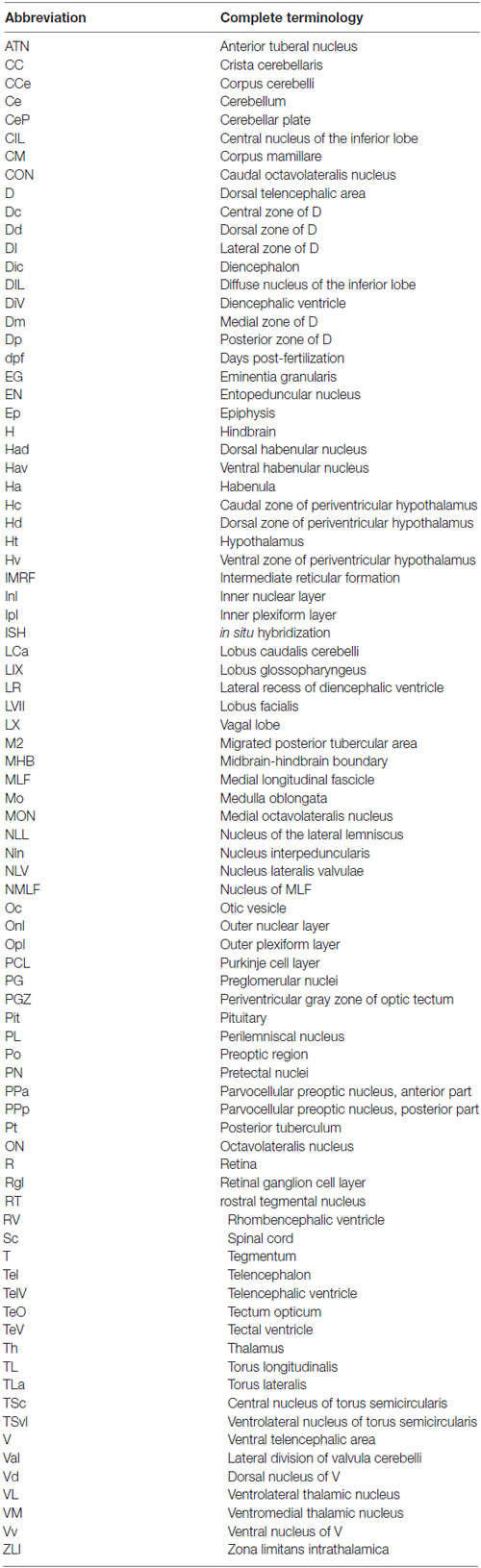
- 1Biocenter, Division of Electron Microscopy, University of Würzburg, Würzburg, Germany
- 2Biocenter, Department of Physiological Chemistry, University of Würzburg, Würzburg, Germany
Septins are a highly conserved family of small GTPases that form cytoskeletal filaments. Their cellular functions, especially in the nervous system, still remain largely enigmatic, but there are accumulating lines of evidence that septins play important roles in neuronal physiology and pathology. In order to further dissect septin function in the nervous system a detailed temporal resolved analysis in the genetically well tractable model vertebrate zebrafish (Danio rerio) is crucially necessary. To close this knowledge gap we here provide a reference dataset describing the expression of selected septins (sept3, sept5a and sept5b) in the zebrafish central nervous system. Strikingly, proliferation zones are devoid of expression of all three septins investigated, suggesting that they have a role in post-mitotic neural cells. Our finding that three septins are mainly expressed in non-proliferative regions was further confirmed by double-stainings with a proliferative marker. Our RNA in situ hybridization (ISH) study, detecting sept3, sept5a and sept5b mRNAs, shows that all three septins are expressed in largely overlapping regions of the developing brain. However, the expression of sept5a is much more confined compared to sept3 and sept5b. In contrast, the expression of all the three analyzed septins is largely similar in the adult brain.
Introduction
Septins, a highly conserved family of small GTPases that form filaments, have various functions in normal physiology as well as pathology (Mostowy and Cossart, 2012; Dolat et al., 2014). For instance, septins are reported to play a role in cytokinesis, axon growth and exocytosis. Furthermore, they are involved in developmental processes, such as left-right-asymmetry regulation, as well as several pathologies, such as cancer, neurodegenerative diseases and psychiatric disorders (Dash et al., 2014; Dolat et al., 2014; Zhai et al., 2014; Marttinen et al., 2015). However, the cellular roles and functions of septins, in particular how they are involved in the physiology of the nervous system, still need to be fully elucidated.
Septins were first discovered in cell division screens in budding yeast (Hartwell et al., 1970) and named after their localization at the septum (Byers and Goetsch, 1976; Sanders and Field, 1994). Interestingly, septin homologs were identified later on in other eukaryotes like fungi, worms, flies, zebrafish and mammals (Cao et al., 2007; Pan et al., 2007; Nishihama et al., 2011). In humans, there are 13 septin genes, which can be divided according to sequence similarity into four homology groups: SEPT2 (SEPT1, SEPT2, SEPT4 and SEPT5), SEPT3/9 (SEPT3, SEPT9 and SEPT12), SEPT6 (SEPT6, SEPT8, SEPT10, SEPT11 and SEPT14), and SEPT7 (SEPT7; Kinoshita, 2003; Dolat et al., 2014). In the zebrafish (Danio rerio) genome, there are currently 17 septin genes annotated (Willis et al., 2016). The increased diversity of septins is likely due to an teleost specific genome duplication event (Postlethwait et al., 2004), so that there are for instance two homologs of SEPT5 (sept5a and sept5b) present in the zebrafish genome (Willis et al., 2016). In addition, genes like zebrafish sept9a and sept9b also have additionally multiple isoforms due to alternative splicing (Landsverk et al., 2010). The overall structure of septins is highly conserved. They share a phosphoinositide-binding polybasic domain, followed by the GTP-binding sites and the Septin Unique Element (SUE). A coiled-coil domain is found in septins belonging to the SEPT2, SEPT6 and SEPT7 groups. Furthermore, septins form heterofilaments consisting of two or more types of septins (Kinoshita, 2003; Mostowy and Cossart, 2012; Dolat et al., 2014).
In the mammalian central nervous system Septin3 and Septin5 have been attributed specific roles. Septin3 (Xue et al., 2004) and Septin5 (Kinoshita et al., 2000) are enriched at presynaptic terminals. Septin5 can inhibit exocytosis and interact with presynaptic proteins (Beites et al., 1999, 2005). Furthermore, Septin5 function has an effect on social behavior of mice (Suzuki et al., 2009; Harper et al., 2012). Septin5 is located in the human genome at position 22q11.2, a region that is the site of the most common micro-deletion syndrome in humans. This deletion is associated with multiple phenotypes including cardiac and palatal anomalies, intellectual disabilities and other disorders affecting the nervous system (Guna et al., 2015). Moreover, for Septin5 a fine-regulated expression at later developmental stages has been reported (Maldonado-Saldivia et al., 2000).
Despite these indications of the important roles of Septin3 and Septin5 in brain function and development, comparably little is known about the temporal expression of these genes during early vertebrate brain development. Previously, it was shown that zebrafish Septin5b is expressed in 1 day post-fertilization (dpf) embryos (Gomez et al., 2012) and Septin3 is generally expressed in neural tissues (Thisse and Thisse, 2004). However, a precise temporal description of septin expression in early vertebrate brain development is currently missing. Such information is necessary and opens up a high resolution view into the cellular role of septins in neurons.
The zebrafish is an established model organism well suited for studies of early vertebrate brain development (Wullimann et al., 1996; Mueller and Wullimann, 2015) and activity (Akerboom et al., 2012). Due to the accessible development and the large number of transparent offspring, zebrafish is a very fitting model for the systematic investigation of gene expression during development using RNA in situ hybridization (ISH; Thisse and Thisse, 2008). In addition to that, zebrafish is very suited for systematic high resolution and functional analysis to shed light on the cellular roles of septins in neurons. The zebrafish is a well-established neurogenetic model with a rich genetic toolbox (Wullimann et al., 1996; Lillesaar, 2011; Mueller and Wullimann, 2015; Willis et al., 2016). These are key advantages of this model, as septins seem to play important roles in synaptic development (Yang et al., 2010).
Here we present a precise description of the expression patterns of septin3, septin5a and septin5b at different developmental stages of zebrafish larvae as well as in adult brains. This study builds the basis for a thorough analysis of the role of these septins in the nervous system using the vertebrate model organism zebrafish (Danio rerio).
Materials and Methods
Zebrafish Culture and Nomenclature
Zebrafish (Danio rerio) larvae and adults (4–6 months) of the wild-type strain (AB) were kept on a day/night cycle of 14 h light and 10 h darkness. Larvae were kept at 28.5°C and staged as reported previously (Kimmel et al., 1995) and were raised in 30% Danieau’s solution (Westerfield, 2000). The pigmentation of zebrafish larvae was inhibited by 0.2 mM 1-phenyl-2-thiourea treatment (Karlsson et al., 2001). The brain structure nomenclature is conforming to reference atlases (Wullimann et al., 1996; Mueller and Wullimann, 2015). All experiments were performed according to the animal welfare regulations of the District Government of Lower Franconia.
Phylogenetic Analyses
The human homologs of zebrafish Sept3 (NP_001019589.1), Sept5a (NP_956282.1), and Sept5b (NP_001003782.1) sequences (Willis et al., 2016) were aligned to Septin3 and Septin5 protein sequences from mouse (Sept3: NP_036019.2; Sept5: NP_998779.2) and human (Sept3 isoform B: NP_061979.3; Sept5 isoform 1: NP_002679.2), as well as to the two known septins from C. elegans (UNC-59: NP_493388.1; UNC-61 isoform a: NP_872156.2). Alignment was performed by the “One Click” Method using the web tool with GBlocks1 (RRID:SCR_010266, Dereeper et al., 2008) combining several algorithms (Castresana, 2000; Guindon and Gascuel, 2003; Edgar, 2004; Anisimova and Gascuel, 2006; Chevenet et al., 2006; Dereeper et al., 2010).
