
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol. , 20 April 2023
Sec. Neurogenetics
Volume 14 - 2023 | https://doi.org/10.3389/fneur.2023.1153509
 Huifang Xu1†
Huifang Xu1† Tongtong Li2,3†
Tongtong Li2,3† Qiyong Gong4
Qiyong Gong4 Haizhen Xu4
Haizhen Xu4 Yongbo Hu4
Yongbo Hu4 Wenqi Lü1
Wenqi Lü1 Xin Yang1
Xin Yang1 Jin Li1
Jin Li1 Wenming Xu2,3*
Wenming Xu2,3* Weihong Kuang1*
Weihong Kuang1*Background: The retrograde endocannabinoid (eCB) pathway is closely associated with the etiology of major depressive disorder (MDD) at both pathophysiological and genetic levels. This study aimed to investigate the potential role of genetic mutations in the eCB pathway and underlying mechanisms in Han Chinese patients with MDD.
Methods: A total of 96 drug-naïve patients with first-episode MDD and 62 healthy controls (HCs) were recruited. Whole-exome sequencing was performed to identify the gene mutation profiles in patients with MDD. Results were filtered to focus on low-frequency variants and rare mutations (minor allele frequencies <0.05) related to depressive phenotypes. Enrichment analyses were performed for 146 selected genes to examine the pathways in which the most significant enrichment occurred. A protein–protein interaction (PPI) network analysis was performed to explore the biological functions of the eCB pathway. Finally, based on current literature, a preliminary analysis was conducted to explore the effect of genetic mutations on the function of this pathway.
Results: Our analysis identified 146 (15.02%) depression-related genetic mutations in patients with MDD when compared with HCs, and 37 of the mutations were enriched in the retrograde eCB signaling pathway. Seven hub genes in the eCB pathway were closely related to mitochondrial function, including Complex I genes (NDUFS4, NDUFV2, NDUFA2, NDUFA12, NDUFB11) and genes associated with protein (PARK7) and enzyme (DLD) function in the regulation of mitochondrial oxidative stress.
Conclusion: These results indicate that genetic mutations in the retrograde eCB pathway represent potential etiological factors associated with the pathogenesis of MDD.
Major depressive disorder (MDD) is a prevalent mental disorder that manifests as a wide spectrum of heterogeneous symptoms. As of 2019, MDD ranked 2nd among the top 25 leading causes of years lived with disability (YLDs) (1). However, the etiology and pathogenesis of MDD remain unclear. Therefore, understanding of the pathological mechanisms underlying MDD is necessary to promote effective treatment.
MDD is a complex disorder influenced by multiple genetic factors. The estimated heritability of MDD ranged from 30 to 50% (2). Although several primary genetic studies have focused on candidate genes that have been associated with MDD—including SLC6A4, BDNF, COMT, HTR2A, TPH1 and TPH2—their results have provided little insight into the impact of these candidate genes on MDD (3). Several previous genome-wide association studies (GWAS) have identified common variations related to MDD. However, the estimated heritability of these common genetic mutations ranges from only 9 to 10% (4). Thus, researchers have shifted their attention to other undiscovered heritability, such as rare gene variations (MAF < 0.5%) and copy number variations (CNVs). Using whole-exome sequencing (WES), several studies have explored the contributions of rare or low-frequency variants to the genetic basis of MDD. One low-coverage whole-genome sequencing study identified the SIRT1 and LEPP genes as risk loci in a sample of 5,303 Han Chinese women with recurrent MDD (5). The authors identified 1,985 variations in 479 MDD-related genes using different approaches and databases, reporting 14 gene mutations that differed between patients with MDD and the general South Asian population (6). A recent study that utilized 16,702 samples from the UK Biobank also highlighted the FOXH1 gene and sphingolipid metabolism pathways as the most significant pathogenic genes for MDD (4). Despite several advances in understanding the genetic mutations underlying MDD, studies are still scarce and have limitations such as small sample sizes and large numbers of Europeans. Significant differences may exist across ethnicities due to the different allele frequencies. Therefore, it is necessary to study the genetic variation of Asians to expand the genetic research.
In the current study, we firstly explored the genetic variation profile of drug-naïve patients with MDD about whole-exome sequencing from China. Meanwhile, we conducted a genetic interaction analysis to infer functional variations via gene enrichment analysis rather than focusing on single-gene mutations. Preliminary explanation of how these genetic mutations affect physiological function were demonstrated through literature review.
A total of 96 first-episode drug-naïve patients with MDD and 62 healthy control (HC) participants were recruited for this study. All patients were recruited from the West China Hospital and had been diagnosed using the Structured Clinical Interview for DSM-IV Disorders (SCID). The inclusion criteria in this study were age of 18–60 years, presence of depressive symptoms for >2 weeks, and no previous exposure to antidepressant treatment. Exclusion criteria were as follows: history of psychosis, significant neurological or medical illness, current electroconvulsive therapy, and any history of alcohol or substance abuse or dependence. All HC participants, who were matched with the MDD group according to age and sex, were recruited from the local community through advertisements. Inclusion criteria for the HC group were as follows: no history of neuropsychiatric illness or brain injury, no family history of any serious mental illness in first-degree relatives.
Age was compared between the groups using nonparametric tests. Sex was compared using the chi-square test. The results are expressed as means ± SEM values and were analyzed by SPSS 26.0 (IBM, Chicago, IL, United States). The value of p < 0.05 is considered to be statistically significant.
DNA samples were extracted and subjected to exome sequencing, including DNA quantification, library construction, exome sequencing, annotation and filtration. Detailed methods and data analysis are described in the Supplementary material.
To better predict the harmfulness of variation, we first utilized the classification system of the American College of Medical Genetics and Genomics (ACMG), which classifies variations as pathogenic, likely pathogenic, of uncertain significance, likely benign, or benign (7). Variations were then screened according to their scores using the SIFT (8), Polyphen (9), MutationTaster (10), and CADD (11) software programs. Potentially deleterious variations were retained if the scores from more than half of the four software programs supported their potential harmfulness (12).
Python scripts were used to extract SNVs and genes associated with a depressive phenotype (HP:0000716). Screening for variants with depressive phenotypes in patients and healthy controls revealed 146 variants that were specific to the patient group. We performed enrichment analyses on this set of 146 genes using clusterProfiler (13) and Metascape1 (14). Significant enrichment was defined as overlap of at least three genes, and the hypergeometric test was used to estimate the significance (p < 0.05). We also used Metascape to analyze the enrichment of mutated genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway to further screen for these functions and interactions.
To identify the biological functions of the selected gene mutations in patients with MDD, 37 genes in the eCB pathway were mapped into the online search tool STRING database.2 A combined score of ≥0.4 was considered significant. Cytoscape software (3version 3.9.1; Institute for Systems Biology, Seattle, WA, United States) was used to construct and visualize the eCB gene variation network. To identify hub genes in the pathway, genes associated with other genes were ranked based on eigenvector centrality (EC) (15) using the CytoNCA v2.1.6 plugin. A survey of the current literature revealed that the top seven genes were related to mitochondrial function.
