- 1Department of Molecular Neuroscience and Integrative Physiology, Graduate School of Medical Sciences, Kanazawa University, Kanazawa, Japan
- 2Department of Cell and Developmental Biology, Wolfson Institute for Biomedical Research, University College London, London, UK
Although, the suprachiasmatic nucleus (SCN) of the hypothalamus acts as the central clock in mammals, the circadian expression of clock genes has been demonstrated not only in the SCN, but also in peripheral tissues and brain regions outside the SCN. However, the physiological roles of extra-SCN circadian clocks in the brain remain largely elusive. In response, we generated Nkx2.1-Bmal1−/− mice in which Bmal1, an essential clock component, was genetically deleted specifically in the ventral forebrain, including the preoptic area, nucleus of the diagonal band, and most of the hypothalamus except the SCN. In these mice, as expected, PER2::LUC oscillation was drastically attenuated in the explants of mediobasal hypothalamus, whereas it was maintained in those of the SCN. Although, Nkx2.1-Bmal1−/− mice were rhythmic and nocturnal, they showed altered patterns of locomotor activity during the night in a 12:12-h light:dark cycle and during subjective night in constant darkness. Control mice were more active during the first half than the second half of the dark phase or subjective night, whereas Nkx2.1-Bmal1−/− mice showed the opposite pattern of locomotor activity. Temporal patterns of sleep-wakefulness and feeding also changed accordingly. Such results suggest that along with mechanisms in the SCN, local Bmal1–dependent clocks in the ventral forebrain are critical for generating precise temporal patterns of circadian behaviors.
Introduction
The circadian oscillator in the suprachiasmatic nucleus (SCN) of the hypothalamus is the central pacemaker in mammals, orchestrating multiple circadian rhythms in organisms (Reppert and Weaver, 2002; Dibner et al., 2010). Each SCN cell has an individual cellular clock driven by autoregulatory transcriptional and translational feedback loops (TTFLs) of clock genes. Notably, such cellular clocks also appear in many peripheral tissues and brain regions outside the SCN (Balsalobre et al., 1998; Yamazaki et al., 2000; Yagita et al., 2001; Abe et al., 2002; Yoo et al., 2004; Guilding et al., 2009; Dibner et al., 2010). Cell type-specific manipulations of clock genes have clarified the roles of peripheral circadian clocks in a variety of tissues (McDearmon et al., 2006; Storch et al., 2007; Lamia et al., 2008; Westgate et al., 2008; Dibner et al., 2010; Marcheva et al., 2010; Sadacca et al., 2011; Fustin et al., 2012; Paschos et al., 2012; Nguyen et al., 2013; Gibbs et al., 2014; Jacobi et al., 2015; Xie et al., 2015; Dudek et al., 2016; Xu et al., 2016). By contrast, knowledge of the physiological roles of brain clocks in extra-SCN regions remains limited (Roybal et al., 2007; Mukherjee et al., 2010; Mieda and Sakurai, 2011; Spencer et al., 2013; Yu et al., 2014; Nakano et al., 2016; Orozco-Solis et al., 2016; Shimizu et al., 2016).
The hypothalamus plays a pivotal role in homeostasis and in controlling multiple bodily functions that exhibit circadian fluctuations, including sleep and wakefulness, body temperature, food intake, and autonomic nervous and endocrine systems. In addition, local brain clocks have been reported in multiple nuclei of the hypothalamus (Abe et al., 2002; Mieda et al., 2006; Guilding et al., 2009; Moriya et al., 2009; Yu et al., 2014; Orozco-Solis et al., 2016). Therefore, it is especially intriguing to pinpoint the roles of extra-SCN brain clocks in the hypothalamus.
To genetically dissect the roles of SCN and extra-SCN clocks, we used mice in which the expression of Cre recombinase was restricted to the hypothalamus outside the SCN. We focused on the developmental regulatory genes involved in the patterning of the neuroepithelium, which are often expressed in domains that give rise to certain brain regions and nuclei in adults. Nkx2.1 is a good candidate because its expression affects a domain that generates the posteroventral hypothalamus, yet is devoid of the Lhx1-positive domain that differentiates into the SCN and its adjacent structures (Puelles and Rubenstein, 2003; Shimogori et al., 2010). Along with the neuroepithelial domain that gives rise to the posteroventral hypothalamus, Nkx2.1 is also expressed developmentally in the medial ganglionic eminence (MGE) and anterior entopeduncular area (AEP) in the ventral forebrain, from which γ-aminobutyric acid (GABA) neurons originate and migrate into the cerebral cortex and basal ganglia, including the striatum and globus pallidus (Puelles and Rubenstein, 2003; Kessaris et al., 2006). The MGE and AEP also generate oligodendrocyte progenitors in developing cortical and subcortical structures, although those cells decline to a very small fraction of all oligodendrocytes in most parts of the adult forebrain (Kessaris et al., 2006).
In our study, we generated mice in which Bmal1, an essential transcription factor of the TTFL, was deleted specifically in cells marked by the developmental expression of Nkx2.1. We demonstrated that although the mice maintained nocturnality and a normal free-running period of behavioral rhythm driven by the intact SCN, the temporal patterns of their nocturnal locomotor activity, sleep-wakefulness cycle, and food intake were impaired.
Methods
Animals
To generate Nkx2.1-Bmal1−/− mice (Nkx2.1-Cre;Bmal1fl/−), Nkx2.1-Cre mice (N > 7 backcrossed to C57BL/6J; Kessaris et al., 2006) were mated with mice carrying a conditional Bmal1 allele (Bmal1fl; Storch et al., 2007) (N > 7 backcrossed to C57BL/6J, The Jackson Laboratory #007668), as well as with mice carrying a null Bmal1 allele (Bmal1−) generated from Bmal1fl mice (Mieda and Sakurai, 2011). Nkx2.1-Bmal1−/− and control mice were further mated with Per2::Luc reporter mice (Yoo et al., 2004) in order to obtain Nkx2.1-Bmal1−/−;Per2::Luc (Nkx2.1-Cre;Bmal1fl/−;Per2::Luc) and Control;Per2::Luc (Bmal1fl/−;Per2::Luc) mice. Mice were maintained under a strict 12-h light:dark (LD) cycle in a temperature- and humidity-controlled room and fed ad libitum. All experimental procedures involving animals were approved by the respective animal care and use committee of Kanazawa University and were in accordance with the guidelines of the National Institutes of Health.
