- 1Department of Neurology, First Affiliated Hospital of Soochow University, Suzhou, China
- 2Department of Neurology, Suzhou Ninth People’s Hospital, Suzhou, China
- 3Department of Neurology, The Affiliated Changzhou NO.2 People’s Hospital of Nanjing Medical University, Changzhou, China
- 4Department of Neurology, Suzhou Guangci Cancer Hospital, Suzhou, China
- 5Department of Neurology, The Dushu Lake Hospital of Soochow University, Suzhou, China
- 6Department of Neurology, The Affiliated Taizhou Second People’s Hospital of Yangzhou University, Yangzhou, China
Background: Previous research has yielded conflicting results on the link between epilepsy risk and lipid-lowering medications. The aim of this study is to determine whether the risk of epilepsy outcomes is causally related to lipid-lowering medications predicted by genetics.
Methods: We used genetic instruments as proxies to the exposure of lipid-lowering drugs, employing variants within or near genes targeted by these drugs and associated with low-density lipoprotein cholesterol (LDL cholesterol) from a genome-wide association study. These variants served as controlling factors. Through drug target Mendelian randomization, we systematically assessed the impact of lipid-lowering medications, including HMG-CoA reductase (HMGCR) inhibitors, proprotein convertase subtilisin/kexin type 9 (PCSK9) inhibitors, and Niemann-Pick C1-like 1 (NPC1L1) inhibitors, on epilepsy.
Results: The analysis demonstrated that a higher expression of HMGCR was associated with an elevated risk of various types of epilepsy, including all types (OR = 1.17, 95% CI:1.03 to 1.32, p = 0.01), focal epilepsy (OR = 1.24, 95% CI:1.08 to 1.43, p = 0.003), and focal epilepsy documented with lesions other than hippocampal sclerosis (OR = 1.05, 95% CI: 1.01 to 1.10, p = 0.02). The risk of juvenile absence epilepsy (JAE) was also associated with higher expression of PCSK9 (OR = 1.06, 95% CI: 1.02 to 1.09, p = 0.002). For other relationships, there was no reliable supporting data available.
Conclusion: The drug target MR investigation suggests a possible link between reduced epilepsy vulnerability and HMGCR and PCSK9 inhibition.
Introduction
Approximately 70 million people worldwide suffer from epilepsy, which has an annual rate ranging from 50.4 to 81.7 per 100,000 people (1, 2). Despite ongoing research, the precise mechanisms underlying epileptic seizures remain incompletely understood. Genetic predispositions, cerebrovascular illness, head traumas, and neurodegenerative disorders are only a few of the recognized causes of epilepsy. According to earlier research, metabolic variables may be very important in the development of epilepsy (3, 4).
Numerous observational studies have to date shown a link between higher circulating lipid levels and an enhanced risk of epilepsy. They have also suggested that the use of lipid-lowering medications might potentially alleviate seizure activity (5–13). Contradictory studies, however, have hypothesized that people with epilepsy could, in general, have lower amounts of circulating blood lipids. Additionally, there are evidence to suggest that increasing circulating blood lipids through the adoption of a ketogenic diet may result in a reduction in seizure frequency (14–16).
HMG-CoA reductase (HMGCR) inhibitors, sometimes referred to as statins, are a very frequently prescribed family of lipid-lowering medications. They offer various advantages, including well-established safety records, cost-effectiveness, and pleiotropic effects. Additionally, the proteins Niemann-Pick C1-like 1 (NPC1L1) and proprotein convertase subtilisin/kexin type 9 (PCSK9) play critical roles in controlling levels of circulating low-density lipoprotein cholesterol (LDL-C) (17, 18).
Genetic epidemiology provides an additional approach to address these questions. Variants located within or near genes responsible for encoding protein drug targets can potentially influence their expression or function. Genetic effects can serve as predictors of drug treatment outcomes (19). In contrast to other kinds of observational epidemiology, genotypes are randomly inherited at conception, much like the treatment allocation in clinical trials; thus, relationships between variations, biomarkers, and illness outcomes are less susceptible to confounding and reverse causation. The methodology known as “Mendelian randomization” (MR) is based on this premise (20). In addition, the general expectation is that genotypes contribute to the variation in traits throughout life. Consequently, by projecting the effects of prolonged therapeutic treatment, MR studies have the potential to inform the validation of pharmacological targets (21). The targets of drugs administered for primary hypercholesterolemia or familial hypercholesterolemia were studied in this study in relation to genetic variations. We also evaluated how they could affect epilepsy risk.
