regulate brain sexual differentiation across these reptilian species. Female painted turtles derived from early exposure to BPA have increased expression of mitochondrial and ribosomal associated genes (Manshack et al., 2017).
While Australian central bearded dragons possess sex chromosomes, GSD can be overridden when eggs are incubated at higher temperatures (Deveson et al., 2017). Females derived from TSD but not those due to GSD have intron retention within jarid2 and jmid, and the same introns are present in the embryonic gonads of TSD alligators and turtles. As introns are not transcribed, this inclusion of introns within these genes might induce sex-dependent epigenetic changes.
This latter study introduces the concept that Sry or other genes regulating brain sexual differentiation may operate via epigenetic changes. Such epigenetic modifications could entail those involving DNA methylation, histone protein modifications, or alteration in miRNAs. Rodent, zebrafish, European sea bass, and Drosophila studies have shown sex-dependent differences in all three types of epigenetic changes (Navarro-Martin et al., 2011; Jasarevic et al., 2012; Sato and Yamamoto, 2014; Shen et al., 2015; Bramble et al., 2016; Chatterjee et al., 2016; Forger, 2016; Kigar et al., 2016; Pfau et al., 2016; Morgan and Bale, 2017; Mosley et al., 2017). Sry may induce several epigenetic changes within the brain (Sekido, 2014). One example as discussed previously is that circSry might act as a “miRNA sponge” by binding to ncRNAs and thereby rendering them inactive, as might occur for miR-148 in brain neuronal cells (Obernosterer et al., 2006; Morton et al., 2008; Eskildsen et al., 2011; Hansen et al., 2013). To address the potential involvement of Sry in altering the brain epigenome, the DNA methylome, histone protein modifications, and miRNA profile should be screened in several brain regions of the FCG mouse model and future models lacking Sry in discrete brain regions or neuronal cell populations. Such approaches might also be considered in the Tokudaia and Ellobius species lacking Sry, monotremes, and fish/reptiles with TSD. By examining several taxa that include those with Sry and those without Sry, common epigenetic changes directing sexual differentiation across taxa might be revealed. In other words, while the emphasis to date has been on identifying genes regulating gonad and brain sexual differentiation, changes within DNA methylation, histone proteins, or miRNAs might instead be the common factor(s) driving gonad and brain sexual differentiation across vertebrate species. Future studies should be directed at using transgenic mice and naturally occurring animal models lacking Sry to address this exciting possibility.
The question posed at the outset was whether SRY is required for brain sexual differentiation. While there is evidence SRY/Sry is expressed and might affect certain pathways in the brain of men and select rodent models, respectively, it is also clear that brain sexual differentiation in therian and prototherian mammals, fish, reptiles, and likely in other vertebrate species occurs in the absence of this gene. However, it is uncertain which genes that presumably reside on autosomal chromosomes underpin brain sexual differentiation in these animals. Sry has been postulated to induce epigenetic changes in the brain, but it remains to be determined which ones are critical in modulating masculinization. Even less is known about the potential sex-dependent epigenetic cascade of events within the brain of species lacking Sry that might orchestrate masculinization or feminization of the brain. Such sexually dimorphic epigenetic modifications might be universal across taxa and possibly even precede the actions of Sry. In sum, a comparative taxa approach that examines atypical animal models where males are devoid of Sry might hold the key to unlocking other genes, pathways, and epigenetic changes essential in sculpting brain sexual differentiation.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was funded by NIH/NIEHS 1R01ES025547-01-A1 to CR.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alam, M. A., Kobayashi, Y., Horiguchi, R., Hirai, T., and Nakamura, M. (2008). Molecular cloning and quantitative expression of sexually dimorphic markers Dmrt1 and Foxl2 during female-to-male sex change in Epinephelus merra. Gen. Comp. Endocrinol. 157, 75–85. doi: 10.1016/j.ygcen.2008.03.018
Arakawa, Y., Nishida-Umehara, C., Matsuda, Y., Sutou, S., and Suzuki, H. (2002). X-chromosomal localization of mammalian Y-linked genes in two XO species of the Ryukyu spiny rat. Cytogenet. Genome Res. 99, 303–309. doi: 10.1159/000071608
Arnold, A. P., and Breedlove, S. M. (1985). Organizational and activational effects of sex steroids on brain and behavior: a reanalysis. Horm. Behav. 19, 469–498. doi: 10.1016/0018-506X(85)90042-X