A total of 96 patients with MDD and 62 HCs were enrolled in this study. Age and sex did not significantly differ between the MDD and HC groups. The demographic and clinical information of the matched groups is presented in Table 1.
The 96 patients with MDD exhibited 15,637 SNPs of variant genes, 972 of which were related to the depressive phenotype. In the MDD group, we also extracted 3,353 SNVs involving 907 genes that were only related to depression phenotypes. Only a small number (146, 15.02%) of depression-associated gene mutations were observed among patients with MDD. The distributions of the 146 genes among patients with MDD are shown in Supplementary material.
Subsequently, enrichment analyses were performed for a set of 146 genes to investigate 17 pathways enriched in the KEGG pathway. Among these, the retrograde eCB signaling pathway was significantly associated with MDD (Gene count = 9, GeneRatio = 0.062, p = 0.000000238, -log10 (value of p) = 6.624). Additional details of 17 pathways were presented in Figure 1 and Supplementary material.
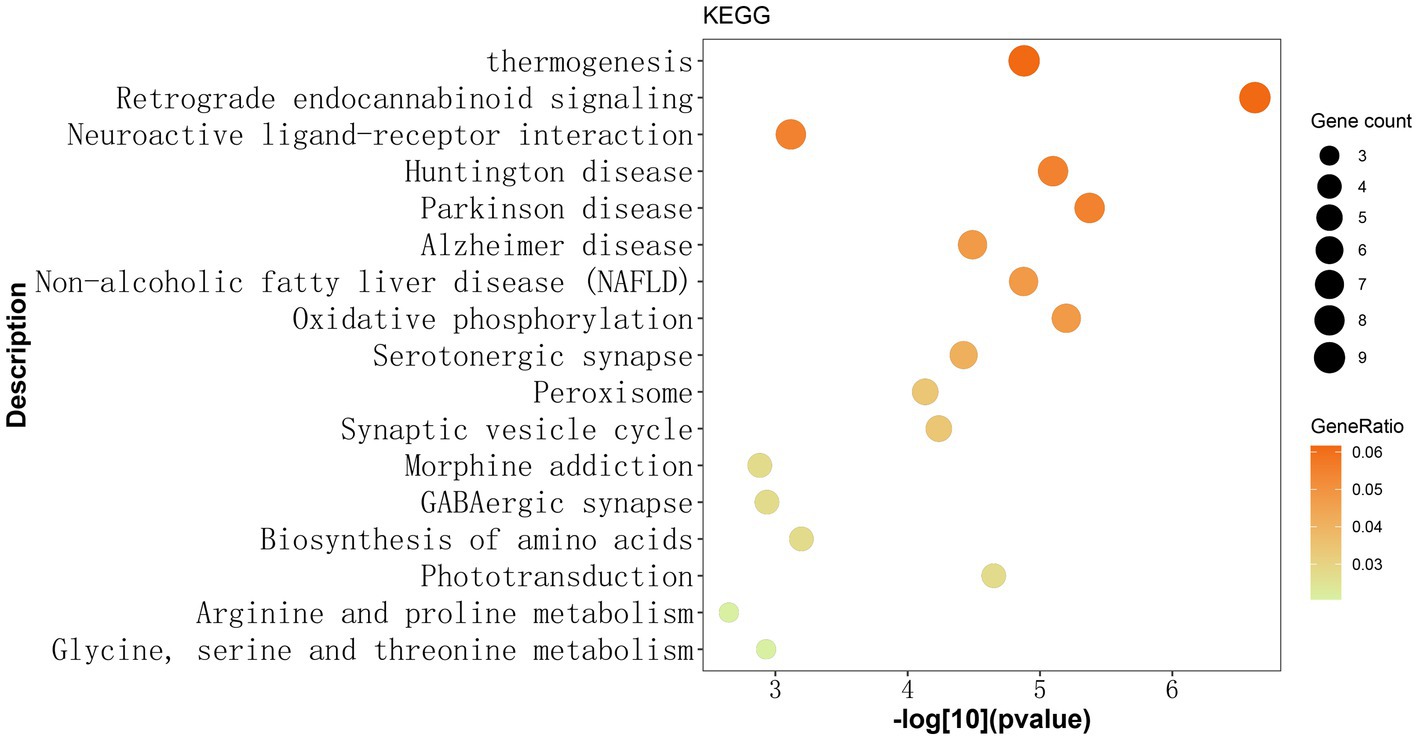
Figure 1. Identified 17 KEGG pathways based on 146 gene mutations observed exclusively in Chinese patients with MDD. The y-axis represents the KEGG pathway term, while the x-axis shows the -log[10](p-value). Each dot on the plot represents the fold enrichment for each KEGG pathway. The size of each dot corresponds to the number of mutated genes related to the depression phenotype, and the color of each dot represents the proportion of enriched genes in that pathway relative to the total number of mutated genes. The darker color indicates a higher proportion of genes. The retrograde endocannabinoid signaling pathway exhibited the most significant enrichment among the 17 KEGG pathways. KEGG: Kyoto Encyclopedia of Genes and Genomes.
There were 37 gene mutations in the retrograde eCB signaling in 55 patients with MDD (Figure 2). Among them, DDC and GLRB mutations were involved in the largest number of patients (five patients). Our PPI study demonstrated that seven hub genes were vitally related to mitochondria function, including Complex I (NDUFS4, NDUFV2, NDUFA2, NDUFA12, NDUFB11) and genes related to protein (PARK7) and enzyme (DLD) function in the eCB pathway, as shown in Figure 3. The details and frequency of the seven genes in the samples are presented in Table 2. In addition, CHCH10 encodes a protein in the mitochondrial intermembrane that regulates mitochondrial function. Other gene variants included those related to synapse metabolism (STXBP1 and VAMP2); protein-regulated neurotransmitters (GNB3, GNAO1, STXBP1, SLC1A3, SLC16A2); and receptors associated with glutamic acid (GLRB), gamma-aminobutyric acid (GABRB2, GABRB3), and 5-hydroxytryptamine (HTR2A). The 37 genes in the retrograde eCB signaling pathway were shown in Supplementary material.
