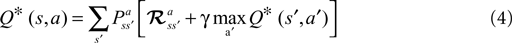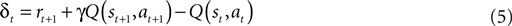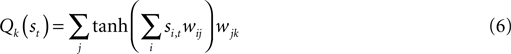In the last decade, there has been an explosion in the number of models and amount of physiological data that can be obtained from either basic neuroscience or computational neuroscience experiments (
Trappenberg, 2002
;
Purves et al., 2004
). The data from these experiments is beginning to be leveraged to address the problem of
reverse-engineering the brain (
Markram, 2006
). This process has been identified by the U.S. National Academy of Engineering as one of the 21st Century’s great challenges for engineering and neuroscience because it has the potential to: (1) impact human health by providing a guide for repairing diseased neural tissue and (2) create innovation in neuromorphic systems based on details of brain function. The challenge arises due to both
anatomical and
behavioral complexity. Human brains contain approximately 10
15 synapses connecting 10
11 neurons of different classes which are hierarchically self-organized across multiple temporal and spatial scales (
Abeles, 1991
). Furthermore, the neuronal properties, structural networks, and functional networks can adapt based on experience and learning.
A prominent project addressing anatomical complexity is the Blue Brain Project which combines realistic neuron models (
Markram, 2006
). Currently, experimental data is fed to a supercomputer (IBM Blue Gene) with 10,000 processors to model a rat neocortical column (one processor per neuron). Despite current analytical and computational limitations, Henry Markram suggests there will be sufficient computational advances to model an entire human brain within 10 years. With a ‘complete’ brain model, behavioral complexity could be investigated through simulated interactions with the environment. An alternative to this approach is to directly investigate behavioral complexity by modeling the relationship between behavior and brain subsystems (e.g. single neurons, neural ensembles)
in vivo. This modeling may be achieved in a typical neurophysiology laboratory given simple models. Notice that both of these approaches only address one area of complexity (anatomical or behavioral).
Alternatively, it would be useful to provide increased computational capacity to facilitate real-time modeling of interactions between multiple brain subsystems, learning, and behaviors. Such interactions may provide insight into existing research questions (e.g. hierarchical vs. connectionist organization) by revealing functional relationships within the brain and between the brain and external world. Recently, increased computation power has been harnessed via a multi-core graphics processing unit for parallel computations (
Wilson and Williams, 2009
). Additionally, the ability for researchers to share (and improve) models has proven successful in projects like
BCI2000 (
Schalk et al., 2004
) and
Physiome (
Asai et al., 2008
). The
Concierge platform at RIKEN provides centralized experimental results and analysis for researchers around the world (
Sakai et al., 2007
). Here we aim to incorporate aspects of all those systems into a modular, user-friendly workstation for
in vivo experiments. We further believe that information technology has an important role to play in encapsulating many details of the laboratory work – translating advanced computational neuroscience methods into accessible tools for experimental neuroscience. The Cyber-Workstation (CW) developed here was designed with those philosophies in mind.
The CW provides a computational infrastructure for real-time neural systems modeling and collection (and storage) of multiple channels of neural and environmental data during behavior. Synchronous modeling and experimentation can provide a window to functional aspects of neural systems that combine sensory, cognitive, and motor processing while interacting with dynamic environments. The CW allows a neuroscientist to expand in vivo paradigms by providing as much computational power as needed.
The CW was developed around and will be demonstrated via a Co-Adaptive Brain-Machine Interface (CABMI). Brain-Machine Interfaces (BMIs) generally create a model of a specific brain subsystem’s (e.g. motor, parietal) interaction with the external world (
Sanchez and Principe, 2007
) such that the BMI can decode user intention to control a prosthetic. Co-adaptive BMIs go a step further by engaging
both the user and a computer model to
learn to control a prosthetic’s movements based on interaction with the environment (
DiGiovanna et al., 2009
). The CW is an ideal platform for CABMI approaches which naturally require concurrent modeling and experimentation. Although neuroscience encompasses many more topics than BMI; implementing a BMI is a microcosm of the challenges in blending computational and experimental neuroscience.
The next section specifies system requirements for BMI research and design choices given our available computational resources. The section ‘Architecture’ describes the system architecture, how the middleware interfaces the software and hardware, and the web-portal user interface. The section ‘Evaluating the CyberWorkstation’ introduces BMI and overviews the CABMI used to demonstrate some CW capabilities. The final section discusses implications of the CW for computational neuroscience.
Functional System Requirements
The practical embodiment of our CW consolidated software and hardware resources across three collaborating labs: Neuroprosthetics Research Group (NRG), Advanced Computing and Information Systems (ACIS) Lab, and Computational NeuroEngineering Laboratory (CNEL) at the University of Florida. The CW enables neurophysiology researchers to setup, carry out, monitor and review experiments
locally that require powerful
remote online and/or offline data processing. The initial design of the CW was centered around the development of BMIs where neural activity was used to directly control the position of a robotic (prosthetic) arm. The CW was designed to improve the efficiency and efficacy of the typical workflow required by closed-loop experiments. Specifically, several aspects of the workflow can be automated, hidden or supported by a cyberinfrastructure that exposes, through a single point of access, only the needed functionality for a neuroscientist to conceptualize, conduct and reason about an experiment. Figure
1
depicts a high-level view of what such an infrastructure might consist of. Neural signals are sampled from behaving animals at NRG and sent across the network for computing. At ACIS (>500 m away from NRG), a variety of computational models process these data on a pool of servers; then results are aggregated and used to control robot movement in real-time at NRG. The architecture of the CW is described in the next section and has general applicability beyond the specific environments of the ACIS, NRG and CNEL laboratories.
Figure 1. Infrastructure for execution of BMI experiments and online/offline analysis of data. Individual components (e.g. experiment management, parallel computing) are discussed in the next section and Figure
2
.
