
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Neuroanat. , 26 July 2022
Volume 16 - 2022 | https://doi.org/10.3389/fnana.2022.937596
This article is part of the Research Topic Sex Differences in the Brain View all 7 articles
Sexual phenotypic differences in the nervous system are one of the most prevalent features across the animal kingdom. The molecular mechanisms responsible for sexual dimorphism throughout metazoan nervous systems are extremely diverse, ranging from intrinsic cell autonomous mechanisms to gonad-dependent endocrine control of sexual traits, or even extrinsic environmental cues. In recent years, the DMRT ancient family of transcription factors has emerged as being central in the development of sex-specific differentiation in all animals in which they have been studied. In this review, we provide an overview of the function of Dmrt genes in nervous system sexual regulation from an evolutionary perspective.
The brain of most animals displays sexual differences at the level of neuronal number, gene expression, synaptic connectivity, and behavior (Trabzuni et al., 2013; Yang and Shah, 2016; McCarthy, 2020; Goodwin and Hobert, 2021). These sexual differences impact innate behaviors, those displayed without any prior learning or training, and as a consequence, innate behaviors have a strong sex bias. More relevant to human pathology, the study of sexual differentiation of the brain is of major significance based on the premise that it might uncover factors involved in the general etiology and sexual skewing of neuropsychiatric disorders (McCarthy, 2016; Zagni et al., 2016).
The molecular mechanisms that determine the sex of an organism are almost as diverse as the species themselves. However, among this wide variety, Doublesex and mab-3 related transcription factors (Dmrt) are unique in their conservation, in organisms as diverse as mammals, fishes, birds, frogs, insects, and nematodes. Their conserved role in gonadal differentiation has been discussed in excellent reviews elsewhere (Matson and Zarkower, 2012; Bellefroid et al., 2013). In this review, we consider the conserved function of Dmrt genes in nervous system sexual differentiation. We comprehensively describe studies in two invertebrate genetic model organisms, Drosophila melanogaster and Caenorhabditis elegans, which have been studied with unprecedented detail. Male and female connectomes are fully reconstructed in C. elegans (White et al., 1986; Cook et al., 2019) and in the Drosophila female (Zheng et al., 2018; Scheffer et al., 2020), and work over the years has revealed several sexual dimorphisms (Goodwin and Hobert, 2021). High genetic access, complemented with stereotyped sex-specific behaviors, has allowed for a comprehensive study of Dmrt genes in sexual differentiation of the nervous system of C. elegans and Drosophila. The first evidence of a Dmrt gene controlling nervous system sexual differentiation was published in 1980, and was related to Drosophila's peripheral nervous system (Baker and Ridge, 1980). The authors found that doublesex (dsx) was required for differentiation of sex combs, a male-specific array of modified mechanosensory bristles needed for mating (Spieth et al., 1952; Cook, 1977; Ng and Kopp, 2008). Likewise, mab-3 was shown to be involved in male-specific sensory organ development and behavior in C. elegans (Shen and Hodgkin, 1988; Yi et al., 2000). However, more than 30 years have passed since these seminal initial works (Baker and Ridge, 1980; Shen and Hodgkin, 1988), and no systematic research in other organisms has been conducted. In the present work, we collect and summarize relevant data about the sexual regulation of the nervous system by the Dmrt genes dsx and dmds (Dmrt nomenclature in C. elegans), as a roadmap for studying the potential conserved function of Dmrt genes in the sexual regulation of the nervous system of other organisms. In addition, we catalog all those animal species in which Dmrt expression in neural tissues has been demonstrated, evidencing Dmrt function, and highlighting its involvement in sex-specific traits (where present) (Table 1).
Dmrt genes are defined by a DNA-binding region called the DM domain. This motif was named after the founding members dsx in Drosophila and mab-3 in C. elegans. Both genes encode transcription factors that perform analogous functions in sexual development. Interestingly, dsx (in its male isoform, see below) can partially substitute for mab-3 in vivo in the peripheral nervous system of C. elegans (Raymond and Zarkower, 1998). The DM domain encodes an intertwined zinc-finger DNA-binding motif, followed by an alpha-helical recognition domain that confers sequence-specific DNA binding through a 13 bp pseudo-palindromic DNA motif (Murphy et al., 2007, 2015). Outside the DM region, Dmrt genes do not share much homology, except for the DMA subfamily (Dmrt3, Dmrt4, and Dmrt5 in mammals, dmrt93B and dmrt99B in Drosophila, or dmd-4 in C. elegans). Although the role of the DMA domain is unclear, recent evidence in C. elegans points to sex-specific protein stabilization through ubiquitin binding to the DMA domain. The human DMRT3 DMA domain is also able to bind ubiquitin, at least in vitro, and when the full protein was ectopically expressed in worm neurons in vivo, it was degraded like the worm DMD-4, suggesting conserved molecular functioning of the DMA domain (Bayer et al., 2020).
DMRT proteins regulate the transcription of target genes by activation or repression. They can form heterodimers on DNA in vitro and in vivo, raising the possibility that they may interact functionally by forming mixed complexes (Murphy et al., 2007). As far as we are aware, most studies on mechanistic aspects of DMRTs derive from DMRT1 function in the mouse gonad, or DSX in Drosophila. DMRT1 functionally collaborates with another key male sex regulator, SOX9. Chromatin immunoprecipitation followed by sequencing (ChIP-Seq) experiments show that they both bind to close sites on target genes, suggesting that they have joint effects on target gene expression to maintain and reprogram sexual cell fate (Rahmoun et al., 2017). Related to this function, DMRT1 appears to act as a pioneer transcription factor, binding “closed” chromatin and opening it to allow binding by other regulators such as SOX9, (Lindeman et al., 2021). Lastly, DMRT1 can bind DNA in a highly unusual type of interaction and can bind with different stoichiometries. However, it is not clear what the functional outcome of different stoichiometry might be. In Drosophila, dsx acts downstream of sex determination cues, cell autonomously establishing most somatic dimorphisms in distinct tissues. dsx is sex-specifically spliced into DSXF and DSXM isoforms. The two proteins share a common region, containing the DM DNA-binding domain and dimerization domains, but differ on sex-specific C-terminal regions (Burtis and Baker, 1989; Erdman and Burtis, 1993; An et al., 1996; Cho and Wensink, 1996; Erdman et al., 1996). These two transcription factors bind largely the same target genes in different tissues in both sexes, but their regulation would depend on the combinatorial activity of other gene-specific factors cooperating with DSX binding (Clough et al., 2014), although such combinatorial action has not yet been described. DSX target genes are involved in signaling (e.g., WNT, ecdysone), transcription and epigenetic regulation (e.g., bric a brac, Drop, grappa), and sex-determination (e.g., fru, Sxl, and dsx itself). Additionally, DSX cross-regulates known transcription factors involved in dsx expression such as Sex comb reduced and Abdominal-B (Williams et al., 2008; Chatterjee et al., 2011; Tanaka et al., 2011; Clough et al., 2014; Neville et al., 2014).
In somatic tissues, Dmrt genes function as developmental regulators that integrate information about sex, position/space, and time, to direct cell populations toward male or female features. This role is perfectly illustrated by the Drosophila sex comb—a strictly male-specific organ. In this case, spatial control is accomplished by HOX genes, and sex combs only develop on the first pair of legs, whereas the sex-specificity is controlled by alternative dsx splicing. Transcription of dsx is induced in the first leg by the HOX gene Sex combs reduced, which is not expressed in the more posterior segments (Tanaka et al., 2011). Similarly, in worms, sexual specificity is controlled by the master regulator TRA-1, and temporal specificity by heterochronic pathway genes. As a consequence, several Dmrt genes (for which sex-specific functions have been found) are expressed in a male-specific manner, but only from the fourth larval stage, when worms become sexually mature. In some cases, it has been shown that space information is assured by neuron-type-specific regulators of neuronal identity, termed terminal selectors (Hobert and Kratsios, 2019). For instance, mab-3 expression in SMD motor neurons is controlled by the SMD identity regulator unc-42, a homeobox gene (Pereira et al., 2019; Berghoff et al., 2021). In this way, the mechanisms that couple developmental timing to sexual maturation are themselves tightly coupled.
Most animal genomes contain multiple Dmrt genes (Figure 1). The short length of the DM domain (~70 amino acids) hampers the construction of phylogenetic relationships among Dmrt paralogs, which is fundamental to unravelling the ancestral developmental functions of this gene family. However, based on several studies that combine syntenic and phylogenic approaches (Ottolenghi et al., 2002; Bellefroid et al., 2013; Wexler et al., 2014; Mawaribuchi et al., 2019) the Dmrt gene family has been classified into eight major subsets: Dmrt1, Dmrt2a/2b (dmrt2, dmrt2a and dmrt2b), Dmrt3, Dmrt4/5 (Dmrt4, Dmrt5 and dmrt99B), Dmrt6, Dmrt7, Dmrt8, and dmrt93B (Figure 1). For instance, in arthropods' genomes there are four Dmrt genes (dsx, dmrt93B, dmrt99B, dmrt11E), while in C. elegans there are ten (mab-3, mab-23, and dmd-3 to dmd-10). In mammals there are seven Dmrt genes, Dmrt1 to Dmrt7 (Ottolenghi et al., 2002; Volff et al., 2003; Bellefroid et al., 2013), and up to three Dmrt-like genes (Dmrt8.1, Dmrt8.2 and Dmrt8.3) that lack a functional DM domain but which are clustered in the X chromosome (Veith et al., 2006b). However, in contrast to all other known DMRT proteins, the DM domain-encoding sequence of DMRT8 is not conserved in humans and mice.
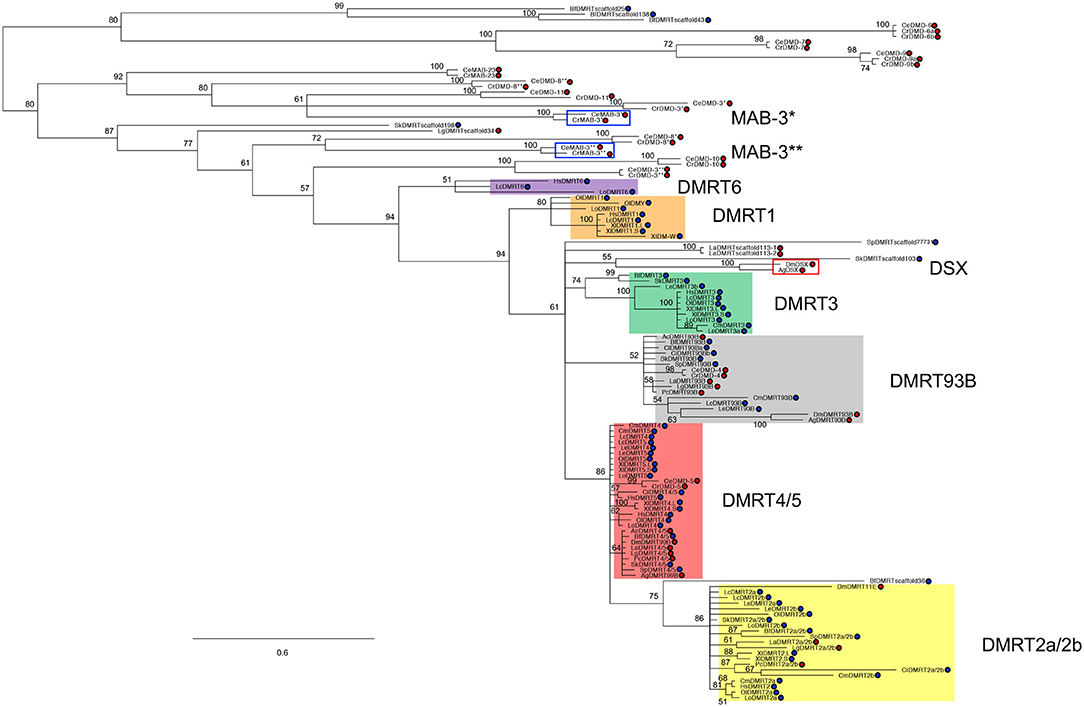
Figure 1. DMRT family phylogeny. Figure adapted from Mawaribuchi et al. (2019). Bayesian tree of bilaterian Dmrt family genes. The tree was constructed using protein sequences of the DM domains from 19 species representing eight different phyla in bilateria. Blue and red circles represent Deuterostomia and Protostomia, respectively. * and ** indicate DM domain regions on 5′ and 3′ sides, respectively. Lingula anatina (La); Aplysia californica (Ac); L. gigantea (Lg); Priapulus caudatus (Pc); Caenorhabditis elegans (Ce); Caenorhabditis remanei (Cr); Anopheles gambiae (Ag); Drosophila melanogaster (Dm); Saccoglossus kowalevskii (Sk); Strongylocentrotus purpuratus (Sp); Ciona intestinalis (Ci); Branchiostoma floridae (Bf); Chondrichthyes, C. milii (Cm); L. erinacea (Le); L. oculatus (Lo); O. latipes (Ol); L. chalumnae (Lc); Xenopus laevis (Xl); Homo sapiens (Hs).
