
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol. , 24 December 2021
Sec. Pediatric Neurology
Volume 12 - 2021 | https://doi.org/10.3389/fneur.2021.804078
This article is part of the Research Topic Genetically Determined Epilepsies: Perspectives in the Era of Precision Medicine View all 19 articles
 Ping Yang1,2*
Ping Yang1,2* Robert Broadbent1,2
Robert Broadbent1,2 Chitra Prasad3
Chitra Prasad3 Simon Levin3
Simon Levin3 Sharan Goobie4
Sharan Goobie4 Joan H. Knoll1,2
Joan H. Knoll1,2 Asuri N. Prasad3
Asuri N. Prasad3Objectives: Mutations in the STXBP1 gene have been associated with epileptic encephalopathy. Previous studies from in vitro neuroblastoma 2A cells showed that haploinsufficiency of STXBP1 is the mechanism for epileptic encephalopathy. In this ex vivo study, STXPB1 DNA mutations and RNA expression were assessed from two patients to help understand the impact of STXBP1 mutations on the disease etiology and mechanism.
Methods: Microarray analysis and DNA sequencing were performed on two children with development delay, one with and one without infantile spasms. Different pathogenic mutations of STXBP1 were identified in the patients and RNA expression of STXPB1 was then performed by RT-Q-PCR on RNA extracted from blood samples of each patient.
Results: Pathogenic deletion [of exons 13–20 and 3′ downstream of STXBP1] and nonsense mutation [c.1663G>T (p.Glu555X) in exon 18 of STXBP1] were detected from the two patients, respectively. RNA analysis showed that 1) the deletion mediated RNA decay, and that 2) no RNA decay was identified for the nonsense mutation at codon 555 which predicts a truncated STXBP1 protein.
Significance: Our RNA expression analyses from the patient blood samples are the first ex vivo studies to support that both haploinsufficiency and truncation of STXBP1 protein (either dominant negative or haploinsufficiency) are causative mechanisms for epileptic encephalopathies, intellectual disability and developmental delay. The RNA assay also suggests that escape from nonsense-mediated RNA decay is possible when the nonsense mutation resides <50 nucleotides upstream of the last coding exon-exon junction even in the presence of additional non-coding exons that are 3′ downstream of the last coding exon.
Missense, nonsense, frame shift mutations and deletions of all or part of the STXBP1 gene have been reported in the literature in more than 280 patients affected with epileptic encephalopathy patients with early infantile onset-4 (OMIM # 612164). All patients with germline pathogenic variants and deletions of STXBP1 have global developmental delay, intellectual disability and cognitive dysfunction. The majority of affected patients present with seizures (1). Mutations or deletions of STXBP1 have been reported primarily in patients affected with Ohtahara syndrome (2, 3), West syndrome (4), and less frequently in patients affected with early myoclonic epileptic encephalopathy (5, 6), Dravet syndrome (7), Lennox-Gaustaut syndrome (8), Angelman/Pitt Hopkins-like syndrome phenotype (9, 10), and atypical Rett/Rett-like phenotypes (11). Ohtahara syndrome is characterized by neonatal onset severe seizures, tonic spasms, burst suppression pattern on EEG, intellectual disability and developmental delay (2). West syndrome is defined by infantile spasm onset between 3 and 12 months of age with atypical hypsarrhythmia (4). Recently, mutations or deletions of STXBP1 have been reported in 14 patients with intellectual disability without epileptic seizures (12, 13). Somatic mutation of STXBP1 was also identified in a patient with focal cortical dysplasia (14).
STXBP1 encodes syntaxin binding protein 1 which is highly expressed in brain and plays a role in neurotransmitter release as part of the synaptic fusion machinery. There are two isoforms of the STXBP1 gene. One isoform (isoform a) contains 20 exons (NM_003165) encoding 603 amino acids (P61764-2) and another isoform (isoform b) has 19 exons (NM_001032221) encoding 594 amino acids (P61764-1), respectively. The difference between the two isoforms at the amino acid sequence level is from positions 576 to 594 or to the C-terminal end (3). Mutations in STXBP1 can be detected by sequencing analysis for 83% of cases, by targeted deletion/duplication analysis for 5% of cases and by microarray analysis for 12% of cases (7). Haploinsufficiency of STXBP1 has been proposed as the mechanism for the epileptic encephalopathies based on expression experiments of mutant STXBP1 proteins in cultured neuroblastoma 2A cells (3). Recently, a dominant negative mechanism has been suggested from missense mutant studies (15).
In this study, we report genomic and RNA findings on two unrelated children with different STXBP1 alterations. The results are presented for each proband in the Genetic Anomalies and STXBP1 Expression sections. Our findings from patient blood samples are the first ex vivo assay to support that both haploinsufficiency and truncation of STXBP1 protein (either dominant negative or haploinsufficiency) are causative mechanisms for epileptic encephalopathies, intellectual disability and developmental delay.
