- Institute of Neuroinformatics, University of Zurich and ETH Zurich, Zurich, Switzerland
Assessment of three-dimensional morphological structure and synaptic connectivity is essential for a comprehensive understanding of neural processes controlling behavior. Different microscopy approaches have been proposed based on light microcopy (LM), electron microscopy (EM), or a combination of both. Correlative array tomography (CAT) is a technique in which arrays of ultrathin serial sections are repeatedly stained with fluorescent antibodies against synaptic molecules and neurotransmitters and imaged with LM and EM (Micheva and Smith, 2007). The utility of this correlative approach is limited by the ability to preserve fluorescence and antigenicity on the one hand, and EM tissue ultrastructure on the other. We demonstrate tissue staining and fixation protocols and a workflow that yield an excellent compromise between these multimodal imaging constraints. We adapt CAT for the study of projection neurons between different vocal brain regions in the songbird. We inject fluorescent tracers of different colors into afferent and efferent areas of HVC in zebra finches. Fluorescence of some tracers is lost during tissue preparation but recovered using anti-dye antibodies. Synapses are identified in EM imagery based on their morphology and ultrastructure and classified into projection neuron type based on fluorescence signal. Our adaptation of array tomography, involving the use of fluorescent tracers and heavy-metal rich staining and embedding protocols for high membrane contrast in EM will be useful for research aimed at statistically describing connectivity between different projection neuron types and for elucidating how sensory signals are routed in the brain and transformed into a meaningful motor output.
Introduction
Acquiring systematic information about synaptic connectivity in the brain is currently one of the greatest challenges in neuroscience. In recent years, significant effort has been invested into developing staining and microscopy techniques to speed up, automate, and make acquisition of large image volumes of brain tissue possible. Different strategies have been pursued, some based on electron microscopy (EM), some on light microcopy (LM), and some on a combination of EM and LM.
The strength of EM-based techniques is their high spatial resolution. Among established EM techniques are serial block-face scanning electron microscopy (SBFSEM), focused ion beam scanning electron microscopy (FIBSEM), serial section transmission electron microscopy (ssTEM), and serial section scanning electron microscopy. In the SBFSEM technique (Denk and Horstmann, 2004), block-face imaging in a scanning electron microscope (SEM) is combined with serial sectioning with a diamond knife of a resin-embedded brain block inside the microscope chamber. SBFSEM allows sectioning of large areas of tissue for hundreds of consecutive sections and is therefore suited for high-resolution imaging of large tissue volumes. An alternative technique for automated cutting and block-face imaging, FIBSEM, relies on using a focused ion beam for milling thin layers of embedded tissue (Knott et al., 2008; Merchan-Perez et al., 2009). The ion beam mills away few nanometer thick layers of tissue, giving rise to close-to-isotropic volumetric data that facilitates the tracking of small neurites through the tissue. The FIBSEM technique is currently limited by the inability to mill areas larger than a few hundreds of square micrometers, because the focused ion beam cannot be deflected arbitrarily. FIBSEM is therefore less suited for imaging of large volumes. Also, block-face techniques such as SBFSEM and FIBSEM have the common disadvantage of section loss because the cut ultrathin sections cannot be collected. It is therefore not possible to repeatedly image a region after the block has been cut, which would be desirable in case that analysis of the imaged volume reveals that higher-resolution images of certain sections are needed. Another disadvantage of block-face EM techniques is that immunohistochemical stainings can only be performed before tissue embedding but not thereafter.
A third EM approach relies on high-throughput ssTEM. Different frameworks have been proposed to automate image acquisition, image tile, and serial section registration, and take advantage of the high imaging speed of transmission electron microscopes (TEM; Anderson et al., 2009; Cardona et al., 2010). Unlike block-face techniques, ssTEM allows for repeated imaging of sections and post-embedding immunolabeling. However ssTEM involves cutting and collecting hundreds of serial sections on small and fragile TEM grids. Preparation necessitates therefore a skillful and trained operator, because section loss can lead to incompleteness in the reconstruction. Moreover, compared to FIBSEM and SBFSEM, ssTEM has the disadvantages that sections with current technology cannot be cut thinner than 30–40 nm, compared to few nanometers with FIBSEM, and that imaged sections need to be registered between each other, since their orientation is lost during preparation.
