
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol., 28 January 2022
Sec. Neurological Biomarkers
Volume 12 - 2021 | https://doi.org/10.3389/fneur.2021.799222
Background: Alterations in the gut microbiota after ischemic stroke have been demonstrated, whereas the effect on stroke outcome remains to be established.
Methods: A total of 132 consecutive patients with acute ischemic stroke were prospectively enrolled. Their gut microbiomes within 24 h of admission were profiled using 16S ribosomal RNA (rRNA) gene (V3–V4 region) sequencing. Microbiota comparisons were made between groups with good outcome (n = 105) and poor outcome (n = 27) based on 3-month modified Rankin Scale scores of 0–2 and 3–6. Propensity score-matching (PSM) analysis was conducted to assess the robustness of our findings. The functional potential was predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt).
Results: Patients in the poor outcome group were characterized by a significant reduction in the alpha diversity (Shannon index, p = 0.025; Simpson index, p = 0.010), an increase in the pathogenic bacteria (e.g., Enterococcaceae and Enterococcus), and a decrease in the short-chain fatty acids (SCFAs)-producing bacteria (e.g., Bacteroidaceae, Ruminococcaceae, and Faecalibacterium) to those with good outcome group (all p < 0.05). Similar results of microbial composition were obtained after PSM. The PICRUSt revealed that the pathway for membrane transport was relatively dominant in patients with poor outcome (p < 0.05).
Conclusion: This study demonstrated that stroke patients with 3-month poor outcome had baseline gut microbiota dysbiosis featured by increased pathogenic bacteria and decreased SCFAs-producing bacteria.
Stroke is one of the leading causes of disability and death, especially in the population aged 50 years and older (1). For acute ischemic stroke, about one-third of patients die or become disabled within the first 3 months (2). Identifying prognostic factors in the acute phase of stroke is important and might provide a novel therapeutic target to improve the stroke outcome.
Evidence is emerging that the gut microbiota is intimately involved in the pathology of a wide range of neurological disorders, including acute ischemic stroke (3). This is supported by the observations that gut microbiota correlates closely with stroke risk factors, such as age, obesity, hypertension, diabetes, dyslipidemia, and atrial fibrillation (4–8). Moreover, recent studies described a consistent pattern of poststroke gut microbiota dysbiosis, which is characterized by an increase in opportunistic pathogens (e.g., Enterobacteriaceae) and a decrease in commensal bacteria (e.g., Fecalibacterium), yielding a pro-inflammatory effect (9–14). Besides, gut microbiota-derived metabolites such as trimethylamine-N-oxide (TMAO) and short-chain fatty acids (SCFAs) might contribute to stroke pathology by modulating inflammation and atherosclerosis (11, 15–18). The SCFAs could regulate the expression of brain-derived neurotrophic factor (BDNF) (16); they could also modulate the effects of brain-invading lymphocytes, leading to an altered level of proinflammatory interleukin-17 (IL-17) and neuroprotective IL-10 (15). Interestingly, previous studies showed that the alterations of gut microbiota after stroke are correlated with the prognosis of stroke (10–13). The enrichment of Parabacteroides, Oscillospira, Enterobacteriaceae, etc., and depletion of Prevotella, Roseburia, Fecalibacterium, etc., were positively correlated with early unfavorable outcomes at discharge in patients with stroke (12). Whereas, the specific groups of bacteria and potential mediators associated with 3-month functional outcome in patients with acute ischemic stroke have not been fully revealed.
Here, we conducted this 16S ribosomal RNA (rRNA) gene sequencing-based study to characterize the gut microbiota in the acute phase of ischemic stroke and to compare the bacteria profiles of patients with different functional outcomes [3-month modified Rankin Scale (mRS) 0–2 vs. 3–6]. We aimed to: (1) reveal the taxonomic dysbiosis of gut microbiota related to unfavorable stroke outcome; (2) infer possible differences of functional composition associated with stroke outcome; (3) explore whether inflammatory cytokines (IL-17 and IL-10), BDNF, and TMAO are associated with gut microbiota and stroke outcome. An additional propensity score-matching (PSM) analysis was performed to correct for sample selection bias.
The participants were consecutively recruited from May 2018 to June 2019 in the Department of Neurology, Nanjing First Hospital (a teaching hospital affiliated with Nanjing Medical University). The inclusion criteria have been described in detail in our previous work (19). In brief, we enrolled patients aged ≥ 50 years with acute ischemic stroke due to anterior cerebral infarction. The exclusion criteria were: radiological evidence of intracerebral hemorrhage; chronic inflammatory or immune diseases (e.g., rheumatoid arthritis, lupus erythematosus, inflammatory bowel disease, or chronic hepatitis); severe comorbidities or an unstable medical condition (e.g., congestive heart failure, respiratory failure, renal failure, or severe liver dysfunction); and consumption of probiotics, antibiotics, or immunosuppressants within 1 month before admission. This study was approved by the Ethics Committee of Nanjing First Hospital with the approval number 20180713-05 and all the participants (or their legal guardians) signed a written informed consent.
Demographic information and medical histories were collected shortly after admission through a face-to-face interview. The baseline National Institutes of Health Stroke Scale (NIHSS) and the Diffusion-Weighted Imaging-Alberta Stroke Program Early CT score (DWI-ASPECTs) were assessed by two neurologists (HS and ZL).
Fecal and fasting blood samples were taken within 24 h after admission. The fecal samples were collected using sterile fecal containers with a spoon and each sample weighed ~2 g. After collection, fresh fecal and serum samples were immediately (within 1 h) stored at −80°C until analysis. The fasting blood samples were collected after overnight fasting and measured at the hospital central laboratory for total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), and fasting blood glucose (FBG).
Functional outcomes were assessed by two neurologists (HS and ZL) through telephone interviews using the mRS score at 3 months after stroke. Three patients were lost to follow-up and were excluded. According to the 3-month mRS scores, participants were divided into the two groups: the good functional outcome group (mRS 0–2) and the poor functional outcome group (mRS 3–6).