Proband 1: A 2 year old male child was referred for microarray testing due to clinical findings of developmental delay, intellectual disability and hypotonia. An intragenic deletion of the STXBP1 gene was detected by microarray analysis. Parental blood samples were obtained for follow-up studies to determine if the deletion was inherited or de novo.
Proband 2: A 6 year old child, with a history of infantile spasms, was tested with a 51-gene infantile epilepsy panel (GeneDx, Gaithersburg, Maryland, USA). A nonsense mutation, c.163G>T (p.Glu555X), was identified in exon 18 of STXBP1.
DNA was extracted from peripheral blood cells from the probands, their parents, and five normal control males using the MagNA pure compact instrument and MagNA Pure LC DNA isolation kit 1 (Roche Diagnostics, Laval, Quebec, Canada). RNA was extracted from blood samples from the probands, and two normal male controls of similar age (1 and 6 years) using the RNeasy mini kit (QIAamp® RNA Blood Mini, Qiagen, Hilden, Germany).
Approximately 200 ng of genomic DNA was used for the ThermoFisher High Resolution CytoScan HD Array studies according to the manufacturer's protocol. Genomic copy number variants (CNVs) and absence of heterozygosity (AOH) were identified using ChAS software from Affymetrix/ThermoFisher Scientific (Waltham, MA, USA).
To confirm the small deletion and determine the inheritance of the deletion, Q-PCR studies were performed on the genomic DNA extracted from proband 1, parents and five normal controls. Two pairs of primers designed from within the hemizygously deleted region and undeleted region were used, respectively for the Q-PCR studies. Primer pair 1 is located in a non-deleted region in STXBP1 intron 1: 5′ GACATTTGCAAAACGGCATC 3′-F and 5′ TGTTGGTGATGAGAAAGGTCA 3′-R (Figure 1A). Primer pair 2 is located within the deleted region in intron 19: 5′TTGCTTGTAACGAGGAAGCT 3′-F and 5′ TGAAGAGTGAACCATTGCCA 3′-R (Figure 1A). A FOXP2 gene amplicon was used as a two-copy reference control for the relative Q-PCR calculation. It was amplified using primer pairs: 5′-TGCTAGAGGAGTGGGACAAGTA 3′-F and 5′ CAAAAGCCACAGCAATCCTT 3′-R (courtesy of TCAG, Hospital for Sick Children, Toronto, Canada). The relative Q-PCR amplification using 9 ng genomic DNA in a total 15 ul reaction volume was performed on the Roche LightCycler®480 real time PCR instrument using the LightCycler® 480 high resolution melting master protocol. Briefly, the DNA was denatured at 95°C for 10 min, amplified at 95°C for 10 s, 60°C for 15 s, 72°C for 15 s for 45 cycles. The PCR products were melted at 95°C for 10 s, 65°C for 1 min, continuous at 95°C. The products were cooled to 37°C for 10 min. The LightCycler®480 software v1.5 using the ΔΔ Ct method determined the ratio of the amplified target sequence (STXBP1) to the amplified reference sequence (FOXP2) and normalized with the five pooled normal control samples (16–18). Each run contained triplicates for each sample and the mean number was obtained from the triplicates. Copy numbers were interpreted as loss (1n), normal (2n), and gain (3n) while the ΔΔ Ct ratios were 0.4–0.7, 0.8–1.2, and 1.3–1.7, respectively.
