
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Neurol. , 06 July 2018
Sec. Stroke
Volume 9 - 2018 | https://doi.org/10.3389/fneur.2018.00514
 Sandro Marini1,2*
Sandro Marini1,2* Umme K. Lena1,2
Umme K. Lena1,2 Katherine M. Crawford1,2
Katherine M. Crawford1,2 Charles J. Moomaw3
Charles J. Moomaw3 Fernando D. Testai4
Fernando D. Testai4 Steven J. Kittner5
Steven J. Kittner5 Michael L. James6
Michael L. James6 Daniel Woo3
Daniel Woo3 Carl D. Langefeld7
Carl D. Langefeld7 Jonathan Rosand1,2,8
Jonathan Rosand1,2,8 Christopher D. Anderson1,2* on behalf of the ERICH investigators
Christopher D. Anderson1,2* on behalf of the ERICH investigatorsBackground: We sought to determine whether a small pool of ancestry-informative DNA markers (AIMs) improves modeling of intracerebral hemorrhage (ICH) risk in heterogeneous populations, compared with self-identified race/ethnicity (SIRE) alone.
Methods: We genotyped 15 preselected AIMs to perform principal component (PC) analysis in the ERICH study (a multi-center case-control study of ICH in whites, blacks, and Hispanics). We used multivariate logistic regression and tests for independent samples to compare associations for genetic ancestry and SIRE with ICH-associated vascular risk factors (VRFs). We then compared the performance of models for ICH risk that included AIMs and SIRE alone.
Results: Among 4,935 subjects, 34.7% were non-Hispanic black, 35.1% non-Hispanic white, and 30.2% Hispanic by SIRE. In stratified analysis of these SIRE groups, AIM-defined ancestry was strongly associated with seven of the eight VRFs analyzed (p < 0.001). Within each SIRE group, regression of AIM-derived PCs against VRFs confirmed independent associations of AIMs across at least two race/ethnic groups for seven VRFs. Akaike information criterion (AIC) (6,294 vs. 6,286) and likelihood ratio test (p < 0.001) showed that genetic ancestry defined by AIMs achieved a better ICH risk modeling compared to SIRE alone.
Conclusion: Genetically-defined ancestry provides valuable risk exposure information that is not captured by SIRE alone. Particularly among Hispanics and blacks, inclusion of AIMs adds value over self-reported ancestry in controlling for genetic and environmental exposures that influence risk of ICH. While differences are small, this modeling approach may be superior in highly heterogeneous clinical poulations. Additional studies across other ancestries and risk exposures are needed to confirm and extend these findings.
Vascular risk factors (VRFs) are often stratified by race and ethnicity. Hypertension has well-documented racial differences in prevalence, with African Americans more frequently hypertensive than whites(1–3). These disparities in hypertension impact both disease risk [the population attributable risk varies from 80.3 to 97.6% (4, 5)] and outcomes (6). Variability in risk exposure by race/ethnicity extends to other VRFs as well. Whites tend to have more unfavorable lipid profiles compared to blacks (7), and diabetes prevalence varies by race/ethnicity from 7.1% in whites to more than 10% in Hispanics and blacks (8). These ethnic and racial differences extend to the epidemiology of intracerebral hemorrhage (ICH), with increased risk in blacks and Hispanics compared with non-Hispanic whites (9, 10). As such, approaches that maximize the capture of racial and ethnic exposures in modeling of disease risk may improve the precision of our risk assessments.
In genome-wide association studies (GWAS), genetic polymorphisms whose allele frequencies vary markedly across different racial and ethnic groups [i.e., ancestry informative markers (AIMs)] are typically used to control for biases resulting from population stratification in their analyses (11). However, outside of GWAS, differences in risk factor prevalence among populations are usually addressed by using self-identified race and ethnicity (SIRE) information. Although SIRE does capture common cultural and social distinctions, it does not capture the full spectrum of genetic and ancestral diversity, nor does it adequately control for varying exposures within ancestral groups (12). Strengths and weaknesses of both approaches have been described elsewhere (13–16), but the use of AIMs to control for ancestry-related exposures remains rare outside of genetic association studies. As such, the potential benefits of leveraging AIMs data to control for racial and ethnic risk factors in epidemiologic studies has not been well-studied (17, 18).
We hypothesized that AIMs can provide value in capturing risk exposures that vary within and between ancestral populations only partially captured by SIRE (19). In testing this hypothesis, we utilized data from the Ethnic/Racial Variations of Intracerebral Hemorrhage (ERICH) study, which contains information on self-reported race and ethnicity from a standardized questionnaire, as well as AIMs data from a small pool of highly-informative variants in ICH cases and stroke-free controls.
