- 1Computational and Evolutionary Biology, Faculty of Life Sciences, The University of Manchester, Manchester, UK
- 2BioScience Project, Wakefield, MA, USA
- 3Institute of Cognitive and Integrative Neuroscience, University of Bordeaux and CNRS, Talence, France
- 4Department of Neurology, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
- 5IFB Adiposity Diseases, Leipzig University Medical Center, Leipzig, Germany
- 6Department of Human Genetics, Donders Institute for Brain, Cognition and Behaviour, Radboud University Medical Center Nijmegen, Nijmegen, Netherlands
- 7German Center for Neurodegenerative Diseases (DZNE), Dresden, Germany
- 8CRTD - Center for Regenerative Therapies Dresden, Genomics of Regeneration, Technische Universität Dresden, Dresden, Germany
- 9Department of Radiology, Center for In Vivo Microscopy, Duke University Medical Center, Durham, NC, USA
Adult neurogenesis, the lifelong production of new neurons in the adult brain, is under complex genetic control but many of the genes involved remain to be identified. In this study, we have integrated publicly available gene expression data from the BXD and CXB recombinant inbred mouse lines to discover genes co-expressed in the adult hippocampus with Nestin, a common marker of the neural precursor cell population. In addition, we incorporated spatial expression information to restrict candidates to genes with high differential gene expression in the hippocampal dentate gyrus. Incorporating data from curated protein-protein interaction databases revealed interactions between our candidate genes and those already known to be involved in adult neurogenesis. Enrichment analysis suggested a link to the Wnt/β-catenin pathway, known to be involved in adult neurogenesis. In particular, our candidates were enriched in targets of Lef1, a modulator of the Wnt pathway. In conclusion, our combination of bioinformatics approaches identified six novel candidate genes involved in adult neurogenesis; Amer3, Eya3, Mtdh, Nr4a3, Polr2a, and Tbkbp1. Further, we propose a role for Lef1 transcriptional control in the regulation of adult hippocampal precursor cell proliferation.
Introduction
In the hippocampal dentate gyrus of many mammalian species, including mice (Kempermann et al., 1997a,b) and humans (Eriksson et al., 1998), there exists a population of neural stem cells that continue to divide and give rise to new granule cell neurons throughout adulthood. The proliferation of these precursor cells is under strong genetic control (Kempermann et al., 1997a, 2006; Kempermann and Gage, 2002); and is modulated by a complex interplay of genetic interactions (Kempermann et al., 2006; Pozniak and Pleasure, 2006; Kempermann, 2011). Although many genes have already been assigned a role in the regulation of proliferation in this system (Overall et al., 2012), many more undoubtedly remain to be identified. In addition, the functional interactions between these genes and their protein products have, in most cases, yet to be established.
Murine genetic reference populations provide excellent tools to address such questions since, firstly, neurogenesis is a well demonstrated phenomenon in mice and, secondly, these sets of strains with fixed, replicable genomes model human genetic complexity and how this influences phenotypes. Recombinant inbred (RI) strain families are genetic reference populations constructed with great experimental control on genotypic variation (Williams et al., 2001), and have been used extensively in quantitative trait loci mapping. Because of their genotypic stability, they provide a platform for phenotype and gene expression data that can be shared between different experimenters, across different time points (Collaborative Cross Consortium, 2012). We identified two such resources for which compatible hippocampal expression data exist (Overall et al., 2009): the BXD RI cross between C57BL/6J and DBA/2J (Taylor, 1978; Taylor et al., 1999; Peirce et al., 2004), and the CXB RI cross between C57BL/6ByJ and BALB/cByJ (Bailey, 1971; Nowakowski, 1984).
We chose to focus on the intermediate filament protein Nestin (Nes), first discovered in neuroepithelial stem cells (Lendahl et al., 1990), and a widely-used marker of the proliferating neural precursor cell population in the adult hippocampal subgranular zone (Reynolds and Weiss, 1992; Yamaguchi et al., 2000). Nestin defines a mixed cell population of proliferating cells, including the type-1 stem cells as well as the type-2a and type-2b transiently amplifying precursor cells (Kempermann et al., 2004). Thus, it would be of great interest to identify related markers which may specify sub-populations to allow more accurate phenotyping of these stages. Also, Nestin, while useful as a marker of precursors, is a structural protein and therefore unlikely to be an upstream modulator of cell fate. It would be of great interest to discover molecules regulating the expression of genes in the precursor cell population. The work presented here has thus examined gene expression profiles correlating with Nestin in two mouse genetic reference panels in order to identify genes potentially regulating proliferation of the neural precursor cell population.
Our results are an example of how a bioinformatics approach, using only information already available in the public domain, can be successfully used to generate novel hypotheses, which can be later tested experimentally at the bench, to help better understand some of the open questions in neurogenomics.
Methods
Correlation with Expression Data
We used GeneNetwork (Chesler et al., 2003, 2005), to gain access to Nestin related traits for BXD and CXB RI families of mice. In the Affymetrix M430v2 microarray platform used (http://www.genenetwork.org/dbdoc/Hippocampus_M430_V2_PDNN_Sept05.html), Nestin is represented by three probesets (1453997_a_at, 1418289_at, 1449022_at). The first principal component of these traits was calculated to create a meta-trait, Nes-PC1. This Nes-PC1 meta-trait was then correlated (Pearson product-moment correlation coefficient) to all probesets in the two hippocampal expression data sets “BXD Hippocampus Consortium M430v2 (Jun06)” normalized by PDNN (GeneNetwork accession: GN112; Overall et al., 2009) and “CXB Hippocampus Consortium M430v2 (Dec05),” normalized by PDNN (GeneNetwork accession: GN99; Overall et al., 2009). The intersection of all probe sets was calculated, using a Pearson product-moment correlation coefficient (r) value greater than 0.8 in both data sets. A correlation coefficient cutoff was used rather than a significance threshold due to the large difference in the number of lines between the two datasets (71 BXD lines, 15 CXB lines). The correlation threshold of 0.8 had a p-value of < 1 × 10 −16 in BXD and ~0.00014 in CXB. Use of a significance value threshold would have resulted in many BXD genes being included which had a very small, but still significant, correlation. The threshold of 0.8 was chosen empirically as a strong correlation, with the intention of selecting for genes highly associated with Nestin. We determined the significance of the intersecting genes by permuting the Nes-PC1 meta-trait data and re-running the analysis 1000 times.
Statistical tests were done using the free and open source software package R (http://www.r-project.org; R Core Team, 2013).
Differential Gene Expression Search Based on the Allen Mouse Brain Atlas
The resulting gene list found to correlate with the Nes-PC1 meta-trait for both BXD and CXB families was further constrained based on in situ hybridization data registered to a common anatomical atlas, available from the Allen Institute for brain sciences (Lein et al., 2007; Allen Institute for Brain Science, 2014; http://www.brain-map.org). These resources allow one to single out genes enriched in areas of interest that are defined in the anatomical atlas. Using a python script, courtesy of Dr. David Feng, we queried the Allen Brain Atlas database to generate a list of possible candidates by examining the differential gene expression patterns for the dentate gyrus, contrasted against the whole gray matter, thresholding the results at a minimum of 2-fold expression enrichment, a threshold that is comparable with values in the literature (Tusher et al., 2001). This produced a list of genes with enhanced expression in the hippocampal dentate gyrus.