Materials and techniques
Research plan
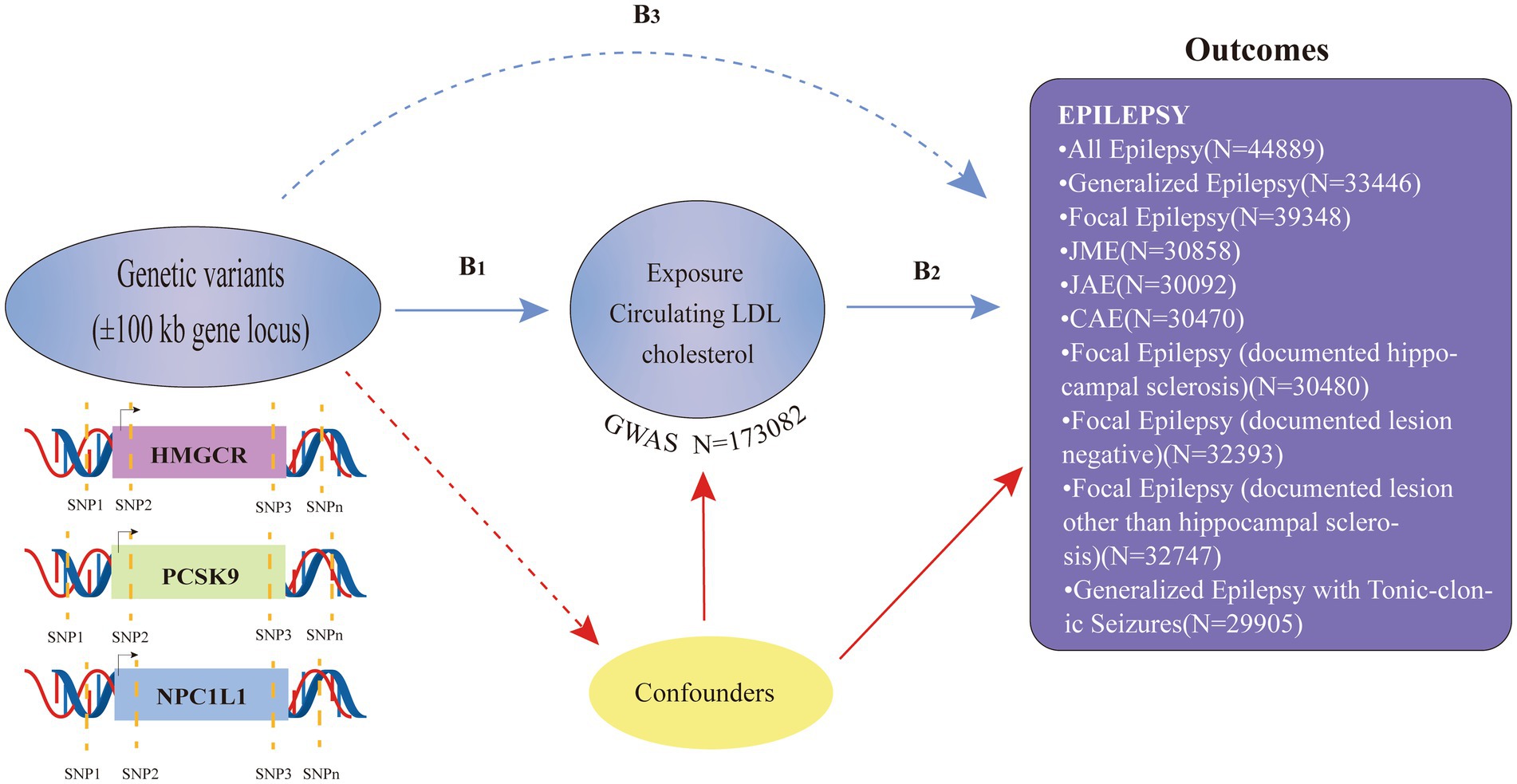
In this two-sample MR analysis, publicly accessible summary-level data from genome-wide association studies (GWASs) were employed (refer to Supplementary Table S1). The following three guiding hypotheses served as the foundation for the MR analysis in this study: (i) the selected genetic variants should demonstrate a significant association with the exposure factor (LDL-C); (ii) the chosen genetic variants exhibit no association with other potential confounding factors.; and (iii) the chosen genetic variations only have an impact on the exposure component (LDL-C), and they have no additional effects on the outcome (epilepsy) (Figure 1). Only publicly available GWAS data were used in this study, and ethical approval and consent to participate could be found in the original GWAS study. The research was performed according to STROBE-MR guidelines (22).
Genetic tool selection
Three kinds of lipid-lowering medications that have received FDA approval were used in this investigation as exposures. HMGCR inhibitors, PCSK9 inhibitors, and NPC1L1 inhibitors are the members of this class.
Our instrument selection procedure chose single-nucleotide polymorphisms (SNPs) located within 100 kb of the gene targeted by each medication, as shown in Supplementary Table S2. These selected SNPs demonstrated a genome-wide association with LDL-C levels (p < 5.0 × 10–8), making them suitable proxies when exposed to lipid-lowering drugs. The analysis relied on GWAS data for LDL-C levels with a sample size of 173,082 sourced from the Global Lipids Genetics Consortium (GLGC) for the identification of these SNPs. The research only took into account frequent SNPs with a minor allele frequency (MAF) higher than 1% (refer to Supplementary Table S2) (23).
Seven SNPs located within 100 kb of the HMGCR gene were chosen to serve as proxies for HMGCR inhibitors. There are 12 SNPs in the PCSK9 gene that have been shown to inhibit PCSK9. Additionally, to represent NPC1L1 inhibitors, three SNPs were selected from the NPC1L1 gene. To enhance the reliability of each instrumental variable for the respective medication, low linkage disequilibrium (R2 < 0.30) between the chosen SNPs was mandated.
Sources of results
We compiled pooled statistics from the GWAS encompassing various forms of epilepsy. These data were sourced from the International League Against Epilepsy (ILAE) Consortium cohort (refer to Supplementary Table S1) (24, 25).
