- Department of Pharmaceutical Sciences, Northeastern University, Boston, MA, USA
Homeostatic synaptic scaling in response to neuronal stimulus or activation, and due to changes in cellular niche, is an important phenomenon for memory consolidation, retrieval, and other similar cognitive functions (Turrigiano and Nelson, 2004). Neurological disorders and cognitive disabilities in autism, Rett syndrome, schizophrenia, dementia, etc., are strongly correlated to alterations in protein expression (both synaptic and cytoplasmic; Cajigas et al., 2010). This correlation suggests that efficient temporal regulation of synaptic protein expression is important for synaptic plasticity. In addition, equilibrium between mRNA processing, protein translation, and protein turnover is a critical sensor/trigger for recording synaptic information, normal cognition, and behavior (Cajigas et al., 2010). Thus a regulatory switch, which controls the lifespan, maturation, and processing of mRNA, might influence cognition and adaptive behavior. Here, we propose a two part novel hypothesis that methylation might act as this suggested coordinating switch to critically regulate mRNA maturation at (1) the pre-transcription level, by regulating precursor-RNA processing into mRNA, via other non-coding RNAs and their influence on splicing phenomenon, and (2) the post-transcription level by modulating the regulatory functions of ribonucleoproteins and RNA binding proteins in mRNA translation, dendritic translocation as well as protein synthesis and synaptic turnover. DNA methylation changes are well recognized and highly correlated to gene expression levels as well as, learning and memory; however, RNA methylation changes are recently characterized and yet their functional implications are not established. This review article provides some insight on the intriguing consequences of changes in methylation levels on mRNA life-cycle. We also suggest that, since methylation is under the control of glutathione anti-oxidant levels (Lertratanangkoon et al., 1997), the redox status of neurons might be the central regulatory switch for methylation-based changes in mRNA processing, protein expression, and turnover. Lastly, we also describe experimental methods and techniques which might help researchers to evaluate the suggested hypothesis.
Methylation Affects Precursor-RNA Processing and mRNA Synthesis
Methylation Based Modulatory Role of MicroRNAs
Precursor-RNA (Pre-RNA) is the immature and the incompletely processed mRNA molecule in the nucleus and which needs to be processed before exporting it into cytoplasm in fully functional mature mRNA form. The pre-RNA processing is an early yet highly regulated event in protein synthesis, wherein regulatory-RNAs (re-RNA) and RNA binding proteins (RNABPs) exert dynamic control. Micro-RNAs (mi-RNA) and other non-coding RNAs [nc-RNA; e.g., long non-coding RNAs (lnc-RNAs)], are the major re-RNAs involved in pre-RNA processing and are capable of inducing alterations in gene expression. A functional complementation exists between the levels of DNA methylation and mi-RNA function (Su et al., 2011) and expression (Saito et al., 2006; Chuang and Jones, 2007). In addition, mi-RNAs, small nucleolar RNA (sn-RNA), and anti-sense RNA can regulate the levels of DNA methylation (Qureshi and Mehler, 2010). Hence, a highly sophisticated dynamic regulatory network exists, which involves intertwined processes, (1) Methylation and (2) multifaceted actions of various non-protein coding RNAs (Weber et al., 2007). This proposal of an interactive loop corresponds to one discussed by authors Bernstein and Allis (2005), wherein the authors indicate the role of nc-RNAs like mi-RNAs, ribosomal-RNA, and transfer-RNA in DNA methylation and transcription, as well as, how some of these RNA molecules are themselves regulated by levels of DNA methylation and methyl binding proteins like MeCP2. (Mehler and Mattick, 2007).
Apart from a DNA methylation role, the mechanism through which mi-RNA or other nc-RNAs regulate pre-RNA processing is not clearly identified. However, experts suggest involvement of mi-RNA and anti-sense RNA to sequester the processing site for mRNA transcripts and not allow normal mRNA binding. This competition between pseudogenes and mRNA for regulation of the processing site by mi-RNA has recently surfaced and is termed as “the competitive endogenous RNA” (ce-RNA) theory of mRNA translation and processing (Salmena et al., 2011). Interestingly, this processing site for regulation of mRNA translation has also been suggested to provide an intrinsic layer of control over expression patterns of mRNA (Rigoutsos and Furnari, 2010). In addition, apart from it’s role in the nucleus mi-RNA also plays a regulatory role at the synapse. This is exemplified via their influence on the function of proteins like Fragile X mental retardation protein (FMRP; Mehler and Mattick, 2007), which is involved in mRNA translation and synaptic transport of about 400 different transcripts (Santoro et al., 2011) including several synaptic proteins and proteins involved in neural development as suggested by Santoro et al. (2011) Reports suggest a correlation between the decline in synaptic localization of proteins and observed neurological disorders, for example: SHANK3 in autism spectrum disorders (ASD; Durand et al., 2007) Neuroligin, neurexin, and PSD-95 (Warren, 2011) in Rett syndrome (Sudhof, 2008) and Fragile X syndrome (Wang et al., 2007). Interestingly, some of these neurological disorders are also associated with the malfunction of FMRP. (Darnell et al., 2011; Santoro et al., 2011).