For many years, Dmrt1, mab-3, and dsx have been postulated as orthologues (Ottolenghi et al., 2002; Clough et al., 2014). However, recent research suggests an independent evolution of Dmrt1, dsx, and mab-3 for sex determination and primary sex differentiation (Wexler et al., 2014; Mawaribuchi et al., 2019) (Figure 1). But, regardless of the independent evolution of Dmrt1 homologues, it seems likely that sexual differentiation regulation is an ancestral function of this gene family (Kopp, 2012; Zarkower and Murphy, 2021). Accordingly, Dmrt genes regulate sexual differentiation in species as divergent as nematodes, flatworms, insects, crustaceans, and vertebrates (Matson and Zarkower, 2012). And remarkably, in most species, Dmrt genes have been detected in somatic tissues such as the central nervous system, the olfactory placode, as well as in somites (see Table 1 and discussion below). However, their roles in the sexual differentiation of somatic tissues, including the nervous system, are best understood in invertebrate model organisms like Drosophila and C. elegans, where sex determination is largely cell autonomous.
In C. elegans, Dmrt genes determine sex-specific neuronal traits, which can be grouped into three major categories in the context of distinct and complementary ways to generate sex-specific circuits: control of cell numbers, cell identity, and connectivity, which are eventually reflected in sexually dimorphic behaviors (recently reviewed in Serrano-Saiz and Isogai, 2021).
C. elegans has two sexes, males and hermaphrodites (which can be considered somatic females capable of producing sperm). The nervous system of males and hermaphrodites is divided into 294 sex-shared neurons (found in both sexes), while each sex additionally contains sex-specific neurons: eight in hermaphrodites (HSN and VCs), involved in egg-laying behavior, and 93 neurons in males, a few of them located in the head and the ventral nerve cord, with most of them in the tail (Sulston et al., 1980; Sulston, 1983; White et al., 1986; Jarrell et al., 2012; Sammut et al., 2015; Cook et al., 2019). As in the hermaphrodite, these male-specific neurons make extensive connections with sex-shared neurons (Cook et al., 2019).
Sex differences in the nervous system of C. elegans occur at multiple levels: sex-specific cell death, sex-specific neurogenesis, and trans-differentiation of glial cells into neurons (Sammut et al., 2015) that lead to sex-specific neuronal numbers. Among the sex-shared neurons (generated in both sexes, positioned in the same manner, and grossly anatomically very similar), sexual differences also appear during sexual maturation at the level of synaptic connectivity across the entire connectome (Cook et al., 2019). Moreover, the systematic mapping of a core identity feature of neurons, their neurotransmitter identity, revealed a number of sexual dimorphisms in neurotransmitter usage as well (Serrano-Saiz et al., 2017b), including sex-specific neurotransmitter switches, and transcriptional scaling. In some cases, these dimorphic features are accompanied by differences in morphology [projections or branching; reviewed by Kim and Kim in this topic (Kim and Kim, 2022)].
The primary sex-determining cue in C. elegans is the ratio of X chromosomes to autosomal chromosomes that converges on a terminal “master regulator” of somatic sexual state, the transcription factor TRA-1. When the ratio is high in XX female embryos, levels of TRA-1 are high, and when the ratio is low in XO male embryos, TRA-1 levels are low. TRA-1 acts cell autonomously to regulate an array of intermediate factors that control more specific aspects of sex-specific development and physiology. In a few cases, TRA-1 has been shown to directly control the expression of effector genes [i.e., the repression of the cell death regulator egl-1 to allow for the survival of HSN hermaphrodite neurons (Conradt and Horvitz, 1999) or the expression of unc-6/netrin, implicated in synaptic development to promote male-specific synaptic connectivity (Weinberg et al., 2018)]. However, many TRA-1 effects are controlled by intermediary factors, the most prominent type being the Dmrt genes. There are 10 Dmrt genes in C. elegans, including the founding member of the family, mab-3. Six of them are dimorphically expressed (mab-3, mab-23, dmd-3, dmd-4, dmd-5, and dmd-10). Most of them are expressed in a male-specific manner in male- and sex-shared neurons. In all cases, Dmrt male-specific expression occurs as a consequence of tra-1 low levels. dmd-4 is an exception, and is degraded in shared neurons in the male tail (Bayer et al., 2020). dmd-4 expression in hermaphrodites also depends on tra-1, however, through a post-transcriptional regulatory mechanism that involves the stabilization of DMD-4 protein. dmd-4 is the only Dmrt across the C. elegans genome that contains a DMA domain, and Bayer et al. described it as a putative ubiquitin-binding domain. The expression of the six Dmrt genes does not cover the entire male-specific nervous system, although their expression pattern is not yet complete, which suggests that other factors might be operating downstream of tra-1. This is the case for another type of transcription factor, the conserved Zn-finger lin-29, which has been found to be expressed in many sex-shared neurons in males (Pereira et al., 2019). Recently, endogenously tagged gfp reporters for mab-3 and dmd-3 have been generated (Pereira et al., 2019). These reagents have served to clarify the molecular mechanisms that position DMRTs at the right moment, at the right site. mab-3 and dmd-3 are precisely timed to begin their expression at the L4 stage when LIN-41, an RNA-binding post-transcriptional regulator, and a key target of the let-7 miRNA, represses their mRNA (Aeschimann et al., 2017). Moreover, mab-3 and dmd-3 are controlled by terminal selectors to be specifically expressed in SMD and PHC neurons, respectively. Finally, the male-specificity of mab-3 and dmd-3 expression in the nervous system is controlled by the TRA-1 transcription factor (Pereira et al., 2019).
Importantly, all sexually dimorphic Dmrt family members have been implicated in controlling sexually dimorphic neuronal identity features. mab-3 is expressed in male-specific and sex-shared neurons. It promotes neurogenesis of male-specific sensory neurons by controlling the expression of a bHLH (basic helix-loop helix) transcription factor. During L3, the atonal-class bHLH transcription factor LIN-32 is activated male-specifically in nine pairs of male-specific ray precursor cells (Zhao and Emmons, 1995). lin-32 expression requires the HOX genes mab-5 and egl-5 (Zhao and Emmons, 1995), as well as mab-3, which indirectly activates lin-32 by repressing its repressor ref-1 (Shen and Hodgkin, 1988; Yi et al., 2000; Ross et al., 2005). In addition to mab-3, both dmd-3 and mab-23 control terminal differentiation programs of male-specific ray neurons, such as their neurotransmitter identities (Yi et al., 2000; Lints and Emmons, 2002; Siehr et al., 2011).
Dmrt genes are also involved in controlling sexually dimorphic neuronal identity features in sex-shared neurons such as projection morphology, gene expression, and dimorphic connectivity, which ultimately impact function. mab-3 is expressed in sex-shared embryonically generated ADF and SMD head motor neurons, but only in males, and only after the L3 stage (Yi et al., 2000; Pereira et al., 2019). However, the role of mab-3 in these neurons is not completely mapped, and it is not known whether it is involved in regulating sexually dimorphic locomotion (Pereira et al., 2019), and loss of mab-3 did not completely eliminate ascaroside attraction mediated by ADF (Fagan et al., 2018). Consequently, mab-3 targets in these neurons are still unknown.
Another interesting case is the sex-shared PHC neuron class that displays highly dimorphic connectivity (Serrano-Saiz et al., 2017a). In hermaphrodites, PHC functions as a sensory neuron, presenting scarce input and output connections. Strikingly, in males, PHC receives heavy input from both male-specific and shared neurons (Jarrell et al., 2012; Cook et al., 2019). Moreover, PHC wiring changes are accompanied by morphological changes and transcriptional scaling in synaptic molecules in order to cope with the increase in the number of connections. All these changes eventually lead to the repurposing of PHC function. dmd-3 orchestrated the entire transformation of PHC. That is, dmd-3 is both required and sufficient to cell autonomously control the male-specific remodeling of the PHC neurons by the L4 stage (Serrano-Saiz et al., 2017a). Although it is not yet known if they are direct targets, the ectopic expression of dmd-3 in PHC neurons in hermaphrodite animals triggered all the gene expression associated changes.
The sex-shared PHB sensory neuron forms synapses with distinct downstream interneurons in males and females, as well as to additional male-specific motor, sensory, and interneurons (Jarrell et al., 2012; Oren-Suissa et al., 2016; Cook et al., 2019). These dimorphisms in connectivity arise through two distinct mechanisms: some connections are formed only in larval males and these persist in adult males (pre-patterning for circuit wiring) while other juvenile connections between two pairs of shared-neurons (PHB>AVA and PHB>AVG) are only eliminated in one sex at L4 (selective pruning). Interestingly, both the sex of the pre-synaptic cells, PHB, and the post-synaptic site are also sufficient to initiate PHB connectivity rewiring. The dimorphic synaptic pruning events depend on the sex-specific activity of several transcription factors, including DMRTs. dmd-5 and dmd-10 (dmd-10 and dmd-11 were previously miss-annotated as two genes; now is dmd-10) mutants display alterations in synaptic wiring, and PHB>AVG male-specific synapses fail to be maintained in males, indicating that dmd-5 and dmd-10 are not required for synapse formation per se, but are specifically involved in controlling sex-specific synapse maintenance by preventing synaptic pruning (Oren-Suissa et al., 2016). Another Dmrt, dmd-4 is expressed in PHA and PHB sensory neurons. In this case, dmd-4 is maintained in hermaphrodites in these neurons, and, in dmd-4 mutants, male specific synapses (PHB>AVG) fail to be pruned in hermaphrodites (Bayer et al., 2020). However, the precise targets of dmd-4 which modulate synaptic connectivity are unknown.
During mating, the male engages in a precise orchestrated series of events to land on a hermaphrodite, find the vulva, and eventually inseminate the mating partner (Liu and Sternberg, 1995; Barr and Garcia, 2006). Male-specific neurons in the tail are responsible for many aspects of copulatory behavior. Therefore, it is not surprising that mutations in Dmrt genes, which affect the generation and specification of male-specific neurons (such as mab-3, dmd-3 or mab-23), have defects in copulation (Yi et al., 2000; Lints and Emmons, 2002; Siehr et al., 2011). Mutations in dmd genes expressed in shared neurons also affect many facets of male mating behavior. In dmd-5;dmd-10 double mutants, vulva location efficiency is affected as a consequence of the loss of proper connectivity between PHB and AVG. Rescue in AVG with either dmd-5 or dmd-10 is sufficient to rescue this behavioral defect (Oren-Suissa et al., 2016). In larval animals, the phasmid neurons PHA, PHB, and PHC function to mediate chemosensory avoidance behavior (Hilliard et al., 2002; Zou et al., 2017), a functional output which is maintained in adult hermaphrodites, but lost in adult males (Oren-Suissa et al., 2016). Similarly, the PHC neuron is important for sensing harsh touch in hermaphrodites, but not in males. Instead, male PHC is involved in mating behavior (Serrano-Saiz et al., 2017a).
In summary, C. elegans powerful model has allowed us to dissect how sexually dimorphic gene expression differences, how reconfiguration of morphology and connectivity rewiring of shared neurons leads to the redeployment of neurons into dimorphic behaviors, and how Dmrts serve as the effectors of many of these scenarios.
In D. melanogaster, sex-specific splicing of dsx and fruitless (fru) is the pivotal event in sexual differentiation of the nervous system. In embryonic development, the ratio of X chromosomes to autosomes causes zygotically transcribed genes to activate the RNA-splicing factor Sex-lethal (Sxl) in females only. SXL protein controls both its own splicing and the splicing of transformer (tra), another RNA-splicing factor. Thus, TRA protein is functional only in females and causes dsx to be spliced, giving rise to the female-specific isoform (DSXF). Moreover, TRA directs a splicing in which a stop codon is introduced into female-specific fru mRNA, and, consequently, it is not translated. In contrast, the absence of SXL and TRA proteins in males allows the production of the male-specific isoform of DSX (DSXM) and FRU (FRUM) (Hoshijima et al., 1991; Ryner and Baker, 1991; Inoue et al., 1992; Ryner et al., 1996; Heinrichs et al., 1998; Usui-Aoki et al., 2000).