Arnold, A. P., and Chen, X. (2009). What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol. 30, 1–9. doi: 10.1016/j.yfrne.2008.11.001
Bagheri-Fam, S., Sreenivasan, R., Bernard, P., Knower, K. C., Sekido, R., Lovell-Badge, R., et al. (2012). Sox9 gene regulation and the loss of the XY/XX sex-determining mechanism in the mole vole Ellobius lutescens. Chromosome Res. 20, 191–199. doi: 10.1007/s10577-011-9269-5
Baumstark, A., Akhverdyan, M., Schulze, A., Reisert, I., Vogel, W., and Just, W. (2001). Exclusion of SOX9 as the testis determining factor in Ellobius lutescens: evidence for another testis determining gene besides SRY and SOX9. Mol. Genet. Metab. 72, 61–66. doi: 10.1006/mgme.2000.3105
Baumstark, A., Hameister, H., Hakhverdyan, M., Bakloushinskaya, I., and Just, W. (2005). Characterization of Pisrt1/Foxl2 in Ellobius lutescens and exclusion as sex-determining genes. Mamm. Genome 16, 281–289. doi: 10.1007/s00335-004-2019-7
Berta, P., Hawkins, J. R., Sinclair, A. H., Taylor, A., Griffiths, B. L., Goodfellow, P. N., et al. (1990). Genetic evidence equating SRY and the testis-determining factor. Nature 348, 448–450. doi: 10.1038/348448a0
Bhandari, R. K., Sadler-Riggleman, I., Clement, T. M., and Skinner, M. K. (2011). Basic helix-loop-helix transcription factor TCF21 is a downstream target of the male sex determining gene SRY. PLoS ONE 6:e19935. doi: 10.1371/journal.pone.0019935
Bohne, A., Sengstag, T., and Salzburger, W. (2014). Comparative transcriptomics in East African cichlids reveals sex- and species-specific expression and new candidates for sex differentiation in fishes. Genome Biol. Evol. 6, 2567–2585. doi: 10.1093/gbe/evu200
Bowers, J. M., Waddell, J., and McCarthy, M. M. (2010). A developmental sex difference in hippocampal neurogenesis is mediated by endogenous oestradiol. Biol. Sex Dif. 1:8. doi: 10.1186/2042-6410-1-8
Bradford, S. T., Hiramatsu, R., Maddugoda, M. P., Bernard, P., Chaboissier, M. C., Sinclair, A., et al. (2009). The cerebellin 4 precursor gene is a direct target of SRY and SOX9 in mice. Biol. Reprod. 80, 1178–1188. doi: 10.1095/biolreprod.108.071480
Bramble, M. S., Roach, L., Lipson, A., Vashist, N., Eskin, A., Ngun, T., et al. (2016). Sex-specific effects of testosterone on the sexually dimorphic transcriptome and epigenome of embryonic neural stem/progenitor cells. Sci. Rep. 6:36916. doi: 10.1038/srep36916
Capel, B., Swain, A., Nicolis, S., Hacker, A., Walter, M., Koopman, P., et al. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030. doi: 10.1016/0092-8674(93)90279-Y
Chakraborty, T., Zhou, L. Y., Chaudhari, A., Iguchi, T., and Nagahama, Y. (2016). Dmy initiates masculinity by altering Gsdf/Sox9a2/Rspo1 expression in medaka (Oryzias latipes). Sci. Rep. 6:19480. doi: 10.1038/srep19480
Chatterjee, A., Lagisz, M., Rodger, E. J., Zhen, L., Stockwell, P. A., Duncan, E. J., et al. (2016). Sex differences in DNA methylation and expression in zebrafish brain: a test of an extended ‘male sex drive’ hypothesis. Gene 590, 307–316. doi: 10.1016/j.gene.2016.05.042
Chen, X., McClusky, R., Chen, J., Beaven, S. W., Tontonoz, P., Arnold, A. P., et al. (2012). The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet. 8:e1002709. doi: 10.1371/journal.pgen.1002709
Chung, W. C., and Auger, A. P. (2013). Gender differences in neurodevelopment and epigenetics. Pflugers Arch. 465, 573–584. doi: 10.1007/s00424-013-1258-4
Cisternas, C. D., Cabrera Zapata, L. E., Arevalo, M. A., Garcia-Segura, L. M., and Cambiasso, M. J. (2017). Regulation of aromatase expression in the anterior amygdala of the developing mouse brain depends on ERbeta and sex chromosome complement. Sci. Rep. 7:5320. doi: 10.1038/s41598-017-05658-6
Cisternas, C. D., Tome, K., Caeiro, X. E., Dadam, F. M., Garcia-Segura, L. M., and Cambiasso, M. J. (2015). Sex chromosome complement determines sex differences in aromatase expression and regulation in the stria terminalis and anterior amygdala of the developing mouse brain. Mol. Cell. Endocrinol. 414, 99–110. doi: 10.1016/j.mce.2015.07.027
Clement, T. M., Bhandari, R. K., Sadler-Riggleman, I., and Skinner, M. K. (2011). SRY directly regulates the neurotrophin 3 promoter during male sex determination and testis development in rats. Biol. Reprod. 85, 277–284. doi: 10.1095/biolreprod.110.090282
Clepet, C., Schafer, A. J., Sinclair, A. H., Palmer, M. S., Lovell-Badge, R., and Goodfellow, P. N. (1993). The human SRY transcript. Hum. Mol. Genet. 2, 2007–2012. doi: 10.1093/hmg/2.12.2007
Cortez, D., Marin, R., Toledo-Flores, D., Froidevaux, L., Liechti, A., Waters, P. D., et al. (2014). Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493. doi: 10.1038/nature13151
Crews, D., Sakata, J., and Rhen, T. (1998). Developmental effects on intersexual and intrasexual variation in growth and reproduction in a lizard with temperature-dependent sex determination. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 119, 229–241. doi: 10.1016/S0742-8413(98)00012-7