Statistical analyses
Primary MR analysis
To determine causal estimates for the primary analysis, we used the inverse variance weighted (IVW) approach to random effects. Given that inhibitors targeting HMGCR, PCSK9, and NC1L1 are commonly used in the treatment of coronary artery disease (CAD), we utilized data from the CARDIoGRAMplusC4D CAD study, which encompasses 60,801 cases and 123,504 control subjects, primarily of European descent. This sample size of 60,801 cases served as a positive control group to assess the validity of the instruments related to HMGCR, PCSK9, and NC1L1 (26, 27). R-version 4.3.1 package was used for the analysis and assessment, and the MR and its MR packages were implemented (28, 29).
Analysis of sensitivity
Using the F-statistic, we evaluated the potency of SNPs used as instruments. To reduce weak instrumental bias, we took care to only include those with an F-statistic greater than 10 (30). The Cochran Q-test was used to assess the heterogeneity of certain SNPs. Potential pleiotropy was suggested by a Cochran Q-derived p-value of 0.05. It is important to note that pleiotropy does not introduce bias to the IVW’s estimates, as long as it remains balanced. We estimated the intercept using MR-Egger regression as a measure of directional pleiotropy to see if the pleiotropy was truly unbalanced. Statistical significance was defined as a value of p of 0.05 (31). If a notable outcome was observed, it would suggest the presence of imbalanced pleiotropy, which prompted us to conduct various sensitivity analyses to validate the findings derived from the IVW estimates. We used a variety of MR methods recognized for their resistance to pleiotropy in many sensitivity studies to confirm the validity of our findings, and Weighted Median Regression was one among the MR methods. It mandates that at least 50% of the weight used in the MR analysis come from reliable devices (32). Additionally, we employed MR-Egger regression, which is capable of detecting and adjusting for directional pleiotropy (33).
Results