The ERICH study is a prospective, multicenter, case-control study of ICH, designed to identify the genetic and epidemiologic factors that affect risk of ICH in a multi-ethnic population. As previously described (20), ERICH recruited approximately equal numbers of self-identified non-Hispanic white (whites), non-Hispanic black (blacks), and Hispanic patients aged ≥18 years with spontaneous ICH (defined as the sudden onset of severe headache, altered level of consciousness, or focal neurologic deficit associated with a focal collection of blood within the brain parenchyma, seen on neuroimaging) across 19 clinical recruitment centers. Cases of peripartum and anticoagulant-associated ICH were included, whereas ICH due to malignancy-associated coagulopathy, dural venous sinus thrombosis, vascular malformations, aneurysms, tumors, or hemorrhagic conversion of a recent ischemic stroke did not qualify as study cases. Controls were identified through random digit dialing to match cases by age (±5 years), sex, race/ethnicity, and geographic area. IRB approval was obtained at all participating centers and informed consent was obtained from all cases or their legally authorized representative.
All cases (or designated proxies) and controls underwent a standardized data collection protocol based on personal interviews. Among the items on baseline interview we focused on variables with established associations with cerebrovascular disease in order to examine their stratification among and within each included race/ethnicity (21). Namely, we analyzed self-reported history of hypertension, diabetes mellitus (DM), hyperlipidemia, coronary artery disease (CAD), carotid artery disease, nephropathy/kidney disease, ischemic stroke, and atrial fibrillation. This approach allowed us to compare cases and controls (that do not have chart abstraction) without over-representing the risk factors in cases. Additionally, to gage whether the prevalence of self-reported hypertension was erroneous, and to exclude systematic reporting bias, we collected also blood pressure readings [treated as dichotomous variable according to the new American College of Cardiology and American Heart Association guidelines (22)]. As in other studies (23), SIRE was classified as: non-Hispanic whites, non-Hispanic blacks, and Hispanics (non-African ancestral Hispanics). Genotype data used to create the ancestral PCs and admixture tables for this analysis are available for download from the International Stroke Genetics Consortium's Cerebrovascular Disease Knowledge Portal (cerebrovascularportal.org) (24). Software tools employed in our analyses were not modified or customized for our use.
For each of the three possible pairs of ancestral populations, we identified markers in which the difference in allele frequency (δ) was at least 0.5 between any two of the recruited ancestral populations. This allele frequency difference was chosen in order to maximize the resolution of ancestral populations with the minimum genotyping requirement. With these restrictions, we identified 15 AIMs that were adequately distributed across the genome (Supplement Table I). Genotyping of ERICH samples was performed using OpenArray TaqMan assay at the University of Miami. The 15 AIMs used in this study were bi-allelic single nucleotide polymorphisms (SNPs) that were selected based on information content for ancestry in the ancestral populations studied. Samples with < 90% call rates, or SNPs with call rates < 95% or deviation from Hardy-Weinberg equilibrium (at P-value < 2.5 × 10−4 for cases and at P-value < 2.5 × 10−2 for controls) were excluded.
To test for association with VRF across SIRE groups, we computed chi-square tests of independence for the baseline characteristics and Mann-Whitney U tests for the ancestry principal components (PC). Similarly, we computed the Mann-Whitney U tests to test for association between the SIRE groups and the ancestral PC. We used logistic regression (adjusted for age and sex) to assess the role of PCs in predicting each VRF, within each self-identified population. To perform PC analysis, we used PLINK v1.9 variance-standardized relationship matrix dimension reduction (25). We combined the results across the SIRE groups using a meta-analysis approach. Specifically, within each SIRE group, we performed backward stepwise regressions (likelihood ratio method) adjusted for age and sex to select the best PC predicting each VRF. We then carried on the absolute values obtained in the inverse variant weighted random effects meta-analysis. We assessed heterogeneity between studies using the Cochran's Q and I2 statistics [derSimonian-Laird estimate (26)].
As a confirmatory approach, we used the software program ADMIXTURE (27) to compute estimates of the three population ancestral proportions (one for each of the three ancestral populations inferred given the PC plot and the Cross-validation errors) using the same autosomal SNP genotypes. We used the three ancestry proportions as the independent variable, similarly to the PC in the previous approach, to test for associations as outlined above.
Finally, VRFs, age and SIRE with and without PCs 1–3 were modeled as predictors of ICH, and Akaike information criterion (AIC) and likelihood ratio (28) were used to compare these regression models. Additional models adjusting for centers of recruitments were performed.