All EM-based techniques provide highly resolved brain volumes. However, they suffer from the inherent drawback of EM to necessitate long imaging and tracing times. Complete EM reconstruction of vertebrate brains requires a prohibitive amount of time (see Helmstaedter et al., 2008 for estimations of reconstruction time). Hence, if long-range projections from and to a specific area cannot be contained in their entirety in the reconstructed volume, they need to be identified using electron-dense tracers (Anderson et al., 1994; da Costa and Martin, 2009). The main disadvantage of electron-dense tracers is that only a limited number of them can be used simultaneously (Smith and Bolam, 1991; Lanciego et al., 1998; Reiner et al., 2000), and therefore only few projection neuron types can be distinguished in a single brain. Therefore, using EM-only methods, it is difficult to describe the connectivity between different brain regions in terms of interactions between different projection neuron types.
In projectomics, the circuitry of inputs and outputs of brain regions is described without necessarily characterizing the local network in similar detail. This approach is different from connectomics, in which a local network is described exhaustively in terms of all synaptic connections present. Whereas connectomics involves a complete EM reconstruction of the region of interest, projectomics requires ways of distinguishing different projections neurons without needing a complete EM reconstruction of the entire brain. LM can overcome the limitation of few distinguishable electron-dense tracers and allows labeling of many more projection neuron populations. These populations can be labeled by injection of different colors into the different brain regions or through expression of fluorescent proteins by viral or transgenic methods.
Different light-based methods have been advertised for circuit reconstruction. The Brainbow is a transgenic technology in which neural circuits are visualized by genetically labeling neurons with different proportions of multiple colors (Livet et al., 2007). Based on proportion differences, neuronal processes belonging to a single cell can be identified in different parts of the brain without needing to completely trace the neuron. Another LM-based technique is array tomography, in which ordered arrays of ultrathin resin-embedded serial sections are repeatedly stained and imaged in the light microscope. In array tomography, large-field volumetric imaging of large numbers of antigens and fluorescent proteins is possible (Micheva and Smith, 2007; Micheva et al., 2010b). Due to small section thickness, the z-resolution of array tomography is comparable to that of EM and much smaller than that of two-photon or confocal imaging. However, the same is not true for the x–y resolution, which in classical LM is smaller than in EM by orders of magnitudes. Recent advances such as stimulated emission depletion (STED) microscopy have increased LM resolution beyond Abbé’s limit, allowing for live imaging and measurement of details including spine neck width and the curvature of the heads of spines (Nagerl et al., 2008; Nagerl and Bonhoeffer, 2010). However STED currently is not an alternative to EM, because exhaustive detection of smaller details such as synaptic vesicles and membrane specializations (necessary for identification of functional synapses) still needs EM today. In addition, with LM it is common to observe signal discontinuities because of partially unlabeled structures. Such discontinuities hinder dense reconstructions of neural circuits. Hence, many dense and high-resolution reconstruction tasks may not be feasible without EM.
Correlative Microscopy for Neural Circuit Reconstruction
In the correlative LM–EM approach, fluorescence LM provides information about neurons and their synapses inside a small subvolume, which is then reconstructed in EM imagery. Multiple neurons or neuronal types are distinguished based on their fluorescence labels and their synapses are detected with EM. As a consequence, there is no need to reconstruct neurons over long distances. Unfortunately, to combine LM and EM is not straightforward in most cases.
Chemical preparation for EM leads to strong fluorescence reduction if the aims are good ultrastructure preservation and high membrane contrast. In addition, during tissue preparation, morphological changes such as tissue shrinkage can occur. Because of such tissue transformations, it is not possible today to directly superpose three-dimensional LM imagery taken prior to embedding with EM micrographs. Although such a correlative microscopy approach would be conceptually very simple and desirable, it is still not feasible. In addition, the correlative pre-embedding LM and EM approach suffers from problem of low z-resolution of LM imagery, rendering it difficult to unambiguously associate synaptic LM labels with the correct synapse seen in the EM.
