- 1Graduate School of Biomedical Sciences, Hiroshima University, Hiroshima, Japan
- 2RIKEN Brain Science Institute, Wako, Saitama, Japan
- 3CREST, Japan Science and Technology Agency, Tokyo, Japan
The circadian oscillation of clock gene expression in mammals is based on the interconnected transcriptional/translational feedback loops of Period (Per) and Bmal1. The Per feedback loop initiates transcription through direct binding of the BMAL1–CLOCK (NPAS2) heterodimer to the E-box of the Per2 promoter region. Negative feedback of PER protein on this promoter subsequently represses transcription. Other circadian transcription regulators, particularly E4BP4 and DEC2, regulate the amplitude and phase of Per2 expression rhythms. Moreover, a direct repeat of E-box-like (EE) elements in the Per2 promoter is required for its cell-autonomous circadian rhythm. However, the detailed mechanism for repression of the two core sequences of the EE element in the Per2 promoter region is unknown. Here, we show that E4BP4 binds to the Per2 EE element with DEC2 to repress transcription and identify the DEC2–E4BP4 heterodimer as a key repressor of the tightly interlocked Per2 feedback loop in the mammalian circadian oscillator. Our results suggest an additional modulatory mechanism for tuning of the phase of cell-autonomous Per2 gene expression cycling.
Introduction
The circadian clock regulates many physiological and behavioral cues, such as hormone secretion, metabolism, body temperature, immune response, and sleep–wake rhythms (1–3). Critical to these processes, the mammalian transcriptional regulators PERIOD (PER), cryptochrome (CRY), BMAL1, and NPAS2/CLOCK play core roles in sustaining interlocked circadian rhythms (4). In particular, BMAL1 forms a heterodimer with either NPAS2 or CLOCK to activate the Per promoter (5, 6). PER and CRY (CRY1 and CRY2), meanwhile, function in negative feedback loops that repress BMAL1–NPAS2/CLOCK, thereby completing a transcription/translation-based autoregulatory feedback loop (7, 8). Three mammalian orthologs of Per, named Per1, Per2, and Per3, have been previously identified (9–14). PER1 and PER2 play important roles for circadian oscillation in the master clock suprachiasmatic nucleus (SCN), whereas PER3 participates in timekeeping in peripheral tissues, such as pituitary and lung (15). Per2 knockout mice exhibit reduction of Per1 and Clock expression levels (16–18), indicating PER2 plays a prominent role in regulating expression of circadian clock genes. Per2 expression mechanisms are different from the classical mechanisms of circadian gene expression in the sense that an E-box-like (EE) (CACGTT) sequence, rather a canonical E-box consensus (CACGTG) sequence, in the Per2 promoter is essential for robust circadian transcription of Per2 (19). The two core sequences of the EE-element in the Per2 promoter region, CACGTT and TATGTG, named E1 and E2, respectively, are required for Per2 rhythmic expressions (20). The E1 site-binding transcription regulators are BMAL1 and CLOCK, while the E2 site-binding transcription regulators remain unidentified (20).
E4BP4, a basic leucine zipper (bZIP) transcription factor, and DEC1/2, basic helix–loop–helix (bHLH) transcription factors, modulate amplitude and phase of Per1/2 expression rhythms (21, 22). Furthermore, Dec1 and Dec2 are expressed in the SCN abundantly. DEC1 represses the CACGTG E-box weakly, whereas DEC2 represses the same E-box more strongly (23). Dec1/2 knockout mice lack normal resetting for phase shift of light–dark cycles and exhibit abnormal amplitude of Per2 expression rhythms in the SCN (22). DEC2 forms a heterodimer with multiple proteins, such as MyoD and retinoid X receptor (RXR), to modulate a wider variety of gene expressions (24, 25). It is thus expected that DEC2 forms heterodimers with hitherto unknown transcription factors to bind the Per2 EE-element and fine-tune the phase of circadian oscillation.
Physiological roles of E4BP4 have been studied in the chicken pineal gland. The light pulse at early subjective night induces E4bp4 expression in the pineal gland, which acts as a light-dependent suppressor of Per2 expression (26, 27). In mammals, E4bp4 is expressed in the SCN in a circadian manner by which the peak of E4bp4 expression occurs at night (28). The E4BP4 binding site (B-site) has been identified, and E4BP4 represses Per2 promoter activity by binding to this B-site, rather than the Per2 EE-element (29). However, E4BP4 regulates the mouse Per2 promoter region (−105 ~ +110), which includes the EE-element but not the B-site (19). E4BP4 may bind to this Per2 promoter region through a binding mechanism different from that of the B-site binding.
In the present study, we investigated how DEC2 and E4BP4 bind to the Per2 EE-element to modulate the phase of circadian oscillation. Here, we report DEC2 and E4BP4 form a heterodimer to repress Per2 promoter activity by binding to the E2 consensus of the Per2 EE-element. Our results suggest that the DEC2–E4BP4 heterodimer plays a crucial role in modulating phase of Per2 circadian rhythms.
Results
DEC2 and E4BP4 Form a Heterodimer
It remains unknown how DEC2 regulates amplitude and phase of circadian rhythms. A homolog of DEC2, Drosophila clockwork orange (CWO), functions as both an activator and a repressor of clock gene expression (30). One possible mechanism for these functions of CWO has been proposed to be the binding of various unknown transcription regulators periodically to regulate circadian rhythms. In fact, DEC2 binds to various transcription regulators, namely BMAL1, MyoD, and RXR (23–25). We therefore hypothesized that DEC2 forms a heterodimer with an unknown transcription regulator to control the Per2 EE-element-driven promoter activity.
