- Department of Neurosurgery, Jinling Hospital, School of Medicine, Nanjing University, Nanjing, Jiangsu, China
Traumatic brain injury (TBI) causes high rates of worldwide mortality and morbidity due to the complex secondary injury cascade. Recently, circular ribonucleic acids (circRNAs) have attracted significant attention in a variety of diseases. However, their expression characteristics in human TBI are still unclear. In this study, we examined brain injury tissues from six severe TBI patients in Jinling Hospital. The TBI tissues and adjacent brain contusion tissues were used to analyze differential expression signatures of circRNAs through full-length transcriptome sequencing, Gene Ontology (GO) analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and ceRNA network construction. Our results found that there were 126 differently expressed circRNAs in TBI. Among them, 64 circRNAs were up-regulated and 62 circRNAs were down-regulated. Moreover, GO and KEGG analyses revealed that the aberrantly expressed circRNAs participated in many pathophysiological processes of TBI, especially regarding microglial cell activation, protein transport, protein processing and inflammation. Furthermore, the ceRNA (circRNA-miRNA-mRNA) network predicted that there existed strong relationship among circRNA, miRNA and mRNA. Taken together, our results indicated for the first time that the expression profiles of circRNAs were different after human TBI. In addition, we found the signaling pathways that were related to circRNAs and predicted a ceRNA network, which provided new insight of circRNAs in human TBI.
1. Introduction
Traumatic brain injury (TBI) is a leading cause of mortality and disability across all age groups, with estimates of more than 50 million people experiencing TBI each year, imposing a tremendous burden on families and society (1). After the initial insult, the primary injury occurs immediately and can lead to cerebral contusion (2). Although primary injury is the main factor determining the prognosis of TBI patients, the biochemical cascade reactions (secondary brain injury), such as oxidative stress, apoptosis and inflammation, will aggravate the brain tissue damage (3). To date, no effective treatments are available to promote functional recovery after TBI. Therefore, novel therapeutics are significant needed to provide new targets for the treatment of TBI.
Circular ribonucleic acids (circRNAs) are non-coding RNAs derived from back-splicing, which are characterized by a single-stranded, covalently closed-loop structure with no 5' end caps or 3' poly (A) tails (4). CircRNAs can locate in the exon, intron or exon-intron boundary. They are abundantly expressed in eukaryotic cells and demonstrate tissue-specific expression patterns (5). CircRNAs were initially thought to be by-products of transcription with no functions (6). However, increasing evidence have indicated that circRNAs possessed a variety of properties and functions such as regulating gene expression and acting as microRNA (miRNA) sponges, protein decoys and protein translators, making circRNAs as the hotspot of non-coding RNAs research field recently (7, 8). The role of circRNAs in TBI has been studied earlier in animal models. For example, Zhao et al. identified 231 significantly and differentially expressed circRNAs in exosomes from the brain extracellular space after TBI. In further studies they found that these differentially expressed circRNAs might be related to the growth and repair of neurons, the development of the nervous system and the transmission of nerve signals. Moreover, the most highly correlated pathways of these circRNAs were involved primarily with glutamatergic synapse and the cyclic guanosine monophosphate-protein kinase G signaling pathway (9). In another study conducted by Xie et al., they observed that a total of 192 circRNAs were differentially expressed in the rat hippocampus following TBI. Furthermore, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis indicated that many messenger RNAs (mRNAs) transcribed from the host genes of altered circRNAs were implicated in brain damage and neural regeneration. These data demonstrated altered circRNA expression pattern in the rat hippocampus after TBI, which may play important roles in post-TBI physiological and pathological processes (10). The role of circRNAs in TBI has also been confirmed in clinical researches. In the blood of humans with TBI, Zheng et al. detected 3,035 differentially expressed circRNAs in the severe TBI group, 2,362 in the moderate group and 433 in the mild group. Moreover, they constructed a ceRNA network and found that the has_circ_0020269 (circHtra1) was significantly up-regulated after brain insults and was correlated with the severity of injury. In addition, circHtra1 could inhibit cell proliferation and promoted apoptosis, and its knockdown reversed these effects (11).
To date, the expression profiles of circRNAs in human TBI brain tissues has not been fully explained. Therefore, we identified the expression profiles of circRNAs from human TBI in our study.
2. Materials and methods
2.1. Patient samples
The TBI tissues were extracted from the center of contusion area, and the adjacent brain contusion tissues were taken from the brain tissues 1.5 cm away from the contusion according to our previous study (12). The specimens were immediately frozen in liquid nitrogen and stored at −80°C.
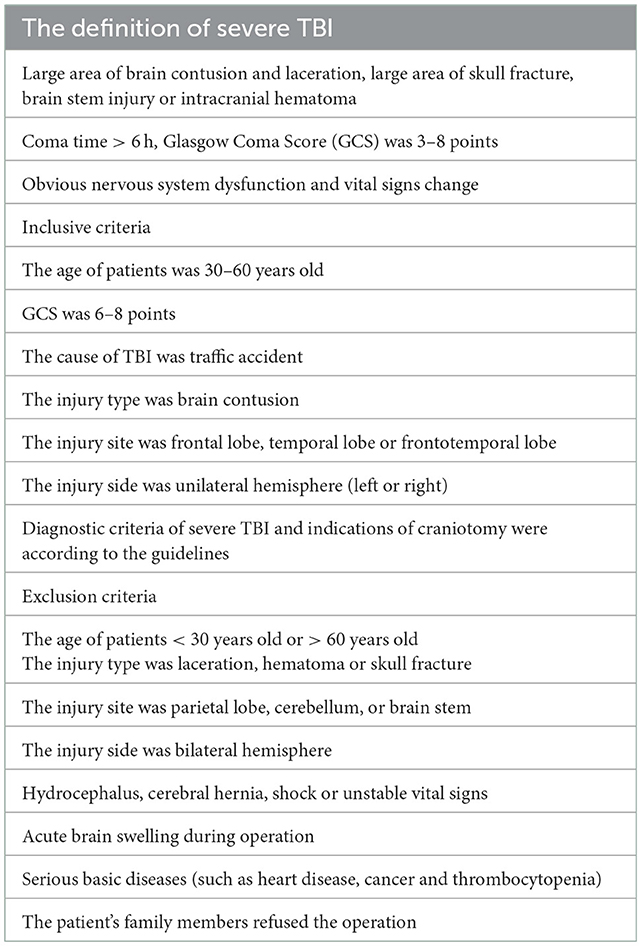
The study protocol was approved by Institutional Review Board of Jinling Hospital (approval number: 2021DZGZR-YBB-082). This study was performed in accordance with the Declaration of Helsinki. A written informed consent to participate and consent for publication were obtained from each patient' surrogates. The clinical data of 6 patients with severe TBI was shown in Table 1. The definition of severe TBI (13), inclusive criteria and exclusion criteria (Brain Trauma Foundation, American Association of Neurological Surgeons and Congress of Neurological Surgeons, 2007) were shown in Table 2.