Under the hood of such interface, middleware would be responsible for supporting:
• Real-time operations that scale with biological responses (<100 ms)
• Parallel processing capability for many multi-input, multi-output models
• Large memory capacity for neurophysiological data streams and model parameterization
• Customizable signal processing models
• Data warehousing for large volumes of neural signals, experiment parameters, and computational results (e.g. prosthetic movements)
• Integrated analysis platform for visualization of modeling and physiological parameters
• Simple yet powerful user interfaces for scientists to manage experiments
These application requirements arise from over three decades of BMI research (
Leuthardt et al., 2006
;
Hatsopoulos and Donoghue, 2009
;
Nicolelis and Lebedev, 2009
). Prior BMI researchers have exploited distributed local computation (
Laubach et al., 2003
;
Wilson and Williams, 2009
) or controlled robots by sending control signals around the world (
Fitzsimmons et al., 2009
). However, to our knowledge, the CW is the first architecture to meet these requirements through distributed, remote computing.
The CW architecture was designed to satisfy all of the application requirements in the prior section. Researchers may combine existing models (e.g. a toolbox) and/or develop novel models via object oriented programming constructs. These models are connected in a block diagram and then adaptive middleware transparently implements the specifications using the full power of the remote hardware. In effect, the middleware deploys an on-demand neuroscience research test-bed that consolidates distributed software and hardware resources to support time-critical and resource-demanding computing.
Multiple interacting and customizable components make it difficult to give a single ‘blueprint’ of the CW. However, the CW is divided into four functional layers in Figure
2
; these layers mirror the key stages of the typical workflow (
Sanchez and Principe, 2007
) of a BMI experiment:
Figure 2. Functional CyberWorkstation layers. The left side of each panel represents entities/actions at a neurophysiology lab and the right side represents their associated events at a computing lab. (A) Physical Access and Setup: The CW is configured to communicate with the user’s specific recording hardware and prosthetic. (B) Model Selection and Composition: The user specifies and connects models and, implicitly, necessary resources to run them. (C) Virtual Resources Request and Reservation: The middleware transparently allocates and configures computing resources and network links (black lines are connected, gray lines not used). (D) Run-time Management: Ensures experiment operates within expected timings and provides user-specified visualizations. Here the ‘real-time’ CW operation loop is shown by green (outgoing) and red (returning) lines.
• Access to experimental test-bed
• Model selection and composition
• Virtual resources request and reservation
• Run-time management of experiments
Each of these four stages is discussed in this section, first in a general context and then as embodied in our CW implementation. Real-time system operation is achieved through the use of low-level communication protocols and interfaces, use of parallel computing to reduce computation times and careful optimization of middleware components in the critical path (closed loop in Figure
1
). These and other IT-related design and implementation details will be reported elsewhere (
Rattanatamrong et al., 2009
).
Access to Experimental Test-Bed
This functionality enables neuroscientists to gain access to a physical system consisting of the subject and all devices needed to enable and monitor subject-environment interaction. Necessarily, such a system is specific to a class of experiments and may change over time (e.g. subject specific differences). If a test-bed can be reused among several researchers, ideally individual researchers could avoid the overhead (time and cost) of interfacing and maintaining different components. Instead, it is desirable to allow researchers to specify needed configurations without having to directly modify individual components, each of which may have its own idiosyncratic programming procedures.
The CW enables both the reuse of existing system components and the ability to integrate with remote resources during experiments through what we labeled as a
physical layer (see Figure
2
A). This physical layer is transparent to the CW users but provides an interface with data acquisition hardware and prosthetic devices. This layer ensures that formatting conventions are respected and network communication between the neurophysiology lab and remote computational resources is stable and reliable. During experimental design, the user must configure the physical layer, i.e. specify the manufacturer and model number such that the CW is aware of all hardware it should interact with. However, this configuration is kept at a block-diagram level. Next we provide an example of basic interfacing in the physical layer and communication to the remote facility.
Data acquisition from neurophysiology hardware
In our physical layer, the CW was configured to sample neural signals from a multi-channel, digital signal-processing (DSP) device. The DSP contains buffers that store estimated firing rates from electrodes implanted in behaving animals at the NRG laboratory. After acquiring neural activity from the brain, the CW packs data into an input data structure and sends it over the campus network to the ACIS compute cluster. Our implementation sampled 32 electrodes (up to 96 neurons on our DSPs) in 8.1 ms (see “Evaluation” section). Expanding this to 256 neurons (assuming eight detected neurons per electrode) with current Tucker-Davis Technologies components would only double the necessary sampling (due to better compression). The theoretical maximum number of electrodes the CW could support involves a tradeoff between other processes in the computation budget and network transfer speed. We believe this sampling time will scale linearly with the number of electrodes for the range of array sizes used in current BMI studies (
Hatsopoulos and Donoghue, 2009
).
Prosthetic control
The physical layer also must be configured to control the prosthetic. At NRG the prosthetic was a robotic arm (Dynaservo, Markham, ON, Canada) using four degrees of freedom. This robot accepts both incremental and point-to-point types of movement commands. Incremental control specifies differential changes for each joint of the robot arm, which must finish within a closed-loop cycle (∼100 ms). Alternatively, point-to-point control specifies absolute endpoints but does not guarantee completion time. The CW could seamlessly communicate with the robot in either mode (and switch between modes) to exploit the advantages of each.
Communication to remote facilities
Once this layer is configured, it is necessary to communicate with the remote compute cluster. A connectionless User Datagram Protocol (RFC 768 Standard) socket was selected to provide low latency communication between the data collection site (NRG) and processing site (ACIS). This protocol provides lower overhead for real-time operation at the expense of reliability. Data loss and unexpected delay could happen at any time during the communication over the campus network because of its unreliable and shared nature; thus, reliable data transfer must be ensured at the application level via middleware to support reliable and real-time experiments. Middleware at the remote facility creates a timeout if it is unlikely that computation results will be available in time to meet the deadline. A circular first-in-first-out (FIFO) buffer is utilized to store newly acquired data samples while retransmitting the previously failed results. The possible delay between neural activity and robot action is limited by the size of the buffer, which is adjustable based on model requirements and user preference.