In Drosophila, nervous system sexual dimorphisms are related to (1) sex-specific neuronal clusters, only present in one sex, (2) sex-specific cell death and proliferation in shared particular clusters producing sex-specific numbers in homologous clusters, and (3) sex-specific neurite projections and synaptic densities. All these features are hypothesized to be responsible for sex-specific behaviors, although evidence of direct links between a given neuronal cluster and a behavioral output are sparse. Male- and female-specific DSX isoforms, together with FRUM, are responsible for most of those dimorphisms, executing distinct—and sometimes antagonistic—functions between the sexes to generate sex-specific neuronal traits and behaviors (Billeter et al., 2006b; Rideout et al., 2007, 2010; Kimura et al., 2008; Sanders and Arbeitman, 2008; Shirangi et al., 2009; Pan et al., 2011; Neville et al., 2014; Pan and Baker, 2014; Ishii et al., 2020).
dsx is expressed in the central nervous system (CNS) of larvae, pupae, and adults (Lee et al., 2002). DSXF is expressed in ~700 neurons; while DSXM is expressed in about 900 neurons, the majority of which also express FRUM (Peng et al., 2021). The ventral nerve cord contains around two-thirds of all dsx-expressing neurons in the CNS, most of them found in the abdominal ganglion (Pan and Baker, 2014). In the CNS, the vast majority of dsx+ cells are cholinergic (excitatory) neurons (Zhou et al., 2014), although dsx expression has also been found in glial cells of both sexes (e.g., the Suboesophageal Lateral Glia, SLG, located in the anteroventral optic cleft) (Robinett et al., 2010). dsx is expressed in ~150 neurons per hemisphere in males, and 30–40 per hemisphere in females. In the entire adult CNS, the number of sex-specific neuronal clusters—only present in one sex—is seven in males (pC2m, pLN, pMN3, P1, SN, TN1, and TN2), and one in females (pMN2); while the number of sexually dimorphic clusters—present in both sexes but presenting different numbers—is four (pCd-1, pC1, pC2l, and abdominal ganglion) (Taylor and Truman, 1992; Nojima et al., 2021). Although, in general, the topology of dsx-axonal projections is similar between both sexes, male neurons notably show higher density of synapses and projections than females ones (Rideout et al., 2010). However, whether dsx has a direct role in the promotion of synaptic density has not yet been described. dsx is also expressed in the peripheral nervous system, e.g., in neuronal cells of gustatory sense organs of the male and female foreleg (Robinett et al., 2010), and in mechanosensory neurons of the male genitalia (Pavlou et al., 2016; Jois et al., 2022). Thus, in the sexually mature fly, dsx is expressed in key nodes of a sexually dimorphic circuitry that produces sex-specific behaviors. As we will explain in greater detail below, these nodes encompass all levels of a neural circuit: the sensory neurons that detect stimuli, the interneurons that process such stimuli, and the motor neurons that materialize the proper behavioral output.
In Drosophila, the main mechanisms to generate sex differences in neuron numbers involve cell proliferation and programmed cell death. Thus, DSXM prolongs cell divisions of four neuroblasts in the abdominal ganglion, resulting in about 20 extra neurons in adult males compared to females (Taylor and Truman, 1992). Furthermore, these terminal neuroblasts do not develop in dsx null flies (Taylor and Truman, 1992), which is consistent with the decreased number of male-specific serotonergic neurons in the abdominal ganglion of dsx null males (Billeter et al., 2006a). These serotonergic neurons control seminal fluid and sperm transfer (Lee and Hall, 2001; Lee et al., 2001), are patterned in two clusters (dorsal and ventral), and most of them co-express FRUM and DSXM (Billeter et al., 2006a). In the posterior part of the protocerebrum, DSXM also promotes additional neuroblast divisions in males, promoting a higher number of neurons in males in two dsx+ neuronal clusters (pC1 and pC2) (Sanders and Arbeitman, 2008; Rideout et al., 2010; Robinett et al., 2010). Likewise, DSXF determines female-specific neuronal numbers configuring the female-specific circuitry. Several studies show that DSXF promotes cell death in different contexts: in sex-specific neuroblasts of the terminal abdominal neuromeres (Birkholz et al., 2013), in the pupal and adult TN1 cluster of the thoracic ventral nerve cord (Sanders and Arbeitman, 2008), and in P1 interneurons of the adult brain (Kimura et al., 2008). TN1 cluster—which disappears completely in females by 48 hours after pupal formation—is a good scenario to illustrate the antagonistic actions of the sex-specific DSX isoforms. While DSXF is necessary for female-specific apoptosis in TN1 progenitors, and no TN1 neurons are generated in females, DSXM promotes additional cell divisions to achieve around 15 ± 0.5 TN1 cell neurons. In fact, XX mutant flies expressing both DSXM and DSXF show an intermediate number of TN1 cells (5 ± 0.3 cells) (Sanders and Arbeitman, 2008). Additionally, DSX also has a role in generating sexual dimorphisms in neuronal processes. In cooperation with FRU, DSX establishes male-specific midline crossing of axons from foreleg gustatory receptor neurons (GRN). In particular, DSXF represses this midline crossing, while DSXM might be promoting it (Mellert et al., 2010). Also, dsx+ aDN neurons display sexually dimorphic arborizations patterns, suggesting that males and females receive information from distinct sensory modalities (Nojima et al., 2021).
Evidence that dsx regulates sexual neuronal circuit differentiation comes initially from behavioral analysis of dsx mutant males and females. Male courtship in D. melanogaster is one of the best-studied animal behaviors (Hall, 1994; Yamamoto and Koganezawa, 2013), although aggression, virgin female receptivity, and female post-mating behaviors in flies are also interesting. All these mentioned behaviors are either sex-specific (e.g., male courtship) or sexually dimorphic (e.g., aggression), and they depend on dsx-expressing neurons (Pan et al., 2011; Kimura et al., 2015; Koganezawa et al., 2016). Since the first evidence of the atypical sexual behavior—performing and eliciting courtship—of XY homozygous mutants for dsx was described in 1985 (McRobert and Tompkins, 1985), a lot of research has been done to better understand the role of dsx in the control of sexual behavior. Courtship consists of a stereotyped sequence of actions displayed by the male to interest the female in copulation. Despite the fact that copulation in dsx null mutants is physically impossible because they do not have external genitalia (Taylor et al., 1994), dsx XY flies can perform, in a qualitatively normal-appearing manner, the courtship steps of following/orientation, tapping, wing extension, licking, and copulation attempts (Villella and Hall, 1996). However, dsx XY mutants exhibit a reduced wing extension time, and some individuals do not perform tapping, licking, or copulation attempts at all, with a high variability in phenotype penetrance (Villella and Hall, 1996). More recent studies demonstrated that the dsx XY fly courtship phenotype is a consequence of defects in the male motor circuit for copulation located in the abdominal ganglion (Pavlou et al., 2016). XX flies expressing constitutively DSXM do not exhibit courtship toward wild-type females (Taylor et al., 1994), suggesting that dsx alone cannot specify courtship behavior, and other regulators such as the fru gene are needed (Rideout et al., 2007; Yamamoto and Koganezawa, 2013).
On the other hand, dsx is fully required for proper courtship singing in males. The courtship song is crucial to stimulate the female to mate, consisting of brief pulses (pulse song) and bouts of humming sound (sine song) (Griffith et al., 1993; Rybak et al., 2002). dsx XY mutants are unable to sing sine song, and they present defects in particular aspects of pulse song (Villella and Hall, 1996; Shirangi et al., 2016). The complete absence of sine song in these mutants is due to the function of dsx in regulating the connectivity between the TN1A interneurons, a subclass of TN1 interneuron, and hg1 motor neurons (Shirangi et al., 2016). The activation or inhibition of some (or all) of the dsx-expressing neurons dramatically alters male courtship and copulatory behaviors. In males, the activation of dsx-expressing neurons triggers all courtship and copulatory behaviors (Pan et al., 2011), while their inhibition blocks them (Rideout et al., 2010).
dsx not only controls the male ability to perform the courtship ritual, but also the capacity of both males and females to elicit courtship in conspecifics. Thus, dsx XY flies elicit courtship in wild-type males at abnormally high levels. Interestingly, dsxF transgene-expressing males—XY flies ectopically expressing the female isoform of dsx—are strongly courted by normal wild-type males and other dsxF transgene males. Presumably, the sex appeal acquired by dsxF transgene-expressing males is due to the concomitant feminization of their synthesis of pheromones (Waterbury et al., 1999). Accordingly, DSXF—but not DSXM—activates the desaturase-F gene, which encodes the enzyme involved in the synthesis of dienes pheromones, crucial for optimal male courtship behavior (Chertemps et al., 2006; Shirangi et al., 2009).
Over the course of the last decade, insights into the function of DSXF in female sexual behavior have finally been investigated. dsx-positive neurons are critical for receptivity to first copulation (pC1 and pCd) in virgin females, for post-mating behaviors such as sperm ejection and storage (Dh44R1 neurons), for oviposition (mechanosensory neurons of the female reproductive system, abdominal ganglion, pMN2, and aDN), and for rejection behavior toward courting males (pC2l, abdominal ganglion) (Rezával et al., 2012, 2014; Zhou et al., 2014; Kimura et al., 2015; Lee et al., 2015; Nojima et al., 2021).
Lastly, competition over territory, food, or mates triggers aggression behavior between pairs of male or female flies (Kravitz and Huber, 2003). It has been demonstrated that the dsx-positive pC1 cluster resolves the males' dilemma between fighting or courting when they encounter conspecific. Thus, pC1 fru-negative sub-fraction acts as an aggression-triggering center, whereas pC1 fru-positive sub-fraction acts as a courtship-triggering center, and these two centers are mutually exclusively activated by two fru-single positive clusters, LC1 and mAL (Koganezawa et al., 2016). Furthermore, the activation of a subset of dsx-expressing pC1 neurons increases aggression behavior in females, but not in males (Palavicino-Maggio et al., 2019). Therefore, once again, we have evidence of dsx-expressing neurons contributing to control a sexually biased behavior.
Once we comprehended how Dmrt genes control sex differentiation of the nervous system in C. elegans and D. melanogaster, we sought to investigate how prevalent Dmrt genes are in nervous systems across Metazoa, with special attention to their potential role in sexual differentiation. Unfortunately, the majority of these studies do not systematically examine both sexes, and in the few cases where Dmrt expression is compared between the sexes, quantitative approaches or enough anatomical resolution are lacking, so dimorphic expressions cannot be unraveled.
Most metazoan studies on Dmrt expression in the nervous system have been conducted in chordates, and among them, studies in fishes stand out for including 11 distinct species. However, studies in DMRT neuronal function have been exclusively focused on model organisms: rat, zebrafish, and Xenopus, but primarily in mouse. For example, in these four species, members of the DMA subfamily (Dmrt3, Dmrt4, and Dmrt5) have been shown to have a conserved role in neurogenesis (Huang et al., 2005; Yoshizawa et al., 2011; Konno et al., 2012, 2019; Kikkawa et al., 2013; Parlier et al., 2013; Saulnier et al., 2013). For excellent reviews see (Bellefroid et al., 2013; Kikkawa and Osumi, 2021).
According to our literature research (summarized in Table 1), Dmrt genes are expressed in neural tissues in a plethora of animal species, including the phyla Chordata, Arthropoda, Platyhelmithes, Onychophora, and Cnidaria. Many of the studies do not reference the sex of the samples analyzed [we indicate this in the table with a (–) in the Dimorphic expression column], or only one sex was analyzed (also indicated in the table). Only 12 of the 53 studies included sex comparisons, and almost all of them deal with fishes or arthropods (Japanese pufferfish, largemouth bass, Medaka, tilapia, platyfish, Atlantic cod, swamp eel, sweetfish, mosquito, bumblebee, silkmoth, and oriental river prawn). Sex differences in Dmrt expression levels or expression patterns have been reported in six studies (highlighted in color in Table 1): sex differences in mRNA levels for dmrt4 were found in the adult telencephalons of tilapia (Cao et al., 2007), and in the adult brains of Atlantic cod (Johnsen and Andersen, 2012); for dmrt5, in the adult brains of the sweetfish (Wang et al., 2014); for dsx, in the adult brains of the silkmoth (Nakata et al., 2021); and for dmrt11E, in the adult abdominal ganglia of the oriental river prawn (Wang et al., 2019). Regarding sex differences in expression patterns, dsx is expressed in different clusters in the antennal lobe and mushroom body of the mosquito Aedes aegypti according to sex (Tomchaney et al., 2014). In the silkmoth, besides the sex differences in dsx expression across development, dsx is expressed ventral to the calyx of the mushroom body (cluster 1) and dorsolateral to the esophagus (cluster 2) in males, while in the female dsx is only expressed in cluster 1 (Nakata et al., 2021).
To investigate sex differences in Dmrt expression, the major limitation of the above mRNA quantifications is their lack of cell resolution. In many studies, whole telencephalon or brain was analyzed, or the authors did not specify whether they dissected a particular region. However, by taking the whole tissue, putative sex differences in particular brain nuclei or cell subpopulations are neglected. In fact, that could be occurring with the similar expression of dmrt1 and dmrt5 observed in the adult brains of males and females in the largemouth bass (Yan et al., 2019) and Atlantic cod (Johnsen and Andersen, 2012), respectively. Interestingly, although not discussed in the article, during gonad transformation in the swamp eel (this animal undergoes sex reversal; first, they have ovaries, then there is an intersex stage between female and male stages in which the animal develops ovotestes, and finally, they develop testes) expression of dmrt2, dmrt2b, dmrt3, dmrt4, and dmrt5 in the brain undergoes concomitant changes, as observed in the differential intensity of bands in cDNA electrophoretic gels (Sheng et al., 2014). There is another study where an evident sex differential expression in the Japanese pufferfish brain for a dmrt exists, but the authors do not mention it (Yamaguchi et al., 2006). Although a quantitative approach was not used, a higher expression of dmrt5 in females can be observed by RT-PCR followed by Southern blot.