RNA Extraction and Reverse Transcription
Zebrafish RNA was isolated from 1 to 4 dpf zebrafish larvae using the RNeasy Mini Kit (Qiagen Gmbh, Düsseldorf, Germany). The RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Braunschweig, Germany) was used to generate cDNA.
Molecular Cloning
Specific PCR primers for sept3 (RefSeq: NM_001024418), sept5a (RefSeq: NM_199988), and the forward primer (fwd) for sept5b (RefSeq: NM_001003782) were generated.
The sept5b reverse primer (rev, CGGTCCTGCTGCTTCGGCTC) was designed according to previously published work (Gomez et al., 2012).
Primers were ordered from Sigma-Aldrich (Munich, Germany):
sept3_fwd (ATGTCAGAAATTGTGCCCCCTGAAGTGA),
sept3_rev (TCACAGGTTGCTTTCTTGTGTATC),
sept5a_fwd (ATGACGACCAACATCCGATACAAGAGCA),
sept5a_rev (TCACTGGTCTTTCTCGTGCATCTG), and
sept5b_fwd (ATGACGAGCAGCGCCAGGTACAAGAGCA).
Septin sequences were amplified from a cDNA mix of 1–4 dpf zebrafish larvae and cloned into the pJET-plasmid using the CloneJET PCR Cloning Kit (Thermo Scientific, Braunschweig, Germany) and verified by sequencing by GATC Biotech AG (Konstanz, Germany). To generate plasmids for RNA ISH probe generation, the septin sequences were cut out of the pJET-plasmids using the restriction enzymes XbaI and XhoI (Thermo Scientific, Braunschweig, Germany) and cloned into the pSC-A-amp/kan plasmid (Agilent Technologies, Santa Clara, CA, USA).
RNA In Situ Hybridization
Digoxigenin-labeled sense and antisense probes were generated using T3 and T7 RNA Polymerases and the DIG RNA Labeling Kit (Roche, Mannheim, Germany). Whole mount RNA ISH was performed on zebrafish larvae and adult brains fixed with 4% PFA as previously published (Thisse et al., 1993; Thisse and Thisse, 2008). After RNA ISH adult brains were embedded in 3% agarose in PBS after stringency washes and 90 μm sections were cut using a TPI Vibratome 1000 Tissue Sectioning System (Technical Products International, Inc. St. Louis, MO, USA). Digoxigenin-labeled probes were detected with an anti-dig-AP-antibody (final dilution 1:5000, Roche, Mannheim, Germany) and revealed with NBT/BCIP (Roche, Mannheim, Germany) or SIGMAFAST Fast Red TR/Naphthol AS-MX Tablets (Sigma-Aldrich, Munich, Germany). Larvae were embedded in Epon (Westerfield, 2000) after RNA ISH and 8 μm sections were cut on a Leica EM UC7 (Leica Microsystems GmbH, Wetzlar, Germany) with a histology diamond knife (Dianova, GmbH, Hamburg, Germany). The Epon sections were counterstained with Hoechst 33342 (Sigma-Aldrich, Munich, Germany, 5 mg/μl stock solution) diluted 1:250.
Immunocytochemistry and Microscopy
Immunocytochemistry was performed after RNA ISH on larvae or on adult brain sections following a previously described protocol (Yamamoto et al., 2011). Primary monoclonal anti-Tyrosine Hydroxylase (TH, clone LNC1, MERCK Millipore, Darmstadt, Germany) mouse antibody purified from PC12 cells was used to identify catecholaminergic neurons (Yamamoto et al., 2011). To identify proliferating cells a monoclonal anti-proliferating cell nuclear antigen (PCNA, Clone PC10, Dako, Glostrup, Denmark) mouse antibody was used (Mueller and Wullimann, 2015). As secondary antibody the donkey anti-mouse Alexa-488 IgG (H + L) antibody (Jackson ImmunoResearch Labratories, Suffolk, UK) was applied. Primary antibodies were used in a dilution of 1:100, the secondary antibody in a dilution of 1:500. Microscopy images were obtained with a Leitz Aristoplan or a Leica TCS SP confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Results
Zebrafish Septin3 and 5 Domain Structure and Conservation
Zebrafish Sept3, Sept5a and Sept5b protein sequences were aligned and show high conservation with their mouse and human homologs (Figure 1). The conserved GTPase domains are present in the zebrafish sequences, specifically the G1 (GXXXXGKS), G3 (DXXG) and G4 (XKXD) motifs described previously (Kinoshita, 2003). Similarly, the S1 (EXXXXR), S2 (DXR(V/I)HXXX(Y/F)F(I/L)XP), S3 (GXXLXXXD), and S4 (WG) motifs are present (Pan et al., 2007; Figure 1). Additionally, the alignment shows that zebrafish Sept5a and Sept5b contain a structured coiled-coil domain at the C-terminus, whereas Sept3 is missing this structure, as reported previously (Kinoshita, 2003).

Figure 1. Sequence alignment. The sequences from C. elegans (CEL UNC-59 and CEL UNC-61 isoform a), mouse (MMU Septin3 and MMU Septin5), human (HSA Septin3 isoform B and HSA Septin5 isoform 1) and zebrafish (DRE Septin3, DRE Septin5a and DRE Septin5b) were aligned with the online tool (http://www.phylogeny.fr/). Similar residues are colored as the most conserved according to BLOSUM62 (cyan: max 3.0 and gray: low 0.5). The G1, G3 and G4 motifs of the GTPase domain (yellow, Kinoshita, 2003) as well as the conserved S1, S2, S3 and S4 motifs (red, Pan et al., 2007) and the coiled-coil domain of the Septin5 group (blue) are highlighted.
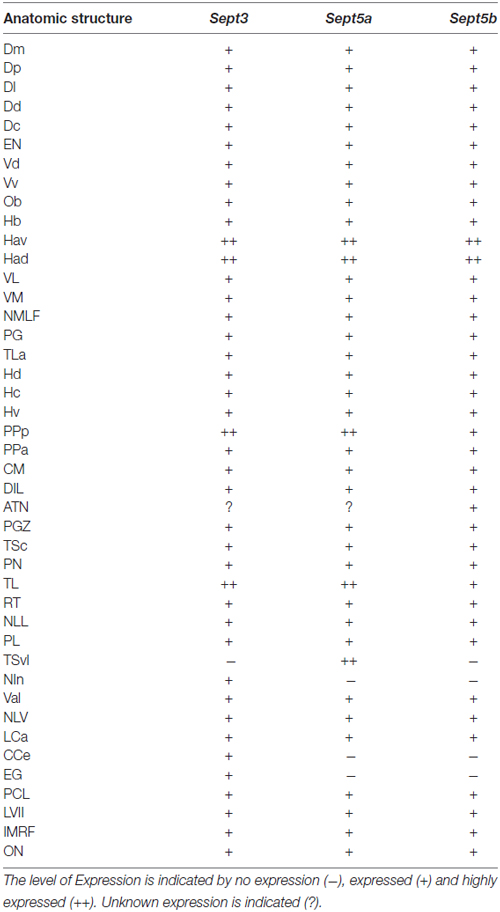
Expression Patterns of sept3, sept5a and sept5b in Zebrafish Larvae during Development
The expression patterns of sept3, sept5a and sept5b, as analyzed by whole mount RNA ISH, in the head region of 1–4 dpf zebrafish larvae are shown in lateral views in Figure 2 and dorsal views in Figure 3. At 1 dpf, sept3 is expressed in the telencephalon (Tel), the diencephalon (Dic), the hindbrain (H), and the spinal cord (Sc; Figures 2A, 3A). Furthermore, sept3 expression is detectable in the habenula (Ha). The tectum opticum (TeO), the mid-hindbrain boundary (MHB), the retina (R) and the periventricular regions are devoid of notable expression. From 2 dpf to 4 dpf, sept3 expression can be found additionally in TeO, cerebellum (Ce), and the retina (Figures 2D,G,J, 3D,G,J). In the ventricular zones (Figures 4, 5) no sept3 expression can be observed during development up to 4 dpf.
