- 1Department of Biological Sciences, Faculty of Pharmaceutical Sciences, Teikyo University, Tokyo, Japan
- 2Department of Molecular Biology, Faculty of Pharmaceutical Sciences, Teikyo University, Tokyo, Japan
Animal personalities are stable, context-dependent behavioral differences. Associations between the personality of birds and polymorphisms in the dopamine receptor D4 (DRD4) gene have been repeatedly observed. In mammals, our understanding of the role of the dopamine (DA) system in higher cognitive functions and psychiatric disorders is improving, and we are beginning to understand the relationship between the neural circuits modulating the DA system and personality traits. However, to understand the phylogenetic continuity of the neural basis of personality, it is necessary to clarify the neural circuits that process personality in other animals and compare them with those in mammals. In birds, the DA system is anatomically and molecularly similar to that in mammals; however, the function of DRD4 remains largely unknown. In this study, we used chicks as model birds to reveal the expression regions of the DA neuron-related markers tyrosine hydroxylase (TH), dopa decarboxylase (DDC), dopamine β-hydroxylase (DBH), and DRD4, as well as other DRDs throughout the forebrain. We found that DRD4 was selectively expressed in the mitral cells of the olfactory bulb (OB). Furthermore, a detailed comparison of the expression regions of DA neurons and DRD4 in the OB revealed a cellular composition similar to that of mammals. Our findings suggest that the animal personality gene DRD4 is important for olfactory information processing in birds, providing a new basis for comparing candidate neural circuits for personality traits between birds and mammals.
1 Introduction
Personality, or animal personality, refers to consistent, genetically-based individual behavioral differences in humans and non-human animals that are stable across time and contexts (Gosling, 2001; van Oers and Mueller, 2010; Wolf and Weissing, 2012). The candidate gene that has been most intensively studied for its association with personality and considered the most promising is the dopamine receptor D4 (DRD4) (Ebstein et al., 1996; Kluger et al., 2002; Munafò et al., 2008). Such associations between personality traits and DRD4 polymorphisms are also known in animal personalities, including those of birds (Fidler et al., 2007), but the results of many studies have been ambiguous across species. To gain a deeper understanding of the phylogenetic continuity of the neural basis of personality, it is necessary to clarify the role of DRD4 in the brain function of each species.
In mammals, the dopamine (DA) system is involved in many physiological and higher cognitive functions and has attracted particular attention because of its involvement in human psychiatric disorders, including Parkinson's disease, schizophrenia, and addiction (Carlsson, 2006; Girault and Greengard, 2004; Iversen and Iversen, 2007; Klein et al., 2019). DA neurons are distributed in clusters in the central nervous system. These clusters are classified into two main groups: the A8-A10 cell groups located in the midbrain and the A11-A16 cell groups located in the forebrain (Dahlström and Fuxe, 1964; Bjorklund and Dunnett, 2007). Midbrain DA neuron cell groups project to a wide area of the forebrain, including the nucleus accumbens (NAc), amygdala, prefrontal cortex, and striatum. Recent research has begun to elucidate how DA influences neural circuits that regulate reward, motivation, and aversion at the cellular level (Hillarp et al., 1966; Bjorklund and Dunnett, 2007; Wise, 2004; Verharen et al., 2020; Yagishita, 2020, 2023). In contrast, forebrain DA neurons have local and spinal projections and are involved in the regulation of many physiological functions, including endocrine regulation (Ben-Jonathan and Hnasko, 2001). The released DA acts on its downstream targets, which is mediated by two families of DA receptors: the D1 and D2 receptor families. The D1 family includes DRD1 and DRD5 (also called DRD1B), and the D2 family includes DRD2, DRD3, and DRD4 (Yamamoto et al., 2015). The DRD genes encoding these DA receptors have different expression distributions in the brain, and each is thought to be involved in the regulation of various brain functions (Callier et al., 2003; Bentivoglio and Morelli, 2005). Studies using knockout mice for each DRD subtype combined with various behavioral assays are actively investigating the specific brain functions regulated by each DRD (Holmes et al., 2004). As for DRD4 knockout mice, findings include increased motor hypersensitivity to ethanol, cocaine, methamphetamine, and methylphenidate (Rubinstein et al., 1997; Keck et al., 2013); reduced novelty seeking (Dulawa et al., 1999); enhanced anxiety-related behavior (Falzone et al., 2002); and effects on memory and learning (Ananth et al., 2019). However, no abnormalities in impulsivity were observed (Helms et al., 2008), and the effects on novelty seeking and anxiety-related behavior varied by sex (Thanos et al., 2015). Notably, behavioral abnormalities in response inhibition were observed in heterozygotes rather than in knockout mice (Young et al., 2011). Thus, although it has not yet been fully resolved, DRD4 appears to be related to novelty seeking, emotionality, and behavioral inhibition in mice.
In birds, the anatomical features of the DA system are similar to those in mammals (Reiner et al., 1994; Smeets and Reiner, 1994; Smeets and González, 2000; Reiner et al., 2004). Furthermore, hodological analyses have revealed that the input-output connections of the midbrain DA nuclei in birds are similar in extent to those in mammals (Csillag, 1999; Durstewitz et al., 1999; Mezey and Csillag, 2002; Balint and Csillag, 2007; Balint et al., 2011). The molecular characteristics of the avian DA system have also been thought to be largely conserved in mammals (Yamamoto and Vernier, 2011). However, recent findings suggest that they are not always highly conserved through vertebrates. For instance, birds have lost the dopamine transporter (DAT) gene from the genome, and the DAT function is compensated for by noradrenaline transporter (NAT) (Lovell et al., 2015; Fujita et al., 2022c). Therefore, the degree of conservation between the avian and mammalian DA systems requires careful re-examination. Although several studies have revealed the expression distribution of avian DRDs in the forebrain (Kubikova et al., 2010; Yamamoto et al., 2013), the region where DRD4 is expressed remains unclear.
The most comprehensive study to date on the expression patterns of DRDs in the avian forebrain was conducted in songbirds. Kubikova et al. (2010) used probes for full-length songbird DRD1, DRD2, DRD3, DRD4, DRD5, and DRD1C (referred to as D1D in their study) to reveal the expression distribution across various songbird brain regions. Additionally, they performed cross-hybridization experiments on adult chicken brains using the same songbird probe sets (Kubikova et al., 2010). To date, no clear expression region for DRD4 has been identified in avian brain regions except for the cerebellum. DRD1E, a member of the D1 family lost in mammals, has been identified in the chicken genome (Yamamoto et al., 2015). Therefore, to reveal the expression distribution of DRDs in the chick forebrain, we developed chicken-specific probes for all DRDs and conducted in situ hybridization (ISH) analysis. Furthermore, the distribution of avian DA neurons in the brain has been elucidated through histochemical and immunohistochemical techniques (Ikeda and Goto, 1971; Dube and Parent, 1981; Guglielmone and Panzica, 1984; Moons et al., 1994; Reiner et al., 1994, 2004). The distribution of cell bodies in the chick midbrain was previously revealed using ISH analysis of DA neuron-“related” markers. DA neurons are typically defined as tyrosine hydroxylase (TH)+/dopa decarboxylase (DDC)+/dopamine β-hydroxylase (DBH)– cell population (Fujita et al., 2022c), but those in the chick forebrain using DA neuron-“related” markers have not yet been clarified.
In this study, to better understand the function of DRDs in avians, we investigated the molecular anatomy of the chick forebrain DA system using ISH. We selected all DA receptor genes—DRD1, DRD2, DRD3, DRD4, DRD5, DRD1C (previously called DRD1D), and DRD1E.—along with the chick orthologs of DA neuron-related marker genes, TH and dopa decarboxylase (DDC), and as well as the noradrenergic neuron-related marker gene [dopamine β-hydroxylase (DBH)].
2 Materials and methods
2.1 Animals
Fertilized eggs of domestic chicks (Gallus gallus domesticus, Cobb strain) were purchased from a domestic company (3-M, Aichi Prefecture, Japan) and incubated at Teikyo University (Kaga, Itabashi-ku, Tokyo). Animal experiments were performed as described by Yamaguchi et al. (2008a,b). All chicks used in this study were 1-day-old chicks [post-hatched on day one (P1)]. All procedures were reviewed and approved by the Committee on Animal Experiments of Teikyo University and performed in accordance with the guidelines of the national regulations for animal welfare in Japan.
2.2 Histology
P1 chicks were anesthetized by intraperitoneal injection of a 1:1 solution of ketamine (10 mg/mL, ketalar-10, Sankyo Co., Tokyo, Japan) and xylazine (2 mg/mL, Sigma, St. Louis, MO, USA) at a dose of 0.40 mL per animal, and then transcardially perfused with 4% paraformaldehyde in 0.1 M phosphate-buffered saline (pH 7.5) (PFA-PBS). After perfusion, whole brains were surgically removed quickly and immediately placed in PFA-PBS, then immersed at 4°C for 1 day. The immersion solution was then changed to an 18% sucrose/PFA-PBS solution for cryoprotection and kept at 4°C for 2 days. The sucrose-substituted brains were then embedded in Tissue-Tek Optimal Cutting Temperature (OCT) compound (Sakura Finetechnical, Tokyo, Japan), rapidly frozen on dry ice, and stored at −80°C until further use. In this study, we used a total of 23 chicks, with the following distribution for each probe: 15 chicks for TH, 12 for DDC, 12 for DBH, 9 for DRD1, 11 for DRD2, 11 for DRD3, 12 for DRD4, 9 for DRD5, 9 for DRD1C, 9 for DRD1E, and 5 for glutamate decarboxylase 2 (GAD2) (Supplementary Table S1). Frozen blocks, including brain samples, were cut into 18 μm-thick sections using a cryostat (Leica CM3050S or Leica CM1850, Leica Biosystems, Nußloch, Germany) and mounted on glass slides. The levels of serial coronal sections (A14.6-A4.6) were consistent with those of the chick brain atlas by Kuenzel and Masson (1988). If necessary, the chick brain atlas by Puelles et al. (2018) was used as a reference.
2.3 Selection of the chick orthologs of mammalian DA system genes
Chick orthologs of specific genes involved in the DA system, which are also present in mammals, were selected. Genes for DA production-related genes (TH, DDC, and DBH) and dopamine receptors (DRD1, DRD2, DRD3, DRD4, DRD5, DRD1C, and DRD1E) were included. The DNA and protein sequence similarities between chick and human genes (or zebrafish) for these DA system genes are presented in Supplementary Table S2. DA neurons are typically defined as neurons that express the rate-limiting enzymes TH and DDC, required for the stepwise synthesis of DA, but do not express DBH, the enzyme responsible for synthesizing noradrenaline from DA. There are two genes encoding TH, TH1, and TH2 in vertebrates other than mammals. Here, TH1 is referred to as TH in this study. Using BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi), we searched for sequence similarities between these DA production-related markers and those of other animals described in a previous study (Fujita et al., 2022c). A summary of these orthologous gene probes is presented in Table 1. When multiple transcript variants of an ortholog were registered in the database, probes were designed to detect all the variants. ISH was performed to analyze the expression patterns of the orthologs in the chick brain.
