
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Neurol., 14 June 2022
Sec. Neurological Biomarkers
Volume 13 - 2022 | https://doi.org/10.3389/fneur.2022.881528
This article is part of the Research TopicPotential Biomarkers in Neurovascular DisordersView all 50 articles
 Akihiko Ueda1*
Akihiko Ueda1* Makoto Nakajima1
Makoto Nakajima1 Yohei Misumi1
Yohei Misumi1 Keiichi Nakahara1
Keiichi Nakahara1 Satoru Shinriki2
Satoru Shinriki2 Masayoshi Tasaki1,3
Masayoshi Tasaki1,3 Hirotaka Matsui2
Hirotaka Matsui2 Mitsuharu Ueda1
Mitsuharu Ueda1This study aimed to evaluate the utility of immunohistochemical staining of vascular Notch3 deposits in biopsied unfixed frozen skin samples from patients with suspected cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). We analyzed vascular Notch3 deposits in unfixed frozen skin biopsy samples obtained from 43 patients with suspected CADASIL by immunohistochemistry using antibodies against the extracellular domain (ECD) of Notch3. We also sequenced the NOTCH3 gene in all patients, as well as evaluated their symptoms and neuroimages. We found granular Notch3 ECD deposits in the vessel walls of unfixed frozen skin biopsy samples in 10 of the 43 suspected patients with CADASIL. All 10 cases with skin Notch3 ECD deposits also carried reported pathogenic variants in the NOTCH3 gene associated with CADASIL. NOTCH3 variants of unknown significance were found in the other four patients without vascular Notch3 ECD or granular osmiophilic material deposits in biopsied skin samples. The remaining 29 cases without vascular Notch3 ECD deposits did not have variants in the NOTCH3 gene. Immunohistochemical evaluation of vascular Notch3 ECD deposits in unfixed frozen biopsied skin samples may be useful for detecting Notch3 deposits in CADASIL.
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is a hereditary cerebrovascular disease caused by mutations in NOTCH3 (1). Migraine, stroke recurrence, and cognitive decline are typical symptoms of CADASIL. Diffuse white matter lesions containing lesions in the temporal pole are characteristic findings of brain magnetic resonance imaging (MRI) in CADASIL. Genetic analysis of NOTCH3 is required for the definitive diagnosis of CADASIL. Most patients with CADASIL have cysteine-related variants in NOTCH3 that lead to an odd number of cysteine residues in epidermal growth factor-like repeats (EGFr) in the Notch3 extracellular domain (ECD). The occurrence of granular osmiophilic material (GOM) and Notch3 ECD deposits have been detected in brain vessels in CADASIL (2, 3).
For the diagnosis of CADASIL, simpler and less invasive detection of vascular Notch3 ECD deposits in the skin is a feasible alternative for detecting vascular pathogenic changes in the brain. Formalin-fixed paraffin-embedded (FFPE) sections, a standard method to fix biopsied samples, have previously been used for the immunohistochemical detection of vascular Notch3 deposits (4–7). However, immunohistochemical detection using FFPE samples has failed to detect Notch3 deposits due to structural alterations during tissue processing. In contrast, unfixed frozen tissue sections, in which the structural conformation of Notch3 ECD is considerably retained, maybe better for the detection of vascular Notch3 ECD deposits in patients with CADASIL (8).
In this retrospective case series, we evaluated the usefulness of immunohistochemical staining for the detection of vascular Notch3 ECD deposits using unfixed frozen skin biopsy samples obtained from 43 patients with suspected CADASIL.
We consulted 380 patients who were suspected of developing CADASIL based on MRI T2 hyperintense lesions in periventricular white matter, deep white matter, and temporal pole white matter each attending doctor between 2008 and 2018 at Kumamoto University Hospital. We enrolled 43 suspected patients with CADASIL with informed consent, who agreed to participate in this study, for the investigation of the diagnostic utility of immunohistochemical detection of vascular Notch3 ECD deposits in the skin of these patients. We did not include patients with CADASIL reported in the previous study (8).
