- Department of Pediatrics, Rady Children's Hospital San Diego, University of California, San Diego, San Diego, CA, United States
Transposable Elements (TE) are mobile DNA elements that can replicate and insert themselves into different locations within the host genome. Their propensity to self-propagate has a myriad of consequences and yet their biological significance is not well-understood. Indeed, retrotransposons have evaded evolutionary attempts at repression and may contribute to somatic mosaicism. Retrotransposons are emerging as potent regulatory elements within the human genome. In the diseased state, there is mounting evidence that endogenous retroelements play a role in etiopathogenesis of inflammatory diseases, with a disposition for both autoimmune and neurological disorders. We postulate that active mobile genetic elements contribute more to human disease pathogenesis than previously thought.
Introduction
Discovered in the context of maize kernel mosaicism (1), transposons are present in virtually all eukaryotes and mobilize from one chromosomal loci to another through either a DNA or RNA intermediate. They parallel viruses in many ways—with regards to their structure and function as they ensure their own survival by way of reintegration (2). Human Endogenous Retroviruses (HERV) and Long-Interspersed Nuclear Element-1 (LINE-1) are two main classes of retrotransposons and are mobilized through a “copy and paste” mechanism. HERV and LINE-1 insertions have accumulated throughout evolution and host genomes have simultaneously coevolved with these mobile elements by employing a variety of factors to suppress aberrant activity (3). LINE-1 somatic retrotransposition has been well-demonstrated to occur in neuronal lineage, however the significance of retroelement activity to normal brain function remains uncertain. Furthermore, the contribution of these elements to the symptomatology of neurodegenerative diseases is a topic of recent exploration. We begin this review with a brief overview of the types of transposable elements and their methods of integration. We will then discuss the consequences of retrotransposon activity and their dynamic relationship with various regulators. Finally, we will review the direct influence of these elements on CNS function and their contribution to disease and neuroinflammation.
Transposable Elements: An Overview
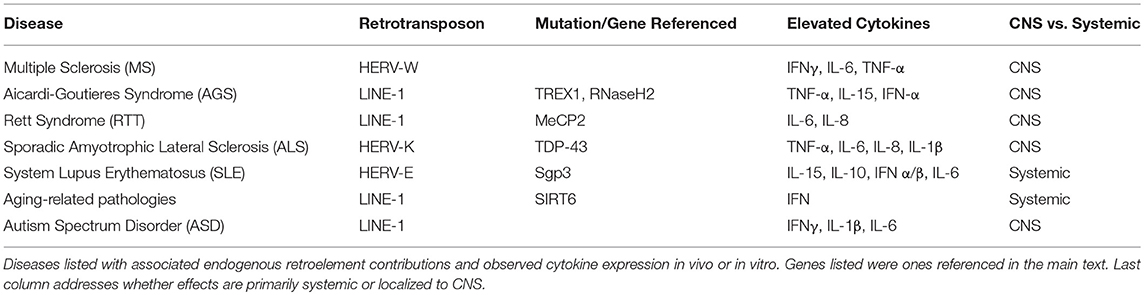
Transposable elements comprise at least 45% of the human genome while coding sequences occupy <3% (4). These highly repetitive strands of “junk” DNA are capable of generating new copies in the human germline and certain somatic tissues. Transposable elements (TE) can be classified as either DNA transposons or retro (RNA) transposons. The mobilization of these elements is referred to as either transposition or retrotransposition. DNA transposons, known as Class I transposons, are flanked by terminal inverted repeats and transpose with a “cut and paste” mechanism whereby the sequence is excised from one region, catalyzed by a transposase enzyme, and integrated into a separate region in the genome [Figure 1; (2, 7)]. DNA transposons constitute 3% of the genome and are no longer active in most mammals (4, 8).
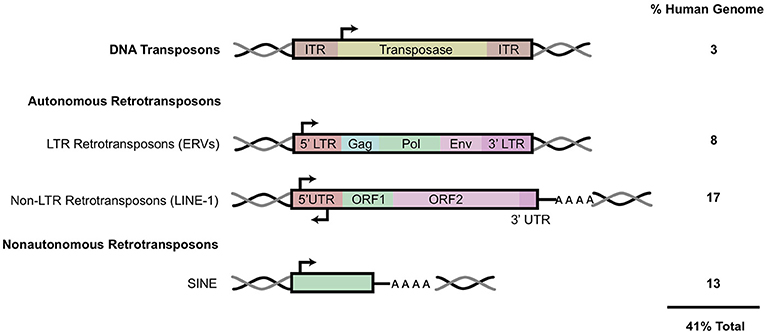
Figure 1. Transposable elements in the human genome. A schematic of the general structure of different classes of mobile elements. On the right is the estimated percentage present within the human genome. ITR, inverted terminal repeat; LTR, long terminal repeat; UTR, untranslated region. HERV genes include gag, which encodes viral structural proteins, pol, which encodes viral enzymes, and env, which encodes for the viral envelope protein. Black arrows indicate sense and antisense promoter regions. Black and gray double helices represent DNA. AAAA, represents polyA tail. This figure has been adapted from several reviews (5, 6).
Retrotransposons, also referred to as Class I transposable elements, integrate into the genome via an RNA intermediate, utilizing a “copy and paste” mechanism; this allows the active retroelements to retain their original location in the genome while accumulating copy numbers elsewhere. These retroelements can be further classified based on the presence of long terminal repeats (LTR) in the sequence. Elements such as Mammalian apparent LTR-retrotransposons (MaLR) and human endogenous retroviruses (HERV) both contain LTR sequences that flank internal coding regions (9). LTR retrotransposons comprise about 8% of the human genome [Figure 1; (10)].
Non-LTR retrotransposons can be further classified into two subtypes: LINE (Long Interspersed Nuclear Elements) and SINE (Short Interspersed Nuclear Elements) (Figure 1). Together, LINE and SINE comprise ~33% of the human genome (4). SINEs, including Alu and SVA elements, are non-autonomous sequences transcribed by RNA Polymerase III (11). Alu sequences are present in over one million copies in the human genome while SINE-R/VNTR/Alu (SVA) elements together constitute more than 10% of the human genome (4). The LINE-1 encoded proteins (ORF1/2p) recognize and bind to non-autonomous SINE sequences in trans to mediate their mobilization.
HERV
HERV copies comprise 5–8% of the human genome, with some lower estimates at 1% (12, 13). HERV elements possess similar genomic organization to that of exogenous retroviruses, such as HIV. Briefly, HERV elements include gag, pol, and env regions that are flanked by LTR sequences on either side (Figure 1). The gag and pol genes encode a retroviral capsid protein and enzymes (protease, reverse transcriptase and integrase) required for viral replication and integration, respectively (12). HERVs also contain the presence of a gene encoding an envelope protein (Env), a remnant of their exogenous retroviral origin prior to their insertion and endogenization into germline cells (14). Functional env proteins have been shown to initiate innate and adaptive immune responses (15).
Transcriptionally active HERV subfamilies have more recently been implicated as pathophysiological contributors to various disorders (Table 1). An initial 1993 study reported that the addition of herpes simplex virus type 1 (HSV-1) to primary leptomeningeal cells isolated from a patient with multiple sclerosis (MS) lead to robust co-expression of retroviral-like particles and reverse transcriptase activity (16). The original multiple sclerosis associated retrovirus (MSRV) sequence identified in the virus-like particles was shown to be highly represented in human DNA but never characterized until MSRV-specific primers defined the previously unknown HERV-W subfamily (17, 18).
This work provided the initial evidence for retroviral-triggered HERV activation as a contributor to the pathology in neurological disorders (16). Similar reports have shown that addition of other exogenous retroviruses, such as HIV-1 and HTLV-1, result in increased expression of HERV-W and HERV-K Env proteins, resulting in a mis-regulated immune response (19). Implications of these elements in neurological diseases sporadic amyotrophic lateral sclerosis (ALS) and multiple sclerosis (MS) will be discussed more extensively later in the review (Table 1).
LINE-1 Retrotransposons
LINE-1 elements are ubiquitous, autonomous retrotransposons with an estimated 500,000 copies contained within the human genome (3, 4). A majority of LINE-1 copies are immobile and unable to retrotranspose, due either to 5′ truncations or inversions introduced into the sequence (20–23). Approximately 80–100 copies are mobile (labeled retrotransposition-competent LINE-1 or RC-L1) (24). Of these, six highly active, or “hot” LINE-1s are responsible for a bulk (84%) of the retrotransposition activity, according to an in-culture cell retrotransposition assay (24).
Full length, RC LINE-1 are 6 kb in length and contain a 5′ untranslated region (5′ UTR), two open reading frames (ORF1, 2), and a 3′ UTR punctuated with a poly-A tract (25) (Figures 1, 2). The LINE-1 promoter region has no TATA-box and displays both sense and antisense activity within the 5′ UTR (25, 26). Additionally, a primate-specific antisense ORF0 of unknown function has also been described within this region (27). The LINE- 1 ORF1 encodes for a protein that has nucleic acid chaperone activities and an RNA binding domain (28, 29). ORF2 encodes for a protein with enzymatic activity strictly required for LINE-1 retrotransposition. ORF2p has both endonuclease (EN) and reverse transcriptase (RT) activities; equally critical for target site cleavage and integration [Figure 2; (30, 31)].