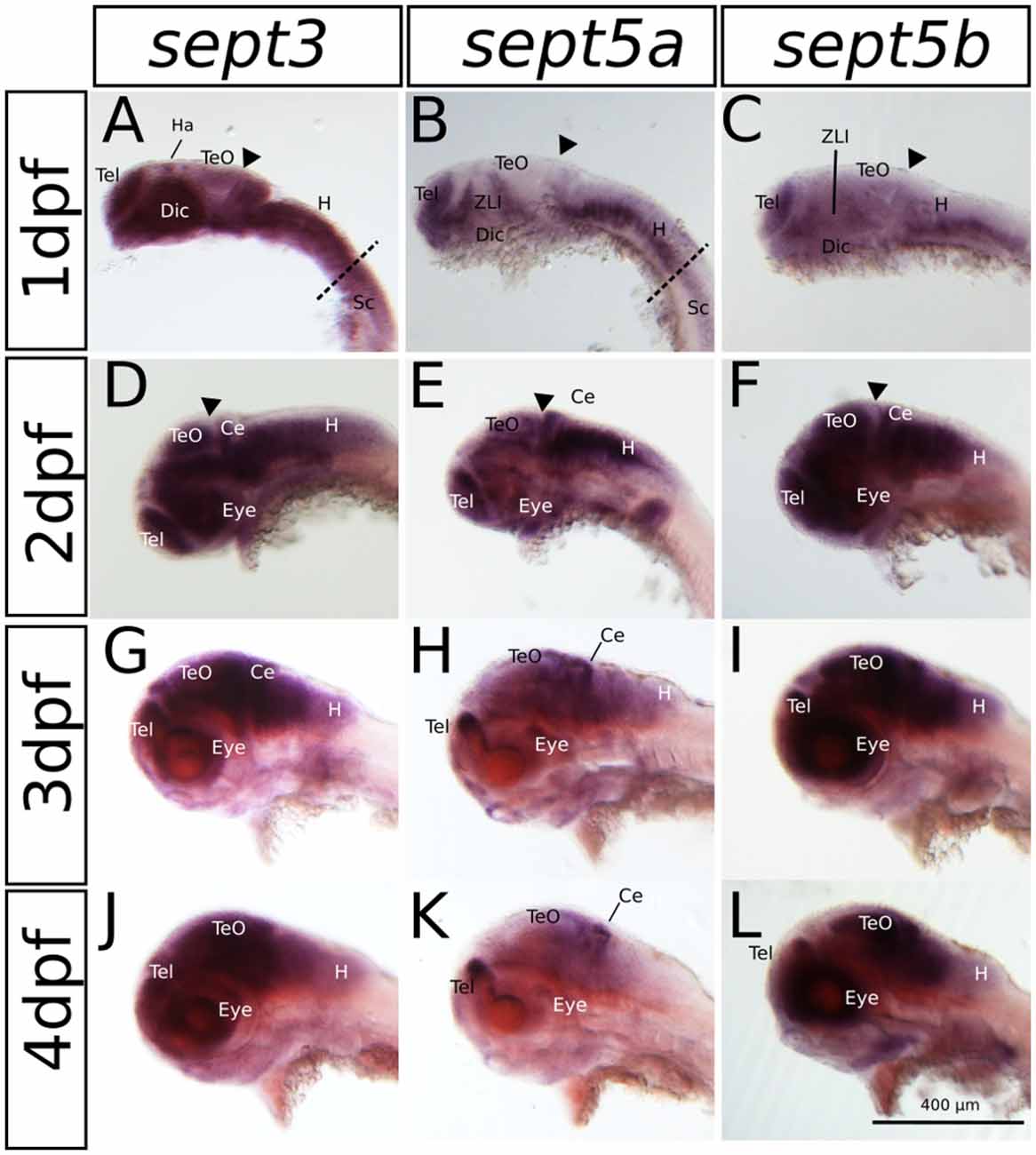
Figure 2. The expression pattern of sept3, sept5a and sept5b in 1–4 dpf zebrafish larvae in a lateral view. Sept3, sept5a and sept5b expression is shown by ISH, the blue color indicates the cells expressing the mRNA of each septin. The expression of sept3 is shown in 1 dpf (A), 2 dpf (D), 3 dpf (G) and 4 dpf (J) zebrafish larvae. Sept5a expression is shown in 1 dpf (B), 2 dpf (E), 3 dpf (H) and 4 dpf (K) larvae. Zebrafish larvae 1–4 dpf stained with the sept5b probe are shown in (C,F,I,L). The telencephalon (Tel), habenula (Ha), diencephalon (Dic), the zona limitans intrathalamica (ZLI), cerebellum (Ce), tectum (TeO), eye, the spinal cord (Sc) as well as the hindbrain (H) are labeled. The border between hindbrain and spinal cord is marked by a dashed line. The midbrain-hindbrain boundary (MHB) is labeled (black arrowhead).
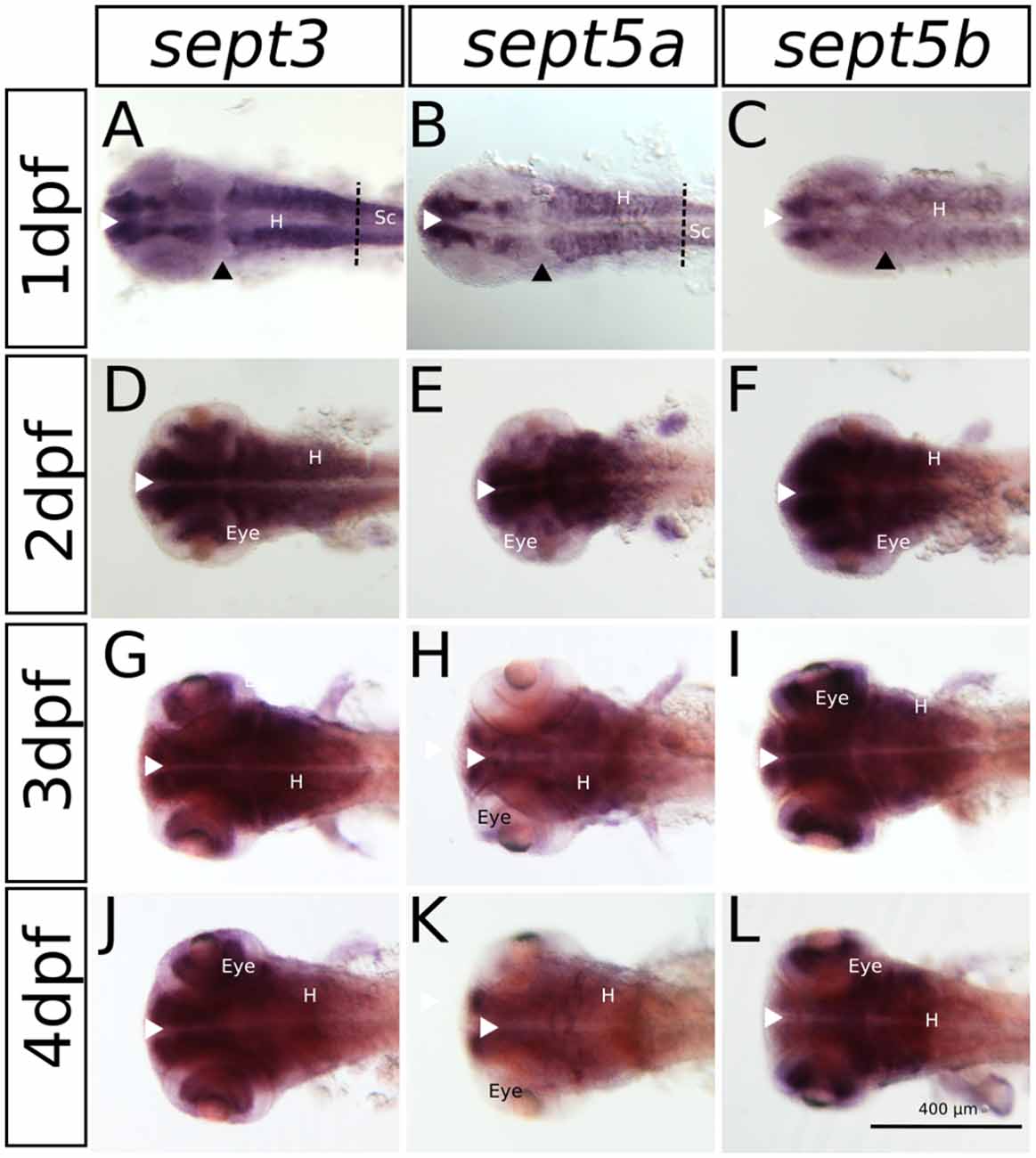
Figure 3. The expression pattern of sept3, sept5a and sept5b in 1–4 dpf zebrafish larvae in a dorsal view. Sept3, sept5a and sept5b expression is shown by ISH, the blue color indicates the cells expressing the mRNA of each septin. The expression of sept3 is shown in 1 dpf (A), 2 dpf (D), 3 dpf (G) and 4 dpf (J) zebrafish larvae. Sept5a expression is shown in 1 dpf (B), 2 dpf (E), 3 dpf (H) and 4 dpf (K) larvae. Zebrafish larvae 1–4 dpf stained with the sept5b probe are shown in (C,F,I,L). The eye, spinal cord (Sc) and hindbrain (H) are marked. The border between hindbrain and spinal cord is marked by a dashed line. Ventricular zones (white arrowhead) and the MHB (black arrowhead) are labeled.
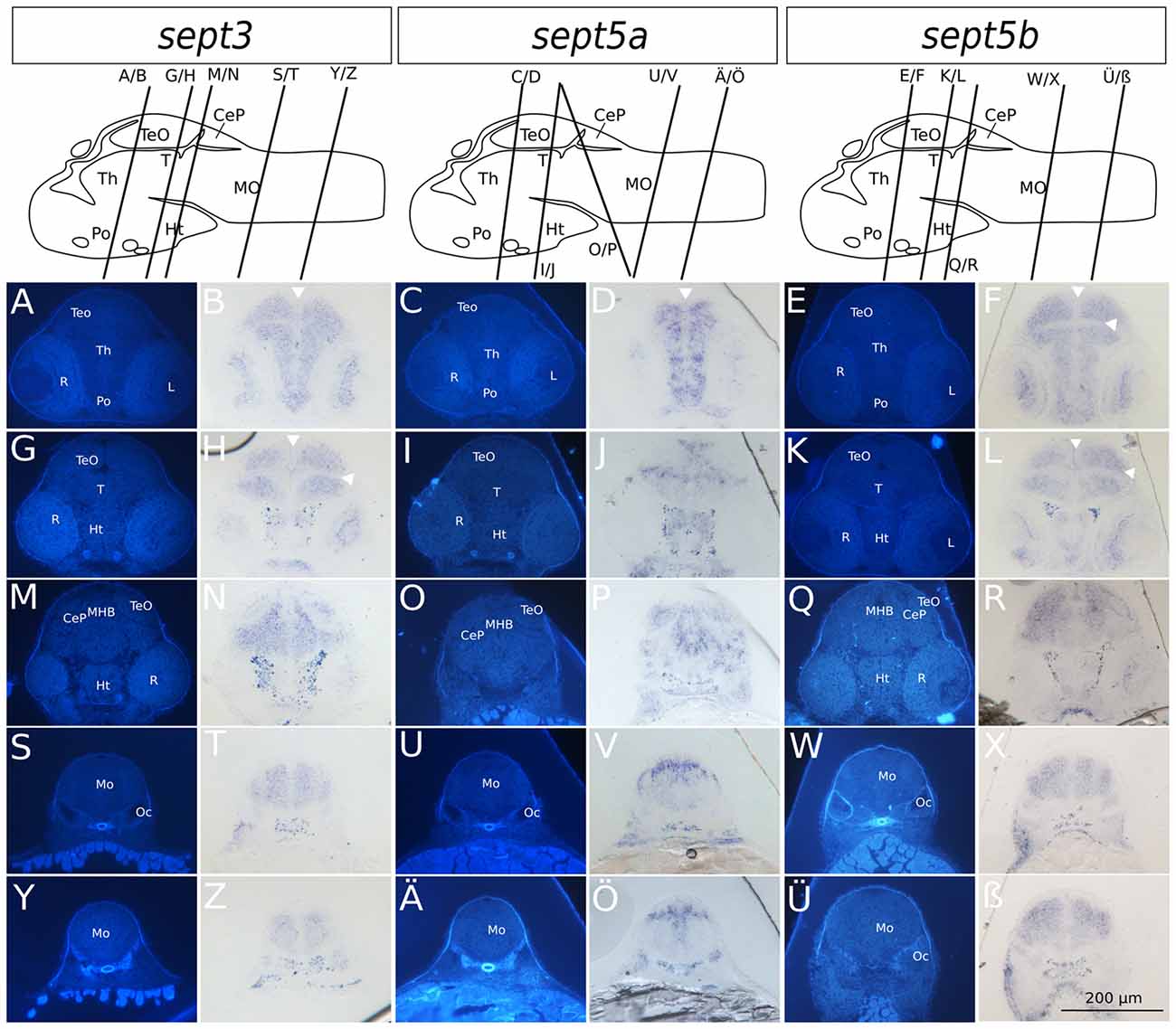
Figure 4. Transverse sections through 2 dpf zebrafish larvae stained with ISH probes against sept3, sept5a and sept5b. 2 dpf zebrafish larvae were stained for sept3 (A,B,G,H,M,N,S,T,Y,Z), sept5a (C,D,I,J,O,P,U,V,Ä,Ö) and sept5b (E,F,K,L,Q,R,W,X,Ü,ß), embedded in epon, and cut in 8 μm thick section. The schemes (modified from Mueller and Wullimann, 2015) are showing the sagittal views of a 2 dpf zebrafish brain with indicated section planes. For better orientation the ISH sections were counterstained with Hoechst (A,G,M,S,Y,C,I,O,U,Ä,E,K,Q,W,Ü). Septin expression is indicated by the blue color (B,H,N,T,Z,D,J,P,V,Ö,F,L,R,X,ß). The retina (R), tectum opticum (TeO), tegmentum (T), preoptic region (Po), thalamus (Th), hypothalamus (Ht), medulla oblongata (Mo), cerebella plate (CeP), MHB and otic capsule (Oc), are marked. The ventricles are indicated with white arrowheads. Schemes for better orientation are modified from Mueller and Wullimann (2015) 2 dpf schemes.