The idea of correlative array tomography (CAT) is to combine LM and EM imagery on the same ultrathin section, thus overcoming the aforementioned problems of low z-resolution and tissue shrinkage. In CAT, endogenous antigens such as tubulin, GABA, SNAP-25, or synapsin, can be labeled with immunofluorescence using staining and fixation protocols also suitable for EM (Micheva and Smith, 2007; Micheva et al., 2010b). It is an open question, however, whether tissue preparation protocols compatible with immunoreactivity and EM also deliver an ultrastructure quality high enough for neural circuit reconstruction. As a step in this direction, we extended the CAT approach by first labeling multiple types of projection neurons using fluorescent tracers and then fixing, staining, and embedding the tissue aiming for high-quality EM ultrastructure, suitable for circuit reconstruction (Oberti et al., 2010). Although our staining and fixation protocols reduce tissue antigenicity and are therefore less suited for detection of endogenous molecules, we show that tracer signal lost during embedding can be recovered using fluorescent antibodies against the tracers, and at the same time synapses can be well resolved in EM. We apply this CAT approach to projection neurons in our animal model, the zebra finch.
The Songbird
The zebra finch is a good animal model to study a complex sensory–motor behavior. Zebra finches are able to imitate the song that they hear sung by their tutors. During a sensory phase, the juvenile animal listens to the tutor and memorizes its song, while in a later sensorimotor phase the juvenile vocalizes and uses auditory feedback to match its own song with the memorized tutor song (Konishi, 1965). Identification of different projection neuron populations is essential to investigate how sensory information enters the brain, how it is processed there, and how a meaningful output is generated. In the zebra finch, projection neurons can be easily labeled because the avian brain is organized in segregated nuclei. Their projection neurons can therefore be labeled by targeted injection of fluorescent tracers of different colors, one for each brain area.
A specialized set of brain areas is involved in song learning and production (Figure 1). One of these is HVC (used as a proper name), a premotor area in the forebrain which drives motor output by a sparse sequence of bursts but also receives auditory information (Nottebohm and Arnold, 1976; Hahnloser et al., 2002, 2008; Long et al., 2010; Roberts et al., 2010). HVC receives input from the nucleus interface of the nidopallium (NIf) and from the thalamic nucleus uveaformis (Uva). HVC contains a population of neurons projecting to the robust nucleus of the arcopallium (RA), which in turns relays information to the motor neurons of the vocal organ and to respiratory areas. Another population of HVC neurons projects to the basal ganglia nucleus Area X, which is involved in generating song variability (Scharff and Nottebohm, 1991; Reiner et al., 2004; Olveczky et al., 2005).
In our strategy, we use fluorescent tracers to label neuronal processes according to their projection target and use EM to visualize the circuit context of the labeled structures (Oberti et al., 2010).
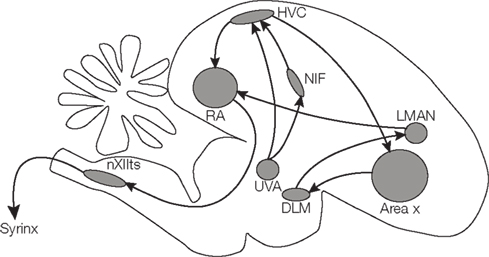
Figure 1. Schematic drawing of the songbird brain, highlighting regions involved in song learning and song production.
Workflow for Correlative Array Tomography Using Neural Tracers in the Songbird
We inject dextran-coupled fluorophores of different colors into the living brain (Figure 2A). After the tracers have diffused, we perfuse the animal with 2% paraformaldehyde and 0.075% glutaraldehyde and slice its brain with a vibratome (Figure 2B). Light microscopy examination of the sections allows us to localize the region of interest, to evaluate quality of labeling, and to proof correctness of the injection sites. We then process the sections for EM. The tissue is stained and fixed with heavy metals (40 min 1.5% potassium ferrocyanide and 1% osmium tetroxide, 40 min 1% osmium tetroxide, 1 h 1% uranyl acetate) dehydrated in a graded series of ethanol dilutions, and finally embedded in an epoxy resin (Durcupan ACM resin, Fluka, Buchs, Switzerland). This preparation results in hardened sections, which, after trimming to smaller pieces, we can cut in ribbons of ultrathin serial sections using a diamond knife (Figure 2C). We put ultrathin sections on various substrates such as pioloform film, glass coated with an electron-conductive substrate, or other electron-conductive materials (such as silicon wafer, as in the examples in this paper).