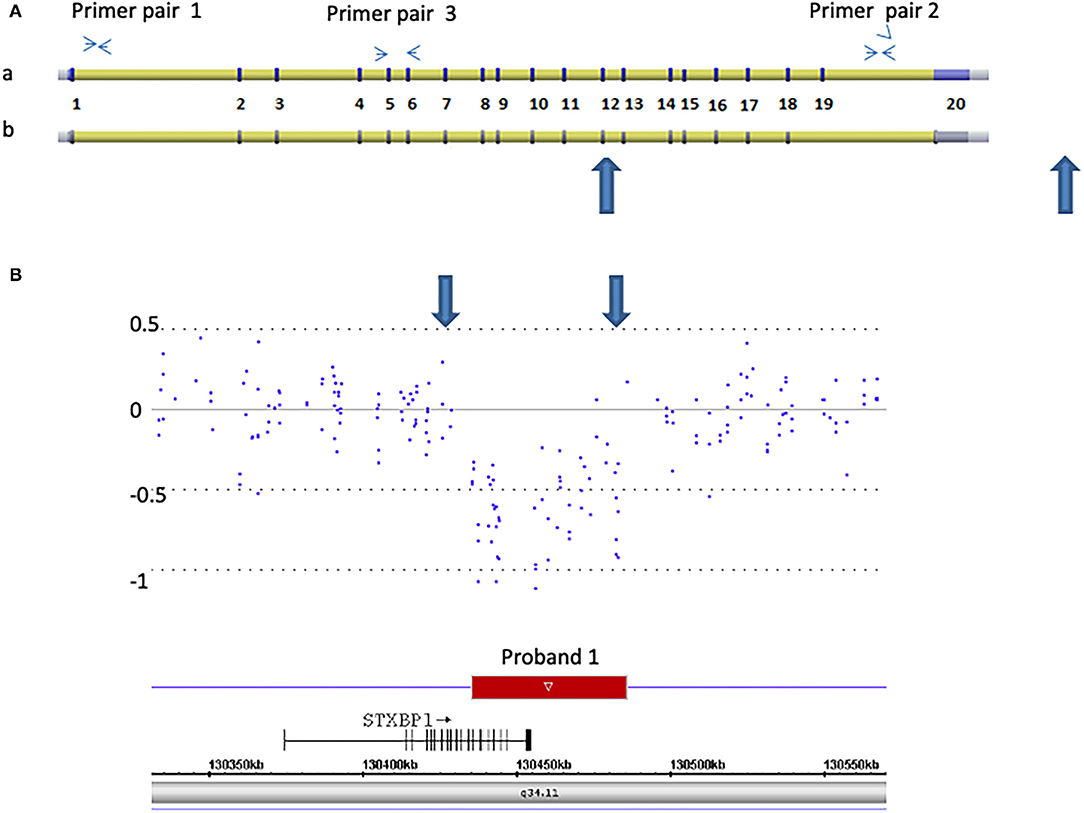
Figure 1. Detection of a 50 kb deletion of STXBP1, exons 13 through 20 and 3′ downstream genomic region in proband 1. (A) Schematic representation of two isoforms of STXBP1 (a: NM_003165. b: NM_001032221) (not scaled). The exons (numbered 1–20) and introns are shown. Primer pairs 1 and 2 are located at introns 1 and 19, respectively. They were used for Q-PCR analysis. Primer pair 3 is located at exons 5 and 6 and was used for RT-Q-PCR analysis. (B) Microarray scan of ~250 kb region encompassing the STXBP1 region. CytoScan HD array probes over the region are represented as dots. The numbers on the Y-axis represent Log2 values. The rectangle represents the region deleted. The STXBP1 gene relative to the deletion (small arrow indicates the orientation from 5′ to 3′ of the gene), genomic location [hg19], and chromosome 9 band is indicated. Large blue arrows demarcate the region deleted in both (A,B).
To determine if the deletion in proband 1 and the nonsense mutation in proband 2 mediate RNA decay, RT-Q-PCR studies were performed on RNA isolated from each proband and similar age- and gender- matched controls. Complementary DNA (cDNA) was obtained from reverse transcription (RT) of 2.5 ug total RNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen/ThermoFisher Scientific, Waltham, MA, USA). One pair of primers, outside of the STXBP1 genomic deletion region, was designed on exons 5 and 6 (primer 3): 5′ TCTCATCAGTGACTTTAAGGACC 3′-F, 5′ AGTTTTGATGACTTTGGCTGCT 3′-R (Figure 1A) and was used for the RT-Q-PCR analysis.
PCR amplification was performed with the Applied Biosystems GeneAmpR PCR System 9700 (Applied Biosystems/ThermoFisher Scientific, Waltham, MA, USA) for 38 Cycles at 95°C denaturation for 10 s, annealing at 59°C for 15 s and extension at 72°C for 15 s. The PCR amplification was completed during the linear phase at cycle 38. The PCR products were separated on the TapeStation 2200 instrument with a High Sensitivity D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany) as per the manufacturer's specifications for analysis of the RT-Q-PCR products. Data were obtained and images were captured using TapeStation Analysis Software (A.02.01) (Agilent Technologies, Waldbronn, Germany). The tests were repeated more than three times.
Consent forms were obtained from each family for this study. Research Ethics Boards (REB) approval was not required by our Institution based on the nature of the study (https://www.uwo.ca/research/_docs/ethics/hsreb_guidelines/Case_report_vs_research.pdf).
Proband 1: This male child presented with feeding difficulty and generalized hypotonia noted in the first few weeks after birth, delayed motor milestones (sitting at 12 months, walking with assistance at 24 months) as well as delays in both fine motor and language domains. At 2 years of age, he displayed behavioral stereotypies (hand flapping) and autistic traits (preoccupation with spinning objects). He has no history of seizures. His growth parameters were: height of 85 cm (10–25th%ile), weight of 11.85 kg (10%ile) and occipital frontal head circumference of 47 cm (3%ile) with a brachycephalic skull shape. Mild bilateral 5th finger clinodactyly was noted on physical examination, but no other significant dysmorphic features. Imaging studies of the brain showed increased periventricular T2 signal suggestive of delayed myelination. At age 3 years he was investigated for staring spells with an electroencephalogram (EEG) that showed mild background slowing, without any evidence of abnormal epileptiform activity. There were no sleep related changes documented on the available record. No other prior EEG recordings were available for review.