As an additional approach to compare the difference between the different models, we also used net reclassification improvement (NRI). We used the cutoff of 30% risk of hemorrhage. This corresponds to a score of >4 points of the ATRIA Bleeding Risk Score, that is the threshold commonly used in clinical settings when assessing eligibility for anticoagulation treatment (29, 30) We performed NRI analyses using the Predictable package in R (31). We used R software, version 3.4.1 (The R Foundation for Statistical Computing) and the statistical package SPSS v. 21, 2012 (www.spss.com). All significance tests were 2-tailed.
The 4,935 subjects with genotype data were 43% female and had a median age of 61 years [interquartile range (IQR): 52–72]. The SIRE groups were relatively balanced between 34.7% non-Hispanic blacks, 35.1% non-Hispanic whites, and 30.2% Hispanics. There were 2,793 ICH cases (median age 60, IQR 51–73; male 59.2%), with the percentage of cases comparable within each SIRE (33.8% for blacks, 32.7% for whites, and 33.5% for Hispanics) (Table 1). Self-reported history of hypertension and blood pressure readings were highly correlated (Spearman's rho 0.138, p < 0.001).
In the assessment of genetic admixture between groups defined by SIRE, there was incomplete overlap between ancestry-based clusters and SIRE (Figure 1) (Supplement Figure I). reflecting considerable admixture between self-identified groups. The result of the PC analysis indicated two primary PCs (Supplement Figure II). Unsupervised reclassification of subjects based on their position in AIM-derived PCs space (Supplement Figure I) showed that 4.1% of whites, 2.6% of blacks, and 2.9% of Hispanics by SIRE would be reclassified to a different racial/ethnic population based on the AIMs-based PC clusters assignment alone (Supplement Table II). Similar results were obtained with the ancestry fractions (AF) obtained from ADMIXTURE (Supplement Figure III). Correlation between AF and PCs are shown in Supplement Table III.
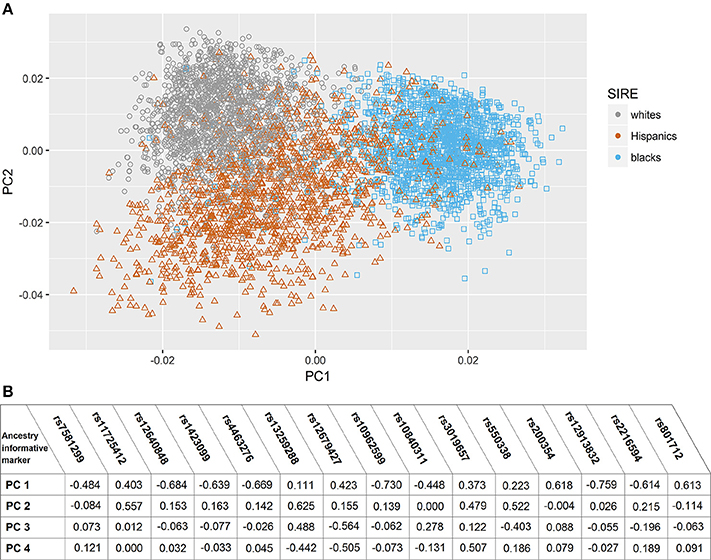
Figure 1. Principal component analysis based on ancestry informative markers. (A) Scatter plot of subjects in Principal Component (PC) space, grouped by their self-identified race/ethnicity. (B) Correlation coefficients between eigenvactors (top 4) and the Ancestry informative markers selected genotypes.
Self-identified race/ethnicity (SIRE) and genetic ancestry determined by AIM-based PCs were both tested for association with reported VRF diagnoses. Prevalence of hypertension, hypercholesterolemia, diabetes, CAD, atrial fibrillation, history of nephropathy, and carotid disease differed across SIRE, while AIM-derived PCs were associated with each of these VRFs (all p < 0.001) (Table 2, Supplement Figure IV). For history of ischemic stroke, there was no association with either PCs or SIRE. When stratified by SIRE, AIMs-based PC remained significantly associated with multiple VRFs (p < 0.001) (Table 3), indicating that genetic ancestry yielded independent information from SIRE relative to these VRFs. For each VRF except hypercholesterolemia, AIMs-based PC were significantly different between affected and unaffected individuals within one or multiple SIRE groups. For example, self-reported blacks demonstrated fine-scale ancestral differences based on hypertension diagnosis. Similarly, a history of CAD was associated with residual genetic ancestry across all three populations studied. These PC-based results were comparable to those obtained when comparing ancestral proportions from ADMIXTURE (see section Methods) in the entire ERICH sample (Supplement Table IV) and within the SIRE groups (Supplement Table V).
