- 1Department of Cellular Neuropathology, Brain Research Institute, Niigata University, Niigata, Japan
- 2International Research Center for Neurointelligence (WPI-IRCN), The University of Tokyo, Tokyo, Japan
Our brain adapts to the environment by optimizing its function through experience-dependent cortical plasticity. This plasticity is transiently enhanced during a developmental stage, known as the “critical period,” and subsequently maintained at lower levels throughout adulthood. Thus, understanding the mechanism underlying critical period plasticity is crucial for improving brain adaptability across the lifespan. Critical period plasticity relies on activity-dependent circuit remodeling through anatomical and functional changes at individual synapses. However, it remains challenging to identify the molecular signatures of synapses responsible for critical period plasticity and to understand how these plasticity-related synapses are spatiotemporally organized within a neuron. Recent advances in genetic tools and genome editing methodologies have enabled single-cell endogenous protein labeling in the brain, allowing for comprehensive molecular profiling of individual synapses within a neuron, namely “single-cell synaptome mapping.” This promising approach can facilitate insights into the spatiotemporal organization of synapses that are sparse yet functionally important within single neurons. In this review, we introduce the basics of single-cell synaptome mapping and discuss its methodologies and applications to investigate the synaptic and cellular mechanisms underlying circuit remodeling during the critical period.
1 Introduction
The brain adapts to the surrounding environment by optimizing its functions in an experience-dependent manner. The neural plasticity that underlies this adaptation peaks at a narrow developmental time window, known as the “critical period” (Hubel and Wiesel, 1970). The brain subsequently reduces its adaptability, maintaining it at lower levels across adulthood (Hübener and Bonhoeffer, 2014). This developmental change enables efficient acquisition of new skills, such as learning a second language in childhood, but also limits recovery from ischemic or traumatic brain damages in adulthood (Bjorklund, 2022). Thus, understanding the mechanisms that govern the opening, maintenance, and closure of the developmental critical period, as well as how the critical period reopens in adulthood, is crucial for advancing our knowledge of brain plasticity and its applications in neurological recovery (Hensch, 2004; Bavelier et al., 2010).
The cellular mechanism underlying critical period plasticity involves activity-dependent circuit remodeling through anatomical and functional changes at individual synapses. The visual cortex, extensively studied as a model of critical period plasticity (Hensch, 2005; Espinosa and Stryker, 2012), requires both Hebbian and non-Hebbian synaptic plasticity for circuit remodeling (Mrsic-Flogel et al., 2007; Yoon et al., 2009; Ranson et al., 2012). Structural changes of dendritic spines, which are protrusions that form excitatory synapses, are enhanced in pyramidal cells during early life (Majewska and Sur, 2003). These synaptic phenotypes vary across individual synapses (Majewska and Sur, 2003; Sun et al., 2019). However, previous studies have often relied on averaged data from a limited number of synapses, potentially overlooking specific synaptic subpopulations that contribute to circuit remodeling. Therefore, comprehensive profiling of individual synapses is essential for a better understanding of the circuit remodeling in critical period plasticity. In this review, we introduce a powerful approach for molecular profiling of individual synapses within a neuron, called “single-cell synaptome mapping.” We also discuss the technical methodologies and applications of this approach to investigate the synaptic and cellular mechanisms underlying circuit remodeling during the critical period in the mouse brain.
2 Single-cell synaptome: comprehensive molecular profiles of individual synapses within a neuron
The term “synaptome” is derived from “synapse” and “ome” (meaning “totality”). It was originally used as a part of the connectome, a comprehensive map of neuronal connections between different neurons or regions within the central nervous system (DeFelipe, 2010). Recently, the concept of the synaptome has expanded to include the synapse proteome, comprising over 1,000 protein species essential for synaptic functions (Koopmans et al., 2019; van Oostrum et al., 2023).