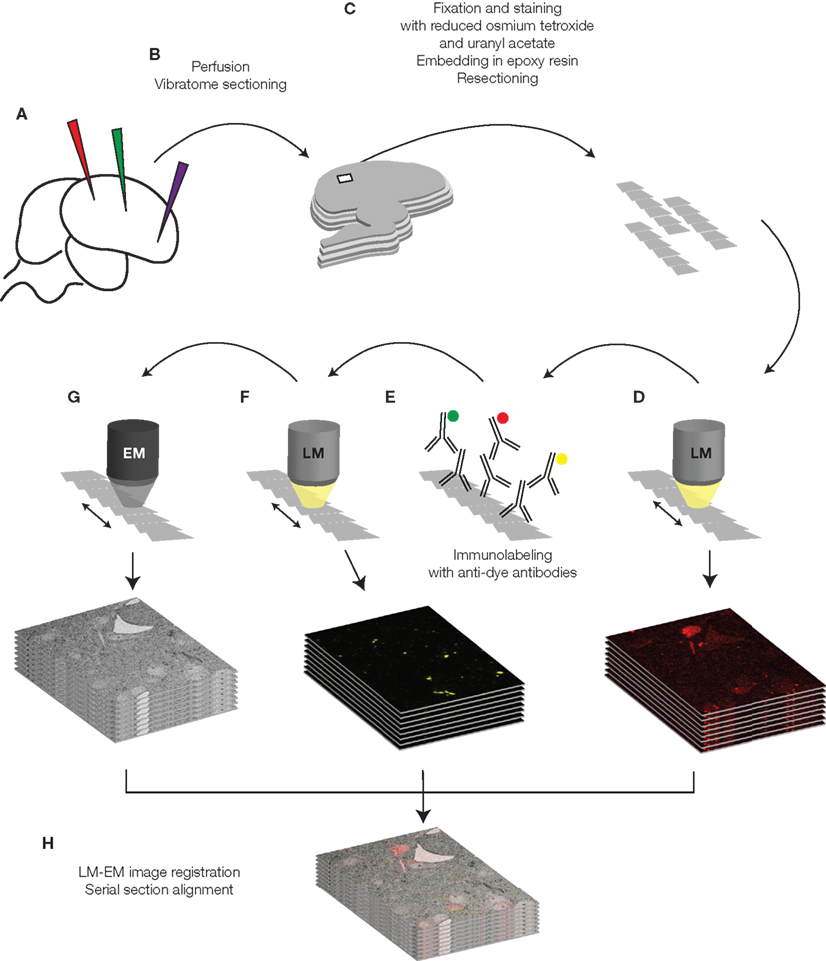
Figure 2. Schematic drawing of the tissue preparation and imaging process. The different steps are described in detail in the text.
We then image the collected ultrathin sections in a conventional wide-field fluorescence microscope (Figure 2D). Exposure times in the order of several seconds are necessary (5–20 s). Preparation of the tissue for EM causes a strong reduction of the fluorescence of the tracers, which varies depending on the fluorophore and can reach in some cases complete quenching. We therefore introduced an additional step to regain fluorescence signal by immunolabeling against the fluorophores (Figure 2E). First the sections are treated with periodic acid and sodium metaperiodate, chemicals which facilitate accessibility and binding of the antibodies to the resin-embedded and osmificated tissue. Subsequently we incubate the sections with primary antibodies that bind to the fluorophores, followed by fluorescent secondary antibodies.
We finally image immunostained sections with LM and EM (Figures 2F,G). In the EM we locate a region of interest previously defined in LM imagery using landmarks such as section borders, blood vessels (visible in some fluorescence channels) and stained cell somata, which can be easily identified in EM images even at low magnification.
After images are acquired with the EM, we align them with LM images using the previously mentioned landmarks or using features such as somata borders or tracer-filled vesicles in retrograde stained cells. The final data consists of multichannel LM pictures superimposed to EM imagery (Figure 2H).