Enrichment Analysis of Candidate Genes
Our candidate genes were investigated using WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt; Zhang et al., 2005; Wang et al., 2013) for enrichment in GeneOntology (GO), KEGG pathways, wikipathways and transcription factor targets. This allowed us to find commonality between our candidate genes. The whole mouse genome was used as the reference set and the Benjamini and Hochberg (1995) method was used to correct for multiple tests.
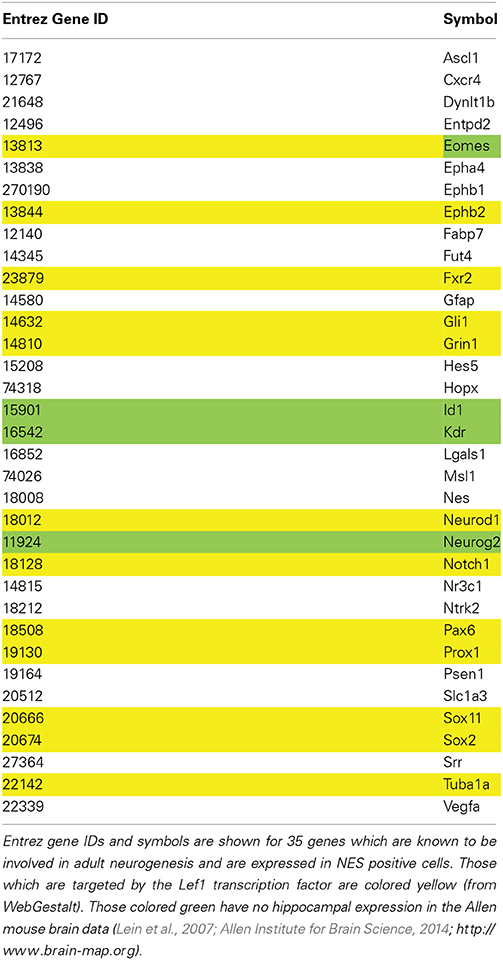
The Mammalian Adult Neurogenesis Gene Ontology, MANGO, is a database of genes known to be involved in adult hippocampal neurogenesis (http://mango.adult-neurogenesis.de; Overall et al., 2012). The MANGO API was used to produce a subset of genes which are expressed in type-1, -2a and -2b cells, i.e., Nestin-positive cells (http://mango.adult-neurogenesis.de/xml/annotations?process=expression&cellstage=t1,t2a,t2b&effect=positive&expression=true).
Predicted and known Interactions with Genes known to be Involved in Adult Neurogenesis
GeneMANIA (http://genemania.org; Mostafavi et al., 2008; Warde-Farley et al., 2010) was used to investigate known and predicted interactions, including protein and genetic interactions, pathways, co-expression, co-localization and protein domain similarity. Our candidate genes, genes known to be expressed in Nestin positive cells and our candidate regulator Lef1 were submitted to the website and a summary network created. Default settings were used.
A summary figure for the role of Lef1 was created using Cytoscape (http://www.cytoscape.org; Saito et al., 2012; Su et al., 2014), incorporating the above transcription factor target and interaction data. Further, IntAct was used to find additional known protein-protein interactions (www.ebi.ac.uk/intact; Orchard et al., 2014).
Correlation Analysis in the Hippocampus of RI Strains
To support links found through protein-protein interactions and enrichment analysis, correlations were carried out between our candidate genes in the BXD and CXB hippocampus microarray data. The probes identified above via the Nes-PC1 meta-trait were correlated against two probes for Lef1 expression (1445568_at and 1454734_at) using the built in functionality of GeneNetwork.
All online analyses and database queries were verified on 24 October 2014.
Results
A Common set of Genes is Associated with Hippocampal Nestin Expression in Different Genetic Reference Populations
Hippocampal expression has been measured previously in two genetic reference populations, BXD and CXB RI strain families, as part of a single experiment (Overall et al., 2009), meaning that the array platform and hybridization handling was common to both. We generated a meta-trait, Nes-PC1, based on the first principal component of the expression profiles for the three probesets targeting Nestin (Figure 1A). For each RI population, we correlated Nes-PC1 against all probesets on the array and selected strong correlations of r > 0.8 (Figure 1B). Of the resulting 450 probesets in BXD and 955 probesets in CXB, 144 probesets (excluding those for Nes itself) were common to both sets (Figure 1C; Supplementary Table 1). No overlapping gene sets of 144 or more members were observed after permutation testing of Nes-PC1 data, with 1000 such permutations. This indicates that the association of these genes with Nes is not due simply to chance.
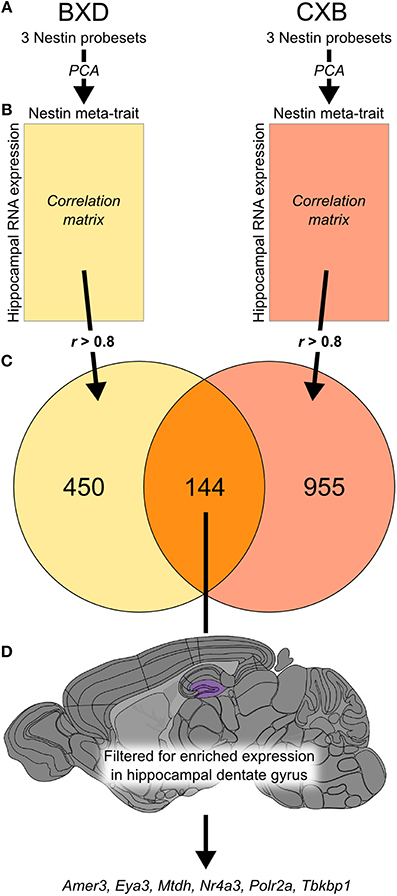
Figure 1. The integration of data from two genetic reference populations revealed a set of 144 genes potentially expressed in adult hippocampal neural precursor cells. Principal component analysis was carried out using three probesets for Nes expression (1453997_a_at, 1418289_at, 1449022_at) in both the BXD and CXB genetic reference populations (A). The first principal component of these traits was used to create a meta-trait, Nes-PC1. This meta-trait was then correlated to all probesets in the two hippocampal expression data sets for the two genetic reference populations (B). The intersection of all probesets (r-value greater than 0.8 in both data sets) revealed 144 probesets in common (C). These genes were then filtered based on enriched expression in the dentate gyrus (highlighted in purple) using the Allen Brain Atlas to yield a subset of 6 candidate genes most likely to be expressed in neural precursor cells (D).