RNABP Methylation and Regulation of Alternative Splicing Patterns
Alternative splicing is one of the earliest phenomena to be identified as a “coding language” used by RNA molecules to generate protein diversity. Alternative splicing occurs by selective and guided skipping of intragenic sequences during transcription and pre-RNA processing. However, over the past decade it has been shown that patterns of alternative splicing are not just involved in protein diversity for evolutionary and developmental purposes, but they also play a major regulatory role in pre-RNA processing. This newly identified role of alternative splicing is especially important in neurons, as it affects cell fate determination, axon guidance, and synaptogenesis (Li et al., 2007). Some experts consider the process of alternative splicing in neurons as an ability of mRNA to adapt and perform differential protein expression in response to local stimulus, neuronal activation, or changes in the neuronal niche, which ultimately promotes homeostasis (Grabowski and Black, 2001; Lipscombe, 2005). Errors of splicing phenomena in neurons have been shown to be involved in several neuromuscular and neurological disorders, including spinal muscular atrophy, fronto-temporal dementia, Fragile X syndrome and Rett syndrome (Li et al., 2007). A similar regulatory role of alternative splicing can be exemplified in FMRP expression and function; for example, in Drosophila melanogaster the short isoform of FMRP (without the glutamine-asparagine domain) is inadequate for participating in short and long term memory formation (Banerjee et al., 2010). Deletion of the homologous region (i.e., the C-terminal domain) in human FMRP does not allow binding of kinesin, and thus consequently inhibits dendritic transport of mRNA molecules (Dictenberg et al., 2008), and affects synaptic plasticity.
The methylation status of the RNA-binding domain of RNABP is believed to regulate splicing pattern on mRNA transcript (Young et al., 2005). Methylcytosine binding protein-2 (MeCP2) recognizes 5-methylcytosine on DNA and is a critical transcription factor implicated in neuro-developmental disorders, including Rett syndrome and autism spectrum disorder (Chahrour et al., 2008). Y-box-binding protein 1 (YBP1) is a RNA binding protein, which interacts with MeCP2, and this conjugation critically regulates splicing, such that mutations in YBP1 or MeCP2, or alterations in MeCP2 levels (as observed in Rett syndrome), can affect mRNA splicing patterns, and cause aberrant gene expression (Young et al., 2005). Thus RNABP methylation status is an important regulator of alternative splicing phenomena.
Effects of Methylation on Post-Transcriptional Regulation of mRNA
Methylation of RNA
Similar to various other post-transcriptional modifications, RNA methylation also occurs on different RNA species like tRNA, rRNA, mRNA, tmRNA, sn-RNA, snoRNA, mi-RNA, and viral RNA (Motorin and Helm, 2011). In fact, RNA methylation occurs at different positions and a variety of RNA-methyltransferases are employed for this process. It is a post-transcriptional modification, dependent on the levels of S-adenosylmethionine (SAM), which serves as the methyl donor (Figure 1A; Martin, 1992). The most common and highly studied RNA methylation is involved in the process of “capping” at the 5′ end. The guanosine nucleotides are methylated and this marking of eukaryotic mRNA allows cells to distinguish host mRNA from other types of RNA molecules including viral mRNA molecules.
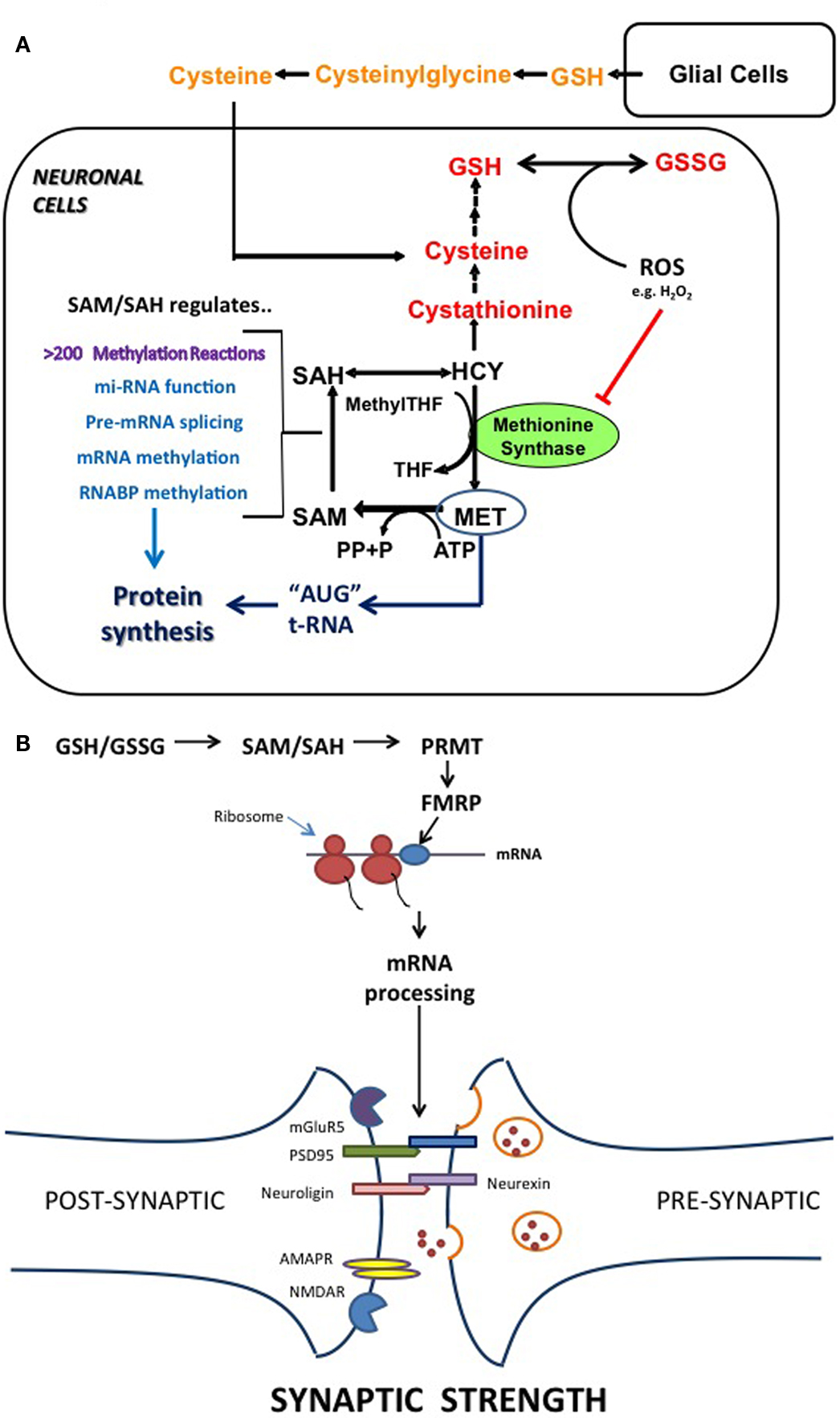