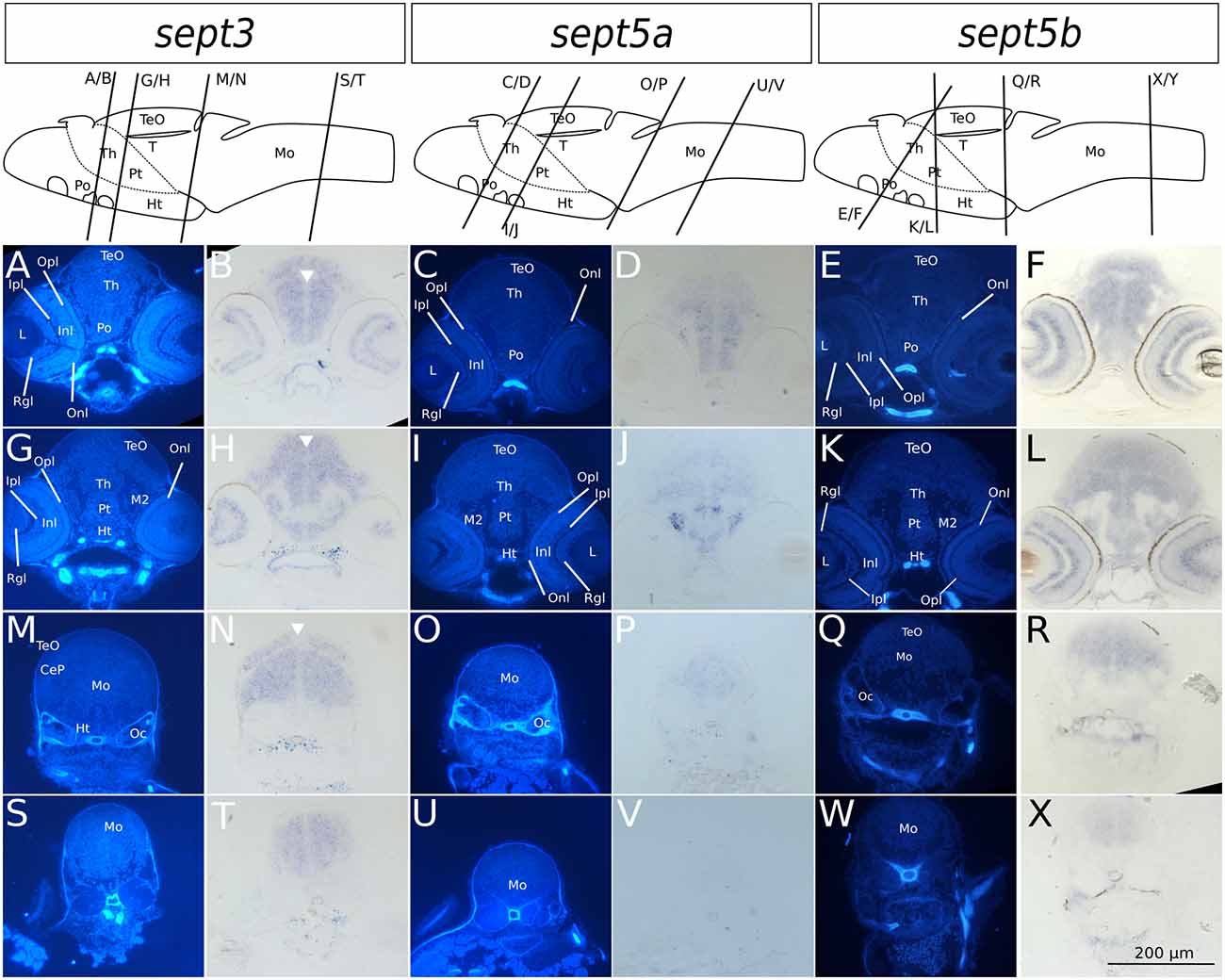
Figure 5. Transverse sections through 4 dpf zebrafish larvae stained with ISH probes against sept3, sept5a and sept5b. 2 dpf zebrafish larvae were stained for sept3 (A,B,G,H,M,N,S,T), sept5a (C,D,I,J,O,P,U,V) and sept5b (E,F,K,L,Q,R,W,X), embedded in epon, and cut in 8 μm thick section. For better orientation, the ISH sections were counterstained with Hoechst (A,G,M,S,C,I,O,U,E,K,Q,W). Septin expression is indicated by the blue color (B,H,N,T,D,J,P,V,F,L,R,X). The tectum (TeO), preoptic region (Po), thalamus (Th), hypothalamus (Ht), cerebellum (Ce), the medulla oblongata (Mo) and the otic capsules (Oc), are marked. Additionally the migrated posterior tubercular area (M2) and the posterior tuberculum (Pt) are annotated. In the eye, the retinal ganglion cell layer (Rgl), the inner and outer nuclear layer (Inl and Onl), as well as the inner and outer plexiform layer (Ipl and Opl) are shown. The ventricles are indicated with white arrowheads. Schemes for better orientation with indicated section planes are modified from Mueller and Wullimann (2015) 5 dpf schemes, as these are structurally very close to the here shown 4 dpf sections.
At 1 dpf, sept5a is expressed in the telencephalon and the diencephalon (Figures 2B, 3B). In the diencephalon the zona limitans intrathalamica (ZLI) remains unstained. Sept5a is further expressed in the rhombomeres of the hindbrain and in the Sc. At 2 dpf sept5a is expressed in the TeO (Figures 2E, 3E). In the retina no sept5a expression is detectable after 2 dpf. From 2 dpf to 4 dpf the expression of sept5a decreases in most brain regions, but is detectable in the telencephalon and the dorsal part of the Ce (Figures 2H,K, 3H,K). No sept5a expression can be detected in the ventricular zones (Figures 4, 5).
At 1 dpf, sept5b is expressed in the telencephalon, diencephalon, and hindbrain (Figures 2C, 3C). In the ZLI and TeO sept5b is not expressed. From 2 dpf to 4 dpf sept5b is additionally detectable in the Ce, the TeO and the retina (Figures 2F,I,L, 3F,I,L). Similar to sept5a, sept5b expression is absent from the ventricular zones (Figures 4, 5).
Summing up, during early development the selected septins are expressed broadly in the brain but proliferative zones, such as ventricular zones in general, and the ZLI and MHB (Figure 4) in particular, seem to be omitted.
Sept3, sept5a and sept5b Expression in Brain Sections of 2 and 4 dpf Zebrafish Larvae
To analyze the developmental expression of the septins in greater spatial detail, cross sections through the head of 2 and 4 dpf larvae were prepared and counterstained with Hoechst (Figures 4, 5). At 2 dpf sept3 is expressed broadly outside the ventricular zones in the TeO (Figures 4A,B,G,H,M,N), the preoptic region (Po; Figures 4A,B), the thalamus (Th; Figures 4A,B), the hypothalamus (Ht; Figures 4G,H,M,N), and the medulla oblongata (Mo; Figures 4S,T,Y,Z), as well as in the developing retina (R; Figures 4A,B,G,H,M,N). While sept5b (Figures 4E,F,K,L,Q,R,W,X,Ü,ß) shows a very similar expression at 2 dpf, sept5a is expressed differently (Figures 4C,D,I,J,O,P,U,V,Ä,Ö). Sept5a is expressed in stripe-like patterns with alternating high and low expressions along the dorso-ventral axis in TeO (Figures 4C,D,I,J,O,P), T (Figures 4I,J), Po (Figures 4C,D), Th (Figures 4C,D), Ht (Figures 4I,J), and Mo (Figures 4U,V,Ä,Ö). The expression of sept5a is more restricted to regions near the ventricles, but there is only patchy expression in the ventricular zone itself. Furthermore, the retina shows faint sept5a expression.