A Subset of Nestin Correlates are Enriched in the Hippocampal Neurogenic Niche
A differential search was performed using the Allen Brain Atlas Resource which produced a list of 2472 genes with enhanced expression in the hippocampal dentate gyrus, contrasted against the whole gray matter of the brain. This was used to narrow the list of 144 genes obtained through correlation with our Nes-PC1 meta-trait down to six candidate genes (Figure 1D). These candidates, correlating with Nes and enriched in the dentate gyrus, are therefore hypothesized to be involved in adult hippocampal neurogenesis. The six candidates are: Amer3 (Fam123c), Eya3, Mtdh, Nr4a3, Polr2a, and Tbkbp1. The ISH data and expression intensity for these 6 genes are presented in Figure 2.
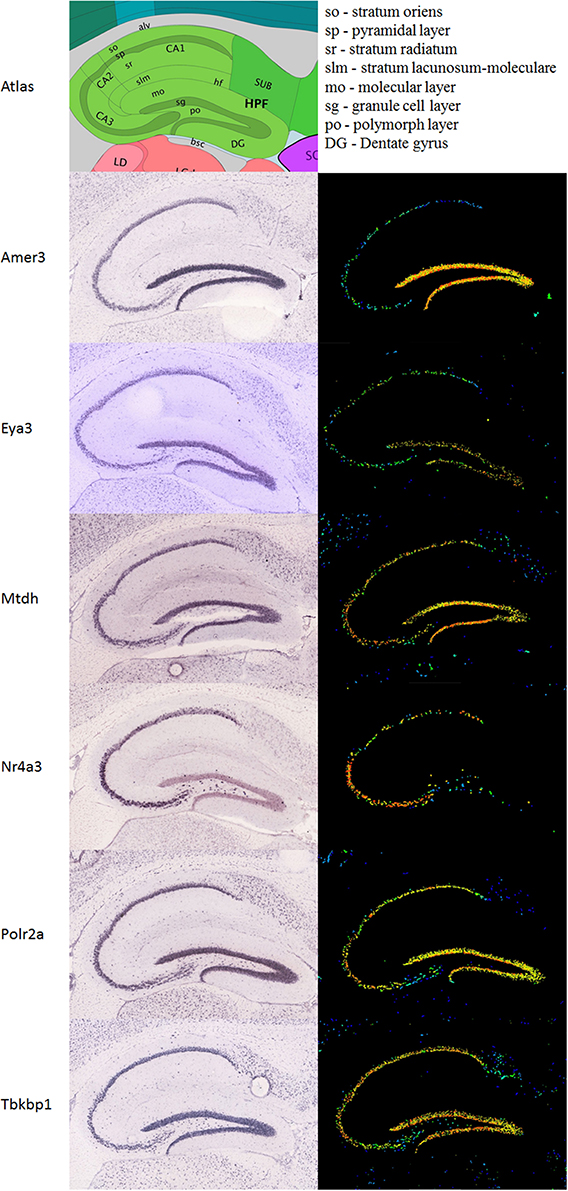
Figure 2. Expression of the six candidate genes in the hippocampus. Immunohistochemistry (ISH) is presented in the left column, the expression in the same sagittal slices in the right column. An atlas is shown above to help orientation. All images are taken from the Allen Mouse brain data (Lein et al., 2007; Allen Institute for Brain Science, 2014; http://www.brain-map.org).
Enrichment Analysis Suggest Lef1 as a Common Regulator in Neural Precursor Cells
Enrichment analysis was carried out using WebGestalt for our six candidate genes, Amer3, Eya3, Mtdh, Nr4a3, Polr2a, and Tbkbp1. There is significant enrichment for several GeneOntology terms related to transcription (Supplementary Table 2). For example “transcription, DNA-dependent,” was significantly enriched (Benjamini and Hochberg adjusted p-value; adjP = 0.0395) due to Nr4a3, Eya3, Mtdh, and Polr2a. In addition, the six candidate genes were enriched (adjP = 0.0001) for the transcription factor Lef1, which targets Nr4a3, Eya3, Amer3, and Polr2a (Table 1).
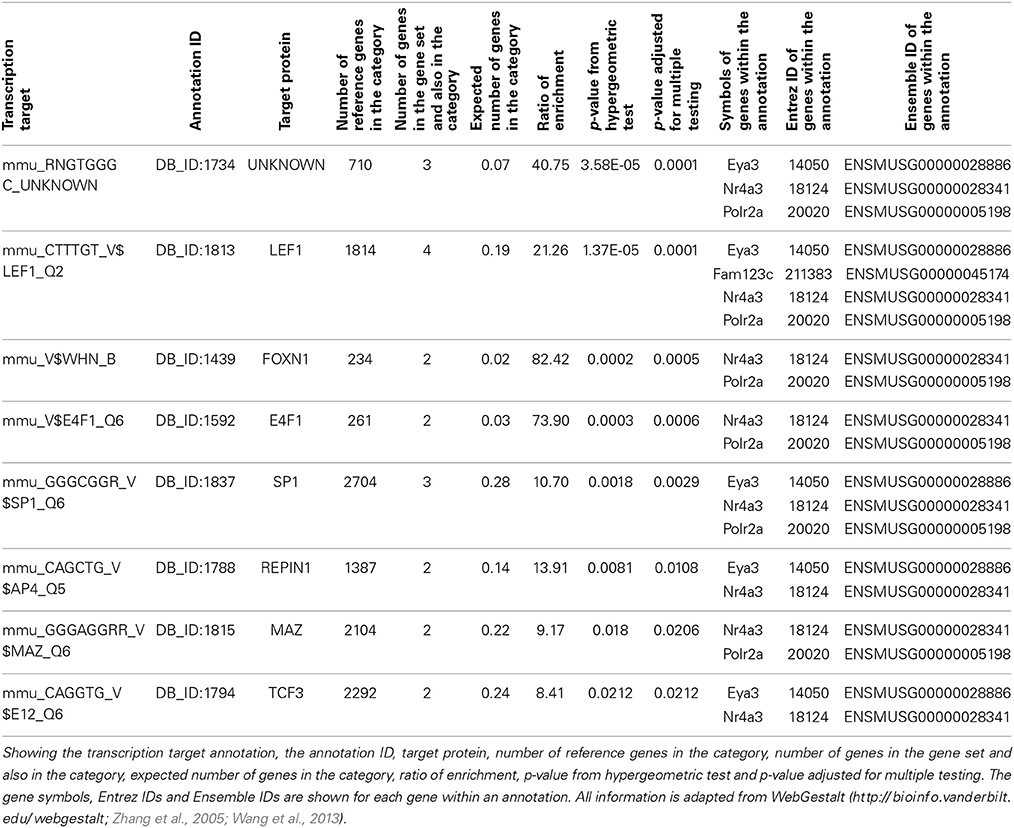
Table 1. Transcription factor target annotation enrichment from WebGestalt for our six candidate genes, Amer3 (Fam123c), Eya3, Mtdh, Nr4a3, Polr2a, and Tbkbp1.
To investigate if this link to Lef1 is common to many adult neurogenesis genes, or just our candidates, a list of genes expressed in Nestin-positive cell stages was retrieved from the MANGO database and was tested for enrichment in transcription factor targets using WebGestalt. This also revealed a significant enrichment for targets of Lef1 (12 / 35 genes, adjP = 2.44 × 10−8).