Correlative Microscopy of Songbird HVC
Figure 3 shows examples of correlative images of zebra finch HVC. The animal was injected with two different tracers in the afferent nucleus Uva (Texas Red dextran) and the efferent nucleus RA (Lucifer Yellow dextran). After resin embedding, we cut ultrathin sections and acquired images of Texas Red direct fluorescence with a LM (Figure 3, left column, red signal). Lucifer Yellow fluorescence was completely quenched during the preparation, but we detected it anew using anti-Lucifer Yellow antibodies and fluorescent secondary antibodies for signal amplification (Figure 3, left column, yellow signal). Sections were subsequently imaged with an SEM using an energy-filtered detector for backscattered electrons (Figure 3, middle column). Finally the two image sets were superimposed (Figure 3, right column, and Figure 4). Texas Red fluorescence that was detected in ultrathin sections without antibody staining survived preparation and was colocalized with retrogradely labeled cell somata (Figures 3C,D, asterisks) and with smaller neuronal processes (Figures 3A,B, asterisks). Lucifer Yellow-dextran tracer, relabeled with antibodies, also colocalized with small neurites (Figures 3C,D, arrows) as well as with presynaptic terminals (Figures 3A,B, arrows). Alignment of the two image sets was based on landmarks such as section borders and blood vessels, and necessitated only rotation, translation, and scaling of the images. Physical deformation of the sections due to the electron beam, as it occurs frequently in TEM, was absent, because the sections were mounted on a silicon wafer, which is a rigid substrate. Optical deformations were measured and found to be minimal in our SEM and negligible in the LM. The reliability of immunostaining signal can be assessed by inspecting the correspondence of immunolabeled structures in consecutive sections (Figure 3). To be conservative, it can be assumed that fluorescence signal present in only individual sections forms false positive staining, because tracer-filled neurons span multiple ultrathin sections. We suspect that our method is more susceptible to false negatives, because small neurites may not be labeled because of insufficient tracer concentration. Tracing small neurites in EM image stacks of several ultrathin sections, however, should allow us to find a region in which the neuron is larger and more likely contains reliably detectable tracer. This tracing should be possible thanks to the good preservation of tissue ultrastructure (Figure 4).
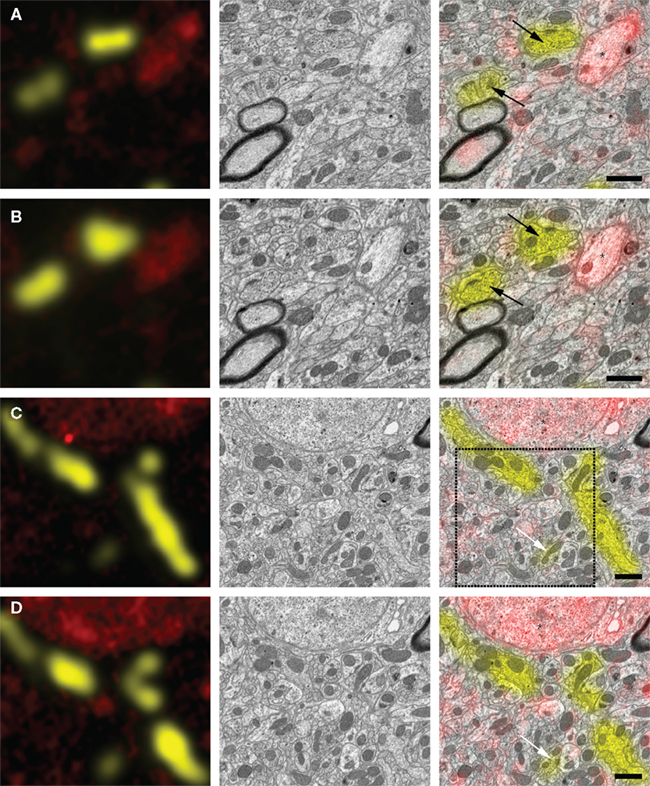
Figure 3. Correlation of EM and LM micrographs of serial ultrathin sections. Left column: superposition of light micrographs of two fluorescence channels associated with two different projection neuron populations (red: Texas Red-dextran direct fluorescence of RA-projecting HVC neruons; yellow: anti-Lucifer Yellow immunolabeling of HVC-projecting Uva neurons). Middle column: SEM micrographs of the same region as in the left column. Right column: immunofluorescence image is superimposed on the SEM micrograph, allowing for classification of synaptic terminals [black arrows in (A,B)], small neural processes [white arrows in (C,D), asterisk in (A,B)], and cell somata [asterisks in (C,D)]. (A,B), as well as (C,D) are images taken from adjacent section pairs: immunofluorescence signal on neighboring sections coincides with the same structures as identified in the EM. The region enclosed by the dashed square is represented in higher magnification in Figure 4. Scale bar 1 μm.