Cui, Z., Liu, Y., Wang, W., Wang, Q., Zhang, N., Lin, F., et al. (2017). Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 7:42213. doi: 10.1038/srep42213
Czech, D. P., Lee, J., Correia, J., Loke, H., Moller, E. K., and Harley, V. R. (2014). Transient neuroprotection by SRY upregulation in dopamine cells following injury in males. Endocrinology 155, 2602–2612. doi: 10.1210/en.2013-2158
Czech, D. P., Lee, J., Sim, H., Parish, C. L., Vilain, E., and Harley, V. R. (2012). The human testis-determining factor SRY localizes in midbrain dopamine neurons and regulates multiple components of catecholamine synthesis and metabolism. J. Neurochem. 122, 260–271. doi: 10.1111/j.1471-4159.2012.07782.x
De Haro, L., and Janknecht, R. (2005). Cloning of the murine ER71 gene (Etsrp71) and initial characterization of its promoter. Genomics 85, 493–502. doi: 10.1016/j.ygeno.2004.12.003
Delbridge, M. L., and Graves, J. A. (1999). Mammalian Y chromosome evolution and the male-specific functions of Y chromosome-borne genes. Rev. Reprod. 4, 101–109. doi: 10.1530/ror.0.0040101
Deveson, I. W., Holleley, C. E., Blackburn, J., Marshall Graves, J. A., Mattick, J. S., Waters, P. D., et al. (2017). Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 3:e1700731. doi: 10.1126/sciadv.1700731
Devlin, R. H., and Nagahama, Y. (2002). Sex determination and sex differentiation in fish: an overview of the genetic, physiological, and environmental influences. Aquaculture 208, 191–364. doi: 10.1016/S0044-8486(02)00057-1
Dewing, P., Chiang, C. W., Sinchak, K., Sim, H., Fernagut, P. O., Kelly, S., et al. (2006). Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 16, 415–420. doi: 10.1016/j.cub.2006.01.017
Dolci, S., Grimaldi, P., Geremia, R., Pesce, M., and Rossi, P. (1997). Identification of a promoter region generating Sry circular transcripts both in germ cells from male adult mice and in male mouse embryonal gonads. Biol. Reprod. 57, 1128–1135. doi: 10.1095/biolreprod57.5.1128
Eskildsen, T., Taipaleenmaki, H., Stenvang, J., Abdallah, B. M., Ditzel, N., Nossent, A. Y., et al. (2011). MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. U.S.A. 108, 6139–6144. doi: 10.1073/pnas.1016758108
Ezaz, T., Sarre, S. D., O'meally, D., Graves, J. A., and Georges, A. (2009). Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet. Genome Res. 127, 249–260. doi: 10.1159/000300507
Ferguson-Smith, M. A., and Rens, W. (2010). The unique sex chromosome system in platypus and echidna. Genetika 46, 1314–1319. doi: 10.1134/S1022795410100054
Forger, N. G. (2016). Epigenetic mechanisms in sexual differentiation of the brain and behaviour. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371:20150114. doi: 10.1098/rstb.2015.0114
Foster, J. W., Brennan, F. E., Hampikian, G. K., Goodfellow, P. N., Sinclair, A. H., Lovell-Badge, R., et al. (1992). Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359, 531–533. doi: 10.1038/359531a0
Fredga, K. (1988). Aberrant chromosomal sex-determining mechanisms in mammals, with special reference to species with XY females. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 322, 83–95. doi: 10.1098/rstb.1988.0116
Ganesh, S., Mohanty, J., and Raman, R. (1997). Male-biased distribution of the human Y chromosomal genes SRY and ZFY in the lizard Calotes versicolor, which lacks sex chromosomes and temperature-dependent sex determination. Chromosome Res. 5, 413–419. doi: 10.1023/A:1018452526903
Gatewood, J. D., Wills, A., Shetty, S., Xu, J., Arnold, A. P., Burgoyne, P. S., et al. (2006). Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J. Neurosci. 26, 2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006
Grafodatskaya, D., Rens, W., Wallis, M. C., Trifonov, V., O'brien, P. C., Clarke, O., et al. (2007). Search for the sex-determining switch in monotremes: mapping WT1, SF1, LHX1, LHX2, FGF9, WNT4, RSPO1 and GATA4 in platypus. Chromosome Res. 15, 777–785. doi: 10.1007/s10577-007-1161-y
Graves, J. A. (2001). From brain determination to testis determination: evolution of the mammalian sex-determining gene. Reprod. Fertil. Dev. 13, 665–672. doi: 10.1071/RD01093
Graves, J. A. (2002). Evolution of the testis-determining gene–the rise and fall of SRY. Novartis Found Symp. 244, 86–97. discussion: 97–101, 203–106, 253–107.
Graves, J. A. (2015). Weird mammals provide insights into the evolution of mammalian sex chromosomes and dosage compensation. J. Genet. 94, 567–574. doi: 10.1007/s12041-015-0572-3
Guiguen, Y., Fostier, A., Piferrer, F., and Chang, C.-F. (2010). Ovarian aromatase and estrogens: a pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 165, 352–366. doi: 10.1016/j.ygcen.2009.03.002
Hacker, A., Capel, B., Goodfellow, P., and Lovell-Badge, R. (1995). Expression of Sry, the mouse sex determining gene. Development 121, 1603–1614.