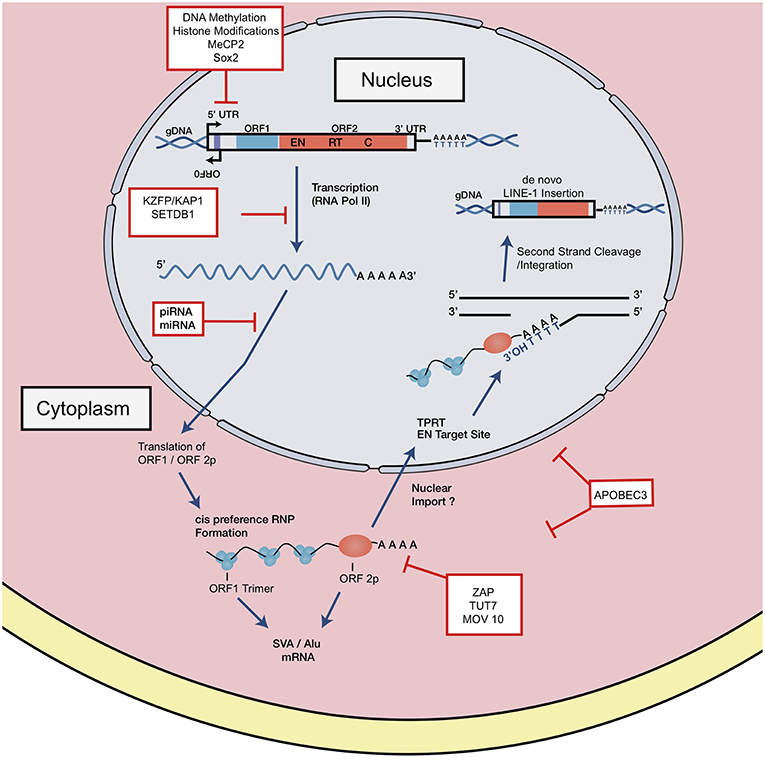
Figure 2. LINE-1 retrotransposition cycle and host factor regulation. Structure of full-length genomic LINE-1. 5′ UTR contains sense and antisense promoter activity. Relative positions of ORF0, ORF1, and ORF2, 3′ UTR and a poly A tail are shown. EN denotes endonuclease, RT denotes reverse transcriptase, and C denotes cysteine-rich domain. RNA polymerase II mediates transcription of retrotransposition-competent LINE-1 sequence. This transcript is exported from the nucleus where it forms an RNP complex with ORF1p and ORF2p. Through a mechanism not well-understood, the RNP is imported into the nucleus to begin reverse transcription and integration through TPRT. The ORF2 EN nicks the bottom strand of the DNA, exposing a 3′ OH, which serves as a primer for the RT to generate the cDNA from the LINE-1 mRNA. How the second strand is synthesized and integrated is a poorly understood mechanism. LINE-1 is regulated at distinct intermediates of retrotransposition, indicated in red boxes.
The LINE-1 transcription start site begins at the RNA polymerase II promoter region, or more precisely, within the first 100 base pairs of the 5′ UTR (25, 32). Once transcribed, the full-length capped and polyadenylated LINE-1 mRNA is exported into the cytoplasm where it combines in cis with ORF1 and ORF2 proteins to form a ribonucleoparticle (RNP) complex (Figure 2) reviewed in Doucet et al. (33), Macia et al. (5), and Elbarbary et al. (34). In a more traditional model, the RNA-protein complex can be imported back into the nucleus by a mechanism not well-understood, where target-primed reverse transcription (TPRT) might take place. The RNP complex binds preferentially to an AT rich consensus target (5′ TTTT/AA 3′ and variants) recognized by the EN (35–37). The EN nicks the bottom DNA strand, exposing a 3′ OH which primes the reverse transcription of the LINE-1 mRNA template into cDNA. Recent observations have challenged this model, suggesting that TPRT might also start in the cytoplasm or either unfinished retrotransposition events might be transported back to the cytoplasm (38). The complete mechanism of second DNA strand cleavage and subsequent cDNA synthesis is unknown, but recent studies have suggested that RNase H2 is required to degrade the RNA:cDNA hybrid generated during this step (39).
Heterogeneity of Insertional Preferences and Distribution of Retrotransposons Across Different Populations
Selective processes and insertion bias impact the distribution of LINE-1 elements in the genome (40). The distribution of retrotransposons within the genome is variable; SINEs are more aptly tolerated by the cell and have been shown to localize in gene-rich (GC-rich) regions whereas pre-existing, static LINEs are highly enriched in intergenic (AT-rich) isochores, likely due to the length of their insertion sequences (34, 41).
A recent meta-analysis of engineered LINE-1 insertions by Flasch and colleagues reported a strong bias for LINE-1 insertional preferences. Wild-type LINE-1 EN preferentially nicks the lagging strand of DNA replication fork, resulting in cDNA insertions into leading strand templates (42). It was determined that varied levels of open chromatin state had only minor influence on LINE-1 insertion preference. A similar study recently published by Sultana and colleagues corroborated these findings. De novo LINE-1 retrotransposition was induced in cultured HeLa S3 cells followed by ATLAS-seq profiling to detect and map integration sites. This study also confirmed minimal association (2–3%) of all insertions in chromatin segments annotated as weak enhancers or with histone modifications characteristic of weak enhancers (H3K4me1) (43). The strongest association was enrichment of LINE-1 insertions within early replicating regions of the genome. Insertion orientation was shown to be influenced by the directionality of the replication machinery; EN cleavage of the bottom strand was highly enriched when replication fork moves leftward. Both studies provide evidence that LINE-1 integration events do not target expressed genes, open chromatin or transcribed regions but instead associate with host DNA replication (42, 43). These findings provide the first clues that LINE-1 integration and DNA replication may be mechanistically linked.
There is heterogeneity in the distribution of endogenous retroelements across different human populations: from the presence/absence of the element to single nucleotide polymorphisms (SNPs) [Figure 3; (20, 44, 45)]. Because RC-L1 mobilize to novel insertion sites, it is logical that individuals will carry differences in the presence or absence of LINE-1 insertions at various loci in their genome. Polymorphisms in retroelement insertions are generated either in the germline or early in embryonic development (Figure 3). Germline insertions are incorporated into all tissue types within the individual and are heritable by the next generation. Somatic de novo LINE-1 insertions, which occur later in development, are not inherited by subsequent generations and are localized to the cell(s) in which the insertion occurred (Figure 3).
Figure 3. Somatic vs. Germline Insertions. Differences between the heritability of germline vs. somatic retrotransposon insertions. (A) Somatic insertions lead to mosaicism within the individual but are not inherited. (B) Generally, germline or early embryonic insertions are incorporated into the three germ layers and are present in all tissue types. (C) Example of polymorphisms across different populations based on the presence or absence of an insertion.
As a consequence, polymorphisms can generate variability in activity levels of retrotransposons. Indeed, allelic variability within the LINE-1 elements was demonstrated to have up to 16-fold differences in activity (44). To study the differential expression of polymorphic LINE-1 elements, Philippe et al. mapped active LINE-1 HS-Ta (human-specific) copies according to their epigenetic signatures in 12 somatic cell lines. A restricted subset of polymorphic LINE-1 loci remain highly active but are differentially regulated according to cell type (46). ORF1p expression was high in neural progenitors and cancer cell types studied while little to no expression was seen in primary fibroblasts, consistent with previous work (47–49). In summary, LINE-1 transcription in somatic cells was shown to be governed by locus- and cell-type- specific determinants (46).
Polymorphisms are observed with HERV elements as well. Next generation sequencing (NGS) characterized unfixed HERV-K insertions across different human populations (14, 50). Scans for polymorphisms in HERV-K loci revealed 17 loci in the individuals studied that were not present in the human reference genome. On average, each individual possessed six loci, often in the heterozygous state, that were not found in the reference genome (50). A separate study analyzed 36 non-reference polymorphic HERV-K proviruses from more than 2,500 globally sampled individuals found insertion frequencies ranging from <0.0005 to >0.75, and varied by population (14).
In summary, a myriad of selective processes influences the distribution and insertional preferences of retrotransposons. This allows for genomic mosaicism within a single individual as well as polymorphisms across different human populations.
Cellular Impact of LINE-1 Retrotransposition
There are innumerable ways in which retrotransposons can influence the genome and impact cellular function (Figure 4). LINE-1 elements generate structural variation/instability within the genome and subsequently interfere with host gene expression. In this section, we review the insertional and transcriptional impact of LINE-1 elements in the human genome.
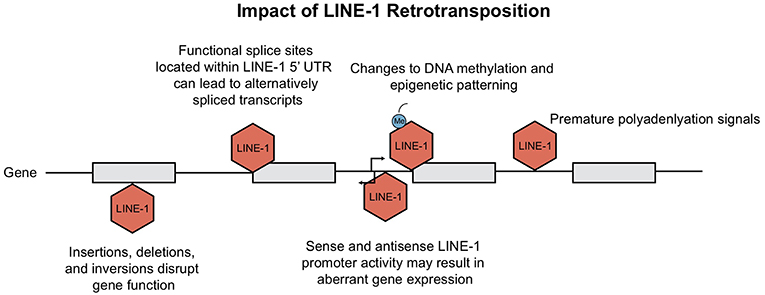
Figure 4. Consequences of LINE-1 retrotransposition. Ongoing retrotransposition generates genomic instability and consequently interferes with host gene expression at many levels. Consequences include disruption of gene function due to LINE-1 insertions, as well as generation of mis-spliced or prematurely truncated transcripts. Insertions have been shown to impact host gene expression through changes in epigenetic patterning and LINE-1 promoter activity. This figure is adapted from Macia et al. (5) and Garcia-Perez et al. (6).
The current rate of LINE-1 retrotransposition has been estimated to occur between 1 out of 20 and 1 out of 200 births, depending upon the method used in the analysis (10). LINE-1 insertional mutagenesis has resulted in over 120 cases of spontaneous or inherited disease; including diseases such as hemophilia A, cystic fibrosis, and breast cancer; reviewed in Chen et al. (51). Apart from mutations caused by insertions, DNA recombination of chimeric LINE-1 can lead to retrotransposition-mediated deletions, duplications, or rearrangements (52, 53).
Insertional mutagenesis is not the only hazard for our cells, the presence of both sense and antisense promoter within the 5′UTR of LINE-1 elements can activate upstream or downstream transcription [(34, 54, 55); Figure 4]. Among all the transcriptionally active elements present in the human genome, the strength of LINE-1 promoters has been shown to be sequence and context-dependent. Lavie et al. generated 5′ UTR constructs to test observable differences in promoter strength. Although there was no clear link between nucleotide variations and transcriptional activity, deletion of 5′ genomic flanking sequences from the constructs resulted in both enhanced and diminished promoter activity, depending on whether the sequence acted as an enhancer or repressor (32). As for the LINE-1 antisense promoter, Matlik and colleagues established its activity as tissue specific; driving transcription of adjacent genes to yield chimeric transcripts of host genes [Figure 4; (56, 57)].