2.2. RNA extraction and qRT-PCR
Total RNA was extracted from the brain contusion tissues and adjacent brain contusion tissues using TRIzol regent (Invitrogen, Carlsbad, CA, USA) according to manual instruction. Total RNA was reverse-transcribed using PrimeScriptTM RT reagent kit with gDNA Eraser.
qRT-PCR was performed using SYBR® Premix Ex TaqTM II and a 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) (14). Relative quantification of gene expression was calculated using the formula: 2−ΔΔCt, ΔΔCt = (Cttargetgene – Ctβ−actin) injury – (Cttargetgene – Ctβ−actin) injury surrounding. Three independent experiments were performed for each condition.
2.3. CircRNA analysis and differential circRNA screening
The Epicentre Ribo-Zero Kit was used to eliminate all ribosomal RNAs from the total RNA (Epicentre). RNase R (Epicentre) digestion was conducted to eliminate all linear RNAs. Finally, pure circRNAs were obtained. After enrichment, the obtained circRNAs were treated with NEBNext® Ultra™ Directional RNA Library preparation kit (Illumina). Finally, the obtained libraries were subjected to paired-end sequencing on an Illumina HiSeq2500 platform (Illumina).
Two algorithms, CIRI2 and CIRCexplorer2 were used to detect circRNAs. The reads were mapped into the human reference genome GRCh37/hg19 (http://genome.ucsc.edu/) by BWA-MEM or Tophat, respectively. CIRI2 detected paired chiastic clipping signals according to the mapping of reads. Systematic filtering in combination with local alignment with BWA-MEM was conducted to eliminate potential false-positive data. CIRCexplorer2 uses Top Hat and Top Hat-Fusion alignment output to detect circRNAs.
If a circRNA was detected by both methods, it was considered to be an identified circRNA. Back-spliced junction reads identified in CIRI2 were integrated and scaled to reads per million (mapped reads, BWA-MEM mapping) to quantify each circRNA. The fold difference of each circRNA was calculated to statistically compare the expression profiles between the TBI tissues and the adjacent brain contusion tissues using Student's t test. The differentially expressed circRNAs were defined as P < 0.05.
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of circRNA
The GO and KEGG pathway analysis were conducted according to previous studies (15). GO enrichment analysis (http://www.geneontology.org) was conducted to explore the potential biological functions of differentially expressed circRNAs. KEGG pathway analysis (http://www.genome.jp/kegg/) was conducted to reveal the signaling pathways of identified circRNAs at the molecular level.
2.5. Bioinformatic analysis
The specific binding of circRNAs to miRNAs was predicted by miRanda (http://www.microrna.org/) and PITA (https://genie.weizmann.ac.il/pubs/mir07/mir07_exe.html). The miRNAs-targeted mRNAs were identified by miRanda (http://www.microrna.org/) and Targetscan (http://www.targetscan.org/).
2.6. ceRNA (circRNA-miRNA-mRNA) network analysis
According to the predicted targeting relationship among circRNA, miRNA, and mRNA, and the correlation expressed in the samples, the negative correlation between circRNA and miRNA and the negative correlation between mRNA and miRNA are taken to construct the endogenous competitive binding ceRNA (circRNA-miRNA-mRNA) network. Three conditions must exist for the ceRNA network to occur (16). Firstly, the relative concentration of ceRNAs and their miRNAs is clearly important. Secondly, the effectiveness of a ceRNA depends on the number of miRNAs that it can “sponge”. Thirdly, not all of the miRNA response elements on ceRNAs are equal. Therefore, we accept only the ceRNA-pair relations that passed filtering measures (p < 0.05). A graph of the circRNA-miRNA-mRNA network was made using Cytoscape software (version 3.5.1) to visualize these relationships.
2.7. Statistical analysis
All statistical analysis was performed with SPSS 19.0 (SPSS Inc., Chicago, IL). Data were presented as mean ± SEM. Differences between the TBI group and control group were analyzed using t-test. A value of p < 0.05 was considered statistically significant.
2.8. Database and accession numbers
The circRNA microarray data in our study were deposited at the NCBI GenBank under the Accession No. BankIt2634779: OP794505-OP79462.