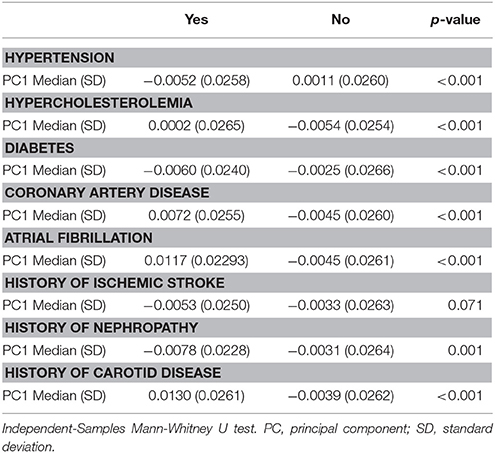
Table 2. Distribution of vascular risk factors across genetic ancestry (Principal Component 1) among affected and non-affected individuals.
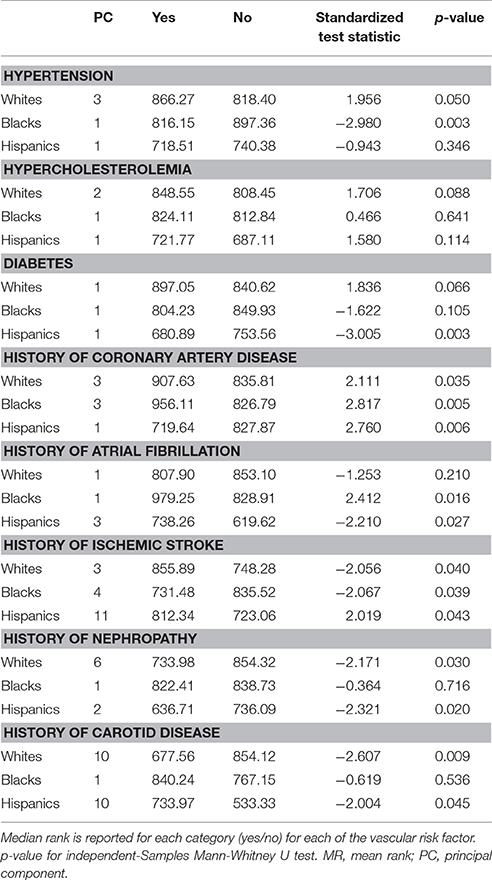
Table 3. PCs differentiate those affected from those unaffected by VRFs, within each self-identified race/ethnicity group.
To further test these associations, we regressed VRF status against the AIM-derived PCs. Logistic regressions for VRFs adjusted for age, sex, and SIRE identified significant independent associations of genetic ancestry with all the VRFs studied except hypercholesterolemia (Supplement Table VI). Inverse variance weighted random effects meta-analysis across SIRE indicated that different PCs were predictive of different VRFs (Supplement Table VII), suggesting that different ancestral subpopulations are enriched for different exposures. In this meta-analysis (Figure 2), risks for hypertension, diabetes, and atrial fibrillation varied significantly among self-reported blacks and Hispanics, when genetic ancestry was taken into account. Similarly, black and Hispanic individuals showed differing risks for ischemic stroke, while genetic ancestry modified risk for nephropathy only within the self-reported white population. Finally, based on ancestry, risk for carotid disease and coronary arthery disease was different in Hispanics and blacks, respectively. Comparable results were obtained when analyzing admixture-based AF and PC-derived ancestry (Supplement Tables VIII, IX).
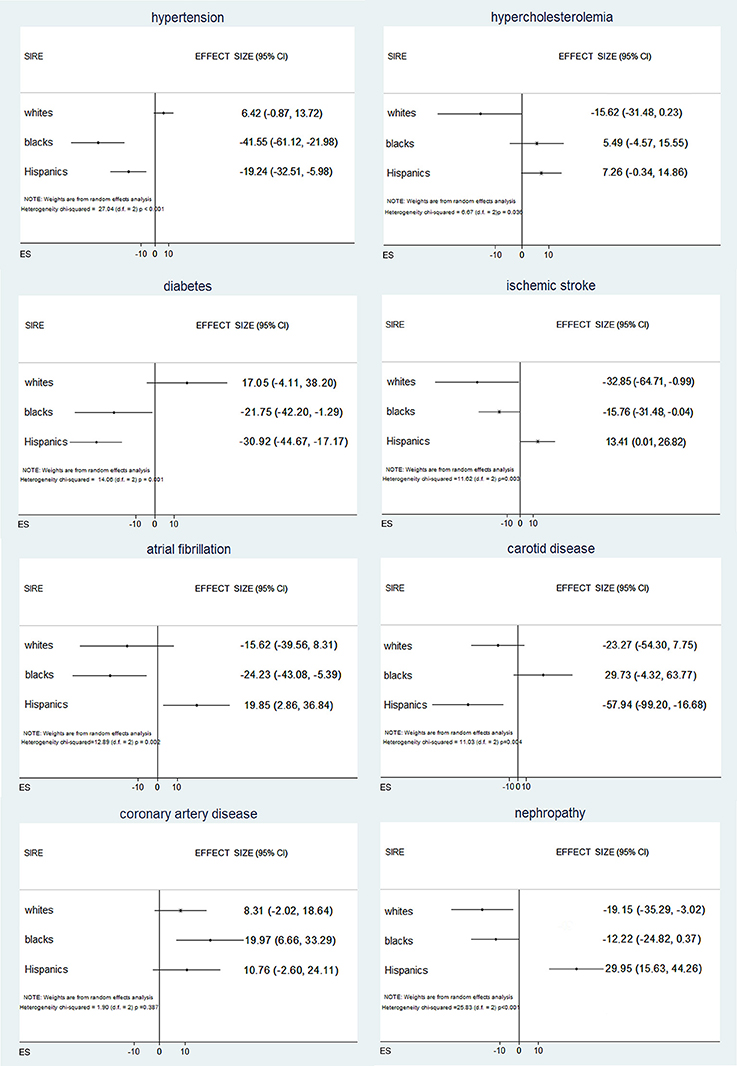
Figure 2. Associations between vascular risk factors and genetic ancestry across self-identified race/ethnicities within each self-identified race/ethnicities (SIRE) group, DNA ancestry is independently associated with vascular risk factor, with differences that vary across SIRE and vascular risk factor.
