- 1 Graduate School of Medicine, Korea University, Seoul, South Korea
- 2 Laboratory of Neuronal and Neuroendocrine Differentiation and Communication, INSERM U982, University of Rouen, Mont-Saint-Aignan, France
The neuropeptides gonadotropin-releasing hormone (GnRH) and kisspeptin (KiSS), and their receptors gonadotropin-releasing hormone receptor (GnRHR) and kisspeptin receptor (KiSSR) play key roles in vertebrate reproduction. Multiple paralogous isoforms of these genes have been identified in various vertebrate species. Two rounds of genome duplication in early vertebrates likely contributed to the generation of these paralogous genes. Genome synteny and phylogenetic analyses in a variety of vertebrate species have provided insights into the evolutionary origin of and relationship between paralogous genes. The paralogous forms of these neuropeptides and their receptors have coevolved to retain high selectivity of the ligand–receptor interaction. These paralogous forms have become subfunctionalized, neofunctionalized, or dysfunctionalized during evolution. This article reviews the evolutionary mechanism of GnRH/GnRHR and KiSS/KiSSR, and the fate of the duplicated paralogs in vertebrates.
Introduction
Neuropeptides serve as messenger molecules to regulate a variety of physiological functions in the nervous and neuroendocrine systems of vertebrates (Hoyle, 1999). In humans, more than 70 genes have been identified to encode approximately 100 short mature peptides that function either at the brain or periphery. Most neuropeptide genes can be categorized into small families consisting of one to four members that originate from a common ancestral gene. These families have diversified through evolutionary mechanisms, like gene/chromosome duplication, followed by gene modification and gene loss (Conlon and Larhammar, 2005; Lee et al., 2009; Kim et al., 2011). Receptors for neuropeptides are generally G protein-coupled receptors (GPCRs) which transmit signals from the cell membrane to the intracellular space through activation of heterotrimeric G proteins (Oh et al., 2006). Similar to neuropeptide ligands, receptors have also diverged evolutionarily to maintain high selectivity for their cognate ligands (Fredriksson et al., 2003; Acharjee et al., 2004; Li et al., 2005; Cho et al., 2007).
Gonadotropin-releasing hormone-1 (GnRH1, also known as mammalian GnRH) and its mammalian type-I receptor (GnRHRm1) are an essential neuropeptide/receptor pair for sexual development and reproductive function in vertebrates (Schally et al., 1971; Millar et al., 2004). GnRH1 is synthesized in the hypothalamus and released into the hypophyseal portal circulation in a pulsatile manner. In the pituitary, binding of GnRH1 to GnRHRm1 regulates secretion of the gonadotropins, luteinizing hormone, and follicle-stimulating hormone. The secreted gonadotropins then stimulate gametogenesis and steroidogenesis in the gonad. Steroid hormones secreted from the gonad exert either negative or positive feedback action on the hypothalamus and pituitary to control GnRH1 and gonadotropin secretion, hence establishing the hypothalamic-pituitary–gonadal (HPG) axis to control the reproductive function in mammals (Hrabovszky et al., 2007; Ohkura et al., 2009). In addition, identification and characterization of kisspeptin, a product of the KiSS1 gene and its receptor gene KiSS1R (or GPR54) indicates that the pair plays a key role in initiating puberty as well as steroid hormone feedback (de Roux et al., 2003; Seminara et al., 2003; d’Anglemont de Tassigny et al., 2007). In mammals, kisspeptin binding to KiSS1R induces GnRH1 secretion from GnRH1 neurons (Messager et al., 2005). Neurons expressing KiSS1 in the anteroventral periventricular nucleus (APVN) and arcuate nucleus (ARC) are the main targets for estrogens to exert either positive or negative feedback on GnRH1 secretion (Oakley et al., 2009; Ohkura et al., 2009). Thus, the KiSS1/KiSS1R pair functions as a molecular gatekeeper of the HPG axis for sexual maturation and reproductive function in mammals.
To date, multiple isoforms of KiSS1 and GnRH1 have been identified in a variety of vertebrates (Um et al., 2010; Kim et al., 2011). For instance, three KiSS isoforms (KiSS1, KiSS1b, and KiSS2) have been identified in Xenopus tropicalis, and three gonadotropin-releasing hormone (GnRH) isoforms (GnRH1, GnRH2, and GnRH3) exist in many fish species. Multiple isoforms for the respective receptors, kisspeptin receptor (KiSSR) and gonadotropin-releasing hormone receptor (GnRHR), have also been identified (Lee et al., 2009; Kim et al., 2011). During evolution, the function of paralogs can diversify. Although the GnRH/kisspeptin system has been well characterized in rodents, which only have one KiSS1 and one GnRH1 ligand–receptor pair, the function of the paralogous genes in other vertebrates remains poorly understood. In addition, some non-mammalian vertebrate species do not have orthologous genes for KiSS1/KiSS1R or GnRH1/GnRHRm1, suggesting that a paralogous set of the genes may substitute for the function of the absent genes. In avian species, the KiSS/KiSSR system is completely absent, indicating that KiSS signaling is not required in birds. During evolution, the KiSS/KiSSR and GnRH/GnRHR genes have been duplicated, generating at least three paralogs whose functions have diverged to some extent. These features make the KiSS and GnRH systems excellent models to understand molecular evolution of peptide ligand and receptor pairs. This article reviews the evolution of the GnRH and KiSS ligand–receptor pairs and the fate of their duplicated paralogous genes in vertebrates.
Two Rounds of Genome Duplication Produced Multiple Isoforms of Neuropeptides and their Receptors
The fact that neuropeptides and receptors group into small families can be accounted for by two rounds of large-scale genome duplication in an early vertebrate ancestor that resulted in up to four copies of each gene (Ohno, 1970; Larhammar et al., 2002). Further, after the divergence of fish and land vertebrates, a teleost fish-specific third round of genome duplication is believed to have produced eight copies of a gene family (Larsson et al., 2008). Genome duplication events often produce paralogous chromosomal regions, also called paralogons (Lundin, 1993). Analysis of gene arrangement within paralogons allows the relationships among and origins of each gene family to be traced (Um et al., 2010).