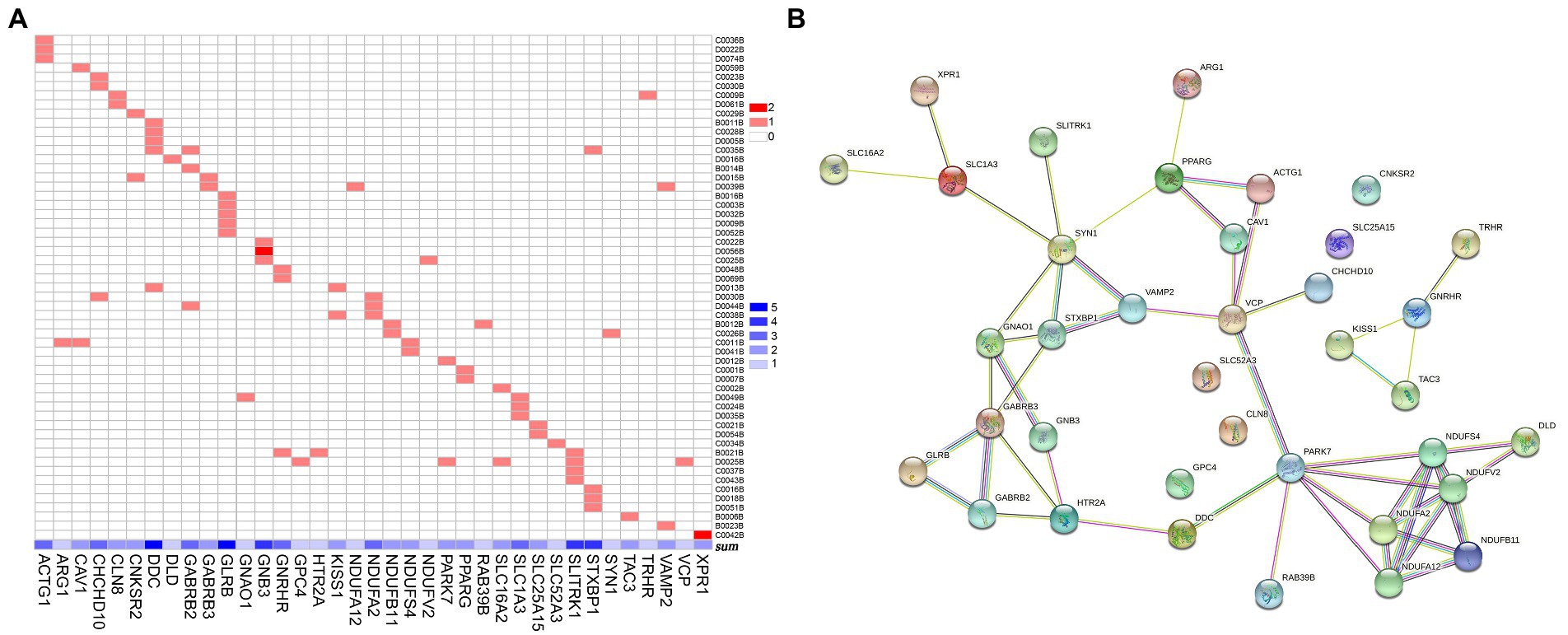
Figure 2. Gene mutations in the retrograde eCB signaling pathway in patients with MDD. (A) The 37 gene mutations in the retrograde eCB signaling pathway observed in 55 patients with MDD. The x-axis shows the 37 gene mutations, with the concentration of blue indicating the number of patients with MDD who carry each mutation. The y-axis shows each patient with MDD. The color in each square reflects the number of genetic mutations. Light red indicates that the gene is a heterozygous mutation, and dark red indicates a homozygous mutation. (B) The biological functions of the genes in the eCB signaling pathway based on the protein–protein interaction network analysis. Each node represents the proteins encoded by the genes, the contents of which are the 3D structures of proteins. Edges represent protein–protein associations contributing to a shared function. Different classifications are indicated by different colors. Known interactions are shown in blue and rose-red, based on information from curated databases and experimental studies, respectively. Predicted interactions are shown in green, red, and deep purple to indicate gene neighborhood, fusions, and co-occurrence, respectively. Yellow, black, and light purple represent interactions based on text mining, co-expression, and protein homology. eCB: endocannabinoid.
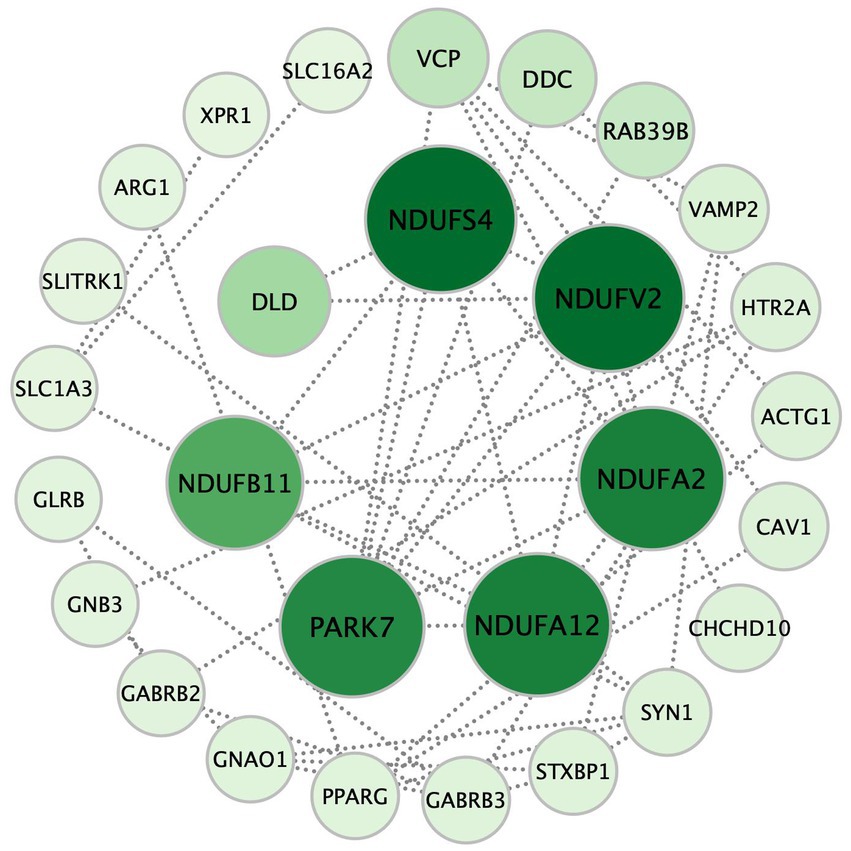
Figure 3. The seven hub genes of the retrograde eCB signaling pathway in patients with MDD based on the PPI analysis. Each circle represents a gene variation, and the sizes and color densities of each circle reflect ranking based on eigenvector centrality (EC). Larger, darker circles indicate the genes that are more significantly enriched in this pathway. The top seven most important genes are in the inner circle, while the others are arranged in the outer circle. eCB: endocannabinoid; PPI: protein–protein interaction.
In the current study we found that 146 depression-associated genes were enrich 17 KEGG pathways and the retrograde eCB signaling pathway was the most notable. The 7 hub genes in this pathway were associated with the mitochondrial function in PPI. Our findings support the relationship between the eCB system and mitochondria in MDD from a genetic and biological function perspective. Figure 4 shows the possible underlying mechanistic pathways of the mutant gene-encoded proteins in mitochondria and the eCB signaling.