Spatial mapping of endogenous proteins at individual synapses is crucial for understanding synaptic protein-based synaptome. Immunohistochemistry is the most common method for labeling endogenous proteins in brain tissues. However, non-specific immunoreactivity and limited antibody penetration often cause challenges to quantitative accuracy (Watanabe et al., 1998), despite recent advances such as computational antibody design (Kim et al., 2023), glyoxal fixation (Konno et al., 2023), and chemical and physical modifications of antibodies and fixed tissues (Yau et al., 2023). Grant and colleagues developed a methodological pipeline for analyzing the molecular profiles of individual synapses across the brain, referred to as the brain-wide synaptome (Figure 1A) (Zhu et al., 2018; Cizeron et al., 2020; Bulovaite et al., 2022; Tomas-Roca et al., 2022; Koukaroudi et al., 2024). This is based on large-scale, high-resolution, quantitative measurements of the expression levels of major endogenous postsynaptic proteins (e.g., postsynaptic scaffolds PSD95 and SAP102) genetically labeled with fluorescent tags across the mouse brain. Mathematical analyses revealed synaptome maps that vary depending on cell types, brain regions, ages, and disease states. However, understanding the intricate synaptome at the cellular level remains a challenge due to the remarkable heterogeneity of neurons within the brain.
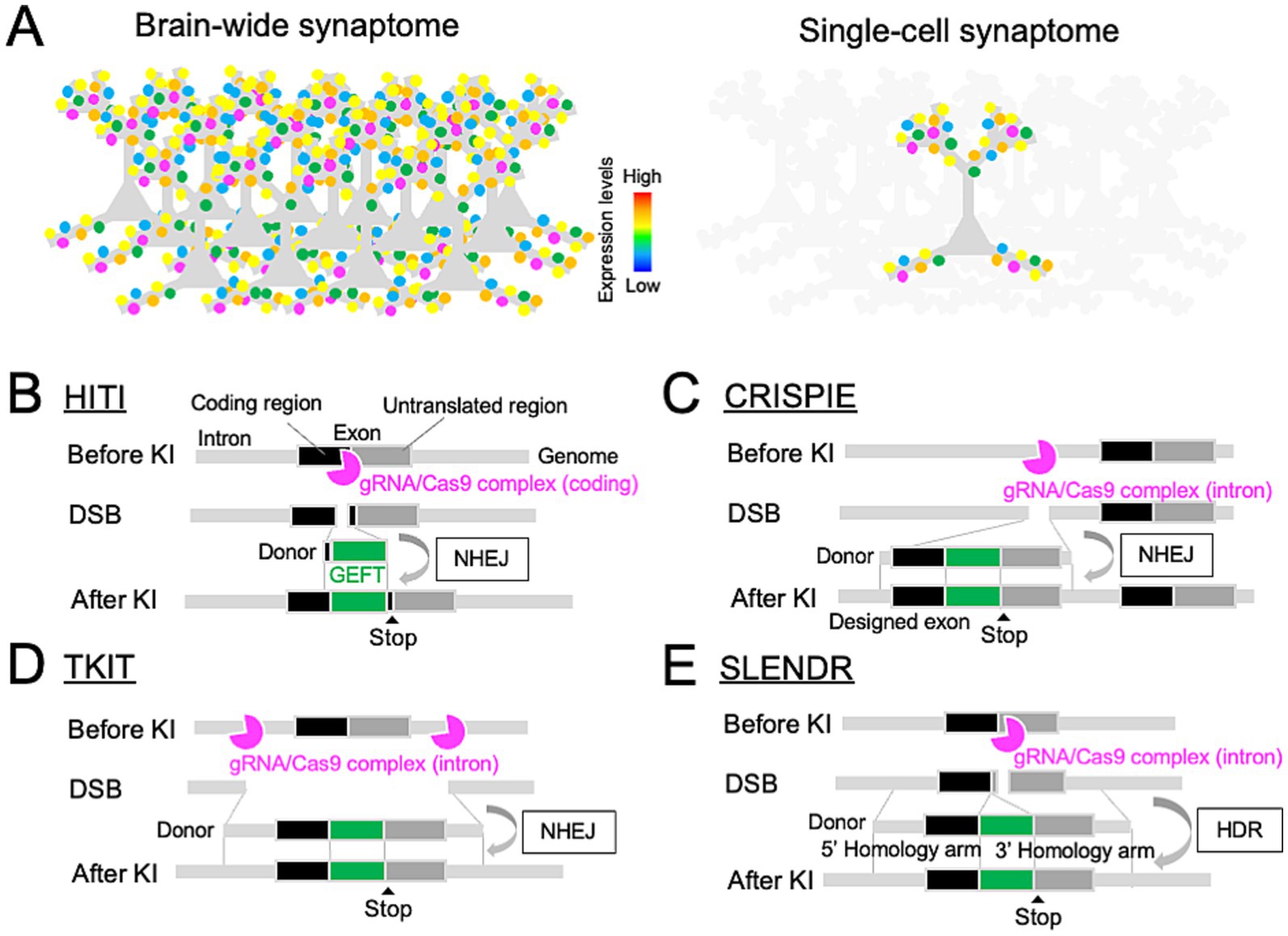
Figure 1. Genome editing toolbox for single-cell endogenous protein labeling in the brain. (A) Comparison of brain-wide (top) and single-cell (bottom) synaptome mapping based on endogenous protein labeling at individual synapses. (B–E) Design of genome editing-mediated GEFT KI through the NHEJ-mediated HITI (B), CRISPIE (C), or TKIT (D), and HDR-mediated SLENDR (E), allowing for single-cell endogenous protein labeling with GEFTs in the brain.