Hansen, T. B., Jensen, T. I., Clausen, B. H., Bramsen, J. B., Finsen, B., Damgaard, C. K., et al. (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. doi: 10.1038/nature11993
Hattori, R. S., Murai, Y., Oura, M., Masuda, S., Majhi, S. K., Sakamoto, T., et al. (2012). A Y-linked anti-Mullerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. U.S.A. 109, 2955–2959. doi: 10.1073/pnas.1018392109
Heule, C., Goppert, C., Salzburger, W., and Bohne, A. (2014). Genetics and timing of sex determination in the East African cichlid fish Astatotilapia burtoni. BMC Genet. 15:140. doi: 10.1186/s12863-014-0140-5
Ito, M., Ishikawa, M., Suzuki, S., Takamatsu, N., and Shiba, T. (1995). A rainbow trout SRY-type gene expressed in pituitary glands. FEBS Lett. 377, 37–40. doi: 10.1016/0014-5793(95)01307-5
Iwata, E., Mikami, K., Manbo, J., Moriya-Ito, K., and Sasaki, H. (2012). Social interaction influences blood cortisol values and brain aromatase genes in the protandrous false clown anemonefish, Amphiprion ocellaris. Zool. Sci. 29, 849–855. doi: 10.2108/zsj.29.849
Jandegian, C. M., Deem, S. L., Bhandari, R. K., Holliday, C. M., Nicks, D., Rosenfeld, C. S., et al. (2015). Developmental exposure to bisphenol A (BPA) alters sexual differentiation in painted turtles (Chrysemys picta). Gen. Comp. Endocrinol. 216, 77–85. doi: 10.1016/j.ygcen.2015.04.003
Jasarevic, E., Geary, D. C., and Rosenfeld, C. S. (2012). Sexually selected traits: a fundamental framework for studies on behavioral epigenetics. ILAR J. 53, 253–269. doi: 10.1093/ilar.53.3-4.253
Jeske, Y. W., Bowles, J., Greenfield, A., and Koopman, P. (1995). Expression of a linear Sry transcript in the mouse genital ridge. Nat. Genet. 10, 480–482. doi: 10.1038/ng0895-480
Jiang, Y., Han, K., Chen, S., Hong, W., Wang, Y., and Zhang, Z. (2015). Molecular cloning, characterization and expression of Lc-Sox11a in large yellow croaker Larimichthys crocea. Gene 574, 287–301. doi: 10.1016/j.gene.2015.08.021
Just, W., Baumstark, A., Hameister, H., Schreiner, B., Reisert, I., Hakhverdyan, M., et al. (2002). The sex determination in Ellobius lutescens remains bizarre. Cytogenet. Genome Res. 96, 146–153. doi: 10.1159/000063031
Just, W., Baumstark, A., Suss, A., Graphodatsky, A., Rens, W., Schafer, N., et al. (2007). Ellobius lutescens: sex determination and sex chromosome. Sex Dev. 1, 211–221. doi: 10.1159/000104771
Just, W., Rau, W., Vogel, W., Akhverdian, M., Fredga, K., Graves, J. A., et al. (1995). Absence of Sry in species of the vole Ellobius. Nat. Genet. 11, 117–118. doi: 10.1038/ng1095-117
Kamiya, T., Kai, W., Tasumi, S., Oka, A., Matsunaga, T., Mizuno, N., et al. (2012). A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8:e1002798. doi: 10.1371/journal.pgen.1002798
Kato, T., Miyata, K., Sonobe, M., Yamashita, S., Tamano, M., Miura, K., et al. (2013). Production of Sry knockout mouse using TALEN via oocyte injection. Sci. Rep. 3:3136. doi: 10.1038/srep03136
Kido, T., Sun, Z., and Lau, Y. C. (2017). Aberrant activation of the human sex-determining gene in early embryonic development results in postnatal growth retardation and lethality in mice. Sci. Rep. 7:4113. doi: 10.1038/s41598-017-04117-6
Kigar, S. L., Chang, L., Hayne, M. R., Karls, N. T., and Auger, A. P. (2016). Sex differences in Gadd45b expression and methylation in the developing rodent amygdala. Brain Res. 1642, 461–466. doi: 10.1016/j.brainres.2016.04.031
Kikuchi, K., and Hamaguchi, S. (2013). Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 242, 339–353. doi: 10.1002/dvdy.23927
Kobayashi, T., Yamada, F., Hashimoto, T., Abe, S., Matsuda, Y., and Kuroiwa, A. (2007). Exceptional minute sex-specific region in the X0 mammal, Ryukyu spiny rat. Chromosome Res. 15, 175–187. doi: 10.1007/s10577-006-1093-y
Konkle, A. T., and McCarthy, M. M. (2011). Developmental time course of estradiol, testosterone, and dihydrotestosterone levels in discrete regions of male and female rat brain. Endocrinology 152, 223–235. doi: 10.1210/en.2010-0607
Kopsida, E., Lynn, P. M., Humby, T., Wilkinson, L. S., and Davies, W. (2013). Dissociable effects of Sry and sex chromosome complement on activity, feeding and anxiety-related behaviours in mice. PLoS ONE 8:e73699. doi: 10.1371/journal.pone.0073699
Kuljis, D. A., Loh, D. H., Truong, D., Vosko, A. M., Ong, M. L., McClusky, R., et al. (2013). Gonadal- and sex-chromosome-dependent sex differences in the circadian system. Endocrinology 154, 1501–1512. doi: 10.1210/en.2012-1921
Kuroiwa, A., Handa, S., Nishiyama, C., Chiba, E., Yamada, F., Abe, S., et al. (2011). Additional copies of CBX2 in the genomes of males of mammals lacking SRY, the Amami spiny rat (Tokudaia osimensis) and the Tokunoshima spiny rat (Tokudaia tokunoshimensis). Chromosome Res. 19, 635–644. doi: 10.1007/s10577-011-9223-6