DEC2 is speculated to have similar functions as E4BP4 for circadian rhythms. Per2 expression levels are repressed by E4BP4 (19, 26, 29), suggesting that E4BP4 potentially modulates the amplitude and phase of circadian rhythms in a manner similar to that of DEC2. Based on this logic, we assessed how E4BP4 interacts with DEC2 to modulate circadian rhythms. Using immunoprecipitation and glutathione S-transferase (GST) pull-down assays, we discovered that DEC2 forms a heterodimer with E4BP4 (Figures 1A,B). Furthermore, given the previous finding that the DEC2 bHLH domain plays a crucial role in forming a heterodimer with MyoD (24), we assessed formations of heterodimer between various DEC2 fragments and E4BP4. DEC2 bHLH domain showed binding with high amount of E4BP4 in the co-immunoprecipitation experiment (Figure 1C). This result indicates that DEC2 bHLH is important for DEC2–E4BP4 heterodimer formation.
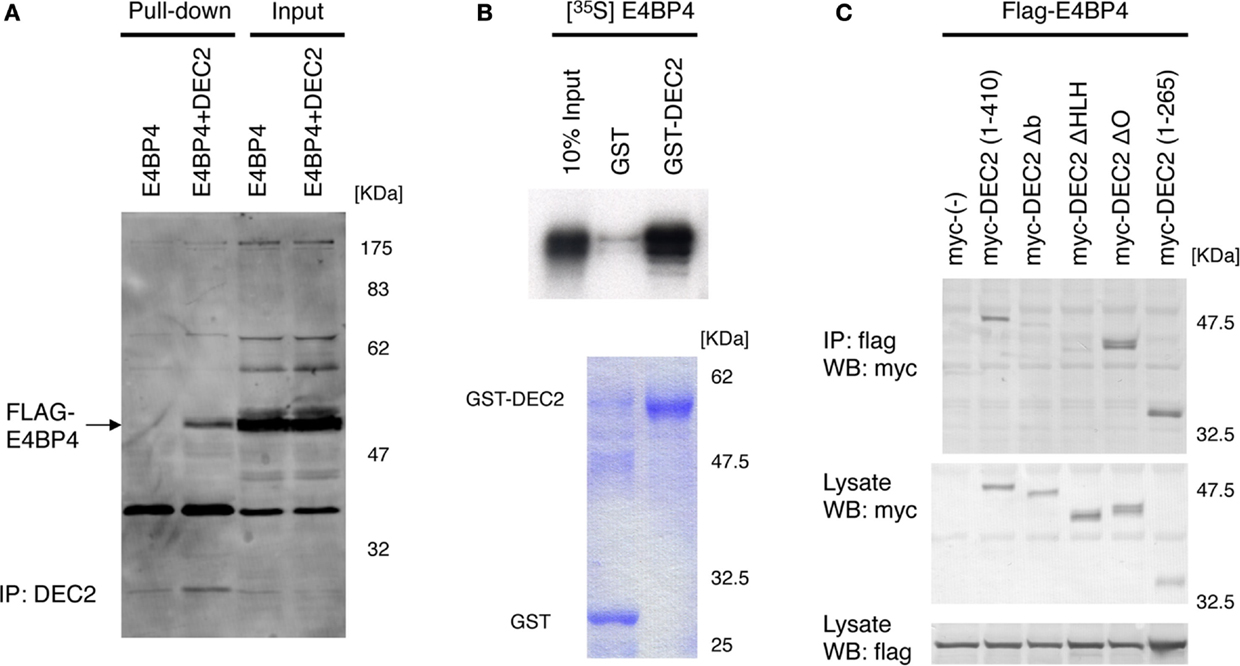
Figure 1. DEC2 forms a heterodimer with E4BP4. (A,B) Immunoprecipitation of E4BP4 and DEC2 complex. Flag-tagged E4BP4–DEC2 heterodimer was pulled down with anti-DEC2 antibody and protein A agarose beads. Flag-tagged E4BP4 protein was detected using immunoblotting with anti-FLAG antibody. Flag-tagged E4BP4–DEC2 heterodimer was detected in NIH3T3 cells co-expressing DEC2 and E4BP4 but not in the cells expressing E4BP4 only (A). The data are a representative result of three independent experiments. Radioisotope labeled S35-E4BP4 was pulled down with GST-tagged DEC2 and glutathione-coupled agarose beads. The bottom panel shows the corresponding SDS-PAGE gel stained with Coomassie blue (B). (C) bHLH domain of DEC2 plays a crucial role in binding of E4BP4. COS-7 cells were transfected with myc-tagged Dec2 expression vectors together with flag-tagged E4BP4 expression vector. Cell lysates were immunoprecipitated with anti-flag antibody and immunoblotted with anti-myc antibody. To confirm expression of E4BP4 and DEC2 proteins, aliquots of the total cell lysates were immunoblotted with the anti-flag and anti-myc antibodies. b, basic region; HLH, helix–loop–helix domain; O, orange domain. Numbers indicate the position of amino acids in the DEC2 protein.
DEC2–E4BP4 Heterodimer Binds to the E2 Consensus of the Per2 EE-Element
Since the Per2 EE-element is essential for normal circadian oscillation (20), we assessed the possibility of DEC2–E4BP4 heterodimer binding to the Per2 EE-element. E4BP4 bound to the oligonucleotide probe of the Per2 EE-element in the presence of DEC2, whereas E4BP4 failed to bind to the Per2 EE-element in the absence of DEC2 (Figure 2A). On the contrary, DEC2 bound to the Per2 EE-element in the absence of E4BP4 (Figure 2B). These results indicate that DEC2 mediates E4BP4 binding to the Per2 EE-element.