Model Selection and Composition
This functionality enables a computational neuroscientist to choose and implement models on the basis of experimental goals. These models may have components involving statistics and machine learning (e.g. adaptive filters, neural networks, Hidden Markov models, etc. (
Trappenberg, 2002
)). Desirably, an investigator should have access to a toolbox of models that could be instantiated easily and composed as necessary to capture the architecture of the neural system under investigation. The toolbox should be easy to extend and share with different researchers. We provided this toolbox in what we labeled an
allocation layer (see Figure
2
B). It uses a template-based approach where a researcher can specify his or her choices of modules for each part of the template. As long as the modules satisfy pre-specified interface requirements, the components instantiated on the template are guaranteed to communicate and interact correctly. In this layer, the user specifies data flow, model types, experiment priority, and visualization parameters. Middleware infers the amount and type of resources that need to be reserved to execute all the models to meet these specifications. Additionally, it checks the software design in the user-created models to ensure there are no logic violations with respect to the expected interfaces. Next we provide more details of basic configuration in the allocation layer.
Data flow
Notice in Figure
2
A that the local
BMI Controller box, which coordinates data acquisition, signal processing, model adaptation, and prosthetic control, is grayed out. The CW makes that component unnecessary in the neurophysiology lab; instead users specify how neuro-physiological data will be processed (e.g. serially or in parallel) at the remote facilities by creating a block diagram to establish the data flow to/from computational models. Figure
2
B illustrates possible connections with gray arrows and possible parallel processing (of model sets) with gray planes. This allows user flexibility and also conveys explicitly what can be executed concurrently in order to exploit the parallel computing capability of remote machines. The data flow partially specifies the overall complexity of the remote processing and can also be used to implicitly identify further parallelism at a finer grain. The actual models used in each block (e.g. Model A, B, or C) also contribute to complexity and are discussed next.
Model selection
After specifying the data flow, the user must also specify lower-level processing, i.e. which computational models will be used for each block. Here the user has the option to select from a toolbox of established signal processing models (e.g. recursive least squares, moving average). This toolbox is advantageous because the models will have been thoroughly error-checked and vetted by other users. Alternatively, the user can upload custom C++ algorithms for each block.
Visualizations
The user also specifies which experimental variables will need to be monitored during the actual experiment, i.e. at run-time. This is also done in the block diagram (see Figure
2
D). The user inserts and connects a
visualization block to an appropriate area to monitor a variable. The user then specifies which type of visualization (e.g. trace, histogram, error rate) is appropriate.
Experiment priority and type
The final decision is the experiment’s priority. The user informs the CW what type of experiment they plan to run, i.e. online, real-time analysis, or offline. An online experiment will run at the user’s lab and has the highest priority – the CW must meet all user-specified deadlines. A real-time analysis experiment piggybacks onto a collaborator’s experiment, e.g. a user at the CNEL lab can run real-time analysis of an online experiment at the NRG lab. An offline experiment has the lowest priority because it does not have any hard deadlines. Offline experiments are useful to try new signal processing techniques on existing data.
Virtual Resources Request and Reservation
Computing BMI models in real-time often requires investigators to properly structure their models into computational tasks that can be executed concurrently and/or use models that have been previously optimized for parallel execution. These tasks then need to be programmed and deployed on computers that could be dedicated to the test bed or part of a shared computer facility that is usually remote. In the latter case, researchers need to figure out the necessary amount of resources, how to request and reserve them, how to connect to these remote resources and how to submit tasks to (and retrieve results from) them. Ideally, investigators should instead only have to do high-level organization that relies on their domain knowledge (i.e. specific connectivity and processing in the allocation layer) rather than having to refashion algorithms and making sure that communications and computations take place properly on available hardware, both of which require IT expertise. Depending on the extent of the computational demands, the need for real-time computation and the complexity of accessing remote resources, this experimental stage can be rather complex and a significant barrier to the successful completion of an experiment.
We provided this functionality in what we labeled the
configuration layer. This transparent layer includes middleware that organizes and allocates virtual resources to the appropriate physical machines based on the needs specified by the allocation layer. An example of user design choices and middleware reservations is shown using black arrows on the left and right side Figure
2
C respectively. The CW uses virtualization technology to create virtual machines (VM) and each VM can be customized with the necessary execution environment, including operating system and libraries, to support seamless deployment of a computational model. Multiple models can run concurrently with dedicated VMs, where resources are dynamically provisioned according to model demands and timing requirements. Virtualization provides efficient utilization and isolation of resources (e.g. for parallel computation).
Communication between different models is realized using the Message Passing Interface (MPI). MPI allows communication with both simplicity and portability across systems (
Snir et al., 1995
). The integrity and consistency of data in the CW is protected through the mechanisms available from communication channels. In addition, we isolate data addition, modification and deletion from different users in the CW using a lock mechanism. Further improvement via transaction-based consistency protocol is expected as the CW becomes available to more users.
Run-Time Management of Experiments
After physical connection, configuration, allocation the experiment is ready to run. All data must properly collected and unanticipated situations properly handled and documented. Beyond collecting data, it is desirable to be able to graphically visualize spatio-temporal data, critical measures, and control experimental parameters (if necessary). The CW includes what we label a
runtime layer (see Figure
2
D) to ensure all specifications are met.