To estimate the changes in the explanatory ability of prediction models that include AIM-derived PCs, two regression models for ICH risk were computed: the first with age, sex, hypertension, and SIRE (model 1), the second adding AIM-derived PCs to the covariates (model 2). Akaike information criterion (AIC) (6,294 for model 1 VS 6,286 for model 2) and likelihood ratio test (p < 0.001) demonstrate superiority of the AIM-inclusive model 2 over model 1. Results showed the same pattern when models were also computed with centers of recruitments and smoking use, as covariates (AIC 6203.9 for model without ancestry VS 6185.0 for model with ancestry).
Our NRI analysis revealed similar results. Reclassifications for subjects with and without ICH events are summarized in Supplement Table X. For 57 subjects who did not experience ICH, classification improved using the model with AIMs, and for nine subjects it became worse. The NRI was estimated at 0.013 (0.004–0.021; p = 0.006), reflecting that individuals with ICH were 1.3% more likely to move up a category than down, compared with individuals without ICH.
In this multi-ethnic study of ICH, SIRE did not fully capture relevant genetic ancestry. Inclusion of AIMs substantially improved modeling of risk factor exposures. While inclusion of AIMs in an ICH prediction model improved model fit over SIRE alone, the overall improvement was modest. Certainly, deploying this approach in different racial and ethnic populations, as well as alternative risk exposures and disease states, are likely to reveal situations in which genetic ancestral modeling results in more dramatic improvements in risk prediction.
Social and cultural aspects such as access or utilization of care undoubtedly affect risk and outcome of disease. SIRE may capture exposures that vary by social background more closely than AIMs alone, and as such there would not appear to be any reason to fully replace self-reported data. However, a better understanding of genetic ancestry could help to understand why racial/ethnic minority groups have a lower risk for short-term all-cause mortality after being hospitalized for ischemic stroke than non-Hispanic whites, or why stratification by race does not improve risk prediction in high-risk hypertensive patients (32, 33). Such differences could be due to biological variation, but could also reflect differences in exposures that vary socioculturally within racial and ethnic groups in ways that are not fully captured by SIRE.
Rather than focusing on the magnitude of improvement with the incorporation of genetic ancestry into ICH prediction models, we submit that our results may serve as a proof of concept of for our approach to racial and ethnic modeling. Our analysis leveraged a limited pool of AIMs. It is likely that as the number of genetic markers increases, the fitness of the model will also improve (34). Further, given the high degree of stratification of VRFs by genetic ancestry in our analysis, we expect that other conditions for which highly stratified risk factors explain an even greater proportion of overall disease risk variance will demonstrate a greater improvement in modeling through inclusion of AIMs. For ICH, changes in AIC were small but reflect the inherent challenges in risk prediction in complex human disease irrespective of racial/ethnic contributions.
Adjusting for population stratification using AIMs improves statistical power in genetic association studies, but less information is available on whether these benefits extend to non-genetic studies (35). There are conflicting reports regarding the ability of SIRE to serve as an accurate predictor of population clusters (36–39). Prior studies have failed to demonstrate a greater predictive power of the genetic based ancestry over the SIRE-based categorization for cardiovascular disease (CVD) risk factors (40, 41). However, in these studies, AIMs were used to construct a continuous PC distribution that was then collapsed into to a categorical variable for analysis. Given the substantial overlap between ancestral population clusters in our study, our approach of using AIM-derived PCs as a continuous variable throughout our analyses may account for the differences in results.