These enrichment analyses suggest a possible novel role of Lef1 as a key transcriptional regulator in proliferating neural precursor cells (Figure 3).
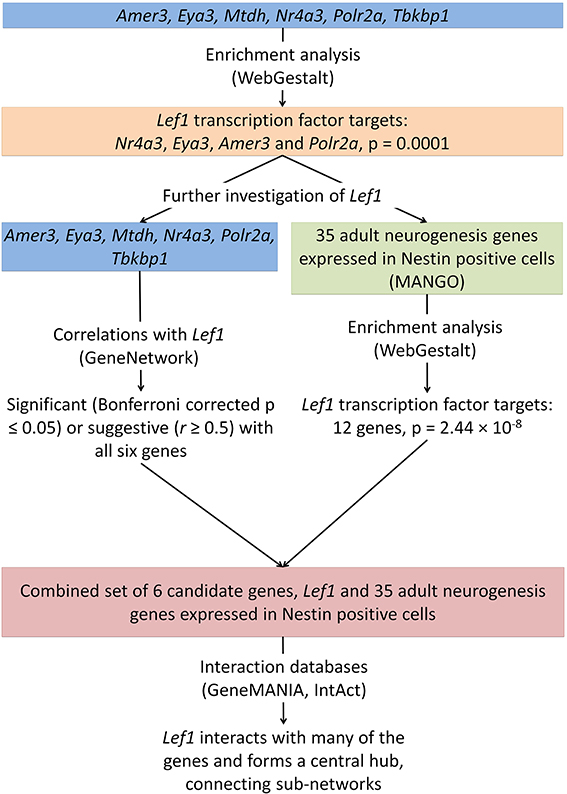
Figure 3. The integration of data from several online tools identifies a novel role for Lef1 in adult hippocampal neurogenesis. Enrichment analysis was carried out on the six candidate genes identified (Figure 1) and for targets of the transcription factor Lef1. This was further investigated using enrichment of genes known to be expressed in Nestin positive cells, correlation analysis between our candidate genes and Lef1, and the use of interaction databases to find links between our candidates.
Hippocampal Expression Patterns of Lef1
To investigate if expression levels of Lef1 correlate with expression of our candidate genes, correlation matrices were produced for the BXD and CXB strains. This shows suggestive correlations between our candidate genes and Lef1 (Table 2). Although the correlation is generally greater (larger r-values) in the CXB set compared to the BXD, the significance is lower (larger p-values), due to far fewer lines being used (n = 71 vs. n = 15). However, in both sets, these correlations are not as strong as the correlations between our candidates and Nes. This suggests that Lef1 is only one of several factors influencing the expression level of our genes of interest.
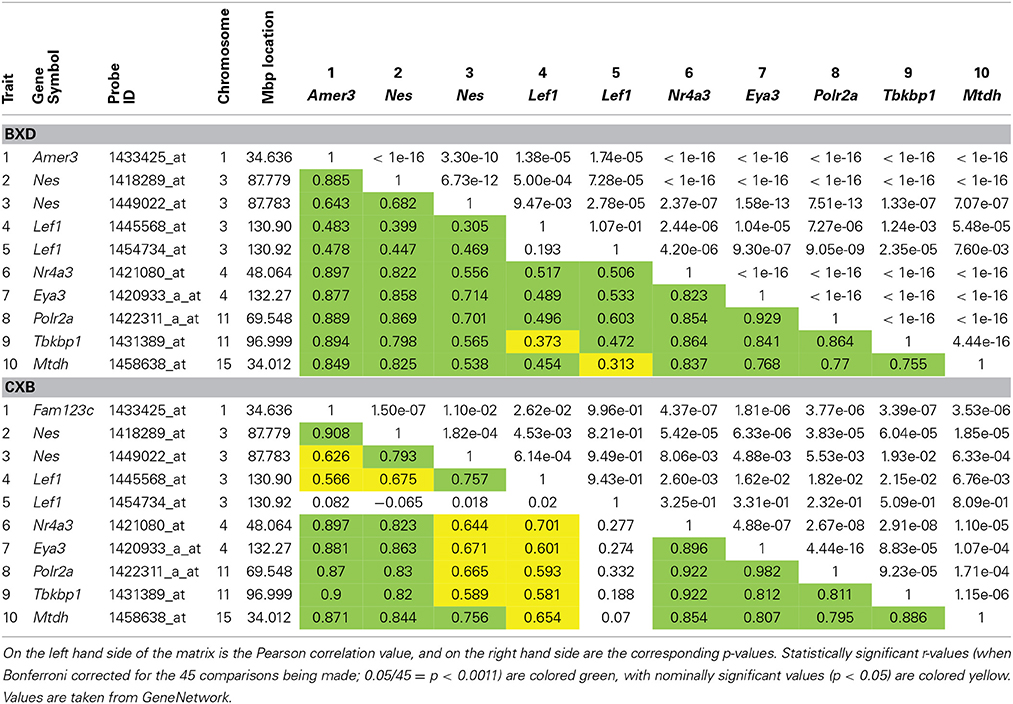
Table 2. Correlation matrices for hippocampal expression of our six candidate genes, Nes and Lef1 in both BXD and CXB genetic reference populations.
The Allen Brain Map showed very little expression of Lef1 in the hippocampus. However, only a small subset of hippocampal cells are proliferative, and Lef1 may only be functioning at one stage in the cell cycle. To investigate whether a strong hippocampal in situ signal is a prerequisite for involvement in neurogenesis, a list of 35 genes from the MANGO database which are known to be expressed in Nestin-positive cells (Table 3) were examined in the Allen Mouse Brain Atlas for hippocampal expression. This showed that four genes known to be involved in adult hippocampal neurogenesis; Id1 (Nam and Benezra, 2009), Neurog2 (Ozen et al., 2007; Roybon et al., 2009), Eomes (Hodge et al., 2008; Azim et al., 2013) and Kdr (Cao et al., 2004; Warner-Schmidt and Duman, 2007; Segi-Nishida et al., 2008; Bernal and Peterson, 2011) all had low expression levels, similar to Lef1. Indeed Nestin itself appears in the Allen Brain Atlas to be poorly expressed, despite it being the dominant marker of the proliferating cell population. This indicates that although high transcript expression in the hippocampus might be supportive of a role in adult neurogenesis, it is certainly not necessary.
Predicted and known Interactions with Genes Involved In Adult Neurogenesis
Our method, outlined above, allowed the identification of six novel candidate genes as being associated with adult hippocampal neurogenesis, and revealed that Lef1 acts as a central hub between these genes. We employed GeneMANIA to predict interactions between the 35 MANGO genes expressed in Nestin positive cells, the six candidate genes and Lef1. This showed numerous interactions among all of the submitted genes (Figure 4A). In particular we saw a large number of interactions between Lef1 and the 35 genes expressed in Nestin-positive cells (Figure 4B). In contrast, Amer3, which also appeared to be a good candidate, showed few interactions (Figure 4C), and none when only physical and predicted physical interactions are used whereas, when only considering the same interactions, the network surrounding Lef1 was essentially unchanged (Figure 4D).