Histological Study
To examine the specificity of Cre-mediated recombination, Nkx2.1-Cre mice were crossed with Rosa26R-lacZ reporter mice (Jackson Laboratory #003474) (Soriano, 1999). Animals were sacrificed by transcardial perfusion with PBS followed by 4% paraformaldehyde fixative. Serial coronal brain sections 30 μm thick were collected in four series, one of which was further stained by β-galactosidase enzymatic labeling as previously described (Kessaris et al., 2006). Immunostaining was performed as previously reported (Mieda et al., 2015) with rabbit anti-BMAL1 (Novus Biologicals, 1:10,000), biotinylated anti-rabbit antibody (Vector Labs, 1:500), and the Vectastain ABC kit (Vector Labs).
Bioluminescence Measurement
Male Nkx2.1-Bmal1−/−;Per2::Luc (n = 6) and control;Per2::Luc mice (Bmal1fl/−;Per2::Luc; n = 5), aged 17–19 weeks, were housed in LD before sampling. PER2::LUC bioluminescence in SCN tissue was measured with a photomultiplier tube (Atto, Kronos Dio) at 10-min intervals with an exposure time of 1 min. Coronal SCN slices of 300 μm were made with a vibratome (Leica, Vi1000S) at approximately ZT8–10. The SCN tissue was dissected at the midrostrocaudal region, and a paired SCN was cultured on a Millicell-CM culture insert (Millipore), as previously described (Mieda et al., 2015). Bilateral MBH explants containing the dorsomedial (DMH), ventromedial (VMH), and arcuate (Arc) nuclei of the hypothalamus, as well as the median eminence (ME) and pars tuberalis (PT) (Guilding et al., 2009) were cultured similarly to the SCN explant. Recorded values were detrended by subtracting 24-h moving average values and were smoothened with a 5-point moving average method. Because luminescence from the SCN of Nkx2.1-Bmal1−/−;Per2::Luc mice was weak and its waveform was noisy, the middle of the time points crossing value 0 upward and downward were defined as acrophases, and the intervals between two adjacent acrophases were calculated for the periods. The average of periods of initial three cycles was calculated for each explant.
Measurements of Locomotor Activity and Food Intake
Male Nkx2.1-Bmal1−/− (Nkx2.1-Cre;Bmal1fl/−; n = 11) and control (Bmal1fl/− and Nkx2.1-Cre;Bmal1+/fl; n = 8 and 6, respectively) mice, aged 9–15 weeks, were housed individually in cages placed in a light-tight chamber with a light intensity of ~50 lux. Spontaneous locomotor activity and food intake were recorded by infrared motion sensors and food intake monitor (O'Hara) in 10-min bins. Actogram, activity profile, and χ2 periodogram analyses were performed using ClockLab (Actimetrics). The free-running period was measured by periodogram for Days 5–21 in constant darkness (DD).
Sleep Recordings
This study used male Nkx2.1-Bmal1−/− (Nkx2.1-Cre;Bmal1fl/−; n = 4) and control (Bmal1fl/− and Nkx2.1-Cre;Bmal1+/fl; n = 3 for each genotype; i.e., 6 control mice in total) mice, aged 17–21 weeks. These mice were different from those used for measurements of locomotor activity and food intake. The implantation of an electroencephalogrammic (EEG) and electromyographic (EMG) electrode was performed as described previously (Sasaki et al., 2011). Following surgery, all animals were housed individually for a recovery period of at least 14 days, after which EEG-EMG recordings were performed on 3 consecutive days in LD. EEG-EMG data were analyzed as previously described (Sasaki et al., 2011).
Statistics
All results are expressed as the mean ± SEM. Comparisons between individuals were analyzed by a two-tailed Student's t-test. In Figure S1, an one-way repeated measure ANOVA was performed, followed by a Tukey-HSD post-hoc analysis. When appropriate, data were analyzed by a two-way repeated measure ANOVA, followed by a Tukey-HSD post-hoc test.
Results
Cre-Mediated Recombination in the Hypothalamus of Nkx2.1-Cre Mice
According to the prosomeric model (Puelles and Rubenstein, 2003), the SCN and its adjacent areas develop from a field dorsal to the anterior basal floor of the secondary prosencephalon, which generates most areas of the hypothalamus and is delineated by the expression of the homeodomain transcription factor Nkx2.1. Therefore, we examined Nkx2.1-Cre mice (Kessaris et al., 2006) as a candidate Cre driver, in which Cre is expressed specifically in the hypothalamus but not the SCN. To localize cells with Cre-mediated recombination, Nkx2.1-Cre mice were crossed with Rosa26R-lacZ reporter mice, which expressed β-Galctosidase (βGal) after the Cre-mediated deletion of a loxP-flanked transcriptional blocker (Soriano, 1999). We mapped βGal+ cells in the entire brain of adult mice (Figure 1). Prominent βGal expression was observed in the medial septum, nucleus of the diagonal band (Figure 1A), medial preoptic area (Figures 1B,G), and most areas of the hypothalamus: the ventromedial (VMH), dorsomedial (DMH), arcuate (Arc) (Figures 1D,J), mammillary nuclei (MN) (Figure 1E), and the lateral hypothalamic area (LHA) at the rostrocaudal level of and posterior to the VMH (Figure 1D). Notably, the SCN, PVH, and adjacent regions were nearly devoid of βGal+ cells (Figures 1C,I). Moreover, βGal expression was detected in cells scattered in the cerebral cortex (Figure 1K), striatum, and globus pallidus (Figure 1H), which were likely GABAergic neurons originated from the Nkx2.1+ MGE and AEP. Such expression was entirely consistent with the developmental expression of Cre recombinase in Nkx2.1-Cre mice (Kessaris et al., 2006). We also detected several βGal+ cells in the dorsal raphe nucleus (Figures 1F,L). Altogether, Nkx2.1-Cre mice may be useful in revealing the roles of extra-SCN clocks in the hypothalamus.