3. Results
3.1. Different express of circRNAs in human TBI tissues and adjacent brain contusion tissues
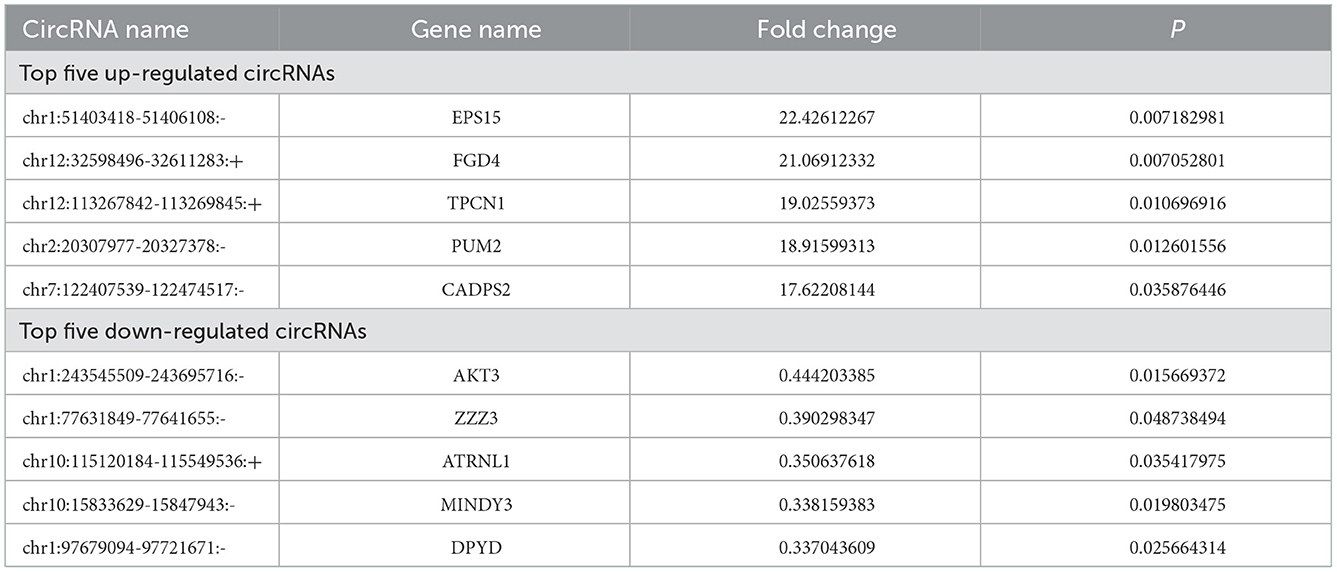
Among the 126 differentially expressed circRNAs (P < 0.05, Figures 1A, B), 64 were up-regulated and 62 were down-regulated. Among them, circRNA epidermal growth factor receptor pathway substrate 15 (EPS15), FYVE, RhoGEF, and PH domain containing four (FGD4), two-pore channel 1 (TPCN1), PUMILIO 2 (PUM2) and calcium-dependent activator protein for secretion 2 (CADPS2) were top five up-regulated circRNAs, while circRNA serine/threonine protein kinase 3 (AKT3), zinc finger ZZ-type containing 3 (ZZZ3), attractin-like 1 (ATRNL1), motif interacting with Ub-containing novel DUB family 3 (MINDY3) and dihydropyrimidine dehydrogenase (DPYD) were top five down-regulated circRNAs (Table 3).
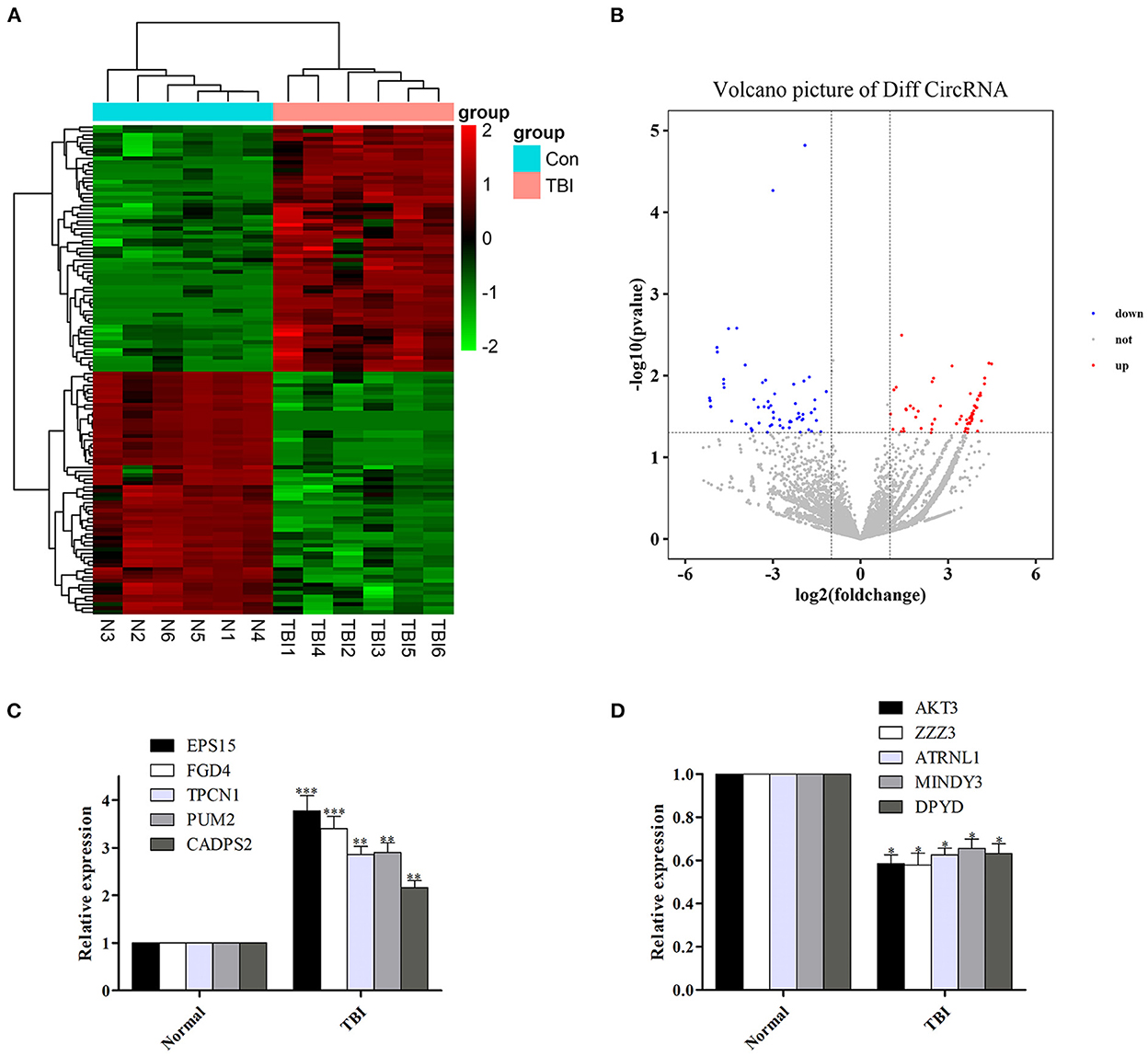
Figure 1. Differentially expressed circRNAs in human TBI tissues, and verify the reliability of sequencing results by qRT-PCR. (A, B) Differentially expressed circRNAs in human TBI tissues. (C) The expression of the top five up-regulated circRNAs was increased in human TBI tissues. (D) The expression of the top five down-regulated circRNAs was decreased in human TBI tissues. n = 6 per group. Data are presented as mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001 vs. adjacent brain contusion group.