Figure 1. Summary of the hypothesis. (A) The relationship between the methionine cycle of methylation and the transulfuration pathway which converts HCY to cysteine. In brain, glial cells are a primary source of released GSH, which is hydrolyzed to cysteine in the extracellular space. (Raps et al., 1989; Hirrlinger et al., 2002) The intracellular availability of cysteine is rate-limiting for GSH synthesis, and the GSH/GSSG-based redox status is regulated through a combination of cysteine uptake and transsulfuration of HCY. Redox status regulates the SAM/SAH level via its influence on methionine synthase. More than 200 methylation reactions are dependent on SAM levels; Key steps in mRNA processing are regulated via SAM-dependent methylation. Levels of intracellular methionine affect protein synthesis, since it is the required amino acid for initiation of translation. (B) An example of a methyltransferase being regulated by redox status which affects neuronal plasticity. PRMT is a SAM-dependent methyltransferase which methylates the RGG domain of FMRP (a RNABP). FMRP is involved in regulation of about 400 different mRNA transcripts, including NLGN3 (neuroligin-3), PSD-95 (post synaptic density protein-95) and the AMPA-type glutamate receptor. Thus redox status, acting via methylation reactions, can control synaptic strength between neurons, thereby providing a potential molecular mechanism for Hebbian learning and memory formation.
Methylation of cytosine (5MeC), well-known for DNA, has also been recently reported for RNA (Rozenski et al., 1999). However, levels of 5MeC in RNA are low, and the major form of methylation in RNA (i.e., about 30–50% of total RNA methylation) is reported to occur at the 6-position on adenine residues (m6A; Martin, 1992). 5MeC has been described in RNA species like rRNA and tRNA (Rozenski et al., 1999), whereas the highly conserved heterogeneous RNAs (hnRNA) show m6A residues (Yu, 2011). However, the methylated 6-adenine (m6A) residue is localized in a general consensus sequence, Gm6AC or Am6AC in almost all RNA transcripts (Wei and Moss, 1977). Some studies discussed in this section also indicate a significant correlation between alterations in m6A levels and subsequent changes in mRNA processing activity. In particular, two separate studies demonstrated about a 1.5 fold elevation in translation of dihydrofolate reductase (DHFR) RNA transcript in correlation with an increase in its level of methylation on mRNA transcripts (Heilman et al., 1996), whereas inhibition of methylation capacity by depleting SAM levels led to a decrease in DHFR transcript processing (Tuck et al., 1999). Levels of m6A also regulate the selection of splicing sites, and supporting evidence shows that SAM depletion disrupts splicing patterns, and decreases cytoplasmic and consequently synaptic localization of mRNA molecules (Caboche and Bachellerie, 1977).
BC1, a small non-coding RNA (snc-RNA) is highly expressed in neurons (Muslimov et al., 2002) and enriched at synapses (Chicurel et al., 1993). It forms a ribonucleoprotein (RNP) with several partners including FMRP and acts as a liaison between FMRP and FMRP’s target mRNA molecules (Zalfa et al., 2003). The tudor domain of FMRP selectively binds to non-methylated BC1 (Zalfa et al., 2003, 2005). Recently, it has also been shown that BC1 in intracellular compartments contains 2′-O-methylation in the FMRP binding domain, whereas if present at synapses, BC1 lacks the 2′-O-methylation mark, which allows its FMRP interaction (Lacoux et al., 2012). Thus, the methylation status of BC1 indirectly regulates translation and mRNA processing at synapses by regulating FMRP (Lacoux et al., 2012). Studies show that hypomethylation of precursors and intermediates of ribosomal-RNA in the nucleus inhibits their cytoplasmic export and prolongs nuclear accumulation, thus inhibiting further RNA processing (Dictenberg et al., 2008). In fact, Vaughan et al. (1967) showed that depriving cells of methionine and limiting the methylation capacity leads to a blockade of ribosome production itself. A similar study with cycloleucine (a reversible inhibitor of nucleic acid methylation) showed that hypomethylation affects the RNA maturation process at different stages and results in altered mRNA levels in a cumulative manner (Caboche and Bachellerie, 1977). All of the above effects can result in alterations in mRNA translation, and in neurons it might lead to protein deprivation at synapses and hence hinder changes in synaptic plasticity. However, these are preliminary results and further proof would be required to support this idea.