We obtained 0.8 × 1.5 cm of skin biopsy samples from the upper arm of the 43 patients with suspected CADASIL. The biopsied skin samples were equally divided into three parts. The first part was fixed in 4% paraformaldehyde solution with 2.5% glutaraldehyde in 0.1-M sodium cacodylate buffer for electron microscopic analysis. The second part was rapidly frozen in isopentane and cooled in nitro liquid to prepare unfixed frozen sections for immunohistochemical staining of vascular Notch3 ECD deposits. The third part was fixed in 4% paraformaldehyde in phosphate buffer solution (PBS) for standard histopathological examinations.
For immunohistochemical staining of vascular Notch3 ECD deposits, we used 10-μm unfixed frozen skin sections. The sections were stained with rabbit antisera against Notch3 ECD (amino acid residues 1,555–1,569), which was prepared according to a previous study (9), overnight at 4°C. The sections were then washed with PBST for 3 h or more. To decrease the non-specific reaction of the primary antibodies, we prolonged the time of washing the sections in this step. The sections were then incubated with horseradish peroxidase (HRP)-conjugated goat secondary antibodies against rabbit immunoglobulin (Agilent, Santa Clara, CA, United States) for 2 h. Then, the sections were washed five times in phosphate-buffered saline with Tween20 (PBST). The sections were then incubated with 0.3 mg/ml diaminobezidine (Dojin Laboratories, Kumamoto, Japan), 0.65 mg/ml of sodium azide, and 100 μl of 30% hydrogen peroxide for 2 min, and counterstained with Victoria blue to visualize the internal elastic lamina.
Electron microscopy was performed as previously described (10). Briefly, the samples were post-fixed in buffered osmium tetroxide, dehydrated in ascending grades of ethanol, and embedded in Epon. Semi-thin sections were cut and stained with toluidine blue to select arteries of the appropriate size for thin sectioning. Thin sections were double-stained with uranyl acetate and lead citrate, and examined by transmission electron microscopy (TH 7700, HITACHI, Tokyo).
We sequenced the NOTCH3 gene in the 43 patients with suspected CADASIL using a next-generation sequencing panel as follows. We had designed a screening panel of genes for use with the Illumina TruSeq Custom Amplicon platform (Illumina, Inc., San Diego, CA, United States). The panel includes amplicons defining all coding exons of the 27 genes whose mutations are known to cause cerebral small vessel diseases including NOTCH3. Sequencing was performed using the MiSeq (Illumina, Inc.). The obtained sequences were aligned to the reference genome (GRCh37hg19) using MiSeq Reporter software (Illumina, Inc.). The generated virtual contact file (VCF) files containing variant calls were reviewed and further filtered. The clinical significance of the NOTCH3 variants detected in the patients was assessed using ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). Pathogenicity of NOTCH3 variants was predicted using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and MutationTaser2021 (https://www.genecascade.org/MutationTaster2021/#transcript). We analyzed the frequencies of each NOTCH3 variant using the Human Genetic Variation Database (HGVD) (https://www.hgvd.genome.med.kyoto-u.ac.jp/about.html) for the Japanese population and the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/) for the general population. Symptoms and MRI findings using the Fazekas scale for white matter lesions were also evaluated.