Proband 2: This 6 year old male presented to the neurogenetic clinic with a history of epileptic spasms with onset at 2 months of age, global developmental delay, poor visual attention (cortical visual impairment) and bilateral clubfeet with no major facial dysmorphism. His EEG findings at age 3 months confirmed the presence of discontinuous suppression burst pattern with multiple independent spike foci, features consistent with an epileptic encephalopathy. Subsequent follow up recordings done at regular intervals showed variable features with slow background rhythms, regional (occipital) expression of spike activity, with further enhancement and activation of spiking in sleep. By age 12 years the EEG recordings had begun to show features of generalized slow spike wave activity (2–2.5 Hz), paroxysmal fast (recruiting rhythms) in keeping with evolution of his epilepsy. Imaging studies showed non-specific features of generalized loss of gray matter volume. Extensive investigative work-up including biochemical studies for inborn errors of metabolism were negative. The patient was initially treated with Vigabatrin titrated to a maximum of 120 mg/kg/day, and further medication changes were dictated by appearance of refractoriness to therapy. Ophthalmological monitoring for retinal toxicity was continued. Valproic acid, and Levetiracetam were added sequentially and withdrawn due to a lack of benefit in terms of seizure control. Eventually, by age 4 years he was weaned off Vigabatrin. He did show a period of seizure remission for about 18 months around age 5 years, after which tonic spasms made a reappearance at age 7 years. At age 12 years, he continues to report nocturnal tonic seizures sometimes in clusters, and is currently maintained on Rufinamide 31 mg/kg/day in three divided doses daily and Clobazam 0.25 mg/kg at bedtime.
Proband 1: A 50 kb deletion including 56 oligonucleotide probes in chromosome region 9q34.11 was detected using CytoScan HD Array and ChAS analysis (Figure 1B). The genome coordinates of the deletion are 9q34.11(130 435 492–130 485 618)x1 [hg19]. This deletion resulted in loss of exons 13 through 20 and 3' downstream of the STXBP1 gene (NM_003165, NM_001032221, OMIM# 602926). The Q-PCR studies confirmed the deletion in the proband and revealed the deletion was de novo (Figures 2A,B and Tables 1, 2). No identical deletion has been previously reported in literature or databases.
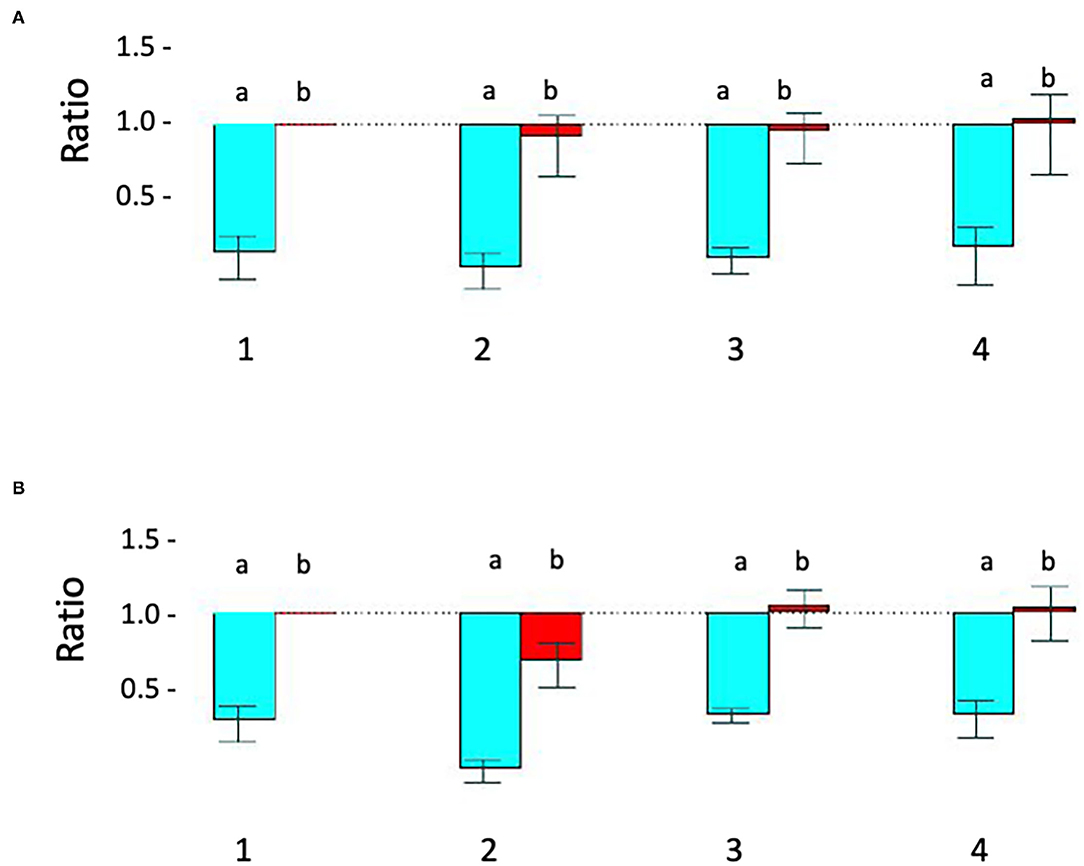
Figure 2. Relative quantitative –PCR [Q-PCR] confirms STXBP1 deletion in proband 1 and demonstrates that it is de novo. (A) Q-PCR product using primer pair 1 (intron 1) did not detect the deletion in the proband 1 (#2), his parents (#3-mother, #4-father) and a pooled normal male sample control (#1). (B) With primer 2 (intron 19), Q-PCR product confirmed the STXBP1 deletion in proband 1 (#2b) detected by microarray. No deletion was detected in the proband's mother (#3) and father (#4) or the control (#1). a = target/reference ratio (i.e., STXBP1/FOXP2); b = normalized ratio (i.e., = proband's a/normal controls a, mother's a/controls a and father's a/controls a). Error bars indicate average standard error of the mean obtained from normalization of triplicates for each sample. The test was reproducible and was repeated more than three times.