Kuroiwa, A., Ishiguchi, Y., Yamada, F., Shintaro, A., and Matsuda, Y. (2010). The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma 119, 519–526. doi: 10.1007/s00412-010-0275-8
Lahr, G., Maxson, S. C., Mayer, A., Just, W., Pilgrim, C., and Reisert, I. (1995). Transcription of the Y chromosomal gene, Sry, in adult mouse brain. Brain Res. Mol. Brain Res. 33, 179–182. doi: 10.1016/0169-328X(95)00136-G
Le Page, Y., Diotel, N., Vaillant, C., Pellegrini, E., Anglade, I., Merot, Y., et al. (2010). Aromatase, brain sexualization and plasticity: the fish paradigm. Eur. J. Neurosci. 32, 2105–2115. doi: 10.1111/j.1460-9568.2010.07519.x
Lephart, E. D. (1996). A review of brain aromatase cytochrome P450. Brain Res. Brain Res. Rev. 22, 1–26. doi: 10.1016/0165-0173(96)00002-1
Manshack, L. K., Conard, C. M., Bryan, S. J., Deem, S. L., Holliday, D. K., Bivens, N. J., et al. (2017). Transcriptomic alterations in the brain of painted turtles (Chrysemys picta) developmentally exposed to bisphenol A or ethinyl estradiol. Physiol. Genomics 49, 201–215. doi: 10.1152/physiolgenomics.00103.2016
Manshack, L. K., Conard, C. M., Johnson, S. A., Alex, J. M., Bryan, S. J., Deem, S. L., et al. (2016). Effects of developmental exposure to bisphenol A and ethinyl estradiol on spatial navigational learning and memory in painted turtles (Chrysemys picta). Horm. Behav. 85, 48–55. doi: 10.1016/j.yhbeh.2016.07.009
Marshall Graves, J. A. (2002). The rise and fall of SRY. Trends Genet. 18, 259–264. doi: 10.1016/S0168-9525(02)02666-5
Matsuda, K. I., Mori, H., and Kawata, M. (2012). Epigenetic mechanisms are involved in sexual differentiation of the brain. Rev. Endocr. Metab. Disord. 13, 163–171. doi: 10.1007/s11154-012-9202-z
Matsuda, M. (2003). Sex determination in fish: lessons from the sex-determining gene of the teleost medaka, Oryzias latipes. Dev. Growth Differ. 45, 397–403. doi: 10.1111/j.1440-169X.2003.00716.x
Matthey, R. (1958). [New type of chromosomal sex determination in the mammals Ellobius lutescens Th. and Microtus (Chilotus) oregoni Bachm. (Muridae, Microtinae)]. Experientia 14, 240–241. doi: 10.1007/BF02159178
Mayer, A., Lahr, G., Swaab, D. F., Pilgrim, C., and Reisert, I. (1998). The Y-chromosomal genes SRY and ZFY are transcribed in adult human brain. Neurogenetics 1, 281–288. doi: 10.1007/s100480050042
Mayer, A., Mosler, G., Just, W., Pilgrim, C., and Reisert, I. (2000). Developmental profile of Sry transcripts in mouse brain. Neurogenetics 3, 25–30. doi: 10.1007/s100480000093
McArthur, S., Mchale, E., and Gillies, G. E. (2007). The size and distribution of midbrain dopaminergic populations are permanently altered by perinatal glucocorticoid exposure in a sex- region- and time-specific manner. Neuropsychopharmacology 32, 1462–1476. doi: 10.1038/sj.npp.1301277
McCarthy, M. M. (2010). How it's made: organisational effects of hormones on the developing brain. J. Neuroendocrinol. 22, 736–742. doi: 10.1111/j.1365-2826.2010.02021.x
McElreavey, K., Vilain, E., Abbas, N., Herskowitz, I., and Fellous, M. (1993). A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proc. Natl. Acad. Sci. U.S.A. 90, 3368–3372. doi: 10.1073/pnas.90.8.3368
McPhie-Lalmansingh, A. A., Tejada, L. D., Weaver, J. L., and Rissman, E. F. (2008). Sex chromosome complement affects social interactions in mice. Horm. Behav. 54, 565–570. doi: 10.1016/j.yhbeh.2008.05.016
Menger, Y., Bettscheider, M., Murgatroyd, C., and Spengler, D. (2010). Sex differences in brain epigenetics. Epigenomics 2, 807–821. doi: 10.2217/epi.10.60
Milsted, A., Serova, L., Sabban, E. L., Dunphy, G., Turner, M. E., and Ely, D. L. (2004). Regulation of tyrosine hydroxylase gene transcription by Sry. Neurosci. Lett. 369, 203–207. doi: 10.1016/j.neulet.2004.07.052
Morgan, C. P., and Bale, T. L. (2017). Sex differences in microRNA-mRNA networks: examination of novel epigenetic programming mechanisms in the sexually dimorphic neonatal hypothalamus. Biol. Sex Differ. 8:27. doi: 10.1186/s13293-017-0149-3
Morris, J. A., Jordan, C. L., and Breedlove, S. M. (2004). Sexual differentiation of the vertebrate nervous system. Nat. Neurosci. 7, 1034–1039. doi: 10.1038/nn1325
Morton, S. U., Scherz, P. J., Cordes, K. R., Ivey, K. N., Stainier, D. Y., and Srivastava, D. (2008). microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 105, 17830–17835. doi: 10.1073/pnas.0804673105
Mosley, M., Weathington, J., Cortes, L. R., Bruggeman, E., Castillo-Ruiz, A., Xue, B., et al. (2017). Neonatal inhibition of DNA methylation alters cell phenotype in sexually dimorphic regions of the mouse brain. Endocrinology 158, 1838–1848. doi: 10.1210/en.2017-00205