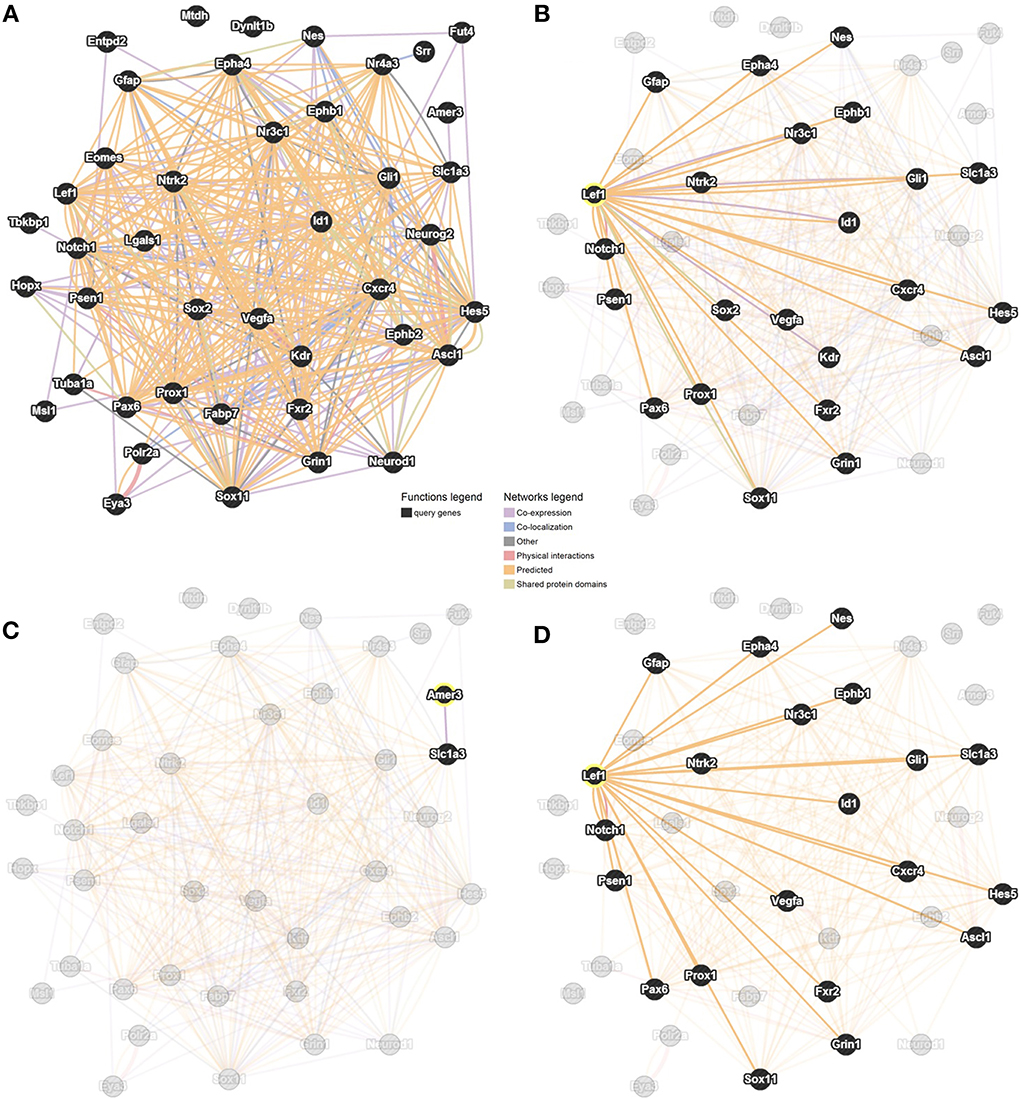
Figure 4. Interaction network between the six candidate genes, Lef1 and 35 MANGO genes expressed in Nestin-positive cells. Adapted from GeneMANIA (http://genemania.org; Mostafavi et al., 2008; Warde-Farley et al., 2010) with links based on co-expression, co-localization, physical and genetic interactions, as well as shared protein domains. The network is shown with all genes and all interactions highlighted (A), only genes with a connection to Lef1 highlighted (B), only genes with a connection to Amer3 highlighted (C), or highlighting restricted to genes with a physical, or predicted physical, interaction with Lef1 (D).
Finally, a network was created incorporating all our evidence for Lef1 as a candidate regulator, including transcription factor targets from WebGestalt and physical interactions from GeneMANIA, as well as additional known protein-protein interactions from IntAct (Figure 5). This shows that Lef1 not only targets many of our genes, but that it connects subsets of physically interacting genes together.
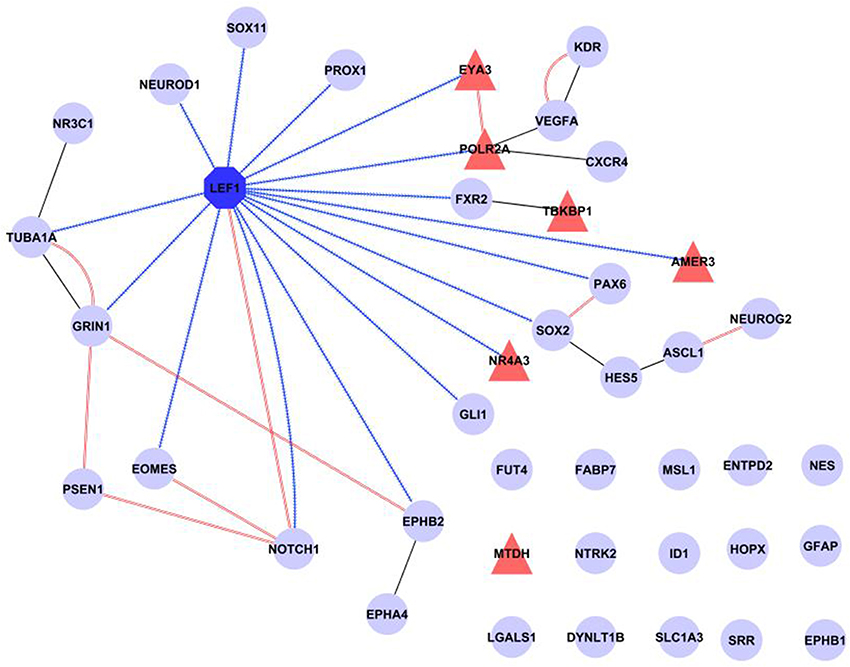
Figure 5. Summary of known interactions shows a central role for Lef1. The transcription factor Lef1 (dark blue octagon) is central in a network comprised of our six candidate genes (red triangles) and 35 genes expressed in Nestin positive cells (pale blue circles). Transcription factor targets (from WebGestalt) are shown as contiguous blue arrows, protein-protein interactions (from IntAct) as black lines and physical interactions (from GeneMANIA) as double red lines. Figure produced in Cytoscape.
Taken together, the results presented above support the hypothesis that Lef1 plays a gene regulatory role in type-1 and type-2 Nestin-positive cells in the adult hippocampal dentate gyrus.