Vascular Notch3 ECD deposits in unfixed frozen skin sections were visualized as granular dots in the arterial walls by immunohistochemical staining in 10 of the 43 patients with suspected CADASIL (Figures 1A–F, Table 1). Although amounts of granular Notch3 deposits were slightly different among patients with CADASIL, we found Notch3 deposits in all the arterioles. We detected Notch3 deposits in 8 (62%) of 13 patients with lesions in the temporal pole and 2 (7%) of 30 patients without lesions in the temporal pole (Table 2). We detected Notch3 deposits in 10 (34%) of 29 patients with Fazekas grade 3 white matter lesions and did not find them in 13 patients with Fazekas grade 1–2 white matter lesions (Table 2). GOM deposits were observed by electron microscopy (Figures 1E,F). The location and morphological features of the vascular Notch3 ECD and the GOM deposits were similar. We found Notch3 ECD deposits in all the three randomly selected CADASIL cases with GOM deposits (Table 1). Moreover, sequencing revealed that all 10 patients with vascular Notch3 ECD deposits had pathogenic or likely pathogenic variants of NOTCH3, such as p.Arg110Cys, p.Tyr258Cys, p Cys408Arg, p.Cys516Phe, p. Trp1003Cys, and p.Tyr1021Cys, which are reportedly associated with CADASIL (Table 1). NOTCH3 p.Arg110Cys and p.Tyr258Cys variants are located in Notch3 EGFr domains 1–6 (case 2 and 10), and NOTCH3 p.Cys408Arg, p.Cys516Phe, p.Trp1003Cys, and p.Tyr1021Cys variants are located in Notch3 EGFr domains 7–34 (cases 1 and 3–9). We did not find significant correlations between the degree of Notch3 deposits and NOTCH3 mutation location. Based on the ClinVar database, the NOTCH3 p.Arg75Gln variant is “likely benign.” The other five variants, such as p.Thr900Pro, p.Leu989Arg, p.Cys1372Gly, p Glu1373Gly, p.Ala1649Thr, and p.Gly1650Ser, were not found in the ClinVar database. According to PolyPhen-2 and MutationTaser2021, p.Leu989Arg, p.Ala1649Thr, and p.Gly1650Ser were predicted to be “benign” and p.Thr900Pro, p.Cys1372Gly, and p Glu1373Gly were predicted to be “probably damaging” and “deleterious” (Table 1).
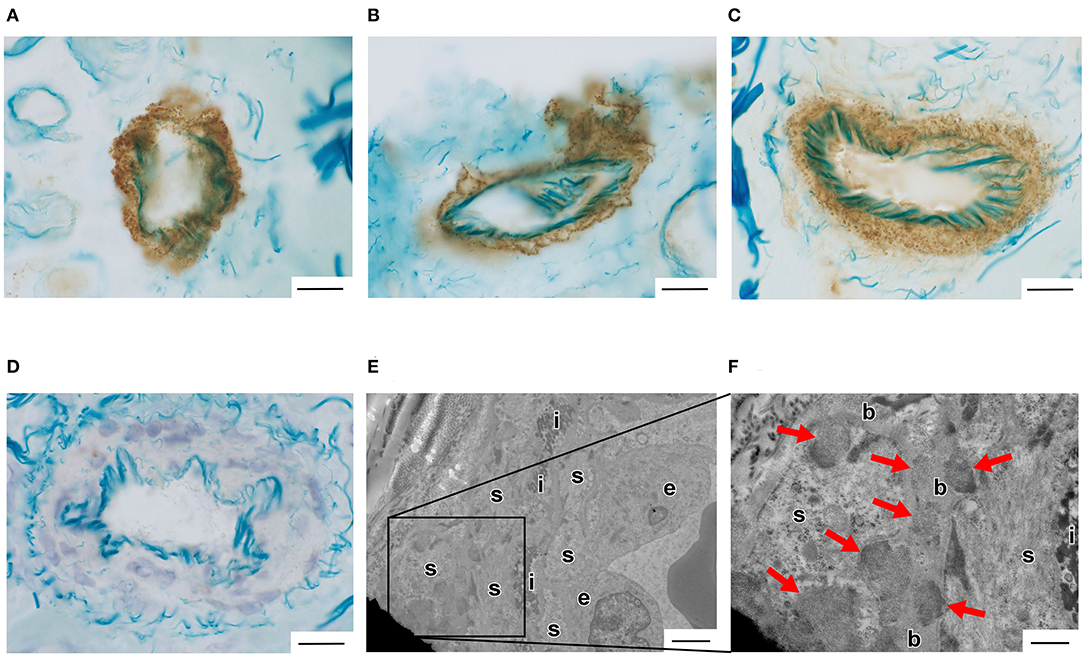
Figure 1. (A–D) Vascular Notch3 ECD deposits were detected by immunohistochemical staining using anti-Notch3 ECD antibodies in an unfixed frozen skin biopsy sample obtained from a patient with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL). (A) Case 2 with the pathogenic NOTCH3 p.Arg110Cys variant. (B) Case 6 with the pathogenic NOTCH3 p.Trp1003Cys variant. (C) Case 9 with the pathogenic NOTCH3 p.Tyr1021Cys variant. (D) Case 35 with the likely benign variants of NOTCH3 p.Arg75Gln (Table 1). Bars = 20 μm. (E,F) Vascular granular osmiophilic material (GOM) deposits were detected by electron microscopic analysis in a biopsied skin sample obtained from a patient with CADASIL (Case 9 with the pathogenic NOTCH3 p.Tyr1021Cys variant, Table 1). Arrows indicate GOM deposits. e, endothelial cells; s, smooth muscle cells; i, internal elastic lamina; b, basal lamina. (E) Bar = 2 μm. (F) Bar = 400 nm.