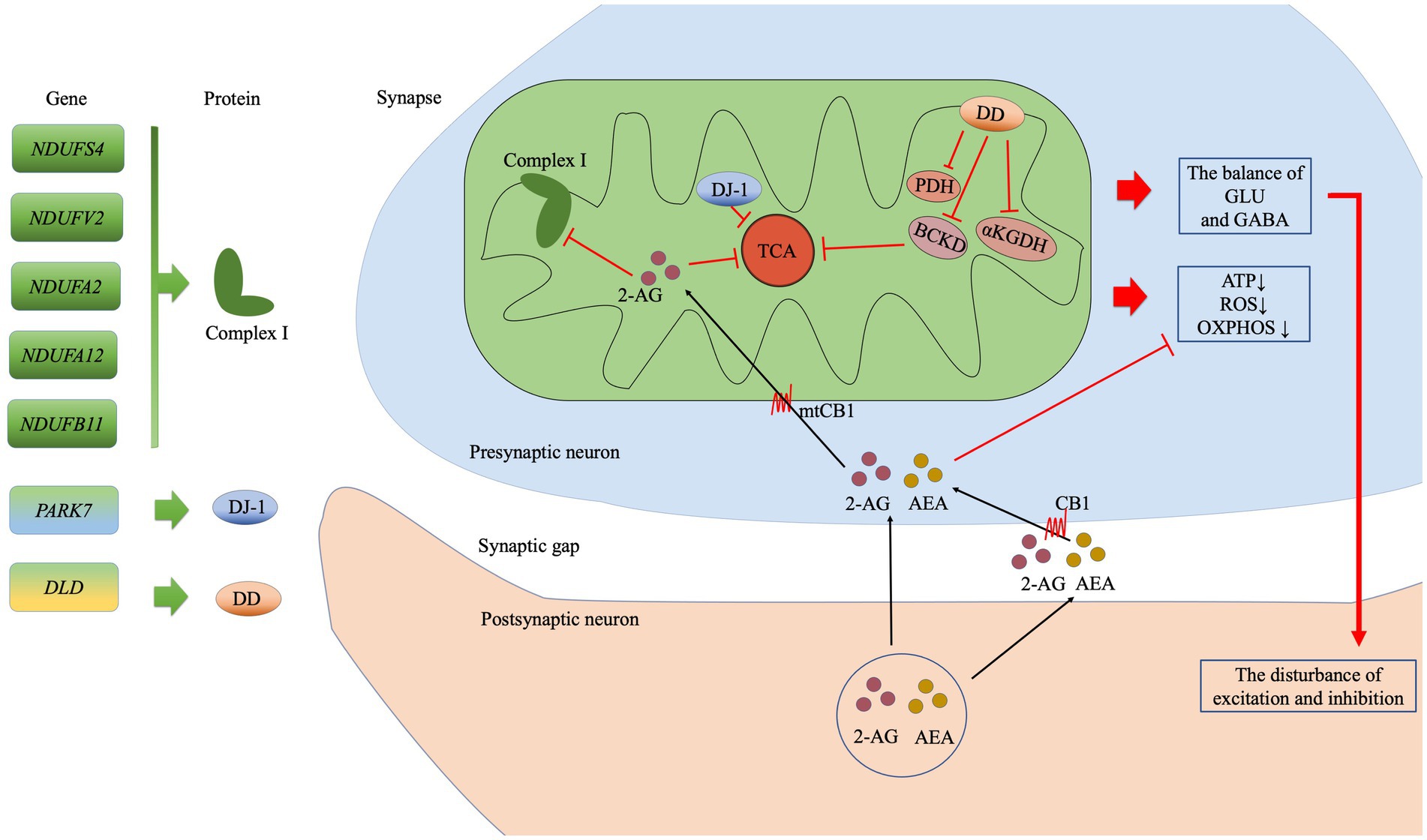
Figure 4. The possible mitochondrial mechanisms associated with the top seven genes in the eCB pathway analysis. The subunits of complex I are encoded by NDUFS4, NDUFV2, NDUFA2, NDUFA12, and NDUFB11. PARK7 encodes the DJ-7 protein, while DLD encodes dihydrolipoamide dehydrogenase (DD), which in turn affects the enzyme complexes such as BCKD, PDH, and αKGDH. The eCBs 2-arachidonoylglycerol (2-AG) and anandamide (AEA) are synthesized in the postsynaptic membrane on demand. Then, 2-AG may enter the presynaptic membrane via CB or simple diffusion, following which it may inhibit oxidative function by acting on CB1 at the mitochondrial membrane (mtCB1) and regulating the subunits of complex I. Ultimately, the effects of this process on the TCA cycle reduced production of ATP, ROS generation, and reduced OXPHOS. In addition, disturbances in the balance of GLU and GABA exert an effect on synaptic function. Abbreviation: complex I: mitochondrial complex I (NADH: ubiquinone-oxidoreductase); BCKD: branched-chain alpha-keto acid dehydrogenase; PDH: pyruvate dehydrogenase; αKGDH: alpha-ketoglutarate dehydrogenase; TCA: tricarboxylic acid; ATP, adenosine triphosphate; ROS: reactive oxygen species; OXPHOS: oxidative phosphorylation; GLU: glutamate; GABA: γ-aminobutyric acid GABA; 2-AG: 2-Arachidonoylglycerol; AEA: anandamide or N-arachidonoylethanolamine; CB1: cannabinoid receptor 1; mtCB1: cannabinoid receptor 1 expressed on the mitochondrial membrane.
We found that the retrograde eCB signaling pathway was significantly correlated with patients with MDD. The retrograde eCB signaling pathway, a component of the endocannabinoid system (ECS), is a widespread neuromodulatory pathway related to a range of physiological and pathological conditions, including the stress response, emotion, cognition, and memory (16). Numerous genetic and metabolomic studies have verified that abnormalities in eCB signaling play an essential role in MDD pathogenesis, impacting neurotransmission as well as the neuroendocrine and neuroimmune systems (17). Analyses based on the Psychiatric Genetic Consortium and UK Biobank have identified 43 differentially expressed genes between MDD and smoking in several neurotransmitter pathways, including the retrograde eCB signaling pathway (18). The retrograde eCB signaling was down-regulated pathway in bipolar disorder type I compared with depressive disorder based on expressed genes (19) and in bipolar II disorder (20). Another study detected 38 hippocampal metabolites related to retrograde eCB signaling in rats with prenatal stress that were associated with depression-like behaviors (21).
Furthermore, the polymorphisms of genes coding for the components of the ECS were related to MDD, such as cannabinoid receptors and the enzymes (22–25). Studies on enrichment analysis of genes involved in MDD also obtained that the neuroactive ligand receptor interaction (26), synaptic structure and neurotransmission (27), hypoxia, epithelial-mesenchymal transition, hedgehog signaling, and reactive oxygen species pathway (28) were achieved significance.