Concerning humans, according to the BrainSpan Atlas of the Developing Human Brain (http://www.brainspan.org/), a transcriptome resource of prenatal and post-natal human specimens of cortical and subcortical structures, DMRT2, DMRTA1/DMRT4, DMRT3 and DMRTA2/DMRT5 are expressed mainly in cortical regions and at higher levels in prenatal stages. In addition to the expression data, two clinical case reports have served to provide insights into how DMRTs operate in the human nervous system. One case report suggests that loss of function of DMRTA2 leads to a severe recessive condition characterized by microcephaly, lissencephaly, and agenesis of the corpus callosum (Urquhart et al., 2016). In fact, Dmrt5−/− mice, among other defects, show a strong reduction of the caudomedial cortex, and they lack corpus callosum (Saulnier et al., 2013), which is coherent with the observed human phenotype. On the other hand, Lerer et al. (2005) reported a case of a familial cerebral palsy with a deletion in the KANK1 gene locus, including a large part of the intergenic region between KANK1 and DMRT1 (KANK1-DMRT1-DMRT3 constitute a synteny in both humans and mice) (Lerer et al., 2005; Kubota et al., 2018). However, this clinical case report does not directly elucidate the role of DMRT3 in cerebral palsy, but it served as a basis for a subsequent study that demonstrates the involvement of DMRT3 via bioinformatics approaches (Kubota et al., 2018). In that study, the authors used comparative genomic and reanalysis of public data for ChIP-Seq and chromatin interaction to postulate an enhancer of DMRT3 as a putative source for spastic cerebral palsy. In addition, this presumed enhancer contains a putative RAR/RXR-binding motif, and Dmrt3 gene expression is upregulated after retinoic acid stimulation in mouse cell culture (Kubota et al., 2018). Since Dmrt3 participates in the specification of spinal cord dI6 interneurons [mutations in the Dmrt3 gene affects locomotion in horses and mice (Andersson et al., 2012)], and dmrt3 is involved in cortical development (Konno et al., 2019), if conserved in these features, DMRT3 could contribute to motor symptoms in cerebral palsy patients as a consequence of impaired development of the motor cortex and dysfunctional locomotion circuits in the spinal cord. Lastly, deletion or duplication of the short arm of chromosome 9, where DMRT1, DMRT2, and DMRT3 are located, is characterized by developmental delay, intellectual disabilities, and craniofacial anomalies (Güneş et al., 2017). In addition, an association of 9p deletion and autistic spectrum disorder has also been reported (Õunap et al., 2004; Vinci et al., 2007; Yang et al., 2012).
One limitation of our search is that we have not systematically looked for neural tissues in animals in which it was demonstrated that Dmrt are not expressed, which would provide more information about Dmrt conservation across the animal kingdom. For instance, some Dmrts are not detected in the adult brains of males and females of several fishes: dmrt6 in rock bream (supplementary material in Li et al., 2021), dmrt4 in Japanese pufferfish (Yamaguchi et al., 2006), dmrt1, dmrt2a, and dmrt3 in Atlantic cod (Johnsen and Andersen, 2012), and dmrt1 in sweetfish (Wang et al., 2014).
As we have exposed previously, the role of Dmrt genes in sexual differentiation and regulation of vertebrate's nervous system has not been systematically approached. However, we would like to delve into potential non-cell autonomous interactions between Dmrt functions in peripheral organs and the brain. In mammals, sexual hormones secreted by the gonad have a fundamental non-cell autonomous role in the nervous system. In the embryo, the coordinated action of fetal Leydig and Sertoli cells is necessary for testosterone release (Shima et al., 2013). Embryonic testosterone is subsequently locally converted to estradiol by aromatase to act on brain cells. According to the organizational-activational hypothesis of brain sexual differentiation (Arnold, 2009), during the organizational phase, several mechanisms will configure dimorphic circuits, such as proliferation, neurogenesis, apoptosis, dendritic growth, synaptogenesis or synaptic pruning among others (Phoenix et al., 1959; Arnold, 2009; McCarthy et al., 2017). Later, in sexually mature adult animals, both testosterone and estradiol (released by adult Leydig cells and by granulosa cells, respectively) will modulate brain activity during the activational phase (McCarthy, 2020; Gegenhuber et al., 2022). Therefore, transcriptional networks—potentially including Dmrt genes—that affect gonadal development and maintenance could indirectly have a major impact on sexual brain differentiation.
Despite profound differences in gonad structure and development among animal phyla, Dmrt genes are specifically expressed in the developing gonads of almost all animals (Matson and Zarkower, 2012). In mice and humans, Dmrt1 expression is sex-specific and developmentally dynamic. In mice (where it has been most extensively studied), Dmrt1 is expressed in germ cells of both sexes through E13.5, disappearing by about E15.5 (Lei et al., 2007). However, Dmrt1 becomes highly male-enriched in somatic Sertoli cells by about E14.5 as testis differentiation initiates, and its expression is maintained thereafter (Raymond et al., 1999; Lei et al., 2007). In males, Dmrt1 is re-expressed in spermatogonia just before birth and maintained in spermatogonia subsequently (Lei et al., 2007; Matson et al., 2010). Human DMRT1 expression is less well-characterized but appears to be similar to that of mice. During fetal development, DMRT1 mRNA is detectable by gestational week 11 (GW11) in both sexes, becoming most abundant in pre-Sertoli cells between GW10 and GW20 (Jorgensen et al., 2012). In adult testes, DMRT1 is expressed both in Sertoli cells and in spermatogonia, as in mice. However, there are discrepancies between the functions ascribed to DMRT1 in humans and mice that are not fully understood yet. In Dmrt1 mutant mice, severe defects are observed in post-natal testes and reveal the importance of Dmrt1 in Sertoli differentiation and male germ cell survival. About two weeks after birth, Dmrt1 mutant immature Sertoli cells acquire expression of female-specific granulosa cell markers (Matson et al., 2011). Interestingly, androgen activity is reduced and estradiol levels are elevated in mutants, suggesting a male-to-female tilt in hormone signaling, which is probably due in part to an elevated expression of the enzyme aromatase. However, as far as we are aware, nervous system sexual differentiation has not been assessed in these mice. Human DMRT1 deletions and microdeletions on the distal part of chromosome 9, where DMRT1 is located, and missense mutations in the DM domain have been associated with Disorders in Sexual Development (DSD), and reported to cause partial or complete 46,XY gonadal dysgenesis (Raymond, 1999; Zarkower and Murphy, 2021) and male infertility cases (Tewes et al., 2014). The degree of DSD can range from male genitalia with small dysgenic testes, to complete sex reversal that will potentially affect testosterone secretion. Moreover, distal 9p monosomy deletions have also been associated with cognitive deficits, developmental delay, and a characteristic array of dysmorphic craniofacial features (Alfi et al., 1976). 9p deletions consistently affect DMRT1. However, most of these deletions also affect other genes, including the adjacent paralogues DMRT3 and DMRT2, which are immediately proximal to DMRT1 on 9p. In mice, Dmrt3 function in gonadal development (and the nearby Dmrt2) has not been reported (Seo et al., 2006; De Clercq et al., 2016; Desmaris et al., 2018).
In vertebrates, most Dmrt family members are expressed in the undifferentiated gonad and in most cases, they are subsequently maintained at higher levels in male as opposed to female gonads (Kim et al., 2003). In mice, at least, mutations in Dmrt4 (Dmrta1), Dmrt6 (Dmrtb1), and Dmrt7 (Dmrtc2) disrupt gonadal development in one or both sexes (Raymond et al., 2000; Balciuniene et al., 2006; Kawamata and Nishimori, 2006; Kim et al., 2007; Zhang et al., 2014). However, these Dmrt genes are expressed in cells that do not affect gonadal hormone secretion. Dmrt7 localizes to spermatocytes (Kawamata and Nishimori, 2006); Dmrt6 and Dmrt7 regulate germ cell differentiation and appear not to be required in any other cell types (Kim et al., 2007; Zhang et al., 2014). A systematic analysis of Dmrt expression in embryonic mouse gonads by RT-PCR revealed that many of them are expressed in male and female gonads at some point of development (Kim et al., 2003). Dmrt3 is expressed in embryonic ovaries and testes, but is undetectable in adults. Conversely, Dmrt4 and Dmrt5 are expressed in adult testes. In the embryo, Dmrt5 is more highly expressed in the ovary, although no gonadal phenotype in Dmrt5 mutants has been described to date, and Dmrt5 null mutants are perinatally lethal and fertility assays have not yet been performed. Dmrt4 is present in the gonads of both embryos and adult animals (Kim et al., 2003; Balciuniene et al., 2006) and while mutant females have polyovular follicles, suggesting a role in folliculogenesis, 25% of mutant males consistently exhibited copulatory behavior toward other males (Balciuniene et al., 2006). However, the origin of this behavior has not yet been fully explored molecularly.
Another interesting site of Dmrt expression that we would like to briefly highlight, is the potential role of Dmrt5 in pituitary gland development. The hypothalamic-pituitary-gonadal axis controls puberty and reproduction (Abreu and Kaiser, 2016). This axis is active in the embryonic and early post-natal stages of life, is subsequently restrained during childhood, and its reactivation culminates in puberty initiation. At that time, the hypothalamus releases gonadotrophin-releasing hormone (GnRH), which can act on pituitary gonadotropes, triggering the release of follicle-stimulating hormone (FSH) and luteinizing hormone (LH). FSH and LH bind to their respective receptors in the ovary, leading to secretion of the sex steroid hormones, estrogen, which stimulates vitellogenesis, and progesterone, which stimulates oocyte meiosis, follicular maturation, and ovulation, even in fish (Yaron and Levavi-Sivan, 2011). Interestingly, in zebrafish pituitaries, dmrt5 controls corticotrope and gonadotrope cell differentiation (Graf et al., 2015). It is tempting to speculate that altered dmrt5 activity could indirectly affect gonadal activity and consequently sexual development because of altered gonadotrope numbers and/or activities. Although this has not been tested, a recent study reported that a deficiency in LH and FSH in zebrafish mutants leads to a significant delay in gonad development and the onset of puberty, as well as infertility and sex reversal (Zhang et al., 2015). Interestingly, Dmrt5 is expressed in the embryonic mouse pituitary (Saulnier et al., 2013), and according to The Human Protein Atlas resource, DMRT5 (DMRTA2) is enriched in pituitary and testes tissues (and not in ovaries) (www.proteinatlas.org).
Regardless of the independent evolutionary origins of Dmrt1, dsx, and mab-3 (Mawaribuchi et al., 2019), research within the last couple of decades has positioned DMRT transcription factors as the most conserved type of protein consistently utilized throughout the animal kingdom to control sex determination and differentiation. We have extensively detailed how Dmrt genes operate in the sexual differentiation of the nervous systems in C. elegans and Drosophila. In light of the similarities between mechanistic aspects of dsx and dmds, it is possible that Dmrt genes control similar target genes to cooperate in the sexual differentiation of the brain in the nervous system of other animals. Indeed, dsx and Dmrt1 have been proposed to regulate similar target genes (Clough et al., 2014). Neurogenesis, cell death, and connectivity—through the control of morphology, neurotransmitter gene expression, or selective synaptic pruning—seem to be the preferred features controlled by Dmrt genes that, ultimately, lead to the generation of sexually dimorphic circuits and behavioral outputs. Significantly, these are recurrent mechanisms that are engaged in the refinement of neuronal circuits at various stages throughout development. It is precisely the fact that Dmrt genes control such mechanisms that situates them in a relevant position in the control of sexually dimorphic circuit configurations.
In all organisms studied so far, the precise role of Dmrts in the specification of sex-specific somatic features, including in the nervous system, results from a dichotomy in their expression pattern in males and females. Up-stream sex-determinant primary cues ensure the sexually dimorphic expression of Dmrt genes: for dsx, a single locus produces two different variants, a female and a male variant deployed in many different cellular contexts; in C. elegans, several dmds are expressed exclusively in males (and dmd-4 in hermaphrodites) as a result of the actions of the master regulator of sex-determination tra-1. In other species, where Dmrts are dimorphically expressed, upstream direct regulators have not yet been identified. However, as we have exposed in this review, not all Dmrt genes are dimorphically expressed (Table 1). One possibility is that the consequences of this loss of dimorphic expression could lead to unisex functions (Bellefroid et al., 2013). However, we hypothesize that as a consequence of the monomorphic expression, in certain cellular contexts, Dmrt genes could have acquired a neofunction to prevent sexual differences from occurring. However, to date, no functional studies have been performed in Dmrt mutants to demonstrate differential outcomes in males and females in Dmrt unisex-expressing regions.
The relatively simple and fully mapped nervous systems of C. elegans and Drosophila in both sexes (in Drosophila, the female neuronal connectivity map has been made available recently, and the male map will soon be released), together with their genetic amenability and access to sexually dimorphic behaviors, has allowed the thorough delineation of the role of Dmrts in sexual differentiation in these animals. In other species, especially in vertebrates, this task might entail a greater challenge due to the greater complexity of their nervous systems and the lack of powerful genetic tools. However, in order to advance our knowledge of the potential functions of Dmrt genes across metazoans (with special focus on the mammalian brain), we would like to highlight a few specific remarks:
• Many of the recent/contemporary studies relied on Dmrt mRNA measurements (either RT-PCR or in situ hybridization). However, by relying solely on these techniques, post-translational regulatory mechanisms will go undetected. This point is especially relevant in Dmrt genes with DMA domain that may undergo sex-specific post-transcriptional regulation, as has been proposed in Bayer et al. (2020).