RNA Binding Protein Methylation
mRNA biogenesis depends upon nuclear formation of a messenger ribonucleoprotein particle (mRNP), which is then exported to the cytoplasmic compartment (Yu, 2011). RNABPs regulate a highly dynamic, yet well-orchestrated molecular organization and recognition pattern for mRNP formation. Arginine methylation, which is a major feature of post-transcriptional regulation, occurs on almost all RNABPs, including heterogeneous RNP (hnRNP) and serine/arginine-rich (SR) proteins (Bedford and Clarke, 2009). Arginine methylation is implicated in various cellular processes including, but not limited, to transcription and RNA processing, which includes nuclear export and synaptic localization of mRNA (Yu, 2011). In mammals, the process of arginine methylation is performed by about 10 known protein arginine methyltransferases (PRMTs), whose activity is dependent on levels of SAM as the methyl group donor (Bedford and Clarke, 2009). Thus depletion of SAM decreases PRMT methylation of FMRP (Santoro et al., 2011), which alters processing of mRNA transcript associated with FMRP.
As mentioned above, RNABP methylation occurs at a specific consensus domain known as “RGG” (arginine flanked by glycine) (Bedford and Clarke, 2009). The RGG domain in RNABP recognizes a particular mRNA transcript and selectively binds to it, which results in mRNA translation and/or transport. Indeed, studies involving FMRP show that alterations in the level of RGG methylation are closely associated with changes in protein-protein and protein-RNA interactions (Dolzhanskaya et al., 2006). In the case of FMRP, it was also suggested that differential RGG methylation levels in FMRP can strongly affect the affinity of FMRP for about 400 different mRNA transcripts, about 95 of which belong to proteins involved in synapse formation, and approximately 28 of these proteins are implicated in autism (Santoro et al., 2011). Preliminary results implicate a similar regulatory role of methylation status of mRNA transcript in the KH2 type RNA-binding domain of RNABP, in RNA-RNABP kissing complex formation (De Boulle et al., 1993). A I304N mutation in the KH2 domain of FMRP blocks its ability to bind to polyribosomes and regulate RNA processing (Feng et al., 1997). Additionally, investigators have generated mouse models for Fragile X syndrome carrying this I304N mutation in the KH2 domain of FMRP, and have showed specifically that lost RNA binding ability (due to mutation) led to decrease in FMRP levels and polyribosome association (Zang et al., 2009). In addition, a similar mutation is observed in the KH2 RNA-binding domain in post-mortem brain samples from Fragile X syndrome patients (De Boulle et al., 1993). Similarly, several other proteins possessing a conserved RGG domain play an important role in pre-RNA processing (e.g., “RGG” methylation in spliceosomal small nuclear ribonucleoproteins (snRNPs) regulates alternative splicing). Hence, binding of RNA to RNABP and formation of mRNPs during transcription is a dynamic yet ordered process, and a number of factors involved in the process, appear to be influenced by methylation capacity and levels of methylation on RNA as well as RNABP.
Regulation of Protein Turnover via Methylation Capacity
Protein arginine methylation, the process of adding monomethyl or dimethyl groups to arginine residues, is a well-known methylation reaction (Gary and Clarke, 1998). About 12 ATPs are required per methylation cycle and evolutionary retention of such an “expensive” system underscores the biological importance of this post-translational modification (Boisvert et al., 2003). Histone arginine methylation and myelin basic protein were the first proteins known to be methylated (Paik and Kim, 1968; Brostoff and Eylar, 1971). At present, more than 200 proteins are known to contain RG-repeats and can be methylated at arginine residue by different classes of PRMT (Boisvert et al., 2003). Most of these proteins are associated with RNA maturation process as mentioned earlier, and are involved in mRNA translation regulation through RNABP (Boisvert et al., 2003). Thus, all these studies support the general concept that protein arginine methylation regulates localization and turnover of synaptic proteins.
Methionine is the initiating amino acid for protein synthesis, as the starting codon sequence “AUG” on any mRNA molecule corresponds to methionine. Hence, intracellular levels of methionine can regulate initiation of protein synthesis. Lower methionine availability (for methionine-loaded MET-tRNA) would result in decreased initiation of translation, affecting a wide range of cellular functions. In neurons this could decrease the rate of synaptic protein synthesis, limiting the ability to dynamically adjust the composition of the proteome in accordance to changes in neuronal niche. Importantly, protein lifespan depends partly upon the ubiquitinylation of exposed lysine residues at their epsilon amino group in a protein which targets these proteins for proteasomal degradation. However, methylation or homocysteinylation of these sites will block ubiquitination and extend protein lifespan, allowing integration with protein synthesis (Shukla et al., 2009; Williamson and Whetton, 2011). Thus, the equilibrium between levels of methionine (MET) and homocysteine (HCY) is important for normal translation, and protein turnover. This would act as a regulatory point for modulating protein homeostasis at synapses, thus regulating synaptic plasticity.