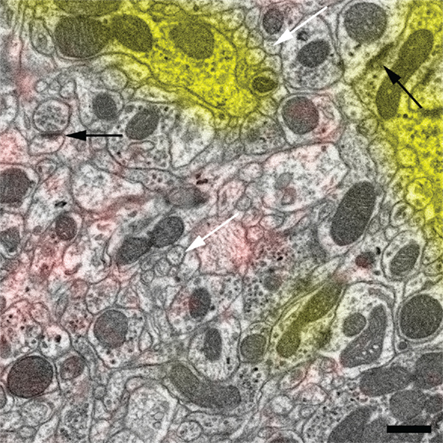
Figure 4. Magnified view of the region in Figure 3C enclosed by the dashed square. The high-quality ultrastructure is illustrated by membranes of small neurites that have no gap artifacts and appear fully closed (white arrows) and by highly contrasted synaptic densities (black arrows). Scale bar 500 nm.
Superposition of the two image sets (Figure 3, right column) allows therefore classification of structures observed in the EM based on LM information. EM-identified synapses can be assigned to a specific projection neuron class (Figures 3A,B, arrows) based on the fluorescence signal in few ultrathin sections, without needing to trace the neuron in a big volume (for additional examples, see Oberti et al., 2010).
Electron microscopy imaging can be done either in the SEM or the TEM. In the first case, sections can be mounted on a variety of electron-conductive substrates, including coated glass and silicon wafers. These substrates allow the collection of long ribbons of serial sections, which are useful in large imaging tasks such as in array tomography (Micheva and Smith, 2007). SEM has the disadvantage of slower imaging speed compared to TEM. With TEM, imaging is typically faster, but sections need to be mounted on a fragile electron-transparent substrate such as pioloform film on a slot grid. Slot grids have a size limited to few millimeters; therefore, only few serial sections can be mounted on each of them. As a consequence TEM imaging is more labor-intensive and error-prone than SEM imaging.
The applicability of multiple tracer types is only limited by the signal to noise ratio of the image data and the availability of antibodies. Immunostaining of multiple tracers on the same section can be done using antibodies raised in different species, or, if the antibodies are all raised in the same species, using consecutive staining and antibody elution rounds (Micheva et al., 2010a). Although the elution protocols developed for array tomography also work when the tissue is embedded in resins other than LR White such as epoxy resins, it remains to be tested whether the excellent ultrastructure and integrity of the ultrathin sections we achieved are affected by the antibody elution. In our work, we embedded the tissue in Durcupan resin because we found the ultrastruture to be better preserved than in LR White. In future work we will seek to find a substrate which on the one hand binds to the Durcupan-embedded sections so that these do not detach due to physical stress during preparation and elution, and on the other hand is inert to the chemicals used for elution and is also electron-conductive for EM imaging.
Alternative Approaches to Correlative Microscopy
Other tissue preparation strategies have been proposed for combining LM and EM. Photooxidation is a technique in which fluorescent dyes are used to oxidize diaminobenzidine into an electron-dense osmiophilic polymer (Maranto, 1982; Deerinck et al., 1994). Photooxidation has the advantage of allowing direct and unambiguous correlation of the same structures in the LM and EM, because the fluorescent labels imaged in the LM are converted into labels visible in the EM. However, this technique has so far not allowed to convert different colors into substrates distinguishable in the EM.
Cryosectioning has also been proposed as a useful technique for correlative microscopy, in particular combined with immunolabeling methods using gold- or FluoroNanogold-labeled antibodies (Takizawa et al., 1998; Slot and Geuze, 2007; van Rijnsoever et al., 2008). Cryosectioning leads to a better antigenicity preservation compared to classical chemical preparation, but its applicability for imaging of large volumes may be limited by section instability – it is almost impossible to reliably collect large numbers of serial sections.
Quantum dots have been used for correlated LM and EM imaging (Giepmans et al., 2005). These small nanocrystals are both fluorescent and electron dense, and they can be discriminated by their color in the LM and their size and shape in the EM, allowing labeling of multiple antigens simultaneously. Unfortunately, to our knowledge quantum dots are not available yet in a form that can be injected into the brain as a neuronal tracer, for example coupled to dextrans.
Our CAT approach to projectomics does not require complete imaging and reconstruction of large volumes. Instead, we expect to gain statistical information about neuron types by reconstructing small, local volumes, in which part of neurons are recognized via their label. Our approach can be applied to different animal models. Connectivity in subcortical regions of the mammalian brain, such as interactions between hypothalamic and brainstem nuclei, could be investigated by targeted injections of fluorescent tracers combined with EM imaging. Moreover, immunostaining against endogenous- or transgenically expressed molecules in interneurons could be used to classify these neurons and their synapses.