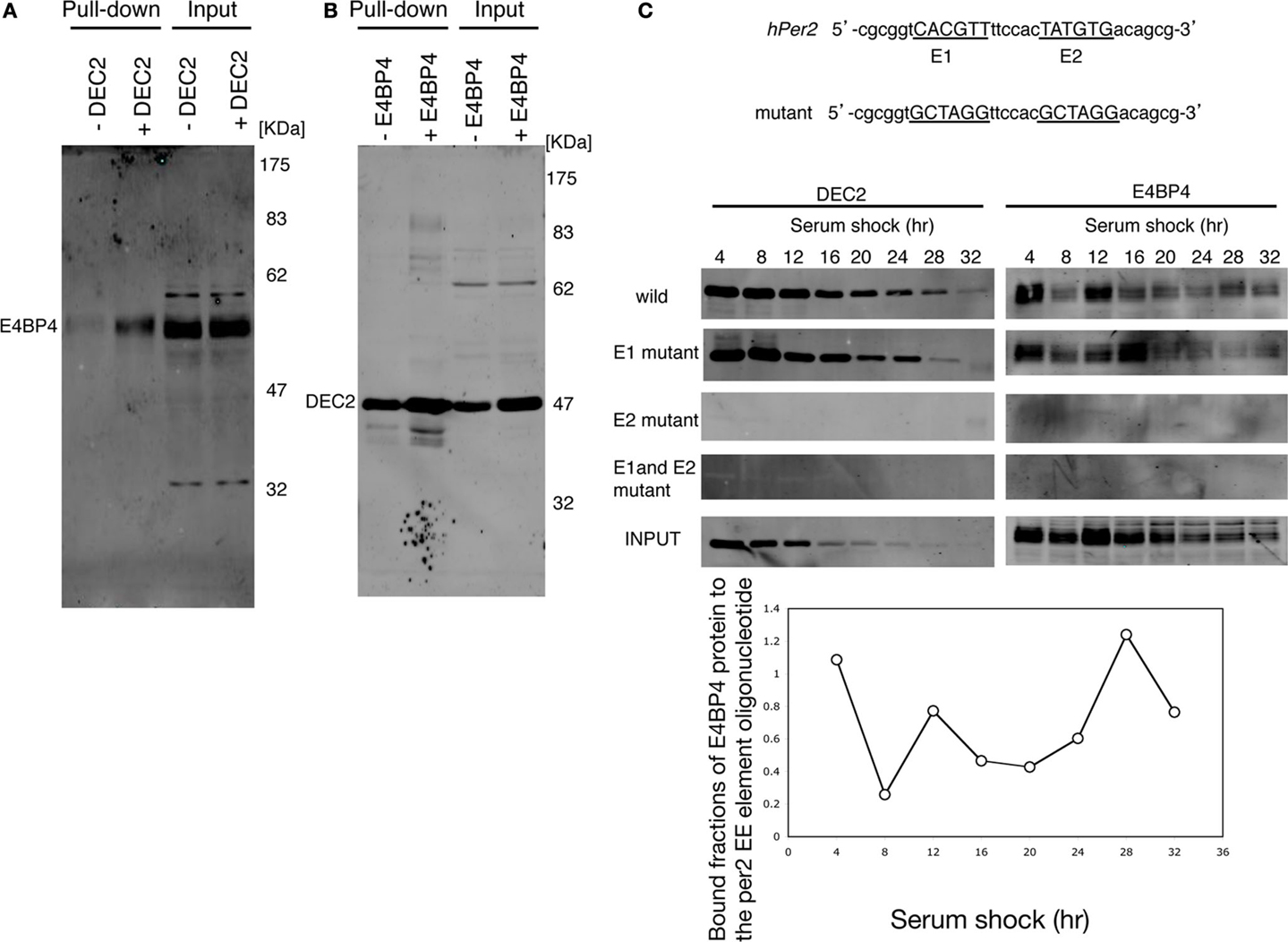
Figure 2. E4BP4 binds to the E2 consensus of Per2 EE element via DEC2. (A) E4BP4 binds to oligonucleotide of the Per2 EE element in the presence of DEC2. The complexes of biotin-labeled Per2 EE-element probe (20) and E4BP4 protein from nuclear extracts of E4BP4-overexpressing NIH3T3 cells were pulled down using streptavidin beads. Detection of E4BP4 was performed using immunoblotting with anti-E4BP4 antibody. E4BP4 was pulled down in the nuclear extracts of NIH3T3 cells co-expressing DEC2 and E4BP4 (indicated with +DEC2). E4BP4 was not pulled down in the nuclear extracts of NIH3T3 cells expressing only E4BP4 (indicated with −DEC2). (B) DEC2 binds to oligonucleotide of the Per2 EE element without E4BP4. Detection of DEC2 was performed using immunoblotting with anti-DEC2 antibody. DEC2 was pulled down in nuclear extracts of NIH3T3 cells both expressing only DEC2 and co-expressing DEC2 and E4BP4, indicated with −E4BP4 and +E4BP4, respectively. (C) DEC2–E4BP4 heterodimer binds the E2 consensus of Per2 EE element. DEC2–E4BP4 heterodimer was pulled down from nuclear extracts of NIH3T3 cells co-expressing DEC2 and E4BP4 every 4-h after cyclohexylamine-shock. Detection of E4BP4 or DEC2 was performed using immunoblotting with DEC2 (left panel) and E4BP4 (right panel). Both DEC2 and E4BP4 were pulled down by wild-type and the E1 mutant probes but not by E2 mutant and E1-E2 mutant probes. Wild-type and mutant sequences of Per2 EE element probe are shown in the upper panel. Representative results of three independent experiments are shown. The bottom figure shows the quantification of bound fractions of E4BP4 to the wild-type per EE element.