Our study complements the genetic mechanism of ECS in MDD as a biomarker and provides a theoretical basis for diagnosing and treating MDD. Activation of the ECS appears rapid-acting treatment for MDD (29). As conventional antidepressant drugs show delayed onset of therapeutic effects, novel treatments for MDD based on the ECS are developing rapidly. The endocannabinoid 2-Arachidonoylglycerol (2-AG) (17), the CB1 and CB2 receptors (30), and the enzyme (31) all have antidepressant pharmacological modulation and are potential new therapeutic targets for the treatment of MDD. Meanwhile, endocannabinoid-related compounds are also in rapid development. N-palmitoylethanolamide (PEA) is an endocannabinoid-like modulator, demonstrating an antidepressant-like effect (32). Cannabidiol, derived from phytocannabinoids, is a non-psychoactive substance that exerts antidepressant effects through multiple targets (33). ECS-based compounds are promising antidepressants in the future.
We identified 7 hub genes in the eCB signaling pathway, which were vitally related to mitochondria function. Among them, there are 5 genes encoding the mitochondrial Complex I (NADH: ubiquinone oxidoreductase), which is the largest inner membrane protein of the respiratory electron chain. The gene mutations in 45 subunits of Complex I have been closely linked to a wide range of neuropsychiatric disorders. NDUFS4 (AQDQ protein) was identified as the most important site in the PPI of eCB in our cohort, as an accessory subunit associated with the assembly and/or stability of Complex I. The NDUFS4 gene regulates the balance between excitatory (glutamate) and inhibitory (γ-aminobutyric acid, GABA) neurotransmission (34). An abnormal balance of glutamate and GABA is also usually found in patients with MDD. NDUFV2 encodes a 24-kDa subunit of the NDUFV2 protein of Complex I. Initial studies have suggested a possible link between the mRNA level of NDUFV2 and the state of bipolar disorder (BD) (35, 36). In addition, a haplotype T-C consisting of NDUFV2 is most likely a protective factor for MDD in the Han Chinese population (37). NDUFA2 encodes a subunit of the hydrophobic protein fraction of Complex 1. The mRNA level of NDUFA2 was most significantly associated with schizophrenia (38) and the remission of psychiatric symptoms (39). NDUFB11 (ESSS protein), located in the short arm of the X-chromosome, is essential for the assembly and activity of Complex I. NDUFB11 may play a role in the mechanism underlying cognitive deficits in children and adolescents born preterm. Notably, cognitive impairment has been identified in more than half of patients with MDD (40). Although there have been no reports on NDUFB11 and MDD, cognitive impairment may be a phenotype that this gene contributes to depressive disorder. NDUFA12 is a small hydrophobic accessory subunit of Complex I, identified as a novel binding partner of the serine/threonine p21-activated kinase that increases susceptibility to type 2 diabetes (41). It is well-known that patients with MDD and diabetes are shared genetic risks (42). The underlying relationship between NDUFA12 and MDD deserves to be explored. PARK7 (DJ-7 protein) is a mitochondrial-associated protein, that exerts a wide range of effects on cellular functions including helping to prevent damage from reactive oxygen species (ROS), maintaining mitochondrial function, and participating in chaperone activity and carbohydrate metabolism (43). DJ-1 protein may act as an antioxidative defense mechanism to regulate mitochondrial dysfunction in the context of depressive disorders (44). DLD encodes a mitochondrial-associated enzyme called dihydrolipoamide dehydrogenase, which forms a subunit of several enzyme complexes, including pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (αKGDHc). Upregulation of DLD in the hippocampus was associated with anxiety-like behavior (45). The genes encoding mitochondria might affect depression-like behavior by regulating synaptic transmission, susceptibility, cognition and antioxidant.
The retrograde eCB signaling is involved in energy metabolism-regulated mitochondrial function via cannabinoid receptor 1 (CB1). CB1 is expressed not only on the cell membrane but also on the mitochondrial membrane (mtCB1) (46). The activity of mtCB1 exerts a great impact on brain mitochondrial physiology-associated bioenergetics, ROS production, and neurotransmitter regulation (47). Studies have indicated that mtCB1 can inhibit soluble adenylate cyclase and protein kinase A (PKA) activity, resulting in reduced PKA-dependent phosphorylation of mitochondrial proteins (46). Further, mtCB1 has been associated with the regulation of synaptic transmission, including glutamate and GABA transmission, via its effects on adenosine triphosphate supply and Ca2+ homeostasis (47). In addition, brain mtCB1 is important for regulating glutamate transmission associated with memory performance. The hippocampus is a vital regulator of memory and learning and is linked to acute mitochondrial activity in the brain. Hippocampal mtCB1 receptors regulate intra-mitochondrial Gαi proteins, resulting in the inhibition of soluble adenylyl cyclase (sAC), leading to a reduction in cAMP levels as well as decreased phosphorylation of PKA and subunit NDUFS2 of Complex I. Eventually, this chain of activities regulates memory processes by decreasing the brain’s mitochondrial energy metabolism (48). Therefore, hippocampal mtCB1 is an important acute regulator of cognitive function. Astrocytes provide energy to the neurons in the brain by regulating cellular glucose metabolism. The activation of astroglial mtCB1 hampers glucose metabolism, reducing the generation of ROS and the phosphorylation of NDUFS4 to destabilize Complex I, eventually impairing neuronal activity and behavioral responses in mice (49). The activation of muscular mtCB1, implicated in the metabolism of the primary tricarboxylic acid (TCA) substrate pyruvate, also participates in the regulation of oxidative activity (50).
Simply, mitochondrial-related genes lead to mitochondrial dysfunction affecting energy metabolism, oxidative stress, neurotransmitter transmission and cognitive function, mediated by mtCB1. Mitochondria have salient biological location in the eCB pathway.
Mitochondria are key organelles for energy production, involved in mechanisms of MDD through neuroimmune and neuroinflammation. The mobilization of energy is very important for dealing with stressful events. Mitochondria are involved in regulating the stress response mitochondrial biology is tissue- and cell-specific, particularly in the immune system (51). Social stress is a risk factor for the development of MDD (52) and contributes to mitochondrial dysfunction, leading to inflammatory disturbances (53). One study of metabolomic signatures reported that mitochondrial oxidative phosphorylation (OXPHOS), morphology, and recycling were crucial elements of the stress response. In addition, the authors reported upregulated protein expression of Complexes I, II, and IV in resilient animals, demonstrating that the electron respiratory chain is positively associated with chronic stress (52). Another study found that the changes in protein levels related to mitochondrial dysfunction were dependent on peripheral inflammation, which regulates the severity of MDD (54). Moreover, plasma levels of inflammatory cytokines such as C-reactive protein (54) and interleukin-6 (55) are regarded as markers of mitochondrial disturbance and are known to modulate the severity of depressive symptoms. A longitudinal study showed of patients with treatment-resistant depression undergoing anti-inflammatory treatment with the tumor necrosis factor antagonist infliximab identified peripheral blood gene transcripts enriched for oxidative stress and mitochondrial degradation, which were related to increases in psychomotor reaction time (55). The inflammatory signaling and metabolic reprogramming in the immune system were associated with inflammation in patients with depressive disorders and may promote psychomotor retardation. In another study, alterations in the dynamic regional homogeneity of the brain were observed between 65 first-episode, treatment-naïve patients with MDD and 66 HCs, which correlated with the 16 gene modules investigated in the weighted gene co-expression network analysis. The expression profiles of the gene modules were enriched for immune, mitochondrial, protein, and synaptic signaling (56).