2.4 cDNA cloning and RNA probe preparations
Total RNA was extracted from a chick brain using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) and subjected to a reverse-transcription (RT) reaction using an oligo (dT) primer with SuperScript III kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RT polymerase chain reaction (RT-PCR) was performed using the following gene-specific primer (forward and reverse) pairs: DRD1:5′-TGGATGGAGAAGGGTTGCTT-3′ and 5′- CTGCTTCTGTTGCCACTTGT-3,' respectively; DRD2:5′-CCCCTGAATCTATCCTGGTACA-3′ and 5′-AGATCTGCACGTACACCAGC-3,' respectively; DRD3:5′-ATGTCATGATGTGCACAGCC-3′ and 5′-GGGCGTTGAGGATGTGAATC-3,' respectively; DRD4:5′-TCGTCCTCATCCTGCTCATC-3′ and 5′-GTGGGCATAAGGGTGGTACT-3,' respectively; DRD5:5′-CATCTTCATCGTGTCGCTGG-3′ and 5′-TGATGGAGGACTTGAGGCTG-3,' respectively; DRD1C: 5′-ACTGGTTTGTGCTGTCGTTG-3′ and 5′-TGACAGAAAGGTAGCAGGCA-3,' respectively; DRD1E: 5′-CAACCCCTTCTGCTACGAGA-3′ and 5′-GGCTGCTTTGTACTCCACTG-3,' respectively; GAD2: 5′-GCACAGAAGTTCACCGGAG-3′ and 5′-GGGAACATCTTGAAACGTGC-3,' respectively. The resulting PCR amplicons were subcloned into a pGEM-T Easy Vector (Promega, Madison, WI, USA). Subcloning of the target sequences was confirmed using Sanger sequencing. For the TH, DDC, and DBH probes, we used plasmids prepared in a previous study (Fujita et al., 2022c). All plasmids containing cDNA fragments were amplified by PCR with M13 primer pairs, and amplicons containing the T7 and SP6 promoter sites were purified using a PCR purification kit (Qiagen, Valencia, CA, USA). The digoxigenin (DIG)-labeled sense and antisense RNA probes were prepared by in vitro transcription using a DIG RNA labeling kit (Roche, Basel, Switzerland).
2.5 ISH
ISH experiments were performed according to the method described by Fujita et al. (2019) with minor modifications. Briefly, brain sections on glass slides were fixed in 4% PFA-PBS. After incubation in 10 μg/ml proteinase K in 10 mM Tris-HCl and 1 mM EDTA, the specimens were post-fixed for 10 min in PFA–PBS, treated with 0.2 M HCl for 10 min, washed in PBS, and treated with 0.25% acetic anhydride in 0.1 M triethanolamine (pH 7.5) for 10 min. Hybridization was performed with DIG-labeled RNA probes at 70°C for 19 h 30 min to 21 h 10 min. The size of the probes used is shown in Table 1. After stringent washing, the hybridized probes were immunohistochemically detected using an alkaline phosphatase-conjugated anti-DIG antibody (1:1,000, Roche, Basel, Switzerland). To visualize the signals, a chromogenic reaction with a nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate was performed at room temperature (~24°C) for 16 h 15 min to 20 h 10 min for all probes. Sense probes were used as negative controls in all the experiments.
2.6 Imaging and data processing
Sections on each slide were semi-automatically acquired as digital slides using a NanoZoomer 2.0HT or NanoZoomer XR system (Hamamatsu Photonics, Shizuoka, Japan). Digital photographs of the regions of interest were manually extracted from digital slides using the NDP.view2 software (Hamamatsu Photonics, Shizuoka, Japan). The entire extracted images were converted to 8-bit images, and the brightness and contrast of the entire extracted images were adjusted using ImageJ (https://imagej.nih.gov/ij/).
3 Results
3.1 Expression of TH, DDC, and DBH in the chick forebrain
We performed ISH analysis to investigate the expression patterns of the DA neuron-related markers—TH, DDC, and DBH—using adjacent sections from A14.6 to A4.6 in the forebrain of chicks on post-hatched day 1(P1). We detected cells showing signals of TH in the A16 (Figures 1A, A'), A15 (Figures 2A, I, A', I'), A14 and A13 (Figures 2A, E, A', E'), A12 and A11 (Figures 3A, E, A', E') regions. DDC-positive cells were detected in the A15 (Figures 2B, J, B', J'), A14 and A13 (Figures 2B, F, B', F'), PVO, A12 and A11 (Figures 3B, F, B', F') regions. We found that the expression patterns of TH and DDC in A15-A11 in the chick were similar, confirming the absence of DBH signals.
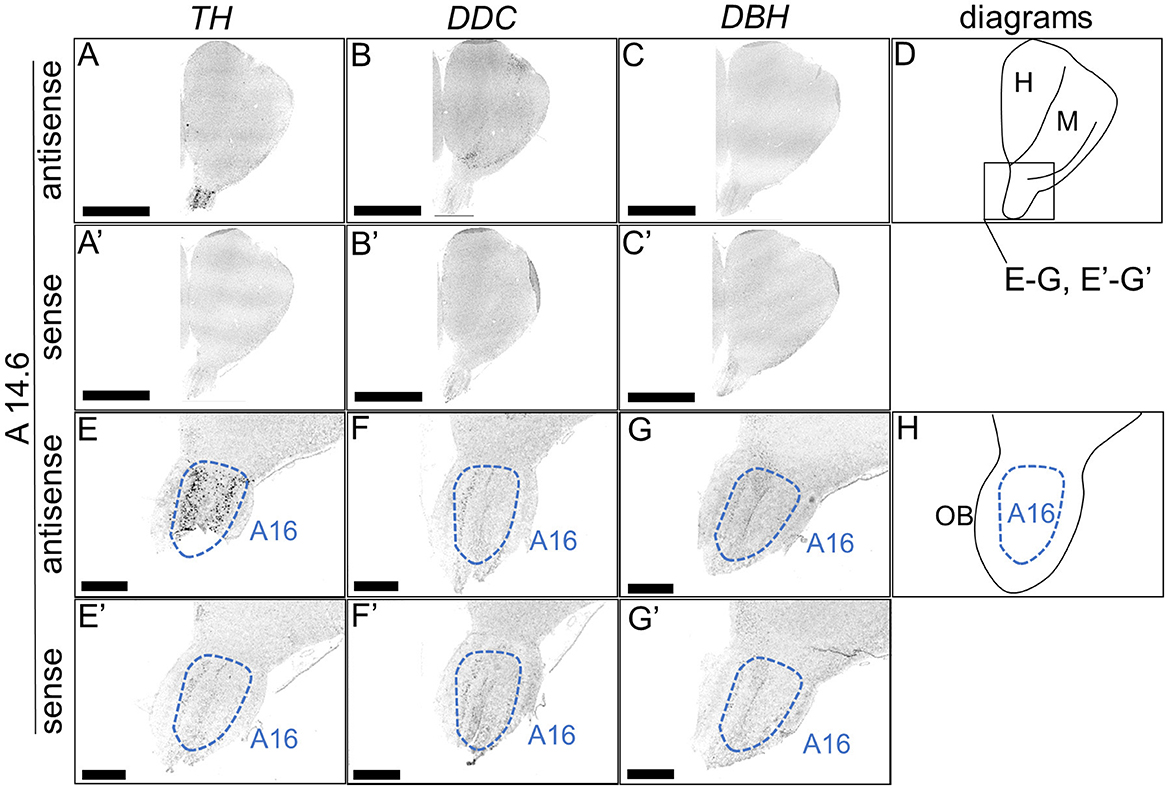
Figure 1. In situ hybridization of TH, DDC, and DBH in the P1 chick rostral forebrain. Digoxigenin-labeled RNA probes, both antisense [TH, (A, E), DDC, (B, F), and DBH, (C, G)] and sense [TH, (A', E'), DDC, (B', F'), and DBH, (C', G')], were used for in situ hybridization in coronal sections of P1 chick forebrains, corresponding level A14.6 in the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility was verified across multiple chicks at the same level, with representative images from neighboring sections provided. (E–G, E'–G') present magnified views of the forebrain region indicated by the box in (D). Diagrams of coronal sections depicted in (A, E) are shown in (D, H), respectively. A16, A16 cell group; H, hyperpallium; M, mesopallium; OB, olfactory bulb. Scale bars = 2.5 mm (A–C, A'–C') and 500 μm (E–G, E'–G').
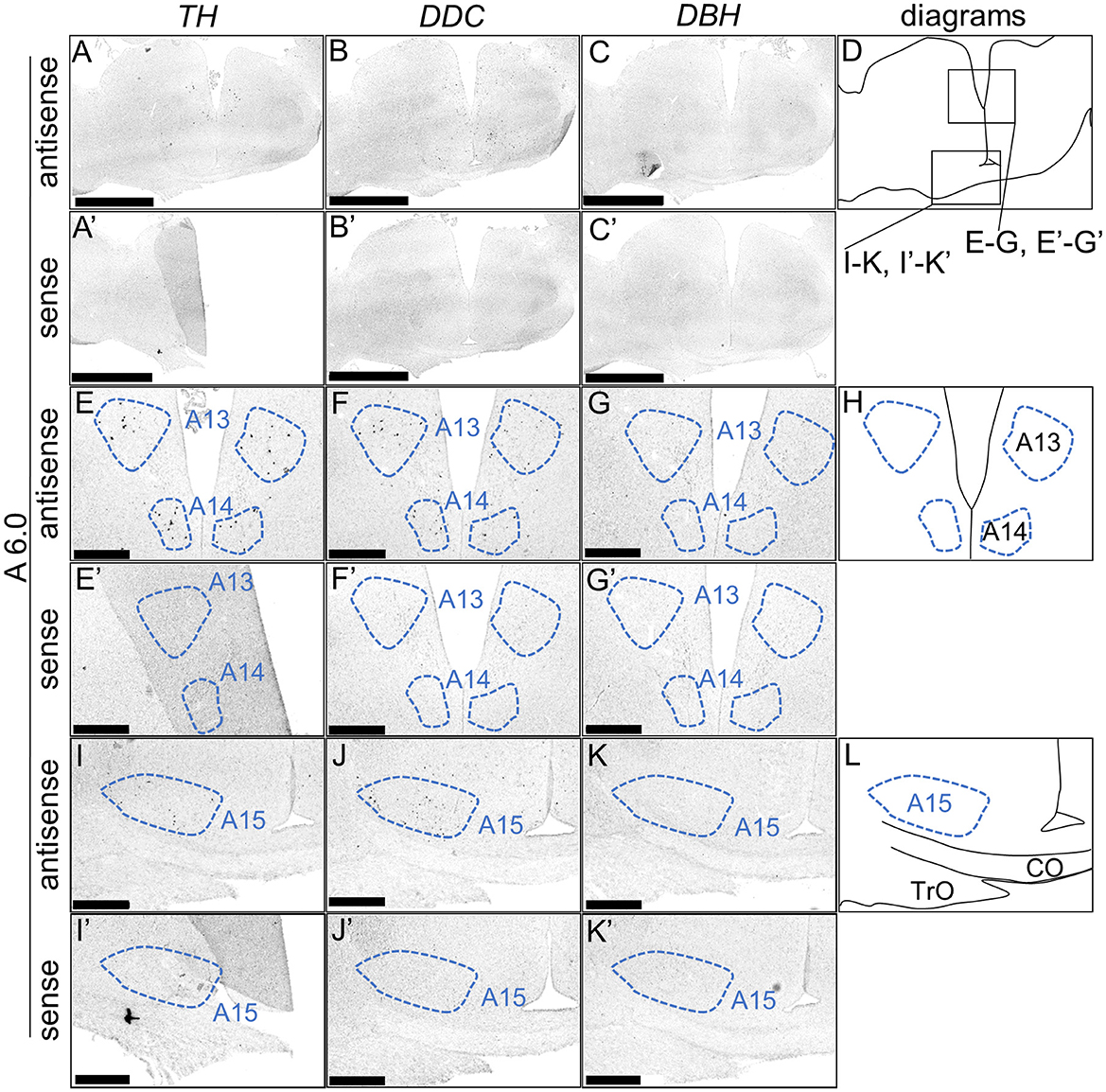
Figure 2. In situ hybridization of TH, DDC, and DBH in the P1 chick diencephalon. Digoxigenin-labeled RNA probes, both antisense [TH, (A, E, I), DDC, (B, F, J), and DBH, (C, G, K)] and sense [TH, (A', E', I'), DDC, (B', F', J'), and DBH, (C', G', K')], were used for in situ hybridization in coronal sections of P1 chick forebrains, corresponding to level A6.0 in the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility was verified across multiple chicks at the same level, with representative images from neighboring sections provided. (E–G, I–K, E'–G', I'–K') present magnified views of the diencephalon region indicated by the box in (D). Diagrams of coronal sections depicted in (A, E, I) are shown in (D, H, L), respectively. A13, A13 cell group; A14, A14 cell group; A15, A15 cell group; CO, chiasma opticum; TrO, tractus opticus. Scale bars = 2.5 mm (A–C, A'–C') and 500 μm (E–G, I–K, E'–G', I'-K').