The Per2 EE-element consists of two consensus sequences, named E1 and E2 (20). We tested whether the DEC2–E4BP4 heterodimer binds to E1 or to E2 through an oligonucleotide pull-down experiment. In Drosophila, circadian transcription regulators bind to the Per promoter dynamically (31). Therefore, we tested time-dependent DEC2 and E4BP4 binding to the Per2 EE-element in serum shocked NIH3T3 cells. DEC2 and E4BP4, which were expressed in the nucleus of the serum shocked NIH3T3 cells, dynamically bound to the oligonucleotide probe with the mutant E1 and the wild-type E2 but failed to bind to the probe with the wild-type E1 and the mutant E2 (Figure 2C). This result indicates that the DEC2–E4BP4 heterodimer binds to the E2 consensus of the Per2 EE-element and not to the E1 consensus sequence.
DEC2–E4BP4 Heterodimer Represses Promoter Activity of the Per2 EE-Element Region
To define the effects of the DEC2–E4BP4 heterodimer on promoter activity of the Per2 EE-element, we measured Per2 EE-element region-driven luciferase activities in the presence of DEC2 and E4BP4. Promoter activities of the Per2 EE-element region were not affected by single expressions of DEC2 or E4BP4, whereas co-expression of E4BP4 and DEC2 repressed promoter activities of the Per2 EE-element (Figures 3A,B). Therefore, it is a heterodimer of DEC2 and E4BP4 that represses promoter activity of the Per2 EE-element region. Furthermore, we quantified the ratio of binding between DEC2 and E4BP4 with exponential fitting. Half-maximal inhibition dosages (IC50) of DEC2 and E4BP4 with respective dimerization partners, E4BP4 100 ng and DEC2 50 g, were estimated to be 101.1 and 56.6 ng, respectively, indicating a 1:1 stoichiochemistry.
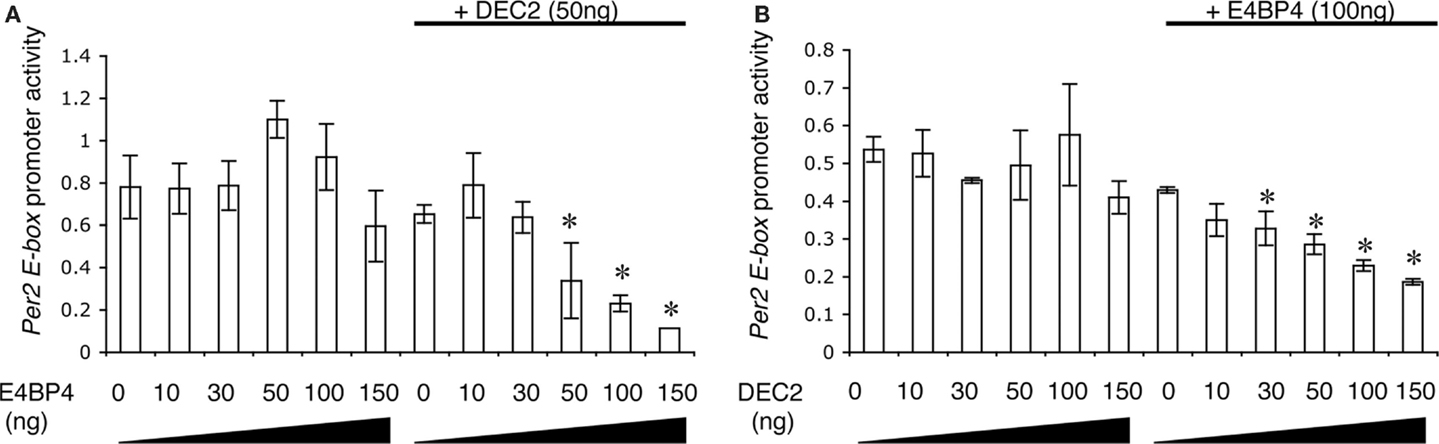
Figure 3. DEC2 represses Per2 EE element promoter activity. (A,B) E4BP4–DEC2 heterodimer represses Per2 EE element promoter activity. NIH3T3 cells were transfected with different amounts of E4bp4/pcDNA3.1 or Dec2/pcDNA3.1 plasmid vector, 100 ng Per2 EE element luciferase/pGL3 plasmid vector, 3 ng of Bmal1/pcDNA3.1 and Clock/pcDNA3.1, and 20 ng pRL-TK plasmid vector. Per2 EE element promoter activity is presented as relative values of the Per2EE promoter-driven luciferase activity compared to internal control luciferase activity (pRL-TK). E4BP4 repressed the Per2 EE element-driven promoter activity in the presence of DEC2 (A). DEC2 repressed the Per2 EE element-driven promoter activity in the presence of E4BP4 (B). Error bars represent SEM from three independent experiments. Asterisk (*), p < 0.05 versus 0 ng of E4BP4 or DEC2.