Central Regulatory Redox Switch
The methylation potential depends on levels of SAM and SAH, as described above and as indicated in Figure 1A. However, levels of SAM are in turn dependent upon the levels of methionine, homocysteine, and activity of the folate and vitamin B12 dependent enzyme methionine synthase (Figure 1A). Most importantly, methionine synthase activity is highly sensitive to cellular redox status and to fluctuations in the major intracellular anti-oxidant, glutathione (GSH; Waly et al., 2004). The methyl group donated by SAM derives from adenosylation of methionine, and during all SAM-dependent methylation reactions, donation of this methyl group results in S-adenosylhomocysteine (SAH), which is an inhibitor of methylation, based upon its competition with SAM for methyltransferase binding (Yi et al., 2000). SAH is reversibly converted to HCY by SAH hydrolase, whose activity affects the rate of methylation reactions (Chiang et al., 1996). Methionine synthase, which forms MET from HCY, also regulates SAM levels, as indicated in Figure 1A. More than 200 methylation reactions (including DNA and RNA methylation) are dependent upon the SAM/SAH ratio (Petrossian and Clarke, 2011). Interestingly, one of the most widely accepted causes of DNA and RNA damage in a cell is oxidative stress, which is induced by a decline in levels of the major anti-oxidant GSH. Levels of GSH in neurons can be maintained by HCY and MET through the intermediates cystathionine and cysteine. Abnormal levels of these metabolites, including cysteine, GSH, SAM, and SAH have been extensively demonstrated in ASD (James et al., 2004; Deth et al., 2008). In addition, redox levels have also been directly linked to regulation of mRNA (Ufer et al., 2010) as well as micro-RNA (Wiesen and Tomasi, 2009; Ufer et al., 2010). However, the reader is asked to seek further literature from the references cited above. Thus, anti-oxidant levels can regulate SAM/SAH-based methylation reactions throughout the cell, with implications for the clinical pathophysiology of neurological and neuro-developmental disorders.
The concept of a redox-based methylation switch for mRNA-related events requires validation through experimental investigation of the hypothesis we put forth. Recent technological advancements allow individual mRNA transcript sequencing as well as whole transcriptome sequencing using the SOLiD™ system (Lao et al., 2009). Hence, methylation changes on mRNA transcripts in animal models of neurological disorders (e.g., Rett syndrome, ASD, etc.), as well as in post-mortem brain samples of patients suffering from these disorders, can be measured at the individual mRNA transcript level. Bisulfite sequencing can be used to measure methylation status in the whole transcriptome, and population-based transcriptome comparisons can be analyzed (Schaefer et al., 2009). In addition, a cause-effect relationship between levels of methylation in mRNA/RNABP or other such effectors, and resulting neurological or behavioral effects, should also be investigated. This is exemplified by the correlative studies described above, involving the FMRP methylating enzyme PRMT and resulting neurological changes observed with its decreased activity (Figure 1B; Santoro et al., 2011). In addition, manipulations of the redox state in neuronal cells can be altered by oxidative insults and/or anti-oxidant interventions (e.g., N-acetylcysteine or GSH). Comparisons of subsequent changes in mRNA methylation patterns across the transcriptome or in individual transcripts can be measured by bisulfite sequencing, as described above, as well as by mass spectrometry (Qiu and McCloskey, 1999).
Some of the experiments suggested above are exemplified from studies performed by researchers in Germany (Hermes et al., 2004). In this study, researchers manipulated redox conditions and investigated subsequent effects on methylation potential (SAM/SAH levels) and alterations in levels of mRNA and DNA methylation. They induced hypoxia in HepG2 cell cultures, which led to increased SAM and decreased SAH levels with about four-fold elevation in methylation potential (Hermes et al., 2004). Real-time PCR amplification quantified specific mRNA transcripts, namely VEGF and erythropoietin. Incorporation of radiolabeled L-[methyl-3[H]-methionine] and 14[C]-uridine into mRNA reported that inhibition of SAH hydrolase led to decreased methylation potential and decreased mRNA methylation, which suggested that increased SAH levels led to probable inhibition of mRNA-methyltransferase, which is consistent with reports from other studies (Backlund et al., 1986). Similar studies could be performed for neuronal cell cultures and with advanced techniques like FRAP (fluorescence recovery after photobleaching), the synaptic localization or transport of proteins could be tested (Antar et al., 2005). Additionally, by keeping animals/cell cultures in a hypobaric or hyperbaric oxygen chamber and using optogenetic tools to selectively stimulate a certain population of cells involved in particular brain function, correlations could be made between brain activity, redox status, and synapse formation. For these purposes, redox status could be evaluated by using magnetic resonance imaging (MRI) or single-photon emission computed tomography (SPECT) to image 99mTechnetium hexamethylpropyleneamine oxime (HMPAO) conjugated to glutathione (Suess et al., 1991). Post-mortem gene expression analysis could then be performed in addition to quantification of mRNA and DNA methylation status using techniques described above. These and other such studies would allow researchers to test the underlying major hypothesis that redox state is the ultimate source of regulatory control over mRNA methylation, mRNA processing, protein synthesis, and protein turnover.