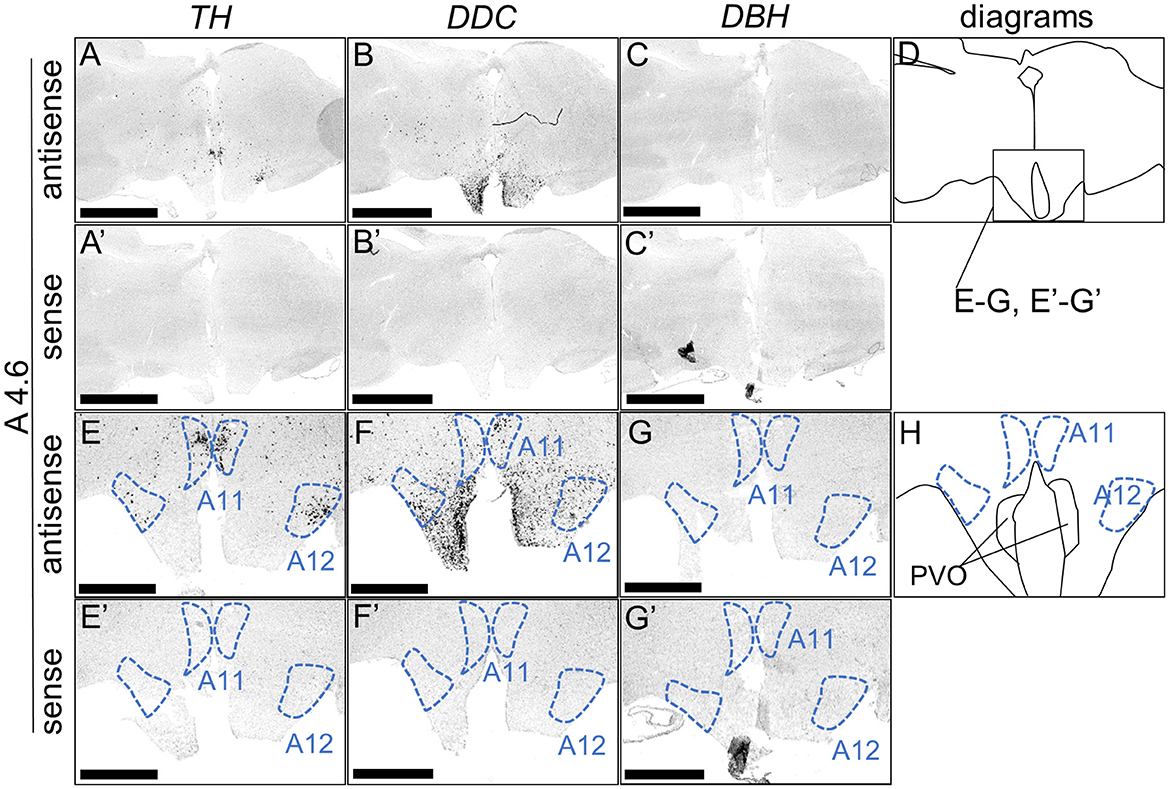
Figure 3. In situ hybridization of TH, DDC, and DBH in the P1 chick hypothalamus. Digoxigenin-labeled RNA probes, both antisense [TH, (A, E), DDC, (B, F), and DBH, (C, G)] and sense [TH, (A', E'), DDC, (B', F'), and DBH, (C', G')], were used for in situ hybridization in coronal sections of P1 chick hypothalamus, corresponding to the A4.6 level in the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility of the signals was confirmed across multiple chicks at the same level, with representative images from neighboring sections provided. (E–G, E'–G') present magnified views of the hypothalamus region indicated by the box in (D). Diagrams of coronal sections depicted in (A, E) are shown in (D, H), respectively. A11, A11 cell group; A12, A12 cell group; PVO, paraventricular organ. Scale bars = 2.5 mm (A–C, A'–C') and 1 mm (E–G, E'–G').
Although typical DA neurons are defined by the expression of TH and DDC but not DBH, neurons exhibiting only some of the characteristics of DA neurons have been identified in mammals (Bjorklund and Dunnett, 2007; Ugrumov, 2009). Specifically, neurons expressing only TH or DDC monoenzymatically, but not DBH, are considered to partially express the DA neuron phenotype. In this study, we detected such neurons in the forebrain of chicks. TH-positive cells were detected in the substantia nigra (SN), medial striatum (MSt), bed nucleus of the stria terminalis, lateral part (BSTL), septum mediale (SM), globus pallidus (GP), olfactory bulb (OB), and periventricular thalamic nuclei (PTM) as shown in Figures 4A, E, I, M, A', E', I', M', 5A, E, I, M, Q, A', E', I', M', Q', and 6A, E, A', E', respectively. Cells with signals of DDC were detected in the BSTL, lateral striatum (LSt), GP, occipito-mesencephalic tract (OM), and PTM (Figures 4B, N, B', N', 5B, J, N, R, B', J', N', R', and 6B, F, B', F'), respectively. No DBH signal was detected in these regions. Additionally, TH and DDC signals were observed in nearby areas, though their expression patterns differed.
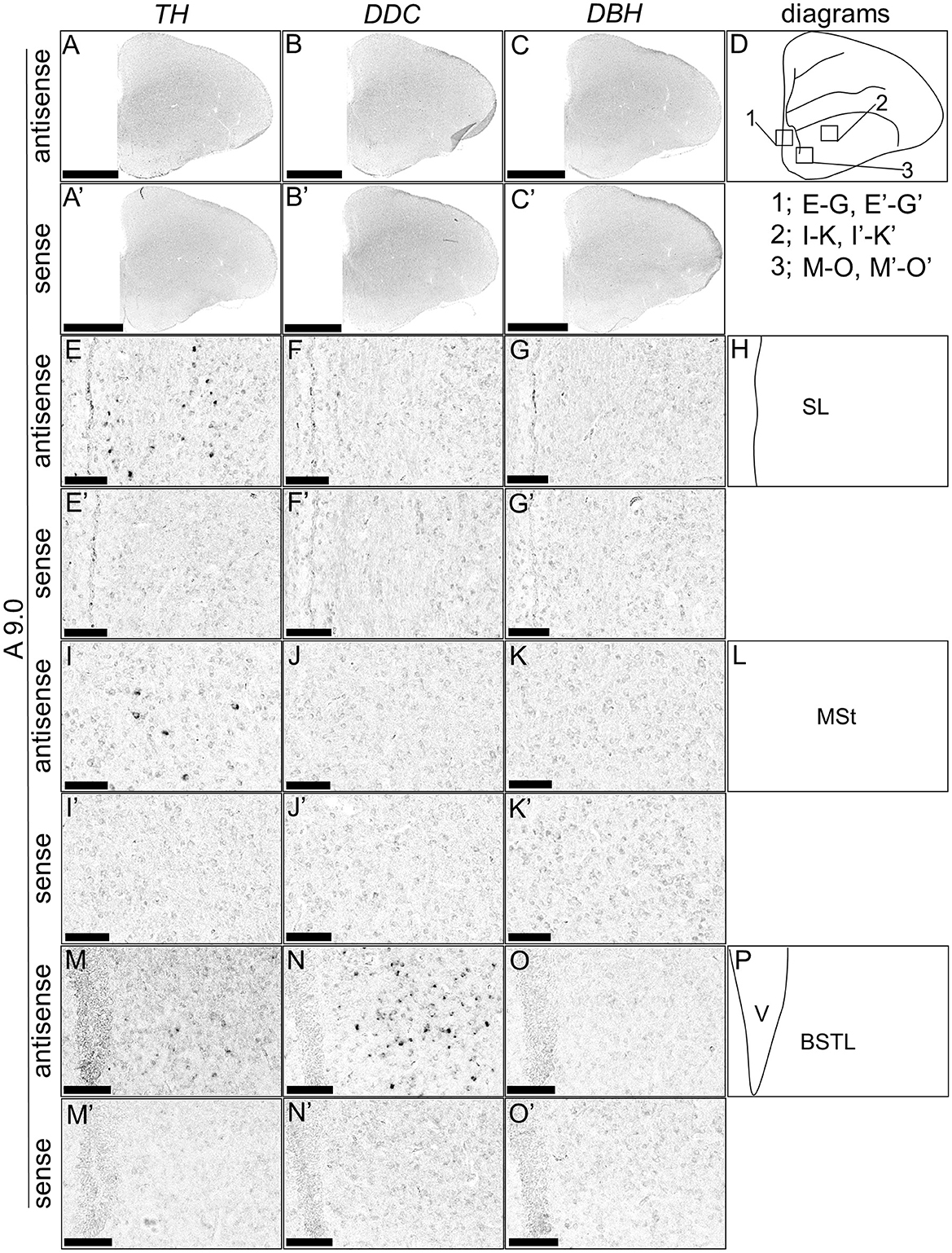
Figure 4. In situ hybridization of TH, DDC, and DBH in the P1 chick A9.0 level forebrain. Digoxigenin-labeled RNA probes, both antisense [TH, (A, E, I, M), DDC, (B, F, J, N), and DBH, (C, G, K, O)] and sense [TH, (A', E', I', M'), DDC, (B', F', J', N'), and DBH, (C', G', K', O')], were used for in situ hybridization in coronal sections of P1 chick forebrains, corresponding to level A9.0 in the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility was verified across multiple chicks at the same level, with representative images from neighboring sections provided. (E–G, I–K, M–O, E'–G', I'–K', M'–O') present magnified views of the forebrains region indicated by the box in (D). Diagrams of coronal sections depicted in (A, E, I, M) are shown in (D, H, L, P), respectively. BSTL, bed nucleus of the stria terminalis; MSt, medial striatum; SL, lateral septal nucleus. Scale bars = 2.5 mm (A–C, A'–C') and 100 μm (E–G, I–K, M–O, E'–G', I'-K', M'-O').