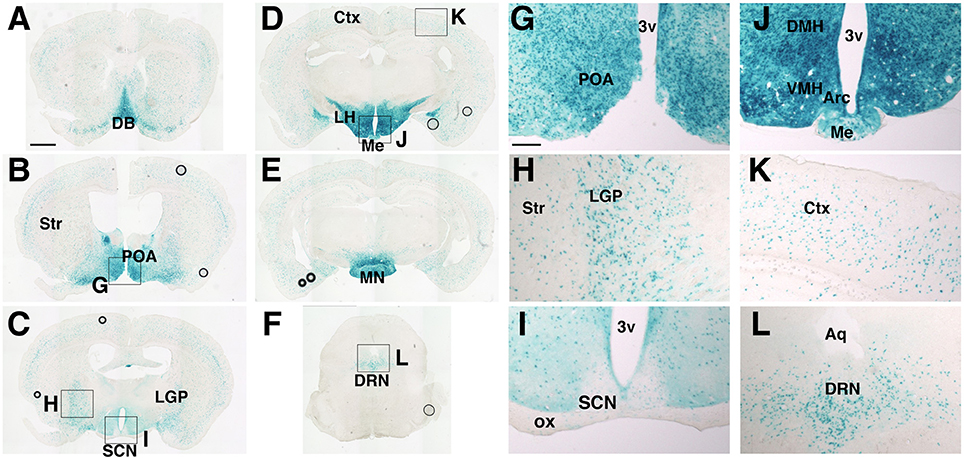
Figure 1. Distribution of cells with Cre-mediated recombination in Nkx2.1-Cre mice crossed with Rosa26R-lacZ reporter mice. Coronal sections from the olfactory bulb to medulla were stained by β-galactosidase enzymatic labeling. (A–F) Representative images with positive cells. (G–L) Magnified images of regions in (B–F). Scale bars, 1 mm for (A–F); 200 μm for (G–L). ac, anterior commissure; 3v, Arc, arcuate hypothalamic nucleus; Aq, aqueduct; Ctx, cerebral cortex; DB, diagonal band; DMH, dorsomedial hypothalamic nucleus; DRN, dorsal raphe nucleus; LGP, lateral globus pallidus; LH, lateral hypothalamus; Me, medial eminence; MN, mammillary nuclei; POA, preoptic area; SCN, suprachiasmatic nucleus; Str, striatum; VMH, ventromedial hypothalamic nucleus (Paxinos and Franklin, 2001).
PER2::LUC Oscillation Attenuated in the Mediobasal Hypothalamus of Nkx2.1-Bmal1−/− Mice
We generated mice without the Bmal1 gene in the hypothalamus, except for in the SCN, by breeding Nkx2.1-Cre mice with mice carrying floxed Bmal1 alleles (Bmal1fl; Storch et al., 2007). To improve the efficiency of Bmal1fl allele deletion, we generated Nkx2.1-Cre;Bmal1fl/− mice to have one floxed and one null allele of the Bmal1 gene (Mieda and Sakurai, 2011), hereafter designated as Nkx2.1-Bmal1−/− mice. We first confirmed in these mice that BMAL1 expression was drastically reduced in the ventral forebrain, including the Arc, VMH, and the DMH, but not in the SCN and adjacent regions (Figure 2A). Before analyzing their behavior in detail, we further crossed Nkx2.1-Cre mice with mice carrying the PER2::LUC reporter (Yoo et al., 2004) to confirm ex vivo that circadian clocks in the mediobasal hypothalamus (MBH; Guilding et al., 2009) were functionally attenuated in Nkx2.1-Bmal1−/− mice. Coronal slices were prepared from adult mice, and PER2::LUC oscillation in the SCN of Nkx2.1-Bmal1−/− mice was similar to that of control mice (amplitude: 6002 ± 435 vs. 5872 ± 559, p = 0.857; Figure 2B). By contrast, PER2::LUC oscillation in the MBH of Nkx2.1-Bmal1−/− mice was far less robust and unstable with reduced amplitude (amplitude: 202 ± 24 vs. 482 ± 113, p = 0.018; Figure 2C). The values of period and first acrophase of PER2::LUC oscillation in control explants were similar to those reported previously in both the SCN and MBH (Figures 2D–F; Yoo et al., 2004; Guilding et al., 2009). Interestingly, the first acrophase of PER2::LUC oscillation in the MBH occurred earlier in Nkx2.1-Bmal1−/− mice than control mice. Thus, local circadian clocks in the MBH were severely impaired in Nkx2.1-Bmal1−/− mice, whereas the SCN central clock remained normal.
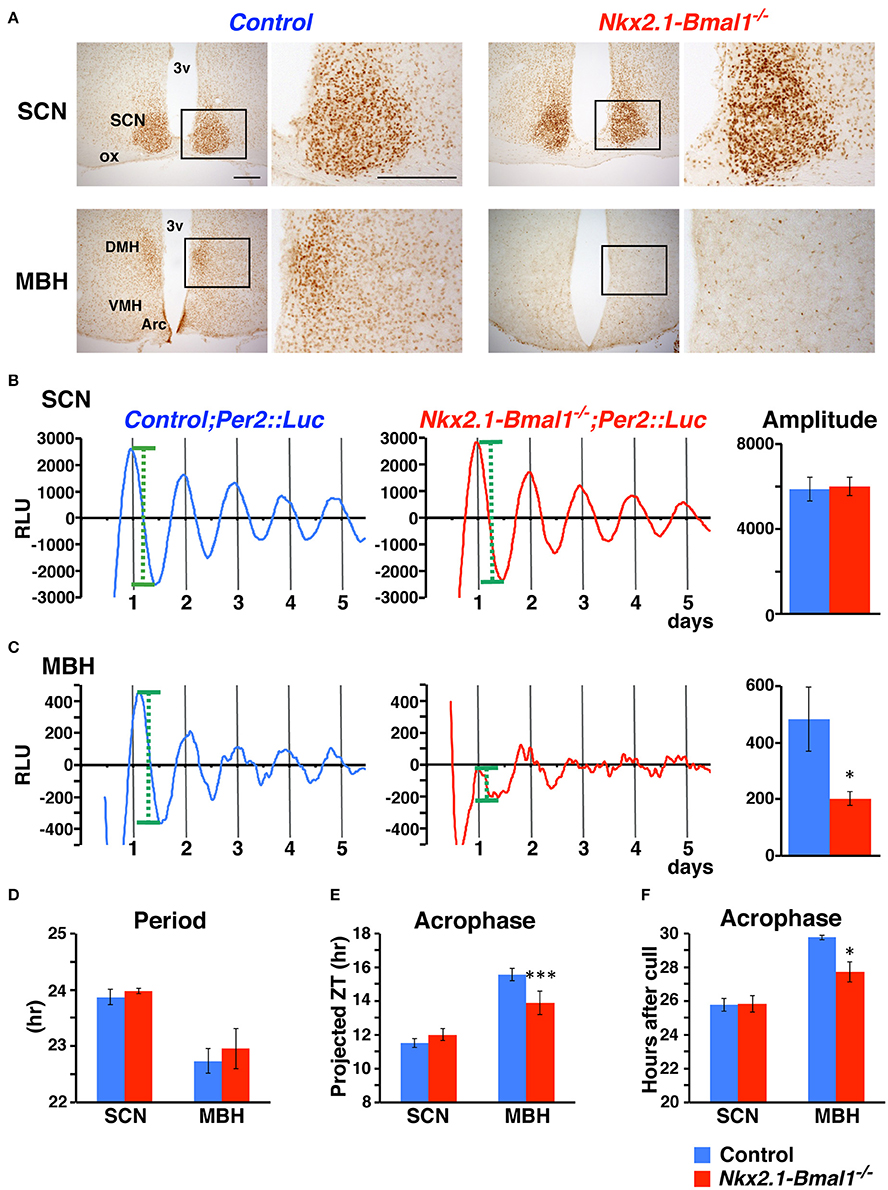
Figure 2. PER2::LUC oscillation is attenuated in the mediobasal hypothalamus but not in the SCN of Nkx2.1-Bmal1−/− mice. (A) BMAL1 expression in the SCN and mediobasal hypothalamus (MBH) of control and Nkx2.1-Bmal1−/− mice. Coronal brain sections were immunostained for BMAL1. The locations of the magnified images are indicated by rectangles in the low-power images. Scale bars, 200 μm. (B,C) Representative circadian rhythm of PER2::LUC in slices of the SCN (B) and MBH (C) of control and Nkx2.1-Bmal1−/− mice housed in LD. Mean amplitudes of PER2::LUC oscillation are shown on the right. Definition of amplitude was demonstrated by green dotted lines in (B,C). (D–F) Mean periods (D) and first acrophases (E,F) of PER2::LUC oscillation. Because PER2::LUC expression in the MBH was reported to be significantly correlated with time of cull (Guilding et al., 2009), the first acrophases were also expressed in hours