Duplications of the KiSS- and KiSSR-Containing Genomes
As X. tropicalis has three forms of KiSS genes, it is presumable that the multiple genes were generated by at least two rounds of genome duplication during early vertebrate evolution (Lee et al., 2009; Um et al., 2010). Genome synteny analysis comparing vertebrate KiSS-containing genome fragments with human chromosomes allowed identification of four genome fragments that share several paralogous genes. These include TEAD1/2/4, K/N/R/HRAS, KCNJ7/8/9/11, CACNA1A/C/S, SOX5/6/13, PLEKHA4/5/6/7, and PIK3C2A/B/G (Figure 1). This observation suggests that these paralogous genes are likely members of four early paralogons, containing KiSS1, KiSS2, KiSS1b, or the fourth form of KiSS. During millions of years of evolution, many paralogous genes have been rearranged within paralogons, translocated to other chromosomes, or lost (Figure 1). The absence of the fourth KiSS gene, despite the presence of the fourth paralogon in vertebrates, suggests that the fourth KiSS gene was lost before or just after the divergence of teleosts and tetrapods. The presence of the KiSS1b gene only in X. tropicalis indicates that the KiSS1b gene has been dysfunctionalized in a wide variety of vertebrates. No teleost-specific KiSS duplication has been observed in fish species.
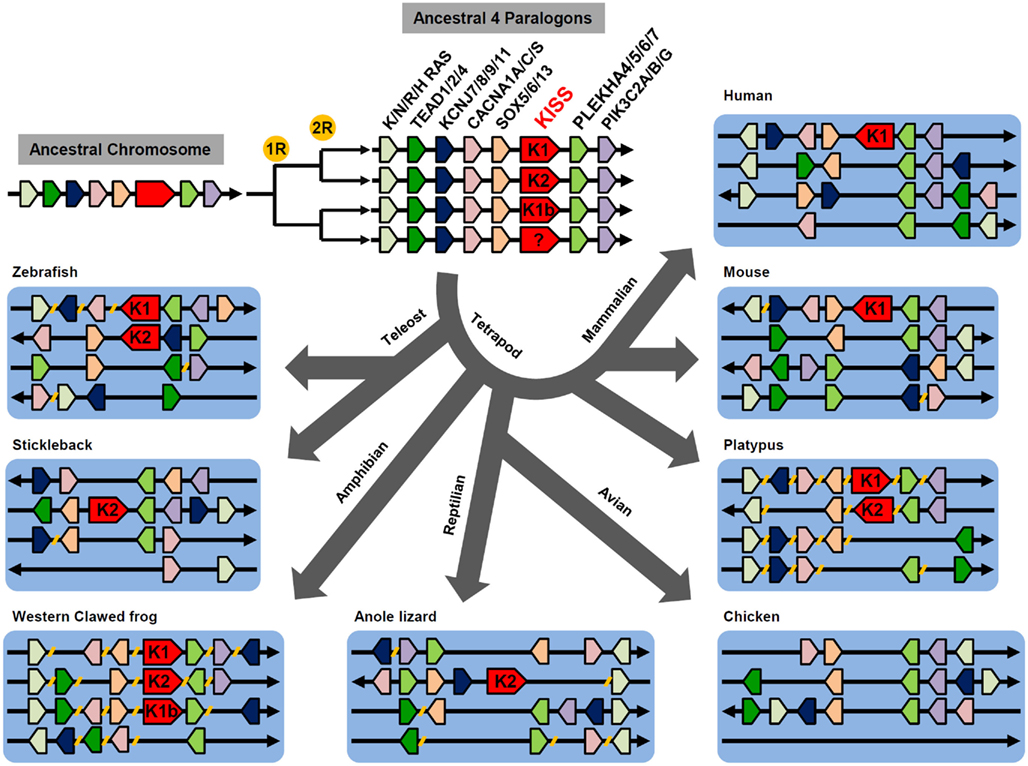
Figure 1. Proposed evolutionary history of KiSS-containing paralogons. Two rounds of genome duplication generated four copies of ancestral paralogons containing the KiSS gene. The KiSS gene in the fourth paralogon likely disappeared before teleost and tetrapod divergence. During vertebrate evolution, some genes, including KiSS, were lost. Genes localized to different genome fragments or chromosomes are indicated by breaks ( // ) on the line.
Comparison of KiSSR-containing genomes in X. tropicalis with other vertebrate species also supports tetraploidization of KiSSR-containing paralogons sharing paralogous genes including NOTCH1/2/3/4, PTPRD/K/S/U, STXBP1/2/3, ARID1A/1B/3A/3C, PALM1/2/D, PTBP1/2/ROD1, and ABCA1/4/7 (unpublished data). The teleost-specific duplication of a KiSSR-containing genome has not been identified. Similar to the KiSS gene, the fourth KiSSR gene is absent in all vertebrates and KiSS1bR is present in only X. tropicalis.
As a result of these genome duplications, vertebrates have various numbers of KiSS/KiSSR pairs in a single species (Figure 2). In the fish species, zebrafish and medaka contain two KiSS1/KiSS1R and KiSS2/KiSS2R pairs, whereas torafugu, green puffer fish, and stickleback have only one KiSS2/KiSS2R pair. In the tetrapod lineage, X. tropicalis preserves all currently identified paralogous isoforms of the KiSS/KiSSR pair and thus has three KiSSs (KiSS1, KiSS1b, and KiSS2) and three KiSSRs (KiSS1R, KiSS1bR, and KiSS2R). On the other hand, the anole lizard has only the KiSS2/KiSS2R pair. Interestingly, in the avian phylum, the chicken and zebra finch do not possess any form of KiSS and KiSSR. Although most mammals, including humans and rodents, contain only KiSS1 and KiSS1R, the platypus, a monotreme species, has KiSS1/KiSS1R and KiSS2/KiSS2R. In all species examined for this article, the KiSS isoform and cognate receptor isoform coexisted (Figure 2).