We retrieved a total of 7, 12, and 3 SNPs from the GWAS summary data of LDL-C levels in the GLGC. These SNPs were found within, close to, or near the HMGCR, PCSK9, and NPC1L1 genes, respectively (refer to Supplementary Table S2). All instrumental variations had F-statistics that were more than 30. This suggests that our study’s efforts to minimize possible weak instrument bias were successful (refer to Supplementary Table S2). The positive control study furnished compelling evidence linking drug exposure to coronary heart disease, utilizing instruments derived from LDL-C GWAS (refer to Supplementary Table S5). This evidence further substantiates the soundness of our chosen genetic instruments.
Primary analysis
We started by lining up all of the outcome datasets with the exposures of the drug target instruments. We performed associated IVW-MR, MR-Egger, and Weighted Median MR experiments in the sections that follow. This process yielded test statistics for heterogeneity and pleiotropy. The standardized correlated MR effect estimates are presented last. According to these estimations, the medication target gene is indirectly inhibited by a one standard deviation (SD) decrease in biomarker LDL-C levels (SD = 38.7 mg/dL).
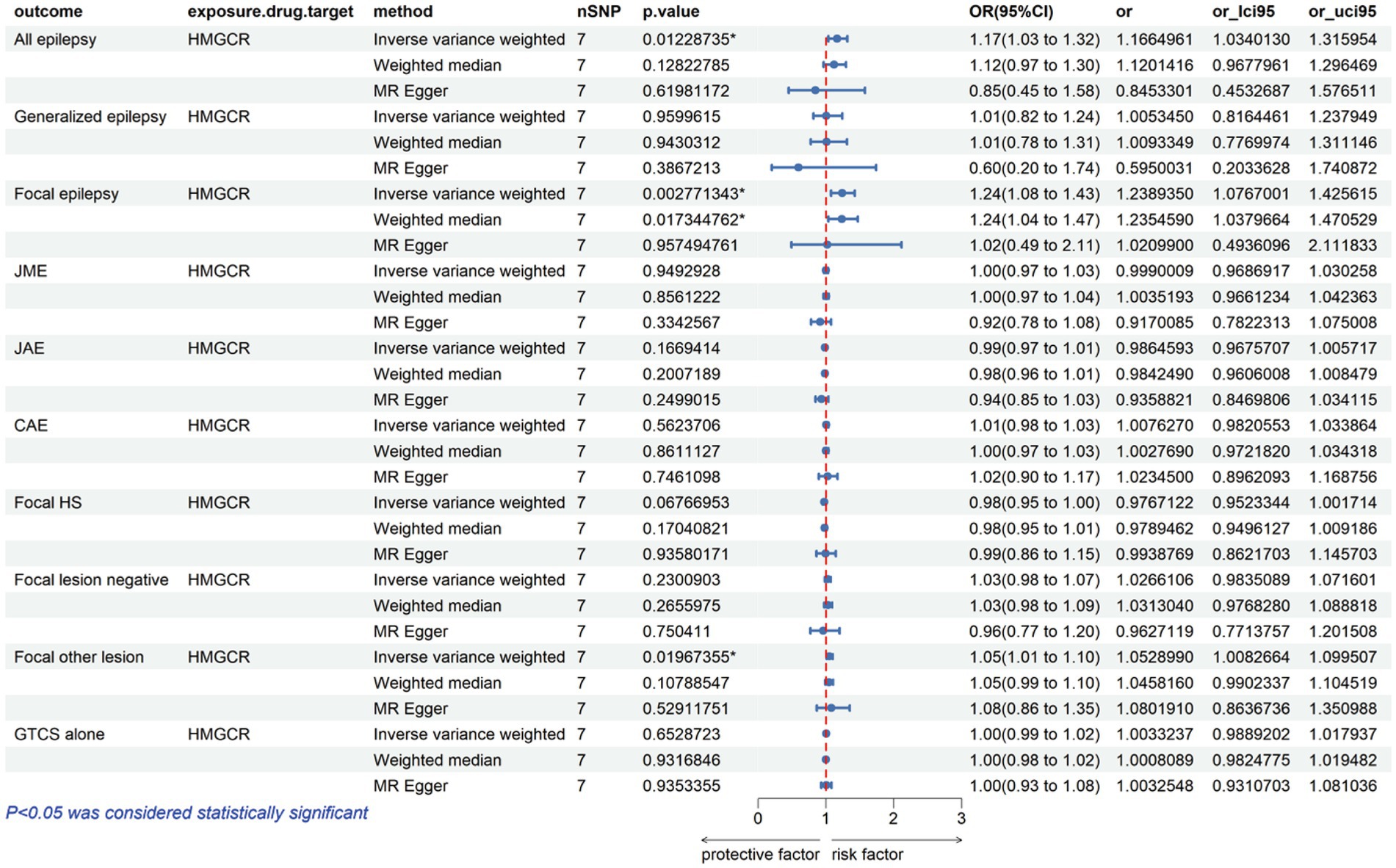
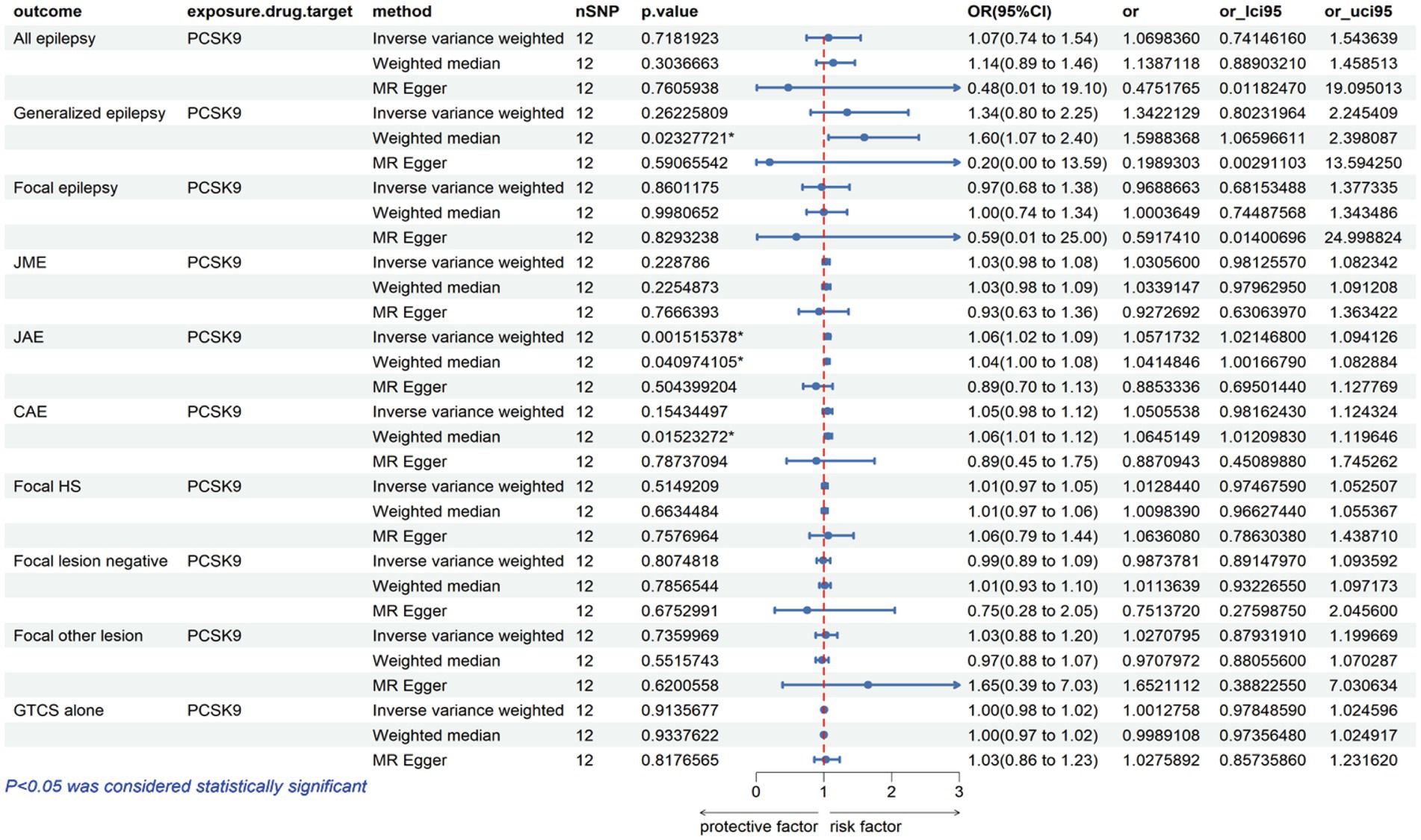
The analysis reveals that a higher expression of HMGCR is associated with an increased risk of various types of epilepsy, including all types, with an odds ratio (OR) of 1.17 with a 95% confidence interval (CI) of 1.03–1.32, accompanied by a corresponding p-value of 0.01. For focal epilepsy, the OR is 1.24 (95% CI = 1.08–1.43, p = 0.003), and for focal epilepsy with documented lesions other than hippocampal sclerosis, the OR is 1.05 (95% CI = 1.01–1.10, p = 0.02) (Figure 2; Supplementary Table S3). Similarly, as indicated in Figure 3 and Supplementary Table S3, for a heightened expression of PCSK9 that was linked with an elevated risk of juvenile absence epilepsy (JAE), the OR is 1.06 (95% CI = 1.02–1.09, p = 0.002). There was no evidence of an association between NPC1L1-mediated LDL-C and the outcome of epilepsy in Supplementary Figure S1 and Supplementary Table S3.