At the compute cluster, control scripts are used to orchestrate computation processes and communication among them. The master process (blue server in Figure
2
D) manages and distributes data to other computers (worker processes) in the cluster (black servers). The worker processes execute the user’s control scheme. The control scheme comprises both
computation of commands from neural signal and continuous
model adaptation. All model set (black planes) results are aggregated at the master process and sent back to NRG for prosthetic control. Model adaptation occurs between neural commands.
Middleware monitors the elapsed time after sending out neural data. The elapsed time includes the segments marked in Figure
2
D with green (outgoing) and red (incoming) arrows. A timeout happens when it detects that it is unlikely to get the computation results back (following the red path) in time to meet the deadline (e.g. 100 ms). In a timeout, the middleware stores the failed data sample in a circular first-in-first-out buffer, and starts a new closed-loop cycle by polling the neurophysiology hardware. During the new cycle, it queues the newly acquired sample in the buffer and retries the transmission of the previously failed sample. A delay between neural activity and robot action will occur after this type of timeout. However, the acceptable extent of this delay is adjustable based on model requirements and user preference.
Web Portal
The prior sections provided an abstract overview of the CW’s functional architecture. Here we describe the Web-based portal where users actually interact with the CW. Through this portal, users can access the CW from anywhere with an Internet connection. Portal functionalities are provided via AJAX-based JSR-168 compliant portlets, which allow flexible interface customization and responsive, asynchronous content update. Users can configure the portal environment to meet their needs. The main portlets developed were model integration, experiment management, and visualization.
Figure
3
illustrates how a user would interact with the web portal to configure the allocation layer (see Model Selection and Composition). After making all physical connections with the CW (e.g. physical layer), the user should be able to interface with all other CW layers through the Web portal. All input, output, and configuration data of this experiment are persistently stored in the compute cluster, so users can retrieve data or replay the experiment later. The
experiment management portlet in the Web portal handles the selection of motor control model sets and model connectivity (i.e. allocation layer). It also ensures that any custom models follow the CW’s model API. Model configuration (e.g. static and dynamic parameters, initializations, etc.) and visualization planning will also be performed in this portlet. Additionally, users can fine-tune dynamic model parameters
on the fly. Beyond the collection of models already provided in the CW, users can add custom models, developed by the CW’s model API, into the CW’s collection via the
integration portlet. This portlet requires an XML (Sperberg-McQueen et al., 2008) model specification file (containing all information regarding the model’s configurable parameters), sample input files, and default parameter values; this information enables the experiment manager portlet to automatically generate a model configuration interface for users.
Figure 3. Flow diagram to configure allocation and configuration layers via the web portal. Orange shapes represent explicit actions the user makes in the CW’s interface, while green shapes represent implicit actions (e.g. decisions, modifications).
A conceptual overview of
visualization portlet is given in Figure
4
, it is a window to the CW’s run-time layer. The visualization portlet supports the monitoring of prosthetic and model behavior via real-time video streaming. When the user terminates the experiment (as scheduled or due to an emergency), they will not lose any results. All details of the experiment can be downloaded as one archived file and examined locally after the experiment is ended. Also, users can perform extensive, remote analysis immediately with the tools (e.g. Matlab) provided in the CW. Offline and analysis experiments are configured similarly (Experiment types detailed in the section ‘Experiment Priority and Type’). If the user would like to adjust the model configuration to study alternative parameters or to improve performance, they can fine-tune dynamic model parameters during the experiment in the experiment management portlet.
Figure 4. Conceptual interface with CW run-time layer.
Evaluating the Cyberworkstation
The CW has been designed to meet seven specifications listed in the section ‘Functional System Requirements’. In the prior sections, we have developed an architecture that is powerful, flexible, and user-friendly. Here, we use an online BMI experiment to benchmark the CW’s effectiveness in meeting each requirement. BMI architectures and learning tasks are briefly overviewed for context. Then we demonstrate CW performance for each of the seven specifications.
Brain-Machine Interfaces
Conceptually, a BMI learns an unknown mapping between neural signals and some prosthetic output (e.g. cursor position, endpoint velocity) to decode user ‘intent’. This mapping is typically initially random but is adapted over time to minimize some error metric (e.g. misalignment, failed trials). There are many BMI implementations (e.g. linear regressors, population vectors, kernel methods, point process models); each is tailored towards a particular application (
Schwartz et al., 2006
;
Sanchez and Principe, 2007
;
Hatsopoulos and Donoghue, 2009
). The CW’s flexible architecture supports a variety of BMI systems whether they use single models [e.g. Recursive Least Squares (RLS)] or combinations of models (e.g.
mixtures of experts). We developed a BMI based on Reinforcement Learning (RL) (
DiGiovanna et al., 2009
); in the next subsections we describe RL, RL-based BMI, and the
learning tasks. This BMI specifically used real-time processing, limited parallel computation, ‘large’ memory capacity, and data warehousing capabilities of the CW. Later, we also address how other CW abilities could be exploited.
Reinforcement learning
The RL paradigm involves an agent that learns to interact with an environment in a fashion that maximizes future rewards (
Sutton and Barto, 1998
). The agent environment interface is modeled as a Markov Decision Process. Agent
actions influence environmental
state and after completing an action, the environment may provide a reward. The agent tries to maximize return
Rt which is simply the discounted (γ≤1) sum of rewards
rn for all future times
n. Equation 1 shows that return is conditional on current state-action pairs. The agent does not know whether selected actions were optimal as they are executed. However, over time it can build an estimate of return – a value function
Q. These value functions (Eq. 2) are recursively consistent such that Eq. 2 can be rearranged into Eq. 3 (please refer to
Sutton and Barto, 1998
for derivation). Given Eq. 3 the agent can start with an initially random
Q and learn the value function from observations of states, actions, and rewards. If the agents learns the optimal value function
Q*, then it can always maximize reward by taking actions which maximize
Q*.