Mathematical Modeling Shows DEC2–E4BP4 Heterodimer has an Impact on Phase of Per2 Expression Cycling
Phases of DEC2 and E4BP4 are opposite in the SCN. Peak points of DEC2 and E4BP4 expression levels are at daytime and midnight (23, 28). Expression levels of DEC2–E4BP4 heterodimer should be high at both subjective dusk and dawn, at which DEC2 and E4BP4 expression levels are equal (Figure 4A). To evaluate the logical consequence of the DEC2–E4BP4 heterodimer repression in Per2 expression cycling, we introduced a mathematical model of circadian oscillation. Since the kinetics of DEC2 and E4BP4 has not been considered in previous mathematical models, we created a hybrid model based on the Becker–Weimann model of the circadian clock (32) by inserting contributions of DEC2 and E4BP4 as exogenous sinusoidal variations as proposed earlier (33). DEC2–E4BP4 heterodimer concentration is given by multiplying the concentration of DEC2 and E4BP4, in accordance with our observation of 1:1 binding (Figure 3). High levels of DEC2–E4BP4-induced phase advances of Per2 expression cycling (Figure 4B). Three times of the tentative norm of the DEC2–E4BP4 level induced an ~1-h phase advance of Per2 expression cycling per day (Figure 4C). The hybrid model test the consequence of rhythmic repression, which shows that modulation of the DEC–E4BP4 heterodimer has an impact on Per2 expression cycling.
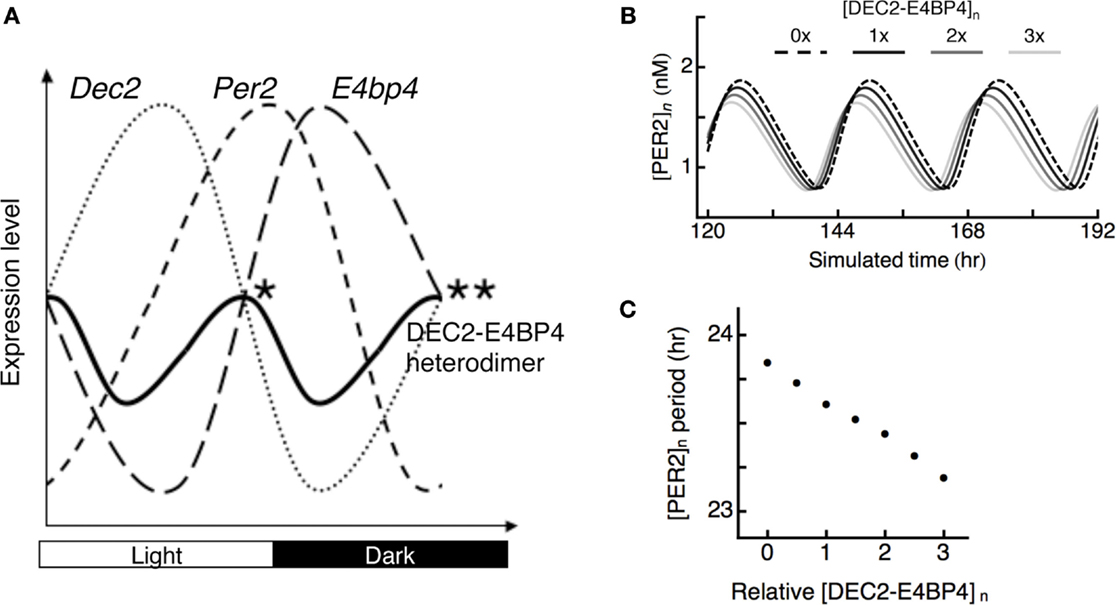
Figure 4. Possible roles of DEC2–E4BP4 heterodimer in the interlocked Per2 oscillation. (A) Phases of clock gene cycling in SCN. Dec2 expression cycling is opposite to E4bp4 expression cycling in the SCN (23, 28). Two peaks of DEC2–E4BP4 heterodimer levels in the SCN should be at subjective dusk (*) and dawn (**), at which DEC2 and E4BP4 expression levels are equal. Per2 expression levels are high at subjective dusk and trough at subjective dawn in SCN (11, 12). DEC2–E4BP4 heterodimer represses Per2 expression levels to determine time points of peak and trough of Per2 expression cycling. (B) Theoretical model of PER2 protein expression cycling under various amounts of DEC2–E4BP4 heterodimer. Oscillations of PER2 expression levels under various amounts of [DEC2–E4BP4]n (n = 0, 1, 2, 3) are indicated with dashed line (n = 0), black solid line (n = 1), gray solid line (n = 2), and light gray line (n = 3). The simulated nuclear concentration of DEC2–E4BP4 heterodimer ([DEC2–E4BP4]n) induces phase advance of PER2 ([PER2]n) expression cycling in a concentration-dependent manner. (C) Periods of the PER2 oscillations under various amount of DEC2–E4BP4 heterodimer in (B). High concentration of DEC2–E4BP4 heterodimer shortens the period of PER2 oscillation.
Discussion
Many circadian transcription regulators dynamically form complexes with other circadian transcription regulators for circadian timekeeping. Here, we found a new complex, the E4BP4–DEC2 heterodimer, which serves as repressor of Per2 promoter activity. E4BP4 and DEC2 are thought to be negative circadian regulators independently (23, 29). In addition to previous reports, we found E4BP4 and DEC2 form a heterodimer to cooperatively regulate the clock function. E4BP4 and DEC2 play important roles in modulating the phase of circadian rhythms. Overexpressing E4BP4 shortens the period length of Per2 oscillation (19). DEC2 is required for normal circadian phase resetting (22). E4BP4 and DEC2 modulate phase of Per2 expression cycling cooperatively through formation of a heterodimer.