Sensitivity assessment
The results consistently aligned across all applied Mendelian Randomization (MR) methods. The results of the Cochran Q-test did not indicate any evidence of heterogeneity in the epilepsy-related endpoints or the HMGCR-mediated LDL-C outcomes. All of the value of ps (refer to Supplementary Table S4) were higher than 0.05. However, some degree of heterogeneity emerged in the results concerning PCSK9-mediated LDL-C, encompassing both all types and focal epilepsy (excluding hippocampal sclerosis lesions) (Supplementary Table S4). Additionally, in the MR-Egger regression analysis, neither intercept term provided substantial evidence of pervasive horizontal pleiotropy (every value of p was higher than 0.05; refer to Supplementary Table S4).
Discussion
Our drug target MR studies have identified a direct correlation between the risk of epilepsy and HMGCR, encompassing all types (OR = 1.17, 95% CI = 1.03–1.32, p = 0.01), focal epilepsy (OR = 1.24, 95% CI = 1.08–1.43, p = 0.003), and focal epilepsy characterized by documented lesions other than hippocampal sclerosis (OR = 1.05, 95% CI = 1.01–1.10, p = 0.02). These results indicate that HMGCR inhibitors might lower the risk of epilepsy. Moreover, our study has provided initial evidence supporting a positive correlation between PCSK9 expression and susceptibility to JAE (OR = 1.06, 95% CI = 1.02–1.09, p = 0.002). This result also suggests a potential risk reduction in epilepsy with the use of PCSK9 inhibitors. It reinforces the notion that these inhibitors could be a valuable consideration in epilepsy prevention. Conversely, we found no association between NPC1L1 expression and epilepsy outcomes.
Epilepsy, with its significant morbidity and mortality, is a prevalent chronic neurological condition. The ILAE describes it as a disorder brought on by either the breakdown of systems that should end seizures or the onset of mechanisms that should end excessively protracted seizures (34). With an estimated yearly prevalence of 800,000 occurrences of vascular epilepsy globally, stroke is also the main cause of acquired epilepsy in elderly people (35, 36). Statins, which are HMGCR inhibitors, have been commonly used for several years in the prevention of both ischemic and hemorrhagic strokes, according to randomized controlled studies and meta-analyses (37, 38). Furthermore, numerous studies have indicated enhanced neurological outcomes and a better acute stroke prognosis with statin administration (39, 40). Since neuroprotective agents have the capacity to influence the onset of epilepsy (41), statins have also been proposed for their potential anticonvulsant properties in epilepsy (42–46). Numerous neurosynaptic transmissions and molecular processes contribute to the antiepileptic properties of statins. PCSK9 inhibitors, serving as a crucial regulator of LDL-C, have become a significant focus in the development of cholesterol-lowering medications (17, 47, 48). Our MR study revealed a previously unreported protective role of PCSK9 inhibitors against JAE. Our study has identified a potential benefit of statins in reducing the risk of epilepsy. This finding suggests that statins could be considered a therapeutic option for patients with post-stroke epilepsy, particularly elderly patients. Additionally, it introduces a novel avenue for drug therapy in both clinical and preclinical studies.
As a genetic epidemiological method, MR overcomes the limitations of conventional observational studies. Previous clinical studies in epilepsy have suffered from varying designs, inconsistent terminology, limited sample sizes, fluctuating durations of follow-up, and uncertainty in the identification and classification of seizures, which has led to inconsistent results. Additionally, there is a limited amount of data available regarding particular subtypes of seizures. Our study effectively circumvents these issues. Previous attempts to establish a causal link between lipids and seizures using two-sample Mendelian randomization techniques yielded conflicting results (49). In this specific investigation, we utilized genetic variants associated with HMGCR-mediated LDL-C levels as instrumental proxies for statin exposure, effectively reducing the potential for confounding bias. This approach harnesses the wealth of currently available genetic information on epilepsy. Furthermore, our study addresses concerns of reverse causality and residual confounding through meticulous MR analysis. We applied various methodologies to validate MR estimates, ensure the consistency of our estimates across different MR models, and check for any violations of MR assumptions.
Genomic analytic techniques such as MR offer early evidence of long-term LDL-C modulation by drug targets; given the recent development of PCSK9 inhibitors, long-term trial data are still lacking. The consistency of our results across different MR methods, accommodating varied assumptions about genetic pleiotropy, significantly strengthens the causal inference of our analysis. Our research indicates that statins demonstrate a risk reduction in the occurrence of various forms of epilepsy, including all types, focal epilepsy, and focal epilepsy with documented lesions other than hippocampal sclerosis. The association between PCSK9 inhibitors and JAE has not been previously documented in existing literature. This unexplored area will be the central focus of our upcoming research endeavors and investigations.