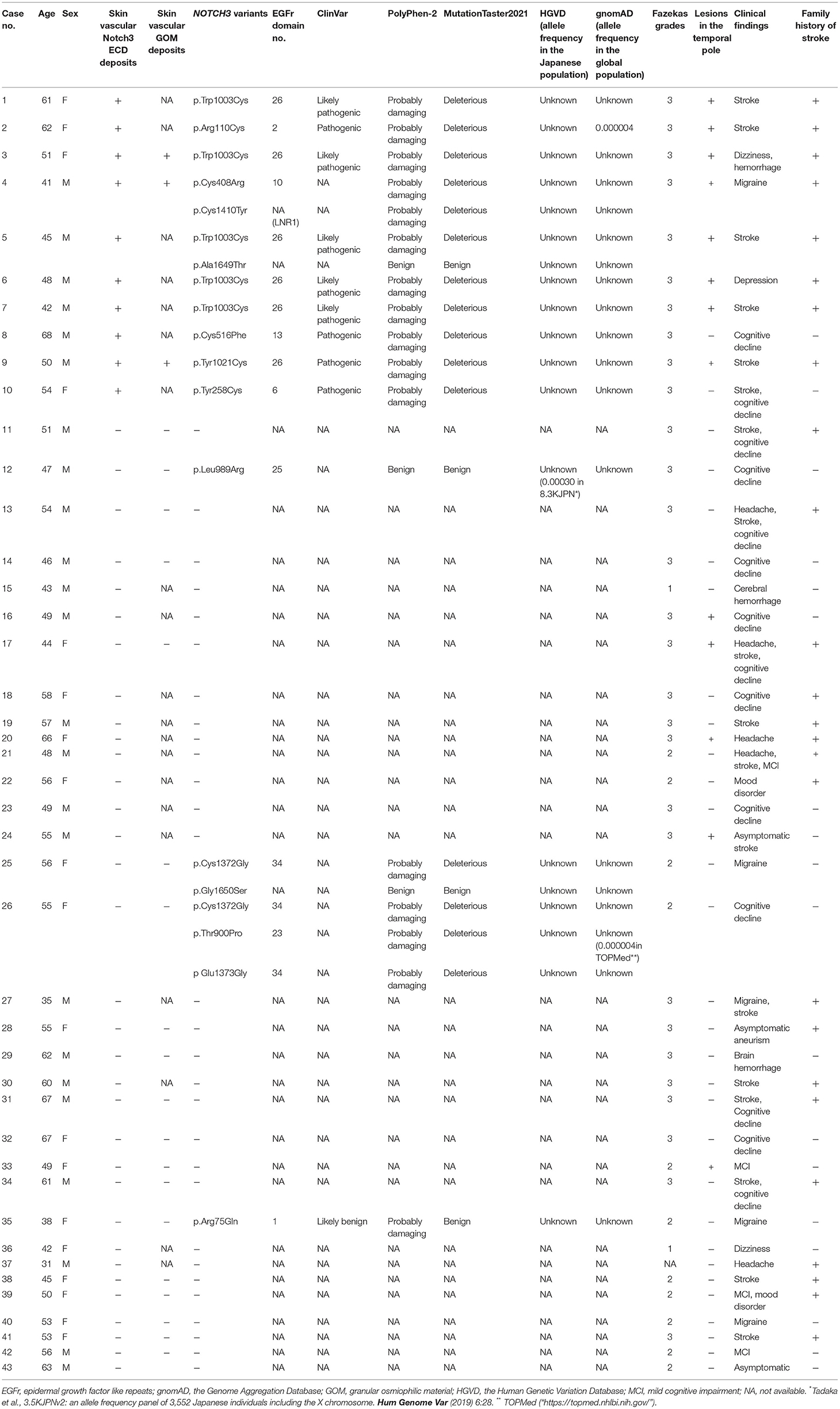
Table 1. Skin vascular Notch3 ECD deposits and NOTCH3 variants in 43 patients with suspected CADASIL.
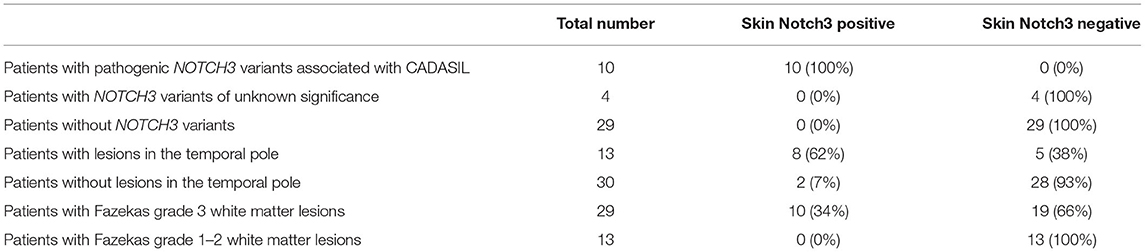
Table 2. Number of patients with and without skin Notch3 deposits among patients with suspected CADASIL.
Four of the 33 patients without vascular Notch3 ECD or GOM deposits in skin biopsy samples (cases 12, 25, 26, and 35) (Table 1) had NOTCH3 variants of unknown significance, such as p.Arg75Gln, p.Thr900Pro, p.Leu989Arg, p.Cys1372Gly, p Glu1373Gly, and p.Gly1650Ser. The remaining 29 patients without vascular Notch3 ECD deposits did not have any NOTCH3 variants. We also investigated vascular GOM deposits in 16 of the 29 patients and found no GOM deposits in any of the 16 patients without vascular Notch3 ECD deposits or NOTCH3 variants (Table 1).
We found lesions in the temporal pole on MRI (Figure 2) and a family history of stroke in eight of the 10 (80%) patients with vascular Notch3 ECD deposits. Two patients (cases 8 and 10) with vascular Notch3 ECD deposits had neither lesion in the temporal pole on MRI nor a family history of stroke (Table 1). In contrast, 5 of the 33 (15%) patients without vascular Notch3 ECD deposits (cases 16, 17, 20, 24, and 33) had lesions in the temporal pole. No NOTCH3 variants were found in these five patients (Table 1).