Discussion
Adult neurogenesis occurs in several mammalian species, although the rate of precursor proliferation and new neuron production vary considerably between species (Kempermann, 2012). It has been widely studied in the mouse, where strain differences are present as well (Kempermann et al., 1997a, 2006; Hayes and Nowakowski, 2002; Clark et al., 2011; Poon and Goldowitz, 2014). Anatomical regions known to support adult neurogenesis are the subventricular zone and the subgranular zone of the hippocampal dentate gyrus, which is the focus of our study. Our understanding of neurogenesis regulatory networks and the functional interactions between genes expressed in this area and their protein products is limited.
Our approach consisted of finding genes that: (a) correlate strongly with Nes gene expression, a defining protein of hippocampal stem cells (Lagace et al., 2007), in two recombinant inbred mouse lines, BXD and CXB; and (b) have enhanced expression in the hippocampal dentate gyrus. In this way, we aimed to leverage existing, publicly available, in silico data to search for novel markers of the Nestin-positive precursor cell population.
The differential search option in the Allen Brain Science Institute's database allowed the identification of enhanced gene expression in a specific brain region. However, data for the subgranular zone, which is located in the immediate vicinity of the granule cell layer, and is only 2–3 cells thick, is not available at the present time. Therefore, the entire dentate gyrus was used as a proxy and compared to the whole brain gray matter. Because the subgranular zone (SGZ) is a thin band of cells located at the boundary of granule cell layer, small inaccuracies in boundary definition or spatial normalization could severely affect the analysis results. Therefore, we decided upon using the whole dentate gyrus as our best candidate for differential search.
Amer3 has a well-defined enhanced expression in the granule cell layer of the hippocampus, and is a particularly good candidate since it has already been linked to embryonic neurogenesis (Comai et al., 2010). Furthermore, it is an Apc membrane recruitment protein, and Apc has already been linked to adult neurogenesis (Imura et al., 2010). Apc inhibits the Wnt signaling pathway via degradation of β-catenin, whereas Amer3 enhances the expression of a β-catenin (Brauburger et al., 2014), suggesting they have antagonistic roles in this important pathway within neurogenesis (Varela-Nallar and Inestrosa, 2013). Amer3 also binds to the Wnt pathway regulator Conductin/Axin2, and unlike Amer1 and Amer2 has been shown to be a positive regulator of the Wnt-β catenin signaling (Brauburger et al., 2014). This suggests that Amer3 is indeed a good candidate for a novel molecular marker of this important cell population.
The reason that we found only a small set of genes using the differential search of Allen Brain Atlas resources is probably due to larger regions being used for our analysis (i.e., the whole hippocampus for Nes correlation and the dentate gyrus of the hippocampus for the differential gene expression search), as small but significant changes within the subgranular cell layer may be obscured by signals from other cell populations. Further refinements of the atlas and availability of a more spatially refined expression data set would likely yield a greater overlap between these two approaches.
Our protein-protein interaction and enrichment analysis revealed that four out of our six candidate genes are targeted by the Lef1 transcription factor, and the remaining two have close relations to it. Lef1 is a downstream effector of the Wnt/β-catenin pathway (Mazumdar et al., 2010), which is important in adult neurogenesis (Machon et al., 2003; Lie et al., 2005; Kuwabara et al., 2009; Wexler et al., 2009; Varela-Nallar and Inestrosa, 2013; Wisniewska, 2013). Lef1 is expressed in cultured hippocampal neural stem cells in response to activation of the Wnt signaling pathway (Cui et al., 2011). Our evidence and the literature both suggest that genes known to be involved in hippocampal adult neurogenesis are targets of Lef1, an important factor in generating granule cells in the dentate gyrus during development (Galceran et al., 2000). The only two genes not targeted by Lef1 can be closely associated with it: Mtdh regulates the expression of Lef1 (Hu et al., 2009; Yoo et al., 2009), and Tbkbp1 physically interacts with a known adult neurogenesis proteins Fxr2, which is expressed in Nestin positive cells and is a Lef1 target (Figure 5).
Combined with the established role of the Wnt signaling pathway in adult hippocampal neurogenesis (Lie et al., 2005) together with the known association between Lef1 and β-catenin (Behrens et al., 1996), our results here suggest that Lef1 is an important part of the Wnt-controlled regulation of neural precursor function in the adult dentate gyrus.
While our approach has limitations, such as relying on one single marker of proliferation, and on spatial proxies for the SGZ, we believe that it proposes a novel method. By repeating a similar procedure, as we followed using Nestin, and then imposing spatial constraints for other markers associated with neurogenesis (e.g., Sox2, Prox1, NeuN, or Dcx), one might expect to produce larger sets of gene candidates and better understand their roles in the various stages of neurogenesis. It is our hope that this can lead to a better understanding of adult neurogenesis and its relationship with its developmental counterpart, as well as give additional insight into the functional relevance of a process that has been demonstrated in several mammalian species (Amrein et al., 2011; Kempermann, 2012; Patzke et al., 2013).
In this study we demonstrate a strategy for finding novel candidate genes linked to adult neurogenesis in the murine hippocampus, using data available in the public domain. By integrating data from several public resources, this method presents an avenue for generating novel hypotheses in silico, and potential gene networks which can be tested in the future in vitro or in vivo.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the International Neuroscience Coordinating Facility (INCF) and the Center for Integrative and Translational Genomics of the University of Tennessee Health Science Center, Memphis, TN, USA for funding the INCF Short Course on Neuroinformatics, Neurogenomics and Brain Disease, which brought us together for this project. We are grateful to Dr. David Feng for his insight into querying the Allen Brain Atlas database, and to Dr. Richard Nowakowski for support in the initial stages of the project. Alexandra Badea acknowledges funding from NIA, through K01 AG041211, and the Center for In Vivo Microscopy, supported by NIBIB through P41 EB015897.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnins.2014.00418/abstract
References
Allen Institute for Brain Science. (2014). Allen Mouse Brain Atlas [Internet]. Available online at: http://mouse.brain-map.org/
Amrein, I., Isler, K., and Lipp, H.-P. (2011). Comparing adult hippocampal neurogenesis in mammalian species and orders: influence of chronological age and life history stage. Eur. J. Neurosci. 34, 978–987. doi: 10.1111/j.1460-9568.2011.07804.x
Azim, K., Zweifel, S., Klaus, F., Yoshikawa, K., Amrein, I., and Raineteau, O. (2013). Early decline in progenitor diversity in the marmoset lateral ventricle. Cereb. Cortex 23, 922–931. doi: 10.1093/cercor/bhs085