Taken together, multiple lines of evidence indicate that stress regulation and neuroimmunology are key areas linking mitochondrial function and MDD etiology. Genetic studies may also provide novel insights into different antioxidants and anti-immunotherapies that are safe for adjunctive treatment of depressive symptoms (57). Based on the available evidence and the current findings, these therapies may be ideal for patients with MDD exhibiting stress-related immune impairments.
Although our findings provide insight into the role of eCB gene mutations in the pathogenesis of MDD, the current study was limited by its small sample size and the use of a literature review to examine potential molecular mechanisms. As this was a cross-sectional study comparing patients with MDD to HCs, additional studies are also required to determine the direction of causality. Moreover, future studies should aim to examine the impact of epigenetic modifications, the expression profile of these gene variants, and real gene–gene interactions in the context of MDD. Given that MDD is a multifactorial disorder influenced by genetic and environmental factors and the interactions between them, these elements should also be included in future studies. Lastly, we were unable to exclude potential confounders known to affect mitochondrial function, such as lifestyle factors, childhood trauma, chronic stress, and suicidal behaviors. The current study should be considered exploratory, and its findings must be verified in a large-scale longitudinal cohort and validated experimental models.
It is noteworthy that MDD is a complex multifactorial disease. Several mechanisms for MDD pathogenesis have been proposed including neuroinflammation, neurotransmitter abnormalities, neuroendocrine dysfunction, mitochondrial abnormalities, and altered stress regulation. No single factor impacts the final effect, as each factor can both modulate and is modulated by other factors, resulting in intricate relationships and complex interactions. The complex relationship between mitochondrial function and the manifestation of MDD remains unclear, and the current findings must be interpreted with caution.
In conclusion, our analysis identified profiles of genetic variation in Han Chinese patients with first-episode MDD and the possible genetic mechanisms by which the retrograde eCB pathway exerts an influence of MDD in these patients. Our preliminary study broadens the current understanding of the complex genetic basis of MDD and highlights genetic mutations in the eCB pathway as potential etiological factors associated with the pathogenesis of MDD. Identifying specific mutations in this pathway may be beneficial for targeted MDD therapy in the future.
The data presented in the study are deposited in the zenodo repository, https://zenodo.org/, accession number 7769978.
This study was approved by the Clinical Trials and Biomedical Ethics Committee of Sichuan University. All participants provided written informed consent.
QG, JL, WX, and WK were designed for current research. HZX, YH, WL, JL, WK, XY, and HFX performed the acquisition of data. HFX and TL analyzed the data and wrote the manuscript. All authors contributed to the discussion of the results and have approved the final manuscript to be published.
This study was supported by the National Natural Science Foundation of China (ZYJC2102981621003 and 2022YFC2009 900/2022YFC2009906), 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC21029) and National Clinical Research Center for Geriatrics, West China Hospital, Sichuan University (Z20191006). We would like to thank Editage (www.editage.cn) for English language editing. Regional Cooperation Project (Sichuan-Guangdong) of Sichuan Science and Technology Programs (2022YFQ0100).
We would like to thank Editage (www.editage.cn) for English language editing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2023.1153509/full#supplementary-material
1. GBD 2019 Mental Disorders Collaborators. Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990-2019: a systematic analysis for the global burden of disease study 2019. Lancet Psychiatry. (2022) 9:137–10. doi: 10.1016/S2215-0366(21)00395-3
2. Kendall, KM, van Assche, E, Andlauer, TFM, Choi, KW, Luykx, JJ, Schulte, EC, et al. The genetic basis of major depression. Psychol Med. (2021) 51:2217–30. doi: 10.1017/S0033291721000441
3. Border, R, Johnson, EC, Evans, LM, Smolen, A, Berley, N, Sullivan, PF, et al. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. AJP. (2019) 176:376–7. doi: 10.1176/appi.ajp.2018.18070881
4. Zhou, W, Chen, L, Jiang, B, Sun, Y, Li, M, Wu, H, et al. Large-scale whole-exome sequencing association study identifies FOXH1 gene and sphingolipid metabolism pathway influencing major depressive disorder. CNS Neurosci Ther. (2021) 27:1425–8. doi: 10.1111/cns.13733
5. Cai, N, Bigdeli, TB, Kretzschmar, W, Li, Y, Liang, J, Song, L, et al. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. (2015) 523:588–1. doi: 10.1038/nature14659
6. Qazi, SR, Irfan, M, Ramzan, Z, Jahanzaib, M, Khan, MZ, Nasir, M, Shakeel, M, and Khan, IA. Identification of putative genetic variants in major depressive disorder patients in Pakistan. Mol Biol Rep. (2022) 49:2283–2292. doi: 10.1007/s11033-021-07050-0
7. Richards, S, Aziz, N, Bale, S, Bick, D, das, S, Gastier-Foster, J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. (2015) 17:405–4. doi: 10.1038/gim.2015.30
8. Kumar, P, Henikoff, S, and Ng, PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. (2009) 4:1073–81. doi: 10.1038/nprot.2009.86
9. Adzhubei, IA, Schmidt, S, Peshkin, L, Ramensky, VE, Gerasimova, A, Bork, P, et al. A method and server for predicting damaging missense mutations. Nat Methods. (2010) 7:248–9. doi: 10.1038/nmeth0410-248
10. Schwarz, JM, Rödelsperger, C, Schuelke, M, and Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. (2010) 7:575–6. doi: 10.1038/nmeth0810-575