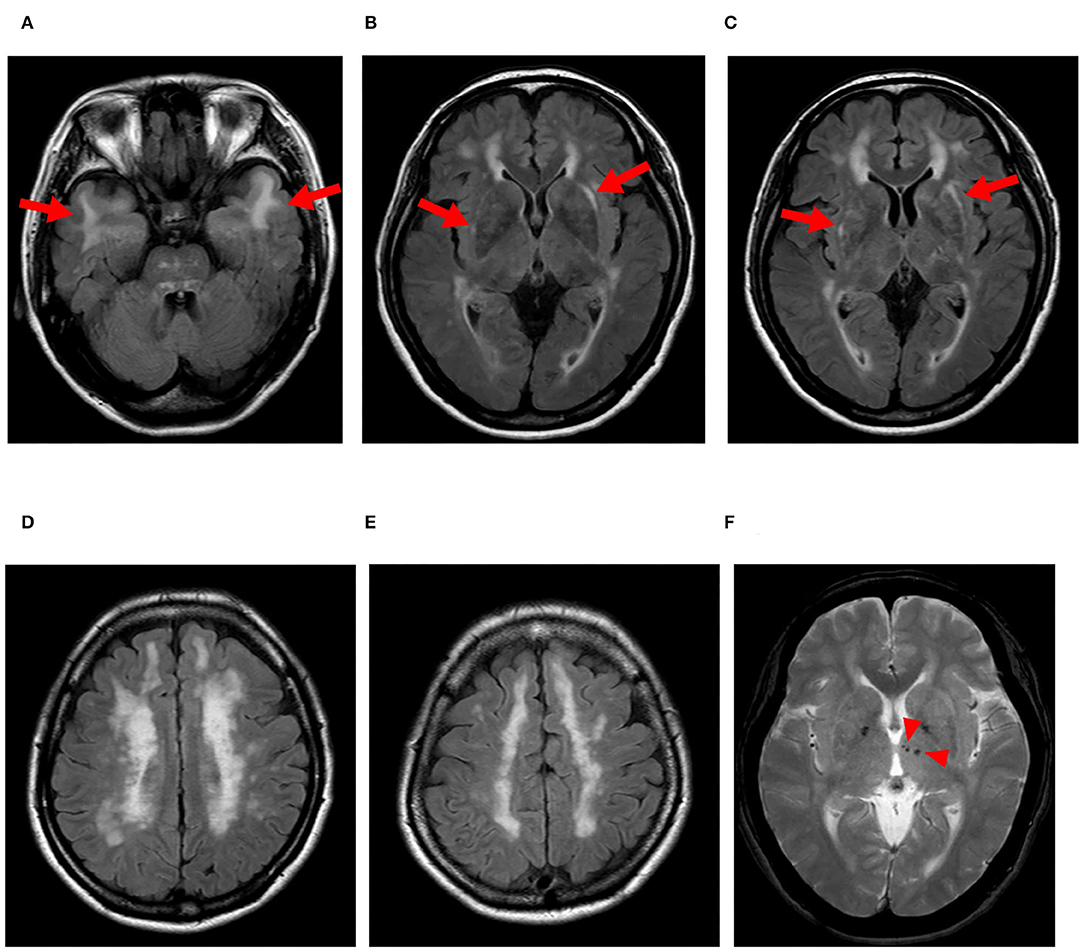
Figure 2. Representative MRI findings of CADASIL. Case 2 with the pathogenic NOTCH3 p.Arg110Cys variant. (A–E) FLAIR images, (F) T2-star weighted image. (A) Arrows indicate lesions in the temporal pole. (B,C) Arrows indicate lesions in the extra capsule of the putamen, and (F) arrowheads indicate microbleeds.
In this case series study of 43 patients with suspected CADASIL, we detected vascular Notch3 ECD deposits in all 10 patients by immunohistochemical staining using unfixed frozen biopsied samples, which were confirmed to have pathogenic NOTCH3 variants causing CADASIL. In contrast, conventional immunohistochemical staining using FFPE tissue samples fails to detect vascular Notch3 deposits in 5–15% of patients with CADASIL with pathogenic variants in NOTCH3 (4–6). Therefore, unfixed frozen biopsied tissue samples may be more suitable than FFPE-biopsied tissue samples for the detection of vascular Notch3 ECD deposits in the skin.