Fecal genomic DNA was extracted from the fecal samples with the QIAamp® DNA Stool Mini Kit (Qiagen, Hilden, Germany), according to the protocol of the manufacturer. The concentration and purity were detected through the Nanodrop, and the integrity was detected through regular 0.8% agarose gel electrophoresis.
The V3–V4 variable regions of the bacterial 16S rRNA gene were amplified by PCR using the forward (5′-CCTACGGGNGGCWGCAG-3′) and reverse (5′-GACTACHVGGGTATCTAATCC-3′) primers. The primary PCR products were checked by agarose gel electrophoresis and pooled to be used as the template for index PCR. The indexed PCR was performed by using index primers for adding the Illumina index to the library. After indexed PCR, the amplification products were checked and purified to be indexed in the 16S V3–V4 library whose quality was assessed on the Qubit@2.0 Fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and Agilent Bioanalyzer 2100 systems. Finally, the pooled library was sequenced on an Illumina MiSeq 250 Sequencer for generating 2 × 250 bp paired-end reads.
The concentration of serum TMAO level was determined using an enzyme-linked immunosorbent assay kit (MyBioSource Incorporation, San Diego, California, USA). The serum IL-17, IL-10, and BDNF levels were determined using enzyme-linked immunosorbent assay kits (R&D Systems Incorporation, Minneapolis, Minnesota, USA).
The raw reads were quality filtered and merged, and the remaining unique reads were chimera checked compared with the gold.fa database (http://drive5.com/uchime/gold.fa). These reads were clustered into operational taxonomic units (OTUs) by UPARSE with a cutoff of 97% similarity. All OTUs were classified based on Ribosomal Database Project by Mothur. Alpha diversity was analyzed by Mothur to describe the abundance and diversity of gut microbiota, including Observed species, Chao1, Abundance-based Coverage Estimator (ACE), Shannon, Simpson, and Coverage index. Beta diversity was utilized to further analyze the between-individual diversity of gut microbiota, using Principal Coordinates Analysis (PCoA) performed by R Project (Vegan package, version 3.3.1). Variation between groups was assessed by permutational multivariate ANOVA (PERMANOVA). Linear discriminant analysis (LDA) coupled with effect size (LEfSe) was conducted to determine the significantly differential taxa between the good outcome group and the poor outcome group. The absolute value of logarithmic LDA score > 2 and p < 0.05 were considered statistically significant. The Spearman correlation analysis was used to detect the associations between biochemical parameters and the relative abundance of microbiota. Functional metabolic profiles of bacterial communities were predicted using the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt).
Statistical analyses were performed with SPSS 22.0 for Windows (SPSS Inc., Chicago, Illinois, USA) and R version 3.3.1 (R Development Core Team, Vienna, Austria). Categorical variables were compared by the chi-squared test and continuous variables were compared by the Student's t-test or the Mann–Whitney U-test. The PSM analysis was performed with age, hypertension, and the baseline NIHSS as covariates. The corresponding propensity score of the grouping variable (mRS 0–2 or mRS 3–6) was calculated for each patient with a 1:1 nearest-neighbor matching algorithm (caliper width 0.2). Matched pairs of samples were obtained for analysis to adjust for sample selection bias. STROBE guidelines were the basis for reporting our results. The resulting p-values were adjusted using the Benjamini–Hochberg false discovery rate correction. A two-sided p < 0.05 was considered as statistically significant.
A total of 132 subjects (89 males and 43 females) aged 50–91 years (mean 64.5 years) were recruited for this study (Supplementary Figure S1). Patients in the mRS 3–6 (n = 27) group were older than those in the mRS 0–2 (n = 105) group (75.2 ± 10.9 vs. 65.9 ± 9.2, p = 0.001). Besides, patients in the poor outcome group had a higher incidence of hypertension and atrial fibrillation, higher baseline NIHSS scores, and lower DWI-ASPECT scores (p < 0.05). No group differences were detected in gender, diabetes mellitus, dyslipidemia, and coronary heart disease. Also, no group differences were observed in medication histories including the use of anticoagulants, antiplatelet, statins, antihypertensive drugs, and antidiabetic agents. Small vessel occlusion was more frequent in the mRS 0–2 group, while the mRS 3–6 group was more likely to have other etiologies (including cardioembolism, the stroke of other determined cause, and stroke of undetermined cause). After PSM, there were no significant differences in demographic and clinical data between the two groups (Table 1).
We obtained a total of 22,167,731 high-quality reads, which were clustered into 2,596 OTUs at 97% identity. The alpha-diversity analyses, including species richness (represented by observed species, Chao1, ACE) and evenness (represented by Shannon index and Simpson index) of the microbial community, showed that the mRS 3–6 group had less diversity than the mRS 0–2 group. There were statistically significant differences in the Shannon and Simpson indices between the two groups (mRS 0–2 vs. mRS 3–6: median Shannon index, 3.56 vs. 3.21, Wilcoxon p = 0.025; median Simpson index, 0.06 vs. 0.10, Wilcoxon p = 0.010). The observed species and Chao1 were lower in the mRS 3–6 group, but the differences between groups did not reach statistical significance (observed species, p = 0.160; Chao1, p = 0.352) (Figure 1A).
