
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol. , 29 July 2021
Sec. Epilepsy
Volume 12 - 2021 | https://doi.org/10.3389/fneur.2021.692412
 Hua Tao1,2
Hua Tao1,2 Zengqiang Chen1
Zengqiang Chen1 Jianhao Wu1
Jianhao Wu1 Jun Chen1
Jun Chen1 Yusen Chen2
Yusen Chen2 Jiawu Fu1
Jiawu Fu1 Chaowen Sun1
Chaowen Sun1 Haihong Zhou1
Haihong Zhou1 Wangtao Zhong1
Wangtao Zhong1 Xu Zhou1,2*
Xu Zhou1,2* Keshen Li2,3*
Keshen Li2,3*Epilepsy is characterized by highly abnormal synchronous discharge of brain neurons, and ion channels are fundamental in the generation and modulation of neural excitability. Considering that abnormal methylation can either activate or repress genes, this study was designed to explore the DNA methylation signature of pathogenic genes encoding ion channels in temporal lobe epilepsy (TLE). In total, 38 TLE patients and 38 healthy controls were enrolled in the study, and genomic DNA and total protein of the lymphocytes were extracted from peripheral blood samples to assess methylation and protein levels. The DNA methylation levels of all 12 genes examined were significantly lower in the TLE group than in the control group. After false-positive correction, 83.3% (10/12) of these genes, namely, gamma-aminobutyric acid type A receptor subunit beta1 (GABRB1), gamma-aminobutyric acid type A receptor subunit beta2 (GABRB2), gamma-aminobutyric acid type A receptor subunit beta1 (GABRB3), glutamate ionotropic receptor NMDA type subunit 1 (GRIN1), glutamate ionotropic receptor NMDA type subunit 2A (GRIN2A), glutamate ionotropic receptor NMDA type subunit 2B (GRIN2B), hyperpolarization activated cyclic nucleotide gated potassium channel 1 (HCN1), potassium voltage-gated channel subfamily A member 2 (KCNA2), potassium voltage-gated channel subfamily B member 1 (KCNB1), and potassium sodium-activated channel subfamily T member 1 (KCNT1), were still differentially expressed. Among these ion channels, HCN1 and KCNA2 were selected to evaluate the effects of DNA methylation, and the levels of these proteins were inversely upregulated in the TLE group compared to the control group. As the genes identified as having differential methylation levels are involved in both excitatory and inhibitory ion channels, this study observed by binary logistic regression that hypermethylated GARAB1 was an independent risk factor for TLE, indicating that the overwhelming effect of ion channels on TLE is probably inhibitory from the perspective of DNA methylation. All these findings support the involvement of DNA methylation in TLE pathologies, but the mechanisms need to be further investigated.
Temporal lobe epilepsy (TLE) is the most common form of human epilepsy and has a high chance of becoming medically intractable. Thus, great efforts have been made to investigate its etiologies and pathologies in attempts to develop reliable strategies for evaluating predisposition toward TLE as well as novel drugs for treatment. TLE is traditionally considered a heterogeneous syndrome resulting from multiple environmental factors, such as perinatal asphyxia and febrile seizures (1), but accumulating evidence in recent decades highlights the influence of single-nucleotide polymorphisms (SNPs) and rare mutations in exons (2). Notably, in past year years, emerging evidence has indicated that epigenetic changes to genomic DNA, such as methylation, regulate gene function via certain modifications to chromatin structure (3).
DNA methylation is a major epigenetic modification that adds a methyl group to the fifth carbon of cytosine to form 5-methylcytosine; when present within cytosine-phosphate-G (CpG) islands proximal to promoters, such modification downregulates genes via transcriptional silencing (4). As it can be inherited by cell division, DNA methylation plays a crucial role in genetic regulation, with important implications for normal biology and disease (5). In one study, methylation analysis of all CpG islands proximal to promoters of the human genome revealed 146 protein-coding genes with an altered DNA methylation status in TLE patients with hippocampal sclerosis compared to those without hippocampal sclerosis, with 81.5% of the promoters displaying hypermethylation (6). Moreover, the expression of DNA methyltransferase 1 and 3a, which facilitate DNA methylation, is increased in the brain tissues of TLE patients. In addition, the status of DNA methylation can predict the risk of drug resistance in TLE patients (7). All this evidence supports the involvement of DNA methylation in seizure susceptibility and maintenance of the disorder.