3.2. Verify the reliability of sequencing results by qRT-PCR
In order to verify the reliability of sequencing results, we detected the expression levels of the top five up-regulated circRNAs (EPS15, FGD4, TPCN1, PUM2, and CADPS2) and down-regulated circRNAs (AKT3, ZZZ3, ATRNL1, MINDY3, DPYD) in the original samples by qRT-PCR. We found that the expression of the top five up-regulated and down-regulated circRNAs was also increased (Figure 1C) and decreased (Figure 1D) in human TBI tissues, suggesting that the results of qRT-PCR were in agreement with the results of sequencing.
3.3. GO and KEGG pathway analyses
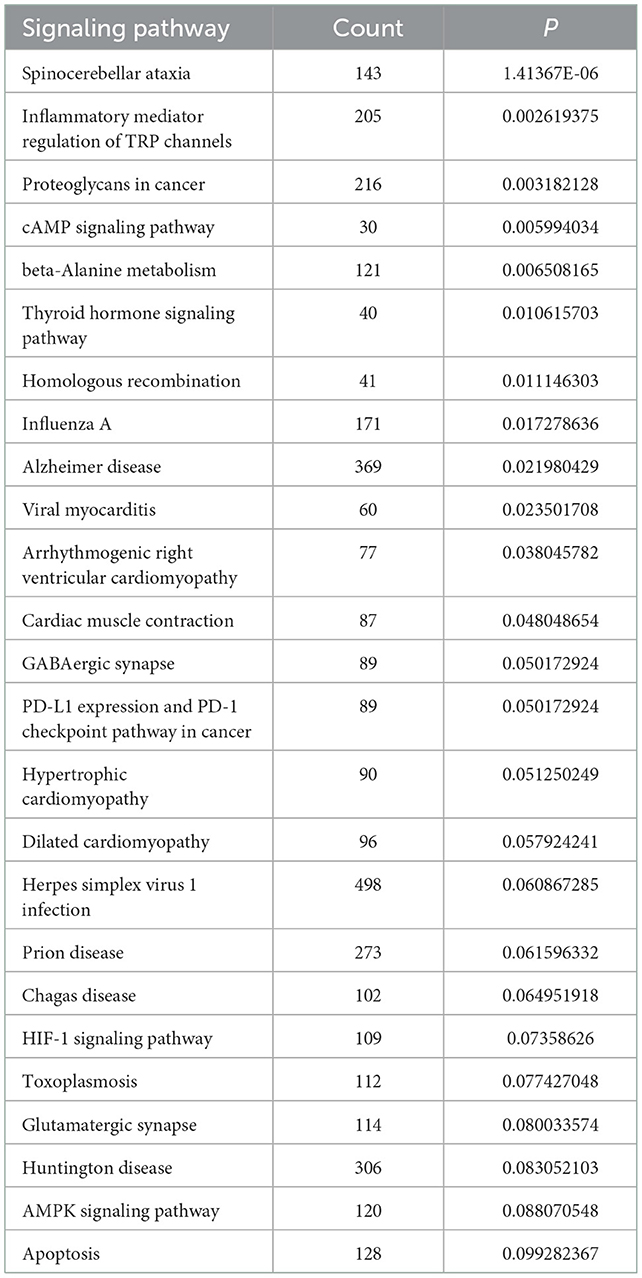
We performed GO analysis on the differentially expressed circRNAs to reveal the changes of biological process after TBI. We found that the GO were mainly enriched in microglial cell activation, protein transport, protein processing, negative regulation of nucleotide-binding oligomerization and inflammation response (Figure 2A). Subsequently, we analyzed KEGG pathway and found that circRNAs were mainly related to spinocerebellar ataxia, inflammatory mediator regulation of transient receptor potential (TRP) channels, proteoglycans in cancer, cyclic adenosine monophosphate (cAMP) signaling pathway and beta-Alanine metabolism (Figure 2B). The top 25 KEGG pathways that related to target genes after TBI were listed in Table 4.
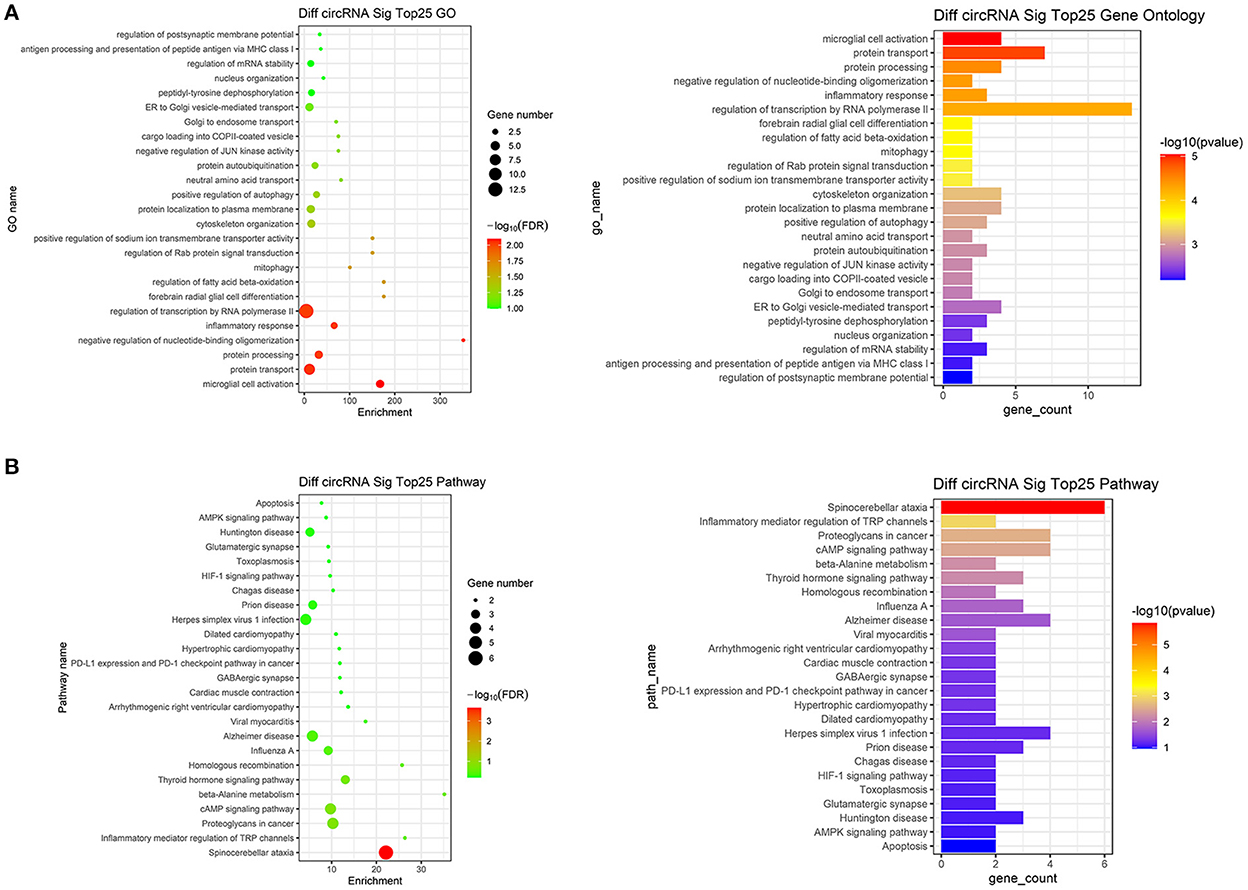
Figure 2. Go analysis and KEGG pathway analysis. (A) The top 25 up-regulated and down-regulated biological processes related to differentially expressed circRNAs after TBI. (B) The top 25 up-regulated and down-regulated pathways related to differentially expressed circRNAs after TBI.