In our animal model, the zebra finch, we expect our method to be useful for statistical quantification of connections between different projection neuron types. In the case of the forebrain area HVC, our goal is to understand how the signal is conveyed by the afferent projection neurons, how it is transformed by the interaction with other neuron populations within HVC, and how it is routed to motor areas by efferent projection neurons. For example we want to quantify the strength of connections of neurons with members of the same class compared to the connections made with other neuron types. This information will help formulate circuit models to better understand how the song-control network can learn and generate a highly stereotyped motor behavior.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Prof. Kevan Martin, Rita Bopp, German Koestinger and Simone Rickauer for technical advice, and the Electron Microscopy Center of ETH Zurich (EMEZ), in particular Dr. Roger Wepf, for support.
Key Concepts
Electron microscopes use an electron beam to illuminate the specimen and reach a resolution higher than light microscopy and sufficient to resolve individual synapses.
Biological tissue prepared for serial section electron microscopy needs to be cut in ultrathin sections. For this purpose the tissue is typically infiltrated with a resin subsequently polymerized so that the hardened tissue can be cut with a diamond knife.
Tracers are chemical compounds that, after injection in the nervous system, are taken up by neurons and transported inside of them. Tracers reveal neuron location and morphology by virtue of labels visible with either LM or EM.
Projectomics is an approach to circuit reconstruction in which mainly afferent and efferent projections of a brain region are reconstructed, without aiming at a complete characterization of the entire local network.
Array tomography is a high-resolution proteomic imaging method based on repeated staining and imaging of ordered arrays of ultrathin, resin-embedded sections (Micheva and Smith, 2007).
References
Anderson, J. C., Douglas, R. J., Martin, K. A., and Nelson, J. C. (1994). Map of the synapses formed with the dendrites of spiny stellate neurons of cat visual cortex. J. Comp. Neurol. 341, 25–38.
Anderson, J. R., Jones, B. W., Yang, J. H., Shaw, M. V., Watt, C. B., Koshevoy, P., Spaltenstein, J., Jurrus, E., Kannan, U. V., Whitaker, R. T., Mastronarde, D., Tasdizen, T., and Marc, R. E. (2009). A computational framework for ultrastructural mapping of neural circuitry. PLoS Biol. 7, e1000074. doi: 10.1371/journal.pbio.1000074
Cardona, A., Saalfeld, S., Preibisch, S., Schmid, B., Cheng, A., Pulokas, J., Tomancak, P., and Hartenstein, V. (2010). An integrated micro- and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biol. 8, e1000502.
da Costa, N. M., and Martin, K. A. (2009). Selective targeting of the dendrites of corticothalamic cells by thalamic afferents in area 17 of the cat. J. Neurosci. 29, 13919–13928.
Deerinck, T. J., Martone, M. E., Lev-Ram, V., Green, D. P., Tsien, R. Y., Spector, D. L., Huang, S., and Ellisman, M. H. (1994). Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J. Cell Biol. 126, 901–910.
Denk, W., and Horstmann, H. (2004). Serial block-face scanning electron microscopy to reconstruct three-dimensional tissue nanostructure. PLoS Biol. 2, e329.
Giepmans, B. N., Deerinck, T. J., Smarr, B. L., Jones, Y. Z., and Ellisman, M. H. (2005). Correlated light and electron microscopic imaging of multiple endogenous proteins using quantum dots. Nat. Methods 2, 743–749.
Hahnloser, R. H., Kozhevnikov, A. A., and Fee, M. S. (2002). An ultra-sparse code underlies the generation of neural sequences in a songbird. Nature 419, 65–70.
Hahnloser, R. H., Wang, C. Z., Nager, A., and Naie, K. (2008). Spikes and bursts in two types of thalamic projection neurons differentially shape sleep patterns and auditory responses in a songbird. J. Neurosci. 28, 5040–5052.
Helmstaedter, M., Briggman, K. L., and Denk, W. (2008). 3D structural imaging of the brain with photons and electrons. Curr. Opin. Neurobiol. 18, 633–641.
Knott, G., Marchman, H., Wall, D., and Lich, B. (2008). Serial section scanning electron microscopy of adult brain tissue using focused ion beam milling. J. Neurosci. 28, 2959–2964.
Konishi, M. (1965). The role of auditory feedback in the control of vocalization in the white-crowned sparrow. Z. Tierpsychol. 22, 770–783.
Lanciego, J. L., Wouterlood, F. G., Erro, E., and Gimenez-Amaya, J. M. (1998). Multiple axonal tracing: simultaneous detection of three tracers in the same section. Histochem. Cell Biol. 110, 509–515.