Research limitations
Our research contains a number of limitations, primarily stemming from the inherent assumptions and constraints of MR. First, while MR is a powerful tool for identifying genetic associations, it cannot fully replace rigorous, long-term randomized controlled trials for potential future gene-editing therapeutics. MR’s conclusions are contingent on the quality and scope of the underlying data. Second, the available data on epilepsy outcomes are relatively small and may lack the power needed to detect potential relationships adequately. It is therefore imperative to replicate these biometric analyses in epilepsy with larger datasets as they become available. Third, it is important to note that statins and PCSK9 inhibitors have limited penetration across the blood–brain barrier. Additionally, their mechanisms of action within the brain might not be reflected in our findings, which may not encompass tissue-specific correlations of statin and PCSK9 expression that could be altered in diseased brain states. Fourth, we admit that, despite our thorough sensitivity analyses that were created to carefully examine the fundamental assumptions of our MR study, there is still a chance of confounding bias and/or horizontal pleiotropy. Finally, it is crucial to note that the majority of the populations of European heritage were included in the GWAS data we used for this study. Consequently, we advise caution when generalizing these findings to other populations.
Conclusion
In this study, we furnish genetic evidence supporting a worldwide as well as a regional link between lipid-lowering drugs such as HMGCR and PCSK9 inhibitors and epilepsy. Grasping this correlation offers potential for pioneering epilepsy treatments. Our MR research establishes a link between the risk of epilepsy and lipid-lowering medications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent from the patients/participants or patients/participants’ legal guardian/next of kin was not required to participate in this study in accordance with the national legislation and the institutional requirements.
Author contributions
SH: Conceptualization, Data curation, Writing – original draft. YL: Formal analysis, Investigation, Methodology, Writing – original draft. YZ: Formal analysis, Project administration, Resources, Writing – original draft. YW: Methodology, Project administration, Software, Writing – original draft. YG: Methodology, Project administration, Software, Writing – original draft. RL: Methodology, Project administration, Writing – original draft. LY: Resources, Software, Validation, Writing – review & editing. XH: Validation, Writing – review & editing. QF: Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the projects Role and mechanism of GluA2-containing-AMPA receptor membrane stability mediated by NSF protein nitrosylation modification in post-stroke depression. National Natural Science Foundation of China, grant no. 82071300, Quantitative EEG Assessment of Cognitive Dysfunction in Elderly Patients with Temporal Lobe Epilepsy. Scientific Research Project on Elderly Health, Jiangsu Provincial Healthcare Commission No. LKM2022019.
Acknowledgments
The authors extend our gratitude to the patients and investigators whose contributions were invaluable to the ILAE Consortium, Global Lipids Genetics Consortium, and CARDIoGRAMplusC4D Consortium. The authors sincerely thank the Physician Scientist Team for their enthusiastic and meticulous teaching and guidance on the Mendelian Randomization study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2024.1331537/full#supplementary-material
References
1. Fisher, RS, Acevedo, C, Arzimanoglou, A, Bogacz, A, Cross, JH, Elger, CE, et al. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. (2014) 55:475–82. doi: 10.1111/epi.12550
2. Collaborators, GBDN. Global, regional, and national burden of neurological disorders, 1990-2016: a systematic analysis for the global burden of disease study 2016. Lancet Neurol. (2019) 18:459–80. doi: 10.1016/S1474-4422(18)30499-X
3. Li, Y, Li, J, Wu, G, Yang, H, Yang, X, Wang, D, et al. Role of SIRT3 in neurological diseases and rehabilitation training. Metab Brain Dis. (2023) 38:69–89. doi: 10.1007/s11011-022-01111-4
4. Lin Lin Lee, V, Kar Meng Choo, B, Chung, YS, Kundap, U, Kumari, Y, and Shaikh, M. Treatment, therapy and Management of Metabolic Epilepsy: a systematic review. Int J Mol Sci. (2018) 19:19. doi: 10.3390/ijms19030871
5. Harnod, T, Chen, H-J, Li, T-C, Sung, F-C, and Kao, C-H. A high risk of hyperlipidemia in epilepsy patients: a nationwide population-based cohort study. Ann Epidemiol. (2014) 24:910–4. doi: 10.1016/j.annepidem.2014.09.008