Epilepsy is characterized by highly abnormal synchronous discharge of brain neurons, and ion channels are fundamental to excitability generation and modulation (8). Most of the variations identified to date in epilepsy patients are also common SNPs in healthy individuals; thus, significant differences between them do not necessarily prove a causal relationship (9). As these variations may be located within flanking, intronic and intergenic regions, they are often considered genetic markers (10–12). In comparison, exon mutations often result in abnormalities in protein structures and function (13). Based on de novo variants of exons in epileptic encephalopathies and additional standards (recurrence of variants in unrelated patients, information on previously identified phenotypes, and data from genetic studies), 17 ion-channel genes are considered to be pathogenic or possibly pathogenic (14). Epileptic encephalopathies are a devastating group of severe epilepsy in infants and children, including Dravet syndrome and West syndrome, which often display some features of TLE in adults (15). Several pathogenic genes with de novo mutations, such as sodium voltage-gated channel alpha subunit 1 (SCN1A) (16), are also involved in TLE, suggesting that de novo variants identified in epileptic encephalopathies might function in TLE.
Overall, increasing evidence indicates that DNA methylation and de novo mutations in exons coexist in many diseases (17–19). Therefore, this study aimed to evaluate the DNA methylation signature of the 17 ion channel genes identified in epileptic encephalopathies via de novo mutations, with the goal of deeply exploring pathologies in TLE patients from an epigenetic perspective.
In total, 38 TLE patients and 38 healthy controls were enrolled from the Affiliated Hospital of Guangdong Medical University. All experimental protocols involving human subjects were approved by the Ethics Committees of the Affiliated Hospital of Guangdong Medical University (18B1018B501). All human participants were Han Chinese, and informed consent was obtained at the time of their enrollment. The diagnosis of TLE was made on the basis of a constellation of clinical, EEG, and MRI criteria, which mainly referred to typical temporal auras or interictal EEG discharges within temporal lobes. Any sign of seizure onset outside the temporal structures by semiology or EEG findings was an exclusion criterion. Before enrollment, healthy controls were free of seizures and any other symptoms involved in neurological disorders, and no disease was diagnosed under routine physical examination items. For the TLE group, age at onset, disease duration and drug response were recorded. According to the definition of drug-resistant epilepsy proposed in 2010 by the commission of the International League Against Epilepsy (20), the responses to anti-epileptic drugs of the TLE patients were classified as follows: patients with drug-resistant epilepsy were determined on the basis of observation, namely, the absence of significant change or reduction in seizure frequency (<60%) or even augmentation after 1-year of treatment, with a schedule of at least two tolerated and properly selected anti-epileptic drugs; the remaining patients were considered to have drug-sensitive epilepsy.
First, CpG islands adjacent to the promoter region (from −2 to 1 kb downstream of the first exon) of the 17 ion channel genes were evaluated based on the following criteria: observed/expected ratio >0.60; percent of alleles C+G > 50.00%; length >200 bp. PCR primers were designed to facilitate multiplexable amplification of bisulfate-converted DNA using Methylation Primer software (Tiangen Biotech, Beijing) and the sequence information for CpG islands (referring to Homo sapiens build CBI37/hg19), and a mixed panel of these primers and their concentrations were further optimized to meet the highest efficiency and specificity.
Peripheral blood samples (3 ml) were collected from each participant, after which genomic DNA was extracted using an automatic nucleic acid extractor (Tiangen Biotech, Beijing). The genomic DNA was subjected to bisulfite treatment using the EZ DNA Methylation-Gold kit following the manufacturer's protocol (Zymo Research, CA, USA) to transform unmethylated allele C to allele U. The bisulfite-treated genomic DNA was amplified using the optimized panel of primer mixtures and the HotStarTaq polymerase kit (TAKARA, Tokyo, Japan). The biotinylated primers were further used for library construction, and the biotinylated products were sequenced using the paired-end sequencing protocol (2*150 bp) in accordance with the manufacturer's protocol (Illumina HiSeq Benchtop Sequencer, CA, USA). After a series of quality controls and data analyses, the levels of CpG methylation were equal to the rate of the reads of C and C+U alleles (Genesky Biotechnologies Inc., Shanghai, China).