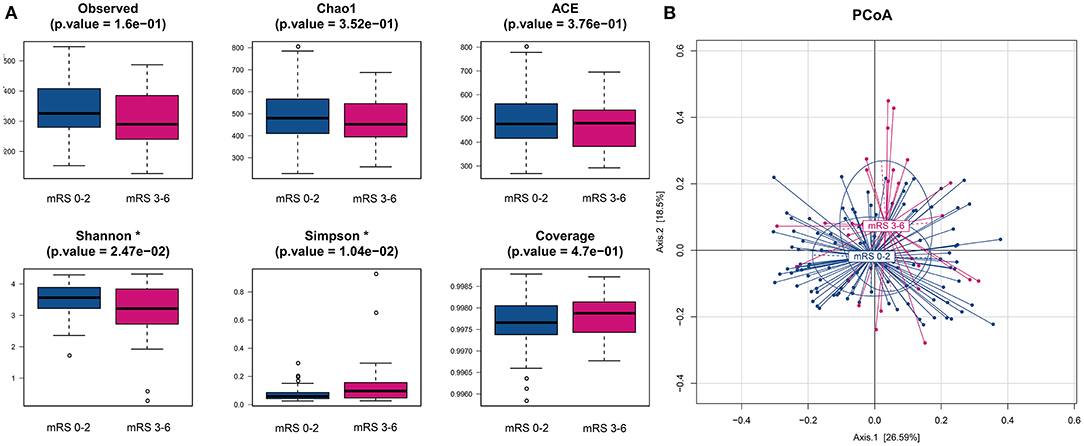
Figure 1. Comparison of alpha- and beta-diversities of gut microbiota between the two groups. (A) Six indices were used to represent the alpha-diversity: observed species, Chao1, Abundance-based Coverage Estimator (ACE), Shannon, Simpson and Coverage. (B) Principal coordinates analysis (PCoA) of the weighted uniFrac distances for the mRS 0–2 and mRS 3–6 subjects. The two components explain 26.6 and 18.5% of the variance, respectively. A significant separation was found between the two groups (p < 0.001, permutation test with pseudo-F ratios). Blue, mRS 0–2 subjects (n = 105); red, mRS 3–6 subjects (n = 27). mRS, modified Rankin Scale.
To compare the differences in microbial structure, PCoA was used (based on weighted-unifrac distance) for beta diversity analysis. The Adonis test was used for statistical significance test and revealed an obvious dissimilarity between the mRS 0–2 and mRS 3–6 groups (Adonis test, p < 0.001, Figure 1B).
The fecal bacteria communities detected in both groups were dominated by 4 main phyla (Supplementary Figure S2A), including Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. The dominant families that account for more than 80% abundance of the total bacteria were Lachnospiraceae, Bacteroidaceae, Ruminococcaceae, Enterobacteriaceae, and Bifidobacteriaceae (Supplementary Figure S2B). The relative abundance of bacteria taxa varied between the two groups. At the phylum level, the gut microbiota in the mRS 3–6 group showed a decrease of beneficial bacteria Bacteroidetes (mean relative abundance, mRS 3–6 group 19.0 vs. mRS 0–2 group 29.8%; p = 0.012, Supplementary Figure S3A). At the family level, beneficial or common core bacteria Bacteroidaceae, Ruminococcaceae, Veillonellaceae, and Streptococcaceae were depleted, while pathogenic bacterium Enterococcaceae was enriched in the mRS 3–6 group, though another opportunistic pathogen Desulfovibrionaceae was depleted (Supplementary Figure S3B). At the genus level, the abundance of six well-known SCFAs-producing genera, Bacteroides, Faecalibacterium, Roseburia, Ruminococcus, Coprococcus, and Butyricicoccus, as well as the other two genera, Streptococcus and Fusicatenibacter were lower in the mRS 3–6 group, while in contrast, the pathogenic bacterium Enterococcus was enriched in this group (Figure 2). Further analysis using LEfSe revealed higher abundances of pathogenic bacteria Enterococcaceae, Enterococcus, and Eggerthella, as well as a lower abundance of SCFAs bacteria Faecalibacterium and Butyricicoccus in the mRS 3–6 group (Figure 3). After PSM, the mRS 3–6 group still showed less abundant SCFAs-producing bacteria of Ruminococcus and Faecalibacterium, and more abundant pathogenic bacterium of Enterococcus (Supplementary Figures S4, S5).
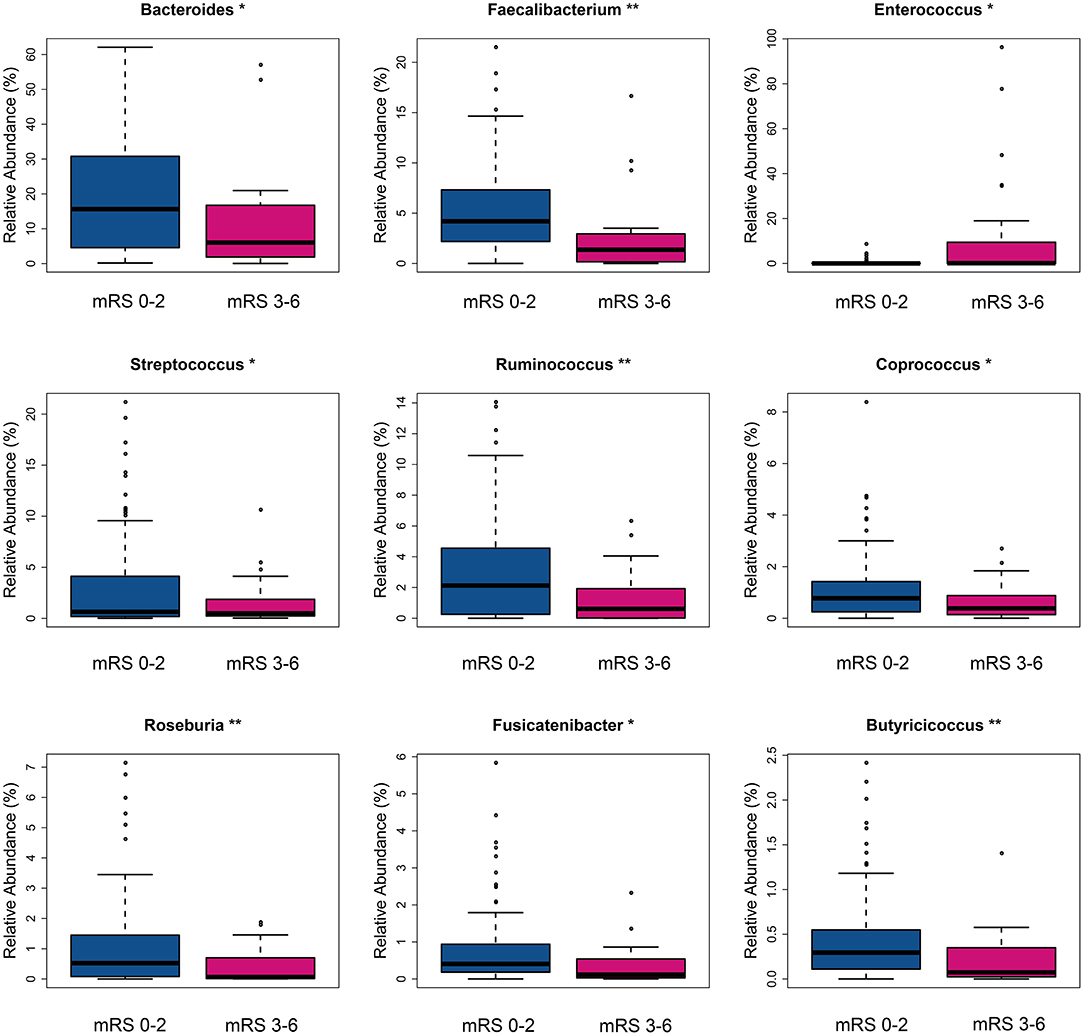
Figure 2. Taxa with significantly different relative abundance between two groups at the genus level. *p < 0.05, **p < 0.01. mRS, modified Rankin Scale.