Single-cell synaptome mapping offers a promising approach for dissecting the complexity of the brain-wide synaptome at cellular levels. In the neocortex, single pyramidal cells receive inputs from other neurons via thousands of synapses on dendritic branches (Iascone et al., 2020). Comprehensive molecular profiling of these synapses within individual neurons, which is referred to as “single-cell synaptome,” enables researchers to explore the molecular diversity of synapses at single-cell levels in the brain (Figure 1A). This approach can help identify synapses that are few in number but functionally impactful within a neuron, and reveal the spatiotemporal organization of individual synaptic profiles that underlies the single-neuron computation. Therefore, single-cell synaptome mapping can gain a deeper insight into the synaptic and cellular mechanisms underlying circuit remodeling during the critical period.
3 Technical methodologies of single-cell synaptome mapping
Single-cell synaptome mapping requires single-cell endogenous protein labeling, volumetric fluorescence imaging, and quantitative signal detection in living or fixed mouse brains. Genetically-encoded fluorescent tags (GEFTs) with brightness and photostability are suitable for these requirements. Indeed, the brain-wide synaptome analysis described by Grant and colleagues is based on genetic labeling of endogenous proteins with GEFTs, such as mEGFP or HaloTag (Zhu et al., 2018; Cizeron et al., 2020; Bulovaite et al., 2022; Tomas-Roca et al., 2022; Koukaroudi et al., 2024). Recent technological advances in genetic tools and genome editing methodologies have revolutionized single-cell endogenous protein labeling. In this section, we provide an overview of several methods for single-cell endogenous labeling with GEFTs in the mammalian brain.
3.1 Intrabody-mediated endogenous protein labeling
Intrabodies are small recombinant antibody-like proteins genetically encoded for the heterologous expression, allowing for efficient access to intact endogenous target proteins within a living neuron transfected with intrabodies (Trimmer, 2022). Different types of intrabodies are available, such as single-chain variable fragments (scFvs), nanobodies (nAbs), and recombinant fibronectin-based molecules (monobodies or FingR). Unlike normal full-length antibodies (IgG) with two pairs of heavy and light chains which contain multiple variable and constant domains, a scFv consists of a single pair of heavy- and light-chain variable domains which tether together via a peptide linker to make a binding site to antigen epitopes (Bird et al., 1988; Huston et al., 1988; Fukata et al., 2013; Kabayama et al., 2020). nAbs are based on the variable domain of camel single-chain antibodies, which are sufficient for binding to antigen epitopes (Hamers-Casterman et al., 1993; Dong et al., 2019; Kilisch et al., 2023; Queiroz Zetune Villa Real et al., 2023). Monobodies or FingR are genetically engineered from human 10th fibronectin type III domains to bind to target proteins via mRNA display (Koide et al., 1998; Karatan et al., 2004; Gross et al., 2013; Mora et al., 2013). For single-cell endogenous protein labeling, these intrabodies are fused with GEFTs, and genetically introduced into specific cells. Since this approach relies on the overexpression of intrabodies, target-binding intrabodies may be outnumbered by unbound intrabodies, which can reduce the signal-to-noise ratio. Transcriptional feedback control can help limit excess unbound intrabodies within the cell (Gross et al., 2013).