According to the manufacturer's protocol, lymphocytes were obtained from the peripheral blood (2 ml) of each participant using lymphocyte separation media (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Total protein was extracted from lymphocytes, and the levels of ion channel proteins, hyperpolarization activated cyclic nucleotide gated potassium channel 1 (HCN1) and potassium voltage-gated channel subfamily A member 2 (KCNA2) were measured using enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems, Minneapolis, USA) following the manufacturer's instructions. Absorbance was measured using an ELISA reader (Bio-Rad Laboratories, Hercules, USA).
Measured data were analyzed with Student's t-test, and the results are provided as the mean ± standard deviation (SD). Count data were analyzed with the chi-square test or Fisher's exact test. Bonferroni correction was employed to adjust false-positive results in multiple statistics. Statistical tests were mainly carried out using SPSS 19.0 (IBM, New York, USA), and p ≤ 0.05 was considered significant.
A total of 76 individuals were recruited, including 38 TLE patients and 38 healthy controls, for the present study. For stratified analysis within the TLE group, age, age at onset and disease duration were grouped according to the dichotomic classification method. The details of sex, age, age at onset, disease duration, and drug response are displayed in Table 1. No significant differences in sex or age were observed between the TLE and healthy control groups (p > 0.05).
First, 17 epilepsy-related ion channel genes were selected to assess their DNA methylation status. However, five of them, sodium voltage-gated channel alpha subunit 2 (SCN2A), glutamate ionotropic receptor NMDA type subunit 2D (GRIN2D), SCN1A, gamma-aminobutyric acid type A receptor subunit alpha1 (GABRA1), and gamma-aminobutyric acid type A receptor subunit gamma2 (GABRG2), were omitted from further testing because no CpG islands were found proximal to their promoters, indicating that genetic regulation of these genes is not modulated by methylation.
The DNA methylation levels of all genes tested were significantly lower in the TLE group than in the control group. After Bonferroni correction, 83.3% (10/12) of these genes, namely, gamma-aminobutyric acid type A receptor subunit beta1 (GABRB1), gamma-aminobutyric acid type A receptor subunit beta2 (GABRB2), gamma-aminobutyric acid type A receptor subunit beta1 (GABRB3), glutamate ionotropic receptor NMDA type subunit 1 (GRIN1), glutamate ionotropic receptor NMDA type subunit 2A (GRIN2A), glutamate ionotropic receptor NMDA type subunit 2B (GRIN2B), HCN1, KCNA2, potassium voltage-gated channel subfamily B member 1 (KCNB1), and potassium sodium-activated channel subfamily T member 1 (KCNT1), were still different between the TLE group and the control group (Table 2). The details of the methylation of CpG islands, CpG sites and related haplotypes of these genes are shown in Table 3. Furthermore, binary logistic regression indicated that among these methylation-related genes, hypermethylated GARAB1 was an independent risk factor for TLE (Table 4).
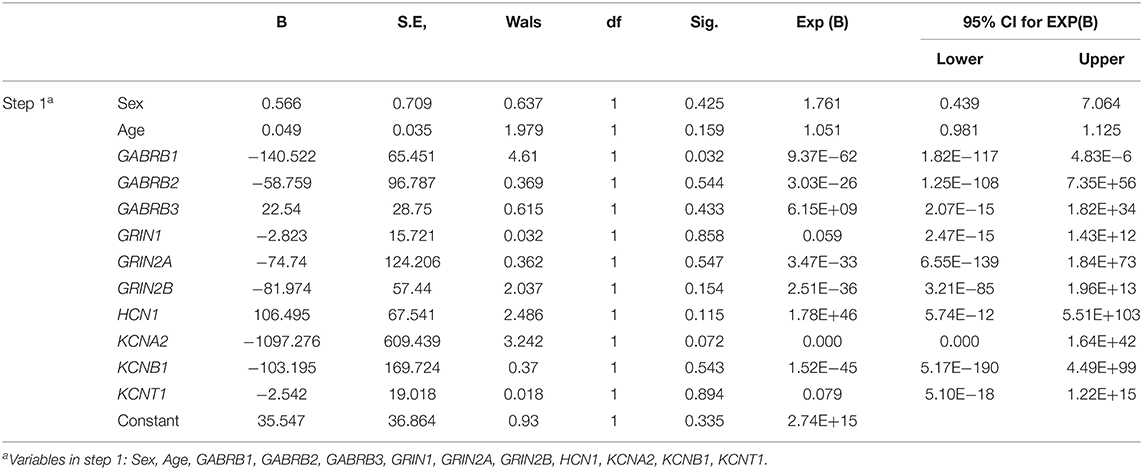
Table 4. Independent risk factors for DNA methylation-related ion channels evaluated by binary logistic regression.