11. Kircher, M, Witten, DM, Jain, P, O'Roak, BJ, Cooper, GM, and Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. (2014) 46:310–5. doi: 10.1038/ng.2892
12. Muona, M, Berkovic, SF, Dibbens, LM, Oliver, KL, Maljevic, S, Bayly, MA, et al. A recurrent de novo mutation in KCNC1 causes progressive myoclonus epilepsy. Nat Genet. (2015) 47:39–46. doi: 10.1038/ng.3144
13. Wu, T, Hu, E, Xu, S, Chen, M, Guo, P, Dai, Z, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb). (2021) 2:100141. doi: 10.1016/j.xinn.2021.100141
14. Zhou, Y, Zhou, B, Pache, L, Chang, M, Khodabakhshi, AH, Tanaseichuk, O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. (2019) 10:1523. doi: 10.1038/s41467-019-09234-6
15. Mistry, D, Wise, RP, and Dickerson, JA. DiffSLC: a graph centrality method to detect essential proteins of a protein-protein interaction network. PLoS One. (2017) 12:e0187091. doi: 10.1371/journal.pone.0187091
16. Navarrete, F, García-Gutiérrez, MS, Jurado-Barba, R, Rubio, G, Gasparyan, A, Austrich-Olivares, A, et al. Endocannabinoid system components as potential biomarkers in psychiatry. Front Psych. (2020) 11:315. doi: 10.3389/fpsyt.2020.00315
17. Gallego-Landin, I, García-Baos, A, Castro-Zavala, A, and Valverde, O. Reviewing the role of the endocannabinoid system in the pathophysiology of depression. Front Pharmacol. (2021) 12:762738. doi: 10.3389/fphar.2021.762738
18. Yao, Y, Xu, Y, Cai, Z, Liu, Q, Ma, Y, Li, AN, et al. Determination of shared genetic etiology and possible causal relations between tobacco smoking and depression. Psychol Med. (2021) 51:1870–9. doi: 10.1017/S003329172000063X
19. Dmitrzak-Weglarz, M, Szczepankiewicz, A, Rybakowski, J, Kapelski, P, Bilska, K, et al. Transcriptomic profiling as biological markers of depression – a pilot study in unipolar and bipolar women. World J Biol Psychiatry. (2021) 22:744–6. doi: 10.1080/15622975.2021.1907715
20. eQTLGen Consortium, BIOS Consortium, the Bipolar Disorder Working Group of the Psychiatric Genomics ConsortiumStahl, EA, Breen, G, Forstner, AJ, McQuillin, A, Ripke, S, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. (2019) 51:793–803. doi: 10.1038/s41588-019-0397-8
21. Zhang, H, He, W, Huang, Y, Zeng, Z, Yang, X, Huang, H, et al. Hippocampal metabolic alteration in rat exhibited susceptibility to prenatal stress. J Affect Disord. (2019) 259:458–7. doi: 10.1016/j.jad.2019.08.002
22. Monteleone, P, Bifulco, M, Maina, G, Tortorella, A, Gazzerro, P, Proto, MC, et al. Investigation of CNR1 and FAAH endocannabinoid gene polymorphisms in bipolar disorder and major depression. Pharmacol Res. (2010) 61:400–4. doi: 10.1016/j.phrs.2010.01.002
23. Kong, X, Miao, Q, Lu, X, Zhang, Z, Chen, M, Zhang, J, et al. The association of endocannabinoid receptor genes (CNR1 and CNR2) polymorphisms with depression: a meta-analysis. Medicine (Baltimore). (2019) 98:e17403. doi: 10.1097/MD.0000000000017403
24. Lazary, J, Eszlari, N, Juhasz, G, and Bagdy, G. Genetically reduced FAAH activity may be a risk for the development of anxiety and depression in persons with repetitive childhood trauma. Eur Neuropsychopharmacol. (2016) 26:1020–8. doi: 10.1016/j.euroneuro.2016.03.003
25. Kim, HK, Zai, G, Müller, DJ, Husain, MI, Lam, RW, Frey, BN, et al. Identification of endocannabinoid predictors of treatment outcomes in major depressive disorder: a secondary analysis of the first Canadian biomarker integration network in depression (CAN-BIND 1) study. Pharmacopsychiatry. (2022) 55:297–3. doi: 10.1055/a-1872-0844
26. Zhang, Y, Li, M, Wang, Q, Hsu, JS, Deng, W, Ma, X, et al. A joint study of whole exome sequencing and structural MRI analysis in major depressive disorder. Psychol Med. (2020) 50:384–5. doi: 10.1017/S0033291719000072
27. Howard, DM, Adams, MJ, Clarke, T-K, Hafferty, JD, Gibson, J, Shirali, M, et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. (2019) 22:343–52. doi: 10.1038/s41593-018-0326-7
28. Qi, D, and Chen, K. Bioinformatics analysis of potential biomarkers and pathway identification for major depressive disorder. Comput Math Methods Med. (2021) 2021:1–11. doi: 10.1155/2021/3036741
29. Sharafi, A, Pakkhesal, S, Fakhari, A, Khajehnasiri, N, and Ahmadalipour, A. Rapid treatments for depression: endocannabinoid system as a therapeutic target. Neurosci Biobehav Rev. (2022) 137:104635. doi: 10.1016/j.neubiorev.2022.104635
30. Morris, G, Walder, K, Kloiber, S, Amminger, P, Berk, M, Bortolasci, CC, et al. The endocannabinoidome in neuropsychiatry: opportunities and potential risks. Pharmacol Res. (2021) 170:105729. doi: 10.1016/j.phrs.2021.105729
31. Rafiei, D, and Kolla, NJ. Elevated brain fatty acid amide hydrolase induces depressive-like phenotypes in rodent models: a review. Int J Mol Sci. (2021) 22:1047. doi: 10.3390/ijms22031047
32. Pinna, G. Biomarkers and treatments for mood disorders encompassing the neurosteroid and endocannabinoid systems. J Neuroendocrinol. (2022):e13226. doi: 10.1111/jne.13226
33. Melas, PA, Scherma, M, Fratta, W, Cifani, C, and Fadda, P. Cannabidiol as a potential treatment for anxiety and mood disorders: molecular targets and epigenetic insights from preclinical research. Int J Mol Sci. (2021) 22:1863. doi: 10.3390/ijms22041863
34. van de Wal, MAE, Adjobo-Hermans, MJW, Keijer, J, Schirris, TJJ, Homberg, JR, Wieckowski, MR, et al. Ndufs4 knockout mouse models of Leigh syndrome: pathophysiology and intervention. Brain. (2022) 145:45–63. doi: 10.1093/brain/awab426
35. Munkholm, K, Peijs, L, Vinberg, M, and Kessing, LV. A composite peripheral blood gene expression measure as a potential diagnostic biomarker in bipolar disorder. Transl Psychiatry. (2015) 5:e614. doi: 10.1038/tp.2015.110
36. Akarsu, S, Torun, D, Erdem, M, Kozan, S, Akar, H, and Uzun, O. Mitochondrial complex I and III mRNA levels in bipolar disorder. J Affect Disord. (2015) 184:160–3. doi: 10.1016/j.jad.2015.05.060