Mulugeta, E., Wassenaar, E., Sleddens-Linkels, E., Van, I. W. F., Heard, E., Grootegoed, J. A., et al. (2016). Genomes of Ellobius species provide insight into the evolutionary dynamics of mammalian sex chromosomes. Genome Res. 26, 1202–1210. doi: 10.1101/gr.201665.115
Murray, H. E., Pillai, A. V., McArthur, S. R., Razvi, N., Datla, K. P., Dexter, D. T., et al. (2003). Dose- and sex-dependent effects of the neurotoxin 6-hydroxydopamine on the nigrostriatal dopaminergic pathway of adult rats: differential actions of estrogen in males and females. Neuroscience 116, 213–222. doi: 10.1016/S0306-4522(02)00578-X
Myosho, T., Otake, H., Masuyama, H., Matsuda, M., Kuroki, Y., Fujiyama, A., et al. (2012). Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191, 163–170. doi: 10.1534/genetics.111.137497
Naftolin, F., Ryan, K. J., Davies, I. J., Reddy, V. V., Flores, F., Petro, Z., et al. (1975). The formation of estrogens by central neuroendocrine tissues. Recent Prog. Horm. Res. 31, 295–319. doi: 10.1016/B978-0-12-571131-9.50012-8
Nagahama, Y. (2005). Molecular mechanisms of sex determination and gonadal sex differentiation in fish. Fish Physiol. Biochem. 31:105. doi: 10.1007/s10695-006-7590-2
Nanda, I., Kondo, M., Hornung, U., Asakawa, S., Winkler, C., Shimizu, A., et al. (2002). A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. U.S.A. 99, 11778–11783. doi: 10.1073/pnas.182314699
Navarro-Martin, L., Galay-Burgos, M., Piferrer, F., and Sweeney, G. (2012). Characterisation and expression during sex differentiation of Sox19 from the sea bass Dicentrarchus labrax. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 163, 316–323. doi: 10.1016/j.cbpb.2012.08.004
Navarro-Martin, L., Galay-Burgos, M., Sweeney, G., and Piferrer, F. (2009). Different sox17 transcripts during sex differentiation in sea bass, Dicentrarchus labrax. Mol. Cell. Endocrinol. 299, 240–251. doi: 10.1016/j.mce.2008.11.013
Navarro-Martin, L., Vinas, J., Ribas, L., Diaz, N., Gutierrez, A., Di Croce, L., et al. (2011). DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 7:e1002447. doi: 10.1371/journal.pgen.1002447
Nugent, B. M., and McCarthy, M. M. (2011). Epigenetic underpinnings of developmental sex differences in the brain. Neuroendocrinology 93, 150–158. doi: 10.1159/000325264
Obernosterer, G., Leuschner, P. J., Alenius, M., and Martinez, J. (2006). Post-transcriptional regulation of microRNA expression. RNA 12, 1161–1167. doi: 10.1261/rna.2322506
Oh, H. J., Li, Y., and Lau, Y. F. (2005). Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 72, 407–415. doi: 10.1095/biolreprod.104.034447
Ohe, K., Lalli, E., and Sassone-Corsi, P. (2002). A direct role of SRY and SOX proteins in pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 99, 1146–1151. doi: 10.1073/pnas.022645899
Otake, T., and Kuroiwa, A. (2016). Molecular mechanism of male differentiation is conserved in the SRY-absent mammal, Tokudaia osimensis. Sci. Rep. 6:32874. doi: 10.1038/srep32874
Pfau, M. L., Purushothaman, I., Feng, J., Golden, S. A., Aleyasin, H., Lorsch, Z. S., et al. (2016). Integrative analysis of sex-specific microRNA networks following stress in mouse nucleus accumbens. Front. Mol. Neurosci. 9:144. doi: 10.3389/fnmol.2016.00144
Phoenix, C. H., Goy, R. W., Gerall, A. A., and Young, W. C. (1959). Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology 65, 369–382. doi: 10.1210/endo-65-3-369
Pontiggia, A., Rimini, R., Harley, V. R., Goodfellow, P. N., Lovell-Badge, R., and Bianchi, M. E. (1994). Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 13, 6115–6124.
Quinnies, K. M., Bonthuis, P. J., Harris, E. P., Shetty, S. R., and Rissman, E. F. (2015). Neural growth hormone: regional regulation by estradiol and/or sex chromosome complement in male and female mice. Biol. Sex Differ. 6, 8. doi: 10.1186/s13293-015-0026-x
Rens, W., O'brien, P. C., Grutzner, F., Clarke, O., Graphodatskaya, D., Tsend-Ayush, E., et al. (2007). The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 8:R243. doi: 10.1186/gb-2007-8-11-r243
Reyhanian Caspillo, N., Volkova, K., Hallgren, S., Olsson, P. E., and Porsch-Hallstrom, I. (2014). Short-term treatment of adult male zebrafish (Danio rerio) with 17alpha-ethinyl estradiol affects the transcription of genes involved in development and male sex differentiation. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 164, 35–42. doi: 10.1016/j.cbpc.2014.04.003
Rosenfeld, C. S., Denslow, N. D., Orlando, E. F., Gutierrez-Villagomez, J. M., and Trudeau, V. L. (2017). Neuroendocrine disruption of organizational and activational hormone programming in poikilothermic vertebrates. J. Toxicol. Environ. Health B Crit. Rev. 20, 276–304. doi: 10.1080/10937404.2017.1370083