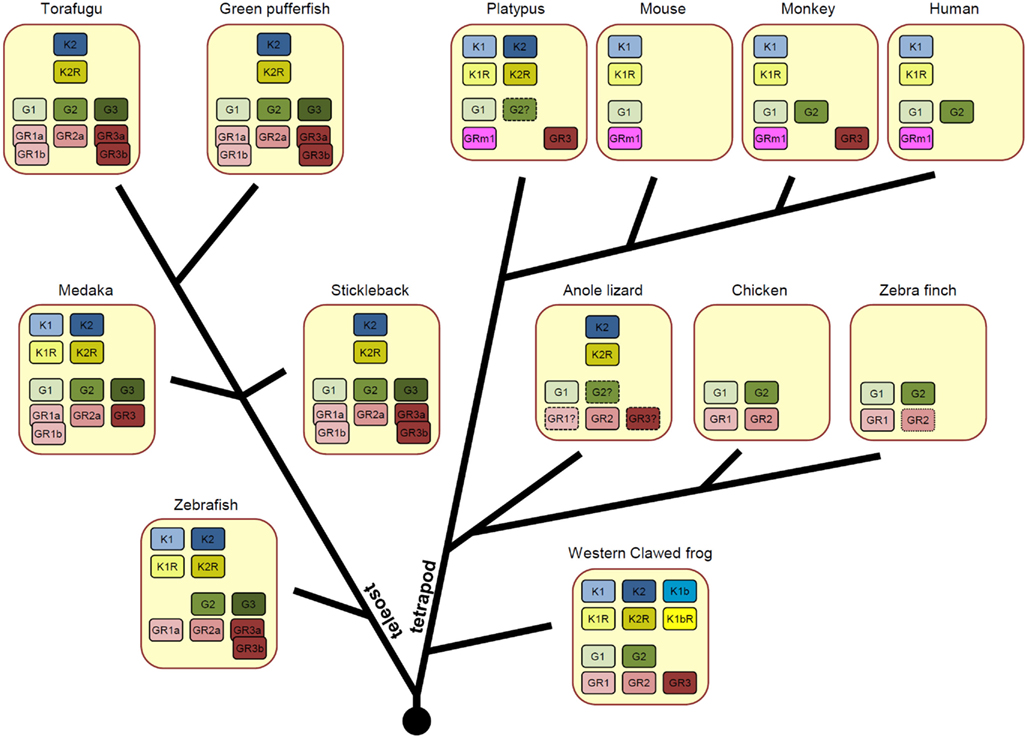
Figure 2. Varied number of KiSS (K), KiSSR (KR), GnRH (G), and GnRHR (GR) genes across vertebrate species. Orthologous genes were categorized in our recent article (Kim et al., 2011) and are presented with the same colors. For teleost fish, the GnRHR genes produced by teleost-specific genome duplication are shown.
Duplications of GnRH- and GnRHR-Containing Genomes
Tetraploidization of GnRH- and GnRHR-containing genomes was recently demonstrated (Kim et al., 2011). Synteny studies of GnRH-containing genomes in tetrapods revealed the presence of four paralogons that contain EBF1/2/3/4, PPP2R2B/D, ADAM12/19, DOCK1/2/5, FAM196A/B, NKX2-3/5/6, STC1/2, CPXM1/2, and PTPRA/E. These studies revealed that the fourth GnRH gene likely disappeared before the divergence of teleosts and tetrapods, and that GnRH3 was lost in the tetrapod lineage after the divergence of teleosts and tetrapods (Kim et al., 2011).
Evolution of the GnRHR gene is more complicated than that of GnRH due to teleost-specific duplications of GnRHR-containing genomes and the inability to resolve the origin of mammalian type-I GnRHR (GnRHRm1). Genome synteny and phylogenetic analyses suggest the presence of four GnRHR paralogs: non-mammalian type-I (GnRHRn1), non-mammalian type-II (GnRHRn2), non-mammalian type-III/mammalian type-II (GnRHRn3/m2), and mammalian type-I (GnRHRm1; Kim et al., 2011). GnRHRn1 and GnRHRn2 were likely generated by a local duplication before the divergence of tetrapods and fish. GnRHRm1 was only identified in mammals and its evolutionary origin and relationship with other GnRHR types is unclear due to a lack of synteny between mammalian genomes containing GnRHRm1 and genomes of any non-mammalian species. In fish, a third round of genome duplication produced a copy of GnRHRn1 and GnRHRn3 (GnRHRn1b and GnRHRn3b, respectively; Kim et al., 2011).
In the five fish species for which genome information is available in ENSEMBL, two or three GnRH isoforms have been identified. Zebrafish has two GnRH genes (GnRH2 and GnRH3), whereas other fish, medaka, stickleback, green puffer fish, and torafugu have all three forms of GnRH (GnRH1, GnRH2, and GnRH3; Figure 2). The literature indicates that salmon species lack GnRH1, while catfish and eel lack GnRH3 (Kah et al., 2007; Okubo and Nagahama, 2008). In the tetrapod lineage, most species except for rodents have two forms of GnRH (GnRH1 and GnRH2). Due to insufficient genetic information investigators were unable to identify the GnRH2 gene in the anole lizard, but believe the gene exists in this species because other reptilian species such as the gecko has two forms of the GnRH gene (Ikemoto and Park, 2003, 2007). Fish have four or five GnRHRs and one or two of them are generated by teleost-specific genome duplication (Kim et al., 2011). Thus, in addition to GnRHRn1, GnRHRn2, and GnRHRn3, torafugu, green puffer fish, and stickleback have two additional isoforms (GnRHRn1b and GnRHRn3b) and medaka and zebrafish each possess one additional isoform (GnRHRn1b and GnRHRn3b, respectively). Amphibians and reptiles likely have three forms of GnRHR (GnRHRn1, GnRHRn2, and GnRHRn3) and avians have two forms of GnRHR (GnRHRn1 and GnRHRn2). In mammals, platypus and monkeys have two GnRHR forms (GnRHRm1 and GnRHRm2), whereas rodents and humans have only one form (GnRHRm1; Figure 2).