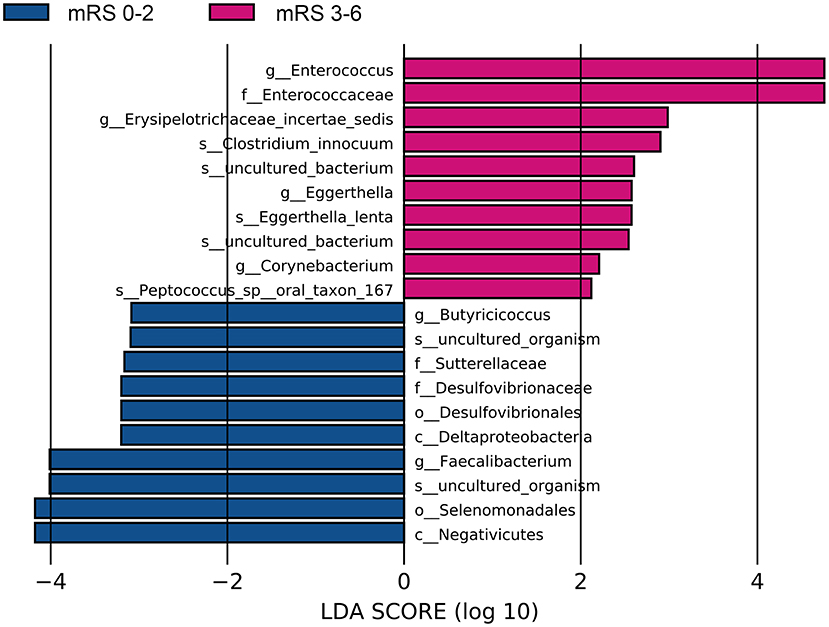
Figure 3. Significantly discriminant taxa between the good outcome and poor outcome patients determined using linear discriminate analysis size effect (LEfSe). LEfSe, linear discriminate analysis size effect; LDA, linear discriminate analysis; mRS, modified Rankin Scale.
As shown in Table 1, The baseline levels of TG, FBG, NIHSS, and DWI-ASPECTs were distributed differently between the two groups. Spearman correlation analyses showed that pathogenic bacterium Clostridium was positively correlated with TG (p = 0.016), and the SCFAs-producing bacterium Roseburia was borderline negatively correlated with FBG (p = 0.050). The pathogenic bacterium Enterococcus was positively correlated with stroke severity (assessed by NIHSS, p < 0.001) and lesion size (estimated by DWI-ASPECTs, p = 0.004), while SCFAs-producing bacteria Bacteroides, Roseburia, and Parabacteroides were negatively correlated with stroke severity (all p < 0.05, Figure 4A). All the above-mentioned bacteria, except for Parabacteroides, had different abundances between the two groups (all p < 0.05).
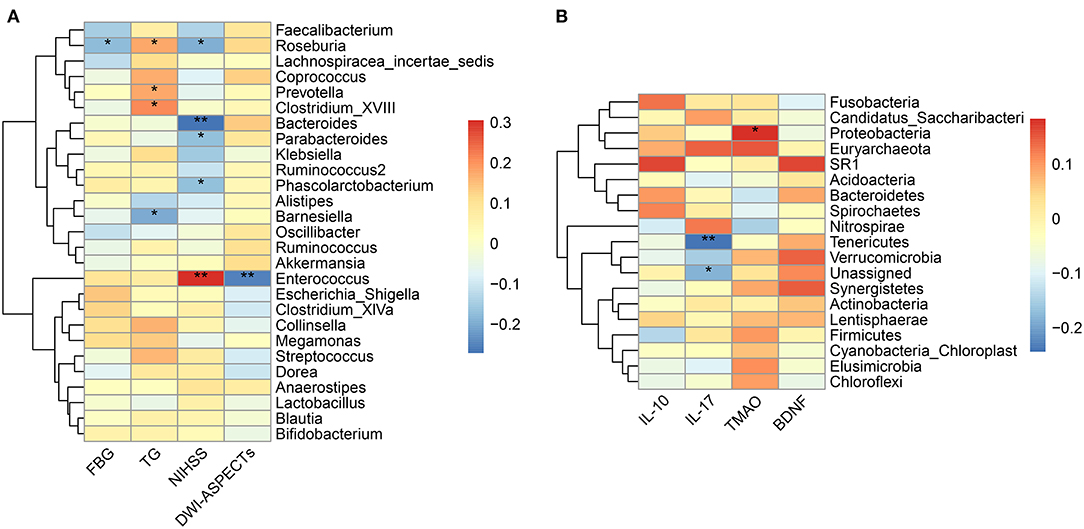
Figure 4. Correlation between the gut microbiota and baseline characteristics. (A) Heatmap of Spearman's correlation analysis between bacteria genera and clinical characteristic parameters. (B) Heatmap of Spearman's correlation analysis between bacteria phyla and laboratory parameters. *p < 0.05, **p < 0.01. FBG, fasting blood glucose; TG, triglyceride; NIHSS, National Institute of Health Stroke Scale; DWI-ASPECTs, DWI-Alberta Stroke Program Early CT score; IL, interleukin; TMAO, trimethylamine N-Oxide; BDNF, brain-derived neurotropic factor.