RL can provide an unbiased estimator of the Bellman Optimality Equation (4) (
Sutton and Barto, 1998
) which can be solved if the state transition
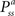
and reward probabilities
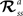
are known and stationary. In situations where these probabilities are unknown and/or non-stationary (e.g. a BMI), RL methods can adapt the value function (Eq. 3)
towards the
Q* based on temporal difference (TD) error (Eq. 5) (TD error is created by rearranging the terms in Eq. 3). Positive error indicates things are better than expected and
Q(
s,
a) should be increased, negative error indicates the opposite. TD learning is a
boot-strapping method using future value estimates to improve current estimates. TD learning is sensitive to the accuracy of
Q because it assumes
Q(
st + 1,
at + 1) is a good approximation of
Rt + 1; hence, it may become unstable. However, TD methods asymptotically converge to
Q* (
Sutton and Barto, 1998
).
RL-based BMI (RLBMI)
We model the interaction of a paralyzed patient (i.e. user) with an intelligent prosthetic controller (agent) to maneuver a robotic arm to a target lever and press it (similar to
reach and grasp) as a cooperative RL task (
DiGiovanna et al., 2009
). A RL framework is useful for a BMI because it creates an agent that
constantly learns from interacting with the world without requiring a ‘desired’ (training) signal (e.g. user movements). We assume relevant control features (e.g. robot position, goal) will be detected by the user and affect the user’s neuronal modulations. To achieve control, the agent must learn both how to detect relevant neuronal states and, given the state, evaluate each possible control action such that it can complete tasks (i.e. earn reward).
Briefly, rats had to use a BMI to maneuver a robot to one of two possible targets (randomly selected). Rats were given cues (e.g. lights, target position, robot position) for the task. In brain control, the rat’s objective was to modulate neurons in primary motor cortex to create a neural state. The agent’s objective was to perform value function estimation on this neural state and select a robot control action every 100 ms. If the agent and rat successfully maneuvered (selected a sequence of actions) the robot endpoint proximal to a target within 4.3 s, both were given a reward and the trial ended successfully. Otherwise, the robot was reset to original position and neither entity was rewarded. Important RL features:
• States: ensemble of neuronal firing rates
• Actions: robot endpoint motions (set of 26)
• Rewards: +1 proximal to correct target, else −0.01
Value function estimation
Neuronal state detection and value function estimation in the above paradigm are both
non-trivial tasks for the agent. The state was (average) 60 dimensional (one firing rate or history per dimension) which is intractable for familiar
look-up table approaches (
Sutton and Barto, 1998
). Instead, an adaptive projection transforms the state into a subspace where segmentation is performed (
DiGiovanna et al., 2007
). Specifically, a Multi-Layer Perceptron (MLP) both segments the state (in the hidden layer) and estimates the value
Q (in the output layer) as:
Q is a function of state and weights (w) with i, j, and k indexing the state, hidden layer, and output layers respectively. An ε-greedy policy typically selects the action corresponding to the
maximum Qk.
There are two computational tasks created here, network
initialization and
adaptation. Initialization refers to rapid adaptation of
initially random MLP weights to estimate
Q based on a sequence of observed states, actions, and rewards. Here the mapping is non-linear and contains many local minima. To increase the probability of converging to a global minimum, multiple networks were created and trained. Networks were adapted by back-propagation of TD(λ) error (similar to TD error described above) as described in (
Sutton and Barto, 1998
). Each network was trained for up to 1000 times (epochs). We initially did this offline due to computational limitations (speed and memory) in our local workstation. Initialization includes many adaptations; however, real-time adaptation is challenge that cannot be addressed offline. After each action (every step) the agent has a new state, action, and reward observation; hence new error.
Q(λ) learning can require up to 42 complete weight updates [see Real-Time Operation that Scale with Biological Responses (<100 ms)] per time-step in this task. This must be completed within 100 ms for adaptation and as soon as possible for initialization.
Experimental Setup
A series of both online and simulated RLBMI experiments was used to benchmark CW performance. A 100-ms deadline was imposed on each closed-loop control cycle, which consists of four phases: data acquisition, network transfer, model computation, and robot control (see Figure
2
D). Neural signals (32 channels) were sampled through a DSP device (online) or from offline data stores (simulated). The data acquisition and robot control server (local) has dual 2.4 GHz Xeon processors and runs Windows Server 2003. Remote computation was conducted on a cluster of VMware ESX server 3.0-based VMs hosted on several dual 3.2 GHz Xeon servers. Each VM has 1 GB RAM and runs Ubuntu Linux 7.04. The experiment was submitted, controlled, and monitored locally via the Web portal.
Evaluation
Real-time operation that scale with biological responses (<100 ms)
In an online RLBMI experiment with 8151 time steps (13 m 22 s), 99% of closed-loop control cycles were completed in less than 10 ms; 100% completed within 100 ms (
Zhao et al., 2008
). This demonstrates the CW provides a high-performance computing environment capable of real-time experiments including BMI adaptation. Table
1
reports timing results in more detail, including the statistics of the entire cycle time as well as individual phase latency (see Figure
2
D for labels).
The data acquisition phase is responsible for a majority of overall variability in closed-loop operation time. Aquiring neural ensemble firing rates from neurophysiology hardware requires 256 bytes (8 bytes per electrode), which is trivial to transfer. However, problems arise because Windows Server 2003 is not a real-time operating system. Although we assign both real-time priority and stop unnecessary services, the Windows Scheduler did not always allow acquisition (or robot control) to start immediately.
The relatively large variance in model computation is related to the specific algorithm used in this example. In
Q(λ) learning, error is back propagated through the MLP at each time step (similar to other neural networks). However, this error includes an approximation of return (Eq. 1), which is not completely available at time
t + 1 (details in
Sutton and Barto, 1998
). Instead the MLP was adapted multiple times; increasingly often as trial length increased. For example, a four-step trial has an average of 1.5 weight updates per time step [(3 + 2 + 1)/4]. If the rat used the maximum trial length (43 steps) then there were 21 average weight updates per time step. Since the rat is free to maneuver the robot along any path (and trials may not be successful), trial length and computation complexity were variable.