Regarding the methylation levels of the 10 differential genes identified above, further stratified analysis within the TLE group was performed, which revealed that 7 genes were downregulated in younger patients, namely, GABRB1, GABRB2, GABRB3, GRIN1, GRIN2A, HCN1, and KCNB1 (Table 5). Furthermore, GABRB1 was downregulated in TLE cases of a long duration compared to those of a short duration (Table 6). Conversely, no difference in methylation level was observed by stratified analysis of sex, age at onset and drug response (data not shown).
Although DNA methylation of most ion channel genes was significantly associated with TLE in this study, it is unknown whether DNA methylation has the potential to regulate protein levels. Hence, further experiments were performed to evaluate the effects of DNA methylation on the protein levels of selected ion-channel genes in peripheral lymphocytes; we did not use brain tissue specimens, which were hard to attain. According to ELISA, the protein levels of HCN1 and KCNA2 were significantly higher in the TLE group than in the control group (Figure 1).
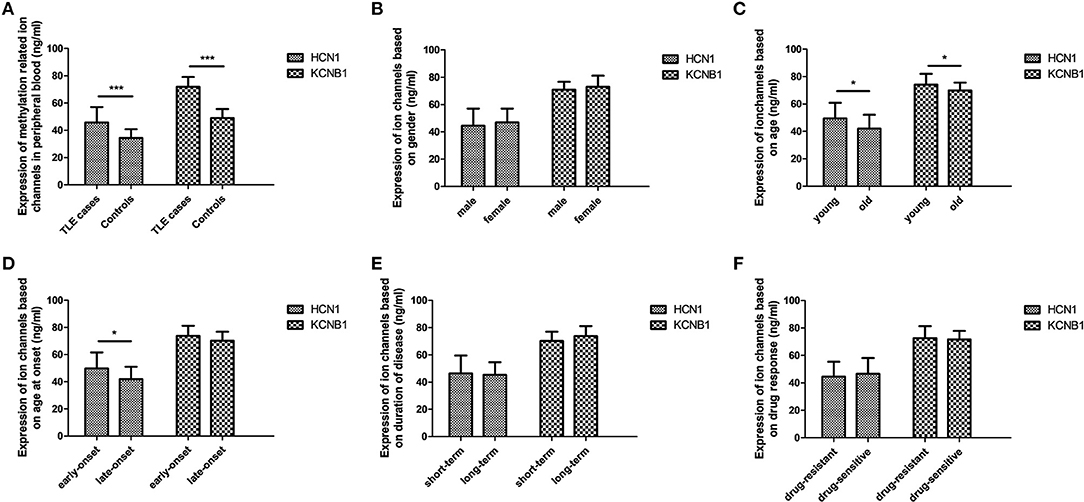
Figure 1. Protein levels of selected ion channel genes in peripheral blood using ELISA. The protein levels of HCN1 and KCNA2 in the TLE group reached a significant level, with a 1.3-fold change and 1.5-fold change in comparison with the control group, respectively (A). Within the TLE group, the levels of HCN1 protein were significantly increased in the young patients and in the early-onset patients (C,D); expression of KCNA2 was also significantly upregulated in young patients compared with old patients (D), but no differences in HCN1 and KCNA2 were observed in accordance with stratified analysis of sex (B), disease duration (E), or drug response (F). *p < 0.05; ***p < 0.001.