6. Banach, M, Czuczwar, SJ, and Borowicz, KK. Statins – are they anticonvulsant? Pharmacol Rep. (2014) 66:521–8. doi: 10.1016/j.pharep.2014.02.026
7. Quintana-Pajaro, LJ, Ramos-Villegas, Y, Joaquim, AF, Agrawal, A, Narvaez-Rojas, AR, Moscote-Salazar, LR, et al. The effect of statins in epilepsy: a systematic review. J Neurosci Rural Pract. (2018) 9:478–86. doi: 10.4103/jnrp.jnrp_110_18
9. Siniscalchi, A. Statins for poststroke seizures the first antiepileptogenic agent? Neurology. (2015) 85:661–2. doi: 10.1212/WNL.0000000000001878
10. Nucera, B, Rinaldi, F, Nardone, R, Lattanzi, S, and Brigo, F. Statins in primary prevention of poststroke seizures and epilepsy: a systematic review. Epilepsy Behav. (2020) 112:107400. doi: 10.1016/j.yebeh.2020.107400
11. Guo, Y, Zhu, L-H, Zhao, K, Guo, X-M, and Yang, M-F. Statin use for the prevention of seizure and epilepsy in the patients at risk: a systematic review and meta-analysis of cohort studies. Epilepsy Res. (2021) 174:106652. doi: 10.1016/j.eplepsyres.2021.106652
12. Das, RR. Statins in epilepsy prime time? Not yet. Neurology. (2010) 75:1490–1. doi: 10.1212/WNL.0b013e3181f962fc
13. McFarland, AJ, Anoopkumar-Dukie, S, Arora, DS, Grant, G, McDermott, C, Perkins, A, et al. Molecular mechanisms underlying the effects of statins in the central nervous system. Int J Mol Sci. (2014) 15:20607–37. doi: 10.3390/ijms151120607
14. Kukuruzovic, M, Basic Kes, V, and Malenica, M. Association between apolipoprotein E polymorphisms and epilepsy in children. Acta Clin Croat. (2021) 60:595–601. doi: 10.20471/acc.2021.60.04.05
15. Elia, M, Klepper, J, Leiendecker, B, and Hartmann, H. Ketogenic diets in the treatment of epilepsy. Curr Pharm Des. (2017) 23:5691–701. doi: 10.2174/1381612823666170809101517
16. Nikolaos, Triantafyllou, Chryssoula, Nikolaou, Irini, Petropoulou, Christos, Manolis, Triantafyllou, Dimitrios, Konstantinos, Patsis, et al. The effect of long-term antiepileptic treatment on serum cholesterol (TC, HDL, LDL) and triglyceride levels in adult epileptic patients on monotherapy. (2004) 10:MT50-52.
17. Sabatine, MS. PCSK9 inhibitors: clinical evidence and implementation. Nat Rev Cardiol. (2019) 16:155–65. doi: 10.1038/s41569-018-0107-8
18. Williams, DM, Finan, C, Schmidt, AF, Burgess, S, and Hingorani, AD. Lipid lowering and Alzheimer disease risk: a mendelian randomization study. Ann Neurol. (2020) 87:30–9. doi: 10.1002/ana.25642
19. Hingorani, A, and Humphries, S. Nature’s randomised trials. Lancet. (2005) 366:1906–8. doi: 10.1016/S0140-6736(05)67767-7
20. Lawlor, DA, Harbord, RM, Sterne, JA, Timpson, N, and Davey, SG. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. (2008) 27:1133–63. doi: 10.1002/sim.3034
21. Swerdlow, DI, Kuchenbaecker, KB, Shah, S, Sofat, R, Holmes, MV, White, J, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. (2016) 45:1600–16. doi: 10.1093/ije/dyw088
22. Skrivankova, VW, Richmond, RC, Woolf, BAR, Davies, NM, Swanson, SA, VanderWeele, TJ, et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. (2021) 375:n2233. doi: 10.1136/bmj.n2233
23. Willer, CJ, Schmidt, EM, Sengupta, S, Peloso, GM, Gustafsson, S, Kanoni, S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. (2013) 45:1274–83. doi: 10.1038/ng.2797
24. International League Against Epilepsy Consortium on Complex E. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. (2018) 9:5269. doi: 10.1038/s41467-018-07524-z
25. Genetic determinants of common epilepsies. A meta-analysis of genome-wide association studies. Lancet Neurol. (2014) 13:893–903. doi: 10.1016/S1474-4422(14)70171-1
26. Gallego-Colon, E, Daum, A, and Yosefy, C. Statins and PCSK9 inhibitors: a new lipid-lowering therapy. Eur J Pharmacol. (2020) 878:173114. doi: 10.1016/j.ejphar.2020.173114
27. Nikpay, M, Goel, A, Won, HH, Hall, LM, Willenborg, C, Kanoni, S, et al. A comprehensive 1, 000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. (2015) 47:1121–30. doi: 10.1038/ng.3396
28. Hemani, G, Zheng, J, Elsworth, B, Wade, KH, Haberland, V, Baird, D, et al. The MR-base platform supports systematic causal inference across the human phenome. eLife. (2018) 7:7. doi: 10.7554/eLife.34408
29. Yavorska, OO, and Burgess, S. Mendelian randomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. (2017) 46:1734–9. doi: 10.1093/ije/dyx034
30. Burgess, S, and Thompson, SG Collaboration CCG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol. (2011) 40:755–64. doi: 10.1093/ije/dyr036
31. Burgess, S, and Thompson, SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. (2017) 32:377–89. doi: 10.1007/s10654-017-0255-x
32. Bowden, J, Davey Smith, G, Haycock, PC, and Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. (2016) 40:304–14. doi: 10.1002/gepi.21965