Patients with CADASIL carrying pathogenic NOTCH3 variants, which were mostly associated with cysteine replacement (11), located in the EGFr domains 7–34, reportedly showed milder phenotypes than those with NOTCH3 variants located in the EGFr domains 1–6 (12). In addition, Gravesteijn et al. (13) recently reported that the amount of vascular Notch3 ECD and GOM deposits in the skin in patients with CADASIL with NOTCH3 variants in EGFr 7–34 was lesser than that in those with NOTCH3 variants in EGFr 1–6. In this case series study, we successfully detected vascular Notch3 ECD deposits in patients with CADASIL with both milder NOTCH3 EGFr 7–34 variants and typical severe NOTCH3 EGFr 1–6 variants by immunohistochemical staining using unfixed frozen biopsied skin samples (Table 1). Therefore, immunohistochemical staining using unfixed frozen biopsied skin samples seems to be suitable for detecting Notch3 deposits in CADASIL regardless of the amount of Notch3 ECD deposits.
Detecting Notch3 and GOM deposits are thought to be helpful for the diagnosis of CADASIL. Brain MRI findings reportedly varied considerably between patients with CADASIL and were dependent on the NOTCH3 genotype (14). While the involvement of the anterior temporal pole and external capsule may be helpful for the diagnosis of CADASIL, these MRI findings were reportedly not sufficient for accurate diagnosis of CADASIL (14). Skin biopsy is less invasive than brain biopsy to directly confirm pathogenic Notch3 and GOM deposits in patients with suspected CADASIL. In this study, 10 of 43 cases were identified as positive of staining of Notch3 and/or GOM. We believe that skin biopsy is useful especially for detecting Notch3 deposits in patients with CADASIL with NOTCH3 variants of unknown significance, while skin biopsy may not be essential for the diagnosis of patients with CADASIL with typical NOTCH3 variants and typical MRI findings in the daily clinical practice.
This study is limited in that it had a small sample size. Large-scale studies including more patients with CADASIL with other genotypes are needed to determine the sensitivity and specificity of this immunohistochemical method in differentiating between CADASIL and other cerebral small vessel diseases.
Immunohistochemical staining of vascular Notch3 ECD deposits in unfixed frozen skin sections may be useful over conventional immunohistochemical staining for detecting Notch3 deposits in CADASIL.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
This study was approved by the Human genome Ethics Committee of Kumamoto University. The patients/participants provided their written informed consent to participate in this study.
AU drafted the manuscript, devised the study concept and design, collected data, and performed the pathological examinations. SS and HM performed the gene analysis. MN, KN, MT, and YM revised the manuscript. MU revised the manuscript, devised the study concept and design, and supervised the study. All authors have contributed to the manuscript and approved the submitted version.
This study was funded by a grant-in-aid for research on intractable diseases from the Japanese Ministry of Health, Labor, and Welfare (Grant No. 21FC1007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank Editage (www.editage.com) for English language editing.
CADASIL, cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy; EGFr, epidermal growth factor-like repeats; ECD, extracellular domain; GOM, granular osmiophilic material; FFPE, formalin-fixed paraffin-embedded sections; PBS, phosphate buffer solution; HRP, horseradish peroxidase; DAB, diaminobenzidine.
1. Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssière C, et al. Strong clustering and stereotyped nature of notch3 mutations in patients with CADASIL. Lancet. (1997) 350:1511–5. doi: 10.1016/S0140-6736(97)08083-5
2. Esters ML, Chimowitz MI, Awad IA, McMahon JT, Furlan AJ, Ratiff NB. Sclerosing vasculopathy of the central nervous system in nonelderly patients. Arch Neurol. (1991) 46:631–6. doi: 10.1001/archneur.1991.00530180087022
3. Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, et al. The ectodomain of the Notch 3 receptor accumulates within the cerebrovascular of patients with CADASIL. J Clin Invest. (2000) 105:597–605. doi: 10.1172/JCI8047
4. Joutel A, Favrole P, Labauge P, Chabriat H, Lescoat C, Andreux F, et al. Skin biopsy immunostaining with a Notch3 monoclonal antibody for CADASIL diagnosis. Lancet. (2001) 358:2049–51. doi: 10.1016/S0140-6736(01)07142-2
5. Lesnik Oberstein SA, van Duinen SG, van den Boom R, Maat-Schieman ML, van Buchem MA, van Houwelingen HC, et al. Evaluation of diagnostic Notch3 immunostaining in CADASIL. Acta Neuropathol. (2003) 106:107–11. doi: 10.1007/s00401-003-0701-6
6. Ampuero I, Alegre-Abarrategui J, Rodal I, España A, Raquel Ros SJLL, et al. On the diagnosis of CADASIL. J Alzhemer's Dis. (2009) 17:787–94. doi: 10.3233/JAD-2009-1112
7. Tikka S, Mykkänen K, Junna M, Bergholm R, Pöyhönen M, Baumann M, et al. Diagnosing Vascular Dementia by Skin Biopsy-Uniqueness of CADASIL. In: Khopkar U, editor. Skin Biopsy-Perspectives. London: Intechopen (2011).
8. Ueda A, Ueda M, Nagatoshi A, Hirano T, Ito T, Arai N, et al. Genotypic and phenotypic spectrum of CADASIL in Japan: the experience at a referral center in Kumamoto university From 1997 to 2014. J Neurol. (2015) 262:1828–36. doi: 10.1007/s00415-015-7782-8
9. Yamamoto Y, Craggs LJ, Watanabe A, Booth T, Attems J, Low RW, et al. Brain microvascular accumulation and distribution of the Notch3 ectodomain and granular osmiophilic material in CADASIL. J Neuropathol Exp Neurol. (2013) 72:416–31. doi: 10.1097/NEN.0b013e31829020b5
10. Tikka S, Mykkänen K, Ruchoux MM, Bergholm R, Junna M, Pöyhönen M, et al. Congruence between Notch3 mutations and GOM in 131 patients with CADASIL. Brain. (2009) 132:933–9. doi: 10.1093/brain/awn364
11. Matsushima T, Conedera S, Tanaka R, Li Y, Yoshino H, Funayama M, et al. Genotype-phenotype correlations of cysteine replacement in CADASIL. Neurobiol Aging. (2017) 169:e7–169. doi: 10.1016/j.neurobiolaging.2016.10.026
12. Rutten JW, Van Eijsden BJ, Duering M, Jouvent E, Opherk C, Pantoni L, et al. The effect of Notch3 pathogenic variant position on CADASIL disease severity: Notch3 EGFr 1–6 pathogenic variant are associated with a more severe phenotype and lower survival compared with EGFr 7–34 pathogenic variant. Genet Med. (2019) 21:676–82. doi: 10.1038/s41436-018-0088-3
13. Gravesteijn G, Hack RJ, Mulder AA, Cerfontaine MN, van Doorn R, Hegeman IM, et al. Notch3 variant position is associated with Notch3 aggregation load in CADASIL vasculature. Neuropathol Appl Neurobiol. (2022) 48:e12751. doi: 10.1111/nan.12751
Keywords: CADASIL, Notch3 deposits, NOTCH3 variants, skin biopsy, immunohistochemistry
Citation: Ueda A, Nakajima M, Misumi Y, Nakahara K, Shinriki S, Tasaki M, Matsui H and Ueda M (2022) Detection of Vascular Notch3 Deposits in Unfixed Frozen Skin Biopsy Sample in CADASIL. Front. Neurol. 13:881528. doi: 10.3389/fneur.2022.881528
Received: 22 February 2022; Accepted: 16 May 2022;
Published: 14 June 2022.
Edited by:
Jinming Han, Capital Medical University, ChinaReviewed by:
Akitoshi Takeda, Osaka City University, JapanCopyright © 2022 Ueda, Nakajima, Misumi, Nakahara, Shinriki, Tasaki, Matsui and Ueda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akihiko Ueda, YWtpaGlrby1zaGltYUBrNC5kaW9uLm5lLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.