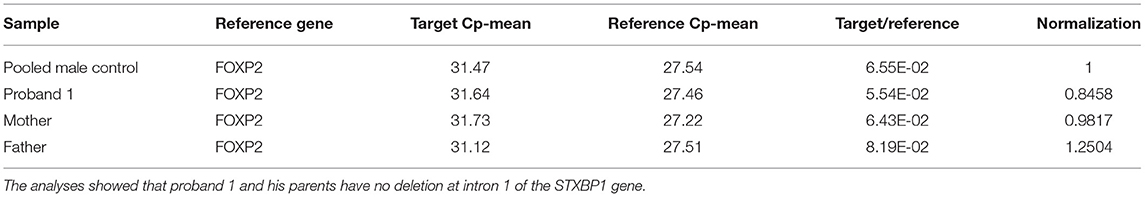
Table 1. Q-PCR analysis results for proband 1, his parents and the five pooled male controls using primer pair 1 at intron 1 of STXBP1.
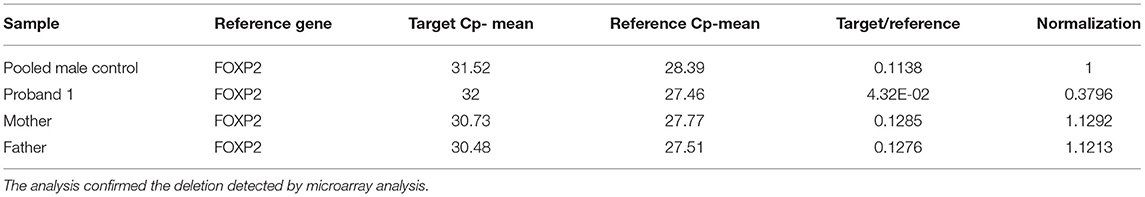
Table 2. Q-PCR analysis results of proband 1, his parents and the five pooled male controls by using primer pair 2 at intron 19 of STXBP1.
Proband 2: Extracted DNA was sent to GeneDx, Inc (Gaithersburg, MD, USA) for testing with their infantile epilepsy 51-gene panel in 2013. A c.1663G>T (p.Glu555X) in exon 18 of STXBP1 was detected. This variant is predicted to result in a premature stop codon at position 555 (nonsense mutation) in domain 2 (13). This variant has not been previously reported in the literature. Parental DNA sequencing studies revealed that this nonsense mutation was de novo.
Proband 1: RT-Q-PCR products from exons 5–6, outside of the deleted genomic region, showed that the expression level of the STXBP1 was reduced to ~half of the expression level of the similar age normal control 2. This indicated that the deletion of exons 13–20 mediated one allele of RNA decay (Figure 3 and Table 3).