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bailey, D. W. (1971). Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11, 325–327. doi: 10.1097/00007890-197103000-00013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Behrens, J., von Kries, J. P., Kühl, M., Bruhn, L., Wedlich, D., Grosschedl, R., et al. (1996). Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382, 638–642. doi: 10.1038/382638a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Bernal, G. M., and Peterson, D. A. (2011). Phenotypic and gene expression modification with normal brain aging in GFAP-positive astrocytes and neural stem cells. Aging Cell 10, 466–482. doi: 10.1111/j.1474-9726.2011.00694.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brauburger, K., Akyildiz, S., Ruppert, J. G., Graeb, M., Bernkopf, D. B., Hadjihannas, M. V., et al. (2014). Adenomatous polyposis coli (APC) membrane recruitment 3, a member of the APC membrane recruitment family of APC-binding proteins, is a positive regulator of Wnt-β-catenin signalling. FEBS J. 281, 787–801. doi: 10.1111/febs.12624
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cao, L., Jiao, X., Zuzga, D. S., Liu, Y., Fong, D. M., Young, D., et al. (2004). VEGF links hippocampal activity with neurogenesis, learning and memory. Nat. Genet. 36, 827–835. doi: 10.1038/ng1395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chesler, E. J., Lu, L., Shou, S., Qu, Y., Gu, J., Wang, J., et al. (2005). Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37, 233–242. doi: 10.1038/ng1518
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chesler, E. J., Wang, J., Lu, L., Qu, Y., Manly, K. F., and Williams, R. W. (2003). Genetic correlates of gene expression in recombinant inbred strains: a relational model system to explore neurobehavioral phenotypes. Neuroinformatics 1, 343–357. doi: 10.1385/NI:1:4:343
Clark, P. J., Kohman, R. A., Miller, D. S., Bhattacharya, T. K., Brzezinska, W. J., and Rhodes, J. S. (2011). Genetic influences on exercise-induced adult hippocampal neurogenesis across 12 divergent mouse strains. Genes. Brain. Behav. 10, 345–353. doi: 10.1111/j.1601-183X.2010.00674.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Collaborative Cross Consortium. (2012). The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190, 389–401. doi: 10.1534/genetics.111.132639
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Comai, G., Boutet, A., Neirijnck, Y., and Schedl, A. (2010). Expression patterns of the Wtx/Amer gene family during mouse embryonic development. Dev. Dyn. 239, 1867–1878. doi: 10.1002/dvdy.22313
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cui, X.-P., Xing, Y., Chen, J.-M., Dong, S.-W., Ying, D.-J., and Yew, D. T. (2011). Wnt/beta-catenin is involved in the proliferation of hippocampal neural stem cells induced by hypoxia. Ir. J. Med. Sci. 180, 387–393. doi: 10.1007/s11845-010-0566-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eriksson, P. S., Perfilieva, E., Björk-Eriksson, T., Alborn, A. M., Nordborg, C., Peterson, D. A., et al. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4, 1313–1317. doi: 10.1038/3305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Galceran, J., Miyashita-Lin, E. M., Devaney, E., Rubenstein, J. L., and Grosschedl, R. (2000). Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127, 469–482.
Hayes, N. L., and Nowakowski, R. S. (2002). Dynamics of cell proliferation in the adult dentate gyrus of two inbred strains of mice. Brain Res. Dev. Brain Res. 134, 77–85. doi: 10.1016/S0165-3806(01)00324-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hodge, R. D., Kowalczyk, T. D., Wolf, S. A., Encinas, J. M., Rippey, C., Enikolopov, G., et al. (2008). Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J. Neurosci. 28, 3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hu, G., Wei, Y., and Kang, Y. (2009). The multifaceted role of MTDH/AEG-1 in cancer progression. Clin. Cancer Res. 15, 5615–5620. doi: 10.1158/1078-0432.CCR-09-0049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Imura, T., Wang, X., Noda, T., Sofroniew, M. V., and Fushiki, S. (2010). Adenomatous polyposis coli is essential for both neuronal differentiation and maintenance of adult neural stem cells in subventricular zone and hippocampus. Stem Cells 28, 2053–2064. doi: 10.1002/stem.524
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G. (2011). Seven principles in the regulation of adult neurogenesis. Eur. J. Neurosci. 33, 1018–1024. doi: 10.1111/j.1460-9568.2011.07599.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G. (2012). New neurons for “survival of the fittest.” Nat. Rev. Neurosci. 13, 727–736. doi: 10.1038/nrn3319
Kempermann, G., Chesler, E. J., Lu, L., Williams, R. W., and Gage, F. H. (2006). Natural variation and genetic covariance in adult hippocampal neurogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 780–785. doi: 10.1073/pnas.0510291103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G., and Gage, F. H. (2002). Genetic influence on phenotypic differentiation in adult hippocampal neurogenesis. Brain Res. Dev. Brain Res. 134, 1–12. doi: 10.1016/S0165-3806(01)00224-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G., Jessberger, S., Steiner, B., and Kronenberg, G. (2004). Milestones of neuronal development in the adult hippocampus. Trends Neurosci. 27, 447–452. doi: 10.1016/j.tins.2004.05.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G., Kuhn, H. G., and Gage, F. H. (1997a). Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc. Natl. Acad. Sci. U.S.A. 94, 10409–10414. doi: 10.1073/pnas.94.19.10409
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kempermann, G., Kuhn, H. G., and Gage, F. H. (1997b). More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495. doi: 10.1038/386493a0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuwabara, T., Hsieh, J., Muotri, A., Yeo, G., Warashina, M., Lie, D. C., et al. (2009). Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 12, 1097–1105. doi: 10.1038/nn.2360
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lagace, D. C., Whitman, M. C., Noonan, M. A., Ables, J. L., DeCarolis, N. A., Arguello, A. A., et al. (2007). Dynamic contribution of nestin-expressing stem cells to adult neurogenesis. J. Neurosci. 27, 12623–12629. doi: 10.1523/JNEUROSCI.3812-07.2007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lein, E. S., Hawrylycz, M. J., Ao, N., Ayres, M., Bensinger, A., Bernard, A., et al. (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176. doi: 10.1038/nature05453
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lendahl, U., Zimmerman, L. B., and McKay, R. D. (1990). CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595. doi: 10.1016/0092-8674(90)90662-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lie, D.-C., Colamarino, S. A., Song, H.-J., Désiré, L., Mira, H., Consiglio, A., et al. (2005). Wnt signalling regulates adult hippocampal neurogenesis. Nature 437, 1370–1375. doi: 10.1038/nature04108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Machon, O., van den Bout, C. J., Backman, M., Kemler, R., and Krauss, S. (2003). Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience 122, 129–143. doi: 10.1016/S0306-4522(03)00519-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mazumdar, J., O'Brien, W. T., Johnson, R. S., LaManna, J. C., Chavez, J. C., Klein, P. S., et al. (2010). O2 regulates stem cells through Wnt/β-catenin signalling. Nat. Cell Biol. 12, 1007–1013. doi: 10.1038/ncb2102
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mostafavi, S., Ray, D., Warde-Farley, D., Grouios, C., and Morris, Q. (2008). GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9(Suppl. 1), S4. doi: 10.1186/gb-2008-9-s1-s4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nam, H., and Benezra, R. (2009). High levels of Id1 expression define B1 type adult neural stem cells. Cell Stem Cell 5, 515–526. doi: 10.1016/j.stem.2009.08.017