Molecular Coevolution of Neuropeptides and Cognate Receptors to Maintain High Affinity Ligand–Receptor Interaction
Although the amino acid sequences of vertebrate neuropeptides and the cognate receptors have diverged from an ancestral system during evolution, core conserved sequences have been retained, allowing for the maintenance of a core structure that is required for ligand–receptor interaction and receptor conformation (Fredriksson et al., 2003; Acharjee et al., 2004; Li et al., 2005; Cho et al., 2007). For instance, sequence alignment of peptides across vertebrate species revealed that core sequences responsible for high affinity binding to the cognate receptor have largely been conserved throughout evolution (Cho et al., 2007). Domains critical for ligand interaction and conformation of the receptor are also well conserved across vertebrate species, indicating high evolutionary selective pressure to maintain high affinity binding to cognate ligands (Wang et al., 2001; Acharjee et al., 2004; Li et al., 2005). Sequence divergence within neuropeptide and receptor families may explain how each paralogous isoform achieved distinct ligand–receptor selectivity. Amino acid changes in a neuropeptide isoform are usually accompanied by a change in the corresponding motif(s) of the cognate receptor (Acharjee et al., 2004; Cho et al., 2007; Moon et al., 2012).
Ligand–Receptor Selectivity in the KiSS–KiSSR System
The KiSS1 peptide sequences differ between non-mammalian species and rodents and humans. Human KiSS1 encodes a 145-amino acid precursor that is enzymatically cleaved into 54-, 14-, 13-, or 10-amino acid peptides sharing a common C-terminal sequence with amidation on phenylalanine (Kotani et al., 2001; Muir et al., 2001; Ohtaki et al., 2001). The KiSS1 peptides in rat and mouse also have a highly conserved C-terminal decapeptide sequence that is amidated on tyrosine. In fact, this C-terminal decapeptide (kisspeptin-10, metastin 45–54) is the minimal sequence required for receptor activation (Kotani et al., 2001; Muir et al., 2001; Ohtaki et al., 2001). The KiSS1 decapeptide sequences from non-mammalian vertebrates vary at position 3 (Leu for fish species, Val for X. tropicalis KiSS1b, and Trp for other species, including KiSS1a from X. tropicalis and KiSS1 from most mammals; Lee et al., 2009). Interestingly, fish KiSS1 and X. tropicalis KiSS1b precursors have a conserved dibasic site followed by a conserved Gln five amino acids upstream of the decapeptide, leading to prediction of 15-amino acid peptide with N-terminal pyroglutamylation. These pentadecapeptides are more potent activators of KiSS1R than decapeptides in fish and X. tropicalis (Lee et al., 2009). Thus, for non-mammalian KiSS1 genes, pyroglutamylated pentadecapeptides are likely the most potent endogenous forms.
The family of the KiSS2 decapeptides exhibits conserved Phe-amidation at the C-terminus, but amino acids at positions 1, 3, and 5 differ from the corresponding residues in the KiSS1 decapeptide. Furthermore, the presence of a conserved basic amino acid three amino acids upstream of the decapeptide suggests that the mature peptide produced from KiSS2 is 12-amino acid long (Lee et al., 2009). The KiSS2 dodecapeptide has been detected in the Xenopus laevis brain using immunoaffinity purification (Lee et al., 2009). Indeed, the KiSS2 dodecapeptides from zebrafish and Xenopus are highly potent activators of KiSS2R expressed in CV-1 cells (Lee et al., 2009). Although some cross-reactivity exists between peptides and receptors, each peptide isoform exhibits relatively high affinity toward its own receptor (Lee et al., 2009), suggesting that KiSS peptides and receptors coevolved to retain high ligand–receptor selectivity. However, the molecular determinants that confer such differential ligand–receptor selectivity remain poorly understood in the KiSS system.
Ligand–Receptor Selectivity in the GnRH–GnRHR System
The amino acid sequence of GnRH1 ([Tyr5, Leu7, Arg8] GnRH) orthologs varies by one or two amino acids at positions 5 and 8 (Powell et al., 1994; Yoo et al., 2000). The GnRH2 ([His5, Trp7, Tyr8] GnRH) sequence is fully conserved in almost all vertebrates and differs from GnRH1 by amino acids at positions 5, 7, and 8 (Fernald and White, 1999). GnRH3 ([Tyr5, Trp7, Leu8] GnRH) is found only in fish species (Powell et al., 1994). Each form of GnRH exhibits a distinct affinity for different GnRHR forms. For instance, GnRH1 generally has a high affinity toward GnRHRm1 but relatively low affinity for GnRHRm2 and other non-mammalian GnRHRs. In contrast, GnRH2 has low affinity for GnRHRm1 but high affinity for other GnRHR forms.
Molecular determinants conferring such ligand–receptor selectivity have been relatively well defined in the GnRH1 and GnRHRm1 system. For instance, the residues in GnRHRm1 (i.e., Asp98, Asn102, Lys121, Asn212, Trp280, Trp289, and Tyr290) that are required for binding to GnRH1 have been identified using mutagenesis and computational modeling (Millar et al., 2004). Interestingly, these residues are similarly conserved in GnRHRn1, n2, and n3/m2 (Millar et al., 2001; Wang et al., 2001). Thus, these residues may not confer differential ligand selectivity toward GnRH1 or GnRH2, but instead are important for the overall ligand–receptor interaction and receptor conformation. In fact, high affinity binding between GnRH1 and GnRHRm1 has been explained by a Ser–Glu/Asp–Pro (SE/DP) motif in extracellular loop 3 (ECL3) of the receptor (Flanagan et al., 1994; Fromme et al., 2001). The positions of Ser and Pro in this motif are known to permit electrostatic interaction with Arg8 of GnRH1 and Glu/Asp in the SE/DP motif (Wang et al., 2004). Thus, altering the Ser and Pro positions (e.g., Pro–Glu–Ser) in GnRHRm1 increased affinity for GnRH2 but reduced affinity for GnRH1. Similarly, mutating a Ser–Gln–Ser (SQS) motif to SEP in bullfrog type-I GnRHR (ortholog of GnRHRn1) increased GnRH2 affinity and decreased GnRH1 affinity (Wang et al., 2004). For the green monkey type-II GnRHR (ortholog of GnRHRn3/m2), multiple residues in ECL3 and transmembrane domain 7 are involved in discriminating between GnRH2 and GnRH1 (Li et al., 2005). Tello et al. (2008) recently demonstrated that zebrafish GnRHR4 (ortholog of GnRHRn1) and GnRHR2 (ortholog of GnRHRn2) exhibit high affinity for both GnRH2 and GnRH3. However, the motifs responsible for such a high affinity binding between these receptors and GnRH3 need to be further clarified.