Sato, K., and Yamamoto, D. (2014). An epigenetic switch of the brain sex as a basis of gendered behavior in Drosophila. Adv. Genet. 86, 45–63. doi: 10.1016/B978-0-12-800222-3.00003-6
Savic, I., Frisen, L., Manzouri, A., Nordenstrom, A., and Linden Hirschberg, A. (2017). Role of testosterone and Y chromosome genes for the masculinization of the human brain. Hum. Brain Mapp. 38, 1801–1814. doi: 10.1002/hbm.23483
Sekido, R. (2010). SRY: a transcriptional activator of mammalian testis determination. Int. J. Biochem. Cell Biol. 42, 417–420. doi: 10.1016/j.biocel.2009.12.005
Sekido, R. (2014). The potential role of SRY in epigenetic gene regulation during brain sexual differentiation in mammals. Adv. Genet. 86, 135–165. doi: 10.1016/B978-0-12-800222-3.00007-3
Sekido, R., and Lovell-Badge, R. (2008). Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature 453, 930–934. doi: 10.1038/nature06944
Seney, M. L., Ekong, K. I., Ding, Y., Tseng, G. C., and Sibille, E. (2013). Sex chromosome complement regulates expression of mood-related genes. Biol. Sex Differ. 4:20. doi: 10.1186/2042-6410-4-20
Shen, E. Y., Ahern, T. H., Cheung, I., Straubhaar, J., Dincer, A., Houston, I., et al. (2015). Epigenetics and sex differences in the brain: a genome-wide comparison of histone-3 lysine-4 trimethylation (H3K4me3) in male and female mice. Exp. Neurol. 268, 21–29. doi: 10.1016/j.expneurol.2014.08.006
Siegel, G., Obernosterer, G., Fiore, R., Oehmen, M., Bicker, S., Christensen, M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11, 705–716. doi: 10.1038/ncb1876
Sinclair, A. H., Berta, P., Palmer, M. S., Hawkins, J. R., Griffiths, B. L., Smith, M. J., et al. (1990). A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 346, 240–244. doi: 10.1038/346240a0
Soullier, S., Hanni, C., Catzeflis, F., Berta, P., and Laudet, V. (1998). Male sex determination in the spiny rat Tokudaia osimensis (Rodentia: Muridae) is not Sry dependent. Mamm. Genome 9, 590–592. doi: 10.1007/s003359900823
Sung, Y. H., Baek, I. J., Kim, D. H., Jeon, J., Lee, J., Lee, K., et al. (2013). Knockout mice created by TALEN-mediated gene targeting. Nat. Biotechnol. 31, 23–24. doi: 10.1038/nbt.2477
Sutou, S., Mitsui, Y., and Tsuchiya, K. (2001). Sex determination without the Y chromosome in two Japanese rodents Tokudaia osimensis osimensis and Tokudaia osimensis spp. Mamm. Genome 12, 17–21. doi: 10.1007/s003350010228
Takada, S., Dinapoli, L., Capel, B., and Koopman, P. (2004). Sox8 is expressed at similar levels in gonads of both sexes during the sex determining period in turtles. Dev. Dyn. 231, 387–395. doi: 10.1002/dvdy.20132
Takamatsu, N., Kanda, H., Tsuchiya, I., Yamada, S., Ito, M., Kabeno, S., et al. (1995). A gene that is related to SRY and is expressed in the testes encodes a leucine zipper-containing protein. Mol. Cell. Biol. 15, 3759–3766. doi: 10.1128/MCB.15.7.3759
Todd, E. V., Liu, H., Muncaster, S., and Gemmell, N. J. (2016). Bending genders: the biology of natural sex change in fish. Sex Dev. 10, 223–241. doi: 10.1159/000449297
Torres Maldonado, L. C., Landa Piedra, A., Moreno Mendoza, N., Marmolejo Valencia, A., Meza Martinez, A., and Merchant Larios, H. (2002). Expression profiles of Dax1, Dmrt1, and Sox9 during temperature sex determination in gonads of the sea turtle Lepidochelys olivacea. Gen. Comp. Endocrinol. 129, 20–26. doi: 10.1016/S0016-6480(02)00511-7
Tsai, H. W., Grant, P. A., and Rissman, E. F. (2009). Sex differences in histone modifications in the neonatal mouse brain. Epigenetics 4, 47–53. doi: 10.4161/epi.4.1.7288
Tsend-Ayush, E., Kortschak, R. D., Bernard, P., Lim, S. L., Ryan, J., Rosenkranz, R., et al. (2012). Identification of mediator complex 26 (Crsp7) gametologs on platypus X1 and Y5 sex chromosomes: a candidate testis-determining gene in monotremes? Chromosome Res. 20, 127–138. doi: 10.1007/s10577-011-9270-z
Uy, B. R., Simoes-Costa, M., Sauka-Spengler, T., and Bronner, M. E. (2012). Expression of Sox family genes in early lamprey development. Int. J. Dev. Biol. 56, 377–383. doi: 10.1387/ijdb.113416bu
Valenzuela, N., Neuwald, J. L., and Literman, R. (2013). Transcriptional evolution underlying vertebrate sexual development. Dev. Dyn. 242, 307–319. doi: 10.1002/dvdy.23897
Vogel, W., Jainta, S., Rau, W., Geerkens, C., Baumstark, A., Correa-Cerro, L. S., et al. (1998). Sex determination in Ellobius lutescens: the story of an enigma. Cytogenet. Cell Genet. 80, 214–221. doi: 10.1159/000014983
Wagner, C. K., Xu, J., Pfau, J. L., Quadros, P. S., De Vries, G. J., and Arnold, A. P. (2004). Neonatal mice possessing an Sry transgene show a masculinized pattern of progesterone receptor expression in the brain independent of sex chromosome status. Endocrinology 145, 1046–1049. doi: 10.1210/en.2003-1219