We did not observe different levels of TMAO, IL-17, IL-10, or BDNF in the mRS 3–6 group when compared to the mRS 0–2 group (Table 1). The results of Spearman correlation analyses showed that the TMAO level was positively correlated with the relative abundances of phylum Proteobacteria and family Enterobacteriaceae (p = 0.050) while negatively correlated with the family Bacillaceae-1 (Figure 4B, Supplementary Figure S6A). At the genus level, TMAO was positively correlated with Escherichia/Shigella (Supplementary Figure S7). The IL-17 level was negatively correlated with phylum Tenericutes and positively correlated with genus Agromyces (Figure 4B, Supplementary Figure S7). The IL-10 level was positively correlated with family Bacteroidaceae and genera Hungatella and Eggerthia (Supplementary Figures S6B, S7). The BDNF level was positively correlated with genus Anaerobacterium (Supplementary Figure S7).
The metagenome functional composition identified by PICRUSt showed that in the gut microbiota of patients with stroke with poor outcome, 5 of the 41 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (level 2) were upregulated, including membrane transport, transcription, metabolism, xenobiotics biodegradation and metabolism, and infectious diseases (Figure 5A). Whereas, in the gut microbiota of patients with stroke with good outcome, 12 KEGG pathways were upregulated, including amino acid metabolism, metabolism of cofactors and vitamins, enzyme families, replication and repair, etc. At KEGG level 3, 34 pathways relating to metabolism showed significant differences between the two groups, including alanine, aspartate and glutamate metabolism, fructose and mannose metabolism, pyruvate metabolism, among others (Supplementary Figure S8).
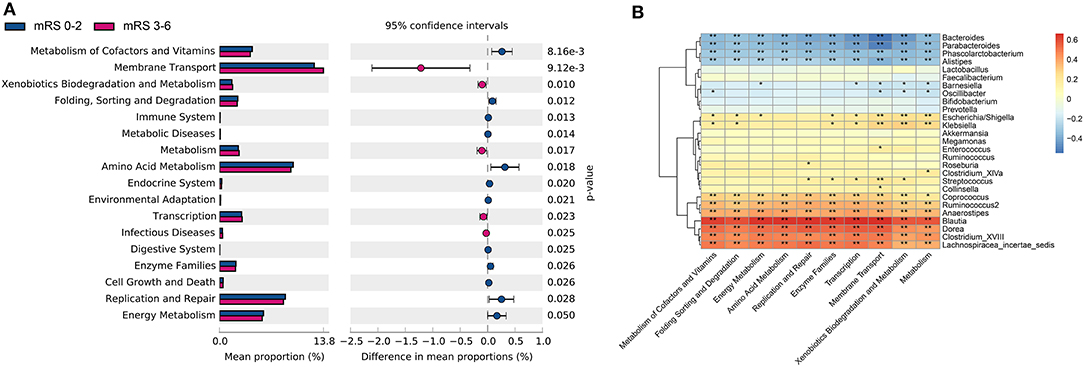
Figure 5. The gut microbiota function differed between the two groups in KEGG pathways and was correlated with bacteria profile. (A) Significantly differed microbiota function in KEGG level 2 pathways. (B) Heatmap of Spearman's correlation analysis between bacteria genera and the KEGG level 2 pathways. KEGG, kyoto encyclopedia of genes and genomes; mRS, modified Rankin Scale.
Next, the Spearman's correlation analyses between the 10 most differently distributed Kyoto Encyclopedia of Genes and Genomes (KEGG) level 2 pathways and 27 bacteria genera with relative abundance > 1% were performed (Figure 5B). Data showed that the genus Enterococcus was positively associated with the membrane transport pathway, while genera Bacteroides and Parabacteroides were negatively correlated with the membrane transport pathway. Besides, SCFAs-producing bacteria Ruminococcus and Coprococcus were positively correlated with the amino acid metabolism pathway.
In this study, we found baseline dysbiosis of gut microbiota in patients with acute ischemic stroke with 3-month unfavorable outcome, featured by decreased alpha diversity, increased opportunistic pathogens (e.g., Enterococcaceae and Enterococcus), and decreased SCFAs-producing bacteria (e.g., Bacteroidaceae, Ruminococcaceae, and Faecalibacterium) compared to those in the favorable outcome group. Although candidate mediators (i.e., serum TMAO, IL-17, IL-10, and BDNF) were comparable in the acute phase of stroke, there were differences in biological functions such as the membrane transport and amino acid metabolism between the two groups. Our findings showed an association between the baseline alterations of gut microbiota and the functional outcome at 3 months after stroke.
The role played by gut microbiota in stroke pathology has been intensively studied in the recent years. Animal studies suggested that stroke would induce dysfunction of the intestinal barrier, such as disruption of tight junction protein (20), dysregulation of the sympathetic nervous system (21), and impairment of gut motility (22), leading to gut microbiota dysbiosis (15, 23). This may reversely impede neuro-regeneration and enlarge stroke core volume, resulting in poor functional outcome (23). In clinical investigations, a consistently altered profile of gut microbiota has been described in patients with stroke, with an increase of pathogenic bacteria and a decrease of common probiotics (10–14). Moreover, a potential relationship with stroke outcome has been suggested for gut microbiota dysbiosis (11, 12) and this study further emphasized that specific bacteria were in association with unfavorable 3-month stroke outcome.