Not only does the activity from the LINE-1 promoter at insertion sites contribute to aberrant host gene expression; LINE-1 insertions can also create splicing variants, generate mis-spliced or prematurely truncated transcripts, promote transcriptional termination or even promote changes to our epigenome (Figure 4). Splicing of pre-mRNAs is a tightly regulated process as it contributes to proteomic diversity and modulates gene expression. RNA extracted from human Ntera2 and Sk-Br-3 cancer cells, which express high levels of LINE-1 transcripts, identified many functional splice sites within the 5′UTR (58). It was demonstrated that several of these LINE-1 splice variants are capable of undergoing retrotransposition. Upon insertion, these active splice sites can disrupt normal gene expression via alternative splicing of mRNA transcripts (58). Similarly, Alu elements harbor consensus sequences that resemble 5′ and 3′ splice site signals, contributing to modifications in pre-mRNA splicing (59, 60). In addition, the presence of LINE-1 adenosine-rich insertions can give rise to new polyadenylation signals, resulting in transcriptional termination [Figure 4; (61)].
LINE-1 elements not only create changes in splicing; LINE-1 loci that contain premature stop codons may still encode for truncated ORF2 proteins with a retained functional EN domain (62). It was suggested that EN activity of ORF2p may generate nicks independent of retrotransposition and could therefore contribute to double stranded breaks (DSBs) formation (63). In fact, the number of observed LINE-1 induced DSBs was demonstrated to be greater than the predicted numbers of successful insertions (63).
Finally, LINE-1 can also alter the epigenome; methylation of LINE and SINE CpG islands leads to “epigenetic patterning” and subsequent silencing of neighboring gene promoters [(64); Figure 4]. Baylin et al. demonstrated that hypermethylation stemming from human Alu elements can silence tumor suppressor genes (65) which has large implications for their role in cancer. Alternatively, hypomethylation of the LINE-1 promoter was shown to directly activate an alternate transcript of the MET oncogene in bladder tumors; inducing ectopic gene expression and possibly altering disease susceptibility (66). In summary, new TE insertions have been shown to have many profound effects on the genome. A contributor to genomic variation, retrotransposons alter host gene expression at the epigenetic, transcriptional and translational level. Host cells have simultaneously evolved many responses in order to repress TE activity, which we review in the next section.
Cellular Responses to Retrotransposition
Intrinsic immune responses to viral pathogens are co-opted to restrict retroviruses and retrotransposons as well. Given that these retroelements can impact the cell in a myriad of ways, the host genome has coevolved to employ a variety of responses to repress aberrant activity.
LINE-1 Transcriptional Repression
Transcriptional repression is a major mechanism of TE regulation and can be achieved with the deposition of repressive epigenetic modifications. DNA methylation, in the form of 5-methylcytosine (5 mC) and N6-methyladenine (6 mA) are widely used chemical modifications in eukaryotes and higher organisms [Figure 2; (67)]. Waves of hypomethylation during embryogenesis are linked with higher rates of retrotransposition. This effect has been replicated in embryonic stem cells (ESC) and induced pluripotent stem cells (iPSCs) (68–70). In preimplantation mouse embryos, which are exceptionally hypomethylated, genomic integrity is maintained with factors such as histone chaperone chromatin assembly factor 1 (CAF-1) (71). CAF−1 was shown to mediate the replacement of Histone variant 3.3 (H3.3) with H3.1/3.2, which serves as a repressive histone modification and subsequently protects the embryo from retrotransposon activity [Figure 2; (71)]. When histone variant H3.3 was deleted from embryonic stem cells, trimethylation of histone 3 on lysine 9 (H3K9me3) was shown to be reduced at HERV sites, establishing an important link between H3.3 and endogenous retrovirus silencing (72).
Transcriptional silencing of retrotransposons can be induced by DNA methylation or histone modifications, but TEs also harbor binding sites for many transcription factors, enabling context-specific transcriptional regulation. The Kruppel-associated box (KRAB)-containing zinc finger proteins (KZFPs) are key regulators of TE activity—often repressing TEs expressed in early embryos (73, 74). Mechanistically, repression is mediated once the C-terminus tandem array of zinc finger motifs binds to target TE sequences and the KRAB domain recruits and tethers to the cofactor KAP-1 (KRAB associated protein 1). KAP-1 then functions as a scaffold for chromatin modifying complexes such as SETDB1 (Set Domain Bifurcated-1) and H3K9 methyl transferase [Figure 2; (75, 76)].
In order for KAP1 to target a specific retroelement, the associating KRAB-ZFP must evolve to bind to that specific regulatory sequence. This is usually followed by mutations in this sequence, such that retrotransposons evade repression; a true evolutionary arms race (77). A recent study by Trono et al. elucidated the role of KZFPs on TEs during embryonic genome activation (EGA), demonstrating that a large proportion of TE-embedded regulatory sequences have been co-opted to serve as lineage- or tissue-specific enhancers of gene expression (78). Authors observed clustered TE sequences of the evolutionarily recent SVA, LTR5Hs-HERVK, and LTR7-HERVH in human embryonic stem cells (hESC) during EGA. These sequences were shown to strongly associate with members of the Kruppel-like factor (KLF) family of transcription factors, notably KLF4. Clustered regularly spaced short palindrome repeats—interference (CRISPRi) targeting of these TE sequences led to up- and downregulation of genes near the target vicinity. For example, CRISPRi of an LTR5Hs-based enhancer resulted in a significant downregulation of PRODH, a neuron-specific gene located 2 kb downstream from the enhancer. Thus, TEs which possess embedded enhancer sequences could subsequently exert large transcriptional influence in hESCs during EGA. By examining the degree of conservation within the zinc fingerprints, evolutionarily recent human KZFPs were shown to target TE subfamilies of similar ages (78). In sum, these recent studies demonstrate the intricate, coevolutionary dynamic between KZFPs and TEs.
However, there is some evidence indicating that this arms-race hypothesis may be too simplistic (79, 80). Imbeault et al. performed a clustering and aging analysis of various KZFPs to estimate their evolutionary ages. These conserved sequences were aligned against various transposable element subfamilies; many transposable element-KZFP pairs were found to be highly conserved long after the transposable elements lost their ability to mobilize (80). In some cases, KZFPs appear much earlier than their TE target, illustrating a complex co-option model where specific regulatory networks are established (78, 80). This is an example whereby the host facilitates the impact TEs can have by regulating their disruptive capacity. In addition, host cells even use conserved TE sequences to their advantage by setting up tissue-specific transcriptional networks.
Post-transcriptional Regulation of LINE-1 Elements
LINE-1 can be regulated post-transcriptionally by small RNAs, like microRNAs (miRNAs) or PIWI-interacting RNAs (piRNAs). Small RNAs can act via targeted RNA degradation, reviewed in Heras et al. (81) and Mita and Boeke (82). The role of RNA interference (RNAi) effectors in regulating TE transcripts is substantial; RNA-induced silencing complex (RISC) pathways are common cellular processes that utilize endonucleolytic cleavage to degrade TE transcripts (83, 84). Mutations to the Dicer protein, a component of the RISC complex, result in elevated transcription of LINE-1 elements (3). The recent discovery of small, non-coding miRNAs also highlighted their critical regulatory role. It was recently shown that miRNA-128 restricts LINE-1 activity via two mechanisms (Figure 2). It can directly target ORF2 RNA for degradation or target the 3′ UTR sequence of required cofactor TNPO1 (nuclear import factor transportin 1) (85). TNPO1 has been proposed to facilitate transport of LINE-1 RNP complex into the nucleus by binding to nuclear localization signals (85). Another recent study identified a separate cellular target of miRNA128-mediated LINE-1 repression (86). hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1) binds to poly(A) sequences in mRNA to facilitate nuclear shuttling (87, 88). hnRNPA1 has been described to interact with LINE-1 ORF1p within the RNP complex and with TNPO1, through its nuclear localization signal (89, 90). miRNA128 was shown to repress hnRNPA1 by directly binding to the coding sequence of hnRNPA1 mRNA, significantly reducing levels of de novo LINE-1 retrotransposition (86). piRNAs are important repressors of TE activity in the germline (Figure 2). They interact with the PIWI subfamily of Argonaute nucleases and have been shown to cleave TE transcripts in cytoplasm as well as recruit repressive histone modifiers to silence transcription (91). Intact piRNA pathway was demonstrated to be necessary for de novo methylation of LINE-1 transgene in male mice testes (92). If the LINE-1 transcript is not targeted for degradation, the cell will employ other host factors to target downstream complexes within the retrotransposition cycle.
Post-translational Mediated Repression
The LINE-1 RNP complex, a retrotransposition intermediate, is commonly targeted for destabilization and degradation (93). The zinc-finger protein ZAP, in addition to targeting several viral families, has been suggested to colocalize with LINE-1 RNA in cytoplasmic stress granules to promote loss of RNP integrity and inhibit LINE and Alu retrotransposition [Figure 2; (94–96)]. Post-transcriptional modifications of LINE-1 mRNAs within the RNP complex offers another way of restricting mobility. TUT7 (terminal uridyl transferase 7) in cooperation with MOV10, transfers uridine residues to LINE-1 mRNA in the cytoplasm. MOV10, a helicase, displaces ORF1p to allow for cytoplasmic 3′ uridylation—ultimately inhibiting ORF2p RT initiation within the nucleus [Figure 2; (97)]. In addition, the APOBEC family of enzymes, specifically APOBEC3G and APOBEC3F, have been shown to work through a process independent of cytosine deamination to selectively inhibit Alu retrotransposition, possibly by destabilizing the RNP complex [Figure 2; (98)]. APOBEC3B/F also strongly interfere with LINE-1 activity; catalytically inactive APOBEC mutants maintained LINE-1 inhibition, also indicating a deamination-independent mechanism (99, 100), while APOBEC3A has been proposed to localize in the nucleus to deaminate the transiently expressed LINE-1 ssDNA that appears during integration and prevent retrotransposition (101). Even with the plethora of host mechanisms put in place to repress endogenous retroelements, de novo insertions still take place within somatic tissues, with substantial LINE-1 retrotransposition occurring in neural lineages.