Conclusion
Thus, evidence from a number of studies indicates that, methylation capacity and methylation levels of mRNA play a major role in it’s maturation and processing, which further affects protein expression and synaptic localization. Any alterations in these key phenomena can trigger an array of effects which might terminally result in neurological disorders. Redox status adds another intricate layer to the sparsely clarified processes mentioned above. However, redox state should be considered as a powerful tool, which can be manipulated to study mRNA regulation and strengthen our current insights of basic biological processes.
The temporospatial localization of proteins is important for synaptic plasticity and a redox-based methylation switch provides modulation of mRNA maturation and lifespan, which eventually influences protein homeostasis at synapses and influences higher-order cognitive functions. A “holonarchy” for synaptic plasticity can be imagined, beginning at mRNA synthesis, transcription, translation, protein turnover, methylation reactions, and at the highest level redox status serves as the central regulatory switch. All these biological processes are individually highly dynamic and complex, yet they are well-coordinated and interrelated processes which provide feedback regulation to each other in order to control and maintain homeostatic synaptic plasticity. However, significant additional evidence supporting this hypothesis is needed, which will not only help in clarifying the functional linkage between key regulatory factors like mi-RNA, RNABPs, GSH, and SAM/SAH, but will also identify potential targets for treating neurological disorders like ASD, Fragile-X syndrome, and other synaptic protein deficiency disorders which can result from defects in mRNA maturation and processing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Antar, L. N., Dictenberg, J. B., Plociniak, M., Afroz, R., and Bassell, G. J. (2005). Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 4, 350–359.
Backlund, P. S., Carotti, D., and Cantoni, G. L. (1986). Effects of the S-adenosylhomocysteine hydrolase inhibitors 3-deazaadenosine and 3-deazaaristeromycin on RNA methylation and synthesis. Eur. J. Biochem. 160, 245–251.
Banerjee, P., Schoenfeld, B. P., Bell, A. J., Choi, C. H., Bradley, M. P., Hinchey, P., Kollaros, M., Park, J. H., McBride, S. M., and Dockendorff, T. C. (2010). Short- and long-term memory are modulated by multiple isoforms of the fragile X mental retardation protein. J. Neurosci. 30, 6782–6792.
Bedford, M. T., and Clarke, S. G. (2009). Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13.
Boisvert, F.-M., Côté, J., Boulanger, M.-C., and Richard, S. (2003). A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 2, 1319–1330.
Brostoff, S., and Eylar, E. H. (1971). Localization of methylated arginine in the A1 protein from myelin. Proc. Natl. Acad. Sci. U.S.A. 68, 765–769.
Caboche, M., and Bachellerie, J. (1977). RNA methylation and control of eukaryotic RNA biosynthesis. Eur. J. Biochem. 74, 19–29.
Cajigas, I. J., Will, T., and Schuman, E. M. (2010). Protein homeostasis and synaptic plasticity. EMBO J. 29, 2746–2752.
Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T., Qin, J., and Zoghbi, H. Y. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320, 1224–1229.
Chiang, P. K., Gordon, R. K., Tal, J., Zeng, G. C., Doctor, B. P., Pardhasaradhi, K., and McCann, P. P. (1996). S-adenosylmethionine and methylation. FASEB J. 10, 471–480.
Chicurel, M. E., Terrian, D. M., and Potter, H. (1993). mRNA at the synapse: analysis of a synaptosomal preparation enriched in hippocampal dendritic spines. J. Neurosci. 13, 4054–4063.
Darnell, J. C., Van Driesche, S. J., Zhang, C., Hung, K. Y., Mele, A., Fraser, C. E., Stone, E. F., Chen, C., Fak, J. J., Chi, S. W., Licatalosi, D. D., Richter, J. D., and Darnell, R. B. (2011). FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146, 247–261.
De Boulle, K., Verkerk, A. J., Reyniers, E., Vits, L., Hendrickx, J., Van Roy, B., Van den Bos, F., de Graaff, E., Oostra, B. A., and Willems, P. J. (1993). A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 3, 31–35.
Deth, R., Muratore, C., Benzecry, J., Power-Charnitsky, V.-A., and Waly, M. (2008). How environmental and genetic factors combine to cause autism: a redox/methylation hypothesis. Neurotoxicology 29, 190–201.
Dictenberg, J. B., Swanger, S. A., Antar, L. N., Singer, R. H., and Bassell, G. J. (2008). A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev. Cell 14, 926–939.
Dolzhanskaya, N., Merz, G., Aletta, J. M., and Denman, R. B. (2006). Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell. Sci. 119, 1933–1946.
Durand, C. M., Betancur, C., Boeckers, T. M., Bockmann, J., Chaste, P., Fauchereau, F., Nygren, G., Rastam, M., Gillberg, I. C., Anckarsäter, H., Sponheim, E., Goubran-Botros, H., Delorme, R., Chabane, N., Mouren-Simeoni, M. C., de Mas, P., Bieth, E., Rogé, B., Héron, D., Burglen, L., Gillberg, C., Leboyer, M., and Bourgeron, T. (2007). Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat. Genet. 39, 25–27.