The decreased alpha diversity in the poor outcome group was observed and similar findings have been reported in patients with other neurological diseases, for instance, Alzheimer's disease and therapy-resistant epilepsy (3). A possible explanation for the decreased alpha diversity observed in this study is that the relative abundance of common core bacteria was decreased due to the “leaky gut” (12, 15, 24), which is characterized by increased intestinal barrier permeability. The stroke mice model found that the supply of SCFAs-producing bacteria increased alpha-diversity and improved outcome (25). Two former studies also found that patients with stroke displayed lower alpha diversity when compared to healthy controls (11, 12), whereas Yin et al. reported elevated alpha diversity, and Li et al. reported no significant difference in alpha diversity between patients with stroke and asymptomatic controls (10, 14). Further studies with a larger sample size might help to verify the prognosis value of alpha diversity in stroke outcome.
In this study, we found the pathogenic bacteria (i.e., Enterococcus and Eggerthella) were increased in the unfavorable outcome group. Likewise, previous reports indicated higher levels of Enterococcus in patients with Parkinson's disease (26) and higher levels of Eggerthella in patients with hypertension (6). The pathogenic bacteria have been reported to be positively correlated with early stroke outcome (12), and this study further confirmed a positive correlation between the pathogen Enterococcus and the 3-month stroke outcome. Enterococcus is a Gram-positive opportunistic pathogen that could cause pneumonia and bacteremia in patients with stroke, worsening functional outcome (20). Mechanically, Enterococcus was reported to have interactions with platelets (27) and could modulate neuronal circuits that mediate stress responses (28). It is noteworthy that the pathogenic bacterium Desulfovibrionaceae, previously reported to be elevated in patients with stroke and associated with stroke severity (9), showed a decrease in the unfavorable outcome group in this study, which gives rise to the complicated patterns of microbial dysbiosis influenced by the stroke that worth further investigations.
Short-chain fatty acids mainly include acetate, propionate, and butyrate and play an important role in maintaining the gut permeability and blood-brain barriers integrity (15, 22, 29, 30). A lower abundance of SCFAs-producing bacteria was previously demonstrated in brain disorders such as Parkinson's disease and metabolic disorders such as type 2 diabetes (31). In patients with stroke, the decrease of SCFAs-producing bacteria was positively associated with poststroke infection (32) and the reduced SCFAs level was closely linked to an increased risk of 3-month poor functional outcomes (11). In the current study, we found the SCFAs-producing bacteria (e.g., Bacteroidaceae, Ruminococcaceae, and Faecalibacterium) were decreased in patients with stroke with poor outcome. This finding was in line with former observations that decreased SCFAs-producing bacteria (e.g., Faecalibacterium, Roseburia, and Coprococcus) predict unfavorable early stroke outcomes (12). Of note, one of the SCFAs, butyrate, which regulates apoptosis and inflammation after stroke (33), may indirectly affect glucose metabolism by interfering with the acetylation of related enzymes (11, 13, 34). Accordingly, we observed an association between the butyrate-producing genus Roseburia and FBG. In total, our findings suggest that the SCFAs-producing bacteria are important for stroke outcome. Further studies focusing on the neuroprotective value of SCFAs-producing bacteria might provide novel therapeutic targets for patients with stroke (23, 25).
Disrupted microbiota composition accompanied by an elevated level of proinflammatory IL-17 and a decreased level of neuroprotective IL-10 were observed in stroke mice (22), and an elevated TMAO level was reported in patients with acute ischemic stroke (35). Nevertheless, this study showed that stroke outcome could not be differentiated by the serum levels of TMAO, IL-17, or IL-10. This result should be interpreted with caution as we collected the blood samples mostly within 72 h after the stroke onset, while the poststroke inflammatory response was reported to peek at 3–5 days (22), and the TMAO might change dynamically after stroke, with an elevation within 24 h and a significant decrease thereafter (36). There is, however, an association between TMAO and the pathogenic bacterium Enterobacteriaceae in the light of previous observations that pathogenic bacteria would worsen disease outcome by regulating TMAO generation (18).
In patients with stroke with poor outcome, the functional alterations of elevated membrane transport suggested intestinal mucosal microbiota disruption (37), and the upregulated enzymatic pathway of xenobiotic metabolism/aromatic biodegradation has been shown to predict poor stroke outcome in the mice model as well (23). On the other hand, the pathway associated with amino acid metabolism, which could be of neuroprotective value (23), was elevated in the favorable outcome group. Furthermore, we found a positive correlation between the genus Enterococcus and the membrane transport pathway, as well as between SCFAs-producing bacteria (i.e., Ruminococcus and Coprococcus) and the amino acid metabolism pathway. Further studies are needed to test the possibility of specific bacteria affecting stroke outcome through these pathways.
This study has several limitations. First, it was a single-center study with a small sample size and is limited to the Chinese Han population. Second, dietary factors were not taken into consideration when comparing the composition of gut microbiota between the two groups. Previous studies have shown that long-term diet affects the fecal microbial community, especially protein and animal fat (Bacteroides) and carbohydrates (Prevotella) (38). However, these hospitalized participants were local residents with similar levels of dietary structure. Third, the participants differed in age, the NIHSS score, and a history of hypertension, which may lead to sampling bias (2, 7). Although we performed PSM analysis to obtain matched pairs, and the results fitted well, further information would be needed from validation cohorts including new patients. Finally, it is widely believed that SCFAs are associated with stroke outcome (23, 24, 30), which we however have not included in the testing of fecal or blood samples.