Is a Genetic Ortholog a Functional Ortholog? Neofunctionalization, Subfunctionalization, and Dysfunctionalization of Paralogs
In mammals, GnRH1 and GnRHRm1 are critical for the onset of puberty and control of reproductive functions. GnRH1-induced activation of the HPG axis is regulated by feedback action from steroid hormones secreted by the gonad. Recently, KiSS1 neurons in the hypothalamus were suggested to mediate this steroid action. KiSS1 neurons are largely distributed in the steroid receptor-enriched APVN of the anterior hypothalamic region and the ARC in the mediobasal hypothalamic region (Kinoshita et al., 2005). The APVN is a potential positive feedback action site for estrogen to control the GnRH1 surge, whereas the ARC is an important negative feedback action area for estrogen to control pulsatile GnRH release (Ohkura et al., 2009).
Although the function of the KiSS/GnRH system in the control of the HPG axis is highly conserved across vertebrate species, there are many examples of KiSS1/GnRH1 paralogs, and not orthologs, functioning in this capacity (Figure 3). In zebrafish, KiSS1 and its receptor KiSS1R are predominantly expressed in the habenula rather than in the hypothalamic area, whereas KiSS2 and KiSS2R are largely distributed in the hypothalamic area (Kitahashi et al., 2009; Servili et al., 2011). This observation suggests that KiSS2 and KiSS2R may be involved in HPG axis regulation. In this species, the KiSS2 peptide is more potent than the KiSS1 peptide at stimulating transcription of GnRH and gonadotropin genes. Furthermore, KiSS2 expression is more sensitive to estrogen feedback than KiSS1 expression (Kitahashi et al., 2009; Servili et al., 2011). Thus, in zebrafish, the functional homologs of mammalian KiSS1 and KiSS1R are likely KiSS2 and KiSS2R (Figure 3). With regard to HPG axis control, zebrafish lacks GnRH1, suggesting that GnRH3 may substitute for this function of GnRH1 (Kah et al., 2007). Salmon species lack GnRH1, whereas catfish and eel lack GnRH3. Because GnRH1 and GnRH3 neurons are both located in the hypothalamus, it is likely that the two complement one another in these species, so that GnRH3 replaces the function of GnRH1 when the latter is lost (e.g., Cypriniformes and most Salmoniformes) and vice versa when GnRH3 is lost (e.g., catfish and eel; Kah et al., 2007; Okubo and Nagahama, 2008).
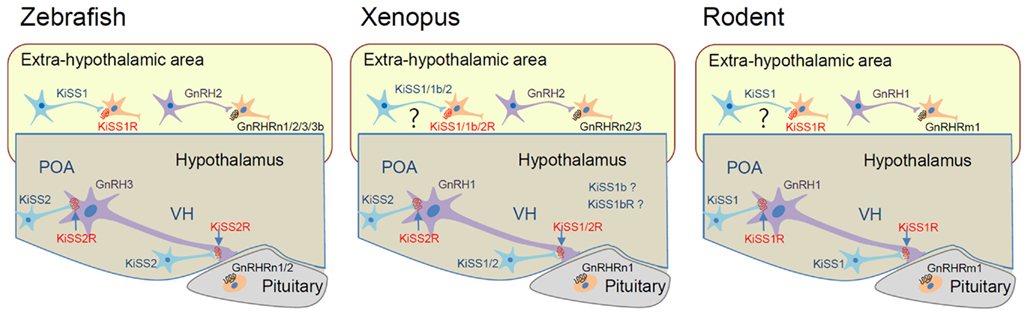
Figure 3. Proposed hypothalamo-pituitary function of GnRH/KiSS and their receptor isoforms in representative vertebrate species. In rodents, KiSS1 neurons in the preoptic area (POA) and ventromedial hypothalamus (VH) control GnRH1 neuronal activity through KiSS1R. GnRH1 action is mediated by GnRHRm1 in the pituitary. In zebrafish, KiSS2 and KiSS2R may be involved in the regulation of GnRH3 neuron in the hypothalamus. KiSS1 and GnRH2 may conduct extrahypothalamic functions through KiSS1R and GnRHRs. GnRH3 action on the pituitary is likely mediated by GnRHRn1 and n2 forms that have relatively high affinity for GnRH3. In Xenopus, both KiSS1 and KiSS2 are likely involved in HPG axis control. Function of KiSS1b and KiSS1bR has not been demonstrated. The GnRHRn1 form in the pituitary may interact with GnRH1 to mediate hypothalamo-pituitary control.
Some fish species, such as stickleback and torafugu, and a reptilian species, the anole lizard, lack KiSS1/KiSS1R. Therefore, KiSS2/KiSS2R in these species may function in a homologous manner to that of mammalian KiSS1/KiSS1R. In Xenopus, KiSS2-expressing neurons are found in the preoptic area and ventromedial hypothalamus, and they send their axons to the median eminence. On the other hand, KiSS1-expressing neurons are present only in the ventromedial hypothalamus (Lee et al., 2009). Thus, both KiSS1 and KiSS2 are likely involved in the control of the HPG axis in this species, with each gene exerting differential control of the GnRH surge and GnRH pulsatile release (Figure 3). Such dual activity suggests the subfunctionalization of KiSS1 and KiSS2 after gene duplication from the ancestral parents. In zebrafish and medaka, KiSS1 and KiSS1R are highly expressed in the habenula, which is not observed in mammals (Kitahashi et al., 2009; Servili et al., 2011), suggesting both genes have become neofunctionalized in these species. Alternatively, KiSS1 and KiSS1R may have become dysfunctionalized in the habenula of mammals.
Interestingly, GnRH1 was not a preferred ligand for any of the non-mammalian GnRHRs, and non-mammalian orthologs of GnRHRm1 have not been definitively identified (Kim et al., 2011). Thus, it has been unclear which isoform of GnRHR is utilized for GnRH1 action in the pituitary of non-mammalian species. Although non-mammalian GnRHRs generally have lower affinity for GnRH1 than GnRH2, some such as bullfrog GnRHR1 (GnRHRn1) and zebrafish GnRHR4 (GnRHRn1) exhibit relatively high affinity for GnRH1. Interestingly, bullfrog GnRHR1 is exclusively expressed in the pituitary (Wang et al., 2001; Tello et al., 2008), suggesting it may interact with GnRH1 to mediate HPG axis control. In zebrafish, GnRHR2 (GnRHRn2) and GnRHR4 exhibit a high affinity for GnRH3 (Tello et al., 2008). Considering GnRH1 is absent in zebrafish, GnRH3 may function like GnRH1 in HPG axis control (Figure 3).