In this study, the DNA methylation levels of all tested epilepsy-related ion-channel genes were significantly downregulated in the TLE group compared to the control group. Moreover, most of these genes were still different after false-positive correction. Among these identified ion channels, HCN1 and KCNA2 were selected to evaluate the effects of DNA methylation, and their protein levels were inversely upregulated in the TLE group compared to the control group. These findings support a key role for DNA methylation in transcriptional silencing of ion channel genes and strongly indicate that DNA methylation of ion channel genes is involved in TLE pathologies.
Nevertheless, the causal relationship between DNA methylation of ion channel genes and epileptic seizures in TLE remains unclear. Interestingly, in further stratified analysis within the TLE group, we found no significant difference in most ion channel genes between patients with long and short durations of TLE or between those with early and late onset, which indicates that recurrent seizures in TLE have no moderating effect on DNA methylation. Considering the association between DNA methylation of ion channel genes and TLE, we conclude that methylation of the ion channel genes identified in this study may be a causal factor for a predisposition toward TLE. An exception is GABRB1, as its methylation level was relatively low in patients with a short duration of TLE, indicating that the pathological effect of methylation of this gene might be enhanced along with a prolonged course of TLE disease.
Interestingly, during long-term observation of a healthy cohort comprising 1919 community-dwelling individuals, the incidence of epileptic seizures increased from 10.6 per 100,000 person-years between age 45 and 59, to 25.8 between age 60 and 74, to 101.1 between age 75 and 89; the cumulative incidence was 0.15% from age 45 to age 60, 0.38% to age 70, 1.01% to age 80, and 1.47% to age 90 (21), indicating an increasing risk of epileptic seizures with aging in the healthy population. In contrast, in a cohort of newly diagnosed epilepsy patients, the cumulative incidence of achieving 2-years seizure remission was 34% at 2 years, 45% at 5 years, 52% at 10 years, and 67% at 20 years (22), which indicates that the incidence of seizures declines with age in epilepsy patients. We performed stratified analysis of age in the TLE group and observed that the methylation levels of seven epilepsy-related ion channel genes, namely, GABRB1, GABRB2, GABRB3, GRIN1, GRIN2A, HCN1, and KCNB1, were relatively low in the young subgroup; this indicates that the methylation of most of the genes tested increases with age in TLE. Considering that these genes do not encode excitatory ion channels such as GRIN1 and GRIN2A or inhibitory ion channels such as GABRB3, interaction between enhanced methylation of related genes and aging in TLE probably occurs and influences the balance of excitation and inhibition, which needs to be further investigated.
Many clinical studies support that females undergo a marginally lower incidence of epileptic seizures than males, which is usually attributed to the greater exposure of male patients to risk factors for brain injury due to symptomatic epilepsy (23). In this study, there was no significant difference in DNA methylation levels between male and female patients, indicating that methylation of epilepsy-related ion channel genes is not involved in such sex differences. In addition, we noted that nearly one-third of epilepsy patients still experienced refractory seizures, even though several new drugs have been introduced in clinical practice in recent decades (24). This study aimed to evaluate the potential of DNA methylation in drug-resistant epilepsy, but no significant difference in the ion channel genes evaluated was observed between TLE patients with drug-sensitive and drug-resistant epilepsy, indicating that methylation of epilepsy-related ion channel genes might not be involved in regulating drug resistance.
Several limitations of this research should be mentioned. First, the total number of participants was relatively small, but differential methylation of almost all of the tested ion-channel genes (10/12) between the TLE patients and controls was confirmed via Bonferroni correction; thus, the findings should be reliable to some extent. Second, most ion channels, such as GABRB1 and GRIN1, play a key role in action potentials and are mainly expressed in the nervous system (25), but some channels, such as HCN1 and KCNA2, also function in non-neuronal cells. Hence, further experiments were performed to evaluate the effect of differential DNA methylation on gene expression levels in peripheral blood, with the aim of indirectly confirming that the effects of DNA methylation in TLE depend on related ion channels. Third, as the genes identified as having differential methylation levels are involved in both excitatory and inhibitory ion channels, it is unclear whether their main effects on epileptic seizures are alleviation or deterioration. Nevertheless, this study observed by binary logistic regression that hypermethylated GARAB1 was an independent risk factor for TLE, indicating that the overwhelming effect of ion channels on TLE is probably inhibitory from the perspective of DNA methylation. In addition, DNA methylation can be modified by postnatal factors and inherited by cell division by the next generation; thus, our findings might be affected by racial and environmental factors, and caution should be exercised before generalizing these findings to other populations.