In 4 dpf zebrafish larvae, sept3 is expressed in the retinal ganglion cell layer (Rgl) and the inner nuclear layer (Inl), (Figures 5A,B,G,H). In the brain proper, sept3 is expressed in the TeO (Figures 5A,B,G,H,M,N), Po (Figures 5A,B), Th (Figures 5A,B,G,H), Ht (Figures 5G,H), and Mo (Figures 5M,N,S,T), as well as strongly in the migrated posterior tubercular area (M2; Figures 5G,H). No sept3 expression can be found in the ventricular zones, or in the posterior tuberculum (Pt; Figures 5G,H). Sept5a is strongly expressed in the Po (Figures 5C,D), M2, Ht and dorsally in the Pt (Figures 5I,J). A weaker expression is detectable in the TeO, Th (Figures 5C,D,I,J), and Mo (Figures 5O,P,U,V). No sept5a expression was detected in the eye or the ventricles. The expression of sept5b resembles the expression pattern of sept3. Sept5b is expressed in the Rgl and Inl of the retina (Figures 5E,F,K,L). In the brain proper, there is strong expression of sept5b in the TeO (Figures 5E,F,K,L), Po (Figures 5E,F), Th (Figures 5E,F,K,L), M2, Ht, Pt (Figures 5K,L), and Mo (Figures 5Q,R,W,X). Interestingly, the expression of both genes is high in the dorsal and low in the ventral regions of the Mo (Figures 5M,N,S,T,Q,R,W,X).
To clarify, if the three septin genes are expressed in proliferation zones, an immunohistochemistry staining was performed on ISH stained 2 dpf zebrafish larvae with an antibody against PCNA (Figure 6), which confirms, that the three septin genes are mostly not expressed in proliferation zones.
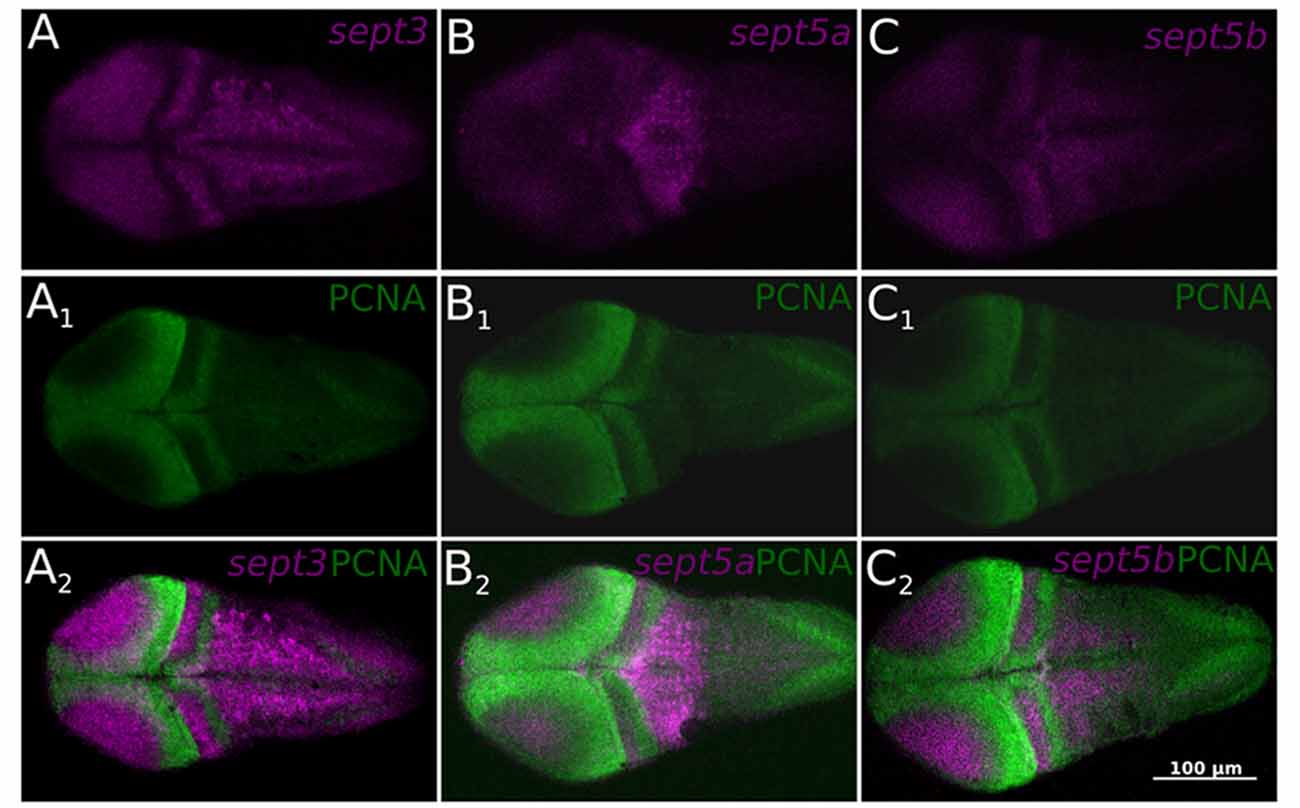
Figure 6. Single confocal section trough 2 dpf zebrafish larvae stained with ISH probes against sept3 (A), sept5a (B), and sept5b (C) from a dorsal view. The larvae were additionally stained with an antibody against Proliferating Cell Nuclear Antigen (PCNA; A1,B1,C1). All three septins (magenta) are mostly not expressed in proliferating cells (green, A2,B2,C2).
To summarize, this more fine resolved analysis on thin sections and co-stainings with a well-established proliferative marker strongly supports the aforementioned notion that sept3, sept5a and sept5b expression is absent from proliferative zones.
Sept3, sept5a and sept5b Expression in Adult Brains
In order to compare septin expression at early stages with the mature nervous system, we analyzed the expression patterns of sept3, sept5a and sept5b in adult brains with ISH (Figures 7, 8). Sept3, sept5a and sept5b are expressed in pallial and subpallial regions of the telencephalon (Figures 7A1,B1,C1, 8A1,B1,C1). In the pallium, transcripts are found in the medial zone of dorsal telencephalic area (Dm) near the ventricle and in the posterior zone of dorsal telencephalic area (Dp; Figures 7A1,B1,C1). Transcripts are also detectable in the lateral zone of dorsal telencephalic area (Dl) and the dorsal zone of dorsal telencephalic area (Dd), but exhibit a more scattered distribution (Figures 7A1,B1,C1). Additionally, septin expression can be found in the central zone of dorsal telencephalic area (Dc; Figures 8A1,B1,C1). In the subpallium, expression is present in the entopeduncular nucleus (EN), dorsal nucleus of the ventral telencephalic area (Vd) and the ventral nucleus of the ventral telencephalic area (Vv; Figures 7A1,B1,C1). In a sagittal view of the telencephalon, expression of sept3, sept5a and sept5b can be found in a stripe in Vd region, as well as in the olfactory bulb (Ob; Figures 8A1,B1,C1). To determine, whether the stripe of cells in the telencephalon are mature dopaminergic neurons (Yamamoto et al., 2011) or part of the differentiating cells in the telencephalon (Grandel et al., 2006), immunohistochemistry staining with an antibody against TH was performed after ISH on adult brain sections (Figures 8D–F). This experiment shows, that sept3, sept5a and sept5b, seem to be not expressed directly within the stream of dopaminergic neurons in the telencephalon but in close proximity.
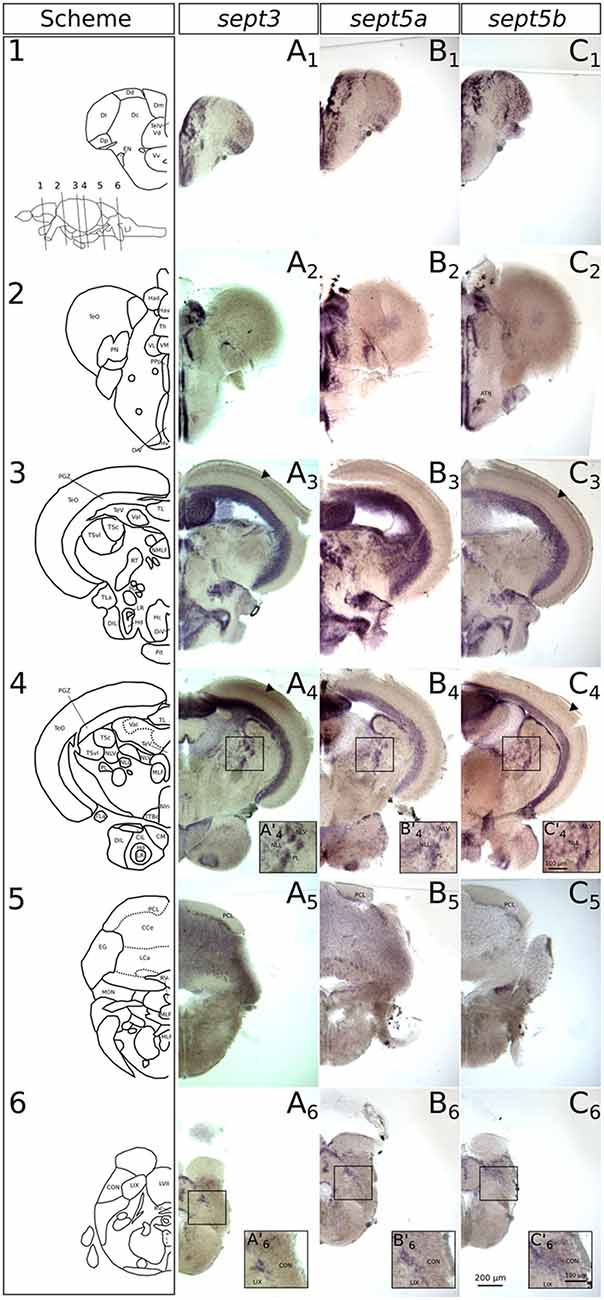
Figure 7. Expression of sept3, sept5a and sept5b in adult zebrafish brains. The expression pattern of sept3 (A1–A6), sept5a (B1–B6) and sept5b (C1–C6) in adult brain sections are shown. The schemes (1–6) show corresponding sections from the zebrafish brain atlas (modified from Wullimann et al., 1996). Abbreviations can be found in Table 1. The anterior tuberal nucleus (ATN) is indicated in (C2). The sept3 and sept5b positive layer in the TeO is labeld with black arrowheads. The inlays in (A’4,B’4,C’4,A’6,B’6,C’6) are showing magnified views of the boxed regions.