LINE-1 in the Developing Brain
A plethora of evidence supports that both endogenous and engineered LINE-1 retrotransposition can occur pre and post-mitotically in the healthy and diseased brain, reviewed in Suarez et al. (102). New retrotransposition events can alter gene expression and ultimately influence cellular phenotype; in the healthy brain this is thought to contribute to neuronal somatic diversification (49, 103). Through the use of an engineered LINE-1 element tagged to a retrotransposition-indicator cassette, retrotransposition events were shown to occur in vitro with adult rat neural progenitor cells (NPCs) and in the brains of mice in vivo (103). This was one of the earliest documented cases of somatic retrotransposition occurrence in vivo (103). A few years later, endogenous LINE-1 mRNA was shown to be detectable in NPCs isolated from human fetal brain cells (47). To investigate endogenous LINE-1 activity, a copy number variant (CNV)-based qPCR assay was performed on genomic DNA extracted from various tissue types of healthy human adults. Interestingly, it was reported that ORF2 content in the hippocampus was consistently higher when compared to heart or liver samples from the same individual (47). Few years later, by using Retrotransposition-capture sequencing (RC-seq), a high throughput sequencing method that targets LINE-1 5′ and 3′ ends, applied to DNA extracted from various tissues again identified significantly higher ORF2 copies and LINE-1 CNV within the hippocampus (49). Separate studies implemented single-cell RC-seq on hippocampal neurons and resulted in an estimated 1.2–13.7 somatic LINE-1 insertions per neuron (104–106), although the frequency of de novo insertions per cell is highly debated (107). The hippocampus, being one of the few brain regions consisting of a neurogenic niche, is ubiquitous with genomic LINE-1 mosaicism. Because LINE-1 retrotransposition events have increased occurrence during neurogenesis (103, 108), it is likely that LINE-1 activity rises in a region like the hippocampus (47). Interestingly, in a recent analysis of 24 hippocampal neurons using RC-seq, whole genome sequencing (WGS) and LINE-1 insertion profiling revealed that somatic insertions which occur during neurodifferentiation in hESCs may occur due to mutations at the Ying Yang 1 (YY1) transcription factor binding site (109). This YYI binding site in the LINE-1 promoter region is known to mediate CpG island methylation and epigenetic repression. Specific loci without an intact YY1 binding site were shown to generate cortical and hippocampal neuron LINE-1 insertions, corroborating an underlying epigenetic mechanism for LINE-1 retrotransposition during neural development (109).
Retrotransposition during neural development may contribute to “genome plasticity” and neuronal diversity by allowing for variation in genomic DNA from cell to cell. By studying the effects of retroelements during neurogenesis, one can examine the early fate choices between different cell lineages (103). Muotri et al. reports a 10-fold increase in LINE-1 promoter during the first 24 h of neuronal induction, consistent with downregulation of the Sox2 promoter (103). It was proposed that subtle changes to LINE-1 promoter methylation may explain the selective activity levels in NPCs; perhaps that the LINE-1 promoter is temporarily released from epigenetic suppression during neurogenesis (47, 49). Indeed, temporal methylation patterning of the LINE-1 promoter during neurogenesis was analyzed in a fairly recent study; researchers applied RC-seq to observe distinct DNA methylation profiles for de novo LINE-1 insertions in an hiPSC line (108). RC-seq performed on cells throughout various timepoints of fibroblast reprogramming and neurodifferentiation identified two well-characterized de novo LINE-1 insertions. General trends showed, on average, ~60% methylated CpG dinucleotides in fibroblasts and mature neurons, while only ~30% were methylated in iPSCs. Surprisingly, they observed a 23% reduction in methylation in day 112 neurons when compared to earlier day 72 neurons. This was followed by a significant 20% increase in methylation at day 156 neurons (108). This study establishes a dynamic temporal patterning of methylation for the LINE-1 promoter during neurodifferentiation. The evidence collected on LINE-1 mobilization in both the developing and adult brain, opens new questions about their contribution to somatic mosaicism, aging and neurological diseases.
Mobile Elements and Neurodegenerative Disorders
There has been an increasing interest in studying endogenous retroelements as their activation has been observed and implicated in a variety of neurological disorders (Table 1). For example, all three HERV-K structural genes (gag, pol, env) have been shown to have increased expression in patients with sporadic ALS when compared to healthy controls (110). ALS is neurodegenerative disease characterized by loss of both upper and lower motor neurons. A number of studies have established the presence of retroviral RT activity in the serum of ALS patients (111, 112). Higher expression of HERV K-Env—a powerful immunopathogenic envelope protein—is observed in cortical pyramidal and spinal neurons in post-mortem brain tissue of ALS patients (110). It is not currently known what triggers expression of HERV-K in adult neurons; however, activation of HERV-K genes was shown to decrease dendritic length, branching, and complexity of transgenic mice motor neurons (110).
Abnormalities in transactive response DNA-binding protein 43 (TDP-43) is likewise observed in the majority of sporadic ALS cases (Table 1). TDP-43 is a dimeric nuclear protein and part of the heterogenous nuclear ribonucleoprotein family (hnRNP) (113). In the CNS, its function is broadly categorized as a regulator of pre and post-transcriptional events as it binds to UG-rich motifs in single-stranded RNA/DNA (113, 114). Crosslinking immunoprecipitation (CLIP-seq) and chromatin immunoprecipitation (ChIP) data exhibits TDP-43 binding broadly to retrotransposon-derived transcripts in human brain tissue and directly to the HERV-K LTR sequence, respectively (110, 115). A study conducted by Lisa Krug et al. addressed whether functionally abnormal TDP-43 expression in Drosophila causes a derepression of retrotransposable elements and if so, whether this contributes to a degenerative phenotype (116). Expression of human TDP-43 (hTDP-43) was shown to induce broad retrotransposon transcript expression in Drosophila neurons and glia. Glial expression of hTDP-43 causes a remarkable reduction of Dicer-2/Argonaute2 mediated silencing while causally inducing DNA-damage mediated cell death (116). In a more recent study, analysis of diseased neuronal nuclei from brain tissue of patients with frontotemporal degeneration ALS (FTD-ALS), provides insight for molecular changes associated with TDP-43 loss. Liu et al. utilized fluorescence-activated cell sorting (FACS) to fractionate diseased neurons followed by ATAC-seq to quantify chromatin accessibility (117). Loss of nuclear TDP-43 was associated with chromatin decondensation around LINE-1 elements. Although it is unclear whether decondensation of LINE-1 elements is a direct result of TDP-43 protein loss, there appeared to be some specificity or preference in decondensation of LINE-1 elements over other repetitive sequences (117). Together, these findings in humans and vertebrate models, suggest that unregulated retroelement expression is somewhat involved in the pathology of ALS, although there is no evidence that it is the primary cause of the syndrome. Potential therapy in the form of antiretroviral RTi is also underway in clinical trials of ALS patients (118, 119).
Sporadic Alzheimer's Disease (SAD) has also been linked to retroelement activation; however, studies have displayed conflicting results on whether LINE-1 sequences are upregulated in patients with SAD (120, 121). “Mosaic genomic recombination events” were observed within the Alzheimer's-related gene, amyloid precursor protein (APP), in neurons of patients with SAD (122). These variants, which lacked intronic sequences, were termed “genomic cDNAs” (gencDNAs). It was hypothesized that they originated from RNA and required endogenous RT activity to insert into double strand breaks. Addition of nucleoside reverse transcriptase inhibitors (nRTi) abacavir (ABC) and azidothymidine (AZT) prevented the production of gencDNAs, further encouraging their therapeutic potential (122). In order to determine the effect of Alzheimer's Tau pathology on TE activity, wild-type and Tau mutant Drosophila were profiled for TE activation including 8 LTR retrotransposons and 4 non-LTR retrotransposons (123). Authors observed a significant increase in expression in three of the 12 TEs assessed in the mutant fly brains (copia, gypsy and het-a) suggesting a possible Tau- associated mechanism for TE activation (123).
Retroelement recombination events are also implicated in early onset Parkinson's Disease (PD). Whole genome sequencing analysis of three families with early onset PD revealed five different structural variations in the PRKN (parkin RBR E3 ubiquitin protein ligase) gene. Structural variation formation is proposed to occur with non-allelic homologous recombination. LTR and non-LTR retrotransposon sequences were identified within two kilobases of the deletion break point, suggesting that the deletions may have originated due to retrotransposition events (124). The link between retroelement activation in neurodegenerative disorders is present but not fully established. We consistently see an upregulation of retroelements in the diseased state, but we do not fully understand how this is initiated or whether it directly contributes to disease pathology. The following section will review the evidence which supports endogenous retroelements as initiators of inflammation and subsequent inflammatory responses in the diseased state.
TEs and Inflammation
The immune system protects against viral infections through coordinated innate and adaptive immune responses and while the contribution of innate immunity to anti-viral defenses has been extensively studied, little is known about the contribution of transposable elements to immune responses exempt of viral infection. When viral DNA is present in the cytoplasm, it triggers activation of the cGAS-STING pathway, subsequently producing interferons to initiate an inflammatory response (Figure 5). The interferon responses elicited during the targeting of virus-infected cells may be mechanistically linked to deregulating retroelement production (125). There is mounting evidence that endogenous retroelements play a large role in initiating neuroinflammation. Endogenous nucleic acid detection by the innate immune system underlies many autoimmune diseases (126) When there is an inflammatory response but no viral infection, what is the role of retroelements? More specifically, how are these engaging the pathophysiological pathways leading to features of the disease pathology?
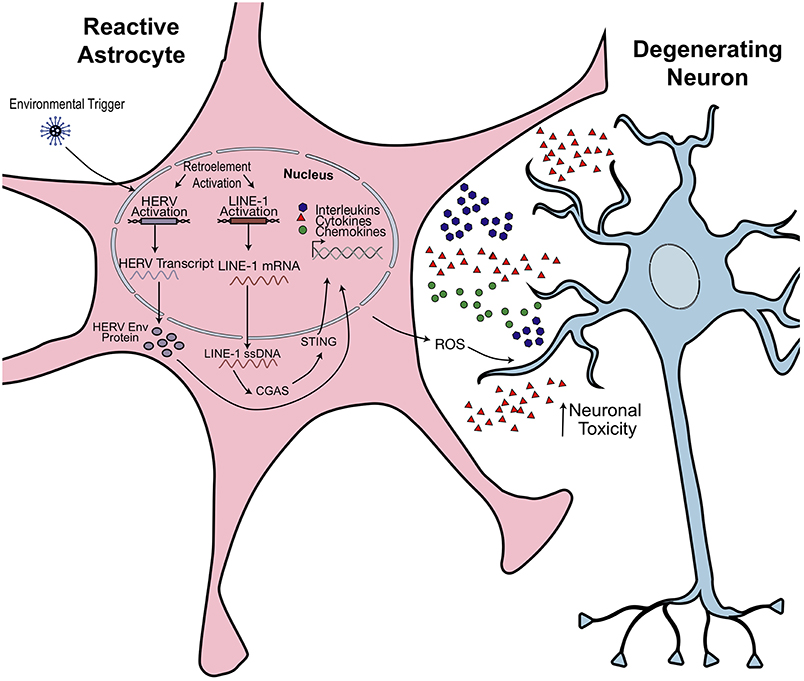
Figure 5. Hypothesized model of retroelement-mediated neuroinflammation. Environmental or cellular triggers may activate retrotransposon transcription. Production of pathogenic HERV Envelope proteins (Env) or cytoplasmic accumulation of LINE-1 ssDNA activates an innate immune response. Astrocytes become activated, releasing proinflammatory cytokines and reactive oxidative species (ROS). This has toxic effects on neighboring neurons and could promote morphological and synaptogenic defects, subsequently promoting neuropathophysiological effects. Figure is adapted from Thomas et al. (38).