3.4. miRNA prediction and ceRNA network construction
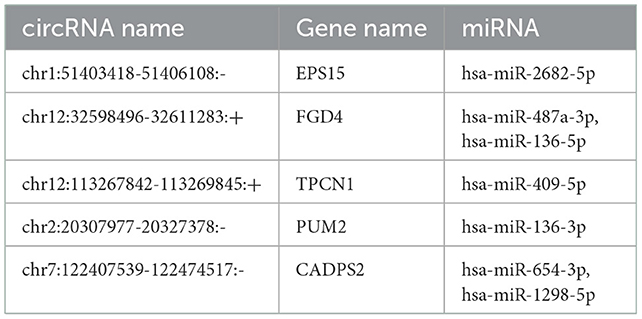
miRanda and PITA were used to predict miRNAs targeted by the differentially expressed circRNAs. The miRNAs predicted to be targeted by the top five significantly differentially expressed circRNAs, which were all contained in the two databases, are presented in Table 5. The miRNA-mRNA regulatory relationships were further identified using miRanda and Targetscan. The circRNA-targeted miRNAs and their regulated mRNAs were further selected for ceRNA network construction (Figure 3).
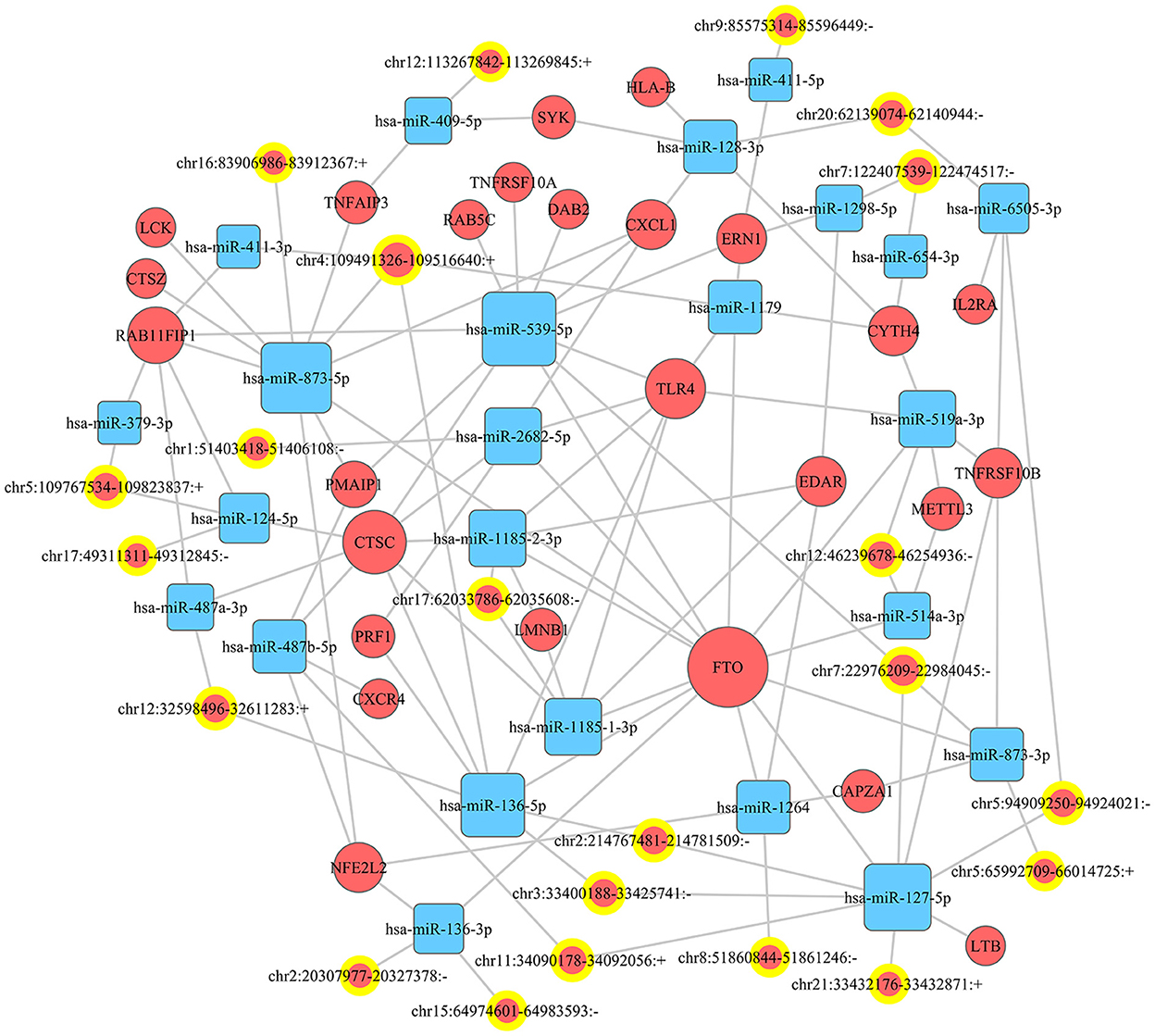
Figure 3. CeRNA network. A ceRNA (circRNA-miRNA-mRNA) network was established to show the relationship among circRNA, miRNA, and mRNA.