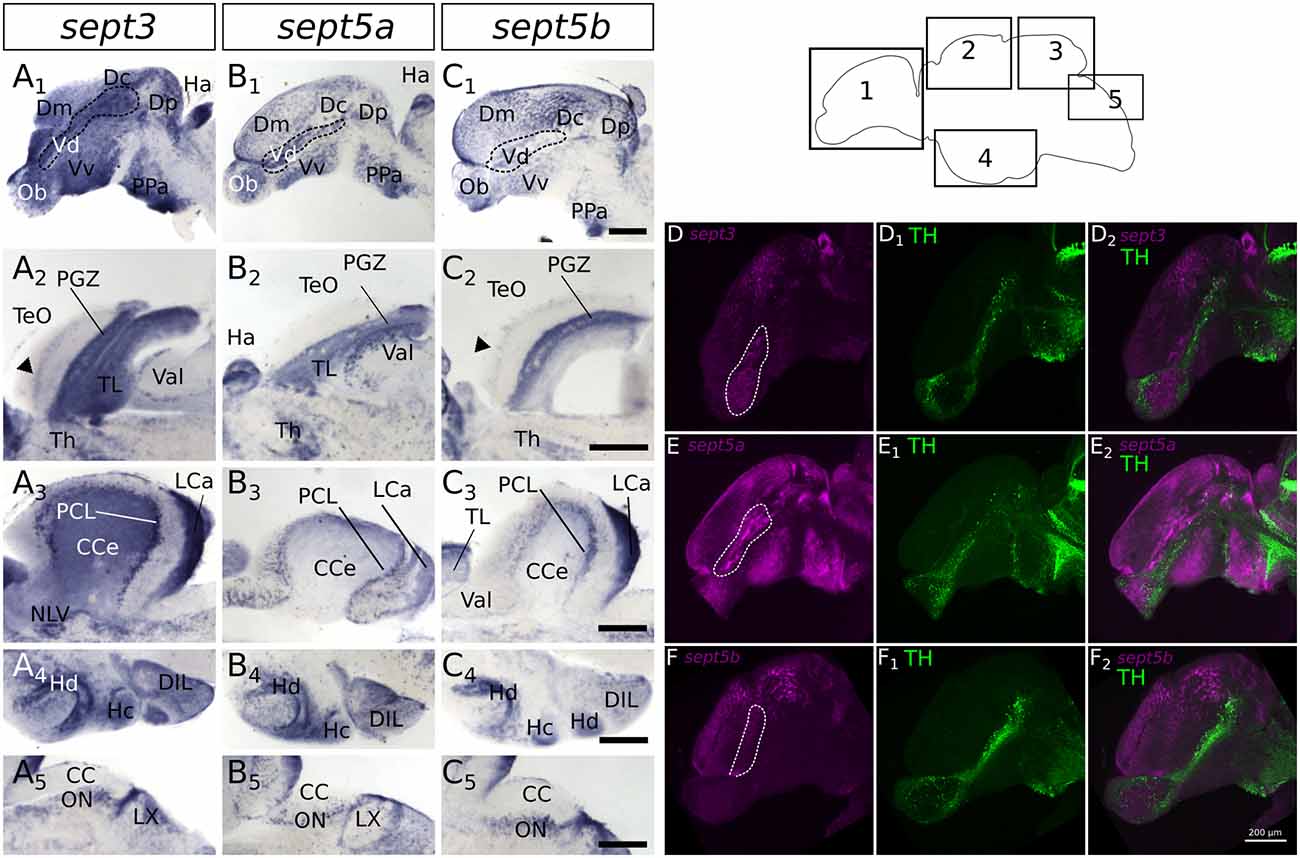
Figure 8. Expression of sept3, sept5a and sept5b in substructures of the adult zebrafish brain. The expression pattern of sept3 (A1–A5), sept5a (B1–B5), and sept5b (C1–C5) is shown in sagittal sections of adult brains. The regions of the brain are indicated in the scheme (1–5). In the telencephalon (1) sept3, sept5a and sept5b are expressed in dorsal telencephalic areas (Dm, Dc and Dp) and ventral telencephalic areas (Vd and Vv). Septins are expressed in a stream in the Vd region (outlined by dotted line). Additionally, sept3, sept5a and sept5b is expressed in the Ob. The sept3 and sept5b positive layer in the TeO is labeld with black arrowheads (A2,C2). In the cerebellum (3,5) sept3, sept5a and sept5b is expressed in the LCa, and in the region of the purkinje cell layer (PCL). Sept3 is additionally expressed in the CCe. Furthermore, hypothalamic sagittal section are shown (4). Abbreviations can be found in Table 1. To identify dopaminergic cells, ISH stained brains (magenta) against sept3 (D), sept5a (E), and sept5b (F) were counterstained with an antibody against tyrosine hydroxylase (TH, green in D1,E1,F1) and analyzed with a confocal microscope and merged (D2,E2,F2). Pictures in (D–F) are maximum projections of confocal slices through adult sagittal brain sections. Scale bars 200 μm.
In the diencephalon (Figures 7A2–4,B2–4,C2–4), all three septins are expressed in nuclei of the habenula (Ha), such as dorsal habenular nucleus (Had) and ventral habenular nucleus (Hav). Furthermore, sept3, sept5a and sept5b are expressed in the pretectal nuclei (PN; Figures 7A2,B2,C2). Additionally, septin expression can be found in thalamic structures like the ventrolateral thalamic nucleus (VL; Figures 7A2,B2,C2), the ventromedial thalamic nucleus (VM; Figures 7A2,B2,C2), anterior and posterior part of the parvocellular preoptic nucleus (PPa; Figures 8A1,B1,C1) and (PPp; Figures 7A2,B2,C2), and the nucleus of the medial longitudinal fascicle (NMLF; Figures 7A3,B3,C3). Septins are also expressed in the preglomerular nuclei (PG; Figures 7A3,B3,C3) and in torus lateralis (TLa; Figures 7A3–4,B3–4,C3–4). In the Ht, septin expression can be found in the dorsal zone of the periventricular hypothalamus (Hd; Figures 7A3–4,B3–4,C3–4, 8A4,B4,C4), caudal zone of the periventricular hypothalamus (Hc; Figures 7A3,B3,C3, 8A4,B4,C4), and ventral zone of the periventricular hypothalamus (Hv; Figures 7A2,B2,C2). Furthermore, the septins are expressed in diffuse nucleus of the inferior lobe (DIL; Figures 7A3–4,B3–4,C3–4, 8A4,B4,C4) and the corpus mamillare (CM; Figures 7A4,B4,C4). Additionally sept5b expression can be found in the anterior tuberal nucleus (ATN; Figure 7C3).
In the mesencephalon (Figures 7A2–4,B2–4,C2–4), expression of all three septins is found in the periventricular gray zone of optic tectum (PGZ; Figures 7A3–4,B3–4,C3–4, 8A2,B2,C2). Further, sept3 and sept5b show clearly two additional fine layers of expression in the TeO (Figures 7A3–4,C3–4, 8A2,B2,C2). Transcripts are also detectable in the central nucleus of torus semicircularis (TSc; Figures 7A3–4,B3–4,C3–4), torus longitudinalis (TL; Figures 7A3–4,B3–4,C3–4, 8A2,B2,C3), rostral tegmental nucleus (RT; Figures 7A3,B3,C3), nucleus of the lateral lemniscus (NLL) and perilemniscal nucleus (PL; Figures 7A4,B4,C4). Interestingly, sept5b is differentially expressed along the anterior-posterior axis of the TL with low expression anteriorly (Figure 7C3) and higher expression posteriorly (Figure 7C4). Additionally, sept5a is strongly expressed in the ventrolateral nucleus of torus semicircularis (TSvl; Figure 7B3). In the nucleus interpeduncularis (Nln; Figures 7A4,B4,C4) sept3 is weakly expressed, whereas sept5a and sept5b are not detectable in this region.
Furthermore, sept3, sept5a, and sept5b are expressed in parts of the rhombencephalon (Figures 7A3–6,B3–6,C3–6, 8A3,A5,B3,B5,C3,C5). Expression is found in the lateral and medial part of the lateral division of valvula cerebelli (Val; Figures 7A3–4,B3–4,C3–4, 8A2,B2,C3) and the nucleus lateralis valvulae (NLV; Figures 7A4,B4,C4). Additionally, sept3, sept5a and sept5b are expressed in the lobus caudalis cerebelli (LCa; Figures 8A3,B3,C3), whereas only sept3 is detectable in the corpus cerebelli (CCe; Figures 7A5,B5,C5, 8A3,B3,C3) and the eminentia granularis (EG; Figures 7A5,B5,C5). Sept3, sept5a and sept5b are expressed in the Purkinje cell layer (PCL; Figures 7A5,B5,C5, 8A3,B3,C3). Moreover, there is expression of sept3, sept5a and sept5b at the superficial layer of the lobus facialis (LVII; Figures 7A6,B6,C6), centrally in LVII and near to the ventricle where cell bodies are enriched and not in projection zones (Yáñez et al., 2017). Furthermore, there is expression of all three septins in a domain of the intermediate reticular formation (IMRF) adjacent to the MLF (Figures 7A6,B6,C6). Finally, the septins are expressed at the border between the caudal octavolateralis nucleus (CON) and the lobus glossopharyngeus (LIX; Figures 7A6,B6,C6). The sagittal sections (Figures 8A5,B5,C5) show, that sept3, sept5a and sept5b expression is detectable in the dorsal part of the octavolateralis nuclei (ON). More specifically, sept5b is expressed in the medial octavolateralis nucleus (MON; Figure 7C5).