There is increasing interest in studying endogenous retroelements as contributors to a variety of inflammatory and neurodegenerative disorders. Diseases such as MS and Aicardi-Goutières syndrome (AGS) have retrotransposon intermediates linked as key effectors of inflammation (16, 38, 127). In addition, there are now several studies that link aging, TEs, and inflammation.
Aging
Mechanisms repressing LINE-1 activity are shown to be less efficient during the aging process. A recent study by De Cecco and colleagues demonstrates increased LINE-1 transcript levels in senescent cells. The accumulation of the cytoplasmic LINE-1 cDNA drives expression of the senescence- associated secretory phenotype (SASP). A type-I interferon (IFN) response is typical with age-associated inflammation in several tissues (Table 1). Liver and adipose tissue of 26-month-old mice showed significant increase in LINE-1 mRNA expression when compared to mice at 5 months. IFN-I and SASP response genes were assessed by RT-qPCR and showed the same trend. This response is antagonized through the use of reverse-transcriptase inhibitors (RTi). Mice treated with RTis demonstrate a reduced IFN response and associated inflammation (128). However, RTis have exhibited an intrinsic anti-inflammatory property, leading some to believe the effects as non-specific (129). The effects of LINE-1 activity in aging were also examined in the mono-ADP-ribosylase/deacetylase protein SIRT6 (Silent Mating Type Information Regulation 2 Homolog 6) deficient mice. SIRT6 KO mice display a severe aging phenotype, with a lifespan of 35 days (130). SIRT6 KO mice demonstrate high levels of LINE-1 expression due to SIRT6's repressive role in ribosylating KAP1 (131, 132). Without SIRT6, LINE-1 cytoplasmic DNA levels increased, triggering cGAS (cyclic GMP-AMP synthase) -mediated IFN response. Treatment with the RTi inhibitors lamivudine and stavudine, significantly expanded the lifespan of the mice while also improving body mass, mobility and behavior phenotypes (130). These results further implicate LINE-1 as a contributor to the pathology of age-related diseases. Although further investigation is needed regarding the mechanisms in which new L1 copies are generated in the cytoplasm, these novel data encourage the potential therapeutic use of RTis for various age-associated conditions.
Multiple Sclerosis
Following the original identification of retroviral-like particles (16), additional research revealed increased RT activity and HERV protein production with Herpesviridae stimulation of various human cells in vitro (133–136). MS is an hyperinflammatory, demyelinating disease of the CNS with no known cure. The disease is characterized by lesions in white matter that lead to blood-brain barrier breakdown and axonal disruption down the spinal cord (137). In addition to genetic and environmental factors, expression of HERVs is now considered a risk factor for MS disease progression (135). The retroviral-like particles isolated from cell culture supernatants of MS patient samples, referred to as MS-associated retroviral agent (MSRV), were demonstrated to originate from HERV elements, as mentioned previously (17, 18, 138). Herpes virus is suggested to upregulate HERV-W expression, which encodes for its own immunopathogenic envelope protein (Env). In vitro stimulation with HERV-W Env proteins displays activation of innate immune responses through pattern recognition receptors TLR4 and CD14, leading to considerable proinflammatory cytokine production (134). Peripheral blood mononuclear cells, isolated from relapse-remitted MS patients and stimulated with the MSRV Env protein, induces elevated IFNγ, IL-6 and TNF- α expression (134, 139). Clinical trials performed on MS patients with the antiretroviral integrase inhibitor, raltegravir, failed to see reduction in lesion count, progression, or inflammatory cytokine levels, suggesting HERV W-Env protein production is not targeted with this integrase (140).
Systemic Lupus Erythematosus
Elevated HERV transcription has been implicated in systemic lupus erythematosus (SLE) pathogenesis. HERV-E mRNA expression levels were found to be higher in lupus CD4+ T cells than in cells from healthy controls (141) but the full contribution of HERV activity to SLE etiology is not known. Deletions in the genes encoding for the KZFP, SNERV1/2 (suppressor of non-ecotropic ERV-1/2) resulted in a 2-fold increase in gene expression of six genes directly overlapping a non-ecotropic ERV sequence (NEERV) (127). The NEERV envelope glycoprotein gp70 is a major immunoantigen and promotes nephritis in murine models (142). SNERV1/2 bind to the gp70-associated loci, Sgp3, and recruit KAP1 to repress transcription. SNERV deletions in New Zealand Black mice resulted in elevated NEERV transcripts and gp70 expression. These results indicate that defects in HERV repression may promote human lupus pathogenesis (127).
Autism Spectrum Disorders
Activity of LINE-1 elements have additionally been implicated in many Autism Spectrum Disorders (ASD) phenotypes. Researchers have found a reduction of methylation and an increase in LINE-1 expression in ASD post-mortem brains (143, 144). ASD is a developmental disorder that impairs communication and behavior; however, little is known about the etiology of the disease. At the cellular level, researchers have found that individuals with ASD frequently show widespread inflammation and elevated brain cytokine expression. Additionally, a growing body of evidence supports the view that a chronic inflammation may contribute to autism symptomatology, with active neuroinflammatory processes being found throughout the brain in both cerebral cortex and cerebellum of patients with autism (145–148). Although new LINE-1 insertions seem to occur frequently in neurons, little is known about the contribution of this element in glial cells. It is becoming increasingly evident that under pathological conditions, there is a non-cell autonomous effect in the CNS, in which glial cells are as vulnerable as neurons (149). Astrocytes are often indicated as the contributors to disease phenotypes and in some instances, the disease initiators (150–152). Accumulating evidence links the cytokine dysregulation and persistent inflammatory phenotypes seen in mice and iPSC-derived models with astrocyte functional abnormalities [Figure 5; (153, 154)]. Indeed, ASD-derived astrocytes secrete elevated cytokines such as interleukin-6 (IL-6), which may interfere with proper neuronal development (155).
Rett Syndrome
Rett Syndrome (RTT), once considered part of ASD, is an X-linked progressive neurodevelopmental disorder with autistic features, characterized predominantly by various mutations in the methyl CpG binding protein-2 (MeCP2) (156). Post-mortem brain tissue samples analyzed showed higher genomic LINE-1 ORF2 sequences in RTT patients when compared to controls (157). Muotri et al. demonstrated that MeCP2 loss of function increases susceptibility for LINE-1 insertions because the 5′UTR sequence within the LINE-1 promoter are targets of MeCP2-mediated transcriptional repression [Figure 2; (157)]. Conditioned media taken from RTT mutant astrocytes had adverse effects on wild type mouse neurons. After just 24 h, neurons grown in RTT astrocyte conditioned media had significantly smaller soma sizes, shorter neurites and less terminal ends (158). Re-expression of MeCP2 specifically in astrocytes improved locomotion, anxiety levels, and prolonged the lifespan of globally deficient MeCP2 mice. Even more, restoration of MeCP2 in astrocytes restored VGlut1 levels and dendritic morphology of neurons in vivo (159). More research must be conducted to confirm whether the abnormally high presence of LINE-1 retroelements seen in RTT contributes to: (1) the inflammatory response seen in astrocytes and (2) disease progression.
Aicardi-Goutières Syndrome
Indeed, it has been found that the accumulation of LINE-1 copies in neurodevelopmental diseases promote inflammatory effects in astrocytes, as is in the case of AGS (38). AGS is a progressive inflammatory disorder that affects newborns and results in severe mental and physical handicap as well as greatly reduced lifespan. AGS can arise from mutations in three-prime repair exonuclease 1 (TREX1). Mutations in TREX1—which functions to degrade dsDNA/ssDNA—result in significant cytoplasmic accumulation of DNA species, which are then sensed as viral or “non-self.” This leads innate immune responses, such as the induction of IFN [Figure 5; (160)]. Thomas et al. demonstrated that a majority of those DNA species consist of cytoplasmic LINE-1 ssDNA. TREX-1 deficient NPCs expressed 70% more LINE-1Hs elements (38). Chronic RTi treatment of TREX1 deficient cell lines was shown to reduce cytoplasmic ssDNA to near control levels as well as improve neurite growth and decrease IFN secretion from astrocytes (38). In a 2018 phase II clinical trial for AGS patients, combinations of RTis were administered for 12 months to observe changes in interferon signaling (161). Across all patients who completed the study, the median interferon score dropped from 9.66 to 5.33. Interferon levels in serum and plasma were also reduced. Strikingly, global interferon-stimulated gene expression (ISG) decreased after 12 months of treatment but then returned to pre-treatment levels 6 months after discontinuing RTi treatment (161). Thus, RTi treatment can prove to be a promising therapeutic alternative for pathologies in which inflammation is a common denominator.
Conclusion
Reviewed here are recent reports which highlight aberrant TE activation as contributors to a variety of neurological, neurodegenerative, and autoimmune pathologies. Activation of retroelements confer genomic and cellular instability as TEs can disrupt coding regions, rewire transcriptional networks, and modify epigenetic and post-transcriptional regulation of gene expression. Indeed, retrotransposons have evaded evolutionary attempts at repression and contribute to somatic mosaicism. In the diseased state, where repression or regulation of retrotransposons is diminished, expression of endogenous nucleic acids are upregulated, promoting a response from the host, similar to the one that occurs upon a viral infection or to environmental triggers. In most cases, the cell will initiate an interferon response. Persistent inflammation leads to functional abnormalities and disease phenotypes; therefore, we speculate that retroelement misregulation impacts human pathogenesis to a larger extent than previously thought.
Author Contributions
AS wrote the manuscript with help from AM and ARM.