In this study, we systematically observed that the DNA methylation levels of 12 epilepsy-related ion channel genes were significantly downregulated in the TLE group compared to the control group. After false-positive correction, 10 genes were still differentially expressed between the groups, indicating that DNA methylation of many ion channel genes is probably involved in TLE pathologies. However, the mechanisms need to be further investigated.
The datasets presented in this article are not readily available because the raw genomic sequencing data is implicated in national safety. Requests to access the datasets should be directed to corresponding authors.
The studies involving human participants were reviewed and approved by Ethics Committees of the Affiliated Hospital of Guangdong Medical University. The patients/participants provided their written informed consent to participate in this study.
HT undertook data analyses and wrote the paper. HT, ZC, JW, and JC performed the biological experiments. YC, JF, CS, HZ, and WZ collected the specimens. XZ and KL conceptualized the hypothesis and designed the study. All authors have read and approved the final manuscript.
In the present study, the collection of blood specimens and ELISA experiments were funded by the Medical Scientific Research Foundation of Guangdong Province of China (Grant No. A2019152) and the National Natural Science Foundation of China (Grant No. 82001368), and the methylation experiments were funded by the National Natural Science Foundation of China (Grant Nos. 81971079 and 8200051442).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate the support of Genesky Biotechnologies Inc., Shanghai, China, for assistance in the methylation experiments.
1. Patterson K, Baram T, Shinnar S. Origins of temporal lobe epilepsy: febrile seizures and febrile status epilepticus. Neurotherapeutics. (2014) 11:242–50. doi: 10.1007/s13311-014-0263-4
2. Salzmann A, Malafosse A. Genetics of temporal lobe epilepsy: a review. Epilep Res Treat. (2012) 2012:863702. doi: 10.1155/2012/863702
3. Jones P. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. (2012) 13:484–92. doi: 10.1038/nrg3230
4. Schübeler D. Function and information content of DNA methylation. Nature. (2015) 517:321–6. doi: 10.1038/nature14192
5. Chen T, Giri M, Xia Z, Subedi Y, Li Y. Genetic and epigenetic mechanisms of epilepsy: a review. Neuropsych Dis Treat. (2017) 13:1841–59. doi: 10.2147/NDT.S142032
6. Miller-Delaney S, Bryan K, Das S, McKiernan R, Bray I, Reynolds J, et al. Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy. Brain. (2015) 138:616–31. doi: 10.1093/brain/awu373
7. Xiao W, Liu C, Zhong K, Ning S, Hou R, Deng N, et al. CpG methylation signature defines human temporal lobe epilepsy and predicts drug-resistant. CNS Neurosci Therap. (2020) 26:1021–30. doi: 10.1111/cns.13394
8. Wei F, Yan L, Su T, He N, Lin Z, Wang J, et al. Ion channel genes and epilepsy: functional alteration, pathogenic potential, and mechanism of epilepsy. Neurosci Bull. (2017) 33:455–77. doi: 10.1007/s12264-017-0134-1
9. Speed D, Hoggart C, Petrovski S, Tachmazidou I, Coffey A, Jorgensen A, et al. A genome-wide association study and biological pathway analysis of epilepsy prognosis in a prospective cohort of newly treated epilepsy. Hum Mol Genet. (2014) 23:247–58. doi: 10.1093/hmg/ddt403
10. Tao H, Si L, Zhou X, Liu Z, Ma Z, Zhou H, et al. Role of glyoxalase I gene polymorphisms in late-onset epilepsy and drug-resistant epilepsy. J Neurol Sci. (2016) 363:200–6. doi: 10.1016/j.jns.2016.01.052
11. Tao H, Zhou X, Xie Q, Ma Z, Sun F, Cui L, et al. SRR intronic variation inhibits expression of its neighbouring SMG6 gene and protects against temporal lobe epilepsy. J Cell Mol Med. (2018) 22:1883–93. doi: 10.1111/jcmm.13473