Feng, Y., Absher, D., Eberhart, D. E., Brown, V., Malter, H. E., and Warren, S. T. (1997). FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell 1, 109–118.
Gary, J. D., and Clarke, S. (1998). RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61, 65–131.
Grabowski, P. J., and Black, D. L. (2001). Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65, 289–308.
Heilman, K. L., Leach, R. A., and Tuck, M. T. (1996). Internal 6-methyladenine residues increase the in vitro translation efficiency of dihydrofolate reductase messenger RNA. Int. J. Biochem. Cell Biol. 28, 823–829.
Hermes, M., Osswald, H., Mattar, J., and Kloor, D. (2004). Influence of an altered methylation potential on mRNA methylation and gene expression in HepG2 cells. Exp. Cell Res. 294, 325–334.
Hirrlinger, J., Schulz, J. B., and Dringen, R. (2002). Glutathione release from cultured brain cells: multidrug resistance protein 1 mediates the release of GSH from rat astroglial cells. J. Neurosci. Res. 69, 318–326.
James, S. J., Cutler, P., Melnyk, S., Jernigan, S., Janak, L., Gaylor, D. W., and Neubrander, J. A. (2004). Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 80, 1611–1617.
Lacoux, C., Di Marino, D., Pilo Boyl, P., Zalfa, F., Yan, B., Ciotti, M. T., Falconi, M., Urlaub, H., Achsel, T., Mougin, A., Caizergues-Ferrer, M., and Bagni, C. (2012). BC1-FMRP interaction is modulated by 2′-O-methylation: RNA-binding activity of the tudor domain and translational regulation at synapses. Nucleic Acids Res. 40, 4086–4096.
Lao, K. Q., Tang, F., Barbacioru, C., Wang, Y., Nordman, E., Lee, C., Xu, N., Wang, X., Tuch, B., Bodeau, J., Siddiqui, A., and Surani, M. A. (2009). mRNA-sequencing whole transcriptome analysis of a single cell on the SOLiDTM system. J. Biomol. Tech. 20, 266–271.
Lertratanangkoon, K., Wu, C. J., Savaraj, N., and Thomas, M. L. (1997). Alterations of DNA methylation by glutathione depletion. Cancer Lett. 120, 149–156.
Li, Q., Lee, J.-A., and Black, D. L. (2007). Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8, 819–831.
Lipscombe, D. (2005). Neuronal proteins custom designed by alternative splicing. Curr. Opin. Neurobiol. 15, 358–363.
Martin, T. T. (1992). The formation of internal 6-methyladenine residues in eucaryotic messenger rna. Int. J. Biochem. 24, 379–386.
Mehler, M. F., and Mattick, J. S. (2007). Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 87, 799–823.
Motorin, Y., and Helm, M. (2011). RNA nucleotide methylation. Wiley Interdiscip. Rev. RNA 2, 611–631.
Muslimov, I. A., Lin, Y., Heller, M., Brosius, J., Zakeri, Z., and Tiedge, H. (2002). A small RNA in testis and brain: implications for male germ cell development. J. Cell. Sci. 115, 1243–1250.
Paik, W. K., and Kim, S. (1968). Protein methylase I purification and properties of the enzymE. J. Biol. Chem. 243, 2108–2114.
Petrossian, T. C., and Clarke, S. G. (2011). Uncovering the human methyltransferasome. Mol. Cell Proteomics 10, M110.000976.
Qiu, F., and McCloskey, J. A. (1999). Selective detection of ribose-methylated nucleotides in RNA by a mass spectrometry-based method. Nucleic Acids Res. 27, e20.
Qureshi, I. A., and Mehler, M. F. (2010). The emerging role of epigenetics in stroke: II. RNA regulatory circuitry. Arch. Neurol. 67, 1435.
Raps, S. P., Lai, J. C., Hertz, L., and Cooper, A. J. (1989). Glutathione is present in high concentrations in cultured astrocytes but not in cultured neurons. Brain Res. 493, 398–401.
Rigoutsos, I., and Furnari, F. (2010). Gene-expression forum: decoy for microRNAs. Nature 465, 1016–1017.
Rozenski, J., Crain, P. F., and McCloskey, J. A. (1999). The RNA modification database: 1999 update. Nucleic Acids Res. 27, 196–197.
Saito, Y., Liang, G., Egger, G., Friedman, J. M., Chuang, J. C., Coetzee, G. A., and Jones, P. A. (2006). Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell 9, 435–443.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358.
Santoro, M. R., Bray, S. M., and Warren, S. T. (2011). Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu. Rev. Pathol. Mech. Dis. 7, 219–245.
Schaefer, M., Pollex, T., Hanna, K., and Lyko, F. (2009). RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 37, e12.
Shukla, A., Chaurasia, P., and Bhaumik, S. R. (2009). Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 66, 1419–1433.