Funding
This work was supported by grants from the National Institutes of Health through the NIH R01MH094753. The work was also supported by the California Institute for Regenerative Medicine (CIRM) award DISC1-08825. AM was supported by a NARSAD Young Investigator grant number 25400.
Conflict of Interest Statement
ARM is a co-founder and has equity interest in TISMOO, a company dedicated to genetic analysis and brain organoid modeling focusing on therapeutic applications customized for autism spectrum disorder and other neurological disorders with genetic origins. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. McClintock B. Controlling elements and the gene. Cold Spring Harb Symp Quant Biol. (1956) 21:197–216. doi: 10.1101/SQB.1956.021.01.017
2. Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. (2011) 12:615–27. doi: 10.1038/nrg3030
3. Goodier JL, Kazazian HH Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. (2008) 135:23–35. doi: 10.1016/j.cell.2008.09.022
4. Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. International Human Genome Sequencing. Initial sequencing and analysis of the human genome. Nature. (2001) 409:860–921. doi: 10.1038/35057062
5. Macia A, Blanco-Jimenez E, Garcia-Perez JL. Retrotransposons in pluripotent cells: impact and new roles in cellular plasticity. Biochim Biophys Acta. (2015) 1849:417–26. doi: 10.1016/j.bbagrm.2014.07.007
6. Garcia-Perez JL, Widmann TJ, Adams IR. The impact of transposable elements on mammalian development. Development. (2016) 143:4101–14. doi: 10.1242/dev.132639
7. Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Curr Genomics. (2010) 11:115–28. doi: 10.2174/138920210790886871
8. Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. (2014) 48:1–27. doi: 10.1146/annurev-genet-120213-092412
9. Smit AF. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. (1993) 21:1863–72. doi: 10.1093/nar/21.8.1863
10. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. (2009) 10:691–703. doi: 10.1038/nrg2640
11. Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. (2003) 35:41–8. doi: 10.1038/ng1223
12. Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, et al. Demystified. Human endogenous retroviruses. Mol Pathol. (2003) 56:11–8. doi: 10.1136/mp.56.1.11
13. Mager DL, Stoye JP. Mammalian endogenous retroviruses. Microbiol Spectr. (2015) 3:MDNA3-0009-2014. doi: 10.1128/microbiolspec.MDNA3-0009-2014
14. Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci USA. (2016) 113:E2326–34. doi: 10.1073/pnas.1602336113
15. Grandi N, Tramontano E. HERV envelope proteins: physiological role and pathogenic potential in cancer and autoimmunity. Front Microbiol. (2018) 9:462. doi: 10.3389/fmicb.2018.00462
16. Perron H, Suh M, Lalande B, Gratacap B, Laurent A, Stoebner P, et al. Herpes simplex virus ICP0 and ICP4 immediate early proteins strongly enhance expression of a retrovirus harboured by a leptomeningeal cell line from a patient with multiple sclerosis. J Gen Virol. (1993) 74 (Pt 1):65–72. doi: 10.1099/0022-1317-74-1-65
17. Perron H, Garson JA, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, et al. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. The collaborative research group on Multiple Sclerosis. Proc Natl Acad Sci USA. (1997) 94:7583–8. doi: 10.1073/pnas.94.14.7583
18. Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, et al. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol. (1999) 73:1175–85.
19. Kury P, Nath A, Creange A, Dolei A, Marche P, Gold J, et al. Human endogenous retroviruses in neurological diseases. Trends Mol Med. (2018) 24:379–94. doi: 10.1016/j.molmed.2018.02.007
20. Boissinot S, Entezam A, Furano AV. Selection against deleterious LINE-1-containing loci in the human lineage. Mol Biol Evol. (2001) 18:926–35. doi: 10.1093/oxfordjournals.molbev.a003893
21. Boissinot S, Furano AV. Adaptive evolution in LINE-1 retrotransposons. Mol Biol Evol. (2001) 18:2186–94. doi: 10.1093/oxfordjournals.molbev.a003765
22. Ostertag EM, Kazazian HH Jr. Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. (2001) 11:2059–2065. doi: 10.1101/gr.205701
23. Pavlicek A, Paces J, Zika R, Hejnar J. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: implications for retrotransposition and pseudogene detection. Gene. (2002) 300:189–94. doi: 10.1016/S0378-1119(02)01047-8
24. Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci USA. (2003) 100:5280–5. doi: 10.1073/pnas.0831042100
25. Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol Cell Biol. (1990) 10:6718–29. doi: 10.1128/MCB.10.12.6718
26. Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. (2001) 21:1973–85. doi: 10.1128/MCB.21.6.1973-1985.2001
27. Denli AM, Narvaiza I, Kerman BE, Pena M, Benner C, Marchetto MC, et al. Primate-specific ORF0 contributes to retrotransposon-mediated diversity. Cell. (2015) 163:583–93. doi: 10.1016/j.cell.2015.09.025
28. Martin SL, Bushman FD. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol Cell Biol. (2001) 21:467–75. doi: 10.1128/MCB.21.2.467-475.2001
29. Martin SL, Cruceanu M, Branciforte DP, Wai-Lun Li, Kwok SC, Hodges RS, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol. (2005) 348:549–61. doi: 10.1016/j.jmb.2005.03.003
30. Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. (1991) 254:1808–1810. doi: 10.1126/science.1722352
31. Feng Q, Moran JV, Kazazian HH Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. (1996) 87:905–16. doi: 10.1016/S0092-8674(00)81997-2
32. Lavie L, Maldener E, Brouha B, Meese EU, Mayer J. The human L1 promoter: variable transcription initiation sites and a major impact of upstream flanking sequence on promoter activity. Genome Res. (2004) 14:2253–60. doi: 10.1101/gr.2745804
33. Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. (2010) 6:e1001150. doi: 10.1371/journal.pgen.1001150
34. Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. (2016) 351:aac7247. doi: 10.1126/science.aac7247
35. Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci USA. (1997) 94:1872–7. doi: 10.1073/pnas.94.5.1872
36. Cost GJ, Boeke JD. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry. (1998) 37:18081–93. doi: 10.1021/bi981858s
37. Monot C, Kuciak M, Viollet S, Mir AA, Gabus C, Darlix JL, et al. The specificity and flexibility of l1 reverse transcription priming at imperfect T-tracts. PLoS Genet. (2013) 9:e1003499. doi: 10.1371/journal.pgen.1003499
38. Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, et al. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell. (2017) 21:319–31 e318. doi: 10.1016/j.stem.2017.07.009
39. Benitez-Guijarro M, Lopez-Ruiz C, Tarnauskaite Z, Murina O, Mian Mohammad M, Williams TC, et al. RNase H2, mutated in Aicardi-Goutieres syndrome, promotes LINE-1 retrotransposition. EMBO J. (2018) 37:e98506. doi: 10.15252/embj.201798506
40. Cheung VG, Spielman RS, Ewens KG, Weber TM, Morley M, Burdick JT. Mapping determinants of human gene expression by regional and genome-wide association. Nature. (2005) 437:1365–9. doi: 10.1038/nature04244
41. Graham T, Boissinot S. The genomic distribution of L1 elements: the role of insertion bias and natural selection. J Biomed Biotechnol. (2006) 2006:75327. doi: 10.1155/JBB/2006/75327
42. Flasch DA, Macia A, Sanchez L, Ljungman M, Heras SR, Garcia-Perez JL, et al. Genome-wide de novo L1 retrotransposition connects endonuclease activity with replication. Cell. (2019) 177:837–51.e28. doi: 10.1016/j.cell.2019.02.050
43. Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, Zine El Aabidine A, et al. The landscape of L1 retrotransposons in the human genome is shaped by pre-insertion sequence biases and post-insertion selection. Mol Cell. (2019) 74:555–70.e7. doi: 10.1016/j.molcel.2019.02.036
44. Lutz SM, Vincent BJ, Kazazian HH Jr, Batzer MA, Moran JV. Allelic heterogeneity in LINE-1 retrotransposition activity. Am J Hum Genet. (2003) 73:1431–7. doi: 10.1086/379744
45. Konkel MK, Wang J, Liang P, Batzer MA. Identification and characterization of novel polymorphic LINE-1 insertions through comparison of two human genome sequence assemblies. Gene. (2007) 390:28–38. doi: 10.1016/j.gene.2006.07.040
46. Philippe C, Vargas-Landin DB, Doucet AJ, van Essen D, Vera-Otarola J, Kuciak M, et al. Activation of individual L1 retrotransposon instances is restricted to cell-type dependent permissive loci. Elife. (2016) 5:e13926. doi: 10.7554/eLife.13926
47. Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. (2009) 460:1127–31. doi: 10.1038/nature08248
48. Belancio VP, Roy-Engel AM, Pochampally RR, Deininger P. Somatic expression of LINE-1 elements in human tissues. Nucleic Acids Res. (2010) 38:3909–22. doi: 10.1093/nar/gkq132
49. Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. (2011) 479:534–7. doi: 10.1038/nature10531
50. Marchi E, Kanapin A, Magiorkinis G, Belshaw R. Unfixed endogenous retroviral insertions in the human population. J Virol. (2014) 88:9529–37. doi: 10.1128/JVI.00919-14
51. Chen JM, Stenson PD, Cooper DN, Ferec C. A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease. Hum Genet. (2005) 117:411–27. doi: 10.1007/s00439-005-1321-0
52. Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. (2002) 110:315–25. doi: 10.1016/S0092-8674(02)00828-0
53. Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. (2007) 8:272–85. doi: 10.1038/nrg2072
54. Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. (2002) 79:628–34. doi: 10.1006/geno.2002.6758
55. Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol. (2011) 31:300–16. doi: 10.1128/MCB.00561-10
56. Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. (2006) 2006:71753. doi: 10.1155/JBB/2006/71753