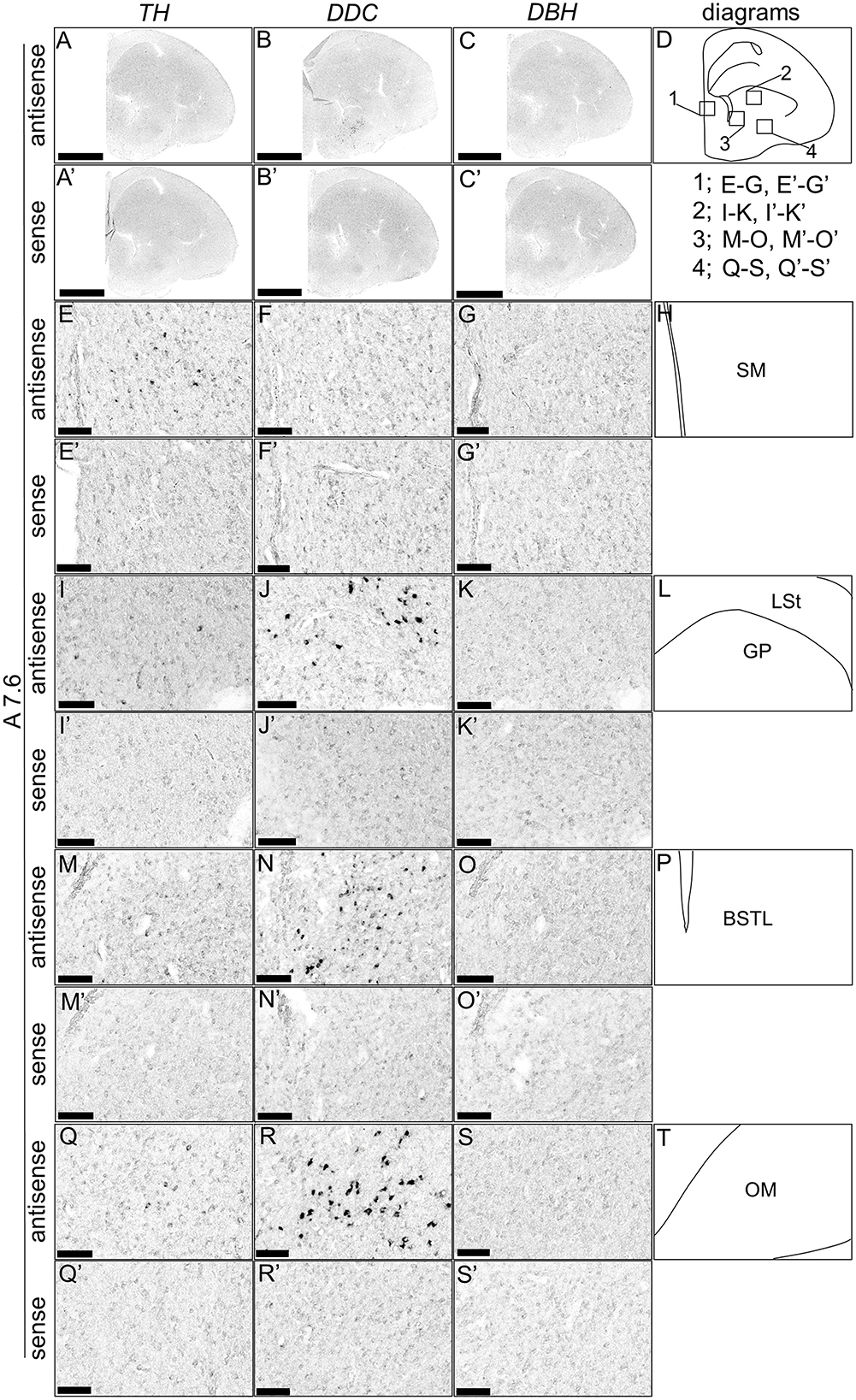
Figure 5. In situ hybridization of TH, DDC, and DBH in the P1 chick A7.6 level forebrain. Digoxigenin-labeled RNA probes, including antisense [TH, (A, E, I, M, Q), DDC, (B, F, J, N, R), and DBH, (C, G, K, O, S)] and sense [TH, (A', E', I', M', Q'), DDC, (B', F', J', N', R'), and DBH, (C', G', K', O', S')] probes, were used for in situ hybridization in coronal sections of P1 chick forebrains, corresponding to the A7.6 level in the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility was verified across multiple chicks at the same level, with representative images from adjacent sections provided. (E–G, I–K, M–O, Q–S, E'–G', I'–K', M'–O', Q'–S') present magnified views of the forebrains region indicated by the box in (D). Diagrams of coronal sections presented in (A, E, I, M, Q) are shown in (D, H, L, P, T), respectively. BSTL, bed nucleus of the stria terminalis; GP, globus pallidus; LSt, lateral striatum; OM, occipito-mesencephalic tract; SM, medial septal nucleus. Scale bars = 2.5 mm (A–C, A'–C') and 100 μm (E–G, I–K, M–O, Q–S, E'–G', I'–K', M'–O', Q'–S').
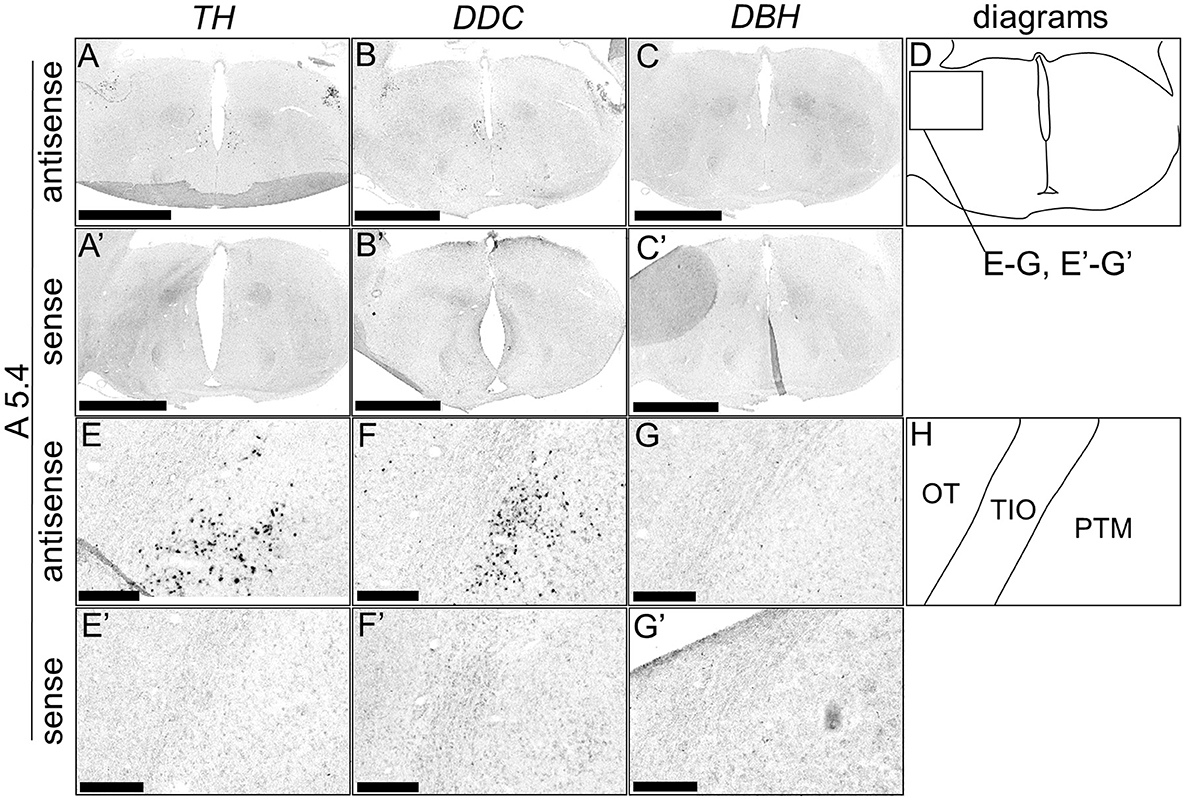
Figure 6. In situ hybridization of TH, DDC, and DBH in the P1 chick A5.4 level forebrain. Digoxigenin-labeled RNA probes, both antisense [TH, (A, E), DDC, (B, F), and DBH, (C, G)] and sense [TH, (A', E'), DDC, (B', F'), and DBH, (C', G')], were used for in situ hybridization in coronal sections of P1 chick forebrains, corresponding to level A5.4 of the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988) was confirmed in multiple chicks at the same level, and representative images of the neighboring sections are shown. (E–G, E'–G') Magnified views of forebrains in the box in (D). (D, H) Diagrams of coronal sections are shown in (A, E). OT, optic tectum; PTM, nucleus pretectalis medialis; TIO, tractus ishmo-opticus. Scale bars = 2.5 mm (A–C, A'–C') and 250 μm (E–G, E'–G').
3.2 Expression of DRD1, DRD2, DRD3, DRD4, DRD5, DRD1C, and DRD1E in the chick forebrain
First, the expression pattern of DRD4 was examined in sections A14.4 to A6.4 in the P1 chick forebrains (Figure 7). Signals were generally poor, consistent with findings from a previous study (Kubikova et al., 2010). However, clear and characteristic signals were detected in the OB (Figures 7A, A', B, B').
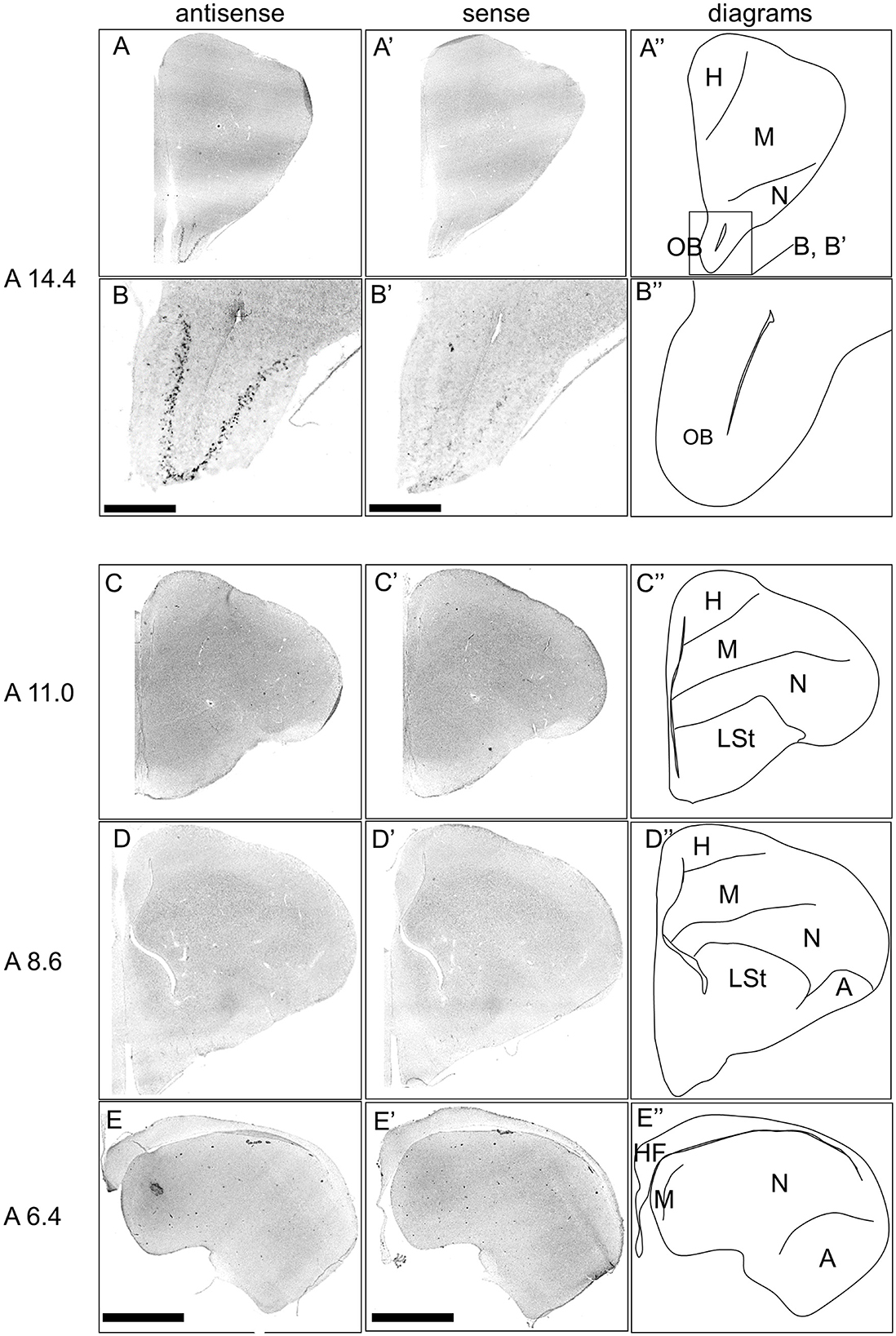
Figure 7. In situ hybridization of DRD4 in P1 chick telencephalons. Digoxigenin-labeled RNA antisense RNA probes, both antisense (A–D) and sense (A'–D') DRD4 probes were used for in situ hybridization in coronal sections of the P1 chick telencephalon. Signal reproducibility was verified across multiple chicks at the same level, with representative images provided. Diagrams of coronal sections are shown in the rightmost (A–D”). Levels of sections (A 14.4 to A 6.4) were in accordance with those mentioned in Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). (B, B') present magnified views of the olfactory bulbs region indicated by the box in (A”). A, arcopallium; H, hyperpallium; HF, hippocampal formation; LSt, lateral striatum; M, mesopallium; N, nidopallium; OB, olfactory bulb. Scale bars = 2.5 mm (A, C–E, A', C'–E') and 500 μm (B, B').