Consistent with prior studies, we found substantial evidence of genetic clustering among the self-identified racial and ethnic populations studied (13). Also, as seen in other studies, the admixed nature of Hispanic populations results in a less compact (37) cluster than white or black populations. In our study, self-identified race generally corresponded well with genetic population clusters, but did not fully overlap. This correspondence is dependent on both the homogeneity within groups as well the sample size. Therefore, cluster analyses based on reported ancestry alone may overlook important components of population structure not captured by SIRE classification.
Beyond our confirmation of previous findings regarding genetic variance captured by SIRE (13) and the improvement of AIMs in defining populations (42), our results demonstrate the promising approach of adding genetic ancestry to the study of epidemiologic risk factors. Ascertainment of a small collection of DNA variants similar to those employed herein could be easily accomplished from blood or saliva in a small direct genotyping assay. Even if the residual benefit to including genetic ancestry remains modest, opmizing patient risk assessment by ancestry may still represent an important step forward in improving health equity as it pertains to providing maximally useful precision risk estimation across representative populations (43).
Strengths of this study include its large sample size, with a geographically diverse population well powered across representative race and ethnic populations. ICH cases and controls were well-characterized, with thorough clinical phenotyping of VRFs and centralized genotyping. Also, our results were confirmed by two distinctive and orthogonal methods for estimating ancestry. Several limitations of our approach deserve mention. As noted, ICH risk model improvements were modest after inclusion of genetic ancestry information. Second, VRF diagnoses were based on subject interview and could be inaccurate, although they were cross-validated and highly structured. Third, our analyses focused on non-Hispanic white, non-Hispanic black, and Hispanic populations. While these racial and ethnic groups are highly prevalent in the US, and are often included in studies of risk factors in US populations, we cannot extrapolate our findings to studies of other racial groups, or in focused studies of highly stratified subpopulations. Future studies in other cardiometabolic traits will be needed to support the generalizability of our approach.
In summary, genetically-defined ancestry provides additional risk exposure information that is not captured by SIRE alone. Inclusion of AIMs reveals additional stratification among risk factors between and within whites, blacks, and Hispanics. While differences in final ICH model specificity are small, incorporation of genetic ancestry into risk models may be superior in highly heterogeneous clinical poulations where risk exposures are highly correlated with race and ethnicity. Additional studies across diverse ancestries and risk exposures are needed to confirm and extend these findings.
This study was carried out in accordance with the recommendations of the Ethical Principles and Guidelines for the Protection of Human Subjects of Research, generally known as the Belmont Report, Partners Human Research Committee. The protocol was approved by the Partners Institutional Review Boards (IRBs).
SM study concept and design, analysis and interpretation of data, drafting of the manuscript. UL and KC analysis and interpretation of data, drafting of the manuscript. CM acquisition of data, analysis and interpretation of data. FT, SK, MJ, and DW critical revision of manuscript for intellectual content, study supervision. CL critical revision of manuscript for intellectual content, acquisition of data, study design, analysis and interpretation of data, study supervision. JR study concept and design, critical revision of manuscript for intellectual content, drafting of the manuscript, study supervision. CA study concept and design, drafting of the manuscript, analysis and interpretation of data, study supervision.