• Dmrt genes may be acting in discrete, scattered, neuronal subpopulations. Within neuronal circuits, subtle changes in synaptic connection affect the communication capabilities of neurons (for instance, by scaling their neurotransmitter levels up or down, or switching post-synaptic partners) and could have major behavioral outcomes. Indeed, this is in line with the notion that homologous circuits between males and females can be reversibly modulated. Searching for neuron-type specific Dmrt expression differences requires the use of technical approaches with high cellular resolution and quantitative characteristics. In this regard, differences in Dmrt level of expression could also be highly relevant.
• If Dmrt genes are expressed in a single neuron type within the circuit, the mutant phenotype may have major behavioral consequences. Therefore, functional studies, based on mutant strains, would be a useful and necessary complement to Dmrt gene expression maps.
• Dmrt genes act as integrators of sex, space, and time (Figure 2). However, the precise molecular mechanisms to integrate these variables are not yet fully understood and still require additional investigation, even in the most-studied invertebrate model organisms. Therefore, when studying Dmrt expression and function, different time points should be considered. In some cases, embryos and adult—sexually mature—animals have been analyzed, but usually a systematic approach throughout the various developmental stages is lacking. For instance, in mammalian brains, Dmrt gene function might be more relevant during the embryonic testosterone surge or during/after puberty. Lastly, many mouse Dmrt mutants are embryonic lethal and have not yet been studied during post-natal stages, when sexual behaviors emerge.
• Lastly, we would like to emphasize that Dmrt functions should be studied using a holistic approach. Dmrt genes are widely expressed in many organs. It may be especially interesting to explore the non-cell autonomous relationships that Dmrt phenotypes in the gonad or the pituitary gland might exert on brain differentiation.
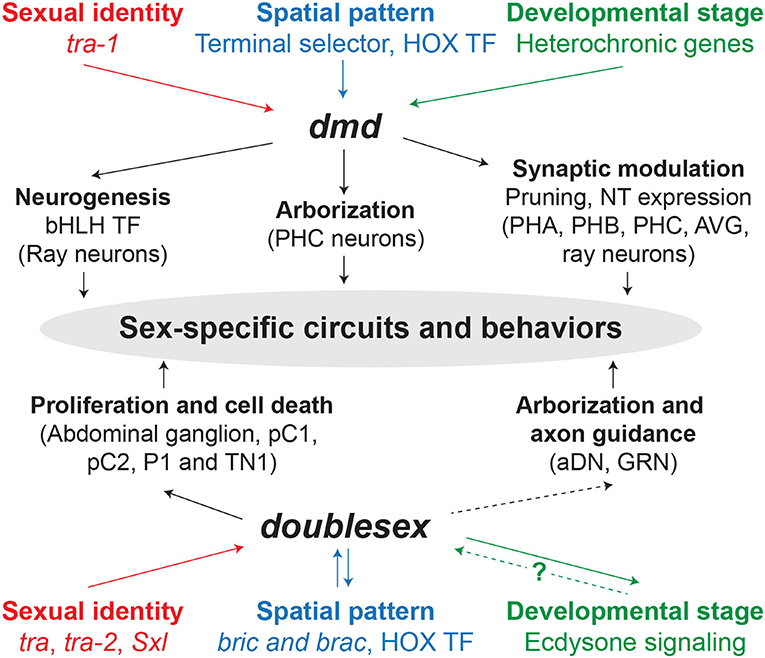
Figure 2. dmd and doublesex mechanisms underlying sex-specific circuits and behaviors. Solid and dashed arrows indicate direct and indirect regulation, respectively. The question mark represents that the mechanism by which ecdysone signaling regulates dsx is unknown. TF, transcription factor; NT, neurotransmitter.
From a human health perspective, DMRTs are excellent genes to be considered as candidate factors related to the etiology and sexual skewing of neuropsychiatric diseases. In the mammalian brain, where sex hormones have a huge impact in establishing sex differences (McCarthy and Arnold, 2011), DMRTs may be acting in parallel in order to create or buffer differences created by gonadal hormones. Hence, DMRTs may either afford protection or generate vulnerability in one sex versus the other for sex-biased mental disorders such as autism spectrum disorder, bipolar disorder, and schizophrenia.
Moreover, given the importance of sex in this review, we feel obliged to remark, regrettably, that most neuroscience research is conducted in males, and even when both sexes are included, the sex is rarely analyzed as a variable (Beery and Zucker, 2011). We hope that this review also serves to raise awareness within the scientific community to more frequently consider sex as a biological variable.
RC-N and ES-S wrote the text. Both authors contributed to the article and approved the submitted version.
This work was supported by Grant PGC2018-101751-A-100, by MCIN/ AEI /10.13039/501100011033/ and by ERDF “A way of making Europe”. RC-N was supported by Ministerio de Universidades - FPU19/02352. ES-S was supported by Grant RYC-2016-20537 funded by MCIN/AEI /10.13039/501100011033 and ESF “Investing in your future”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are thankful to Dr. Mawaribuchi and Dr. Ito for giving us permission to include the DMRT phylogenetic tree from Mawaribuchi et al. (2019).
Abreu, A. P., and Kaiser, U. B. (2016). Pubertal development and regulation. Lancet Diabetes Endocrinol. 4, 254–264. doi: 10.1016/S2213-8587(15)00418-0
Aeschimann, F., Kumari, P., Bartake, H., Gaidatzis, D., Xu, L., Ciosk, R., et al. (2017). LIN41 post-transcriptionally silences mRNAs by two distinct and position-dependent mechanisms. Mol. Cell 65, 476–489.e4. doi: 10.1016/j.molcel.2016.12.010
Alfi, O. S., Donnell, G. N., Allderdice, P. W., and Derencsenyi, A. (1976). The 9p- syndrome. Ann. Genet. 19, 11–6.
An, W., Cho, S., Ishii, H., and Wensink, P. C. (1996). Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol. Cell. Biol. 16, 3106–3111. doi: 10.1128/MCB.16.6.3106
Anand, A., Patel, M., Lalremruata, A., Singh, A. P., Agrawal, R., Singh, L., et al. (2008). Multiple alternative splicing of Dmrt1 during gonadogenesis in Indian mugger, a species exhibiting temperature-dependent sex determination. Gene 425, 56–63. doi: 10.1016/j.gene.2008.08.005
Andersson, L. S., Larhammar, M., Memic, F., Wootz, H., Schwochow, D., Rubin, C.-J., et al. (2012). Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 488, 642–646. doi: 10.1038/nature11399
Arnold, A. P. (2009). The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm. Behav. 55, 570–578. doi: 10.1016/j.yhbeh.2009.03.011
Baker, B. S., and Ridge, K. A. (1980). Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94, 383–423. doi: 10.1093/genetics/94.2.383
Balciuniene, J., Bardwell, V. J., and Zarkower, D. (2006). Mice mutant in the DM domain gene Dmrt4 are viable and fertile but have polyovular follicles. Mol. Cell. Biol. 26, 8984–8991. doi: 10.1128/MCB.00959-06
Barr, M. M., and Garcia, L. R. (2006). Male mating behavior. WormBook. 1–11. doi: 10.1895/wormbook.1.78.1
Bayer, E. A., Stecky, R. C., Neal, L., Katsamba, P. S., Ahlsen, G., Balaji, V., et al. (2020). Ubiquitin-dependent regulation of a conserved DMRT protein controls sexually dimorphic synaptic connectivity and behavior. Elife 9, e59614. doi: 10.7554/eLife.59614.sa2
Beery, A. K., and Zucker, I. (2011). Sex bias in neuroscience and biomedical research. Neurosci. Biobehav. Rev. 35, 565–572. doi: 10.1016/j.neubiorev.2010.07.002
Bellefroid, E. J., Leclère, L., Saulnier, A., Keruzore, M., Sirakov, M., Vervoort, M., et al. (2013). Expanding roles for the evolutionarily conserved Dmrt sex transcriptional regulators during embryogenesis. Cell. Mol. Life Sci. 70, 3829–3845. doi: 10.1007/s00018-013-1288-2
Berghoff, E. G., Glenwinkel, L., Bhattacharya, A., Sun, H., Varol, E., Mohammadi, N., et al. (2021). The Prop1-like homeobox gene unc-42 specifies the identity of synaptically connected neurons. Elife 10, 1–37. doi: 10.7554/eLife.64903
Billeter, J., Rideout, E. J., Dornan, A. J., and Goodwin, S. F. (2006b). Control of male sexual behavior in Drosophila by the sex determination pathway. Curr. Biol. 16, 766–776. doi: 10.1016/j.cub.2006.08.025
Billeter, J. C., Villella, A., Allendorfer, J. B., Dornan, A. J., Richardson, M., Gailey, D. A., et al. (2006a). Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr. Biol. 16, 1063–1076. doi: 10.1016/j.cub.2006.04.039
Birkholz, O., Rickert, C., Berger, C., Urbach, R., and Technau, G. M. (2013). Neuroblast pattern and identity in the Drosophila tail region and role of doublesex in the survival of sex-specific precursors. Development 140, 1830–1842. doi: 10.1242/dev.090043
Burtis, K. C., and Baker, B. S. (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997–1010. doi: 10.1016/0092-8674(89)90633-8
Cao, J., Cao, Z., and Wu, T. (2007). Generation of antibodies against DMRT1 and DMRT4 of Oreochromis aurea and analysis of their expression profile in Oreochromis aurea tissues. J. Genet. Genomics 34, 497–509. doi: 10.1016/S1673-8527(07)60055-1
Chatterjee, S. S., Uppendahl, L. D., Chowdhury, M. A., Ip, P. L., and Siegal, M. L. (2011). The female-specific Doublesex isoform regulates pleiotropic transcription factors to pattern genital development in Drosophila. Development 138, 1099–1109. doi: 10.1242/dev.055731
Chertemps, T., Duportets, L., Labeur, C., Ueyama, M., and Wicker-Thomas, C. (2006). A female-specific desaturase gene responsible for diene hydrocarbon biosynthesis and courtship behaviour in Drosophila melanogaster. Insect Mol. Biol. 15, 465–473. doi: 10.1111/j.1365-2583.2006.00658.x
Cho, S., and Wensink, P. C. (1996). Purification and physical properties of the male and female double sex proteins of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 93, 2043–2047. doi: 10.1073/pnas.93.5.2043
Chong, T., Iii, J. J. C., Brubacher, J. L., Zarkower, D., and Newmark, P. A. (2013). A sex-specific transcription factor controls male identity in a simultaneous hermaphrodite. Nat. Commun. 4, 1812–1814. doi: 10.1038/ncomms2811
Clough, E., Jimenez, E., Kim, Y. A., Whitworth, C., Neville, M. C., Hempel, L. U., et al. (2014). Sex- and tissue-specific functions of drosophila doublesex transcription factor target genes. Dev. Cell 31, 761–773. doi: 10.1016/j.devcel.2014.11.021
Conradt, B., and Horvitz, H. R. (1999). The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 98, 317–327. doi: 10.1016/S0092-8674(00)81961-3
Cook, R. M. (1977). Behavioral role of the sexcombs in Drosophila melanogaster and Drosophila simulans. Behav. Genet. 7, 349–357. doi: 10.1007/BF01077448
Cook, S. J., Jarrell, T. A., Brittin, C. A., Wang, Y., Bloniarz, A. E., Yakovlev, M. A., et al. (2019). Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71. doi: 10.1038/s41586-019-1352-7
Cui, Y., Behura, S. K., and Franz, A. W. E. (2022). Cellular diversity and gene expression profiles in the male and female brain of Aedes aegypti. BMC Genomics 23, 119. doi: 10.1186/s12864-022-08327-9
De Clercq, S., Keruzore, M., Desmaris, E., Pollart, C., Assimacopoulos, S., Preillon, J., et al. (2016). DMRT5 together with DMRT3 directly controls hippocampus development and neocortical area map formation. Cereb. Cortex 28, 493–509. doi: 10.1093/cercor/bhw384
del Pozo, A., Manuel, R., Iglesias Gonzalez, A. B., Koning, H. K., Habicher, J., Zhang, H., et al. (2020). Behavioral characterization of dmrt3a mutant zebrafish reveals crucial aspects of vertebrate locomotion through phenotypes related to acceleration. eNeuro 7, ENEURO.0047-20.2020. doi: 10.1523/ENEURO.0047-20.2020
Desmaris, E., Keruzore, M., Saulnier, A., Ratié, L., Assimacopoulos, S., De Clercq, S., et al. (2018). DMRT5, DMRT3, and EMX2 cooperatively repress GSX2 at the pallium–subpallium boundary to maintain cortical identity in dorsal telencephalic progenitors. J. Neurosci. 38, 9105–9121. doi: 10.1523/JNEUROSCI.0375-18.2018
Erdman, S. E., and Burtis, K. C. (1993). The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 12, 527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x
Erdman, S. E., Chen, H. J., and Burtis, K. C. (1996). Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics 144, 1639–1652. doi: 10.1093/genetics/144.4.1639
Fagan, K. A., Luo, J., Lagoy, R. C., Schroeder, F. C., Albrecht, D. R., and Portman, D. S. (2018). A single-neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Curr. Biol. 28, 902–914.e5. doi: 10.1016/j.cub.2018.02.029
Galazo, M. J., Emsley, J. G., and Macklis, J. D. (2016). Corticothalamic projection neuron development beyond subtype specification: Fog2 and intersectional controls regulate intraclass neuronal diversity. Neuron 91, 90–106. doi: 10.1016/j.neuron.2016.05.024
Gegenhuber, B., Wu, M. V., Bronstein, R., and Tollkuhn, J. (2022). Gene regulation by gonadal hormone receptors underlies brain sex differences. Nature. doi: 10.1038/s41586-022-04686-1