Overall, and in contrast to early expression patterns of these three septin genes, where sept5a stood out, the adult brain shows an even more unified expression. The expression is summarized in Table 2.
Discussion
Evidence is accumulating that septins play important roles in neuronal function (Mostowy and Cossart, 2012; Dolat et al., 2014; Marttinen et al., 2015). In order to dissect the neuronal roles of septins, a reference dataset describing the expression of selected neuronally-expressed septins in a tractable vertebrate model like zebrafish is necessary. This work fills this knowledge gap and shows a detailed description of the expression patterns of zebrafish sept3, sept5a and sept5b during stages of early brain development as well as in the adult brain now providing a framework for the functional analysis of the roles of these septins in the nervous system of a tractable vertebrate model organism.
Characteristic Differences in Early Septin Expression Patterns
While sept3 and sept5b are similarly expressed during early development, a unifying principle of their expression patterns is that proliferative zones, in particular ventricular zones and the organizers at the ZLI and the MHB, stay free of sept3 and sept5b expression. The expression of these two genes in the developing brain is largely overlapping with zones where the expression of the Hu protein has been described (Mueller and Wullimann, 2015), which hints to a role of sept3 and sept5b in neuronal differentiation. While sept5a is mostly not expressed in the ventricular zones it seems to be expressed in a more specific pattern during early development. The expression pattern of sept5a in 2 dpf zebrafish larvae is more restricted and is downregulated in large parts of the brain during development to 4 dpf larvae, where it is strongly expressed in cells of the M2 area. Furthermore, in contrast to sept3 and sept5b, no sept5a expression is detectable in the retina at 4 dpf. The observed expression of sept5b in the zebrafish retina is in accordance with immunohistochemistry studies of SEPT5 in human eye tissues (Pache et al., 2005; Xin et al., 2007), which suggests an important conserved role of Sept5 in retinal development (Xin et al., 2007). The sept5b expression differs from the expression of sept3 in the retina. While both septins are expressed in the retinal ganglion layer, sept3 is only expressed in the Inl in the region of the amacrine cells. Additional sept5b expressing cells can be found near to the Opl, where horizontal cells can be found (Gramage et al., 2014). In contrast to the defined differences during early development the expression of the three studied septin genes is much more similar in adult zebrafish brains. All three septin genes are expressed in regions with high proliferation activity in the adult brain (Grandel et al., 2006), but are not expressed in the proliferating zones themselves (Figure 6), which indicates a more specific role in neuronal differentiation processes, as it has been shown for Sept5 for rodent synaptogenesis (Yang et al., 2010).
Interestingly, the sept5a expressing cell clusters, especially in the thalamus of the zebrafish larvae (Figures 4, 5), resemble the distribution of GABAergic neurons (Mueller and Wullimann, 2015). Furthermore, this expression pattern is very similar to the expression of the metabotropic glutamate receptor (mglur1a). Additionally, sept5a and mglur1a are enriched in cells of the cerebellum in a similar pattern (Haug et al., 2013), hinting to a specific role of sept5a in a special subset of neurons.
Comparison of Zebrafish and Mammalian sept3, sept5a and sept5b Expression
The human ortholog of Sept5 (HCDCREL-1) is expressed in neuronal tissues (Yagi et al., 1998). Furthermore, developing mouse embryos show strong neuronal and retinal expression of CDCrel-1 (Sept5; Maldonado-Saldivia et al., 2000). This expression seems to be more similar to the widely expressed sept5b in zebrafish than to the more restricted sept5a expression. The different expression patterns of the two sept5 genes in zebrafish might be explained by subfunctionalization of the paralogs sept5a and sept5b after a specific genome duplication event in teleost fish (Postlethwait et al., 2004), where the paralogous genes were then free to diverge in their expression patterns. Interestingly differential expression of Sept5 isoforms is described in different regions of mouse brains (Asada et al., 2010). In adult mouse, Sept3 and Sept5 are broadly expressed in the whole brain2, showing a particular intense expression in the cerebellum in the region of the PCL. Similar findings were described in immunoreactivity studies for Sept3 in rat brain (Xue et al., 2004) and are in line with the observed specific expression at the PCL in adult zebrafish brains for sept3, sept5a and sept5b.
The Cellular Role in Neurons of sept3, sept5a and sept5b
Notably, the orthologous proteins Sept3 and Sept5 are located at presynaptic terminals in rodent brains, indicating a key role for synaptic function (Kinoshita et al., 2000; Xue et al., 2004; Yang et al., 2010; Tsang et al., 2011). Due to the highly specific Septin5 expression in the brain and, in particular, its localization at nerve terminals (Kinoshita et al., 2000; Xue et al., 2004; Yang et al., 2010; Tsang et al., 2011) the involvement of Sept5 in exocytosis (Beites et al., 1999, 2005), as well as the interaction of Sept5 with PARK2 (Zhang et al., 2000) has linked this particular Septin to diseases of the nervous system like, schizophrenia, epilepsy and degenerative diseases like Parkinson’s and Alzheimer’s (Dolat et al., 2014; Marttinen et al., 2015). In the same line, the overexpression of Sept5 leads to a degeneration of dopaminergic neurons (Dong et al., 2003), which provides further indications to important roles of septins in brain function and development, but the specific cellular roles are not yet fully elucidated. Interestingly, in C. elegans, where only the two septins unc-59 and unc-61 are described in the genome, both septins have been shown to be involved in the development of the nervous system. Mutants of these genes show several behavioral defects like uncoordinated movement and abnormal egg laying (Nguyen et al., 2000; Finger et al., 2003). Moreover, a mouse knock out of Sept3 and Sept5 shows no obvious neuronal defects during development (Tsang et al., 2008), but further detailed behavioral studies show, that Sept5 has an influence on mouse affective, cognitive and social behavior (Suzuki et al., 2009; Harper et al., 2012). With our expression study of sept3, sept5a and sept5b in the zebrafish brain, we provide now a detailed analysis for further investigations into the functions of septins in vertebrates from the cellular level to behavior.
Author Contributions
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. FH, CL and CS: study concept and design; analysis and interpretation of data; drafting of the manuscript; critical revision of the manuscript for important intellectual content; FH and CL: acquisition of data; CS: study supervision.
Funding
This work was supported by Universitätsbund Würzburg (AZ14-48). CL is funded by the Program Chancengleichheit für Frauen in Forschung und Lehre from the Bayerische Gleichstellungsförderung (BGF) and University of Würzburg. This publication was funded by the German Research Foundation (DFG) and the University of Würzburg in the funding programme Open Access Publishing.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We kindly thank M. Schartl for the generous possibility to use the fish facility. Furthermore we are very thankful to G. Krohne, A. Wehman, D. Liedtke, S. Markert and C. Berger for supportive comments on the manuscript and fruitful discussions throughout the project. We further thank M. Stepf for imaging support.
Footnotes
References
Akerboom, J., Chen, T.-W., Wardill, T. J., Tian, L., Marvin, J. S., Mutlu, S., et al. (2012). Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 32, 13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012
Anisimova, M., and Gascuel, O. (2006). Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55, 539–552. doi: 10.1080/10635150600755453
Asada, A., Takahashi, J., Taniguchi, M., Yamamoto, H., Kimura, T., Saito, T., et al. (2010). Neuronal expression of two isoforms of mouse septin 5. J. Neurosci. Res. 88, 1309–1316. doi: 10.1002/jnr.22294
Beites, C. L., Campbell, K. A., and Trimble, W. S. (2005). The septin sept5/CDCrel-1 competes with α-SNAP for binding to the SNARE complex. Biochem. J. 385, 347–353. doi: 10.1042/bj20041090
Beites, C. L., Xie, H., Bowser, R., and Trimble, W. S. (1999). The septin CDCrel-1 binds syntaxin and inhibits exocytosis. Nat. Neurosci. 2, 434–439. doi: 10.1038/8100
Byers, B., and Goetsch, L. (1976). A highly ordered ring of membrane-associated filaments in budding yeast. J. Cell Biol. 69, 717–721. doi: 10.1083/jcb.69.3.717
Cao, L., Ding, X., Yu, W., Yang, X., Shen, S., and Yu, L. (2007). Phylogenetic and evolutionary analysis of the septin protein family in metazoan. FEBS Lett. 581, 5526–5532. doi: 10.1016/j.febslet.2007.10.032
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. doi: 10.1093/oxfordjournals.molbev.a026334
Chevenet, F., Brun, C., Bañuls, A.-L., Jacq, B., and Christen, R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7:439. doi: 10.1186/1471-2105-7-439
Dash, S. N., Lehtonen, E., Wasik, A. A., Schepis, A., Paavola, J., Panula, P., et al. (2014). Sept7b is essential for pronephric function and development of left-right asymmetry in zebrafish embryogenesis. J. Cell Sci. 127, 1476–1486. doi: 10.1242/jcs.138495