Su, Z., Xia, J., and Zhao, Z. (2011). Functional complementation between transcriptional methylation regulation and post-transcriptional microRNA regulation in the human genome. BMC Genomics 12, S15. doi:10.1186/1471-2164-12-S5-S15
Sudhof, T. C. (2008). Neuroligins and neurexins link synaptic function to cognitive disease. Nature 455, 903–911.
Suess, E., Malessa, S., Ungersböck, K., Kitz, P., Podreka, I., Heimberger, K., Hornykiewicz, O., and Deecke, L. (1991). Technetium-99m-d,1-hexamethylpropyleneamine oxime (HMPAO) uptake and glutathione content in brain tumors. J. Nucl. Med. 32, 1675–1681.
Tuck, M. T., Wiehl, P. E., and Pan, T. (1999). Inhibition of 6-methyladenine formation decreases the translation efficiency of dihydrofolate reductase transcripts. Int. J. Biochem. Cell Biol. 31, 837–851.
Turrigiano, G. G., and Nelson, S. B. (2004). Homeostatic plasticity in the developing nervous system. Nat. Rev. Neurosci. 5, 97–107.
Ufer, C., Wang, C. C., Borchert, A., Heydeck, D., and Kuhn, H. (2010). Redox control in mammalian embryo development. Antioxid. Redox Signal. 13, 833–875.
Vaughan, M. H., Soeiro, R., Warner, J. R., and Darnell, J. E. (1967). The effects of methionine deprivation on ribosome synthesis in HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 58, 1527–1534.
Waly, M., Olteanu, H., Banerjee, R., Choi, S. W., Mason, J. B., Parker, B. S., Sukumar, S., Shim, S., Sharma, A., Benzecry, J. M., Power-Charnitsky, V. A., and Deth, R. C. (2004). Activation of methionine synthase by insulin-like growth factor-1 and dopamine: a target for neurodevelopmental toxins and thimerosal. Mol. Psychiatry 9, 358–370.
Wang, W., van Niekerk, E., Willis, D. E., and Twiss, J. L. (2007). RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev. Neurobiol. 67, 1166–1182.
Warren, S. T. (2011). Trinucleotide repeat syndromes (fragile X). Annu. Rev. Pathol. 7, 110301101326014.
Weber, B., Stresemann, C., Brueckner, B., and Lyko, F. (2007). Methylation of human microRNA genes in normal and neoplastic cells. Cell Cycle 6, 1001–1005.
Wei, C., and Moss, B. (1977). 5’-Terminal capping of RNA by guanylyltransferase from HeLa cell nuclei. Proc. Natl. Acad. Sci. U.S.A. 74, 3758–3761.
Wiesen, J. L., and Tomasi, T. B. (2009). Dicer is regulated by cellular stresses and interferons. Mol. Immunol. 46, 1222–1228.
Williamson, A. J. K., and Whetton, A. D. (2011). The requirement for proteomics to unravel stem cell regulatory mechanisms. J. Cell. Physiol. 226, 2478–2483.
Yi, P., Melnyk, S., Pogribna, M., Pogribny, I. P., Hine, R. J., and James, S. J. (2000). Increase in plasma homocysteine associated with parallel increases in plasma S-adenosylhomocysteine and lymphocyte DNA hypomethylation. J. Biol. Chem. 275, 29318–29323.
Young, J. I., Hong, E. P., Castle, J. C., Crespo-Barreto, J., Bowman, A. B., Rose, M. F., Kang, D., Richman, R., Johnson, J. M., Berget, S., and Zoghbi, H. Y. (2005). Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. U.S.A. 102, 17551–17558.
Yu, M. C. (2011). The role of protein arginine methylation in mRNP dynamics. Mol. Biol. Int. 2011, 1–10.
Zalfa, F., Adinolfi, S., Napoli, I., Kühn-Hölsken, E., Urlaub, H., Achsel, T., Pastore, A., and Bagni, C. (2005). Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 280, 33403–33410.
Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B., and Bagni, C. (2003). The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327.
Zang, J. B., Nosyreva, E. D., Spencer, C. M., Volk, L. J., Musunuru, K., Zhong, R., Stone, E. F., Yuva-Paylor, L. A., Huber, K. M., Paylor, R., Darnell, J. C., and Darnell, R. B. (2009). A mouse model of the human fragile X syndrome I304N mutation. PLoS Genet. 5, e1000758. doi:10.1371/journal.pgen.1000758
Keywords: alternative splicing, FMRP, glutathione, homeostatis, redox status, Rett syndrome, S-adenosylmethionine, synaptic scaling
Citation: Trivedi MS and Deth RC (2012) Role of a redox-based methylation switch in mRNA life cycle (pre- and post-transcriptional maturation) and protein turnover: implications in neurological disorders. Front. Neurosci. 6:92. doi: 10.3389/fnins.2012.00092
Received: 31 January 2012; Accepted: 06 June 2012;
Published online: 26 June 2012.
Edited by:
Jernej Ule, University of Cambridge, UKReviewed by:
Elizabeth A. Thomas, Scripps Research Institute, USAMichaela Frye, University of Cambridge, UK
Copyright: © 2012 Trivedi and Deth. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Malav S. Trivedi, Department of Pharmaceutical Sciences, Northeastern University, 140 The Fenway, Boston, MA 02115, USA. e-mail:bTN2ZWRpMTk4NkBnbWFpbC5jb20=