Gennet, N., Gale, E., Nan, X., Farley, E., Takacs, K., Oberwallner, B., et al. (2011). Doublesex and mab-3-related transcription factor 5 promotes midbrain dopaminergic identity in pluripotent stem cells by enforcing a ventral-medial progenitor fate. Proc. Natl. Acad. Sci. U. S. A. 108, 9131–9136. doi: 10.1073/pnas.1016679108
Gennet, N., Tamburini, C., Nan, X., and Li, M. (2016). FolR1: a novel cell surface marker for isolating midbrain dopamine neural progenitors and nascent dopamine neurons. Sci. Rep. 6, 32488. doi: 10.1038/srep32488
Goodwin, S. F., and Hobert, O. (2021). Molecular mechanisms of sexually dimorphic nervous system patterning in flies and worms. Annu. Rev. Cell Dev. Biol. 37, 519–547. doi: 10.1146/annurev-cellbio-120319-115237
Graf, M., Teo Qi-Wen, E.-R., Sarusie, M. V., Rajaei, F., and Winkler, C. (2015). Dmrt5 controls corticotrope and gonadotrope differentiation in the zebrafish pituitary. Mol. Endocrinol. 29, 187–199. doi: 10.1210/me.2014-1176
Griffith, L. C., Verselis, L. M., Aitken, K. M., Kyriacou, C. P., Danho, W., and Greenspan, R. J. (1993). Inhibition of calcium/calmodulin-dependent protein kinase in Drosophila disrupts behavioral plasticity. Neuron 10, 501–509. doi: 10.1016/0896-6273(93)90337-Q
Gruzin, M., Mekheal, M., Ruhlman, K., Winkowski, M., and Petko, J. (2020). Developmental expression of doublesex-related transcripts in the common house spider, Parasteatoda tepidariorum. Gene Expr. Patterns 35, 119101. doi: 10.1016/j.gep.2020.119101
Güneş, S., Ekinci, Ö., Ekinci, N., and Toros, F. (2017). Coexistence of 9p deletion syndrome and autism spectrum disorder. J. Autism Dev. Disord. 47, 520–521. doi: 10.1007/s10803-016-2943-x
Guo, Y., Li, Q., Gao, S., Zhou, X., He, Y., Shang, X., et al. (2004). Molecular cloning, characterization, and expression in brain and gonad of Dmrt5 of zebrafish. Biochem. Biophys. Res. Commun. 324, 569–575. doi: 10.1016/j.bbrc.2004.09.085
Heinrichs, V., Ryner, L. C., and Baker, B. S. (1998). Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 18, 450–458. doi: 10.1128/MCB.18.1.450
Hilliard, M. A., Bargmann, C. I., and Bazzicalupo, P. (2002). C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr. Biol. 12, 730–734. doi: 10.1016/S0960-9822(02)00813-8
Hobert, O., and Kratsios, P. (2019). Neuronal identity control by terminal selectors in worms, flies, and chordates. Curr. Opin. Neurobiol. 56, 97–105. doi: 10.1016/j.conb.2018.12.006
Hoshijima, K., Inoue, K., Higuchi, I., Sakamoto, H., and Shimura, Y. (1991). Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252, 833–836. doi: 10.1126/science.1902987
Huang, X., Hong, C.-S., O'Donnell, M., and Saint-Jeannet, J.-P. (2005). The doublesex-related gene, XDmrt4, is required for neurogenesis in the olfactory system. Proc. Natl. Acad. Sci. U. S. A. 102, 11349–11354. doi: 10.1073/pnas.0505106102
Imai, K. S., Levine, M., Satoh, N., and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183–1187. doi: 10.1126/science.1123404
Inoue, K., Hoshijima, K., Higuchi, I., Sakamoto, H., and Shimura, Y. (1992). Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc. Natl. Acad. Sci. U. S. A. 89, 8092–8096. doi: 10.1073/pnas.89.17.8092
Ishii, K., Wohl, M., Desouza, A., and Asahina, K. (2020). Sex-determining genes distinctly regulate courtship capability and target preference via sexually dimorphic neurons. eLife. 9, e52701. doi: 10.7554/eLife.52701
Jarrell, T. A., Wang, Y., Bloniarz, A. E., Brittin, C. A., Xu, M., Thomson, J. N., et al. (2012). The connectome of a decision-making neural network. Science 337, 437–444. doi: 10.1126/science.1221762
Johnsen, H., and Andersen, Ø. (2012). Sex dimorphic expression of five dmrt genes identified in the Atlantic cod genome. The fish-specific dmrt2b diverged from dmrt2a before the fish whole-genome duplication. Gene 505, 221–232. doi: 10.1016/j.gene.2012.06.021
Jois, S., Chan, Y.-B., Fernandez, M. P., Pujari, N., Janz, L. J., Parker, S., et al. (2022). Sexually dimorphic peripheral sensory neurons regulate copulation duration and persistence in male Drosophila. Sci. Rep. 12, 6177. doi: 10.1038/s41598-022-10247-3
Jorgensen, A., Nielsen, J. E., Blomberg Jensen, M., Graem, N., and Rajpert-De Meyts, E. (2012). Analysis of meiosis regulators in human gonads: a sexually dimorphic spatio-temporal expression pattern suggests involvement of DMRT1 in meiotic entry. Mol. Hum. Reprod. 18, 523–534. doi: 10.1093/molehr/gas030
Kasahara, R., Aoki, F., and Suzuki, M. G. (2018). Deficiency in dmrt99B ortholog causes behavioral abnormalities in the silkworm, Bombyx mori. Appl. Entomol. Zool. 53, 381–393. doi: 10.1007/s13355-018-0569-5
Kawamata, M., and Nishimori, K. (2006). Mice deficient in Dmrt7 show infertility with spermatogenic arrest at pachytene stage. FEBS Lett. 580, 6442–6446. doi: 10.1016/j.febslet.2006.10.066
Kikkawa, T., Obayashi, T., Takahashi, M., Fukuzaki-Dohi, U., Numayama-Tsuruta, K., and Osumi, N. (2013). Dmrta1 regulates proneural gene expression downstream of Pax6 in the mammalian telencephalon. Genes to Cells 18, 636–649. doi: 10.1111/gtc.12061
Kikkawa, T., and Osumi, N. (2021). Multiple functions of the dmrt genes in the development of the central nervous system. Front Neurosci. 15, 789583. doi: 10.3389/fnins.2021.789583
Kikkawa, T., Sakayori, N., Yuuki, H., Katsuyama, Y., Matsuzaki, F., Konno, D., et al. (2020). Dmrt genes participate in the development of Cajal-Retzius cells derived from the cortical hem in the telencephalon. Dev. Dyn. 249, 698–710. doi: 10.1002/dvdy.156
Kim, D., and Kim, B. (2022). Anatomical and functional differences in the sex-shared neurons of the nematode C. elegans. Front. Neuroanat. 16, 906090. doi: 10.3389/fnana.2022.906090
Kim, S., Kettlewell, J. R., Anderson, R. C., Bardwell, V. J., and Zarkower, D. (2003). Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr. Patterns 3, 77–82. doi: 10.1016/S1567-133X(02)00071-6
Kim, S., Namekawa, S. H., Niswander, L. M., Ward, J. O., Lee, J. T., Bardwell, V. J., et al. (2007). A mammal-specific doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 3, e62. doi: 10.1371/journal.pgen.0030062
Kimura, K., ichi Hachiya, T., Koganezawa, M., Tazawa, T., and Yamamoto, D. (2008). Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron 59, 759–769. doi: 10.1016/j.neuron.2008.06.007
Kimura, K., Sato, C., Koganezawa, M., and Yamamoto, D. (2015). Drosophila ovipositor extension in mating behavior and egg deposition involves distinct sets of brain interneurons. PLoS ONE 10, e0126445. doi: 10.1371/journal.pone.0126445
Koganezawa, M., Kimura, K., and Yamamoto, D. (2016). The neural circuitry that functions as a switch for courtship versus aggression in Drosophila males. Curr. Biol. 26, 1395–1403. doi: 10.1016/j.cub.2016.04.017
Kondo, M., Froschauer, A., Kitano, A., Nanda, I., and Hornung, U. (2002). Molecular cloning and characterization of DMRT genes from the medaka Oryzias latipes and the platyfish Xiphophorus maculatus. Gene 295, 213–222. doi: 10.1016/S0378-1119(02)00692-3
Konno, D., Iwashita, M., Satoh, Y., Momiyama, A., Abe, T., Kiyonari, H., et al. (2012). The mammalian DM domain transcription factor Dmrta2 is required for early embryonic development of the cerebral cortex. PLoS ONE 7, e46577. doi: 10.1371/journal.pone.0046577
Konno, D., Kishida, C., Maehara, K., Ohkawa, Y., Kiyonari, H., Okada, S., et al. (2019). Dmrt factors determine the positional information of cerebral cortical progenitors via differential suppression of homeobox genes. Development 146, dev174243. doi: 10.1242/dev.174243
Kopp, A. (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175–184. doi: 10.1016/j.tig.2012.02.002
Kravitz, E. A., and Huber, R. (2003). Aggression in invertebrates. Curr. Opin. Neurobiol. 13, 736–743. doi: 10.1016/j.conb.2003.10.003
Kubota, N., Yokoyama, T., Hoshi, N., and Suyama, M. (2018). Identification of a candidate enhancer for DMRT3 involved in spastic cerebral palsy pathogenesis. Biochem. Biophys. Res. Commun. 496, 133–139. doi: 10.1016/j.bbrc.2018.01.011
Lee, G., and Hall, J. C. (2001). Abnormalities of male-specific FRU protein and serotonin expression in the CNS of fruitless mutants in Drosophila. J. Neurosci. 21, 513–526. doi: 10.1523/JNEUROSCI.21-02-00513.2001
Lee, G., Hall, J. C., and Park, J. A. E. H. (2002). Doublesex gene expression in the central nervous system of Drosophila melanogaster. J. Neurogenet. 16, 229–248. doi: 10.1080/01677060216292
Lee, G., Villella, A., Taylor, B. J., and Hall, J. C. (2001). New reproductive anomalies infruitless-mutant Drosophila males: extreme lengthening of mating durations and infertility correlated with defective serotonergic innervation of reproductive organs. J. Neurobiol. 47, 121–149. doi: 10.1002/neu.1021
Lee, K., Daubnerová, I., Isaac, R. E., Zhang, C., Choi, S., Chung, J., et al. (2015). A neuronal pathway that controls sperm ejection and storage in female Drosophila. Curr. Biol. 25, 790–797. doi: 10.1016/j.cub.2015.01.050
Lei, N., Hornbaker, K. I., Rice, D. A., Karpova, T., Agbor, V. A., and Heckert, L. L. (2007). Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol. Reprod. 77, 466–475. doi: 10.1095/biolreprod.106.058784
Lerer, I., Sagi, M., Meiner, V., Cohen, T., Zlotogora, J., and Abeliovich, D. (2005). Deletion of the ANKRD15 gene at 9p24. 3 causes parent-of-origin-dependent inheritance of familial cerebral palsy. Hum Mol Genet 14, 3911–3920. doi: 10.1093/hmg/ddi415
Li, H., Zhu, Q., Chen, R., Liu, M., and Xu, D. (2021). Identification and characterization of dimorphic expression of sex-related genes in rock bream, a fish with multiple sex chromosomes. Front. Genet. 12, 1–11. doi: 10.3389/fgene.2021.791179
Li, Q., Zhou, X., Guo, Y., Shang, X., Chen, H., Lu, H., et al. (2008). Nuclear localization, DNA binding and restricted expression in neural and germ cells of zebrafish Dmrt3. Biol Cell 100, 453–463. doi: 10.1042/BC20070114
Li, S., Li, F., Yu, K., and Xiang, J. (2018). Identification and characterization of a doublesex gene which regulates the expression of insulin-like androgenic gland hormone in Fenneropenaeus chinensis. Gene 649, 1–7. doi: 10.1016/j.gene.2018.01.043
Lindeman, R. E., Murphy, M. W., Agrimson, K. S., Gewiss, R. L., Bardwell, V. J., Gearhart, M. D., et al. (2021). The conserved sex regulator DMRT1 recruits SOX9 in sexual cell fate reprogramming. Nucleic Acids Res. 49, 6144–6164. doi: 10.1093/nar/gkab448
Lints, R., and Emmons, S. W. (2002). Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 16, 2390–2402. doi: 10.1101/gad.1012602
Liu, K. S., and Sternberg, P. W. (1995). Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron 14, 79–89. doi: 10.1016/0896-6273(95)90242-2
Matson, C. K., Murphy, M. W., Griswold, M. D., Yoshida, S., Bardwell, V. J., and Zarkower, D. (2010). The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev. Cell 19, 612–624. doi: 10.1016/j.devcel.2010.09.010
Matson, C. K., Murphy, M. W., Sarver, A. L., Griswold, M. D., Bardwell, V. J., and Zarkower, D. (2011). DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104. doi: 10.1038/nature10239
Matson, C. K., and Zarkower, D. (2012). Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163–174. doi: 10.1038/nrg3161
Matsushita, Y., Oshima, Y., and Nakamura, M. (2007). Expression of DMRT genes in the gonads of Rana rugosa during sex determination. Zoolog. Sci. 24, 95–99. doi: 10.2108/zsj.24.95
Mawaribuchi, S., Ito, Y., and Ito, M. (2019). Independent evolution for sex determination and differentiation in the DMRT family in animals. Biol. Open 8, 1–8. doi: 10.1242/bio.041962
McCarthy, M. M. (2016). Multifaceted origins of sex differences in the brain. Philos. Trans. R. Soc. B Biol. Sci. 371, 20150106. doi: 10.1098/rstb.2015.0106