The absence of any KiSS and KiSSR genes in birds raises a fundamental question about the importance of KiSS/KiSSR signaling in vertebrate sexual development and reproduction. A recent finding that ablation of either KiSS1- or KiSS1R-expressing neurons in early development results in the normal onset of puberty in mice suggests these genes may not be required for this activity (Mayer and Boehm, 2011). However, acute ablation of KiSS1- but not KiSSR-expressing cells in adult mice caused an absence of the estrous cycle (Mayer and Boehm, 2011). These observations suggest that KiSS/KiSSR signaling is not the sole system inducing puberty, and that a complementary system can be established in the absence of KiSS1- or KiSSR-expressing cells during puberty. Such a complementary system cannot be established, however, when KiSS1-expressing cells are ablated in the adult period. Whether a complementary system replaces KiSS function in birds needs to be further investigated. The lack of any KiSS/KiSSR genes in chicken and zebra finch indicates that the KiSS/KiSSR system may be completely dysfunctionalized in birds.
Conclusion
The function of GnRH and GnRHR is critical for sexual maturation and reproduction. The existence of GnRH- and GnRHR-like molecules in early vertebrates and invertebrates tells us that regulation of reproduction is highly conserved in animal phyla (Tello and Sherwood, 2009; Lindemans et al., 2011). Further, comparative analyzes of ligand–receptor interaction among various forms of GnRH and GnRHR provide insights into how this ligand–receptor pair coevolved in invertebrates and vertebrates (Tello and Sherwood, 2009). In vertebrates, chromosome/gene duplications have generated multiple paralogous isoforms of GnRH and GnRHR. The presence of lamprey-specific GnRH-I and -III (GnRH4 group), however, reveals more complicated evolutionary scenario of the GnRH gene during early vertebrate evolution (Silver et al., 2004; Kavanaugh et al., 2008). This issue will be further disclosed when genome data of Agnathan and Chondrichthyan species are available. The occurrence of multiple KiSS and KiSSR isoforms has made the regulation of sexual maturation and reproduction in vertebrates even more diverse and complex. Although the amino acid sequences in GnRH and KiSS paralogs have diverged, accompanying amino acid changes have occurred in the cognate receptors, thereby maintaining high selectivity of the ligand–receptor interaction. Some paralogous isoforms acquired a new function, whereas others preserved the ancestral one. In other cases, each paralogous isoform took on a distinct subset of the ancestral functions, complementing each other to fulfill the ancestral function. Finally, some paralogs have been functionally lost, becoming pseudogenes or disappearing completely from the genome. Thus, after duplications of the KiSS and GnRH systems in early vertebrates, the paralogs of KiSS/KiSSR and GnRH/GnRHR became neofunctionalized, subfunctionalized, or dysfunctionalized, and the function related to regulation of reproduction was adopted by either of the KiSS/GnRH isoforms during evolution.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants (2011K00277) from the Brain Research Center of the Twenty-First Century Frontier Research Program, the Brain Research Program of the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0019205), and Science and Technology Medical Research Program to Jae Young Seong and Hubert Vaudry.
Abbreviations
APVN, anteroventral periventricular nucleus; ARC, arcuate nucleus; ECL, extracellular loop; GnRH, gonadotropin-releasing hormone; GnRHR, gonadotropin-releasing hormone receptor; GPCR, G protein-coupled receptor; HPG, hypothalamic-pituitary–gonadal; KiSS, kisspeptin; KiSSR, kisspeptin receptor.
References
Acharjee, S., Do-Rego, J. L., Oh, D. Y., Ahn, R. S., Choe, H., Vaudry, H., Kim, K., Seong, J. Y., and Kwon, H. B. (2004). Identification of amino acid residues that direct differential ligand selectivity of mammalian and nonmammalian V1a type receptors for arginine vasopressin and vasotocin. Insights into molecular coevolution of V1a type receptors and their ligands. J. Biol. Chem. 279, 54445–54453.
Cho, H. J., Acharjee, S., Moon, M. J., Oh, D. Y., Vaudry, H., Kwon, H. B., and Seong, J. Y. (2007). Molecular evolution of neuropeptide receptors with regard to maintaining high affinity to their authentic ligands. Gen. Comp. Endocrinol. 153, 98–107.
Conlon, J. M., and Larhammar, D. (2005). The evolution of neuroendocrine peptides. Gen. Comp. Endocrinol. 142, 53–59.
d’Anglemont de Tassigny, X., Fagg, L. A., Dixon, J. P., Day, K., Leitch, H. G., Hendrick, A. G., Zahn, D., Franceschini, I., Caraty, A., Carlton, M. B., Aparicio, S. A., and Colledge, W. H. (2007). Hypogonadotropichypogonadism in mice lacking a functional Kiss1 gene. Proc. Natl. Acad. Sci. U.S.A. 104, 10714–10719.
de Roux, N., Genin, E., Carel, J. C., Matsuda, F., Chaussain, J. L., and Milgrom, E. (2003). Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. U.S.A. 100, 10972–10976.
Fernald, R. D., and White, R. B. (1999). Gonadotropin-releasing hormone genes: phylogeny, structure, and functions. Front. Neuroendocrinol. 20, 224–240.
Flanagan, C. A., Becker, II Davidson, J. S., Wakefield, I. K., Zhou, W., Sealfon, S. C., and Millar, R. P. (1994). Glutamate 301 of the mouse gonadotropin-releasing hormone receptor confers specificity for arginine 8 of mammalian gonadotropin-releasing hormone. J. Biol. Chem. 269, 22636–22641.
Fredriksson, R., Lagerstrom, M. C., Lundin, L. G., and Schioth, H. B. (2003). The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272.
Fromme, B. J., Katz, A. A., Roeske, R. W., Millar, R. P., and Flanagan, C. A. (2001). Role of aspartate7.32(302) of the human gonadotropin-releasing hormone receptor in stabilizing a high-affinity ligand conformation. Mol. Pharmacol. 60, 1280–1287.