57. Weber B, Kimhi S, Howard G, Eden A, Lyko F. Demethylation of a LINE-1 antisense promoter in the cMet locus impairs Met signalling through induction of illegitimate transcription. Oncogene. (2010) 29:5775–84. doi: 10.1038/onc.2010.227
58. Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. (2006) 34:1512–21. doi: 10.1093/nar/gkl027
59. Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. (2007) 13:1603–8. doi: 10.1261/rna.682507
60. Shen S, Lin L, Cai JJ, Jiang P, Kenkel EJ, Stroik MR, et al. Widespread establishment and regulatory impact of Alu exons in human genes. Proc Natl Acad Sci USA. (2011) 108:2837–42. doi: 10.1073/pnas.1012834108
61. Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. (2003) 35:363–6. doi: 10.1038/ng1269
62. Kines KJ, Sokolowski M, deHaro DL, Christian CM, Belancio VP. Potential for genomic instability associated with retrotranspositionally-incompetent L1 loci. Nucleic Acids Res. (2014) 42:10488–502. doi: 10.1093/nar/gku687
63. Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J Mol Biol. (2006) 357:1383–93. doi: 10.1016/j.jmb.2006.01.089
64. Estecio MR, Gallegos J, Dekmezian M, Lu Y, Liang S, Issa JP. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. (2012) 10:1332–42. doi: 10.1158/1541-7786.MCR-12-0351
65. Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. (1998) 72:141–96. doi: 10.1016/S0065-230X(08)60702-2
66. Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. (2010) 6:e1000917. doi: 10.1371/journal.pgen.1000917
67. Deniz O, Frost JM, Branco MR. Author correction: regulation of transposable elements by DNA modifications. Nat Rev Genet. (2019) 20:432. doi: 10.1038/s41576-019-0117-3
68. Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. (2007) 16:1569–77. doi: 10.1093/hmg/ddm105
69. Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. (2010) 466:769–73. doi: 10.1038/nature09209
70. Klawitter S, Fuchs NV, Upton KR, Munoz-Lopez M, Shukla R, Wang J, et al. Author correction: reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun. (2018) 9:5398. doi: 10.1038/s41467-018-07917-0
71. Hatanaka Y, Inoue K, Oikawa M, Kamimura S, Ogonuki N, Kodama EN, et al. Histone chaperone CAF-1 mediates repressive histone modifications to protect preimplantation mouse embryos from endogenous retrotransposons. Proc Natl Acad Sci USA. (2015) 112:14641–6. doi: 10.1073/pnas.1512775112
72. Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. (2015) 522:240–4. doi: 10.1038/nature14345
73. Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. (2017) 18:71–86. doi: 10.1038/nrg.2016.139
74. Guo G, von Meyenn F, Rostovskaya M, Clarke J, Dietmann S, Baker D, et al. Correction: epigenetic resetting of human pluripotency (10.1242/dev.146811). Development. (2018) 145:dev166397. doi: 10.1242/dev.166397
75. Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mob DNA. (2015) 6:17. doi: 10.1186/s13100-015-0050-8
76. Yang P, Wang Y, Macfarlan TS. The role of KRAB-ZFPs in transposable element repression and mammalian evolution. Trends Genet. (2017) 33:871–81. doi: 10.1016/j.tig.2017.08.006
77. Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. (2014) 516:242–5. doi: 10.1038/nature13760
78. Pontis J, Planet E, Offner S, Turelli P, Duc J, Coudray A, et al. Hominoid-specific transposable elements and KZFPs facilitate human embryonic genome activation and control transcription in naive human ESCs. Cell Stem Cell. (2019) 24:724–35.e5. doi: 10.1016/j.stem.2019.03.012
79. Ecco G, Imbeault M, Trono D. KRAB zinc finger proteins. Development. (2017) 144:2719–29. doi: 10.1242/dev.132605
80. Imbeault M, Helleboid PY, Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. (2017) 543:550–4. doi: 10.1038/nature21683
81. Heras SR, Macias S, Caceres JF, Garcia-Perez JL. Control of mammalian retrotransposons by cellular RNA processing activities. Mob Genet Elements. (2014) 4:e28439. doi: 10.4161/mge.28439
82. Mita P, Boeke JD. How retrotransposons shape genome regulation. Curr Opin Genet Dev. (2016) 37:90–100. doi: 10.1016/j.gde.2016.01.001
83. Yang N, Kazazian HH Jr. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. (2006) 13:763–71. doi: 10.1038/nsmb1141
84. Bodak M, Yu J, Ciaudo C. Regulation of LINE-1 elements by miR-128 is not conserved in mouse embryonic stem cells. Front Genet. (2018) 9:683. doi: 10.3389/fgene.2018.00683
85. Idica A, Sevrioukov EA, Zisoulis DG, Hamdorf M, Daugaard I, Kadandale P, et al. MicroRNA miR-128 represses LINE-1 (L1) retrotransposition by down-regulating the nuclear import factor TNPO1. J Biol Chem. (2017) 292:20494–508. doi: 10.1074/jbc.M117.807677
86. Fung L, Guzman H, Sevrioukov E, Idica A, Park E, Bochnakian A, et al. miR-128 restriction of LINE-1 (L1) retrotransposition is dependent on targeting hnRNPA1 mRNA. Int J Mol Sci. (2019) 20:1955. doi: 10.3390/ijms20081955
87. Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. (1992) 355:730–2. doi: 10.1038/355730a0
88. Izaurralde E, Jarmolowski A, Beisel C, Mattaj IW, Dreyfuss G, Fischer U. A role for the M9 transport signal of hnRNP A1 in mRNA nuclear export. J Cell Biol. (1997) 137:27–35. doi: 10.1083/jcb.137.1.27
89. Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. (1995) 129:551–60. doi: 10.1083/jcb.129.3.551
90. Goodier JL, Cheung LE, Kazazian HH Jr. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res. (2013) 41:7401–19. doi: 10.1093/nar/gkt512
91. Toth KF, Pezic D, Stuwe E, Webster A. The piRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol. (2016) 886:51–77. doi: 10.1007/978-94-017-7417-8_4
92. Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, et al. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci USA. (2017) 114:E5635–E5644. doi: 10.1073/pnas.1701069114
93. Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. (2005) 14:3237–48. doi: 10.1093/hmg/ddi354
94. Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr. LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. (2007) 27:6469–6483. doi: 10.1128/MCB.00332-07
95. Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH Jr. The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. (2015) 11:e1005252. doi: 10.1371/journal.pgen.1005252
96. Moldovan JB, Moran JV. The zinc-finger antiviral protein ZAP inhibits LINE and alu retrotransposition. PLoS Genet. (2015) 11:e1005121. doi: 10.1371/journal.pgen.1005121
97. Warkocki Z, Krawczyk PS, Adamska D, Bijata K, Garcia-Perez JL, Dziembowski A. Uridylation by TUT4/7 restricts retrotransposition of human LINE-1s. Cell. (2018) 174:1537–48 e1529. doi: 10.1016/j.cell.2018.07.022
98. Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. (2007) 390:199–205. doi: 10.1016/j.gene.2006.08.032
99. Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. (2006) 281:22161–72. doi: 10.1074/jbc.M601716200
100. Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. (2006) 281:16837–41. doi: 10.1074/jbc.M602367200
101. Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV. APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife. (2014) 3:e02008. doi: 10.7554/eLife.02008
102. Suarez NA, Macia A, Muotri AR. LINE-1 retrotransposons in healthy and diseased human brain. Dev Neurobiol. (2018) 78:434–55. doi: 10.1002/dneu.22567
103. Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. (2005) 435:903–10. doi: 10.1038/nature03663
104. Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. (2012) 151:483–96. doi: 10.1016/j.cell.2012.09.035
105. Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. (2015) 161:228–39. doi: 10.1016/j.cell.2015.03.026
106. Erwin JA, Paquola ACM, Singer T, Gallina I, Novotny M, Quayle C, et al. Author correction: L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat Neurosci. (2018) 21:1016. doi: 10.1038/s41593-018-0131-3
107. Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. (2010) 33:345–54. doi: 10.1016/j.tins.2010.04.001
108. Salvador-Palomeque C, Sanchez-Luque FJ, Fortuna PRJ, Ewing AD, Wolvetang EJ, Richardson SR, et al. Dynamic methylation of an L1 transduction family during reprogramming and neurodifferentiation. Mol Cell Biol. (2019) 39(7). doi: 10.1128/MCB.00499-18
109. Sanchez-Luque FJ, Kempen MHC, Gerdes P, Vargas-Landin DB, Richardson SR, Troskie R-L, et al. LINE-1 evasion of epigenetic repression in humans. Mol Cell. (2019) 75:P590–604.E12. doi: 10.1016/j.molcel.2019.05.024
110. Li W, Lee MH, Henderson L, Tyagi R, Bachani M, Steiner J, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. (2015) 7:307ra153. doi: 10.1126/scitranslmed.aac8201
111. Andrews WD, Tuke PW, Al-Chalabi A, Gaudin P, Ijaz S, Parton MJ, et al. Detection of reverse transcriptase activity in the serum of patients with motor neurone disease. J Med Virol. (2000) 61:527–532. doi: 10.1002/1096-9071(200008)61:4<527::AID-JMV17>3.0.CO;2-A
112. Steele AJ, Al-Chalabi A, Ferrante K, Cudkowicz ME, Brown RH Jr, Garson JA. Detection of serum reverse transcriptase activity in patients with ALS and unaffected blood relatives. Neurology. (2005) 64:454–8. doi: 10.1212/01.WNL.0000150899.76130.71
113. Hergesheimer RC, Chami AA, de Assis DR, Vourc'h P, Andres CR, Corcia P, et al. The debated toxic role of aggregated TDP-43 in amyotrophic lateral sclerosis: a resolution in sight? Brain. (2019) 142:1176–94. doi: 10.1093/brain/awz078
114. Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. (2009) 37:1799–808. doi: 10.1093/nar/gkp013
115. Li W, Jin Y, Prazak L, Hammell M, Dubnau J. Transposable elements in TDP-43-mediated neurodegenerative disorders. PLoS ONE. (2012) 7:e44099. doi: 10.1371/journal.pone.0044099
116. Krug L, Chatterjee N, Borges-Monroy R, Hearn S, Liao WW, Morrill K, et al. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. (2017) 13:e1006635. doi: 10.1371/journal.pgen.1006635