37. Zhang, Z, Ni, J, Zhang, J, Tang, W, Li, X, Wu, Z, et al. A haplotype in the 5′-upstream region of the NDUFV2 gene is associated with major depressive disorder in Han Chinese. J Affect Disord. (2016) 190:329–2. doi: 10.1016/j.jad.2015.10.034
38. Sun, L, Min, L, Li, M, Shao, F, and Wang, W. Transcriptomic analysis reveals oxidative phosphorylation activation in an adolescent social isolation rat model. Brain Res Bull. (2018) 142:304–2. doi: 10.1016/j.brainresbull.2018.08.013
39. Ota, VK, Santoro, ML, Spindola, LM, Pan, PM, Simabucuro, A, Xavier, G, et al. Gene expression changes associated with trajectories of psychopathology in a longitudinal cohort of children and adolescents. Transl Psychiatry. (2020) 10:99. doi: 10.1038/s41398-020-0772-3
40. Ambaw, A, and Desalegn, GT. Magnitude and correlates of cognitive impairment among major depressive disorder patients in Addis Ababa: institution based cross-sectional study. BMC Res Notes. (2019) 12:135. doi: 10.1186/s13104-019-4184-5
41. Goutzelas, Y, Kontou, P, Mamuris, Z, Bagos, P, and Sarafidou, T. Meta-analysis of gene expression data in adipose tissue reveals new obesity associated genes. Gene. (2022) 818:146223. doi: 10.1016/j.gene.2022.146223
42. Liu, D, McIntyre, RS, Li, R, Yang, M, Xue, Y, and Cao, B. Genetic association between major depressive disorder and type 2 diabetes mellitus: shared pathways and protein networks. Prog Neuro-Psychopharmacol Biol Psychiatry. (2021) 111:110339. doi: 10.1016/j.pnpbp.2021.110339
43. Heremans, IP, Caligiore, F, Gerin, I, Bury, M, Lutz, M, Graff, J, et al. Parkinson’s disease protein PARK7 prevents metabolite and protein damage caused by a glycolytic metabolite. Proc Natl Acad Sci USA. (2022) 119:e2111338119. doi: 10.1073/pnas.2111338119
44. Głombik, K, Stachowicz, A, Trojan, E, Olszanecki, R, Ślusarczyk, J, Suski, M, et al. Evaluation of the effectiveness of chronic antidepressant drug treatments in the hippocampal mitochondria – a proteomic study in an animal model of depression. Prog Neuro-Psychopharmacol Biol 03 Psychiatry. (2017) 78:51–60. doi: 10.1016/j.pnpbp.2017.05.014
45. Tang, M, Huang, H, Li, S, Zhou, M, Liu, Z, Huang, R, et al. Hippocampal proteomic changes of susceptibility and resilience to depression or anxiety in a rat model of chronic mild stress. Transl Psychiatry. (2019) 9:260. doi: 10.1038/s41398-019-0605-4
46. Oláh, A, Alam, M, Chéret, J, Kis, NG, Hegyi, Z, Szöllősi, AG, et al. Mitochondrial energy metabolism is negatively regulated by cannabinoid receptor 1 in intact human epidermis. Exp Dermatol. (2020) 29:616–2. doi: 10.1111/exd.14110
47. Djeungoue-Petga, H-CE, Hebert-Chatelain, E, and Transmission, S. Linking mitochondria and synaptic Transmission: the CB1 receptor. BioEssays. (2017) 39:1700126. doi: 10.1002/bies.201700126
48. Hebert-Chatelain, E, Desprez, T, Serrat, R, Bellocchio, L, Soria-Gomez, E, Busquets-Garcia, A, et al. A cannabinoid link between mitochondria and memory. Nature. (2016) 539:555–9. doi: 10.1038/nature20127
49. Jimenez-Blasco, D, Busquets-Garcia, A, Hebert-Chatelain, E, Serrat, R, Vicente-Gutierrez, C, Ioannidou, C, et al. Glucose metabolism links astroglial mitochondria to cannabinoid effects. Nature. (2020) 583:603–8. doi: 10.1038/s41586-020-2470-y
50. Mendizabal-Zubiaga, J, Melser, S, Bénard, G, Ramos, A, Reguero, L, et al. Cannabinoid CB1 receptors are localized in striated muscle mitochondria and regulate mitochondrial respiration. Front Physiol. (2016) 7:476. doi: 10.3389/fphys.2016.00476
51. Brasanac, J, Gamradt, S, Otte, C, Milaneschi, Y, Monzel, AS, Picard, M, et al. Cellular specificity of mitochondrial and immunometabolic features in major depression. Mol Psychiatry. (2022) 27:2370–1. doi: 10.1038/s41380-022-01473-2
52. Brivio, P, Audano, M, Gallo, MT, Gruca, P, Lason, M, Litwa, E, et al. Metabolomic signature and mitochondrial dynamics outline the difference between vulnerability and resilience to chronic stress. Transl Psychiatry. (2022) 12:87. doi: 10.1038/s41398-022-01856-7
53. Hollis, F, Pope, BS, Gorman-Sandler, E, and Wood, SK. Neuroinflammation and mitochondrial dysfunction link social stress to depression In: KA Miczek and R Sinha, editors. Neuroscience of social stress. Cham: Springer International Publishing (2022)
54. Scaini, G, Mason, BL, Diaz, AP, Jha, MK, Soares, JC, Trivedi, MH, et al. Dysregulation of mitochondrial dynamics, mitophagy and apoptosis in major depressive disorder: does inflammation play a role? Mol Psychiatry. (2022) 27:1095–102. doi: 10.1038/s41380-021-01312-w
55. Bekhbat, M, Goldsmith, DR, Woolwine, BJ, Haroon, E, Miller, AH, and Felger, JC. Transcriptomic signatures of psychomotor slowing in peripheral blood of depressed patients: evidence for Immunometabolic reprogramming. Mol Psychiatry. (2021) 26:7384–92. doi: 10.1038/s41380-021-01258-z
56. Xue, K, Liang, S, Yang, B, Zhu, D, Xie, Y, Qin, W, et al. Local dynamic spontaneous brain activity changes in first-episode, treatment-naïve patients with major depressive disorder and their associated gene expression profiles. Psychol Med. (2022) 52:2052–61. doi: 10.1017/S0033291720003876
Keywords: major depressive disorder, whole exome sequencing, genetic mutation, retrograde endocannabinoid signaling, mitochondrial function
Citation: Xu H, Li T, Gong Q, Xu H, Hu Y, Lü W, Yang X, Li J, Xu W and Kuang W (2023) Genetic variations in the retrograde endocannabinoid signaling pathway in Chinese patients with major depressive disorder. Front. Neurol. 14:1153509. doi: 10.3389/fneur.2023.1153509
Received: 02 February 2023; Accepted: 20 March 2023;
Published: 20 April 2023.
Edited by:
Manabu Funayama, Juntendo University, JapanReviewed by:
Wenzhi Hao, Jinan University, ChinaCopyright © 2023 Xu, Li, Gong, Xu, Hu, Lü, Yang, Li, Xu and Kuang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenming Xu, eHV3ZW5taW5nQHNjdS5lZHUuY24=; Weihong Kuang, a3doaGxqQHNjdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.