We examined the expression pattern of DRD1 in sections A14.0 to A6.2 in the P1 chick forebrains (Supplementary Figure S1). Signals were detected in the striatum (Supplementary Figures S1B, C, B', C'), lateral nidopallium, dorsal arcopallium (Supplementary Figures S1D, D'), and the hippocampal formation (HF), particularly in the dorsolateral region (Supplementary Figures S1D, D'). We examined the expression pattern of DRD2 in sections A14.4 to A5.8 in the P1 chick forebrains (Supplementary Figure S2), and signals were detected in the striatum, except for GP (Supplementary Figures S2B, C, B', C'). We examined the expression pattern of DRD5 in sections A14.0 to A6.4 in the P1 chick forebrains (Supplementary Figure S3). Signals were detected in the mesopallium (Supplementary Figures S3A, B, C, A', B', C'), striatum (Supplementary Figures S3B, C, B', C'), and HF, especially the V-shape region (Supplementary Figures S3D, D'). We examined the expression pattern of DRD1C in sections A14.0 to A5.8 in the P1 chick forebrains (Supplementary Figure S4). Signals were detected in the mesopallium (Supplementary Figures S4B, C, D, B', C', D'), hyperpallium (Supplementary Figures S4C, C'), and HF, especially the V-shape region (Supplementary Figures S4C, D, C', D'). The DRD1, DRD2, DRD5, and DRD1C expression sites were consistent with those of previous studies (Kubikova et al., 2010; Yamamoto et al., 2013), suggesting that the expression sites of these receptors are generally complete in P1 chicks.
Next, we examined the expression pattern of DRD3 in sections A14.0 to A6.4 in the P1 chick forebrains (Figure 8). Signals were detected in the dorsal and ventral mesopallium (Figures 8A, B, D, E, A', B', D', E'), hyperpallium, especially the interstitial nucleus of the hyperpallium apicale (IHA) (Figures 8A, B, A', B'), and the intermediate arcopallium (Figures 8D, D'), which is consistent with a previous study (Kubikova et al., 2010). Moreover, we detected very sparse signals in the entopallium (Figures 8B, C, B', C') and the HF, especially the V-shape region (Figures 8E, F, E', F').
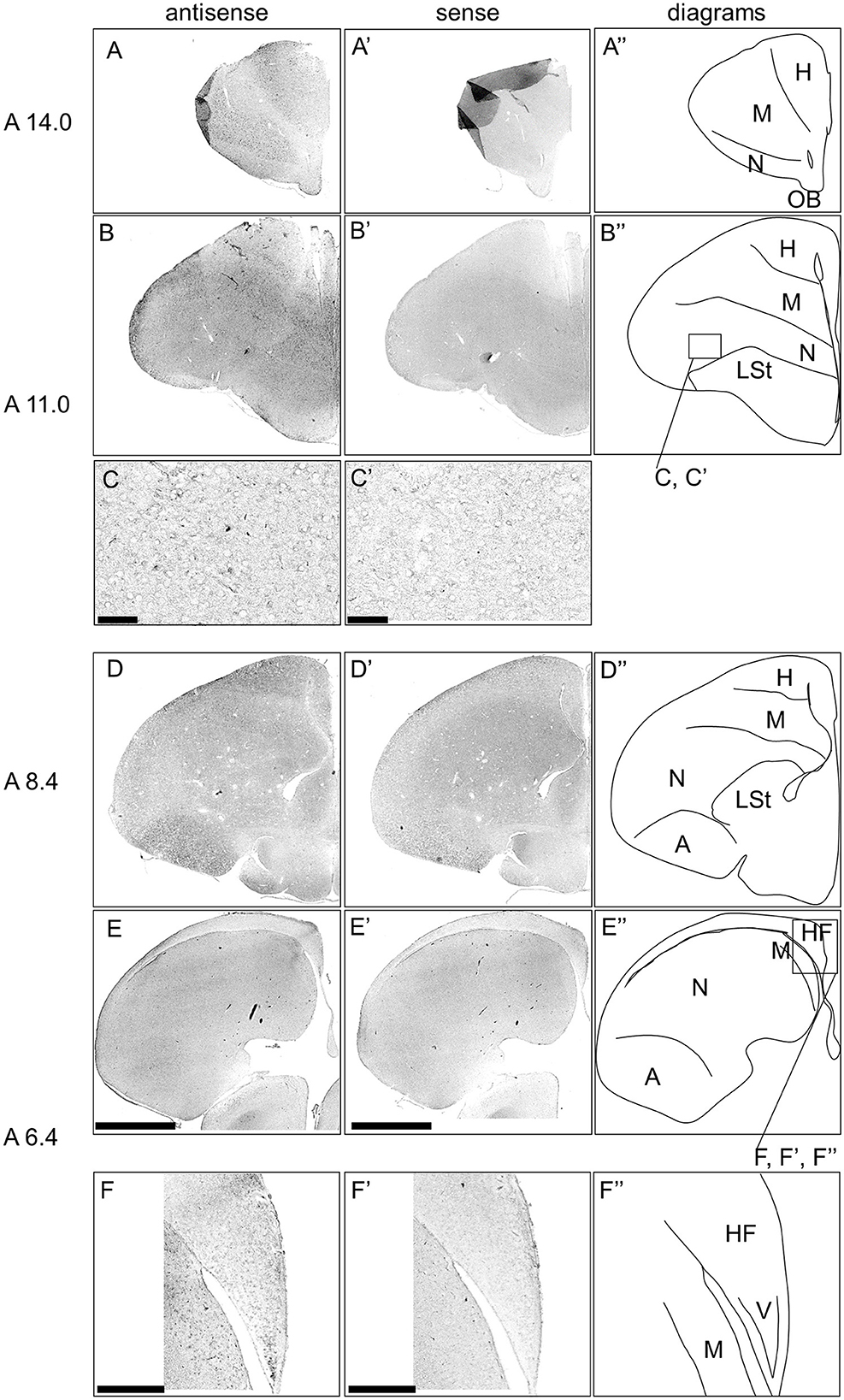
Figure 8. In situ hybridization of DRD3 in P1 chick telencephalons. Digoxigenin-labeled RNA antisense (A–F) and sense (A'–F') DRD3 probes were used for in situ hybridization in coronal sections of the P1 chick telencephalon. Reproducibility of signals was confirmed in multiple chicks at the same level, and representative images are shown. Diagrams of coronal sections are shown in the rightmost (A”–F”). The levels of sections (A 14.0–A 6.4) were in accordance with those mentioned in Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). (C, C') show magnified views of the entopallium (C, C') regions indicated by the box in (B”), and (F, F') show magnified views of the HF (F, F') regions indicated by the box in (E”). A, arcopallium; H, hyperpallium; HF, hippocampal formation; LSt, lateral striatum; M, mesopallium; N, nidopallium; OB, olfactory bulb; V, V-shape region. Scale bars = 2.5 mm (A–D, A'–D'), 100 μm (E, E'), and 500 μm (F, F').
Finally, we examined the expression pattern of DRD1E in sections A14.0 to A6.4 in the P1 chick forebrains and did not obtain clear signals (Supplementary Figure S5).
3.3 Comparison of the TH, DRD4, and GAD2 in the chick OB
We detected signals of TH and DRD4 in the OB. In mammals, the DA neurons in the OB are known to be γ-aminobutyric acid (GABA) ergic interneurons (Cave and Baker, 2009). To further explore the DA system in the avian OB, we compared the expression patterns of TH, DRD4, and GAD2, a marker for GABAergic neurons that encodes the enzyme for GABA synthesis (Figure 9). GAD2 signals were densely distributed in the glomerular layer (GrO), moderately in the inner plexiform layer (IPL), and sparsely in the external granular layer (EPL) (Figures 9A, E, A', E'). DRD4 signals were specifically detected in the mitral cell layer (ML) (Figures 9B, F, B', F'), while TH signals were moderately distributed in the GrO and IPL and sparsely in the EPL (Figures 9C, G, C', G').
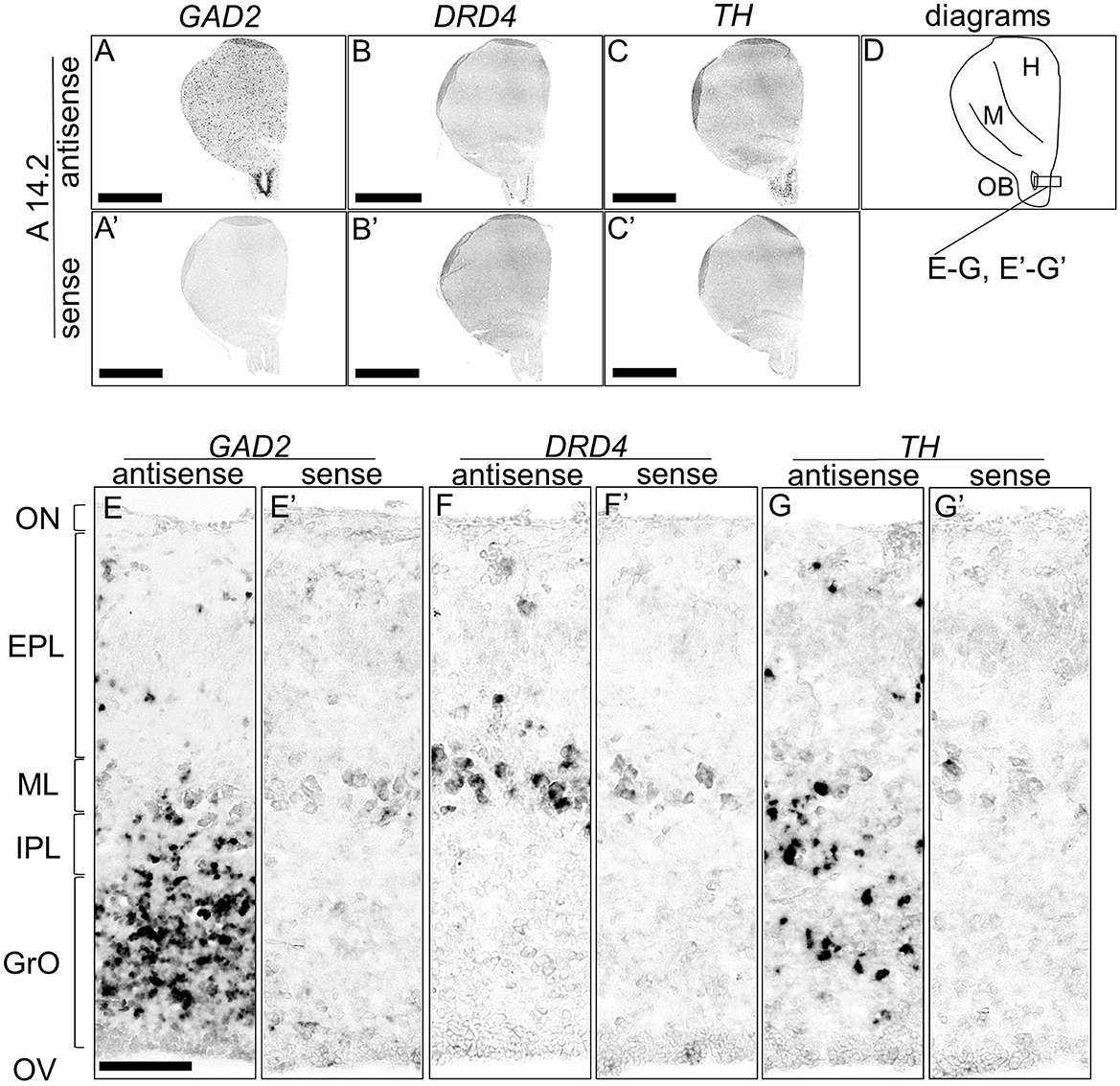
Figure 9. Comparison of GAD2, DRD4, and TH expression patterns in the olfactory bulb in the P1 chick rostral forebrain. Digoxigenin-labeled RNA probes, both antisense [GAD2, (A, E), DRD4, (B, F), and TH, (C, G)] and sense [GAD2, (A', E'), DRD4, (B', F'), and TH, (C', G')], were used for in situ hybridization in coronal sections of P1 chick rostral forebrains, corresponding to level A14.2 of the Kuenzel and Masson's chick atlas (Kuenzel and Masson, 1988). Signal reproducibility was confirmed across multiple chicks at the same level. Representative images of neighboring sections are shown. (E–G, E'–G') present magnified views of the olfactory bulb region indicated by the box in (D). Diagram of coronal sections depicted in (A) is shown in (D). EPL, external plexiform layer; GrO, granule cell layer of the olfactory bulb; H, hyperpallium; IPL, internal plexiform layer; M, mesopallium; ML, mitral cell layer; OB, olfactory bulb; ON, olfactory nerve layer; OV, olfactory ventricle. Scale bars = 2.5 mm (A–C, A'–C') and 100 μm (E–G, E'–G').