Many intrabodies have been developed to target various synaptic proteins, such as CaMKIIα (Mora et al., 2013), PSD95 (Fukata et al., 2013; Gross et al., 2013; Rimbault et al., 2024), Gephyrin (Gross et al., 2013; Dong et al., 2019), and Synaptotagmin-1 (Kabayama et al., 2020; Queiroz Zetune Villa Real et al., 2023). Furthermore, co-expression of these intrabodies fused to distinct GEFTs allows for multi-color imaging of different synaptic proteins within the same neurons (Son et al., 2016). However, the intrabody-based approach has some caveats. Currently, specific intrabodies are available for only a limited number of synaptic proteins. Generating new intrabodies against a specific protein is costly, time-consuming, and labor-intensive, hindering the expansion of the library of intrabodies for various synaptic proteins. Even with available synaptic protein-targeting intrabodies, insufficient penetration into densely packed synaptic structures can impair accurate synaptic protein quantification. Furthermore, intrabody-based labeling of synaptic proteins can cause abnormal synaptic functions within a living neuron by disrupting the molecular conformation or interaction of these proteins (Marchionni et al., 2009). Therefore, individual intrabodies require careful validations before use.
3.2 Single-cell KI-mediated endogenous protein labeling
Endogenous protein tagging using GEFT knock-in (KI) is another powerful approach for quantitative mapping of endogenous proteins in brain tissue, as demonstrated in brain-wide synaptome analyses by Zhu et al. (2018), Cizeron et al. (2020), Bulovaite et al. (2022), Tomas-Roca et al. (2022), and Koukaroudi et al. (2024). Direct fusion of GEFTs to target proteins via genetic KI allows for spatially precise and quantitative measurements of fluorescent signals from GEFT-labeled proteins, with expression levels controlled by endogenous promoters. Although GEFT labeling of target proteins may potentially affect their native functions, previous studies have validated some tagging sites for each protein with minimal effects on its function. Artificial intelligence-based predictions of protein structures, such as the AlphaFold, may also be helpful (Jumper et al., 2021). Currently, two major approaches are available for single-cell GEFT KI in the brain.
3.2.1 Conditional KI mouse
Cre-loxP-mediated conditional KI mice enable single-cell endogenous protein tagging with GEFTs in the brain. A duplicated exon strategy (called “ENABLED”) facilitates the generation of conditional KI mouse lines for GEFT tagging of target proteins at the C-terminus (Fortin et al., 2014). This method duplicates the stop-codon-containing exon into floxed/untagged and GEFT-tagged ones, allowing for a Cre-dependent transcription of the GEFT-tagged exon. Zhong and colleagues generated a conditional KI mouse line in which the C-terminus of PSD95 was tagged with GEFTs in a Cre-dependent manner (Fortin et al., 2014). A sparse introduction of the Cre gene into this mouse line provides single-cell labeling of PSD95 with GEFTs in both living and fixed brains.
A couple of technical limitations have disrupted applications of the ENABLED strategy to other proteins. First, the generation of conditional KI mouse lines is lower-throughput compared with that of knockout models. This productivity might be enhanced by the improved genome editing via oviductal nucleic acid delivery (i-GONAD) (Gurumurthy et al., 2016; Abe et al., 2023). Second, it may be difficult to apply the ENABLED-based KI design to regions of target proteins other than the C-terminus. Duplicated exons of target genes can be both spliced in at mRNA levels, consequently leading to an unnecessary amino acid insertion or frame-shift-mediated pre-mature termination at protein levels. Third, additional exons involved in the ENABLED strategy can cause abnormal transcription and translation of target genes in the absence of Cre recombinase. Indeed, PSD95-ENABLED mice exhibited the down-regulation of untagged PSD95 at both mRNA and protein levels (Fortin et al., 2014).