12. Al-Eitan L, Al-Dalalah I, Elshammari A, Khreisat W, Almasri A. The impact of potassium channel gene polymorphisms on antiepileptic drug responsiveness in Arab patients with epilepsy. J Personal Med. (2018) 8:37. doi: 10.3390/jpm8040037
13. Helbig K, Farwell Hagman K, Shinde D, Mroske C, Powis Z, Li S, et al. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet Med. (2016) 18:898–905. doi: 10.1038/gim.2015.186
14. He N, Lin Z, Wang J, Wei F, Meng H, Liu X, et al. Evaluating the pathogenic potential of genes with de novo variants in epileptic encephalopathies. Genet Med. (2019) 21:17–27. doi: 10.1038/s41436-018-0011-y
15. Agari T, Kobayashi K, Watanabe K, Date I, Ohtsuka Y. Cryptogenic West syndrome and subsequent mesial temporal lobe epilepsy. Epil Disord. (2012) 14:334–9. doi: 10.1684/epd.2012.0518
16. Tiefes A, Hartlieb T, Tacke M, von Stülpnagel-Steinbeis C, Larsen L, Hao Q, et al. Mesial temporal sclerosis in SCN1A-related epilepsy: two long-term EEG case studies. Clin EEG Neurosci. (2019) 50:267–72. doi: 10.1177/1550059418794347
17. Hernández-Valero M, Rother J, Gorlov I, Frazier M, Gorlova O. Interplay between polymorphisms and methylation in the H19/IGF2 gene region may contribute to obesity in Mexican-American children. J Dev Orig Health Dis. (2013) 4:499–506. doi: 10.1017/S204017441300041X
18. Mukherjee N, Lockett G, Merid S, Melén E, Pershagen G, Holloway J, et al. DNA methylation and genetic polymorphisms of the Leptin gene interact to influence lung function outcomes and asthma at 18 years of age. Int J Mol Epidem Genet. (2016) 7:1–17.
19. Verloes A, Spits C, Vercammen M, Geens M, LeMaoult J, Sermon K, et al. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. (2017) 19:118–27. doi: 10.1016/j.scr.2017.01.005
20. Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, et al. Definition of drug resistant epilepsy: consensus proposal by the ad hoc task force of the ILAE commission on therapeutic strategies. Epilepsia. (2010) 51:1069–77. doi: 10.1111/j.1528-1167.2009.02397.x
21. Hussain S, Haut S, Lipton R, Derby C, Markowitz S, Shinnar S. Incidence of epilepsy in a racially diverse, community-dwelling, elderly cohort: results from the Einstein aging study. Epilep Res. (2006) 71:195–205. doi: 10.1016/j.eplepsyres.2006.06.018
22. Giussani G, Canelli V, Bianchi E, Erba G, Franchi C, Nobili A, et al. Long-term prognosis of epilepsy, prognostic patterns and drug resistance: a population-based study. Europ J Neurol. (2016) 23:1218–27. doi: 10.1111/ene.13005
23. McHugh J, Delanty N. Epidemiology and classification of epilepsy: gender comparisons. Int Rev Neurob. (2008) 83:11–26. doi: 10.1016/S0074-7742(08)00002-0
24. Dalic L, Cook M. Managing drug-resistant epilepsy: challenges and solutions. Neuropsych Dis Treat. (2016) 12:2605–16. doi: 10.2147/NDT.S84852
Keywords: DNA methylation, epigenetic, temporal lobe epilepsy, genetic susceptibility, ion channels
Citation: Tao H, Chen Z, Wu J, Chen J, Chen Y, Fu J, Sun C, Zhou H, Zhong W, Zhou X and Li K (2021) DNA Methylation Signature of Epileptic Encephalopathy-Related Pathogenic Genes Encoding Ion Channels in Temporal Lobe Epilepsy. Front. Neurol. 12:692412. doi: 10.3389/fneur.2021.692412
Received: 08 April 2021; Accepted: 23 June 2021;
Published: 29 July 2021.
Edited by:
Dinesh Upadhya, Manipal Academy of Higher Education, IndiaReviewed by:
Shama Prasada Kabekkodu, Manipal Academy of Higher Education, IndiaCopyright © 2021 Tao, Chen, Wu, Chen, Chen, Fu, Sun, Zhou, Zhong, Zhou and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xu Zhou, MTIwMTg1MTdAcXEuY29t; Keshen Li, bGlrZXNoZW4xOTcxQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.