In conclusion, this study suggested the gut microbiota in the acute phase of stroke had an association with the functional outcome at 3 months. Patients with stroke with 3-month unfavorable outcome are characterized by an increase of pathogenic bacteria and a decrease of SCFAs-producing bacteria, as well as the upregulated membrane transport and xenobiotics biodegradation pathways. Additional multicenter studies with larger sample sizes are needed to validate our findings, as well as to verify if the conclusions are confined to a specific region or are general.
The datasets presented in this study can be found in online repositories. The name of the repository and accession number can be found at: Genome Sequence Archive (GSA), https://ngdc.cncb.ac.cn/gsa/, CRA005254.
The studies involving human participants were reviewed and approved by the Ethical Review Board of Nanjing First Hospital (Nanjing, China). The patients/participants provided their written informed consent to participate in this study.
HS, MG, and XC conceived the study. HS and MG analyzed the data. HS, MG, and ZL interpreted the data. HS wrote the manuscript. XC and JZ revised the manuscript.
This study was supported by the National Nature Science Foundation of China (81701064), the Medical Science and Technology Program of Nanjing (JQX20007) and National Science and Technology Innovation 2030 – Major program of “Brain Science and Brain-Inspired Intelligence Research” (2021ZD0201807).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We would like to thank all the study participants, clinical, and research staff from the Nanjing First Hospital for their contributions to this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2021.799222/full#supplementary-material
1. Vos T, Lim SS, Abbafati C, Abbas KM, Abbasi M, Abbasifard M, et al. Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. (2020) 396:1204–22. doi: 10.1016/S0140-6736(20)30925-9
2. Wu SW, Wu B, Liu M, Chen Z, Wang W, Anderson CS, et al. Stroke in China: advances and challenges in epidemiology, prevention, and management. Lancet Neurol. (2019) 18:394–405. doi: 10.1016/S1474-4422(18)30500-3
3. Cryan JF, O'Riordan KJ, Sandhu K, Peterson V, Dinan TG. The gut microbiome in neurological disorders. Lancet Neurol. (2020) 19:179–94. doi: 10.1016/S1474-4422(19)30356-4
4. Horne RG, Yu Y, Zhang R, Abdalqadir N, Rossi L, Surette M, et al. High Fat-High Fructose Diet-Induced Changes In The Gut Microbiota Associated With Dyslipidemia In Syrian Hamsters. Nutrients. (2020) 12:3557. doi: 10.3390/nu12113557
5. Jiang Z, Sun TY, He Y, Gou W, Zuo LS, Fu Y, et al. Dietary fruit and vegetable intake, gut microbiota, and type 2 diabetes: results from two large human cohort studies. BMC Med. (2020) 18:371. doi: 10.1186/s12916-020-01842-0
6. Silveira-Nunes G, Durso DF, Jr LRAO, Cunha EHM, Maioli TU, Vieira AT, et al. Hypertension is associated with intestinal microbiota dysbiosis and inflammation in a Brazilian population. Front Pharmacol. (2020) 11:258. doi: 10.3389/fphar.2020.00258
7. Spychala MS, Venna VR, Jandzinski M, Doran SJ, Durgan DJ, Ganesh BP, et al. Age-related changes in the gut microbiota influence systemic inflammation and stroke outcome. Ann Neurol. (2018) 84:23–36. doi: 10.1002/ana.25250
8. Zuo K, Yin X, Li K, Zhang J, Wang P, Jiao J, et al. Different types of atrial fibrillation share patterns of gut microbiota dysbiosis. mSphere. (2020) 5:2. doi: 10.1128/mSphere.00071-20
9. Bonsack B, Jiang RH, Borlongan CV. A gut feeling about stroke reveals gut-brain axis' active role in homeostasis and dysbiosis. J Cereb Blood Flow Metab. (2019) 40:1132–4. doi: 10.1177/0271678X19900037
10. Li N, Wang X, Sun C, Wu X, Lu M, Si Y, et al. Change of intestinal microbiota in cerebral ischemic stroke patients. BMC Microbiol. (2019) 19:191. doi: 10.1186/s12866-019-1552-1
11. Tan C, Wu Q, Wang H, Gao X, Xu R, Cui Z, et al. Dysbiosis of gut microbiota and short-chain fatty acids in acute ischemic stroke and the subsequent risk for poor functional outcomes. JPEN J Parenter Enteral Nutr. (2020) 45:32–42. doi: 10.1002/jpen.1861
12. Xia GH, You C, Gao XX, Zeng XL, Zhu JJ, Xu KY, et al. Stroke Dysbiosis Index (SDI) in gut microbiome are associated with brain injury and prognosis of stroke. Front Neurol. (2019) 10:397. doi: 10.3389/fneur.2019.00397
13. Yamashiro K, Tanaka R, Urabe T, Ueno Y, Yamashiro Y, Nomoto K, et al. Gut dysbiosis is associated with metabolism and systemic inflammation in patients with ischemic stroke. PLoS One. (2017) 12:e0171521. doi: 10.1371/journal.pone.0171521
14. Yin J, Liao SX, He Y, Wang S, Xia GH, Liu FT, et al. Dysbiosis of gut microbiota with reduced Trimethylamine-N-Oxide level in patients with large-artery atherosclerotic stroke or transient ischemic attack. J Am Heart Assoc. (2015) 4:e002699. doi: 10.1161/JAHA.115.002699
15. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. (2013) 504:451–455. doi: 10.1038/nature12726
16. Maqsood R, Stone TW. The gut-brain axis, BDNF. NMDA and CNS disorders. Neurochem Res. (2016) 41:2819–35. doi: 10.1007/s11064-016-2039-1