4 Discussion
There is a lot of evidence showing the association between DRD4 polymorphism and animal personality and physical condition in birds (Fidler et al., 2007; Flisikowski et al., 2009; Korsten et al., 2010; Gillingham et al., 2012; Mueller et al., 2013; Garamszegi et al., 2014; Mueller et al., 2014; Timm et al., 2015; van Dongen et al., 2015; Holtmann et al., 2016). DRD4 polymorphism was also detected in chickens (Sugiyama et al., 2004). However, the evidence is inconsistent depending on the bird population and the conditions of behavior measurement (Mueller et al., 2013; Edwards et al., 2015; Riyahi et al., 2015, 2017; Timm et al., 2019; Mai et al., 2023). Recently, Silva et al. demonstrated, through a behavioral pharmacological study on D1 and D2 receptors, that personality-related behaviors in birds can be influenced by the manipulation of DA signaling (Silva et al., 2020). However, since no drugs with high selectivity for DRD4 alone among D2 receptors are available, the neural functions of DRD4 in birds are unknown; therefore, the mechanism by which DRD4 affects animal personality in birds is unclear. In the present study, we clarified the distribution of DA neurons and the expression regions of DRDs in the chick forebrain to obtain clues about the function of the animal personality gene DRD4 in the avian brain.
First, we revealed the expression patterns of the DA neuron–related markers TH, DDC, and DBH in the chick forebrain. Typically, DA neurons can be defined as a TH+/DDC+/DBH– cell population. We detected neuron populations with such characteristics in the chick A11 to A15 regions. This characteristic is similar to that observed in the avian forebrain using anti-DA or anti-TH antibodies (Bailhache and Balthazart, 1993; Moons et al., 1994; Reiner et al., 1994). In contrast, TH+/DDC–/DBH– neurons were detected in A16 region. Generally, neurons with these characteristics may be immature and have only partial properties of DA neurons or maybe a DOPAergic neuron population. Nevertheless, previous studies using anti-DA antibodies in other vertebrates have shown that DA is abundant in the OB, and because there is known species variation in DDC expression, these TH+/DDC–/DBH– neuron populations are proposed to be DA neurons (Reiner et al., 1994; Cave and Baker, 2009). TH–/DDC+/DBH– neurons were detected in the PVO (Figure 3). This population is absent in mammals. Previous studies have revealed DA immunopositivity and TH immunonegativity, suggesting that these neurons do not synthesize DA but rather accumulate it (Smeets and Reiner, 1994). In contrast, recent studies have shown that in vertebrates other than mammals, there are two genes encoding TH, TH1 and TH2, which have different immunogenicities. However, TH2 has been lost from the mammalian genome (Yamamoto et al., 2010). Furthermore, TH2 is expressed in the corresponding PVO population in fish, amphibians, and birds (chicken), strongly suggesting that this population has the ability to synthesize and secrete DA (Yamamoto et al., 2011; Xavier et al., 2017). Our data (a TH–/DDC+/DBH– neuron population in the PVO) support the latter possibility (Figure 3). Future studies should focus on the biochemical characterization of TH2 enzyme activity in chicken PVO. Taken together, our data are consistent with the previous findings on avian DA systems.
In addition to the classical DA population, regions with TH+/DDC–/DBH– and TH–/DDC+/DBH– characteristics were detected in chick forebrain (Figures 4–6). Many such monoenzymatic DA neuron populations are well-documented in mammals, with distribution across various regions, including the striatum and BSTL (Ugrumov, 2009; Bupesh et al., 2014). Our results demonstrate that monoenzymatic DA neuron populations are distributed in multiple brain regions, including the striatum and BSTL, in the chick forebrain. This suggests that the presence of monoenzymatic DA neurons in these regions may be conserved between mammals and birds. Previous studies have suggested that these populations contain immature DA neurons and that in the rat arcuate nucleus, TH+/DDC– and TH–/DDC+ monoenzymatic neuron populations are present in close proximity to each other, suggesting that they synthesize and secrete DA in a coordinated manner (Ugrumov, 2009). In our data, we observed such a tendency, for example, in the TH+/DDC–/DBH– and TH–/DDC+/DBH– populations in the chick PTM (Figure 6). The existence of such populations in the forebrain may not only be evolutionarily conserved between birds and mammals but also in birds, where such a cooperative relationship exists.
Next, we investigated the expression patterns of all DRDs, including DRD1, DRD2, DRD3, DRD4, DRD5, and DRD1C, in the chick forebrain. Our findings clearly demonstrate, for the first time, that DRD4 is specifically expressed in the mitral cells of the olfactory bulb, a region where its expression was previously unknown (Figures 7, 9). In mammals, mitral cells are output neurons that transmit information from the olfactory nerve to outside the OB and are modulated by DA (Cave and Baker, 2009). DA modulation of mitral cells via DRD4 may also play an important role in olfactory information processing in birds. The relationship between olfactory information processing and animal personality is currently unknown, and neither how mitral cells process external information may reflect animal personality. Birds have sophisticated sensory systems, and their visual and auditory systems in particular have been the subject of intensive research (Wylie et al., 2015; Iwaniuk and Wylie, 2020). For instance, DRD4 is present in the retina (Macisaac et al., 2024; Klitten et al., 2008), where it contributes to light adaptation (Flood and Eggers, 2021). The olfactory system in birds is important not only for food location but also for navigation in homing pigeons (Gagliardo and Bingman, 2024), nest recognition, predator avoidance, reproductive control, including social interaction (Balthazart and Taziaux, 2009), and behavioral ecology (Corfield et al., 2015). There is evidence that birds such as seabirds (Nevitt et al., 1995), penguins (Amo et al., 2013), and turkey vultures (Smith et al., 2002; Grigg et al., 2017) use their sense of olfaction in their foraging behavior. In this study, we revealed that the A16 DA neuron population is present in the GrO, which is rich in GABAergic interneurons, in chicks, similar to the cellular composition of the mammalian OB. We also found that DRD4, a member of the D2 family, is expressed in chick mitral cells, similar to that in mammalian mitral cells. These findings suggest that the neural circuits in the OB that process olfactory information are likely to be well-conserved between birds and mammals. Physiological confirmation using avian OB is required in the future.
We also examined the expression patterns of the other DRDs in the chick forebrain; however, no signal was detected for DRD1E. The expression patterns of DRD1, DRD2, DRD5, and DRD1C were consistent with those reported in previous studies (Kubikova et al., 2010; Yamamoto et al., 2013), while previously unknown expression regions were identified for DRD3 and DRD4. Specifically, DRD3 was observed in the IHA, entopallium, and V-shape region in the HF, suggesting that DA modulation via DRD3 may play an important role in these regions. We previously revealed that in the avian HF, different subtypes of receptors for serotonin, a different monoamine neuromodulator, are expressed in different populations in each subregion (Fujita et al., 2020, 2022a) and that they are expressed in serotonergic neurons (Fujita et al., 2022b). A very small population expressing 5-HTR1A, 5-HTR1B, and 5-HTR3A was observed in the V-shape region, indicating that serotonergic modulation of the avian HF is characteristic and important (Fujita et al., 2023). Previous studies have established that DRD5 and DRD1C, both D1 family receptors, are expressed in the V-shape region (Kubikova et al., 2010; Yamamoto et al., 2013). In contrast, our study identifies the expression of DRD3, a D2 family receptor, further complicating the understanding of dopamine modulation in this region. Therefore, both D1 and D2 receptors are widely expressed in the V-shape region of birds, suggesting that dopamine, in conjunction with serotonin, plays a pivotal role in the functional regulation of this area.
In conclusion, we comprehensively described the expression distribution of DA neuron-related markers in the chick forebrain, revealing that monoenzymatic DA neurons are also distributed in the avian brain. These findings suggest that the presence of monoenzymatic DA neurons may represent a conserved feature of the vertebrate DA system. Furthermore, we found that DRD4, an avian personality gene, is highly selectively expressed in the mitral cells of the OB and revealed expression sites for other DRDs, including previously undescribed sites. Our findings will enhance our understanding of DA regulation in the avian forebrain and provide insight into how the personality gene DRD4 contributes to the regulation of brain functions in the avian brain.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by the Committee on Animal Experiments of Teikyo University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
TF: Conceptualization, Investigation, Writing – original draft, Writing – review & editing, Formal analysis, Methodology, Validation. NA: Methodology, Writing – original draft, Writing – review & editing, Resources. CM: Methodology, Writing – original draft, Writing – review & editing, Software. KH: Writing – original draft, Writing – review & editing. SY: Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Investigation, Supervision, Validation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Teikyo University Research Encouragement Grant (TF), ACRO Incubation Grants of Teikyo University (TF, SY), Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (SY, 24590096, 15K07945, 18K06667; NA, 24790089, 20K06915; CM, 20K16472 and KH, 26440182, 17K07492, 20K06747), Molecular Profiling Committee, Grant-in-Aid for Transformative Research Areas “Advanced Animal Model Support (AdAMS)” from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (22H04922; to KH), Fund for the Promotion of Joint International Research [Fostering Joint International Research (B)] (TF, KH, 19KK0211), the Uehara Memorial Foundation (SY), the Sagawa Foundation for Promotion of Cancer Research (SY), a Grant-in-Aid for Scientific Research on Innovative Areas “Memory dynamism” (26115522), “Adaptive circuit shift” (15H01449) and “Evolinguistics” (20H05012) from the Ministry of Education, Culture, Sports, Science and Technology (KH), the Naito Foundation (KH), and the Japan Foundation for Applied Enzymology (KH).
Acknowledgments
We thank C. Miyamoto, K. Murata, N. Saito, A. Sakai, and N. Wakahara (Faculty of Pharmaceutical Sciences, Teikyo University) for their technical assistance. We thank Editage (www.editage.com) for the English language editing.
Conflict of interest
The authors declare that this study was conducted in the absence of any commercial or financial relationships that could be construed as potential conflicts of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnana.2025.1531200/full#supplementary-material
References
Amo, L., Rodríguez-Gironés, M. Á., and Barbosa, A. (2013). Olfactory detection of dimethyl sulphide in a krill-eating Antarctic penguin. Mar. Ecol. Prog. Ser. 474, 277–285. doi: 10.3354/meps10081
Ananth, M., Hetelekides, E. M., Hamilton, J., and Thanos, P. K. (2019). Dopamine D4 receptor gene expression plays important role in extinction and reinstatement of cocaine-seeking behavior in mice. Behav. Brain Res. 365, 1–6. doi: 10.1016/j.bbr.2019.02.036
Bailhache, T., and Balthazart, J. (1993). The catecholaminergic system of the quail brain: immunocytochemical studies of dopamine β-hydroxylase and tyrosine hydroxylase. J. Compar. Neurol. 329, 230–256. doi: 10.1002/cne.903290206
Balint, E., and Csillag, A. (2007). Nucleus accumbens subregions: hodological and immunohistochemical study in the domestic chick (Gallus domesticus). Cell Tissue Res. 327, 221–230. doi: 10.1007/s00441-006-0295-0
Balint, E., Mezey, S., and Csillag, A. (2011). Efferent connections of nucleus accumbens subdivisions of the domestic chicken (Gallus domesticus): an anterograde pathway tracing study. J. Comp. Neurol. 519, 2922–2953. doi: 10.1002/cne.22672
Balthazart, J., and Taziaux, M. (2009). The underestimated role of olfaction in avian reproduction? Behav. Brain Res. 200, 248–259. doi: 10.1016/j.bbr.2008.08.036
Ben-Jonathan, N., and Hnasko, R. (2001). Dopamine as a prolactin (PRL) inhibitor. Endocrine Rev. 22, 724–763. doi: 10.1210/edrv.22.6.0451
Bentivoglio, M., and Morelli, M. (2005). “Chapter I The organization and circuits of mesencephalic dopaminergic neurons and the distribution of dopamine receptors in the brain,” in Handbook of chemical neuroanatomy (Amsterdam: Elsevier), 1–107.