Parallel processing capability for multi-input, multi-output models
In addition to closed-loop (online) mode, the CW excels in offline BMI training. A modest example is shown in Figure
5
for the real world problem of RLBMI initialization (also common to other BMI). Specifically, value function estimation (see Value Function Estimation) is not guaranteed to converge to a global optimum, so multiple MLPs (planes in Figure
2
) are initialized and trained to find the best performer. The CW allocates VMs to initialize and train each MLP. The CW outperforms local computation if at least 2 VM (each training 18 MLPs) are used. If 18 VM (each training 2 MLPs) are used, initialization time is reduced by nearly a factor of 5. This creates the opportunity to fully initialize (or re-initialize) the BMI with the CW between normal trials. This contrasts sharply with our prior solution of offline batch training in (
DiGiovanna et al., 2009
) or the common practice of disjoint
train and
testing phases (
Hatsopoulos and Donoghue, 2009
).
Figure 5. Cyberworkstation performance as a function of number of virtual machines used. The time required (error bars are standard deviation in 10 trials) to initialize a RLBMI using rat data from (
DiGiovanna et al., 2009
). The CW is faster (mean) than the local computer (AMD Turion 64 X2 [1.8 GHz dual core], 4 Gb RAM) when more than one VM is used (single VM time was 4883 ± 33 s). Additionally, the local computer is free for other processing tasks.
The CW provides the necessary computational performance in experiments where scalability and code reusability are critical. A remote, resource-rich computing center makes it possible to provision the CW with large numbers of virtual/physical machines. This removes resource impediments to the scaling of models and data sets leaving only algorithm parallelization constraints. Two interesting aspects of Figure
5
are that the CW is slower than local processing when a single VM is used and computation time increases from 18 to 36 VMs. Overheads in virtualization, communication, resource sharing, parallelization and middleware execution that are not present in a local computer cause the first issue. Using only a single remote VM creates this overhead without any of the benefits; hence, the local workstation is faster. Using additional VM creates speed increases up to a certain point (here >18) when the number of VMs becomes sufficiently large such that the communication and sharing behavior (all VMs write to the same set of files) between VMs exceed the computational savings gained from parallel computation of RLBMI networks.
Large memory for neurophysiological data streams and model parameterization
Brain-machine interfaces can present variable challenges to a cyber workstation depending which models (or model sets) are being employed. For example, a linear regression from neural signals to hand position using least mean squares (common in BMI) has complexity O(N) (where N is the number of neural inputs) for each output dimension, D. If a designer wanted to use Gaussian Processes to transform the neural data before filtering, they could add another model but this would have complexity O(M3) where M is the number of training samples. Neurophysiological data often is repeatedly processed in a BMI; hence it may require large, dynamically allocated memory blocks that must scale with N and time of control. Admittedly, RLBMI is parsimonious in parameters relative to other BMI; we expect CW strengths will be better showcased by other BMI – some of which scale exponentially with
N or D.
Customizable signal processing models
The aspect of the CW with the greatest research potential is its inherent flexibility. Both data flow and signal processing are totally customizable, e.g. in Figure
2
B the CW user is presented with a fully interconnected set of three processing models between the input and output. Figure
2
C shows how the user could configure the data flow. Specifically, the input goes directly to Model A. Model A has reciprocal connections with B and an outgoing connection to C. Model C has outgoing connections to B and the BMI output. The remaining gray arrows were not utilized. Additionally, the user has selected multiple parallel BMI implementations that may or may not have the same data flow as described above (the user can customize each parallel implementation). The models (i.e. A, B, C) in Figures
2
B,C are customizable. Users can either select models from an established model
toolbox (e.g. least-squares, RLS) or create their own models in C++ and upload them to the CW.
Model topology and configuration will depend on the particular task attempted. One interesting combination that could be applied to a state-based controller (e.g. RLBMI) involves Gaussian Process models (
Deisenroth et al., 2009
) to segment neural state, linear filters to calculate state value, state/reward transition models to estimate the future, models that aggregate learning from experience and prediction, and finally inverse kinematics optimization models to find robot actuations that achieve selected actions. It may also be useful to have a
mixture of experts, which is parallel implementation of some of these models such that there is an
expert (e.g. value function estimator) for different areas of the state space. We are actively pursuing this topic using the CW.
Integrated analysis platform for visualization of modeling and physiological parameters
The CW also facilitates integrated analysis both for the local (neurophysiology) lab and collaborators elsewhere. A conceptual overview of this integration is shown in Figure
6
where local CW users are running a standard experiment with a single visualization. Additionally, collaborators are simultaneously analyzing (processing represented by dashed lines) the same experiment. The remote lab can perform additional analysis and communicate with the local lab through an instant messaging client. This is advantageous for the local lab because they get feedback and assistance from the remote lab. The remote lab can implement advanced analysis metrics and even alternative models without causing the local lab to divert their own attention from ensuring the BMI user’s safety, the prosthetic function, and their own analysis. If the remote lab develops a model that exhibits better performance (prosthetic control), then ideally their model could be selected and put online locally. The remote lab also benefits because they are given access to a closed-loop experiment without having to invest in access to BMI users and neurophysiology hardware.
Figure 6. Integrated analysis platform for real-time modeling and collaboration.
Data warehousing
The CW also provides vast data warehousing capability. Before the CW, computational neuroscience experiments at the NRG lab generated 1–2 Gb of data per hour. Storage was scaled via external hard drives; however, drive failures could be catastrophic, switching between users required switching drives, and these drives were not easily incorporated into a data back-up system. The CW simplifies and centralizes this storage to a Redundant Array of Inexpensive Disks (RAID) high-capacity storage server in the ACIS lab.