117. Liu EY, Russ J, Cali CP, Phan JM, Amlie-Wolf A, Lee EB. Loss of nuclear TDP-43 is associated with decondensation of LINE retrotransposons. Cell Rep. (2019) 27:1409–21 e1406. doi: 10.1016/j.celrep.2019.04.003
118. Douville R, Liu J, Rothstein J, Nath A. Identification of active loci of a human endogenous retrovirus in neurons of patients with amyotrophic lateral sclerosis. Ann Neurol. (2011) 69:141–51. doi: 10.1002/ana.22149
119. Alfahad T, Nath A. Retroviruses and amyotrophic lateral sclerosis. Antiviral Res. (2013) 99:180–7. doi: 10.1016/j.antiviral.2013.05.006
120. Bollati V, Galimberti D, Pergoli L, Dalla Valle E, Barretta F, Cortini F, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun. (2011) 25:1078–83. doi: 10.1016/j.bbi.2011.01.017
121. Protasova MS, Gusev FE, Grigorenko AP, Kuznetsova IL, Rogaev EI, Andreeva TV. Quantitative analysis of L1-retrotransposons in Alzheimer's disease and aging. Biochemistry. (2017) 82:962–71. doi: 10.1134/S0006297917080120
122. Lee MH, Siddoway B, Kaeser GE, Segota I, Rivera R, Romanow WJ, et al. Publisher correction: somatic APP gene recombination in Alzheimer's disease and normal neurons. Nature. (2019) 566:E6. doi: 10.1038/s41586-019-0905-0
123. Guo C, Jeong HH, Hsieh YC, Klein HU, Bennett DA, De Jager PL, et al. Tau activates transposable elements in Alzheimer's disease. Cell Rep. (2018) 23:2874–80. doi: 10.1016/j.celrep.2018.05.004
124. Bravo P, Darvish H, Tafakhori A, Azcona LJ, Johari AH, Jamali F, et al. Molecular characterization of PRKN structural variations identified through whole-genome sequencing. Mol Genet Genomic Med. (2018) 6:1243–8. doi: 10.1002/mgg3.482
125. Macia A, Tejwani L, Mesci P, Muotri A, Garcia-Perez JL. Activity of retrotransposons in stem cells and differentiated cells. In: Cristofari G, editor. Human Retrotransposons in Health and Disease. Cham: Springer (2017). doi: 10.1007/978-3-319-48344-3_6
126. Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol. (2014) 15:415–22. doi: 10.1038/ni.2872
127. Treger RS, Pope SD, Kong Y, Tokuyama M, Taura M, Iwasaki A. The lupus susceptibility locus Sgp3 encodes the suppressor of endogenous retrovirus expression SNERV. Immunity. (2019) 50:334–47 e339. doi: 10.1016/j.immuni.2018.12.022
128. De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. (2019) 566:73–8. doi: 10.1038/s41586-018-0784-9
129. Fowler BJ, Gelfand BD, Kim Y, Kerur N, Tarallo V, Hirano Y, et al. Nucleoside reverse transcriptase inhibitors possess intrinsic anti-inflammatory activity. Science. (2014) 346:1000–3. doi: 10.1126/science.1261754
130. Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab. (2019) 29:871–85 e875. doi: 10.1016/j.cmet.2019.02.014
131. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. (2006) 124:315–29. doi: 10.1016/j.cell.2005.11.044
132. Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. (2014) 5:5011. doi: 10.1038/ncomms6011
133. Christensen T, Dissing Sorensen P, Riemann H, Hansen HJ, Munch M, Haahr S, et al. Molecular characterization of HERV-H variants associated with multiple sclerosis. Acta Neurol Scand. (2000) 101:229–38. doi: 10.1034/j.1600-0404.2000.101004229.x
134. Rolland A, Jouvin-Marche E, Viret C, Faure M, Perron H, Marche PN. The envelope protein of a human endogenous retrovirus-W family activates innate immunity through CD14/TLR4 and promotes Th1-like responses. J Immunol. (2006) 176:7636–44. doi: 10.4049/jimmunol.176.12.7636
135. Perron H, Bernard C, Bertrand JB, Lang AB, Popa I, Sanhadji K, et al. Endogenous retroviral genes, herpesviruses and gender in multiple sclerosis. J Neurol Sci. (2009) 286:65–72. doi: 10.1016/j.jns.2009.04.034
136. Gottle P, Forster M, Gruchot J, Kremer D, Hartung HP, Perron H, et al. Rescuing the negative impact of human endogenous retrovirus envelope protein on oligodendroglial differentiation and myelination. Glia. (2019) 67:160–70. doi: 10.1002/glia.23535
137. Dendrou CA, Fugger L, Friese MA. Immunopathology of multiple sclerosis. Nat Rev Immunol. (2015) 15:545–58. doi: 10.1038/nri3871
138. Morandi E, Tanasescu R, Tarlinton RE, Constantinescu CS, Zhang W, Tench C, et al. The association between human endogenous retroviruses and multiple sclerosis: a systematic review and meta-analysis. PLoS ONE. (2017) 12:e0172415. doi: 10.1371/journal.pone.0172415
139. Saresella M, Rolland A, Marventano I, Cavarretta R, Caputo D, Marche P, et al. Multiple sclerosis-associated retroviral agent (MSRV)-stimulated cytokine production in patients with relapsing-remitting multiple sclerosis. Mult Scler. (2009) 15:443–7. doi: 10.1177/1352458508100840
140. Gold J, Marta MC, Meier UC, Christensen T, Miller D, Altmann D, et al. Phase 2 baseline versus treatment clinical trial of the HIV drug raltegravir in patients with active relapsing remitting multiple sclerosis: the INSPIRE study biomarker outcome results. Mult Scler J. (2016) 22:188–9. doi: 10.1016/j.msard.2018.06.002
141. Wu Z, Mei X, Zhao D, Sun Y, Song J, Pan W, et al. DNA methylation modulates HERV-E expression in CD4+ T cells from systemic lupus erythematosus patients. J Dermatol Sci. (2015) 77:110–6. doi: 10.1016/j.jdermsci.2014.12.004
142. Baudino L, Yoshinobu K, Morito N, Kikuchi S, Fossati-Jimack L, Morley BJ, et al. Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J Immunol. (2008) 181:2846–54. doi: 10.4049/jimmunol.181.4.2846
143. Shpyleva S, Melnyk S, Pavliv O, Pogribny I, Jill James S. Overexpression of LINE-1 Retrotransposons in Autism Brain. Mol Neurobiol. (2018) 55:1740–9. doi: 10.1007/s12035-017-0421-x
144. Tangsuwansri C, Saeliw T, Thongkorn S, Chonchaiya W, Suphapeetiporn K, Mutirangura A, et al. Investigation of epigenetic regulatory networks associated with autism spectrum disorder (ASD) by integrated global LINE-1 methylation and gene expression profiling analyses. PLoS One. (2018) 13:e0201071. doi: 10.1371/journal.pone.0201071
145. Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol. (2005) 57:67–81.
146. Thacker MA, Clark AK, Marchand F, McMahon SB. Pathophysiology of peripheral neuropathic pain: immune cells and molecules. Anesth Analg. (2007) 105:838–47.
147. Morgan JT, Chana G, Pardo CA, Achim C, Semendeferi K, Buckwalter J, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. (2010) 68:368–76.
148. Kern JK, Geier DA, Sykes LK, Geier MR. Relevance of neuroinflammation and encephalitis in autism. Front Cell Neurosci. (2015) 9:519. doi: 10.3389/fncel.2015.00519
149. Okabe Y, Takahashi T, Mitsumasu C, Kosai K, Tanaka E, Matsuishi T. Alterations of gene expression and glutamate clearance in astrocytes derived from an MeCP2-null mouse model of Rett syndrome. PLoS ONE. (2012) 7:e35354. doi: 10.1371/journal.pone.0035354
150. Di Giorgio FP, Carrasco MA, Siao MC, Maniatis T, Eggan K. Non-cell autonomous effect of glia on motor neurons in an embryonic stem cell-based ALS model. Nat Neurosci. (2007) 10:608–14.
151. Lobsiger CS, Cleveland DW. Glial cells as intrinsic components of non-cell-autonomous neurodegenerative disease. Nat Neurosci. (2007) 10:1355–60.
152. Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. (2009) 187:761–72. doi: 10.1083/jcb.200908164
153. Cortelazzo A, De Felice C, Guerranti R, Signorini C, Leoncini S, Pecorelli A, et al. Subclinical inflammatory status in Rett syndrome. Mediators Inflamm. (2014) 2014:480980. doi: 10.1155/2014/480980
154. Cortelazzo A, De Felice C, De Filippis B, Ricceri L, Laviola G, Leoncini S, et al. Persistent unresolved inflammation in the Mecp2-308 female mutated mouse model of rett syndrome. Mediators Inflamm. (2017) 2017:9467819. doi: 10.1155/2017/9467819
155. Russo FB, Freitas BC, Pignatari GC, Fernandes IR, Sebat J, Muotri ARB, et al. (2018). Modeling the interplay between neurons and astrocytes in autism using human induced pluripotent stem cells. Biol Psychiatry. 83:569–78. doi: 10.1016/j.biopsych.2017.09.021
156. Zhao B, Wu Q, Ye AY, Guo J, Zheng X, Yang X, et al. Somatic LINE-1 retrotransposition in cortical neurons and non-brain tissues of Rett patients and healthy individuals. PLoS Genet. (2019) 15:e1008043. doi: 10.1371/journal.pgen.1008043
157. Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. (2010) 468:443–6. doi: 10.1038/nature09544
158. Williams EC, Zhong X, Mohamed A, Li R, Liu Y, Dong Q, et al. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum Mol Genet. (2014) 23:2968–80. doi: 10.1093/hmg/ddu008
159. Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, et al. A role for glia in the progression of Rett's syndrome. Nature. (2011) 475:497–500. doi: 10.1038/nature10214
160. Crow YJ, Rehwinkel J. Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet. (2009) 18:R130–6. doi: 10.1093/hmg/ddp293
Keywords: LINE-1, HERV, retrotransposition, CNS, inflammation, reverse transcriptase inhibitors
Citation: Saleh A, Macia A and Muotri AR (2019) Transposable Elements, Inflammation, and Neurological Disease. Front. Neurol. 10:894. doi: 10.3389/fneur.2019.00894
Received: 27 April 2019; Accepted: 02 August 2019;
Published: 20 August 2019.
Edited by:
Avindra Nath, National Institute of Neurological Disorders and Stroke, United StatesReviewed by:
Hervé Perron, Independent Researcher, Geneva, SwitzerlandSantiago Morell, University of Cambridge, United Kingdom
Carmen Salvador-Palomeque, The University of Queensland, Australia
Copyright © 2019 Saleh, Macia and Muotri. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alysson R. Muotri, bXVvdHJpQHVjc2QuZWR1
 Aurian Saleh
Aurian Saleh Angela Macia
Angela Macia Alysson R. Muotri
Alysson R. Muotri