after cull (F). Two-way repeated measures ANOVAs revealed statistical significances in the effect of brain region for period [F(1, 8) = 17.16, p = 0.003], in the effects of genotype [F(1, 8) = 619.19, p < 0.001] and region [F(1, 8) = 230.96, p < 0.001], as well as in the interaction between genotype and region [F(1, 8) = 339.05, p < 0.001] for first acrophase (projected ZT), and in the effects of genotype [F(1, 8) = 6.92, p = 0.030] and region [F(1, 8) = 22.27, p = 0.002] for first acrophase (hours after cull). Values are mean ± SEM. *p < 0.05; ***p < 0.001; comparison between genotypes by a Tukey-HSD post-hoc. n = 5 for control;Per2::Luc, n = 6 for Nkx2.1-Bmal1−/−;Per2::Luc mice.
Altered Temporal Pattern of Nocturnal Locomotor Activity in Nkx2.1-Bmal1−/− Mice
We next measured the circadian rhythm of spontaneous locomotor activity in Nkx2.1-Bmal1−/− (Nkx2.1-Cre;Bmal1fl/−) mice. We examined two strains of mice, Bmal1fl/− and Nkx2.1-Cre;Bmal1+/fl, as control mice. All three lines demonstrated a clear nocturnal pattern of behavioral rhythm in LD (Figure 3 and Figure S1). The free-running periods of behavioral rhythm in DD were similar between Nkx2.1-Bmal1−/− and two strains of control mice (Figure 3C and Figure S1C). However, the activity was substantially reduced in Nkx2.1-Bmal1−/− and Nkx2.1-Cre;Bmal1+/fl mice compared to that of Bmal1fl/− mice in LD (Figures S1A,E). We observed a similar attenuation of activity during the subjective night in those two strains free-running in DD (Figures S1A,D,E). Such results suggest that having the Nkx2.1-Cre allele caused a reduction of nocturnal activity regardless of the expression of Bmal1. Notably, the temporal pattern of nocturnal locomotor activity was clearly altered in Nkx2.1-Bmal1−/− mice compared to both control strains (Figure S1A). Such an alteration was more obvious when daily activity profiles were adjusted in accordance with activity levels (Figure 3B and Figure S1B). In both LD and DD, control mice were more active during the first than the second half of the dark phase or subjective night. By contrast, Nkx2.1-Bmal1−/− mice were more active during the second half of the dark phase or subjective night. The amplitude of the free-running rhythm of behavior, for which we calculated Qp values by periodogram analyses, was significantly lower in Nkx2.1-Bmal1−/− mice than in control mice (Figure 3D and Figure S1D). Thus, Nkx2.1-Bmal1−/− mice remained nocturnal, but did not maintain the precise temporal pattern of nocturnal activity. Because the function of SCN remained normal in those mice, peripheral circadian clocks in the extra-SCN hypothalamus might play a role in delineating the activity profile during the dark phase and subjective night.
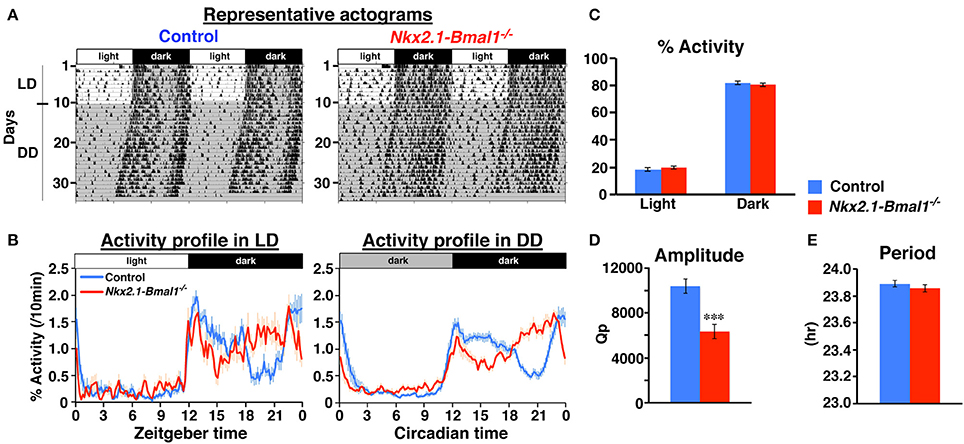
Figure 3. Temporal pattern of nocturnal locomotor activity is altered in Nkx2.1-Bmal1−/−. (A) Representative locomotor activity of control and Nkx2.1-Bmal1−/− mice. Animals were initially housed in 12:12-h light/dark (LD) conditions and later transferred to constant darkness (DD). Gray shading indicates when lights were off. (B) Daily profile of locomotor activity in LD or DD. Activity counts are expressed as percentage of daily total. (C) Relative activity during 12-h light phase (Light) and 12-h dark phase (Dark). No significant difference was detected between genotypes. (D,E) Mean free-running period and amplitude (Qp values of periodogram) in DD. Values for activity in DD were calculated for data on Days 5–19 in DD. Values are mean ± SEM. n = 14 for control, n = 11 for Nkx2.1-Bmal1−/− mice. ***p < 0.001 by a two-tailed Student's t-test.