Wallis, M. C., Delbridge, M. L., Pask, A. J., Alsop, A. E., Grutzner, F., O'brien, P. C., et al. (2007a). Mapping platypus SOX genes; autosomal location of SOX9 excludes it from sex determining role. Cytogenet. Genome Res. 116, 232–234. doi: 10.1159/000098192
Wallis, M. C., Waters, P. D., and Graves, J. A. (2008). Sex determination in mammals–before and after the evolution of SRY. Cell. Mol. Life Sci. 65, 3182–3195. doi: 10.1007/s00018-008-8109-z
Wallis, M. C., Waters, P. D., Delbridge, M. L., Kirby, P. J., Pask, A. J., Grutzner, F., et al. (2007b). Sex determination in platypus and echidna: autosomal location of SOX3 confirms the absence of SRY from monotremes. Chromosome Res. 15, 949–959. doi: 10.1007/s10577-007-1185-3
Wang, H., Hu, Y. C., Markoulaki, S., Welstead, G. G., Cheng, A. W., Shivalila, C. S., et al. (2013). TALEN-mediated editing of the mouse Y chromosome. Nat. Biotechnol. 31, 530–532. doi: 10.1038/nbt.2595
Wang, R., Cheng, H., Xia, L., Guo, Y., Huang, X., and Zhou, R. (2003). Molecular cloning and expression of Sox17 in gonads during sex reversal in the rice field eel, a teleost fish with a characteristic of natural sex transformation. Biochem. Biophys. Res. Commun. 303, 452–457. doi: 10.1016/S0006-291X(03)00361-9
Wapstra, E., and Warner, D. A. (2010). Sex allocation and sex determination in squamate reptiles. Sex Dev. 4, 110–118. doi: 10.1159/000272459
Watson, J., and Adkins-Regan, E. (1989a). Activation of sexual behavior by implantation of testosterone propionate and estradiol benzoate into the preoptic area of the male Japanese quail (Coturnix japonica). Horm. Behav. 23, 251–268. doi: 10.1016/0018-506X(89)90065-2
Watson, J., and Adkins-Regan, E. (1989b). Testosterone implanted in the preoptic area of male Japanese quail must be aromatized to activate copulation. Horm. Behav. 23, 432–447. doi: 10.1016/0018-506X(89)90055-X
Wei, L., Yang, C., Tao, W., and Wang, D. (2016). Genome-wide identification and transcriptome-based expression profiling of the sox gene family in the nile tilapia (Oreochromis niloticus). Int. J. Mol. Sci. 17:270. doi: 10.3390/ijms17030270
Western, P. S., Harry, J. L., Graves, J. A., and Sinclair, A. H. (1999a). Temperature-dependent sex determination in the American alligator: AMH precedes SOX9 expression. Dev. Dyn. 216, 411–419. doi: 10.1002/(SICI)1097-0177(199912)216:4/5<411::AID-DVDY9>3.0.CO;2-Y
Western, P. S., Harry, J. L., Graves, J. A., and Sinclair, A. H. (1999b). Temperature-dependent sex determination: upregulation of SOX9 expression after commitment to male development. Dev. Dyn. 214, 171–177. doi: 10.1002/(SICI)1097-0177(199903)214:3<171::AID-AJA1>3.0.CO;2-S
Wu, J. B., Chen, K., Li, Y., Lau, Y. F., and Shih, J. C. (2009). Regulation of monoamine oxidase A by the SRY gene on the Y chromosome. FASEB J. 23, 4029–4038. doi: 10.1096/fj.09-139097
Yano, A., Guyomard, R., Nicol, B., Jouanno, E., Quillet, E., Klopp, C., et al. (2012). An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22, 1423–1428. doi: 10.1016/j.cub.2012.05.045
Yatsu, R., Miyagawa, S., Kohno, S., Parrott, B. B., Yamaguchi, K., Ogino, Y., et al. (2016). RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genomics 17:77. doi: 10.1186/s12864-016-2396-9
Yatsu, R., Miyagawa, S., Kohno, S., Saito, S., Lowers, R. H., Ogino, Y., et al. (2015). TRPV4 associates environmental temperature and sex determination in the American alligator. Sci. Rep. 5:18581. doi: 10.1038/srep18581
Yokoi, H., Kobayashi, T., Tanaka, M., Nagahama, Y., Wakamatsu, Y., Takeda, H., et al. (2002). Sox9 in a teleost fish, medaka (Oryzias latipes): evidence for diversified function of Sox9 in gonad differentiation. Mol. Reprod. Dev. 63, 5–16. doi: 10.1002/mrd.10169
Zhang, L., Zhu, T., Lin, D., Zhang, Y., and Zhang, W. (2010). A second form of Sox11 homologue identified in the orange-spotted grouper Epinephelus coioides: analysis of sequence and mRNA expression patterns. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 157, 415–422. doi: 10.1016/j.cbpb.2010.09.004
Keywords: steroid hormones, sexual dimorphism, neuroscience, mammals, therian, prototherian, Sox9 transcription factor, dopaminergic neurons
Citation: Rosenfeld CS (2017) Brain Sexual Differentiation and Requirement of SRY: Why or Why Not? Front. Neurosci. 11:632. doi: 10.3389/fnins.2017.00632
Received: 18 August 2017; Accepted: 30 October 2017;
Published: 16 November 2017.
Edited by:
Ishwar Parhar, Monash University Malaysia, MalaysiaReviewed by:
Elaine M. Hull, Florida State University, United StatesMarco Segatto, University of Milan, Italy
Copyright © 2017 Rosenfeld. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cheryl S. Rosenfeld, cm9zZW5mZWxkY0BtaXNzb3VyaS5lZHU=
 Cheryl S. Rosenfeld
Cheryl S. Rosenfeld