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nowakowski, R. S. (1984). The mode of inheritance of a defect in lamination in the hippocampus of BALB/c mice. J. Neurogenet. 1, 249–258. doi: 10.3109/01677068409107090
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Orchard, S., Ammari, M., Aranda, B., Breuza, L., Briganti, L., Broackes-Carter, F., et al. (2014). The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363. doi: 10.1093/nar/gkt1115
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Overall, R. W., Kempermann, G., Peirce, J., Lu, L., Goldowitz, D., Gage, F. H., et al. (2009). Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front. Neurosci. 3:55. doi: 10.3389/neuro.15.003.2009
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Overall, R. W., Paszkowski-Rogacz, M., and Kempermann, G. (2012). The mammalian adult neurogenesis gene ontology (MANGO) provides a structural framework for published information on genes regulating adult hippocampal neurogenesis. PLoS ONE 7:e48527. doi: 10.1371/journal.pone.0048527
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ozen, I., Galichet, C., Watts, C., Parras, C., Guillemot, F., and Raineteau, O. (2007). Proliferating neuronal progenitors in the postnatal hippocampus transiently express the proneural gene Ngn2. Eur. J. Neurosci. 25, 2591–2603. doi: 10.1111/j.1460-9568.2007.05541.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patzke, N., Spocter, M. A., Karlsson, K. A., Bertelsen, M. F., Haagensen, M., Chawana, R., et al. (2013). In contrast to many other mammals, cetaceans have relatively small hippocampi that appear to lack adult neurogenesis. Brain Struct. Funct. doi: 10.1007/s00429-013-0660-1. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Peirce, J. L., Lu, L., Gu, J., Silver, L. M., and Williams, R. W. (2004). A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 5:7. doi: 10.1186/1471-2156-5-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poon, A., and Goldowitz, D. (2014). Identification of genetic loci that modulate cell proliferation in the adult rostral migratory stream using the expanded panel of BXD mice. BMC Genomics 15:206. doi: 10.1186/1471-2164-15-206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pozniak, C. D., and Pleasure, S. J. (2006). Genetic control of hippocampal neurogenesis. Genome Biol. 7:207. doi: 10.1186/gb-2006-7-3-207
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
R Core Team. (2013). R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. Available online at: http://www.r-project.org/
Reynolds, B. A., and Weiss, S. (1992). Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710. doi: 10.1126/science.1553558
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Roybon, L., Hjalt, T., Stott, S., Guillemot, F., Li, J.-Y., and Brundin, P. (2009). Neurogenin2 directs granule neuroblast production and amplification while NeuroD1 specifies neuronal fate during hippocampal neurogenesis. PLoS ONE 4:e4779. doi: 10.1371/journal.pone.0004779
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076. doi: 10.1038/nmeth.2212
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Segi-Nishida, E., Warner-Schmidt, J. L., and Duman, R. S. (2008). Electroconvulsive seizure and VEGF increase the proliferation of neural stem-like cells in rat hippocampus. Proc. Natl. Acad. Sci. U.S.A. 105, 11352–11357. doi: 10.1073/pnas.0710858105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Su, G., Morris, J. H., Demchak, B., and Bader, G. D. (2014). Biological network exploration with cytoscape 3. Curr. Protoc. Bioinformat. 47, 8.13.1–8.13.24. doi: 10.1002/0471250953.bi0813s47
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Taylor, B. A. (1978). “Recombinant inbred strains: use in genetic mapping,” in Origins of Inbred Mice, ed H. Morse (New York, NY: Academic Press), 423–438.
Taylor, B. A., Wnek, C., Kotlus, B. S., Roemer, N., MacTaggart, T., and Phillips, S. J. (1999). Genotyping new BXD recombinant inbred mouse strains and comparison of BXD and consensus maps. Mamm. Genome 10, 335–348. doi: 10.1007/s003359900998
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tusher, V. G., Tibshirani, R., and Chu, G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121. doi: 10.1073/pnas.091062498
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varela-Nallar, L., and Inestrosa, N. C. (2013). Wnt signaling in the regulation of adult hippocampal neurogenesis. Front. Cell. Neurosci. 7:100. doi: 10.3389/fncel.2013.00100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, J., Duncan, D., Shi, Z., and Zhang, B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. doi: 10.1093/nar/gkt439
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–W220. doi: 10.1093/nar/gkq537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warner-Schmidt, J. L., and Duman, R. S. (2007). VEGF is an essential mediator of the neurogenic and behavioral actions of antidepressants. Proc. Natl. Acad. Sci. U.S.A. 104, 4647–4652. doi: 10.1073/pnas.0610282104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wexler, E. M., Paucer, A., Kornblum, H. I., Palmer, T. D., Plamer, T. D., and Geschwind, D. H. (2009). Endogenous Wnt signaling maintains neural progenitor cell potency. Stem Cells 27, 1130–1141. doi: 10.1002/stem.36
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, R. W., Gu, J., Qi, S., and Lu, L. (2001). The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol. 2, research0046–research0046.18. doi: 10.1186/gb-2001-2-11-research0046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wisniewska, M. B. (2013). Physiological role of β-Catenin/TCF signaling in neurons of the adult brain. Neurochem. Res. 38, 1144–1155. doi: 10.1007/s11064-013-0980-9
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamaguchi, M., Saito, H., Suzuki, M., and Mori, K. (2000). Visualization of neurogenesis in the central nervous system using nestin promoter-GFP transgenic mice. Neuroreport 11, 1991–1996. doi: 10.1097/00001756-200006260-00037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yoo, B. K., Emdad, L., Su, Z., Villanueva, A., Chiang, D. Y., Mukhopadhyay, N. D., et al. (2009). Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J. Clin. Invest. 119, 465–477. doi: 10.1172/JCI36460
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhang, B., Kirov, S., and Snoddy, J. (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748. doi: 10.1093/nar/gki475
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: adult neurogenesis, BXD, CXB, neuroinformatics, recombinant inbred mice, systems genetics, Lef1, Wnt pathway
Citation: Ashbrook DG, Delprato A, Grellmann C, Klein M, Wetzel R, Overall RW and Badea A (2014) Transcript co-variance with Nestin in two mouse genetic reference populations identifies Lef1 as a novel candidate regulator of neural precursor cell proliferation in the adult hippocampus. Front. Neurosci. 8:418. doi: 10.3389/fnins.2014.00418
Received: 19 September 2014; Accepted: 27 November 2014;
Published online: 12 December 2014.
Edited by:
Robert W. Williams, University of Tennessee Health Science Center, USAReviewed by:
Sulev Kõks, University of Tartu, EstoniaCamron D. Bryant, Boston University School of Medicine, USA
Susan E. Bergeson, Texas Tech University Health Sciences Center, USA
Copyright © 2014 Ashbrook, Delprato, Grellmann, Klein, Wetzel, Overall and Badea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra Badea, Department of Radiology, Center for In Vivo Microscopy, Duke University Medical Center, Research Drive, Durham, NC 27710, USA e-mail:YWxleGFuZHJhLmJhZGVhQGR1a2UuZWR1
 David G. Ashbrook
David G. Ashbrook Anna Delprato
Anna Delprato Claudia Grellmann
Claudia Grellmann Marieke Klein
Marieke Klein Richard Wetzel
Richard Wetzel Rupert W. Overall
Rupert W. Overall Alexandra Badea
Alexandra Badea