Altered Temporal Sleep-Wakefulness Pattern Parallel to Locomotor Activity in Nkx2.1-Bmal1−/− Mice
We also examined whether the temporal pattern of daily sleep-wakefulness was altered in Nkx2.1-Bmal1−/− mice. As expected, Nkx2.1-Bmal1−/− mice remained nocturnal, but the hourly patterns of wakefulness, NREM sleep, and REM sleep were all altered similarly to that of locomotor activity (Figure 4). Time spent in wakefulness during 24-h of day or during the 12-h dark phase were significantly reduced (whole day: 722.1 ± 10.7 vs. 654.6 ± 21.8 m, p = 0.015; dark phase: 505.1 ± 7.8 vs. 455.0 ± 21.6 m, p = 0.034), while those in NREM sleep were significantly increased in Nkx2.1-Bmal1−/− mice (whole day: 642.3 ± 10.5 vs. 701.5 ± 25.9 m, p = 0.041; dark phase: 200.7 ± 7.5 vs. 249.4 ± 20.9 m, p = 0.034).
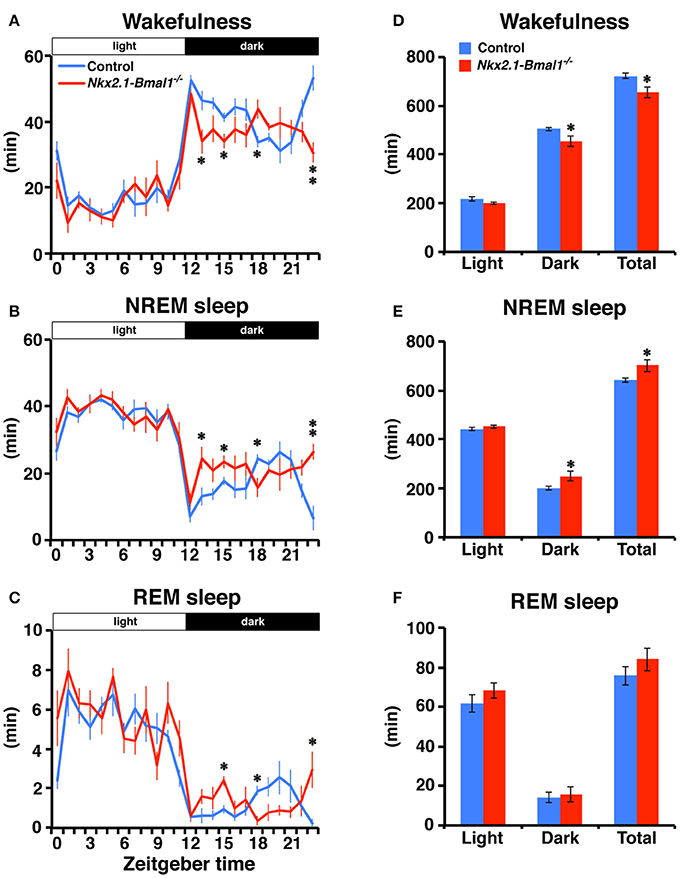
Figure 4. Temporal pattern of sleep-wakefulness during the dark phase is altered in Nkx2.1-Bmal1−/ − mice. (A–C) Hourly plots of time spent in wakefulness (A), NREM sleep (B), or REM sleep (C) in LD. (D–F) Time spent in wakefulness (D), NREM sleep (E), or REM sleep (F) during 12-h light phase (Light), 12-h dark phase (Dark), and 24-h day (Total). Values are mean ± SEM. n = 6 for control, n = 4 for Nkx2.1-Bmal1−/− mice. *p < 0.05; **p < 0.01 by a Tukey-HSD post-hoc. The p-values calculated by two-way repeated measures ANOVAs are: (A) the effect of genotype, F(1, 184) = 9.52, p = 0.015; the effect of time, F(23, 184) = 31.71, p < 0.001; and the interaction between genotype and time, F(23, 184) = 2.60, p < 0.001; (B) F(1, 184) = 5.94, p = 0.041; F(23, 184) = 29.30, p < 0.001; F(23, 184) = 2.60, p < 0.001; (C) F(23, 184) = 24.36, p < 0.001; F(23, 184) = 2.26, p = 0.02; (D) the effect of genotype, F(1, 8) = 9.52, p = 0.015; the effect of time (Light vs. Dark), F(1, 8) = 559.33, p < 0.001; and the interaction between genotype and time, F(1, 8) = 2.04, p = 0.191; (E) F(1, 8) = 5.94, p = 0.041; F(1, 8) = 567.63, p < 0.001; F(1, 8) = 4.17, p = 0.075; (F) F(1, 8) = 1.24, p = 0.299; F(1, 8) = 155.87, p < 0.001; F(1, 8) = 0.41, p = 0.538.
Altered Temporal Pattern of Nighttime Food Intake in Nkx2.1-Bmal1−/− Mice
We further examined the hourly food intake of Nkx2.1-Bmal1−/− mice. As with the patterns of locomotor activity and sleep-wakefulness, the temporal pattern of food intake during the dark phase was altered in Nkx2.1-Bmal1−/− mice (Figure 5). Daily food intake according to body weight did not differ between control and Nkx2.1-Bmal1−/− mice (0.119 ± 0.003 vs. 0.119 ± 0.004 g/g body weight, p = 0.960). Intriguingly, food intake in the light phase was significantly increased in Nkx2.1-Bmal1−/− mice compared to control mice (0.024 ± 0.002 vs. 0.017 ± 0.001 g/g body weight, p = 0.002), especially in the second half of the light phase. This increase might be correlated with a similar increase of locomotor activity in the latter half of the light phase (Figure 3A). As such, locomotor activity, sleep-wakefulness, and food intake demonstrated similar changes in their temporal patterns during the night in Nkx2.1-Bmal1−/− mice.