With respect to cycling phases of Dec2, Per2, and E4bp4 expression levels, we speculate that two peaks of DEC2–E4BP4 heterodimer levels should be around subjective dusk and dawn, at which points Dec2 and E4bp4 expression levels are the same in the SCN (Figure 4A). As for the significance of these two peaks of E4BP4-heterodimer in Per2 expression cycling, we propose the following model: at subjective dusk, the DEC2–E4BP4 heterodimer determines the peak time point of Per2 expression levels through repression of Per2 expression induction. At subjective dawn, the DEC2–E4BP4 heterodimer determines the trough point of Per2 expression cycling through further encouraging the decrease of Per2 expression. This dynamic binding eventually fine-tunes the phase of Per2 expression cycling. This idea is supported by a previous report that showed the overexpression of E4BP4 induces a shorter period length for Per2 oscillation (19). Furthermore, the mathematical model supports the idea that DEC2–E4BP4 modulates the phase of Per2 expression cycling by shortening the length of period (Figures 4B,C), which is consistent with the recent report (34). Increasing DEC2–E4BP4 heterodimer enhances phase advances of Per2 expression cycling in this model. Additionally, the mathematical model can explain the mechanism of phase advance of Per2 expression cycling by light at early subjective night. Light induces E4BP4 expression in the avian circadian system (26). The induced E4BP4 forms a heterodimer with DEC2 and binds to the Per2 EE-element to repress Per2 promoter activities. The oscillatory acceleration through this suppression eventually creates phase advance of Per2 expression cycling.
In this study, we showed that the E4BP4–DEC2 heterodimer binds to the E2 consensus to repress Per2 promoter activity (Figures 2 and 3). Considering that E4BP4 and DEC2 are modulators of clock functions rather than drivers of clock functions, such as BMAL1 and CLOCK, we can propose that the E2 consensus is responsible for modulating the phase and amplitude of circadian oscillation. EE-elements are also located on other promoter regions of many BMAL1 target genes, such as Dbp, Nr1d1, and Tef (20, 35, 36). E4BP4–DEC2 heterodimer can thus regulate the expression rhythms of these genes as well, though further experiments are needed.
The dynamic binding of E4BP4 to the Per2 EE-element we observed, in addition to single E4BP4 binding to the B-box in the Per2 exon, likely contributes to controlling Per2 expression (29). DEC2 is required for E4BP4 binding to the Per2 EE-element. DEC2 is able to bind to various proteins, MyoD, RXR, histone deacetylase, and Sirtuin 2 (24). Understanding how a complex of DEC2 and these proteins regulates the Per2 EE-element activities will shed light on mechanisms to modulate phases of circadian oscillators.
Materials and Methods
Plasmids
The bacterial expression vector for full-length Dec2 that fused with GST was described previously (24). Mammalian expression vectors for myc-tagged full-length and deletion mutants of DEC2 were described previously (24).
Cell Culture and DNA Transfection
NIH3T3 cells were cultured as described (37). DNA transfection was performed according to the manufacturer’s instructions (Lifetechnologies).
Immunoprecipitation
One microgram of pcDNA3.1-Dec2 (38) and pcDNA3.1-FLAG-E4bp4 (19) were transfected into NIH3T3 cells. After 28 h, the cells were harvested and homogenized using ultrasonication in buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.3% Triton X-100, 1 mM PMSF, 1 mg/ml leupeptin, and 1 mg/ml aprotinin. The homogenate was centrifugated (15,000 × g, 5 min, 4°C). Supernatants were incubated with Protein A agarose beads (Roche) and anti-DEC2 antibody (24) (dilution 1:200) for overnight, 4°C. The immunoprecipitated FLAG tagged E4BP4 was detected on Immunoblotting using anti-FLAG antibody (Sigma-Aldrich; dilution 1:500). The interactions of E4BP4 with myc-tagged DEC2 mutant proteins were also examined. The co-immunoprecipitation assay was performed according to a previously described protocol (24).
GST-Pull Assay
The BL21 strain of Escherichia coli was transformed with plasmid pET-41-Dec2 for expression of GST-DEC2 fusion protein or, as a control, pET-41 for expression of GST. GST-pull down assay was performed using purified GST or GST-DEC2 fusion proteins, as described previously (24). [35S] methionine-labeled proteins were synthesized in vitro using the TNT Quick Coupled Transcription/Translation System (Promega, WI, USA).
Luciferase Reporter Assay
Different amounts of pcDNA3.1-Dec2 and pcDNA3.1-FLAG-E4bp4 were cotransfected with 20 ng pRL and 100 ng Per2 EE-element promoter-driven luciferase (20) into NIH3T3T cells. Transfected cells were assayed for luciferase activity in a luminometer (Turner Designs, CA, USA) by using the dual-luciferase reporter assay system (Promega, WI, USA).
Oligonucleotide Probe Pull-Down Experiment
Three micrograms of pcDNA3-Dec2 and pcDNA3-e4bp4 were transfected into NIH3T3T cells. At 23 h after transfection, the medium was exchanged for 100 nM cyclohexylamine containing medium, and 2 h later, the medium was replaced with 10% FBS containing medium. The cells were harvested every 4 h and used for oligonucleotide probe pull-down experiment as described (20). The pull-downed E4BP4 and DEC2 were detected on Immunoblot using anti-E4BP4 antibody (39, 40) and anti-DEC2 antibody.