Dereeper, A., Audic, S., Claverie, J.-M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Dolat, L., Hu, Q., and Spiliotis, E. T. (2014). Septin functions in organ system physiology and pathology. Biol. Chem. 395, 123–141. doi: 10.1515/hsz-2013-0233
Dong, Z., Ferger, B., Paterna, J.-C., Vogel, D., Furler, S., Osinde, M., et al. (2003). Dopamine-dependent neurodegeneration in rats induced by viral vector-mediated overexpression of the parkin target protein, CDCrel-1. Proc. Natl. Acad. Sci. U S A 100, 12438–12443. doi: 10.1073/pnas.2132992100
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Finger, F. P., Kopish, K. R., and White, J. G. (2003). A role for septins in cellular and axonal migration in C. elegans. Dev. Biol. 261, 220–234. doi: 10.1016/s0012-1606(03)00296-3
Gomez, G., Lee, J.-H., Veldman, M. B., Lu, J., Xiao, X., and Lin, S. (2012). Identification of vascular and hematopoietic genes downstream of etsrp by deep sequencing in zebrafish. PLoS One 7:e31658. doi: 10.1371/journal.pone.0031658
Gramage, E., Li, J., and Hitchcock, P. (2014). The expression and function of midkine in the vertebrate retina. Br. J. Pharmacol. 171, 913–923. doi: 10.1111/bph.12495
Grandel, H., Kaslin, J., Ganz, J., Wenzel, I., and Brand, M. (2006). Neural stem cells and neurogenesis in the adult zebrafish brain: origin, proliferation dynamics, migration and cell fate. Dev. Biol. 295, 263–277. doi: 10.1016/j.ydbio.2006.03.040
Guindon, S., and Gascuel, O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Guna, A., Butcher, N. J., and Bassett, A. S. (2015). Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J. Neurodev. Disord. 7:18. doi: 10.1186/s11689-015-9113-x
Harper, K. M., Hiramoto, T., Tanigaki, K., Kang, G., Suzuki, G., Trimble, W., et al. (2012). Alterations of social interaction through genetic and environmental manipulation of the 22q11.2 gene sept5 in the mouse brain. Hum. Mol. Genet. 21, 3489–3499. doi: 10.1093/hmg/dds180
Hartwell, L. H., Culotti, J., and Reid, B. (1970). Genetic control of the cell-division cycle in yeast, I. Detection of mutants. Proc. Natl. Acad. Sci. U S A 66, 352–359. doi: 10.1073/pnas.66.2.352
Haug, M. F., Gesemann, M., Mueller, T., and Neuhauss, S. C. F. (2013). Phylogeny and expression divergence of metabotropic glutamate receptor genes in the brain of zebrafish (Danio rerio). J. Comp. Neurol. 521, 1533–1560. doi: 10.1002/cne.23240
Karlsson, J., von Hofsten, J., and Olsson, P.-E. (2001). Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Mar. Biotechnol. 3, 522–527. doi: 10.1007/s1012601-0053-4
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kinoshita, A., Noda, M., and Kinoshita, M. (2000). Differential localization of septins in the mouse brain. J. Comp. Neurol. 428, 223–239. doi: 10.1002/1096-9861(20001211)428:2<223::AID-CNE3>3.0.CO;2-M
Landsverk, M. L., Weiser, D. C., Hannibal, M. C., and Kimelman, D. (2010). Alternative splicing of sept9a and sept9b in zebrafish produces multiple mRNA transcripts expressed throughout development. PLoS One 5:e10712. doi: 10.1371/journal.pone.0010712
Lillesaar, C. (2011). The serotonergic system in fish. J. Chem. Neuroanat. 41, 294–308. doi: 10.1016/j.jchemneu.2011.05.009
Maldonado-Saldivia, J., Funke, B., Pandita, R. K., Schüler, T., Morrow, B. E., and Schorle, H. (2000). Expression of Cdcrel-1 (Pnutl1), a gene frequently deleted in velo-cardio-facial syndrome/DiGeorge syndrome. Mech. Dev. 96, 121–124. doi: 10.1016/s0925-4773(00)00370-1
Marttinen, M., Kurkinen, K. M. A., Soininen, H., Haapasalo, A., and Hiltunen, M. (2015). Synaptic dysfunction and septin protein family members in neurodegenerative diseases. Mol. Neurodegener. 10:16. doi: 10.1186/s13024-015-0013-z
Mostowy, S., and Cossart, P. (2012). Septins: the fourth component of the cytoskeleton. Nat. Rev. Mol. Cell Biol. 13, 183–194. doi: 10.1038/nrm3284
Mueller, D. T., and Wullimann, M. (2015). Atlas of Early Zebrafish Brain Development: A Tool for Molecular Neurogenetics. Amsterdam: Elsevier.
Nguyen, T. Q., Sawa, H., Okano, H., and White, J. G. (2000). The C. elegans septin genes, unc-59 and unc-61, are required for normal postembryonic cytokineses and morphogenesis but have no essential function in embryogenesis. J. Cell Sci. 113, 3825–3837.
Nishihama, R., Onishi, M., and Pringle, J. R. (2011). New insights into the phylogenetic distribution and evolutionary origins of the septins. Biol. Chem. 392, 681–687. doi: 10.1515/BC.2011.086
Pache, M., Zieger, B., Bläser, S., and Meyer, P. (2005). Immunoreactivity of the septins SEPT4, SEPT5 and SEPT8 in the human eye. J. Histochem. Cytochem. 53, 1139–1147. doi: 10.1369/jhc.4a6588.2005
Pan, F., Malmberg, R. L., and Momany, M. (2007). Analysis of septins across kingdoms reveals orthology and new motifs. BMC Evol. Biol. 7:103. doi: 10.1186/1471-2148-7-103
Postlethwait, J., Amores, A., Cresko, W., Singer, A., and Yan, Y.-L. (2004). Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 20, 481–490. doi: 10.1016/j.tig.2004.08.001
Sanders, S. L., and Field, C. M. (1994). Cell division: septins in common? Curr. Biol. 4, 907–910. doi: 10.1016/S0960-9822(00)00201-3
Suzuki, G., Harper, K. M., Hiramoto, T., Sawamura, T., Lee, M., Kang, G., et al. (2009). Sept5 deficiency exerts pleiotropic influence on affective behaviors and cognitive functions in mice. Hum. Mol. Genet. 18, 1652–1660. doi: 10.1093/hmg/ddp086
Thisse, B., and Thisse, C. (2004). “Fast release clones: a high throughput expression analysis,” in ZFIN Direct Data Submission. Available online at: http://zfin.org
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Thisse, C., Thisse, B., Schilling, T. F., and Postlethwait, J. H. (1993). Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203–1215.
Tsang, C. W., Estey, M. P., DiCiccio, J. E., Xie, H., Patterson, D., and Trimble, W. S. (2011). Characterization of presynaptic septin complexes in mammalian hippocampal neurons. Biol. Chem. 392, 739–749. doi: 10.1515/BC.2011.077
Tsang, C. W., Fedchyshyn, M., Harrison, J., Xie, H., Xue, J., Robinson, P. J., et al. (2008). Superfluous role of mammalian septins 3 and 5 in neuronal development and synaptic transmission. Mol. Cell. Biol. 28, 7012–7029. doi: 10.1128/MCB.00035-08
Westerfield, M. (2000). The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). 4th Edn. Eugene: University of Oregon Press.
Willis, A., Mazon-Moya, M., and Mostowy, S. (2016). Investigation of septin biology in vivo using zebrafish. Methods Cell Biol. 136, 221–241. doi: 10.1016/bs.mcb.2016.03.019
Wullimann, M. F., Rupp, B., and Reichert, H. (1996). Neuroanatomy of the Zebrafish Brain. Basel: Birkhäuser Basel. Available online at: http://link.springer.com/10.1007/978-3-0348-8979-7 [Accessed December 11, 2015].
Xin, X., Pache, M., Zieger, B., Bartsch, I., Prünte, C., Flammer, J., et al. (2007). Septin expression in proliferative retinal membranes. J. Histochem. Cytochem. 55, 1089–1094. doi: 10.1369/jhc.7a7188.2007
Xue, J., Tsang, C. W., Gai, W.-P., Malladi, C. S., Trimble, W. S., Rostas, J. A. P., et al. (2004). Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J. Neurochem. 91, 579–590. doi: 10.1111/j.1471-4159.2004.02755.x
Yagi, M., Zieger, B., Roth, G. J., and Ware, J. (1998). Structure and expression of the human septin gene HCDCREL-1. Gene 212, 229–236. doi: 10.1016/s0378-1119(98)00146-2
Yamamoto, K., Ruuskanen, J. O., Wullimann, M. F., and Vernier, P. (2011). Differential expression of dopaminergic cell markers in the adult zebrafish forebrain. J. Comp. Neurol. 519, 576–598. doi: 10.1002/cne.22535
Yáñez, J., Souto, Y., Piñeiro, L., Folgueira, M., and Anadón, R. (2017). Gustatory and general visceral centers and their connections in the brain of adult zebrafish: a carbocyanine dye tract-tracing study. J. Comp. Neurol. 525, 333–362. doi: 10.1002/cne.24068
Yang, Y.-M., Fedchyshyn, M. J., Grande, G., Aitoubah, J., Tsang, C. W., Xie, H., et al. (2010). Septins regulate developmental switching from microdomain to nanodomain coupling of Ca2+ influx to neurotransmitter release at a central synapse. Neuron 67, 100–115. doi: 10.1016/j.neuron.2010.06.003
Zhai, G., Gu, Q., He, J., Lou, Q., Chen, X., Jin, X., et al. (2014). Sept6 is required for ciliogenesis in Kupffer’s vesicle, the pronephros, and the neural tube during early embryonic development. Mol. Cell. Biol. 34, 1310–1321. doi: 10.1128/MCB.01409-13
Zhang, Y., Gao, J., Chung, K. K., Huang, H., Dawson, V. L., and Dawson, T. M. (2000). Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc. Natl. Acad. Sci. U S A 97, 13354–13359. doi: 10.1073/pnas.240347797
Keywords: septin, RNA in situ hybridization, neuronal development, retinal development, sept3, sept5a, sept5b
Citation: Helmprobst F, Lillesaar C and Stigloher C (2017) Expression of sept3, sept5a and sept5b in the Developing and Adult Nervous System of the Zebrafish (Danio rerio). Front. Neuroanat. 11:6. doi: 10.3389/fnana.2017.00006
Received: 06 October 2016; Accepted: 25 January 2017;
Published: 17 February 2017.
Edited by:
Gonzalo Alvarez-Bolado, Heidelberg University, GermanyReviewed by:
Steffen Scholpp, Karlsruhe Institute of Technology, GermanyMario F. Wullimann, Ludwig Maximilian University of Munich, Germany
Copyright © 2017 Helmprobst, Lillesaar and Stigloher. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution and reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Stigloher, Y2hyaXN0aWFuLnN0aWdsb2hlckB1bmktd3VlcnpidXJnLmRl
 Frederik Helmprobst
Frederik Helmprobst Christina Lillesaar2
Christina Lillesaar2 Christian Stigloher
Christian Stigloher