McCarthy, M. M. (2020). A new view of sexual differentiation of mammalian brain. J. Comp. Physiol. A 206, 369–378. doi: 10.1007/s00359-019-01376-8
McCarthy, M. M., and Arnold, A. P. (2011). Reframing sexual differentiation of the brain. Nat. Neurosci. 14, 677–683. doi: 10.1038/nn.2834
McCarthy, M. M., Nugent, B. M., and Lenz, K. M. (2017). Neuroimmunology and neuroepigenetics in the establishment of sex differences in the brain. Nat. Rev. Neurosci. 18, 471–484. doi: 10.1038/nrn.2017.61
McRobert, S. P., and Tompkins, L. (1985). The effect of transformer, doublesex and intersex mutations on the sexual behavior of Drosophila melanogaster. Genetics 111, 89–96. doi: 10.1093/genetics/111.1.89
Mellert, D. J., Knapp, J.-M., Manoli, D. S., Meissner, G. W., and Baker, B. S. (2010). Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development 137, 323–332. doi: 10.1242/dev.045047
Muralidharan, B., Keruzore, M., Pradhan, S. J., Roy, B., Shetty, A. S., Kinare, V., et al. (2017). Dmrt5, a novel neurogenic factor, reciprocally regulates Lhx2 to control the neuron-glia cell fate switch in the developing hippocampus. J. Neurosci. 37, 11245–11254. doi: 10.1523/JNEUROSCI.1535-17.2017
Murphy, M. W., Lee, J. K., Rojo, S., Gearhart, M. D., Kurahashi, K., Banerjee, S., et al. (2015). An ancient protein-DNA interaction underlying metazoan sex determination. Nat. Struct. Mol. Biol. 22, 442–451. doi: 10.1038/nsmb.3032
Murphy, M. W., Zarkower, D., and Bardwell, V. J. (2007). Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol. Biol. 8, 58. doi: 10.1186/1471-2199-8-58
Nakata, M., Kikuchi, Y., Iwami, M., Takayanagi-Kiya, S., and Kiya, T. (2021). Identification and characterization of sexually dimorphic neurons that express the sex-determining gene doublesex in the brain of silkmoth Bombyx mori. Insect Biochem. Mol. Biol. 129, 103518. doi: 10.1016/j.ibmb.2021.103518
Neville, M. C., Nojima, T., Ashley, E., Parker, D. J., Walker, J., Southall, T., et al. (2014). Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr. Biol. 24, 229–241. doi: 10.1016/j.cub.2013.11.035
Ng, C. S., and Kopp, A. (2008). Sex combs are important for male mating success in Drosophila melanogaster. Behav. Genet. 38, 195–201. doi: 10.1007/s10519-008-9190-7
Nojima, T., Rings, A., Allen, A. M., Billeter, J., Neville, M. C., Goodwin, S. F., et al. (2021). A sex-specific switch between visual and olfactory inputs underlies adaptive sex differences in behavior. Curr. Biol. 31, 1175–1191.e6. doi: 10.1016/j.cub.2020.12.047
Oren-Suissa, M., Bayer, E. A., and Hobert, O. (2016). Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 533, 206–211. doi: 10.1038/nature17977
Ottolenghi, C., Fellous, M., Barbieri, M., and McElreavey, K. (2002). Novel paralogy relations among human chromosomes support a link between the phylogeny of doublesex -related genes and the evolution of sex determination. Genomics 79, 333–343. doi: 10.1006/geno.2002.6711
Õunap, K., Uibo, O., Zordania, R., Kiho, L., Ilus, T., Õiglane-Shlik, E., et al. (2004). Three patients with 9p deletions including DMRT1 and DMRT2: A girl with XY complement, bilateral ovotestes, and extreme growth retardation, and two XX females with normal pubertal development. Am. J. Med. Genet. Part A 130A, 415–423. doi: 10.1002/ajmg.a.30269
Palavicino-Maggio, C. B., Chan, Y., McKellar, C., and Kravitz, E. A. (2019). A small number of cholinergic neurons mediate hyperaggression in female Drosophila. Proc. Natl. Acad. Sci. U. S. A. 116, 17029–17038. doi: 10.1073/pnas.1907042116
Pan, Y., and Baker, B. S. (2014). Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell 156, 236–248. doi: 10.1016/j.cell.2013.11.041
Pan, Y., Robinett, C. C., and Baker, B. S. (2011). Turning males on: activation of male courtship behavior in Drosophila melanogaster. PLoS ONE 6, e21144. doi: 10.1371/journal.pone.0021144
Panara, V., Budd, G. E., and Janssen, R. (2019). Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front. Zool. 16, 23. doi: 10.1186/s12983-019-0322-0
Parlier, D., Moers, V., Van Campenhout, C., Preillon, J., Leclère, L., Saulnier, A., et al. (2013). The Xenopus doublesex-related gene Dmrt5 is required for olfactory placode neurogenesis. Dev. Biol. 373, 39–52. doi: 10.1016/j.ydbio.2012.10.003
Pavlou, H. J., Lin, A. C., Neville, M. C., Nojima, T., Diao, F., Chen, B. E., et al. (2016). Neural circuitry coordinating male copulation. Elife 5, 1–26. doi: 10.7554/eLife.20713
Peng, Q., Chen, J., and Pan, Y. (2021). From fruitless to sex : On the generation and diversification of an innate behavior. Genes Brain Behav. 20, e12772. doi: 10.1111/gbb.12772
Pereira, L., Aeschimann, F., Wang, C., Lawson, H., Serrano-Saiz, E., Portman, D. S., et al. (2019). Timing mechanism of sexually dimorphic nervous system differentiation. Elife 8, 1–31. doi: 10.7554/eLife.42078
Phoenix, C. H., Goy, R. W., Gerall, A. A., and Young, W. C. (1959). Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology 65, 369–382. doi: 10.1210/endo-65-3-369
Rahmoun, M., Lavery, R., Laurent-Chaballier, S., Bellora, N., Philip, G. K., Rossitto, M., et al. (2017). In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res. 45, 7191–7211. doi: 10.1093/nar/gkx328
Ratié, L., Desmaris, E., García-Moreno, F., Hoerder-Suabedissen, A., Kelman, A., Theil, T., et al. (2020). Loss of Dmrt5 affects the formation of the subplate and early corticogenesis. Cereb. Cortex 30, 3296–3312. doi: 10.1093/cercor/bhz310
Raymond, C. (1999). A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 8, 989–996. doi: 10.1093/hmg/8.6.989
Raymond, C. S., Kettlewell, J. R., Hirsch, B., Bardwell, V. J., and Zarkower, D. (1999). Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 215, 208–220. doi: 10.1006/dbio.1999.9461
Raymond, C. S., Murphy, M. W., O'Sullivan, M. G., Bardwell, V. J., and Zarkower, D. (2000). Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14, 2587–2595. doi: 10.1101/gad.834100
Raymond, C. S., and Zarkower, D. (1998). Evidence for evolutionary conservation of sex-determining genes. Nature 391, 1251–1253. doi: 10.1038/35618
Rezával, C., Nojima, T., Neville, M. C., Lin, A. C., and Goodwin, S. F. (2014). Sexually dimorphic octopaminergic neurons modulate female postmating behaviors in Drosophila. Curr. Biol. 24, 725–730. doi: 10.1016/j.cub.2013.12.051
Rezával, C., Pavlou, H. J., Dornan, A. J., Chan, Y., and Kravitz, E. A. (2012). Neural circuitry underlying Drosophila female postmating behavioral responses. Curr. Biol. 22, 1155–1165. doi: 10.1016/j.cub.2012.04.062
Rideout, E. J., Billeter, J. C., and Goodwin, S. F. (2007). The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr. Biol. 17, 1473–1478. doi: 10.1016/j.cub.2007.07.047
Rideout, E. J., Dornan, A. J., Neville, M. C., Eadie, S., and Goodwin, S. F. (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat. Neurosci. 13, 458–466. doi: 10.1038/nn.2515
Robinett, C. C., Vaughan, A. G., Knapp, J., and Baker, B. S. (2010). Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 8, e1000365. doi: 10.1371/journal.pbio.1000365
Ross, J. M., Kalis, A. K., Murphy, M. W., and Zarkower, D. (2005). The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev. Cell 8, 881–892. doi: 10.1016/j.devcel.2005.03.017
Rybak, F., Aubin, T., Moulin, B., and Jallon, J. (2002). Acoustic communication in Drosophila melanogaster courtship: are pulse- and sine-song frequencies important for courtship success? Can. J. Zool. 80, 987–996. doi: 10.1139/z02-082
Ryner, L. C., and Baker, B. S. (1991). Regulation of doublesex pre-mRNA processing occurs by 3'-splice site activation. Genes Dev. 5, 2071–2085. doi: 10.1101/gad.5.11.2071
Ryner, L. C., Goodwin, S. F., Castrillon, D. H., Anand, A., Villella, A., Baker, B. S., et al. (1996). Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 87, 1079–1089. doi: 10.1016/S0092-8674(00)81802-4
Sammut, M., Cook, S. J., Nguyen, K. C. Q., Felton, T., Hall, D. H., Emmons, S. W., et al. (2015). Glia-derived neurons are required for sex-specific learning in C. elegans. Nature 526, 385–390. doi: 10.1038/nature15700
Sanders, L. E., and Arbeitman, M. N. (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev. Biol. 320, 378–390. doi: 10.1016/j.ydbio.2008.05.543
Satou, C., Kimura, Y., Hirata, H., Suster, M. L., and Kawakami, K. (2013). Transgenic tools to characterize neuronal properties of discrete populations of zebrafish neurons. Development 140, 3927–3931. doi: 10.1242/dev.099531
Satou, C., Sugioka, T., Uemura, Y., Shimazaki, T., Zmarz, P., and Kimura, Y. (2020). Functional diversity of glycinergic commissural inhibitory neurons in larval zebrafish. Cell Rep. 30, 3036-3050.e4. doi: 10.1016/j.celrep.2020.02.015
Saulnier, A., Keruzore, M., De Clercq, S., Bar, I., Moers, V., Magnani, D., et al. (2013). The doublesex homolog Dmrt5 is required for the development of the caudomedial cerebral cortex in mammals. Cereb. Cortex. 23, 2552–2567.
Scheffer, L. K., Xu, C. S., Januszewski, M., Lu, Z., Takemura, S., Hayworth, K. J., et al. (2020). A connectome and analysis of the adult Drosophila central brain. Elife 9, 1–74. doi: 10.7554/eLife.57443
Seo, K. W., Wang, Y., Kokubo, H., Kettlewell, J. R., Zarkower, D. A., and Johnson, R. L. (2006). Targeted disruption of the DM domain containing transcription factor Dmrt2 reveals an essential role in somite patterning. Dev. Biol. 290, 200–210. doi: 10.1016/j.ydbio.2005.11.027
Serrano-Saiz, E., and Isogai, Y. (2021). Single-cell molecular and developmental perspectives of sexually dimorphic circuits underlying innate social behaviors. Curr. Opin. Neurobiol. 68, 159–166. doi: 10.1016/j.conb.2021.03.010
Serrano-Saiz, E., Oren-Suissa, M., Bayer, E. A., and Hobert, O. (2017a). Sexually dimorphic differentiation of a C. elegans hub neuron is cell autonomously controlled by a conserved transcription factor. Curr. Biol. 27, 199–209. doi: 10.1016/j.cub.2016.11.045
Serrano-Saiz, E., Pereira, L., Gendrel, M., Aghayeva, U., Battacharya, A., Howell, K., et al. (2017b). A neurotransmitter atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics 206, 1251–1269. doi: 10.1534/genetics.117.202127
Shen, M. M., and Hodgkin, J. (1988). mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell 54, 1019–1031. doi: 10.1016/0092-8674(88)90117-1
Sheng, Y., Chen, B., and Zhang, L. (2014). Identification of Dmrt genes and their up-regulation during gonad transformation in the swamp eel (Monopterus albus). Mol. Biol. Rep. 41, 1237–1245. doi: 10.1007/s11033-013-2968-6
Shima, Y., Miyabayashi, K., Haraguchi, S., Arakawa, T., Otake, H., Baba, T., et al. (2013). Contribution of Leydig and Sertoli cells to testosterone production in mouse fetal testes. Mol. Endocrinol. 27, 63–73. doi: 10.1210/me.2012-1256
Shirangi, T. R., Dufour, H. D., Williams, T. M., and Carroll, S. B. (2009). Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 7, e1000168. doi: 10.1371/journal.pbio.1000168
Shirangi, T. R., Wong, A. M., Truman, J. W., and Stern, D. L. (2016). Doublesex regulates the connectivity of a neural circuit controlling Drosophila male courtship song. Dev. Cell 37, 533–544. doi: 10.1016/j.devcel.2016.05.012
Siehr, M. S., Koo, P. K., Sherlekar, A. L., Bian, X., Bunkers, M. R., Miller, R. M., et al. (2011). Multiple doublesex-related genes specify critical cell fates in a C. elegans male neural circuit. PLoS ONE 6, e26811. doi: 10.1371/journal.pone.0026811
Smith, C. A., Hurley, T. M., McClive, P. J., and Sinclair, A. H. (2002). Restricted expression of DMRT3 in chicken and mouse embryos. Mech. Dev. 119, S73–S76. doi: 10.1016/S0925-4773(03)00094-7
Spieth, H. T., The, O. F., Museum, A., and Natural, O. F. (1952). Mating behavior within the genus Drosophila (Diptera). Bull. Am. Mus. Nat. Hist. 99, 395–474.