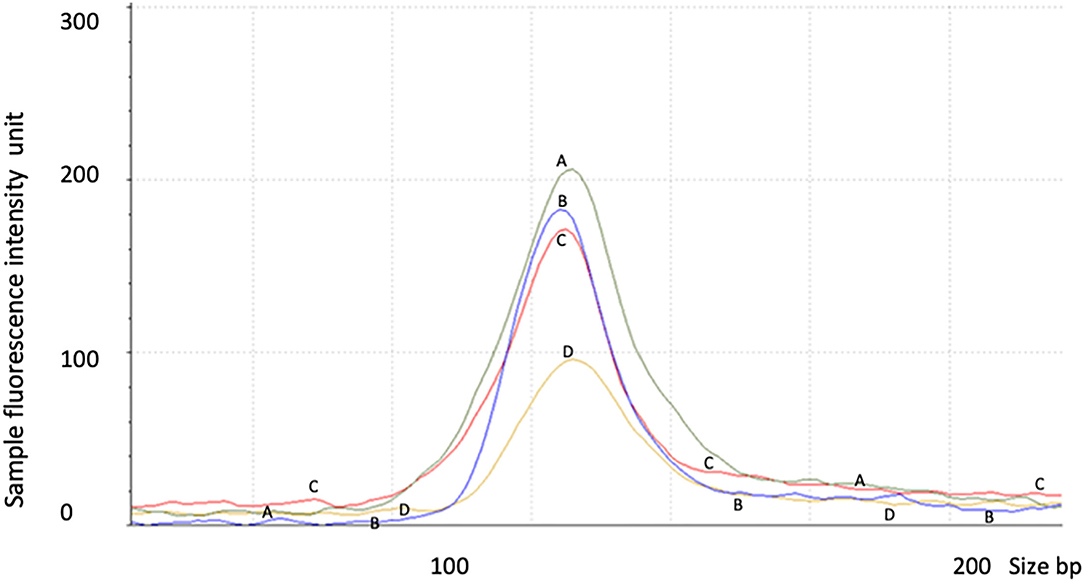
Figure 3. RNA decay observed in proband 1 with STXBP1 deletion not observed in proband 2 with nonsense mutation. Control 1 (A) = 6 year old male, control 2 (C) = 1 year old male, proband 1 (D) = 2 year old male with partial deletion of STXBP1, and proband 2 (B) = a 6 year old male with c.1663G>T nonsense mutation of STXBP1. The data were averaged from triplicates for each sample. The test was repeated for at least three times.
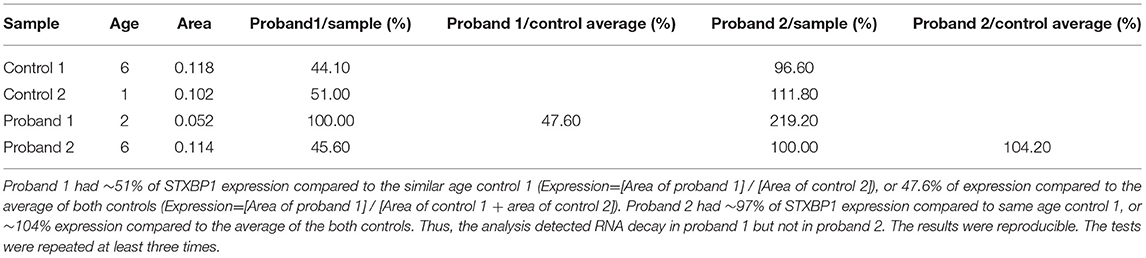
Table 3. RT-Q-PCR analyses of RNA expression from probands 1 and 2 relative to age-matched controls using primer pair 3 located at exons 5 and 6 of STXBP1.
Proband 2: The expression level of STXBP1 was not reduced (Figure 3 and Table 3). It is predicted to result in a truncated STXBP1 protein product with stop codon at amino acid position 555 in domain 2.
STXBP1 protein plays an important role in regulating the release of neurotransmitters during synaptic docking and fusion of the vesicle at the synaptic membrane. Mutations of STXBP1 have been associated with autosomal dominant patterns associated with diverse neurodevelopmental phenotypes that include autism, intellectual disability, developmental delay and early infantile epileptic encephalopathy (7, 14). Approximately 136 point mutations (including splice, nonsense, missense), small indels (<100 kb, including insertions, deletions and duplications) and 26 cases with large copy number variants (CNVs) affecting STXBP1 have been reported in the literature (14). Estimated incidence rate is 3.3–3.8 per 100,000 births (14). The mutations were distributed throughout all domains (domains 1 through 3) of the STXBP1 protein, and either reduced the amount of functional protein (haploinsufficiency) or caused an abnormal structural protein. An abnormal amount or structure of STXBP1 protein impairs the release of neurotransmitters that lead to uncontrolled excitation of neurons and seizures. However, the cause of developmental delay, intellectual disability and other phenotypes by altered STXBP1 protein is unknown. No established genotype and phenotype correlation has been established (15).