17. Zhai Q, Wang X, Chen C, Tang Y, Wang Y, Tian J, et al. Prognostic value of plasma Trimethylamine N-Oxide levels in patients with acute ischemic stroke. Cell Mol Neurobiol. (2019) 39:1201–6. doi: 10.1007/s10571-019-00714-3
18. Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. (2016) 165:111–24. doi: 10.1016/j.cell.2016.02.011
19. Gu M, Chen N, Sun H, Li Z, Chen X, Zhou J, et al. Roseburia abundance associates with severity, evolution and outcome of acute ischemic stroke. Front Cell Infect Microbiol. (2021) 11:669322. doi: 10.3389/fcimb.2021.669322
20. Stanley D, Mason LJ, Mackin KE, Srikhanta YN, Lyras D, Prakash MD, et al. Translocation and dissemination of commensal bacteria in post-stroke infection. Nat Med. (2016) 22:1277–84. doi: 10.1038/nm.4194
21. Mracsko E, Liesz A, Karcher S, Zorn M, Bari F, Veltkamp R. Differential effects of sympathetic nervous system and hypothalamic-pituitary-adrenal axis on systemic immune cells after severe experimental stroke. Brain Behav Immun. (2014) 41:200–9. doi: 10.1016/j.bbi.2014.05.015
22. Singh V, Roth S, Llovera G, Sadler R, Garzetti D, Stecher B, et al. Microbiota dysbiosis controls the neuroinflammatory response after stroke. J Neurosci. (2016) 36:7428–40. doi: 10.1523/JNEUROSCI.1114-16.2016
23. Benakis C, Poon C, Lane D, Brea D, Sita G, Moore J, et al. Distinct commensal bacterial signature in the gut is associated with acute and long-term protection from ischemic stroke. Stroke. (2020) 51:1844–54. doi: 10.1161/STROKEAHA.120.029262
24. Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. (2014) 20:779–86. doi: 10.1016/j.cmet.2014.07.003
25. Chen R, Xu Y, Wu P, Zhou H, Lasanajak Y, Fang Y, et al. Transplantation of fecal microbiota rich in short chain fatty acids and butyric acid treat cerebral ischemic stroke by regulating gut microbiota. Pharmacol Res. (2019) 148:104403. doi: 10.1016/j.phrs.2019.104403
26. Li W, Wu X, Hu X, Wang T, Liang S, Duan Y, et al. Structural changes of gut microbiota in Parkinson's disease and its correlation with clinical features. Sci China Life Sci. (2017) 60:1223–33. doi: 10.1007/s11427-016-9001-4
27. Nallapareddy SR, Sillanpaa J, Mitchell J, Singh KV, Chowdhury SA, Weinstock GM, et al. Conservation of Ebp-type pilus genes among Enterococci and demonstration of their role in adherence of Enterococcus faecalis to human platelets. Infect Immun. (2011) 79:2911–20. doi: 10.1128/IAI.00039-11
28. Wu WL, Adame MD, Liou CW, Barlow JT, Lai TT, Sharon G, et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature. (2021) 595:409–14. doi: 10.1038/s41586-021-03669-y
29. Bailey MT, Cryan JF. The microbiome as a key regulator of brain, behavior and immunity: commentary on the 2017 named series. Brain Behav Immun. (2017) 66:18–22. doi: 10.1016/j.bbi.2017.08.017
30. Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. (2016) 165:1332–45. doi: 10.1016/j.cell.2016.05.041
31. Silva YP, Bernardi A, Frozza RL. The role of short-chain fatty acids from gut microbiota in gut-brain communication. Front Endocrinol. (2020) 11:25. doi: 10.3389/fendo.2020.00025
32. Haak BW, Westendorp WF, van Engelen TSR, Brands X, Brouwer MC, Vermeij JD, et al. Disruptions of anaerobic gut bacteria are associated with stroke and post-stroke infection: a prospective case-control study. Transl Stroke Res. (2021) 12:581–92. doi: 10.1007/s12975-020-00863-4
33. Zhou Z, Xu N, Matei N, McBride DW, Ding Y, Liang H, et al. Sodium butyrate attenuated neuronal apoptosis via GPR41/Gbetagamma/PI3K/Akt pathway after MCAO in rats. J Cereb Blood Flow Metab. (2020) 41:267–81. doi: 10.1177/0271678X20910533
34. Bourassa MW, Alim I, Bultman SJ, Ratan RR. Butyrate, neuroepigenetics and the gut microbiome: can a high fiber diet improve brain health? Neurosci Lett. (2016) 625:56–63. doi: 10.1016/j.neulet.2016.02.009
35. Schneider C, Okun JG, Schwarz KV, Hauke J, Zorn M, Nurnberg C, et al. Trimethylamine-N-oxide (TMAO) is elevated in the acute phase after ischemic stroke and decreases within the first days. Eur J Neurol. (2020) 27:1596–603. doi: 10.1111/ene.14253
36. Tan C, Wang H, Gao X, Xu R, Zeng X, Cui Z, et al. Dynamic changes and prognostic value of gut microbiota-dependent Trimethylamine-N-Oxide in acute ischemic stroke. Front Neurol. (2020) 11:29. doi: 10.3389/fneur.2020.00029
37. Stanley D, Moore RJ, Wong CHY. An insight into intestinal mucosal microbiota disruption after stroke. Sci Rep. (2018) 8:568. doi: 10.1038/s41598-017-18904-8
Keywords: gut microbiota, 16S rRNA, dysbiosis, ischemic stroke, functional outcome
Citation: Sun H, Gu M, Li Z, Chen X and Zhou J (2022) Gut Microbiota Dysbiosis in Acute Ischemic Stroke Associated With 3-Month Unfavorable Outcome. Front. Neurol. 12:799222. doi: 10.3389/fneur.2021.799222
Received: 21 October 2021; Accepted: 30 December 2021;
Published: 28 January 2022.
Edited by:
Ren-You Gan, Chinese Academy of Agricultural Sciences (CAAS), ChinaReviewed by:
Zhifeng Fang, Jiangnan University, ChinaCopyright © 2022 Sun, Gu, Li, Chen and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangliang Chen, Y2hlbnhsQG5qbXUuZWR1LmNu; Junshan Zhou, emhqc2gzMzNAMTI2LmNvbQ==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.