4. Discussion
In the present study, we found that: (1) there existed differentially expressed circRNAs between human TBI tissues and adjacent brain contusion tissues; (2) GO and KEGG analyses indicated that the aberrantly expressed circRNAs mainly participated in microglial cell activation and inflammation; (3) the circRNAs-targeted miRNAs and miRNAs-targeted mRNAs were predicted; (4) a ceRNA network was established.
It has been shown that circRNAs participated in the pathophysiological process of CNS diseases (5, 17). However, the role of circRNAs in TBI was unknown. Some circRNAs have been proposed which may explain the effects of circRNAs in TBI. In a mouse TBI model, circHtra1 could promote neuronal loss and immune deficiency in TBI (11). Moreover, knockdown of circRNA_009194 improved neurological outcomes in TBI (18). Furthermore, inhibition of circIgfbp2 alleviated mitochondrial dysfunction and oxidative stress-induced synapse dysfunction after TBI (19). In our study, we examined circRNAs expression profiles from six TBI patients. A total of 126 circRNAs (64 up-regulated and 62 down-regulated) were differentially expressed between server TBI tissues and adjacent brain contusion tissues. Furthermore, we found that the five top up-regulated circRNAs were EPS15, FGD4, TPCN1, PUM2 and CADPS2, and the five top down-regulated circRNAs were AKT3, ZZZ3, ATRNL1, MINDY3 and DPYD. Among them, circRNA EPS15 owned the maximized up-regulated fold change. CircRNA EPS15 is derived from the parent gene Eps15. Eps15 is transcribed from ch38. p13, which encodes a component of the epidermal growth factor receptor (EGFR) pathway, exists in the pits of the reticular protein coating, and participates in the receptor-mediated endocytosis of EGF (20). To date, the top five up/down-regulated circRNAs have not been reported in previous studies. We would confirm the role of these circRNAs in TBI in our future researches.
In order to understand these differentially expressed circRNAs, the possible biological functions of their parent gene were analyzed by GO and KEGG analysis. GO analysis showed that the mostly enriched GO were located in the microglial cell activation, protein transport, protein processing, negative regulation of nucleotide-binding oligomerization and inflammation response. Based on KEGG results, the most enriched pathways were spinocerebellar ataxia, inflammatory mediator regulation of TRP channels, proteoglycans in cancer, cAMP signaling pathway and beta-Alanine metabolism. From GO and KEGG analysis, we found that microglial cell activation-induced inflammation response was very critical in TBI. Microglial cells, derived from myelogenous cells, support physiological processes by secreting neurotrophic factors and assisting with the formation and elimination of synapses (21). Moreover, microglial cells provide immune responses in respond to a number of stimuli (22). After TBI, microglia are activated, the activated microglia secrete a large number of neuroimmune inflammatory factors such as interleukin-1β (IL-1β), IL-10 and tumor necrosis factor-α (TNF-α), thus exacerbating the inflammatory response and leading to a series of brain injuries, including cognitive impairment, blood-brain barrier (BBB) disruption and brain edema (23, 24). Recently, Eps15, the parent gene of circRNA EPS15, has also been suggested to regulate microglial cell activation and inflammation. It has been found that in intracerebral hemorrhage (ICH), microglial activation marker CD68 was co-located with EHD2, a member of the Eps15 homology domain (EH domain) family (25). Moreover, the Eps15 homology domain-containing protein 1 (EHD1) could mediate the formation of functional IL-6 receptor complexes through D4-positive raft (26). In addition, knockdown of Eps15 significantly decreased endocytosed particulate matter (PM) and reduced the PM-induced production of IL-6 (27). Moreover, cAMP pathway is another significant signaling cascade in TBI. cAMP signaling cascade is one of the most essential and best-understood cellular pathways which mediate the action of a wide variety of hormones, neurotransmitters, and growth factors (28). Besides, the presence of cAMP is essential for the control and regulation of gene expression, growth, cell differentiation as well as proliferation and apoptosis (29). The versatility of cAMP-related effects depends on the expression of many factors such as adenylyl cyclase isoform, phosphodiesterase and A kinase-anchoring proteins (AKAP) (30).
A great number of evidence have suggested that circRNAs can serve as ceRNAs for miRNAs, namely, as miRNA sponges. A circRNA may contain multiple miRNA binding sites and may have adsorptive and suppressive effects on miRNAs. For example, in acute lymphoblastic leukemia (ALL), circ_0000745 knockdown restrained cell cycle progression, and triggered cell apoptosis and ferroptosis. Circ_0000745 acted as a molecular sponge for miR-494-3p in ALL cells and miR-494-3p silencing partly diminished circ_0000745 knockdown-induced anti-tumor effects (31). In addition, in prostate cancer (PC), circPDHX deficiency attenuated PC cell proliferation, migration and fatty acid metabolites. Moreover, circPDHX could bind to miR-497-5p to act as ceRNA (32). In our study, the potential target genes of circRNAs were examined to investigate the functions of these circRNAs. We firstly predicted miRNA candidates based on MiRanda and PITA, and listed the miRNA candidates for the top 5 differentially expressed circRNAs (EPS15, FGD4, TPCN1, PUM2 and CADPS2). Then, the target-binding mRNAs for miRNA candidates were predicted via miRanda and Targetscan. In our study, the ceRNA (circRNA-miRNA-mRNA) network was constructed, which presented a large interaction network for bioinformatic analysis. These results together lay the groundwork for future research into the specific ceRNA network, which is beneficial for investigating the role of circRNAs in regulating the expression of target genes. From the ceRNA network, wo found that fat mass and obesity-associated protein (FTO) was the key mRNA that could be regulated by a lot of miRNAs and circRNAs. FTO is the first discovered demethylase that belongs to the alpha-ketoglutarate-dependent dioxygenase B (ALKB) family. It is an oxygenase dependent on Fe2+ and 2-oxoglutarate, which can catalyze the demethylation of nucleotides (33). FTO-mediated demethylation process is gradually completed. M6A is first converted to hm6A, then transformed to fm6A, and finally reduced to original adenylate (34, 35). FTO is enriched in brain, especially in neurons, and plays an important regulatory role in central nervous system (CNS), including brain development and function (36). Dysregulation of FTO may participate in CNS injuries such as TBI. Yu et al. found that the expression of FTO was significantly down-regulated in rat cerebral cortex after TBI. Moreover, functional FTO was necessary to repair TBI-induced neurological damage (37).