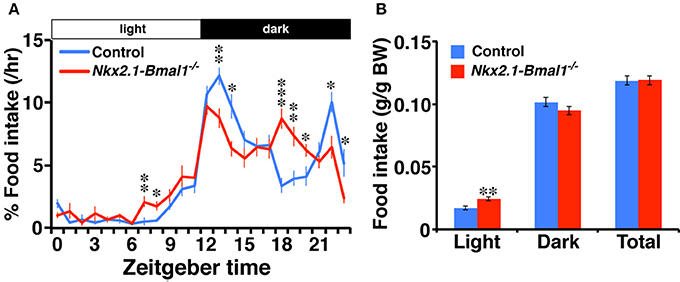
Figure 5. Temporal pattern of food intake during the dark phase is altered in Nkx2.1-Bmal1−/− mice. (A) Hourly plots of food intake, expressed as percentage of daily total. (B) Food intake per body weight during 12-h light phase (Light), 12-h dark phase (Dark), and 24-h day (Total). Values are mean ± SEM. n = 6 for control, n = 4 for Nkx2.1-Bmal1−/− mice. *p < 0.05; **p < 0.01; ***p < 0.001 by a Tukey-HSD post-hoc. The p-values calculated by two-way repeated measures ANOVAs are: (A) the effect of genotype, F(1, 529) > 1000, p < 0.001; the effect of time, F(23, 529) = 53.22, p < 0.001; and the interaction between genotype and time, F(23, 529) = 5.65, p < 0.001; (B) the effect of genotype, F(1, 23) = 0.003, p = 0.960; the effect of time (Light vs. Dark), F(1, 23) = 559.33, p < 0.001; and the interaction between genotype and time, F(1, 23) = 2.04, p = 0.026.
Discussion
We demonstrated that the SCN central clock alone cannot precisely delineate the locomotor activity pattern in the dark phase. The SCN grants mice nocturnality and roughly determines when they are active, whereas Bmal1-dependent local clocks in regions defined by developmental Nkx2.1-Cre expression might fine-tune the temporal pattern of nocturnal activity.
A remarkable feature of Nkx2.1-Cre mice in our study was that Cre expression occurred in most of the hypothalamus, but not in the SCN or its adjacent areas. Such a feature made this Cre driver line suitable for studying local circadian clocks in the hypothalamus without affecting the central clock in the SCN. During development, the SCN and PVH originate from a region immediately dorsal to an area marked by the expression of Nkx2.1 and that generates many hypothalamic structures, including the VMH, DMH, ARC, and MN (Puelles and Rubenstein, 2003; Shimogori et al., 2010). In addition to the hypothalamus, Nkx2.1 is developmentally expressed in the AEP and MGE, from which inhibitory neurons in the cortex and basal ganglia originate (Puelles and Rubenstein, 2003; Kessaris et al., 2006). Therefore, the distribution of βGal+ cells in adult Nkx2.1-Cre;Rosa26R-lacZ mice matched well with developmental Nkx2.1 expression, except for those in the DRN, which might be due to an ectopic expression of Cre in Nkx2.1-Cre mice (Puelles and Rubenstein, 2003; Kessaris et al., 2006; Shimogori et al., 2010).
In contrast to those addressing peripheral clocks in peripheral organs, a limited number of reports address what local brain clocks do in the regulation of behavior and physiology. Dudley et al. initially reported that the lack of NPAS2, a paralog of CLOCK expressed in the forebrain, altered temporal patterns of locomotor activity and sleep-wakefulness in the active phase, meaning that a brief break in circadian locomotor activity beginning midway through the active phase was attenuated (Dudley et al., 2003). Pioneered by McClung et al. the contribution of local clocks in the ventral tegmental area and nucleus accumbens, which constitute the mesolimbic dopaminergic pathway, in mood regulation has been well-characterized (McClung et al., 2005; Roybal et al., 2007; Mukherjee et al., 2010; Spencer et al., 2013; Chung et al., 2014). More recently, Shimizu et al. reported that the suprachiasmatic nucleus circadian oscillatory protein (SCOP) mediates the circadian regulation of long-term memory formation by local clocks in the hippocampus (Shimizu et al., 2016), and Nakano et al., found that the same molecule regulates the circadian expression of anxiety-like behavior under the control of local clocks in the basolateral amygdala (Nakano et al., 2016). On another front, Yu et al. reported that Bmal1-dependent local clock in histaminergic neurons of tuberomammillary nucleus (TMN) regulate sleep architecture, likely via transcriptional control of the histidine decarboxylase gene, which encodes a histamine-synthesizing enzyme, but is not involved in regulating circadian rhythms (Yu et al., 2014). Furthermore, Orozco-Solis et al. revealed that the Bmal1-dependent local clock of VMH Sf1 neurons controls circadian energy expenditure through the rhythmic activation of BAT's metabolism, yet plays little role in delineating circadian activity and feeding rhythms (Orozco-Solis et al., 2016).
It is notable that circadian rhythms of locomotor activity, sleep-wakefulness, and food intake were all altered in parallel in Nkx2.1-Cre mice. Hypothalamic areas originating from Nkx2.1-positive neuroepithelium include several nuclei and neurons implicated in the regulation of locomotor activity, sleep-wakefulness, and feeding. For instance, orexinergic neurons in the perifornical area and histaminergic neurons in the TMN are wake-active and promote wakefulness, while a population of GABAergic neurons in the POA is sleep-active and increases sleep (Mieda and Sakurai, 2009; Saper et al., 2010). The basal forebrain also contains multiple types of neurons that control sleep-wakefulness (Saper et al., 2010; Xu et al., 2015). The Arc, VMH, DMH, and LHA play important roles in the regulation of feeding and energy metabolism (Gautron et al., 2015). As mentioned previously, mice without Bmal1 specifically in histaminergic or VMH Sf1 neurons showed phenotypes unlike those of Nkx2.1-Bmal1−/− mice (Yu et al., 2014; Orozco-Solis et al., 2016). Such observations suggest that Bmal1-dependent local clocks in histaminergic or Sf1 neurons are not solely responsible for altering nocturnal activity patterns in Nkx2.1-Bmal1−/− mice. Orexinergic neurons may be well-placed to regulate multiple rhythms consistently because these neurons have been suggested to control locomotor activity, wakefulness, and food intake cooperatively (Mieda and Sakurai, 2009). Alternatively, multiple local brain clocks may differentially fine-tune these three rhythms.
Although, Bmal1 is the sole non-redundant factor of cellular clocks, the impairment caused by loss of Bmal1 may not be necessarily due to disruption of clock function. Indeed, several defects observed in standard Bmal1 knockout mice, such as reduced life span, fertility, and body weight, were not detected in inducible Bmal1 knockout mice that expressed the gene during embryogenesis but not after birth (Yang et al., 2016). Because Nkx2.1 is expressed in the neuroepithelium during embryogenesis, the possibility that the impairments of circadian rhythms in Nkx2.1-Bmal1−/− might reflect the function of Bmal1 unrelated to the cellular clock cannot be excluded.
In conclusion, we demonstrated that Bmal1-dependent circadian clocks in the SCN and extra-SCN brain regions cooperate to delineate the precise daily patterns of locomotor activity, sleep-wakefulness, and food intake. However, the location of relevant local clocks remains unidentified and thus a task for future research.