Mathematical Modeling
An expected time course of the DEC2–E4BP4 heterodimer is inserted directly into as an inhibitory term in the Michaelis–Menten equation (33) describing Per2 transcription (y1). The dynamic DEC2–E4BP4 heterodimer concentration is given by
In this equation, the first cosine describes the DEC2 time course, the second describes the E4BP4 time course, and n determines the relative concentration of the DEC2–E4BP4 heterodimer. We inserted this equation into the equations of the Becker–Weimann model (32) as follows:
The time t is in hours and all concentrations are in nanomolar. y1 is the concentration of Per2/Cry mRNA, y2 is cysolic PER2/CRY protein complex, y3 is nuclear PER2/CRY protein complex, y4 is Bmal1 mRNA, y5 is cytosolic BMAL1, y6 is nuclear BMAL1, and y7 is nuclear of BMAL1/CLK heterodimer. All parameter values are unchanged from the original model: v1b = 9 nM/h, k1b = 1 nM, k1i = 0.56 nM, c = 0.01 nM, p = 8, k1d = 0.12/h, k2b = 0.3 nM/h, q = 2, k2d = 0.05/h, k2t = 0.24/h. k3t = 0.02/h, k3d = 0.12/h, v4b = 3.6 nM/h, k4b = 2.16 nM, r = 3, k4d = 0.75/h, k5b = 0.24/h, k5d = 0.06/h, k5t = 0.45/h, k6t = 0.06/h, k6d = 0.12/h, k6a = 0.09/h, k7a = 0.003/h, and k7d = 0.09/h.
The differential equations were solved using commercial software, Mathematica 7 (Wolfram Research, Champaign, IL, USA).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all technicians of the Takumi Laboratory for their assistance and Jamasb Sayadi for comments on the manuscript. This work was supported in part by Ministry of Education, Culture, Sports, Science and Technology in Japan Grants-in-Aid for Scientific Research, CREST, and Strategic International Coorperative Program from the Japan Science and Technology Agency, Intramural Research Grant for Neurological and Psychiatric Disorders of NCNP and the Takeda Science Foundation.
Abbreviations
Dbp, D-binding protein; Dec1, differentiated embryonic chondrocyte-expressed 1; Dec2, differentiated embryonic chondrocyte-expressed 2; E4BP4, E-promoter-binding protein 4; Nr1d1, nuclear receptor subfamily 1, group D; RXR, retinoid X receptor; SCN, suprachiasmatic nuclei; Tef, thyrotroph embryonic factor; ZT, zeitgeber time.
References
1. Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science (2010) 330:1349–54. doi:10.1126/science.1195027
2. Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron (2012) 74:246–60. doi:10.1016/j.neuron.2012.04.006
3. Brown SA, Kowalska E, Dallmann R. (Re)inventing the circadian feedback loop. Dev Cell (2012) 22:477–87. doi:10.1016/j.devcel.2012.02.007
4. Bozek K, Relogio A, Kielbasa SM, Heine M, Dame C, Kramer A, et al. Regulation of clock-controlled genes in mammals. PLoS One (2009) 4:e4882. doi:10.1371/journal.pone.0004882
5. Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science (1998) 280:1564–9. doi:10.1126/science.280.5369.1564
6. Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science (2001) 293:506–9. doi:10.1126/science.1060699
7. Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, et al. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron (1998) 21:1101–13. doi:10.1016/S0896-6273(00)80627-3
8. Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell (1999) 98:193–205. doi:10.1016/S0092-8674(00)81014-4
9. Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell (1997) 91:1055–64. doi:10.1016/S0092-8674(00)80495-X
10. Sun ZS, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee CC. RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell (1997) 90:1003–11. doi:10.1016/S0092-8674(00)80366-9
11. Takumi T, Matsubara C, Shigeyoshi Y, Taguchi K, Yagita K, Maebayashi Y, et al. A new mammalian period gene predominantly expressed in the suprachiasmatic nucleus. Genes Cells (1998) 3:167–76. doi:10.1046/j.1365-2443.1998.00178.x
12. Takumi T, Taguchi K, Miyake S, Sakakida Y, Takashima N, Matsubara C, et al. A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J (1998) 17:4753–9. doi:10.1093/emboj/17.16.4753
13. Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature (1997) 389:512–6. doi:10.1038/39086
14. Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron (1998) 20:1103–10. doi:10.1016/S0896-6273(00)80492-4
15. Pendergast JS, Friday RC, Yamazaki S. Distinct functions of Period2 and Period3 in the mouse circadian system revealed by in vitro analysis. PLoS One (2010) 5:e8552. doi:10.1371/journal.pone.0008552
16. Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron (2001) 30:525–36. doi:10.1016/S0896-6273(01)00302-6
17. Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, et al. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature (1999) 400:169–73. doi:10.1038/22118
18. Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell (2001) 105:683–94. doi:10.1016/S0092-8674(01)00380-4
19. Akashi M, Ichise T, Mamine T, Takumi T. Molecular mechanism of cell-autonomous circadian gene expression of Period2, a crucial regulator of the mammalian circadian clock. Mol Biol Cell (2006) 17:555–65. doi:10.1091/mbc.E05-05-0396