33. Bowden, J, Davey Smith, G, and Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. (2015) 44:512–25. doi: 10.1093/ije/dyv080
34. Manford, M. Recent advances in epilepsy. J Neurol. (2017) 264:1811–24. doi: 10.1007/s00415-017-8394-2
35. Group GBDNDC. Global, regional, and national burden of neurological disorders during 1990-2015: a systematic analysis for the global burden of disease study 2015. Lancet Neurol. (2017) 16:877–97. doi: 10.1016/S1474-4422(17)30299-5
36. Galovic, M, Dohler, N, Erdelyi-Canavese, B, Felbecker, A, Siebel, P, Conrad, J, et al. Prediction of late seizures after ischaemic stroke with a novel prognostic model (the SeLECT score): a multivariable prediction model development and validation study. Lancet Neurol. (2018) 17:143–52. doi: 10.1016/S1474-4422(17)30404-0
37. Ni Chroinin, D, Asplund, K, Asberg, S, Callaly, E, Cuadrado-Godia, E, Diez-Tejedor, E, et al. Statin therapy and outcome after ischemic stroke: systematic review and meta-analysis of observational studies and randomized trials. Stroke. (2013) 44:448–56. doi: 10.1161/STROKEAHA.112.668277
38. McKinney, JS, and Kostis, WJ. Statin therapy and the risk of intracerebral hemorrhage: a meta-analysis of 31 randomized controlled trials. Stroke. (2012) 43:2149–56. doi: 10.1161/STROKEAHA.112.655894
39. Cappellari, M. The THRombolysis and STatins (THRaST) study. Neurology. (2013) 80:655–61. doi: 10.1212/WNL.0b013e318281cc83
40. Flint, AC. Inpatient statin use predicts improved ischemic stroke discharge disposition. Neurology. (2012) 78:1678–83. doi: 10.1212/WNL.0b013e3182575142
41. Fernandes MJdS. Neuroprotective agents and modulation of temporal lobe epilepsy. Front Biosci. (2015) 7:90–106. doi: 10.2741/719
42. Moazzami, K, Emamzadeh-Fard, S, and Shabani, M. Anticonvulsive effect of atorvastatin on pentylenetetrazole-induced seizures in mice: the role of nitric oxide pathway. Fundam Clin Pharmacol. (2013) 27:387–92. doi: 10.1111/j.1472-8206.2012.01038.x
43. Moezi, L, Shafaroodi, H, Hassanipour, M, Fakhrzad, A, Hassanpour, S, and Dehpour, AR. Chronic administration of atorvastatin induced anti-convulsant effects in mice: the role of nitric oxide. Epilepsy Behav. (2012) 23:399–404. doi: 10.1016/j.yebeh.2012.02.001
44. Shafaroodi, H, Moezi, L, Fakhrzad, A, Hassanipour, M, Rezayat, M, and Dehpour, AR. The involvement of nitric oxide in the anti-seizure effect of acute atorvastatin treatment in mice. Neurol Res. (2012) 34:847–53. doi: 10.1179/1743132812Y.0000000080
45. Pugh, MJ, Knoefel, JE, Mortensen, EM, Amuan, ME, Berlowitz, DR, and Van Cott, AC. New-onset epilepsy risk factors in older veterans. J Am Geriatr Soc. (2009) 57:237–42. doi: 10.1111/j.1532-5415.2008.02124.x
46. Etminan, M. Statin use and risk of epilepsy a nested case-control study. Neurology. (2010) 75:1496–500. doi: 10.1212/WNL.0b013e3181f96253
47. Mullard, A. Nine paths to PCSK9 inhibition. Nat Rev Drug Discov. (2017) 16:299–301. doi: 10.1038/nrd.2017.83
48. Shapiro, MD, Tavori, H, and Fazio, S. PCSK9: from Basic science discoveries to clinical trials. Circ Res. (2018) 122:1420–38. doi: 10.1161/CIRCRESAHA.118.311227
49. Liang, Z, Zhao, L, Lou, Y, and Liu, S. Causal effects of circulating lipids and lipid-lowering drugs on the risk of epilepsy: a two-sample Mendelian randomization study. QJM. (2023) 116:421–8. doi: 10.1093/qjmed/hcad048
Glossary
Keywords: lipid-lowering medications, epilepsy, drug target Mendelian randomization, causal, genetic, lipid metabolism
Citation: Huang S, Liu Y, Zhang Y, Wang Y, Gao Y, Li R, Yu L, Hu X and Fang Q (2024) Analyzing the causal relationship between lipid-lowering drug target genes and epilepsy: a Mendelian randomization study. Front. Neurol. 15:1331537. doi: 10.3389/fneur.2024.1331537
Edited by:
Kette D. Valente, University of São Paulo, BrazilReviewed by:
Silvia Vincentiis, University of São Paulo, BrazilVenkataraghavan Ramamoorthy, Baptist Health South Florida, United States
Copyright © 2024 Huang, Liu, Zhang, Wang, Gao, Li, Yu, Hu and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Fang, ZmFuZ3FpXzAwOEAxMjYuY29t
†These authors share first authorship
‡These authors share last authorship
§ORCID: Shicun Huang, https://orcid.org/0000-0002-2810-5723
Yuan Liu, https://orcid.org/0000-0001-8316-3806
Yi Zhang, https://orcid.org/0000-0001-8625-2054
Yiqing Wang, https://orcid.org/0000-0003-0576-3624
Ya Gao, https://orcid.org/0009-0000-4910-7931
Runnan Li, https://orcid.org/0000-0002-9833-6105
Lidong Yu, https://orcid.org/0009-0005-5505-5724
Xiaowei Hu, https://orcid.org/0000-0002-3744-4436
Qi Fang, https://orcid.org/0009-0004-9006-3554
 Shicun Huang1†§
Shicun Huang1†§ Qi Fang
Qi Fang