Our RT-Q-PCR assay demonstrated that the nonsense mutation detected at amino acid position 555 of STXBP1 in proband 2 did not lead to RNA decay. This mutation is located in exon 18, the second to last exon and ~40 nucleotides upstream of the last exon (exon 20) of STXBP1 in isoform b and is consistent with the mechanism of escape from nonsense-mediated RNA decay where it is <50 nucleotides upstream of the last exon-exon junction (19). For isoform a, the mutation is located at the third last exon and approximately 37 nucleotides upstream of the second last exon (exon 19). However, the exon 20 in isoform a is not a coding exon, thus exon 19 is the last coding exon. It indicates that escape from nonsense-mediated RNA decay is possible where the nonsense mutation resides <50 nucleotides upstream of the last coding exon-exon junction even in the presence of additional non-coding exons at the 3′ downstream end of the coding exon. The escape of nonsense-mediated RNA decay predicts that truncated STXBP1 proteins could be produced. The predicted truncated proteins are short of 39 and 48 amino acids at the C-terminal end in domain 2 for isoforms b and a, respectively. This indicates that domain 2 is critical for the normal function of STXBP1. It has been proposed that missense mutations result in STXBP1 protein destabilization and are prone to misfolding, aggregation and degradation (20). Based on missense mutations that can cause symptoms as severe as frameshift and heterozygous deletion mutations, it was proposed that missense mutants likely act to deplete the normal SYXBP1 protein in a dominant negative way (14). Similarly, the truncated STXBP1 protein predicted in proband 2 may work in a dominant negative way that could lead to normal protein destabilization or inappropriate binding with syntaxin-1 protein or other SNARE (soluble N-ehtylmaleimide-sensitive factor attachment receptor) proteins required in synaptic vesicle docking, priming and fusion (20). Alternatively, the truncated STXBP1 protein in proband 2 may lose its function completely and result in haploinsufficiency as discussed in a patient with truncated STXBP1 with atypical Rett/Rett-like phenotypes (11). More studies of structural and functional assays on the truncated STXBP1 protein will be needed to clarify whether dominant negative or haploinsufficiency is the causative mechanism for epileptic encephalopathies.
In proband 1, who had a deletion of exons 13 through 20, our RT-Q-PCR assay suggested mediated RNA decay, expected to result in haploinsufficiency of STXBP1 protein. This finding demonstrates the haploinsufficiency mechanism for STXBP1 related disorders directly from RNA extracted from patient blood samples.
Proband 1 and proband 2 have common clinical features of developmental delay and intellectual disability. However, proband 2 presented with infantile spasms which were not noted in proband 1. Whether the difference is due to the different nature of mutations in STXBP1 (deletion vs. nonsense mutation) remains unclear. Several studies revealed no correlation between the type of mutations or position within the gene with respect to epilepsy, cognitive abilities and other phenotypes (7, 20). Thus, the differences may have contribution from each individual's genetic background. Epistasis is a possible explanation for this, which the phenotypes depend on the presence of one or more modifier genes (21).
We believe our RT-Q-PCR assay represents the first ex vivo assay to demonstrate that both truncation and deletion of STXBP1, resulting in truncated STXBP1 protein and reduced protein product, cause STXBP1 related disorders. Domain 2 of STXBP1 protein is critical for the normal function. The findings support haploinsufficiency and truncation/structure anomaly (either acts as dominant negative or haploinsufficiency) as mechanisms for the disorders. Our RNA assay also indicates that escape from nonsense-mediated RNA decay is possible where the nonsense mutation resides <50 nucleotides upstream of the last coding exon-exon junction even in the presence of additional non-coding exons at the 3' downstream of the coding exon. STXBP1 RNA and protein is highly expressed in brain, pancreas, soft tissue and lowly expressed in white blood cells (https://www.genecards.org/cgi-bin/carddisp.pl?gene=STXBP1). Our RT-Q-PCR assay supported that STXPB1 RNA is also expressed in peripheral blood, and this provides a much more accessible way for ex vivo study.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/clinvar/variation/932281/.
Ethical review and approval was not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
PY contributed to conception, design of the study, and wrote the first draft of the manuscript. ANP wrote clinical sections of the manuscript. ANP, CP, and SL contributed clinical and treatment data and edited the manuscript. SG contributed clinical data. RB conducted the experiments and analyzed the data. JHK discussed the study and edited the manuscript. All authors contributed to manuscript, read, and approved the submitted version.
Both patients were funded for the clinical testing through Ontario Ministry of Health and Long-Term Care, Canada.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the two families for participating in this study. We also thank the staff at the Clinical Cytogenetics Laboratory, Department of Pathology and Laboratory Medicine, London Health Science Center, London, Ontario, Canada.
1. Saitsu H, Kato M, Mizuguchi T, Hamada K, Osaka H, Tohyama, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nature Genet. (2008) 40:782–8. doi: 10.1038/ng.150
2. Saitsu H, Kato M, Shimono M, Senju A, Tanabe S, Kimura T, et al. Association of genomic deletions in the STXBP1 gene with Ohtahara syndrome. Clin Genet. (2012) 81:399–402. doi: 10.1111/j.1399-0004.2011.01733.x
3. Saitsu H, Kato M, Matsumoto N. Haploinsufficiency of STXBP1 and Ohtahara syndrome. In: Jasper's Basic Mechanisms of the Epilepsies. Bethesda, MD: National Center for Biotechnology Information (US). (2012) p. 1–12.