Simple user interfaces
As previously described in the section ‘Architecture’, the CW was designed with a focus on creating a simple and user-friendly interface to allow BMI developers to focus on research instead of low-level technical details. In order to use available models in the CW, the user does not need to know the specific details of the models; he or she only needs to understand the concepts. Help is provided via in-line mouse-over tips to be able to work with the models in short amount of time. The user interface also integrates multiple terminals (portlets) while conducting online experiments to give users a unified control interface.
We have developed here a new framework to seamlessly bridge computational resources with in vivo experiments to better study the functional aspects of brain systems operating in dynamic environments. Online and offline BMI experiments execute in a closed-loop manner that includes in vivo (for online experiments) data acquisition, reliable network transfer, parallel model computation, and real-time robot control. Scientists can conveniently deploy their algorithms and control structures on the cyber-infrastructure and conduct research through its Web portal. We have shown proof of concept that BMI control schemes (specifically RLS and RLBMI) could be implemented and tested on the CW. Additionally; we demonstrated that the CW met each requirement on the BMI designer’s wish list.
This CW potentially dissolves analysis barriers in neurophysiology laboratories while further systematizing both experimental and computational neurophysiological investigations. The CW achieves this functionality by linking available hardware with user-friendly software architecture via a powerful middleware layer that manages their interaction. This novel integration approach makes the CW powerful and customizable but also hides complexity with easy-to-use interfaces for users to conduct research. This simple interface makes the CW accessible to a variety of users, e.g. scientists, engineers, and clinicians. We hope this accessibility catalyzes collaborative research.
It is often the case that data from BMI or general neuroscience experiments are used for follow-up studies which require access to the experimental data according to specific formats and execution of processing tasks to analyze, mine, or identify patterns of interest in the data. One hour of experiments can easily generate gigabytes of data. The CW securely preserves this data in persistent storage, yet keeps it easily available for online processing and visualization. Additionally, analysis code can be centrally stored to ensure reported results are reproducible.
The CW provides the necessary resources for future studies including models to estimate future neural states, environmental rewards, and user’s internal reward (
Mahmoudi et al., 2009
). This modification would allow a BMI to learn from both
experience (
DiGiovanna et al., 2009
) and model
prediction of possible environmental interactions; thus facilitating faster learning. Additionally, finding explicit functional relationships between brain states, prosthetic actions, and rewards could provide insight into how the different brain areas process and share information. Even biologically reduced models (reductionist approach) can be analyzed to reveal features (e.g. parameter sensitivity, hidden states) which correspond to neural or behavioral decision variables (
Corrado and Doya, 2007
).
The capability to include extra models is also useful for prosthetic control, specifically for finding differential commands for each robot joint based on predicted endpoint positions. Determining appropriate commands requires inverse kinematics, but this calculation for a redundant four degree-of-freedom robot (non-unique endpoint to joint angles mapping) does not have a closed form solution (
Craig, 1989
). Joint angles can be found via optimization, but it is not guaranteed to find a feasible solution. Error checking and repeated optimizations consume the computing budget. Parallel computation increases the probability of at least one feasible solution in minimal time.
The CW does face a number of possible limitations. The major impediment is that researchers have developed their own models and are not able to devote resources to convert them to C++ compatible with the CW’s API. This could limit both the number of available models in the toolbox and number of CW users (to refine said models). Future CW development may expand support for models written in other languages (e.g. Matlab, Python); however, CW adoption may be too time consuming for some labs. Modeling flexibility could be helpful for users who do use the CW; however, middleware is only responsible for checking that users’ code includes a set of data structures for communication – there is no check whether the code works. Users will need to validate models on similar processing architectures before uploading them to the CW. Additionally, transmitting neurophysiological data from animals or humans across the Internet could raise privacy and security concerns [e.g. compliance with Health Insurance Portability and Accountability Act (HIPAA) laws]. Finally, real-time communication cannot be guaranteed for arbitrary Internet connections whose communication latencies are excessive.
Overall, this solution for distributed BMIs could lay the ground for scalable middleware techniques that, in the long run, can support increasingly elaborate neurophysiologic research test beds in which subjects can carry out more complex tasks. These advanced test beds will be essential for the development and optimization of the computational components that can be implemented on future workstations with multiple multi-core processors which, collectively, will be able to provide the necessary resources for deeper study of neural coding and function. This new computational platform is transformative by providing: (1) access to user friendly interfaces to dynamically manage and analyze experiments, (2) unrestricted computer power for simulation, signal processing of brain signals and experimental control, (3) huge storage for data and (4) real-time and closed-loop subject feedback, 24/7, anywhere in the world.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was funded by the NSF under Grant #CNS-0540304. We thank Loris Marchal for his development of RLS algorithms for pilot testing of the CW.
Abeles, M. (1991). Corticonics: Neural Circuits of the Cerebral Cortex. New York, Cambridge University Press.
Asai, Y., Suzuki, Y., Kido, Y., Oka, H., Heien, E., Nakanishi, M., Urai, T., Hagihara, K., Kurachi, Y., and Nomura, T. (2008). Specifications of insilicoML 1.0: a multilevel biophysical model description language.
J. Physiol. Sci. 58, 447–458.
Corrado, G., and Doya, K. (2007). Understanding neural coding through the model-based analysis of decision making.
J. Neurosci. 27, 8178–8180.
Craig, J. J. (1989). Introduction to Robotics: Mechanics and Control, 2nd Edn. Reading, Addison-Wesley Publishing Co., Inc.
Deisenroth, M. P., Rassmussen, C. E., and Peters, J. (2009). Gaussian process dynamic programming.