There were some limitations in our study. Firstly, we did not confirm the regulatory relationship of circRNA, miRNA and mRNA via gain and loss experiment in our study. Secondly, we did not examine the role of circRNA in animal TBI models, which we would further study in our future experiments. Thirdly, we did not validate whether the signaling pathways found in our study by KEGG was involved in animal TBI models. Fourthly, the six severe TBI patients were chosen only in Jinling Hospital of China, and nothing about other countries, so our study might not be representative.
In conclusion, our study indicated that the expression of circRNAs was significantly different in TBI. Moreover, microarray, proteomic and metabolomic analyses of the downstream moleculars of circRNAs may offer new avenues for restoring normal neuronal network and blocking the vital nodes promoting brain damage. We considered that circRNAs can to be attractive therapeutic targets for patients suffering from TBI. Continued discoveries in this field will bring novel insights on circRNAs involved in biological functions and disease progression. Ultimately, circRNAs may hold promise for clinical challenges.
Data availability statement
The data presented in the study are deposited in the NCBI GenBank repository, accession number BankIt2634779: OP794505-OP79462.
Ethics statement
The studies involving human participants were reviewed and approved by Jinling Hospital (approval number: 2021DZGZR-YBB-082). The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
ZL was responsible for the data collection and manuscript writing. YL was responsible for the clinical specimen collection and proofreading. LM was responsible for the literature collection and manuscript review. LZ was responsible for the design of the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82101461) from ZL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Reddi S, Thakker-Varia S, Alder J, Giarratana AO. Status of precision medicine approaches to traumatic brain injury. Neural Regen Res. (2022) 17:2166–71. doi: 10.4103/1673-5374.335824
2. Ambunda P, Lourens A. Severe traumatic brain injuries secondary to motor vehicle crashes in two Namibian regions: a retrospective review. Afr J Emerg Med. (2022) 12:225–30. doi: 10.1016/j.afjem.2022.04.009
3. Kalra S, Malik R, Singh G, Bhatia S, Al-Harrasi A, Mohan S, et al. Pathogenesis and management of traumatic brain injury (TBI): role of neuroinflammation and anti-inflammatory drugs. Inflammopharmacology. (2022) 30:1153–66. doi: 10.1007/s10787-022-01017-8
4. Sun P, Hamblin MH, Yin KJ. Non-coding RNAs in the regulation of blood-brain barrier functions in central nervous system disorders. Fluids Barriers CNS. (2022) 19:27. doi: 10.1186/s12987-022-00317-z
5. Zhang L, Li Z, Mao L, Wang H. Circular RNA in acute central nervous system injuries: a new target for therapeutic intervention. Front Mol Neurosci. (2022) 15:816182. doi: 10.3389/fnmol.2022.816182
6. Yang R, Xu B, Yang B, Fu J, Chen H, Wang X. Non-coding RNAs: the extensive and interactive regulators of the blood-brain barrier permeability. RNA Biol. (2021) 18:108–16. doi: 10.1080/15476286.2021.1950465
7. Chen M, Lai X, Wang X, Ying J, Zhang L, Zhou B, et al. Long Non-coding RNAs and circular RNAs: insights into microglia and astrocyte mediated neurological diseases. Front Mol Neurosci. (2021) 14:745066. doi: 10.3389/fnmol.2021.745066
8. Gu Q, Liu H, Ma J, Yuan J, Li X, Qiao L. A Narrative review of circular rnas in brain development and diseases of preterm infants. Front Pediatr. (2021) 9:706012. doi: 10.3389/fped.2021.706012
9. Zhao RT, Zhou J, Dong XL Bi CW, Jiang RC, Dong JF, et al. Circular ribonucleic acid expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice. J Neurotrauma. (2018) 35:2056–66. doi: 10.1089/neu.2017.5502
10. Xie BS, Wang YQ, Lin Y, Zhao CC, Mao Q, Feng JF, et al. Circular RNA expression profiles alter significantly after traumatic brain injury in rats. J Neurotrauma. (2018) 35:1659–66. doi: 10.1089/neu.2017.5468
11. Zheng P, Shu L, Ren D, Kuang Z, Zhang Y, Wan J. circHtra1/miR-3960/GRB10 axis promotes neuronal loss and immune deficiency in traumatic brain injury. Oxid Med Cell Longev. (2022) 2022:3522492. doi: 10.1155/2022/3522492
12. Zhang L, Tian M, Wang H, Mao L. Expression characteristics of long noncoding RNA and messenger RNA in human traumatic brain injury. Neuroreport. (2022) 33:90–100. doi: 10.1097/WNR.0000000000001756
13. Hawryluk GWJ, Aguilera S, Buki A, Bulger E, Citerio G, Cooper DJ, et al. A management algorithm for patients with intracranial pressure monitoring: the Seattle International Severe Traumatic Brain Injury Consensus Conference (SIBICC). Intensive Care Med. (2019) 45:1783–94. doi: 10.1007/s00134-019-05805-9
14. Wang S, Liao L, Wang M, Zhou H, Huang Y, Wang Z, et al. Pin1 promotes regulated necrosis induced by glutamate in rat retinal neurons via CAST/Calpain2 pathway. Front Cell Neurosci. (2017) 11:425. doi: 10.3389/fncel.2017.00425
15. Dong Z, Deng L, Peng Q, Pan J, Wang Y. CircRNA expression profiles and function prediction in peripheral blood mononuclear cells of patients with acute ischemic stroke. J Cell Physiol. (2020) 235:2609–18. doi: 10.1002/jcp.29165
16. Qin C, Liu CB, Yang DG, Gao F, Zhang X, Zhang C, et al. Circular RNA expression alteration and bioinformatics analysis in rats after traumatic spinal cord injury. Front Mol Neurosci. (2018) 11:497. doi: 10.3389/fnmol.2018.00497