4. Otsuka M, Oguni H, Liang JS, Ikeda H, Imai K, Hirasawa K, et al. STXBP1 mutations cause not only Ohtahara syndrome but also West syndrome–result of Japanese cohort study. Epilepsia. (2010) 51:2449–52. doi: 10.1111/j.1528-1167.2010.02767.x
5. Deprez L, Weckhuysen S, Holmgren P, Suls A, Van Dyck T, Goossens D, et al. Clinical spectrum of early-onset epileptic encephalopathies associated with STXBP1 mutations. Neurology. (2010) 75:1159–65. doi: 10.1212/WNL.0b013e3181f4d7bf
6. Milh M, Villeneuve N, Chouchane M, et al. Epileptic and nonepileptic features in patients with early onset epileptic encephalopathy and STXBP1 mutations. Epilepsia. (2011) 52:1828–34. doi: 10.1111/j.1528-1167.2011.03181.x
7. Carvill GL, Weckhuysen S, McMahon JM, Hartmann C, Moller RS, Hjalgrim H, et al. GABRA1 and STXBP1: novel genetic causes of Dravet syndrome. Neurology. (2014) 82:1245–53. doi: 10.1212/WNL.0000000000000291
8. Khaikin Y, Mercimek-Mahmutoglu S. STXBP1 Encephalopathy with Epilepsy. In: GeneReviews. Seattle, WA: University of Washington, Seattle. (2016) p. 1993–2021.
9. Olson HE, Tambunan D, LaCoursiere C, Goldenberg M, Pinsky R, Martin E, et al. Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A. (2015) 167A:2017–25. doi: 10.1002/ajmg.a.37132
10. Lopes F, Barbosa M, Ameur A, Soares G, de Sá J, Dias A, et al. Identification of novel genetic causes of Rett syndrome-like phenotypes. J Med Genet. (2016) 53:190–9. doi: 10.1136/jmedgenet-2015-103568
11. Cogliati F, Giorgini V, Masciadri M, Bonati MT, Marchi M, Cracco I, et al. Pathogenic variants in STXBP1 and in genes for GABAa receptor subunities cause atypical Rett/Rett-like phenotypes. Int J Mol Sci. (2019) 20:3621. doi: 10.3390/ijms20153621
12. Mastrangelo M, Peron A, Spaccini L, Novara F, Scelsa B, Introvini P et al. Neonatal suppression-burst without epileptic seizures: expanding the electroclinical phenotype of STXBP1-related, early-onset encephalopathy. Epileptic Disord. (2013) 15:55–61. doi: 10.1684/epd.2013.0558
13. Hamdan FF, Gauthier J, Dobrzeniecka S, Lortie A, Mottron L, Vanasse M et al. Intellectual disability without epilepsy associated with STXBP1 disruption. Eur J Hum Genet. (2011) 19:607–9. doi: 10.1038/ejhg.2010.183
14. Uddin M, Woodbury-Smith M, Chan A, Brunga L, Lamoureux S, Pellecchia G, et al. Germ line and somatic mutations in STXBP1 with diverse neurodevelopmental phenotypes. Neurol Genet. (2017) 3:e199; doi: 10.1212/NXG.0000000000000199
15. Abramov D, Guiberson NGL, Burré J. STXBP1 encephalopathies: Clinical spectrum, disease mechanisms, therapeutic strategies. J Neurochem. (2020) 157:165–78. doi: 10.1111/jnc.15120
16. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. (2008) 3:1101–8. doi: 10.1038/nprot.2008.73
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
18. Haimes J, Kelley M. Demonstration of a ΔΔCq Calculation Method to Compute Relative Gene Expression from qPCR Data. Tech Note. Lafayette, CO: Dharmacon, A Horizon Discovery Group Company (dharmacon.horizondiscovery.com) (2013).
19. Kurosaki T, Maquat YE. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci. (2016) 129:461–7. doi: 10.1242/jcs.181008
Keywords: STXBP1, haploinsufficiency, dominant negative, nonsense mutation, RNA expression
Citation: Yang P, Broadbent R, Prasad C, Levin S, Goobie S, Knoll JH and Prasad AN (2021) De novo STXBP1 Mutations in Two Patients With Developmental Delay With or Without Epileptic Seizures. Front. Neurol. 12:804078. doi: 10.3389/fneur.2021.804078
Received: 28 October 2021; Accepted: 29 November 2021;
Published: 24 December 2021.
Edited by:
Mario Mastrangelo, Umberto 1 Polyclinic, ItalyReviewed by:
Bruria Ben-Zeev, Sheba Medical Center, IsraelCopyright © 2021 Yang, Broadbent, Prasad, Levin, Goobie, Knoll and Prasad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Yang, cGluZ3k3OEBob3RtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.