Bjorklund, A., and Dunnett, S. B. (2007). Dopamine neuron systems in the brain: an update. Trends Neurosci. 30, 194–202. doi: 10.1016/j.tins.2007.03.006
Bupesh, M., Vicario, A., Abellán, A., Desfilis, E., and Medina, L. (2014). Dynamic expression of tyrosine hydroxylase mRNA and protein in neurons of the striatum and amygdala of mice, and experimental evidence of their multiple embryonic origin. Brain Struct. Funct. 219, 751–776. doi: 10.1007/s00429-013-0533-7
Callier, S., Snapyan, M., Le Crom, S., Prou, D., Vincent, J. D., Vernier, P., et al. (2003). Evolution and cell biology of dopamine receptors in vertebrates. Biol. Cell 95, 489–502. doi: 10.1016/S0248-4900(03)00089-3
Carlsson, A. (2006). The neurochemical circuitry of schizophrenia. Pharmacopsychiatry 39, 10–14. doi: 10.1055/s-2006-931483
Cave, J. W., and Baker, H. (2009). Dopamine systems in the forebrain. Dev. Eng. Dopamine Neur. 15–35. doi: 10.1007/978-1-4419-0322-8_2
Corfield, J. R., Price, K., Iwaniuk, A. N., Gutiérrez-Ibáñez, C., Birkhead, T., Wylie, D. R., et al. (2015). Diversity in olfactory bulb size in birds reflects allometry, ecology, and phylogeny. Front. Neuroanat. 9:102. doi: 10.3389/fnana.2015.00102
Csillag, A. (1999). Striato-telencephalic and striato-tegmental circuits: relevance to learning in domestic chicks. Behav. Brain Res. 98, 227–236. doi: 10.1016/S0166-4328(98)00088-6
Dahlström, A., and Fuxe, K. (1964). Evidence for the existence of monoamine-containing neurons in the central nervous system. I. Demonstration of monoamines in the cell bodies of brain stem neurons. Acta Physiol. Scand. Suppl. 232, 1–55.
Dube, L., and Parent, A. (1981). The monoamine-containing neurons in avian brain: I. A study of the brain stem of the chicken (Gallus domesticus) by means of fluorescence and acetylcholinesterase histochemistry. J. Comp. Neurol. 196, 695–708. doi: 10.1002/cne.901960413
Dulawa, S. C., Grandy, D. K., Low, M. J., Paulus, M. P., and Geyer, M. A. (1999). Dopamine D4 receptor-knock-out mice exhibit reduced exploration of novel stimuli. J. Neurosci. 19, 9550–9556. doi: 10.1523/JNEUROSCI.19-21-09550.1999
Durstewitz, D., Kroner, S., and Gunturkun, O. (1999). The dopaminergic innervation of the avian telencephalon. Prog. Neurobiol. 59, 161–195. doi: 10.1016/S0301-0082(98)00100-2
Ebstein, R. P., Novick, O., Umansky, R., Priel, B., Osher, Y., Blaine, D., et al. (1996). Dopamine D4 receptor (D4DR) exon III polymorphism associated with the human personality trait of novelty seeking. Nat. Genet. 12, 78–80. doi: 10.1038/ng0196-78
Edwards, H. A., Hajduk, G. K., Durieux, G., Burke, T., and Dugdale, H. L. (2015). No association between personality and candidate gene polymorphisms in a wild bird population. PLoS ONE 10:e0138439. doi: 10.1371/journal.pone.0138439
Falzone, T. L., Gelman, D. M., Young, J. I., Grandy, D. K., Low, M. J., Rubinstein, M., et al. (2002). Absence of dopamine D4 receptors results in enhanced reactivity to unconditioned, but not conditioned, fear. Eur. J. Neurosci. 15, 158–164. doi: 10.1046/j.0953-816x.2001.01842.x
Fidler, A. E., Van Oers, K., Drent, P. J., Kuhn, S., Mueller, J. C., Kempenaers, B., et al. (2007). Drd4 gene polymorphisms are associated with personality variation in a passerine bird. Proc. Royal Soc. B Biol. Sci. 274, 1685–1691. doi: 10.1098/rspb.2007.0337
Flisikowski, K., Schwarzenbacher, H., Wysocki, M., Weigend, S., Preisinger, R., Kjaer, J. B., et al. (2009). Variation in neighbouring genes of the dopaminergic and serotonergic systems affects feather pecking behaviour of laying hens. Anim. Genet. 40, 192–199. doi: 10.1111/j.1365-2052.2008.01821.x
Flood, M. D., and Eggers, E. D. (2021). Dopamine D1 and D4 receptors contribute to light adaptation in ON-sustained retinal ganglion cells. J. Neurophysiol. 126, 2039–2052. doi: 10.1152/jn.00218.2021
Fujita, T., Aoki, N., Fujita, E., Matsushima, T., Homma, K. J., Yamaguchi, S., et al. (2019). The chick pallium displays divergent expression patterns of chick orthologues of mammalian neocortical deep layer-specific genes. Sci. Rep. 9:20400. doi: 10.1038/s41598-019-56960-4
Fujita, T., Aoki, N., Mori, C., Fujita, E., Matsushima, T., Homma, K. J., et al. (2020). The dorsal arcopallium of chicks displays the expression of orthologs of mammalian fear related serotonin receptor subfamily genes. Sci. Rep. 10:21183. doi: 10.1038/s41598-020-78247-9
Fujita, T., Aoki, N., Mori, C., Fujita, E., Matsushima, T., Homma, K. J., et al. (2022a). Chick hippocampal formation displays subdivision- and layer-selective expression patterns of serotonin receptor subfamily genes. Front. Physiol. 13:882633. doi: 10.3389/fphys.2022.882633
Fujita, T., Aoki, N., Mori, C., Fujita, E., Matsushima, T., Homma, K. J., et al. (2022b). Serotonergic neurons in the chick brainstem express various serotonin receptor subfamily genes. Front. Physiol. 12:815997. doi: 10.3389/fphys.2021.815997
Fujita, T., Aoki, N., Mori, C., Homma, K. J., and Yamaguchi, S. (2023). Molecular biology of serotonergic systems in avian brains. Front. Mol. Neurosci. 16:1226645. doi: 10.3389/fnmol.2023.1226645
Fujita, T., Aoki, N., Mori, C., Serizawa, S., Kihara-Negishi, F., Homma, K. J., et al. (2022c). Dopaminergic nuclei in the chick midbrain express serotonin receptor subfamily genes. Front. Physiol. 13:1030621. doi: 10.3389/fphys.2022.1030621
Gagliardo, A., and Bingman, V. P. (2024). The avian olfactory system and hippocampus: complementary roles in the olfactory and visual guidance of homing pigeon navigation. Curr. Opin. Neurobiol. 86:102870. doi: 10.1016/j.conb.2024.102870
Garamszegi, L. Z., Mueller, J. C., Markó, G., Szász, E., Zsebok, S., Herczeg, G., et al. (2014). The relationship between DRD 4 polymorphisms and phenotypic correlations of behaviors in the collared flycatcher. Ecol. Evol. 4, 1466–1479. doi: 10.1002/ece3.1041
Gillingham, M. A., Bechet, A., Geraci, J., Wattier, R., Dubreuil, C., Cezilly, F., et al. (2012). Genetic polymorphism in dopamine receptor D4 is associated with early body condition in a large population of greater flamingos, Phoenicopterus roseus. Mol. Ecol. 21, 4024–4037. doi: 10.1111/j.1365-294X.2012.05669.x
Girault, J-.A., and Greengard, P. (2004). The neurobiology of dopamine signaling. Arch. Neurol. 61, 641–644. doi: 10.1001/archneur.61.5.641
Gosling, S. D. (2001). From mice to men: what can we learn about personality from animal research? Psychol. Bull. 127, 45–86. doi: 10.1037//0033-2909.127.1.45
Grigg, N. P., Krilow, J. M., Gutierrez-Ibanez, C., Wylie, D. R., Graves, G. R., Iwaniuk, A. N., et al. (2017). Anatomical evidence for scent guided foraging in the turkey vulture. Sci. Rep. 7:17408. doi: 10.1038/s41598-017-17794-0
Guglielmone, R., and Panzica, G. C. (1984). Typology, distribution and development of the catecholamine-containing neurons in the chicken brain. Cell Tissue Res. 237, 67–79. doi: 10.1007/BF00229201
Helms, C. M., Gubner, N. R., Wilhelm, C. J., Mitchell, S. H., and Grandy, D. K. (2008). D4 receptor deficiency in mice has limited effects on impulsivity and novelty seeking. Pharmacol. Biochem. Behav. 90, 387–393. doi: 10.1016/j.pbb.2008.03.013
Hillarp, N. A., Fuxe, K., and Dahlstrom, A. (1966). Demonstration and mapping of central neurons containing dopamine, noradrenaline, and 5-hydroxytryptamine and their reactions to psychopharmaca. Pharmacol. Rev. 18, 727–741.
Holmes, A., Lachowicz, J. E., and Sibley, D. R. (2004). Phenotypic analysis of dopamine receptor knockout mice; recent insights into the functional specificity of dopamine receptor subtypes. Neuropharmacology 47, 1117–1134. doi: 10.1016/j.neuropharm.2004.07.034
Holtmann, B., Grosser, S., Lagisz, M., Johnson, S. L., Santos, E. S. A., Lara, C. E., et al. (2016). Population differentiation and behavioural association of the two ‘personality'genes DRD 4 and SERT in dunnocks (Prunella modularis). Mol. Ecol. 25, 706–722. doi: 10.1111/mec.13514
Ikeda, H., and Goto, J. (1971). Distribution of monoamine-containing cells in the central nervous system of the chicken. Jpn. J. Pharmacol. 21, 763–784. doi: 10.1016/S0021-5198(19)36177-3
Iversen, S. D., and Iversen, L. L. (2007). Dopamine: 50 years in perspective. Trends Neurosci. 30, 188–193. doi: 10.1016/j.tins.2007.03.002
Iwaniuk, A. N., and Wylie, D. R. (2020). Sensory systems in birds: what we have learned from studying sensory specialists. J. Comp. Neurol. 528, 2902–2918. doi: 10.1002/cne.24896
Keck, T. M., Suchland, K. L., Jimenez, C. C., and Grandy, D. K. (2013). Dopamine D4 receptor deficiency in mice alters behavioral responses to anxiogenic stimuli and the psychostimulant methylphenidate. Pharmacol. Biochem. Behav. 103, 831–841. doi: 10.1016/j.pbb.2012.12.006
Klein, M. O., Battagello, D. S., Cardoso, A. R., Hauser, D. N., Bittencourt, J. C., Correa, R. G., et al. (2019). Dopamine: functions, signaling, and association with neurological diseases. Cell. Mol. Neurobiol. 39, 31–59. doi: 10.1007/s10571-018-0632-3
Klitten, L. L., Rath, M. F., Coon, S. L., Kim, J-. S., Klein, D. C., Møller, M., et al. (2008). Localization and regulation of dopamine receptor D4 expression in the adult and developing rat retina. Exp. Eye Res. 87, 471–477. doi: 10.1016/j.exer.2008.08.004
Kluger, A. N., Siegfried, Z., and Ebstein, R. P. (2002). A meta-analysis of the association between DRD4 polymorphism and novelty seeking. Mol. Psychiatry 7, 712–717. doi: 10.1038/sj.mp.4001082
Korsten, P., Mueller, J. C., Hermannstädter, C., Bouwman, K. M., Dingemanse, N. J., Drent, P. J., et al. (2010). Association between DRD4 gene polymorphism and personality variation in great tits: a test across four wild populations. Mol. Ecol. 19, 832–843. doi: 10.1111/j.1365-294X.2009.04518.x
Kubikova, L., Wada, K., and Jarvis, E. D. (2010). Dopamine receptors in a songbird brain. J. Comp. Neurol. 518, 741–769. doi: 10.1002/cne.22255
Kuenzel, W. J., and Masson, M. (1988). A Stereotaxic Atlas of the Brain of the Chick (Gallus domesticus). Baltimore, MD: Johns Hopkins University Press.