The study was funded by NIH/NINDS grants: K23NS086873, U01NS069763, R01NS093870, and R01NS103924. SM is supported by the AHA/ASA postdoctoral fellowship 18POST34080063.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fneur.2018.00514/full#supplementary-material
1. Wang X, Poole JC, Treiber FA, Harshfield GA, Hanevold CD, Snieder H. Ethnic and gender differences in ambulatory blood pressure trajectories: results from a 15-year longitudinal study in youth and young adults. Circulation (2006) 114:2780–7. doi: 10.1161/CIRCULATIONAHA.106.643940
2. Kramer H, Han C, Post W, Goff D, Diez-Roux A, Cooper R, et al. Racial/ethnic differences in hypertension and hypertension treatment and control in the multi-ethnic study of atherosclerosis (MESA). Am J Hypertens (2004) 17: 963–70. doi: 10.1016/j.amjhyper.2004.06.001
3. Koch S, Elkind MSV, Testai FD, Brown WM, Martini S, Sheth KN, et al. Racial-ethnic disparities in acute blood pressure after intracerebral hemorrhage. Neurology (2016) 87:786–91. doi: 10.1212/WNL.0000000000002962
4. Ariesen MJ, Claus SP, Rinkel GJE, Algra A. Risk factors for intracerebral hemorrhage in the general population: a systematic review. Stroke (2003) 34:2060–5. doi: 10.1161/01.STR.0000080678.09344.8D
5. O'donnell MJ, Chin SL, Rangarajan S, Xavier D, Liu L, Zhang H, et al. Global and regional eff ects of potentially modifi able risk factors associated with acute stroke in 32 countries (INTERSTROKE): a case-control study. Lancet (2016) 388:761–75. doi: 10.1016/S0140-6736(16)30506-2
6. Lackland DT. Racial differences in hypertension: implications for high blood pressure management. Am J Med Sci. (2014) 348:135–8. doi: 10.1097/MAJ.0000000000000308
7. Morrison JA, Khoury P, Mellies M, Kelly K, Horvitz R, Glueck CJ. Lipid and lipoprotein distributions in black adults. The cincinnati lipid research clinic's princeton school study. JAMA (1981) 245:939–42. doi: 10.1001/jama.1981.03310340029022
8. Chandler R, Monnat S. Racial/ethnic differences in use of health care services for diabetes management. Heal Educ Behav. (2015) 42:783–92. doi: 10.1002/nbm.3369.Three
10. Labovitz DL, Halim A, Boden-Albala B, Hauser WA, Sacco RL. The incidence of deep and lobar intracerebral hemorrhage in whites, blacks, and Hispanics. Neurology (2005) 65:518–22. doi: 10.1212/01.wnl.0000172915.71933.00
11. Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff S, et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics (2005) 2:81–9. doi: 10.1186/1479-7364-2-2-81
12. Bamshad M, Wooding S, Salisbury BA, Stephens JC. Deconstructing the relationship between genetics and race. Nat Rev Genet. (2004) 5:598. doi: 10.1038/nrg1401
13. Tang H, Quertermous T, Rodriguez B, Kardia SLR, Zhu X, Brown A, et al. Genetic structure, self-identified race/ethnicity, and confounding in case-control association studies. Am J Hum Genet. (2005) 76:268–75. doi: 10.1086/427888
14. Braun L. Race, ethnicity, and health: can genetics explain disparities? Perspect Biol Med. (2002) 45:159–74. doi: 10.1353/pbm.2002.0023
15. Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet (2003) 361:598–604. doi: 10.1016/S0140-6736(03)12520-2
16. Enoch M-A, Shen P-H, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. (2006) 20:19–26. doi: 10.1177/1359786806066041
17. Via M, Ziv E, Burchard E. Recent advances of genetic ancestry testing in biomedical research and direct to consumer testing. Clin Genet. (2009) 76:225–35. doi: 10.1111/j.1399-0004.2009.01263.x.RECENT
18. Royal CD, Novembre J, Fullerton SM, Goldstein DB, Long JC, Bamshad MJ, et al. Inferring genetic ancestry: opportunities, challenges, and implications. Am J Hum Genet. (2010) 86:661–73. doi: 10.1016/j.ajhg.2010.03.011
19. Mersha TB, Abebe T. Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Hum Genomics (2015) 9:1. doi: 10.1186/s40246-014-0023-x
20. Woo D, Rosand J, Kidwell C, Mccauley JL, Osborne J, Brown MW, et al. The ethnic/racial variations of intracerebral hemorrhage (ERICH) study protocol. Stroke (2013) 44:1–7. doi: 10.1161/STROKEAHA.113.002332
21. Martini SR, Flaherty ML, Brown WM, Haverbusch M, Comeau ME, Sauerbeck LR, et al. Risk factors for intracerebral hemorrhage differ according to hemorrhage location. Neurology (2012) 79:2275–82. doi: 10.1212/WNL.0b013e318276896f
22. Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Dennison Himmelfarb C, et al. ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults. J Am Coll Cardiol. (2018) 71:e127–e248. doi: 10.1016/j.jacc.2017.11.006
23. Chen C, Brown WM, Moomaw CJ, Langefeld CD, Osborne J, Worrall BB, et al. Alcohol use and risk of intracerebral hemorrhage. Neurology (2017) 1–10. doi: 10.1212/WNL.0000000000003952