Author Contributions
MM conceived and performed experiments, wrote the manuscript, and secured funding. EH performed experiments. MM and NK provided resources. TS provided expertise and feedback.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported in part by Grants-in-Aid for Scientific Research (B) (24390052 and 16H05120 to MM) and for Scientific Research on Innovative Areas (25126708, 16H01608 to MM and 15H01425 to TS) from JSPS; by the Cabinet Office, Government of Japan, through its “Funding Program for Next Generation World-Leading Researchers” (LS048 to TS); and by European Research Council (ERC) Starting Grant (207807 to NK). We thank for P. Soriano for the Rosa26R-lacZ reporter mice; C. J. Weitz for the Bmal1fl/f mouse; and J. Takahashi for the Per2::Luc reporter mouse.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fnins.2017.00055/full#supplementary-material
References
Abe, M., Herzog, E. D., Yamazaki, S., Straume, M., Tei, H., Sakaki, Y., et al. (2002). Circadian rhythms in isolated brain regions. J. Neurosci. 22, 350–356.
Balsalobre, A., Damiola, F., and Schibler, U. (1998). A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell 93, 929–937. doi: 10.1016/S0092-8674(00)81199-X
Chung, S., Lee, E. J., Yun, S., Choe, H. K., Park, S. B., Son, H. J., et al. (2014). Impact of circadian nuclear receptor REV-ERBalpha on midbrain dopamine production and mood regulation. Cell 157, 858–868. doi: 10.1016/j.cell.2014.03.039
Dibner, C., Schibler, U., and Albrecht, U. (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu. Rev. Physiol. 72, 517–549. doi: 10.1146/annurev-physiol-021909-135821
Dudek, M., Gossan, N., Yang, N., Im, H. J., Ruckshanthi, J. P., Yoshitane, H., et al. (2016). The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J. Clin. Invest. 126, 365–376. doi: 10.1172/JCI82755
Dudley, C. A., Erbel-Sieler, C., Estill, S. J., Reick, M., Franken, P., Pitts, S., et al. (2003). Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301, 379–383. doi: 10.1126/science.1082795
Fustin, J. M., Doi, M., Yamada, H., Komatsu, R., Shimba, S., and Okamura, H. (2012). Rhythmic nucleotide synthesis in the liver: temporal segregation of metabolites. Cell Rep. 1, 341–349. doi: 10.1016/j.celrep.2012.03.001
Gautron, L., Elmquist, J. K., and Williams, K. W. (2015). Neural control of energy balance: translating circuits to therapies. Cell 161, 133–145. doi: 10.1016/j.cell.2015.02.023
Gibbs, J., Ince, L., Matthews, L., Mei, J., Bell, T., Yang, N., et al. (2014). An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20, 919–926. doi: 10.1038/nm.3599
Guilding, C., Hughes, A. T., Brown, T. M., Namvar, S., and Piggins, H. D. (2009). A riot of rhythms: neuronal and glial circadian oscillators in the mediobasal hypothalamus. Mol. Brain 2:28. doi: 10.1186/1756-6606-2-28
Jacobi, D., Liu, S., Burkewitz, K., Kory, N., Knudsen, N. H., Alexander, R. K., et al. (2015). Hepatic bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 22, 709–720. doi: 10.1016/j.cmet.2015.08.006
Kessaris, N., Fogarty, M., Iannarelli, P., Grist, M., Wegner, M., and Richardson, W. D. (2006). Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat. Neurosci. 9, 173–179. doi: 10.1038/nn1620
Lamia, K. A., Storch, K. F., and Weitz, C. J. (2008). Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177. doi: 10.1073/pnas.0806717105
Marcheva, B., Ramsey, K. M., Buhr, E. D., Kobayashi, Y., Su, H., Ko, C. H., et al. (2010). Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631. doi: 10.1038/nature09253
McClung, C. A., Sidiropoulou, K., Vitaterna, M., Takahashi, J. S., White, F. J., Cooper, D. C., et al. (2005). Regulation of dopaminergic transmission and cocaine reward by the Clock gene. Proc. Natl. Acad. Sci. U.S.A. 102, 9377–9381. doi: 10.1073/pnas.0503584102
McDearmon, E. L., Patel, K. N., Ko, C. H., Walisser, J. A., Schook, A. C., Chong, J. L., et al. (2006). Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 314, 1304–1308. doi: 10.1126/science.1132430
Mieda, M., Ono, D., Hasegawa, E., Okamoto, H., Honma, K., Honma, S., et al. (2015). Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron 85, 1103–1116. doi: 10.1016/j.neuron.2015.02.005
Mieda, M., and Sakurai, T. (2009). Integrative physiology of orexins and orexin receptors. CNS Neurol. Disord. Drug Targets 8, 281–295. doi: 10.2174/187152709788921663
Mieda, M., and Sakurai, T. (2011). Bmal1 in the nervous system is essential for normal adaptation of circadian locomotor activity and food intake to periodic feeding. J. Neurosci. 31, 15391–15396. doi: 10.1523/JNEUROSCI.2801-11.2011
Mieda, M., Williams, S. C., Richardson, J. A., Tanaka, K., and Yanagisawa, M. (2006). The dorsomedial hypothalamic nucleus as a putative food-entrainable circadian pacemaker. Proc. Natl. Acad. Sci. U.S.A. 103, 12150–12155. doi: 10.1073/pnas.0604189103
Moriya, T., Aida, R., Kudo, T., Akiyama, M., Doi, M., Hayasaka, N., et al. (2009). The dorsomedial hypothalamic nucleus is not necessary for food-anticipatory circadian rhythms of behavior, temperature or clock gene expression in mice. Eur. J. Neurosci. 29, 1447–1460. doi: 10.1111/j.1460-9568.2009.06697.x
Mukherjee, S., Coque, L., Cao, J. L., Kumar, J., Chakravarty, S., Asaithamby, A., et al. (2010). Knockdown of Clock in the ventral tegmental area through RNA interference results in a mixed state of mania and depression-like behavior. Biol. Psychiatry 68, 503–511. doi: 10.1016/j.biopsych.2010.04.031
Nakano, J. J., Shimizu, K., Shimba, S., and Fukada, Y. (2016). SCOP/PHLPP1beta in the basolateral amygdala regulates circadian expression of mouse anxiety-like behavior. Sci. Rep. 6:33500. doi: 10.1038/srep33500
Nguyen, K. D., Fentress, S. J., Qiu, Y., Yun, K., Cox, J. S., and Chawla, A. (2013). Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 341, 1483–1488. doi: 10.1126/science.1240636
Orozco-Solis, R., Aguilar-Arnal, L., Murakami, M., Peruquetti, R., Ramadori, G., Coppari, R., et al. (2016). The circadian clock in the ventromedial hypothalamus controls cyclic energy expenditure. Cell Metab. 23, 467–478. doi: 10.1016/j.cmet.2016.02.003
Paschos, G. K., Ibrahim, S., Song, W. L., Kunieda, T., Grant, G., Reyes, T. M., et al. (2012). Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat. Med. 18, 1768–1777. doi: 10.1038/nm.2979
Paxinos, G., and Franklin, K. B. (2001). The Mouse Brain in Stereotaxic Coordinates. San Diego, CA: Academic Press.