Lovell, P. V., Kasimi, B., Carleton, J., Velho, T. A., and Mello, C. V. (2015). Living without DAT: loss and compensation of the dopamine transporter gene in sauropsids (birds and reptiles). Sci. Rep. 5:14093. doi: 10.1038/srep14093
Macisaac, A. R., Wellington, A. J., Filicetti, K., and Eggers, E. D. (2024). Impaired dopamine signaling in early diabetic retina: insights from D1R and D4R agonist effects on whole retina responses. Exp. Eye Res. 247:110049. doi: 10.1016/j.exer.2024.110049
Mai, S., Wittor, C., Merker, S., and Woog, F. (2023). DRD4 allele frequencies in greylag geese vary between urban and rural sites. Ecol. Evol. 13:e9811. doi: 10.1002/ece3.9811
Mezey, S., and Csillag, A. (2002). Selective striatal connections of midbrain dopaminergic nuclei in the chick (Gallus domesticus). Cell Tissue Res. 308, 35–46. doi: 10.1007/s00441-002-0514-2
Moons, L., van Gils, J., Ghijsels, E., and Vandesande, F. (1994). Immunocytochemical localization of L-dopa and dopamine in the brain of the chicken (Gallus-domesticus). J. Comp. Neurol. 346, 97–118. doi: 10.1002/cne.903460107
Mueller, J. C., Edelaar, P., Carrete, M., Serrano, D., Potti, J., Blas, J., et al. (2014). Behaviour-related DRD 4 polymorphisms in invasive bird populations. Mol. Ecol. 23, 2876–2885. doi: 10.1111/mec.12763
Mueller, J. C., Korsten, P., Hermannstaedter, C., Feulner, T., Dingemanse, N. J., Matthysen, E., et al. (2013). Haplotype structure, adaptive history and associations with exploratory behaviour of the DRD4 gene region in four great tit (Parus major) populations. Mol. Ecol. 22, 2797–2809. doi: 10.1111/mec.12282
Munafò, M. R., Yalcin, B., Willis-Owen, S. A., and Flint, J. (2008). Association of the dopamine D4 receptor (DRD4) gene and approach-related personality traits: meta-analysis and new data. Biol. Psychiatry 63, 197–206. doi: 10.1016/j.biopsych.2007.04.006
Nevitt, G. A., Veit, R. R., and Kareiva, P. (1995). Dimethyl sulphide as a foraging cue for Antarctic procellariiform seabirds. Nature 376, 680–682. doi: 10.1038/376680ao
Puelles, L., Martinez-De-La-Torre, M., Martinez, S., Watson, C., and Paxinos, G. (2018). The Chick Brain in Stereotaxic Coordinates and Alternate Stains: Featuring Neuromeric Divisions and Mammalian Homologies. Cambridge, MA: Academic Press.
Reiner, A., Perkel, D.J., Bruce, L.L., Butler, A.B., Csillag, A., Kuenzel, W., et al. (2004). Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol. 473, 377–414. doi: 10.1002/cne.20118
Reiner, A., Karle, E., Anderson, K., and Medina, L. (1994). Catecholaminergic perikarya and fibers in the avian nervous system. Phyl. Dev. Catechol. Syst. CNS Vert. 135–181.
Riyahi, S., Björklund, M., Mateos-Gonzalez, F., and Senar, J. C. (2017). Personality and urbanization: behavioural traits and DRD4 SNP830 polymorphisms in great tits in Barcelona city. J. Ethol. 35, 101–108. doi: 10.1007/s10164-016-0496-2
Riyahi, S., Sánchez-Delgado, M., Calafell, F., Monk, D., and Senar, J. C. (2015). Combined epigenetic and intraspecific variation of the DRD4 and SERT genes influence novelty seeking behavior in great tit Parus major. Epigenetics 10, 516–525. doi: 10.1080/15592294.2015.1046027
Rubinstein, M., Phillips, T. J., Bunzow, J. R., Falzone, T. L., Dziewczapolski, G., Zhang, G., et al. (1997). Mice lacking dopamine D4 receptors are supersensitive to ethanol, cocaine, and methamphetamine. Cell 90, 991–1001. doi: 10.1016/S0092-8674(00)80365-7
Silva, P. A., Trigo, S., Marques, C. I., Cardoso, G. C., and Soares, M. C. (2020). Experimental evidence for a role of dopamine in avian personality traits. J. Exp. Biol. 223:jeb216499. doi: 10.1242/jeb.216499
Smeets, W. J. A. J., and González, A. (2000). Catecholamine systems in the brain of vertebrates: new perspectives through a comparative approach. Brain Res. Rev. 33, 308–379. doi: 10.1016/S0165-0173(00)00034-5
Smeets, W., and Reiner, A. (1994). Part III: Catecholamines in the CNS of vertebrates: current concepts of evolution and functional significance. Phyl. Dev. Catechol. Syst. CNS Vertebr. 463. University Press of Cambridge.
Smith, H. R., Degraaf, R. M., and Miller, R. S. (2002). Exhumation of food by turkey vulture. J. Raptor Res. 36, 144–145.
Sugiyama, A., Inoue-Murayama, M., Miwa, M., Ohashi, R., Kayang, B. B., Mizutani, M., et al. (2004). Polymorphism of dopamine receptor D4 exon I corresponding region in chicken. Zool. Sci. 21, 941–946. doi: 10.2108/zsj.21.941
Thanos, P. K., Roushdy, K., Sarwar, Z., Rice, O., Ashby Jr, C. R., Grandy, D. K., et al. (2015). The effect of dopamine D 4 receptor density on novelty seeking, activity, social interaction, and alcohol binge drinking in adult mice. Synapse 69, 356–364. doi: 10.1002/syn.21822
Timm, K., Mägi, M., Telve, K., and Tilgar, V. (2019). The behavioural response of great tits to novel environment and handling is affected by the DRD4 gene. Ibis 161, 91–100. doi: 10.1111/ibi.12604
Timm, K., Tilgar, V., and Saag, P. (2015). DRD4 gene polymorphism in great tits: gender-specific association with behavioural variation in the wild. Behav. Ecol. Sociobiol. 69, 729–735. doi: 10.1007/s00265-015-1887-z
Ugrumov, M. V. (2009). Non-dopaminergic neurons partly expressing dopaminergic phenotype: distribution in the brain, development and functional significance. J. Chem. Neuroanat. 38, 241–256. doi: 10.1016/j.jchemneu.2009.08.004
van Dongen, W. F. D., Robinson, R. W., Weston, M. A., Mulder, R. A., and Guay, P-. J. (2015). Variation at the DRD4 locus is associated with wariness and local site selection in urban black swans. BMC Evol. Biol. 15, 253. doi: 10.1186/s12862-015-0533-8
van Oers, K., and Mueller, J. C. (2010). Evolutionary genomics of animal personality. Philos. Transac. Royal Soc. B Biol. Sci. 365, 3991–4000. doi: 10.1098/rstb.2010.0178
Verharen, J. P. H., Zhu, Y., and Lammel, S. (2020). Aversion hot spots in the dopamine system. Curr. Opin. Neurobiol. 64, 46–52. doi: 10.1016/j.conb.2020.02.002
Wise, R. A. (2004). Dopamine, learning and motivation. Nat. Rev. Neurosci. 5, 483–494. doi: 10.1038/nrn1406
Wolf, M., and Weissing, F. J. (2012). Animal personalities: consequences for ecology and evolution. Trends Ecol. Evol. 27, 452–461. doi: 10.1016/j.tree.2012.05.001
Wylie, D. R., Gutiérrez-Ibáñez, C., and Iwaniuk, A. N. (2015). Integrating brain, behavior, and phylogeny to understand the evolution of sensory systems in birds. Front. Neurosci. 9:281. doi: 10.3389/fnins.2015.00281
Xavier, A. L., Fontaine, R., Bloch, S., Affaticati, P., Jenett, A., Demarque, M., et al. (2017). Comparative analysis of monoaminergic cerebrospinal fluid-contacting cells in Osteichthyes (bony vertebrates). J. Comp. Neurol. 525, 2265–2283. doi: 10.1002/cne.24204
Yagishita, S. (2020). Transient and sustained effects of dopamine and serotonin signaling in motivation-related behavior. Psy. Clin. Neurosci. 74, 91–98. doi: 10.1111/pcn.12942
Yagishita, S. (2023). Cellular bases for reward-related dopamine actions. Neurosci. Res. 188, 1–9. doi: 10.1016/j.neures.2022.12.003
Yamaguchi, S., Fujii-Taira, I., Katagiri, S., Izawa, E., Fujimoto, Y., Takeuchi, H., et al. (2008a). Gene expression profile in cerebrum in the filial imprinting of domestic chicks (Gallus gallus domesticus). Brain Res. Bull. 76, 275–281. doi: 10.1016/j.brainresbull.2008.02.002
Yamaguchi, S., Fujii-Taira, I., Murakami, A., Hirose, N., Aoki, N., Izawa, E., et al. (2008b). Up-regulation of microtubule-associated protein 2 accompanying the filial imprinting of domestic chicks (Gallus gallus domesticus). Brain Res. Bull. 76, 282–288. doi: 10.1016/j.brainresbull.2008.02.010
Yamamoto, K., Fontaine, R., Pasqualini, C., and Vernier, P. (2015). Classification of dopamine receptor genes in vertebrates: nine subtypes in osteichthyes. Brain Behav. Evol. 86, 164–175. doi: 10.1159/000441550
Yamamoto, K., Mirabeau, O., Bureau, C., Blin, M., Michon-Coudouel, S., Demarque, M., et al. (2013). Evolution of dopamine receptor genes of the D1 class in vertebrates. Mol. Biol. Evol. 30, 833–843. doi: 10.1093/molbev/mss268
Yamamoto, K., Ruuskanen, J. O., Wullimann, M. F., and Vernier, P. (2010). Two tyrosine hydroxylase genes in vertebrates: new dopaminergic territories revealed in the zebrafish brain. Mol. Cell. Neurosci. 43, 394–402. doi: 10.1016/j.mcn.2010.01.006
Yamamoto, K., Ruuskanen, J. O., Wullimann, M. F., and Vernier, P. (2011). Differential expression of dopaminergic cell markers in the adult zebrafish forebrain. J. Comp. Neurol. 519, 576–598. doi: 10.1002/cne.22535
Yamamoto, K., and Vernier, P. (2011). The evolution of dopamine systems in chordates. Front. Neuroanat. 5:21. doi: 10.3389/fnana.2011.00021
Keywords: personality gene, dopamine receptor, dopamine neuron, DRD4, chick, dopamine, olfactory bulb, mitral cell
Citation: Fujita T, Aoki N, Mori C, Homma KJ and Yamaguchi S (2025) Molecular characterization of chicken DA systems reveals that the avian personality gene, DRD4, is expressed in the mitral cells of the olfactory bulb. Front. Neuroanat. 19:1531200. doi: 10.3389/fnana.2025.1531200
Received: 20 November 2024; Accepted: 02 January 2025;
Published: 15 January 2025.
Edited by:
Andras Csillag, Semmelweis University, HungaryReviewed by:
Lubica Kubikova, Slovak Academy of Sciences, SlovakiaCatherine Montagnese, Semmelweis University, Hungary
Copyright © 2025 Fujita, Aoki, Mori, Homma and Yamaguchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinji Yamaguchi, c2hpbmppLXlAcGhhcm0udGVpa3lvLXUuYWMuanA=
 Toshiyuki Fujita
Toshiyuki Fujita Naoya Aoki
Naoya Aoki Chihiro Mori
Chihiro Mori Koichi J. Homma
Koichi J. Homma Shinji Yamaguchi
Shinji Yamaguchi