Livet, J., Weissman, T. A., Kang, H., Draft, R. W., Lu, J., Bennis, R. A., Sanes, J. R., and Lichtman, J. W. (2007). Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62.
Long, M. A., Jin, D. Z., and Fee, M. S. (2010). Support for a synaptic chain model of neuronal sequence generation. Nature 468, 394–399.
Maranto, A. R. (1982). Neuronal mapping: a photooxidation reaction makes Lucifer yellow useful for electron microscopy. Science 217, 953–955.
Merchan-Perez, A., Rodriguez, J. R., Alonso-Nanclares, L., Schertel, A., and Defelipe, J. (2009). Counting synapses using FIB/SEM microscopy: a true revolution for ultrastructural volume reconstruction. Front. Neuroanat. 3:18.
Micheva, K. D., O’Rourke, N., Busse, B., and Smith, S. J. (2010a). Array tomography: immunostaining and antibody elution. Cold Spring Harb. Protoc. 2010, pdb prot5525.
Micheva, K. D., Busse, B., Weiler, N. C., O’Rourke, N., and Smith, S. J. (2010b). Single-synapse analysis of a diverse synapse population: proteomic imaging methods and markers. Neuron 68, 639–653.
Micheva, K. D., and Smith, S. J. (2007). Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron 55, 25–36.
Nagerl, U. V., and Bonhoeffer, T. (2010). Imaging living synapses at the nanoscale by STED microscopy. J. Neurosci. 30, 9341–9346.
Nagerl, U. V., Willig, K. I., Hein, B., Hell, S. W., and Bonhoeffer, T. (2008). Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. U.S.A. 105, 18982–18987.
Nottebohm, F., and Arnold, A. P. (1976). Sexual dimorphism in vocal control areas of the songbird brain. Science 194, 211–213.
Oberti, D., Kirschmann, M. A., and Hahnloser, R. H. (2010). Correlative microscopy of densely labeled projection neurons using neural tracers. Front. Neuroanat. 4:24.
Olveczky, B. P., Andalman, A. S., and Fee, M. S. (2005). Vocal experimentation in the juvenile songbird requires a basal ganglia circuit. PLoS Biol. 3, e153.
Reiner, A., Laverghetta, A. V., Meade, C. A., Cuthbertson, S. L., and Bottjer, S. W. (2004). An immunohistochemical and pathway tracing study of the striatopallidal organization of area X in the male zebra finch. J. Comp. Neurol. 469, 239–261.
Reiner, A., Veenman, C. L., Medina, L., Jiao, Y., Del Mar, N., and Honig, M. G. (2000). Pathway tracing using biotinylated dextran amines. J. Neurosci. Methods 103, 23–37.
Roberts, T. F., Tschida, K. A., Klein, M. E., and Mooney, R. (2010). Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature 463, 948–952.
Scharff, C., and Nottebohm, F. (1991). A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: implications for vocal learning. J. Neurosci. 11, 2896–2913.
Smith, Y., and Bolam, J. P. (1991). Convergence of synaptic inputs from the striatum and the globus pallidus onto identified nigrocollicular cells in the rat: a double anterograde labelling study. Neuroscience 44, 45–73.
Takizawa, T., Suzuki, K., and Robinson, J. M. (1998). Correlative microscopy using FluoroNanogold on ultrathin cryosections. Proof of principle. J. Histochem. Cytochem. 46, 1097–1102.
Keywords: correlative microscopy, projectomics, songbird, array tomography, neural tracers
Citation: Oberti D, Kirschmann MA and Hahnloser RH, (2011) Projection neuron circuits resolved using correlative array tomography. Front. Neurosci. 5:50. doi:10.3389/fnins.2011.00050
Received: 21 January 2011; Paper pending published: 01 March 2011;
Accepted: 28 March 2011; Published online: 12 April 2011.
Edited by:
Kristen M. Harris, The University of Texas at Austin, USAReviewed by:
Brian Antonsen, Marshall University, USAGraziella Dicristo, University of Montreal, Canada
Copyright: © 2011 Oberti, Kirschmann and Hahnloser. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Richard H. R. Hahnloser, University of Zurich/ETH Zurich, Institute of Neuroinformatics, Zurich, Switzerland,cmljaEBpbmkucGh5cy5ldGh6LmNo