Hoyle, C. H. (1999). Neuropeptide families and their receptors: evolutionary perspectives. Brain Res. 848, 1–25.
Hrabovszky, E., Kallo, I., Szlavik, N., Keller, E., Merchenthaler, I., and Liposits, Z. (2007). Gonadotropin-releasing hormone neurons express estrogen receptor-beta. J. Clin. Endocrinol. Metab. 92, 2827–2830.
Ikemoto, T., and Park, M. K. (2003). Identification and characterization of the reptilian GnRH-II gene in the leopard gecko, Eublepharis macularius, and its evolutionary considerations. Gene 316, 157–165.
Ikemoto, T., and Park, M. K. (2007). Comparative analysis of the pituitary and ovarian GnRH systems in the leopard gecko: signaling crosstalk between multiple receptor subtypes in ovarian follicles. J. Mol. Endocrinol. 38, 289–304.
Kah, O., Lethimonier, C., Somoza, G., Guilgur, L. G., Vaillant, C., and Lareyre, J. J. (2007). GnRH and GnRH receptors in metazoa: a historical, comparative, and evolutive perspective. Gen. Comp. Endocrinol. 153, 346–364.
Kavanaugh, S. I., Nozaki, M., and Sower, S. A. (2008). Origins of gonadotropin-releasing hormone (GnRH) in vertebrates: identification of a novel GnRH in a basalvertebrate, the sealamprey. Endocrinology 149, 3860–3869.
Kim, D. K., Cho, E. B., Moon, M. J., Park, S., Hwang, J. I., Kah, O., Sower, S. A., Vaudry, H., and Seong, J. Y. (2011). Revisiting the evolution of gonadotropin-releasing hormones and their receptors in vertebrates: secrets hidden in genomes. Gen. Comp. Endocrinol. 170, 68–78.
Kinoshita, M., Tsukamura, H., Adachi, S., Matsui, H., Uenoyama, Y., Iwata, K., Yamada, S., Inoue, K., Ohtaki, T., Matsumoto, H., and Maeda, K. (2005). Involvement of central metastin in the regulation of preovulatory luteinizing hormone surge and estrous cyclicity in female rats. Endocrinology 146, 4431–4436.
Kitahashi, T., Ogawa, S., and Parhar, I. S. (2009). Cloning and expression of kiss2 in the zebrafish and medaka. Endocrinology 150, 821–831.
Kotani, M., Detheux, M., Vandenbogaerde, A., Communi, D., Vanderwinden, J. M., Le Poul, E., Brezillon, S., Tyldesley, R., Suarez-Huerta, N., Vandeput, F., Blanpain, C., Schiffmann, S. N., Vassart, G., and Parmentier, M. (2001). The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J. Biol. Chem. 276, 34631–34636.
Larhammar, D., Lundin, L. G., and Hallbook, F. (2002). The human Hox-bearing chromosome regions did arise by block or chromosome (or even genome) duplications. Genome Res. 12, 1910–1920.
Larsson, T. A., Olsson, F., Sundstrom, G., Lundin, L. G., Brenner, S., Venkatesh, B., and Larhammar, D. (2008). Early vertebrate chromosome duplications and the evolution of the neuropeptide Y receptor gene regions. BMC Evol. Biol. 8, 184. doi:10.1186/1471-2148-8-184
Lee, Y. R., Tsunekawa, K., Moon, M. J., Um, H. N., Hwang, J. I., Osugi, T., Otaki, N., Sunakawa, Y., Kim, K., Vaudry, H., Kwon, H. B., Seong, J. Y., and Tsutsui, K. (2009). Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology 150, 2837–2846.
Li, J. H., Choe, H., Wang, A. F., Maiti, K., Wang, C., Salam, A., Chun, S. Y., Lee, W. K., Kim, K., Kwon, H. B., and Seong, J. Y. (2005). Extracellular loop 3 (EL3) and EL3-proximal transmembrane helix 7 of the mammalian type I and type II gonadotropin-releasing hormone (GnRH) receptors determine differential ligand selectivity to GnRH-I and GnRH-II. Mol. Pharmacol. 67, 1099–1110.
Lindemans, M., Janssen, T., Beets, I., Temmerman, L., Meelkop, E., and Schoofs, L. (2011). Gonadotropin-releasing hormone and adipokinetic hormone signaling systems share a common evolutionary origin. Front. Endocrinol. 2:16. doi:10.3389/fendo.2011.00016
Lundin, L. G. (1993). Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16, 1–19.
Mayer, C., and Boehm, U. (2011). Female reproductive maturation in the absence of kisspeptin/GPR54 signaling. Nat. Neurosci. 14, 704–710.
Messager, S., Chatzidaki, E. E., Ma, D., Hendrick, A. G., Zahn, D., Dixon, J., Thresher, R. R., Malinge, I., Lomet, D., Carlton, M. B., Colledge, W. H., Caraty, A., and Aparicio, S. A. (2005). Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc. Natl. Acad. Sci. U.S.A. 102, 1761–1766.
Millar, R., Lowe, S., Conklin, D., Pawson, A., Maudsley, S., Troskie, B., Ott, T., Millar, M., Lincoln, G., Sellar, R., Faurholm, B., Scobie, G., Kuestner, R., Terasawa, E., and Katz, A. (2001). A novel mammalian receptor for the evolutionarily conserved type II GnRH. Proc. Natl. Acad. Sci. U.S.A. 98, 9636–9641.
Millar, R. P., Lu, Z. L., Pawson, A. J., Flanagan, C. A., Morgan, K., and Maudsley, S. R. (2004). Gonadotropin-releasing hormone receptors. Endocr. Rev. 25, 235–275.
Moon, M. J., Kim, H. Y., Park, S., Kim, D. K., Cho, E. B., Park, C. R., You, D. Y., Hwang, J.-I., Kim, K., Choe, H., and Seong, J. Y. (2012). Evolutionarily conserved residues at the GLP-1 receptor core confer ligand-induced receptor activation. J. Biol. Chem. doi: 10.1074/jbc.mill.276808. [Epub ahead of print].