20. Nakahata Y, Yoshida M, Takano A, Soma H, Yamamoto T, Yasuda A, et al. A direct repeat of E-box-like elements is required for cell-autonomous circadian rhythm of clock genes. BMC Mol Biol (2008) 9:1. doi:10.1186/1471-2199-9-1
21. Nakashima A, Kawamoto T, Honda KK, Ueshima T, Noshiro M, Iwata T, et al. DEC1 modulates the circadian phase of clock gene expression. Mol Cell Biol (2008) 28:4080–92. doi:10.1128/MCB.02168-07
22. Rossner MJ, Oster H, Wichert SP, Reinecke L, Wehr MC, Reinecke J, et al. Disturbed clockwork resetting in Sharp-1 and Sharp-2 single and double mutant mice. PLoS One (2008) 3:e2762. doi:10.1371/journal.pone.0002762
23. Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature (2002) 419:841–4. doi:10.1038/nature01123
24. Fujimoto K, Hamaguchi H, Hashiba T, Nakamura T, Kawamoto T, Sato F, et al. Transcriptional repression by the basic helix-loop-helix protein Dec2: multiple mechanisms through E-box elements. Int J Mol Med (2007) 19:925–32. doi:10.3892/ijmm.19.6.925
25. Cho Y, Noshiro M, Choi M, Morita K, Kawamoto T, Fujimoto K, et al. The basic helix-loop-helix proteins differentiated embryo chondrocyte (DEC) 1 and DEC2 function as corepressors of retinoid X receptors. Mol Pharmacol (2009) 76:1360–9. doi:10.1124/mol.109.057000
26. Doi M, Nakajima Y, Okano T, Fukada Y. Light-induced phase-delay of the chicken pineal circadian clock is associated with the induction of cE4bp4, a potential transcriptional repressor of cPer2 gene. Proc Natl Acad Sci U S A (2001) 98:8089–94. doi:10.1073/pnas.141090998
27. Hatori M, Hirota T, Iitsuka M, Kurabayashi N, Haraguchi S, Kokame K, et al. Light-dependent and circadian clock-regulated activation of sterol regulatory element-binding protein, X-box-binding protein 1, and heat shock factor pathways. Proc Natl Acad Sci U S A (2011) 108:4864–9. doi:10.1073/pnas.1015959108
28. Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev (2001) 15:995–1006. doi:10.1101/gad.873501
29. Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res (2007) 35:648–55. doi:10.1093/nar/gkl868
30. Richier B, Michard-Vanhee C, Lamouroux A, Papin C, Rouyer F. The clockwork orange Drosophila protein functions as both an activator and a repressor of clock gene expression. J Biol Rhythms (2008) 23:103–16. doi:10.1177/0748730407313817
31. Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M. Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev (2010) 24:358–67. doi:10.1101/gad.1883910
32. Becker-Weimann S, Wolf J, Herzel H, Kramer A. Modeling feedback loops of the Mammalian circadian oscillator. Biophys J (2004) 87:3023–34. doi:10.1529/biophysj.104.040824
33. Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, et al. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol (2005) 25:3232–46. doi:10.1128/MCB.25.8.3232-3246.2005
34. Yamajuku D, Shibata Y, Kitazawa M, Katakura T, Urata H, Kojima T, et al. Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. FEBS Lett (2011) 585:2217–22. doi:10.1016/j.febslet.2011.05.038
35. Hatanaka F, Matsubara C, Myung J, Yoritaka T, Kamimura N, Tsutsumi S, et al. Genome-wide profiling of the core clock protein BMAL1 targets reveals a strict relationship with metabolism. Mol Cell Biol (2010) 30:5636–48. doi:10.1128/MCB.00781-10
36. Goriki A, Hatanaka F, Myung J, Kim JK, Yoritaka T, Tanoue S, et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol (2014) 12:e1001839. doi:10.1371/journal.pbio.1001839
37. Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol (2005) 12:441–8. doi:10.1038/nsmb925
38. Hamaguchi H, Fujimoto K, Kawamoto T, Noshiro M, Maemura K, Takeda N, et al. Expression of the gene for Dec2, a basic helix-loop-helix transcription factor, is regulated by a molecular clock system. Biochem J (2004) 382:43–50. doi:10.1042/BJ20031760
39. Ikushima S, Inukai T, Inaba T, Nimer SD, Cleveland JL, Look AT. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci U S A (1997) 94:2609–14. doi:10.1073/pnas.94.6.2609
Keywords: PER2, DEC2, E4BP4, circadian rhythm, E-box
Citation: Tanoue S, Fujimoto K, Myung J, Hatanaka F, Kato Y and Takumi T (2015) DEC2–E4BP4 heterodimer represses the transcriptional enhancer activity of the EE element in the Per2 promoter. Front. Neurol. 6:166. doi: 10.3389/fneur.2015.00166
Received: 02 June 2015; Accepted: 09 July 2015;
Published: 23 July 2015
Edited by:
Urs Albrecht, University of Fribourg, SwitzerlandReviewed by:
Jürgen A. Ripperger, University of Fribourg, SwitzerlandFilippo Tamanini, Imperial College, UK
Megumi Hatori, Keio University, Japan
Copyright: © 2015 Tanoue, Fujimoto, Myung, Hatanaka, Kato and Takumi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Toru Takumi, RIKEN Brain Science Institute, 2-1 Hirosawa, Wako, Saitama 351-0198, Japan,dG9ydS50YWt1bWlAcmlrZW4uanA=
 Shintaro Tanoue
Shintaro Tanoue Katsumi Fujimoto
Katsumi Fujimoto Jihwan Myung
Jihwan Myung Fumiyuki Hatanaka
Fumiyuki Hatanaka Yukio Kato
Yukio Kato Toru Takumi
Toru Takumi