Su, L., Zhou, F., Ding, Z., Gao, Z., Wen, J., Wei, W., et al. (2015). Transcriptional variants of Dmrt1 and expression of four Dmrt genes in the blunt snout bream, Megalobrama amblycephala. Gene 573, 205–215. doi: 10.1016/j.gene.2015.07.044
Sulston, J. E. (1983). Neuronal cell lineages in the nematode Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 48, 443–452. doi: 10.1101/SQB.1983.048.01.049
Sulston, J. E., Albertson, D. G., and Thomson, J. N. (1980). The Caenorhabditis elegans male: postembryonic development of nongonadal structures. Dev. Biol. 78, 542–576. doi: 10.1016/0012-1606(80)90352-8
Tanaka, K., Barmina, O., Sanders, L. E., Arbeitman, M. N., and Kopp, A. (2011). Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 9, e1001131. doi: 10.1371/journal.pbio.1001131
Taylor, B. J., and Truman, J. W. (1992). Commitment of abdominal neuroblasts in Drosophila to a male or female fate is dependent on genes of the sex-determining hierarchy. Development 114, 625–642. doi: 10.1242/dev.114.3.625
Taylor, B. J., Villella, A., Ryner, L. C., Baker, B. S., and Hall, J. C. (1994). Behavioral and neurobiological implications of sex-determining factors in Drosophila. Dev. Genet. 296, 275–296. doi: 10.1002/dvg.1020150309
Tewes, A.-C., Ledig, S., Tüttelmann, F., Kliesch, S., and Wieacker, P. (2014). DMRT1 mutations are rarely associated with male infertility. Fertil. Steril. 102, 816–820.e3. doi: 10.1016/j.fertnstert.2014.05.022
Tomchaney, M., Mysore, K., Sun, L., Li, P., Emrich, S. J., and Severson, D. W. (2014). Examination of the genetic basis for sexual dimorphism in the Aedes aegypti (dengue vector mosquito) pupal brain. Biol. Sex Differ. 5, 1–22. doi: 10.1186/s13293-014-0010-x
Trabzuni, D., Ramasamy, A., Imran, S., Walker, R., Smith, C., Weale, M. E., et al. (2013). Widespread sex differences in gene expression and splicing in the adult human brain. Nat. Commun. 4, 2771. doi: 10.1038/ncomms3771
Tresser, J., Chiba, S., Veeman, M., El-nachef, D., Newman-smith, E., Horie, T., et al. (2010). Doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development 137, 2197–2203. doi: 10.1242/dev.045302
Ugajin, A., Matsuo, K., Kubo, R., Sasaki, T., and Ono, M. (2016). Expression profile of the sex determination gene doublesex in a gynandromorph of bumblebee, Bombus ignitus. Sci. Nat. 103, 17. doi: 10.1007/s00114-016-1342-7
Urquhart, J. E., Beaman, G., Byers, H., Roberts, N. A., Chervinsky, E., O'Sullivan, J., et al. (2016). DMRTA2 (DMRT5) is mutated in a novel cortical brain malformation. Clin. Genet. 89, 724–727. doi: 10.1111/cge.12734
Usui-Aoki, K., Ito, H., Ui-Tei, K., Takahashi, K., Lukacsovich, T., Awano, W., et al. (2000). Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2, 500–506. doi: 10.1038/35019537
Veith, A., Schäfer, M., Klüver, N., Schmidt, C., Schultheis, C., Schartl, M., et al. (2006a). Tissue-specific expression of dmrt genes in embryos and adults of the platyfish Xiphophorus maculatus. Zebrafish 3, 325–337. doi: 10.1089/zeb.2006.3.325
Veith, A.-M., Klattig, J., Dettai, A., Schmidt, C., Englert, C., and Volff, J.-N. (2006b). Male-biased expression of X-chromosomal DM domain-less Dmrt8 genes in the mouse. Genomics 88, 185–195. doi: 10.1016/j.ygeno.2006.01.003
Villella, A., and Hall, J. C. (1996). Courtship anomalies caused by doublesex mutations in Drosophila melanogaster. Genetics 143, 331–344. doi: 10.1093/genetics/143.1.331
Vinci, G., Chantot-Bastaraud, S., El Houate, B., Lortat-Jacob, S., Brauner, R., and McElreavey, K. (2007). Association of deletion 9p, 46, XY gonadal dysgenesis and autistic spectrum disorder. Mol. Hum. Reprod. 13, 685–689. doi: 10.1093/molehr/gam045
Volff, J.-N., Zarkower, D., Bardwell, V. J., and Schartl, M. (2003). Evolutionary dynamics of the DM domain gene family in metazoans. J. Mol. Evol. 57, S241–S249. doi: 10.1007/s00239-003-0033-0
Wagner, E., and Levine, M. (2012). FGF signaling establishes the anterior border of the Ciona neural tube. Development 139, 2351–2359. doi: 10.1242/dev.078485
Wan, H., Zhong, J., Zhang, Z., Zou, P., Zeng, X., and Wang, Y. (2021). Discovery of the Dmrt gene family members based on transcriptome analysis in mud crab Scylla paramamosain. Gene 784, 145576. doi: 10.1016/j.gene.2021.145576
Wang, J., Miao, L., Li, M., Guo, X., Pan, N., and Chen, Y. (2014). Cloning the Dmrt1 and DmrtA2 genes of ayu (Plecoglossus altivelis) and mapping their expression in adult, larval, and embryonic stages. Zool. Res. 35, 99–107. doi: 10.11813/j.issn.0254-5853.2014.2.099
Wang, Y., Jin, S., Fu, H., Qiao, H., Sun, S., Zhang, W., et al. (2019). Identification and characterization of the DMRT11E gene in the oriental river prawn Macrobrachium nipponense. Int. J. Mol. Sci. 20, 1734. doi: 10.3390/ijms20071734
Waterbury, J. A., Jackson, L. L., and Schedl, P. (1999). Analysis of the doublesex female protein in Drosophila melanogaster: role in sexual differentiation and behavior and dependence on intersex. Genetics 152, 1653–1667. doi: 10.1093/genetics/152.4.1653
Weinberg, P., Berkseth, M., Zarkower, D., and Hobert, O. (2018). Sexually dimorphic unc-6/netrin expression controls sex-specific maintenance of synaptic connectivity. Curr. Biol. 28, 623–629.e3. doi: 10.1016/j.cub.2018.01.002
Wen, A., You, Æ. F., and Tan, Æ. X. (2009). Expression pattern of dmrt4 from olive flounder (Paralichthys olivaceus) in adult gonads and during embryogenesis. Fish Physiol. Biochem. 35, 421–433. doi: 10.1007/s10695-008-9267-5
Wexler, J. R., Plachetzki, D. C., and Kopp, A. (2014). Pan-metazoan phylogeny of the DMRT gene family: a framework for functional studies. Dev. Genes Evol. 224, 175–181. doi: 10.1007/s00427-014-0473-0
White, J. G., Southgate, E., Thomson, J. N., and Brenner, S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. London. B, Biol. Sci. 314, 1–340. doi: 10.1098/rstb.1986.0056
Williams, T. M., Selegue, J. E., Werner, T., Gompel, N., Kopp, A., and Carroll, S. B. (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. doi: 10.1016/j.cell.2008.06.052
Winkler, C., Hornung, U., Kondo, M., Neuner, C., Duschl, J., Shima, A., et al. (2004). Developmentally regulated and non-sex-specific expression of autosomal dmrt genes in embryos of the Medaka fish (Oryzias latipes). Mech. Dev. 121, 997–1005. doi: 10.1016/j.mod.2004.03.018
Yamaguchi, A., Hoon, K., Fujimoto, H., and Kadomura, K. (2006). Expression of the DMRT gene and its roles in early gonadal development of the Japanese pufferfish Takifugu rubripes B. Comp. Biochem. Physiol. Part D Genomics Proteomics 1, 59–68. doi: 10.1016/j.cbd.2005.08.003
Yamamoto, D., and Koganezawa, M. (2013). Genes and circuits of courtship behaviour in Drosophila males. Nat. Rev. Neurosci. 14, 681–692. doi: 10.1038/nrn3567
Yan, N., Hu, J., Li, J., Dong, J., Sun, C., and Ye, X. (2019). Genomic organization and sexually dimorphic expression of the Dmrt1 gene in largemouth bass (Micropterus salmoides). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 234, 68–77. doi: 10.1016/j.cbpb.2019.05.005
Yang, T., and Shah, N. M. (2016). Molecular and neural control of sexually dimorphic social behaviors. Curr. Opin. Neurobiol. 38, 89–95. doi: 10.1016/j.conb.2016.04.015
Yang, Y., Wang, C., Wang, F., Zhu, L., Liu, H., and He, X. (2012). Novel chromosomal translocation t(11;9)(p15;p23) involving deletion and duplication of 9p in a girl associated with autism and mental retardation. Gene 502, 154–158. doi: 10.1016/j.gene.2012.04.036
Yaron, Z., and Levavi-Sivan, B. (2011). “Hormonal control of reproduction and growth: Endocrine regulation of fish reproduction,” in Encyclopedia of Fish Physiology, ed A. P. Farrell (San Diego, CA: Academic Press), 1500–8. doi: 10.1016/B978-0-12-374553-8.00058-7
Yi, W., Ross, J. M., and Zarkower, D. (2000). Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development 127, 4469–4480. doi: 10.1242/dev.127.20.4469
Yoshizawa, A., Nakahara, Y., Izawa, T., Ishitani, T., Tsutsumi, M., Kuroiwa, A., et al. (2011). Zebrafish Dmrta2 regulates neurogenesis in the telencephalon. Genes Cells 16, 1097–1109. doi: 10.1111/j.1365-2443.2011.01555.x
Zagni, E., Simoni, L., and Colombo, D. (2016). Sex and gender differences in central nervous system-related disorders. Neurosci. J. 2016, 1–13. doi: 10.1155/2016/2827090
Zarkower, D., and Murphy, M. W. (2021). DMRT1: An ancient sexual regulator required for human gonadogenesis. Sex Dev. 1, 1–14. doi: 10.1159/000518272
Zhang, T., Murphy, M. W., Gearhart, M. D., Bardwell, V. J., and Zarkower, D. (2014). The mammalian Doublesex homolog DMRT6 coordinates the transition between mitotic and meiotic developmental programs during spermatogenesis. Development 141, 3662–3671. doi: 10.1242/dev.113936
Zhang, Z., Zhu, B., and Ge, W. (2015). Genetic analysis of zebrafish gonadotropin (FSH and LH) functions by TALEN-mediated gene disruption. Mol. Endocrinol. 29, 76–98. doi: 10.1210/me.2014-1256
Zhao, C., and Emmons, S. W. (1995). A transcription factor controlling development of peripheral sense organs in C. elegans. Nature 373, 74–78. doi: 10.1038/373074a0
Zheng, Z., Lauritzen, J. S., Perlman, E., Robinson, C. G., Nichols, M., Milkie, D., et al. (2018). A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell 174, 730–743.e22. doi: 10.1016/j.cell.2018.06.019
Zhou, C., Pan, Y., Robinett, C. C., Meissner, G. W., and Baker, B. S. (2014). Central brain neurons expressing doublesex regulate female receptivity in Drosophila. Neuron 83, 149–163. doi: 10.1016/j.neuron.2014.05.038
Keywords: Dmrt, sexual differentiation, nervous system, doublesex, dmd, conservation
Citation: Casado-Navarro R and Serrano-Saiz E (2022) DMRT Transcription Factors in the Control of Nervous System Sexual Differentiation. Front. Neuroanat. 16:937596. doi: 10.3389/fnana.2022.937596
Received: 06 May 2022; Accepted: 15 June 2022;
Published: 26 July 2022.
Edited by:
Luis Miguel Garcia-Segura, Spanish National Research Council (CSIC), SpainReviewed by:
Byunghyuk Kim, Dongguk University Seoul, South KoreaCopyright © 2022 Casado-Navarro and Serrano-Saiz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esther Serrano-Saiz, ZXN0aGVyLnNlcnJhbm8uc2FpekBnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.