Muir, A. I., Chamberlain, L., Elshourbagy, N. A., Michalovich, D., Moore, D. J., Calamari, A., Szekeres, P. G., Sarau, H. M., Chambers, J. K., Murdock, P., Steplewski, K., Shabon, U., Miller, J. E., Middleton, S. E., Darker, J. G., Larminie, C. G., Wilson, S., Bergsma, D. J., Emson, P., Faull, R., Philpott, K. L., and Harrison, D. C. (2001). AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J. Biol. Chem. 276, 28969–28975.
Oakley, A. E., Clifton, D. K., and Steiner, R. A. (2009). Kisspeptin signaling in the brain. Endocr. Rev. 30, 713–743.
Oh, D. Y., Kim, K., Kwon, H. B., and Seong, J. Y. (2006). Cellular and molecular biology of orphan G protein-coupled receptors. Int. Rev. Cytol. 252, 163–218.
Ohkura, S., Uenoyama, Y., Yamada, S., Homma, T., Takase, K., Inoue, N., Maeda, K., and Tsukamura, H. (2009). Physiological role of metastin/kisspeptin in regulating gonadotropin-releasing hormone (GnRH) secretion in female rats. Peptides 30, 49–56.
Ohtaki, T., Shintani, Y., Honda, S., Matsumoto, H., Hori, A., Kanehashi, K., Terao, Y., Kumano, S., Takatsu, Y., Masuda, Y., Ishibashi, Y., Watanabe, T., Asada, M., Yamada, T., Suenaga, M., Kitada, C., Usuki, S., Kurokawa, T., Onda, H., Nishimura, O., and Fujino, M. (2001). Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature 411, 613–617.
Okubo, K., and Nagahama, Y. (2008). Structural and functional evolution of gonadotropin-releasing hormone in vertebrates. Acta Physiol. (Oxf) 193, 3–15.
Powell, J. F., Zohar, Y., Elizur, A., Park, M., Fischer, W. H., Craig, A. G., Rivier, J. E., Lovejoy, D. A., and Sherwood, N. M. (1994). Three forms of gonadotropin-releasing hormone characterized from brains of one species. Proc. Natl. Acad. Sci. U.S.A. 91, 12081–12085.
Schally, A. V., Arimura, A., Kastin, A. J., Matsuo, H., Baba, Y., Redding, T. W., Nair, R. M., Debeljuk, L., and White, W. F. (1971). Gonadotropin-releasing hormone: one polypeptide regulates secretion of luteinizing and follicle-stimulating hormones. Science 173, 1036–1038.
Seminara, S. B., Messager, S., Chatzidaki, E. E., Thresher, R. R., Acierno, J. S. Jr., Shagoury, J. K., Bo-Abbas, Y., Kuohung, W., Schwinof, K. M., Hendrick, A. G., Zahn, D., Dixon, J., Kaiser, U. B., Slaugenhaupt, S. A., Gusella, J. F., O’Rahilly, S., Carlton, M. B., Crowley, W. F. Jr., Aparicio, S. A., and Colledge, W. H. (2003). The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 349, 1614–1627.
Servili, A., Le Page, Y., Leprince, J., Caraty, A., Escobar, S., Parhar, I. S., Seong, J. Y., Vaudry, H., and Kah, O. (2011). Organization of two independent kisspeptin systems derived from evolutionary-ancient kiss genes in the brain of zebrafish. Endocrinology 152, 1527–1540.
Silver, M. R., Kawauchi, H., Nozaki, M., and Sower, S. A. (2004). Cloning and analysis of the lamprey GnRH-III cDNA from eight species of lamprey representing the three families of Petromyzontiformes. Gen. Comp. Endocrinol. 139, 85–94.
Tello, J. A., and Sherwood, N. M. (2009). Amphioxus: beginning of vertebrate and end of invertebrate type GnRH receptor lineage. Endocrinology 150, 2847–2856.
Tello, J. A., Wu, S., Rivier, J. E., and Sherwood, N. M. (2008). Four functional GnRH receptors in zebrafish: analysis of structure, signaling, synteny and phylogeny. Integr. Comp. Biol. 48, 570–587.
Um, H. N., Han, J. M., Hwang, J. I., Hong, S. I., Vaudry, H., and Seong, J. Y. (2010). Molecular coevolution of kisspeptins and their receptors from fish to mammals. Ann. N. Y. Acad. Sci. 1200, 67–74.
Wang, C., Yun, O., Maiti, K., Oh, D. Y., Kim, K. K., Chae, C. H., Lee, C. J., Seong, J. Y., and Kwon, H. B. (2004). Position of Pro and Ser near Glu7.32 in the extracellular loop 3 of mammalian and nonmammalian gonadotropin-releasing hormone (GnRH) receptors is a critical determinant for differential ligand selectivity for mammalian GnRH and chicken GnRH-II. Mol. Endocrinol. 18, 105–116.
Wang, L., Bogerd, J., Choi, H. S., Seong, J. Y., Soh, J. M., Chun, S. Y., Blomenrohr, M., Troskie, B. E., Millar, R. P., Yu, W. H., McCann, S. M., and Kwon, H. B. (2001). Three distinct types of GnRH receptor characterized in the bullfrog. Proc. Natl. Acad. Sci. U.S.A. 98, 361–366.
Keywords: kisspeptin, gonadotropin-releasing hormone, G protein-coupled receptors, comparative genomics, evolution
Citation: Kim D-K, Cho EB, Moon MJ, Park S, Hwang J-I, Do Rego J-L, Vaudry H and Seong JY (2012) Molecular coevolution of neuropeptides gonadotropin-releasing hormone and kisspeptin with their cognate G protein-coupled receptors. Front. Neurosci. 6:3. doi: 10.3389/fnins.2012.00003
Received: 22 November 2011;
Accepted: 05 January 2012;
Published online: 24 January 2012.
Edited by:
Billy K. C. Chow, University of Hong Kong, ChinaReviewed by:
Jack Falcon, Université Pierre et Marie Curie, FranceLeo T. O. Lee, The University of Hong Kong, Hong Kong
Copyright: © 2012 Kim, Cho, Moon, Park, Hwang, Do Rego, Vaudry and Seong. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Jae Young Seong, Graduate School of Medicine, Korea University, Seoul 136-705, South Korea. e-mail:anlzZW9uZ0Brb3JlYS5hYy5rcg==
 Dong-Kyu Kim1
Dong-Kyu Kim1