17. He L, Zhang F, Zhu Y, Lu M. A crosstalk between circular RNA, microRNA, and messenger RNA in the development of various brain cognitive disorders. Front Mol Neurosci. (2022) 15:960657. doi: 10.3389/fnmol.2022.960657
18. Huang XJ, Su GJ, Wu CW, Sha XS, Zou JF, Liu XS, et al. Knockdown of rno_circRNA_009194 improves outcomes in traumatic brain injury rats through inhibiting voltage-gated sodium channel Nav1.3. J Neurotrauma. (2022) 39:196–210. doi: 10.1089/neu.2020.7520
19. Du M, Wu C, Yu R, Cheng Y, Tang Z, Wu B, et al. A novel circular RNA, circIgfbp2, links neural plasticity and anxiety through targeting mitochondrial dysfunction and oxidative stress-induced synapse dysfunction after traumatic brain injury. Mol Psychiatry. (2022) 27:4575–89. doi: 10.1038/s41380-022-01711-7
20. Jiang B, Tian M, Li G, Sadula A, Xiu D, Yuan C, et al. circEPS15 overexpression in hepatocellular carcinoma modulates tumor invasion and migration. Front Genet. (2022) 13:804848. doi: 10.3389/fgene.2022.804848
21. Bajwa E, Klegeris A. Neuroinflammation as a mechanism linking hypertension with the increased risk of Alzheimer's disease. Neural Regen Res. (2022) 17:2342–6. doi: 10.4103/1673-5374.336869
22. Long HZ, Zhou ZW, Cheng Y, Luo HY Li FJ, Xu SG, et al. The role of microglia in Alzheimer's disease from the perspective of immune inflammation and iron metabolism. Front Aging Neurosci. (2022) 14:888989. doi: 10.3389/fnagi.2022.888989
23. Shao F, Wang X, Wu H, Wu Q, Zhang J. Microglia and neuroinflammation: crucial pathological mechanisms in traumatic brain injury-induced neurodegeneration. Front Aging Neurosci. (2022) 14:825086. doi: 10.3389/fnagi.2022.825086
24. Doganyigit Z, Erbakan K, Akyuz E, Polat AK, Arulsamy A, Shaikh MF. The role of neuroinflammatory mediators in the pathogenesis of traumatic brain injury: a narrative review. ACS Chem Neurosci. (2022) 13:1835–48. doi: 10.1021/acschemneuro.2c00196
25. Ke K, Rui Y, Li L, Zheng H, Xu W, Tan X, et al. Upregulation of EHD2 after intracerebral hemorrhage in adult rats. J Mol Neurosci. (2014) 54:171–80. doi: 10.1007/s12031-014-0271-1
26. Woo JH, Park SJ, Park SM, Joe EH, Jou I. Interleukin-6 signaling requires EHD1-mediated alteration of membrane rafts. FEBS J. (2022) 289:5914–32. doi: 10.1111/febs.16458
27. Wu YF Li ZY, Dong LL Li WJ, Wu YP, Wang J, et al. Inactivation of MTOR promotes autophagy-mediated epithelial injury in particulate matter-induced airway inflammation. Autophagy. (2020) 16:435–50. doi: 10.1080/15548627.2019.1628536
28. Pan Z, Wu X, Zhang X, Hu K. Phosphodiesterase 4B activation exacerbates pulmonary hypertension induced by intermittent hypoxia by regulating mitochondrial injury and cAMP/PKA/p-CREB/PGC-1alpha signaling. Biomed Pharmacother. (2022) 158:114095. doi: 10.1016/j.biopha.2022.114095
29. Gong C, Xu D, Sun D, Kang J, Wang W, Xu JR, et al. FgSnt1 of the Set3 HDAC complex plays a key role in mediating the regulation of histone acetylation by the cAMP-PKA pathway in Fusarium graminearum. PLoS Genet. (2022) 18:e1010510. doi: 10.1371/journal.pgen.1010510
30. Kilanowska A, Ziolkowska A, Stasiak P, Gibas-Dorna M. cAMP-dependent signaling and ovarian cancer. Cells. (2022) 11:23. doi: 10.3390/cells11233835
31. Yang X, Li Y, Zhang Y, Liu J. Circ_0000745 promotes acute lymphoblastic leukemia progression through mediating miR-494-3p/NET1 axis. Hematology. (2022) 27:11–22. doi: 10.1080/16078454.2021.2008590
32. Chen J, Xie Q, Miao W, Fan J, Zhou X, Li M. CircPDHX promotes prostate cancer cell progression in vitro and tumor growth in vivo via miR-497-5p/ACSL1 axis. Biochem Biophys Res Commun. (2022) 620:35–41. doi: 10.1016/j.bbrc.2022.06.012
33. Li L, Xu N, Liu J, Chen Z, Liu X, Wang J. m6A methylation in cardiovascular diseases: from mechanisms to therapeutic potential. Front Genet. (2022) 13:908976. doi: 10.3389/fgene.2022.908976
34. Kumari R, Ranjan P, Suleiman ZG, Goswami SK Li J, Prasad R, et al. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res. (2022) 118:1680–92. doi: 10.1093/cvr/cvab160
35. Song N, Cui K, Zhang K, Yang J, Liu J, Miao Z, et al. The role of m6A RNA methylation in cancer: implication for nature products anti-cancer research. Front Pharmacol. (2022) 13:933332. doi: 10.3389/fphar.2022.933332
36. Chang R, Huang Z, Zhao S, Zou J, Li Y, Tan S. Emerging roles of FTO in neuropsychiatric disorders. Biomed Res Int. (2022) 2022:2677312. doi: 10.1155/2022/2677312
Keywords: human traumatic brain injury, circular RNA, biological processes, KEGG pathway, ceRNA
Citation: Li Z, Lin Y, Mao L and Zhang L (2023) Expression characteristics of circular RNA in human traumatic brain injury. Front. Neurol. 13:1086553. doi: 10.3389/fneur.2022.1086553
Received: 01 November 2022; Accepted: 28 December 2022;
Published: 11 January 2023.
Edited by:
Tao Huang, Shanghai Institute of Nutrition and Health (CAS), ChinaReviewed by:
Jinjie Zhong, Zhejiang University, ChinaZhu Lei, Shanghai Changzheng Hospital, China
Hongrong Zhang, First Affiliated Hospital of Chongqing Medical University, China
Junchi He, First Affiliated Hospital of Chongqing Medical University, China
Chen Rui, University of South China, China
Copyright © 2023 Li, Lin, Mao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Zhang,  emhhbmdsaTIwMDdqc0AxMjYuY29t
emhhbmdsaTIwMDdqc0AxMjYuY29t
 Zhenxing Li
Zhenxing Li Yixing Lin
Yixing Lin Lei Mao
Lei Mao Li Zhang
Li Zhang