Puelles, L., and Rubenstein, J. L. (2003). Forebrain gene expression domains and the evolving prosomeric model. Trends Neurosci. 26, 469–476. doi: 10.1016/S0166-2236(03)00234-0
Reppert, S. M., and Weaver, D. R. (2002). Coordination of circadian timing in mammals. Nature 418, 935–941. doi: 10.1038/nature00965
Roybal, K., Theobold, D., Graham, A., DiNieri, J. A., Russo, S. J., Krishnan, V., et al. (2007). Mania-like behavior induced by disruption of CLOCK. Proc. Natl. Acad. Sci. U.S.A. 104, 6406–6411. doi: 10.1073/pnas.0609625104
Sadacca, L. A., Lamia, K. A., deLemos, A. S., Blum, B., and Weitz, C. J. (2011). An intrinsic circadian clock of the pancreas is required for normal insulin release and glucose homeostasis in mice. Diabetologia 54, 120–124. doi: 10.1007/s00125-010-1920-8
Saper, C. B., Fuller, P. M., Pedersen, N. P., Lu, J., and Scammell, T. E. (2010). Sleep state switching. Neuron 68, 1023–1042. doi: 10.1016/j.neuron.2010.11.032
Sasaki, K., Suzuki, M., Mieda, M., Tsujino, N., Roth, B., and Sakurai, T. (2011). Pharmacogenetic modulation of orexin neurons alters sleep/wakefulness states in mice. PLoS ONE 6:e20360. doi: 10.1371/journal.pone.0020360
Shimizu, K., Kobayashi, Y., Nakatsuji, E., Yamazaki, M., Shimba, S., Sakimura, K., et al. (2016). SCOP/PHLPP1[β] mediates circadian regulation of long-term recognition memory. Nat. Commun. 7:12926 doi: 10.1038/ncomms12926
Shimogori, T., Lee, D. A., Miranda-Angulo, A., Yang, Y., Wang, H., Jiang, L., et al. (2010). A genomic atlas of mouse hypothalamic development. Nat. Neurosci. 13, 767–775. doi: 10.1038/nn.2545
Soriano, P. (1999). Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21, 70–71. doi: 10.1038/5007
Spencer, S., Falcon, E., Kumar, J., Krishnan, V., Mukherjee, S., Birnbaum, S. G., et al. (2013). Circadian genes Period 1 and Period 2 in the nucleus accumbens regulate anxiety-related behavior. Eur. J. Neurosci. 37, 242–250. doi: 10.1111/ejn.12010
Storch, K. F., Paz, C., Signorovitch, J., Raviola, E., Pawlyk, B., Li, T., et al. (2007). Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell 130, 730–741. doi: 10.1016/j.cell.2007.06.045
Westgate, E. J., Cheng, Y., Reilly, D. F., Price, T. S., Walisser, J. A., Bradfield, C. A., et al. (2008). Genetic components of the circadian clock regulate thrombogenesis in vivo. Circulation 117, 2087–2095. doi: 10.1161/CIRCULATIONAHA.107.739227
Xie, Z., Su, W., Liu, S., Zhao, G., Esser, K., Schroder, E. A., et al. (2015). Smooth-muscle BMAL1 participates in blood pressure circadian rhythm regulation. J. Clin. Invest. 125, 324–336. doi: 10.1172/JCI76881
Xu, C., Ochi, H., Fukuda, T., Sato, S., Sunamura, S., Takarada, T., et al. (2016). Circadian clock regulates bone resorption in mice. J. Bone Miner. Res. 31, 1344–1355. doi: 10.1002/jbmr.2803
Xu, M., Chung, S., Zhang, S., Zhong, P., Ma, C., Chang, W. C., et al. (2015). Basal forebrain circuit for sleep-wake control. Nat. Neurosci. 18, 1641–1647. doi: 10.1038/nn.4143
Yagita, K., Tamanini, F., van Der Horst, G. T., and Okamura, H. (2001). Molecular mechanisms of the biological clock in cultured fibroblasts. Science 292, 278–281. doi: 10.1126/science.1059542
Yamazaki, S., Numano, R., Abe, M., Hida, A., Takahashi, R., Ueda, M., et al. (2000). Resetting central and peripheral circadian oscillators in transgenic rats. Science 288, 682–685. doi: 10.1126/science.288.5466.682
Yang, G., Chen, L., Grant, G. R., Paschos, G., Song, W. L., Musiek, E. S., et al. (2016). Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 8, 324ra316. doi: 10.1126/scitranslmed.aad3305
Yoo, S. H., Yamazaki, S., Lowrey, P. L., Shimomura, K., Ko, C. H., Buhr, E. D., et al. (2004). PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. U.S.A. 101, 5339–5346. doi: 10.1073/pnas.0308709101
Keywords: bmal1, hypothalamus, Nkx2.1, circadian rhythm, sleep-wakefulness regulation, food intake
Citation: Mieda M, Hasegawa E, Kessaris N and Sakurai T (2017) Fine-Tuning Circadian Rhythms: The Importance of Bmal1 Expression in the Ventral Forebrain. Front. Neurosci. 11:55. doi: 10.3389/fnins.2017.00055
Received: 14 October 2016; Accepted: 25 January 2017;
Published: 09 February 2017.
Edited by:
Etienne Challet, CNRS UPR3212, University of Strasbourg, FranceReviewed by:
Andrew Coogan, Swansea University, UKFeillet Celine, University of Nice Sophia Antipolis, France
Copyright © 2017 Mieda, Hasegawa, Kessaris and Sakurai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michihiro Mieda, bWllZGFAbWVkLmthbmF6YXdhLXUuYWMuanA=
†Present Address: Emi Hasegawa and Takeshi Sakurai, Faculty of Medicine/International Institute for Integrative Sleep Medicine (WPI-IIIS)/Life Science Center for Tsukuba Advanced Research Alliance, University of Tsukuba, Tsukuba, Japan
 Michihiro Mieda
Michihiro Mieda Emi Hasegawa
Emi Hasegawa Nicoletta Kessaris
Nicoletta Kessaris Takeshi Sakurai
Takeshi Sakurai