24. Crawford KM, Gallego-Fabrega C, Kourkoulis C, Miyares L, Marini S, Flannick J, et al. Cerebrovascular disease knowledge portal: an open-access data resource to accelerate genomic discoveries in stroke. Stroke (2018) 49:470–475. doi: 10.1161/STROKEAHA.117.018922
25. Woo D, Falcone GJ, Devan WJ, Brown WM, Biffi A, Howard TD, et al. Meta-analysis of genome-wide association studies identifies 1q22 as a susceptibility locus for intracerebral hemorrhage. Am J Hum Genet. (2014) 94:511–21. doi: 10.1016/j.ajhg.2014.02.012
26. DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials (2015) 45:139–45. doi: 10.1016/j.cct.2015.09.002
27. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. (2009) 19:1655–64. doi: 10.1101/gr.094052.109
29. Lip GYH, Frison L, Halperin JL, Lane DA. Comparative validation of a novel risk score for predicting bleeding risk in anticoagulated patients with atrial fibrillation. J Am Coll Cardiol. (2011) 57:173–80. doi: 10.1016/j.jacc.2010.09.024
30. Fang MC, Go AS, Chang Y, Borowsky LH, Pomernacki NK, Udaltsova N, et al. A new risk scheme to predict warfarin-associated hemorrhage. J Am Coll Cardiol. (2011) 58:395–401. doi: 10.1016/j.jacc.2011.03.031
31. Kundu S, Aulchenko YS, Van Duijn CM, Janssens ACJW. PredictABEL: an R package for the assessment of risk prediction models. Eur J Epidemiol. (2011) 26:261–4. doi: 10.1007/s10654-011-9567-4
32. Drawz PE, Baraniuk S, Davis BR, Brown CD, Colon PJ, Cujyet AB, et al. Cardiovascular risk assessment: addition of CKD and race to the Framingham equation. Am Heart J. (2012) 164:925–31.e2. doi: 10.1016/j.ahj.2012.09.003
33. Qian F, Fonarow GC, Smith EE, Xian Y, Pan W, Hannan EL, et al. Racial and ethnic differences in outcomes in older patients with acute ischemic stroke. Circ Cardiovasc Qual Outcomes (2013) 6:284–92. doi: 10.1161/CIRCOUTCOMES.113.000211
34. Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL. The genetic ancestry of African Americans, Latinos, and European Americans across the United States. Am J Hum Genet. (2018) 96:37–53. doi: 10.1016/j.ajhg.2014.11.010
35. Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. (2008) 17:143–50. doi: 10.1093/hmg/ddn268
36. Haga SB, Venter JC. Genetics. FDA races in wrong direction. Science (2003) 301:466. doi: 10.1126/science.1087004
37. Stephens JC. Haplotype variation and linkage disequilibrium in 313 human genes. Science (2001) 293:489–93. doi: 10.1126/science.1059431
38. Bamshad MJ, Wooding S, Watkins WS, Ostler CT, Batzer M, Jorde LB. Human population genetic structure and inference of group membership. Am J Hum Genet. (2003) 72:578–89. doi: 10.1086/368061
39. Marcus GM, Alonso A, Peralta CA, Lettre G, Vittinghoff E, Lubitz SA, et al. European ancestry as a risk factor for atrial fibrillation in African Americans. Circulation (2010) 122:2009–15. doi: 10.1161/CIRCULATIONAHA.110.958306
40. Halder I, Kip KE, Mulukutla SR, Aiyer AN, Marroquin OC, Huggins GS, et al. Biogeographic ancestry, self-identified race, and admixture-phenotype associations in the Heart SCORE Study. Am J Epidemiol. (2012) 176:146–55. doi: 10.1093/aje/kwr518
41. Basu A, Tang H, Lewis CE, North K, Curb JD, Quertermous T, et al. Admixture mapping of quantitative trait loci for blood lipids in African-Americans. Hum Mol Genet. (2009) 18:2091–8. doi: 10.1093/hmg/ddp122
42. Yaeger R, Avila-Bront A, Abdul K, Nolan P, Grann V, Birchette M, et al. Comparing genetic ancestry and self-described race in African Americans Born in the United States and in Africa. Cancer Epidemiol Biomarkers Prev. (2008) 17:1329–38. doi: 10.1017/S1368980009991996.Validation
Keywords: genetics population, precision medicine, vascular diseases, risk factors, genetics, intracranial hemorrhage, race and ethnicity
Citation: Marini S, Lena UK, Crawford KM, Moomaw CJ, Testai FD, Kittner SJ, James ML, Woo D, Langefeld CD, Rosand J and Anderson CD (2018) Comparison of Genetic and Self-Identified Ancestry in Modeling Intracerebral Hemorrhage Risk. Front. Neurol. 9:514. doi: 10.3389/fneur.2018.00514
Received: 17 April 2018; Accepted: 11 June 2018;
Published: 06 July 2018.
Edited by:
Antonio Arauz, Instituto Nacional de Neurología y Neurocirugía (INNN), MexicoReviewed by:
Miguel A. Barboza, University of Costa Rica, Costa RicaCopyright © 2018 Marini, Lena, Crawford, Moomaw, Testai, Kittner, James, Woo, Langefeld, Rosand and Anderson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sandro Marini, c21hcmluaTFAbWdoLmhhcnZhcmQuZWR1
Christopher D. Anderson, Y2RhbmRlcnNvbkBwYXJ0bmVycy5vcmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.