Neurocomputing 72, 1508–1524.
DiGiovanna, J., Mahmoudi, B., Fortes, J., Principe, J. C., and Sanchez, J. C. (2009). Co-adaptive brain-machine interface via reinforcement learning.
IEEE Trans. Biomed. Eng. 56, 54–64.
DiGiovanna, J., Mahmoudi, B., Mitzelfelt, J., Sanchez, J. C., and Principe, J. C. (2007). Brain-machine interface control via reinforcement learning. In IEEE EMBS Conference on Neural Engineering. Kohala Coast.
Fitzsimmons, N. A., Lebedev, M. A., Peikon, I. D., and Nicolelis, M. A. L. (2009). Extracting kinematic parameters for monkey bipedal walking from cortical neuronal ensemble activity.
Front. Integr. Neurosci. 3. doi:10.3389/neuro.07.003.2009.
Hatsopoulos, N. G., and Donoghue, J. P. (2009). The science of neural interface systems.
Annu. Rev. Neurosci. 32, 249–266.
Laubach, M., Arieh, Y., Luczak, A., Oh, J., and Xu, Y. (2003). A cluster of workstations for on-line analyses of neurophysiological data.
IEEE Bioengineering Conference.
Leuthardt, E. C., Schalk, G., Moran, D., and Ojemann, J. G. (2006). The emerging world of motor neuroprosthetics: a neurosurgical perspective.
Neurosurgery 59, 1–13.
Mahmoudi, B., Principe, J. C., and Sanchez, J. C. (2009). An actor-critic architecture and simulator for goal-directed brain-machine interfaces.
IEEE Eng. Med. Biol. Conference (pp. 3365–3368), Minneapolis.
Markram, H. (2006). The blue brain project.
Nat. Rev. Neurosci. 7, 153–160.
Nicolelis, M. A. L., and Lebedev, M. A. (2009). Principles of neural ensemble physiology underlying the operation of brain-machine interfaces.
Nat. Rev. Neurosci. 10, 530–540.
Purves, D., Augustine, G. J., Fitzpatrick, D., Hall, W. C., LaMantia, A.-S., McNamara, J. O., and Williams, S. M. (eds). (2004). Neuroscience, 3rd Edn. Sunderland, MA, Sinauer Associates, Inc.
Rattanatamrong, P., Zhao, M., DiGiovanna, J., Mahmoudi, B., Principe, J. C., Sanchez, J. C., Figueiredo, R., Hermer-Vazquez, L., and Fortes, J. (2009). Design and Implementation of a Cyber-Workstation for Computational Neuroscience (No. TR-ACIS-09-003). Gainesville, University of Florida.
Sakai, H., Aoyama, T., Yamaji, K., and Usui, S. (2007). Concierge: personal database software for managing digital research resources.
Front. Neuroinformatics 1, 5. doi: 10.3389/neuro.11.005.2007.
Sanchez, J. C., and Principe, J. C. (2007). Brain Machine Interface Engineering: Morgan and Claypool.
Schalk, G., McFarland, D. J., Hinterberger, T., Birbaumer, N., and Wolpaw, J. R. (2004). BCI2000: a general-purpose brain-computer interface (BCI) system.
IEEE Trans. Biomed. Eng. 51, 1034–1043.
Schwartz, A., Cui, X. T., Weber, D. J., and Moran, D. W. (2006). Brain-controlled interfaces: movement restoration with neural prosthetics.
Neuron 52, 205–220.
Snir, M., Otto, S., Huss-Lederman, S., Walker, D., and Dongarra, J. (1995). MPI: The Complete Reference. Cambridge, MA, MIT Press.
Sutton, R. S., and Barto, A. G. (1998). Reinforcement Learning: An Introduction. Cambridge, MIT Press.
Trappenberg, T. P. (2002). Fundamentals of Computational Neuroscience. New York, Oxford University Press.
Wilson, J. A., and Williams, J. C. (2009). Massively parallel signal processing using the graphics processing unit for real-time brain-computer interface feature extraction.
Front. Neuroeng. 2:11, doi: 10. 3389/neuro.16.011.2009.
Zhao, M., Rattanatamrong, P., DiGiovanna, J., Mahmoudi, B., Figueiredo, R., Sanchez, J. C., Principe, J. C., and Fortes, J. (2008). BMI Cyberworkstation: enabling dynamic data-driven brain-machine interface research through cyberinfrastructure.
IEEE Eng. Med. Biol. Vancouver.




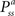 and reward probabilities
and reward probabilities 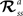 are known and stationary. In situations where these probabilities are unknown and/or non-stationary (e.g. a BMI), RL methods can adapt the value function (Eq. 3) towards the Q* based on temporal difference (TD) error (Eq. 5) (TD error is created by rearranging the terms in Eq. 3). Positive error indicates things are better than expected and Q(s, a) should be increased, negative error indicates the opposite. TD learning is a boot-strapping method using future value estimates to improve current estimates. TD learning is sensitive to the accuracy of Q because it assumes Q(st + 1, at + 1) is a good approximation of Rt + 1; hence, it may become unstable. However, TD methods asymptotically converge to Q* (Sutton and Barto, 1998
).
are known and stationary. In situations where these probabilities are unknown and/or non-stationary (e.g. a BMI), RL methods can adapt the value function (Eq. 3) towards the Q* based on temporal difference (TD) error (Eq. 5) (TD error is created by rearranging the terms in Eq. 3). Positive error indicates things are better than expected and Q(s, a) should be increased, negative error indicates the opposite. TD learning is a boot-strapping method using future value estimates to improve current estimates. TD learning is sensitive to the accuracy of Q because it assumes Q(st + 1, at + 1) is a good approximation of Rt + 1; hence, it may become unstable. However, TD methods asymptotically converge to Q* (Sutton and Barto, 1998
).