3.2.2 In vivo genome editing-mediated KI
In vivo genome editing-mediated KI is also a promising approach for single-cell endogenous protein labeling. This technique leverages the cell-intrinsic DNA repair machinery in response to target-specific DNA double-strand breaks (DSBs), which are introduced by a Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) complex composed of a target-specific guide RNA (gRNA) and the Cas9 endonuclease protein (Jinek et al., 2012; Cong et al., 2013; Ran et al., 2013; Doudna and Charpentier, 2014). Two major DNA repair mechanisms, non-homologous end joint (NHEJ) and homology-directed repair (HDR), are frequently utilized for genome editing-mediated KI of GEFTs in neuronal tissues.
3.2.2.1 NHEJ-mediated KI
NHEJ directly ligates pairs of free DNA ends at DSB sites (Lieber, 2010). Since the NHEJ machinery is active during both mitotic and non-mitotic phases of the cell cycle, it can repair CRISPR/Cas9-induced DNA DSBs with high efficiency in non-dividing neurons. Leveraging this DNA repair machinery, donor DNA fragments encoding tag sequences without any homology arms can be inserted at CRISPR/Cas9-induced DSB sites, enabling highly-efficient GEFT KI in neurons through NHEJ-mediated repair. This method is known as homology-independent targeted integration (HITI, Figure 1B) (Suzuki et al., 2016). The HITI technique is based on a DNA vector- or adeno-associated virus (AAV)-mediated delivery of target gene-specific gRNA and in-frame donor sequences into primary neuronal cultures, organotypic slices, or living brains. To enhance the generalizability of the HITI technique, a series of validated gRNA sequences have been demonstrated for mouse genes encoding 15 and 38 neuronal proteins as part of the HITI-based genome editing toolboxes, HiUGE and ORANGE, respectively (Table 1) (Gao et al., 2019; Willems et al., 2020). The HiUGE toolbox further offers a universal design for donor sequences that can be integrated into virtually any genomic target loci.
HITI-based genome editing requires higher on-target and lower off-target activity of CRISPR/Cas9 complex. This can be achieved through optimized gRNA design (Doench et al., 2016; Xiang et al., 2021) and improved Cas9 derivatives, such as HiFi Cas9 and eSpCas9 (Slaymaker et al., 2016; Vakulskas et al., 2018). Since NHEJ is an error-prone DNA repair machinery, it can cause unfavorable DNA insertions or deletions (INDELs) at DSB sites (Lieber, 2010). Consequently, the HITI can cause INDELs within the coding sequence of the target genes, leading to nonsense or frame-shift mutations at protein levels. To mitigate this issue, the HITI has been modified to target CRISPR/Cas9-mediated DSBs in non-coding regions of the target genes. The CRISPR-mediated insertion of Exon (CRISPIE) inserts a designed exon-like donor, which consists of an exon encoding a tag sequence flanked by introns, into an intronic site in the target gene (Figure 1C) (Zhong et al., 2021). INDELs involved in the insertion of CRISPIE donors can be spliced out, leading to no INDELs at mRNA levels. However, an amino acid position to insert a tag sequence in the target protein depends on the exon-intron structure of the corresponding gene. Alternatively, Targeted Knock-In with Two guides (TKIT) makes CRISPR/Cas9-mediated DSBs at two intronic or non-coding sites, flanking a coding exon of the target gene to be replaced with a tagged one (Figure 1D) (Fang et al., 2021). This allows for an efficient, scarless tag KI at any target genomic loci. For instance, the TKIT provided a precise insertion of pH-sensitive fluorescent proteins a few amino acids downstream of the signal sequence of ionotropic glutamate receptors (Table 1). However, this strategy could cut off the target coding region flanked by two non-coding DSB sites or insert a designed exon at either DSB site (Danner et al., 2021), potentially resulting in a knockout of the target gene at the protein level. Another caveat of NHEJ-mediated genome editing is its poor compatibility with multiplex targeting of different genes in the same cells due to possible crosstalk between target gene-specific donors. Inducible Cre-mediated sequential targeting of different genes, which is named the Conditional Activation of KI Expression (CAKE), might solve this issue, while thorough optimizations of experimental conditions would be necessary for effective multiple targeting of distinct genes (Droogers et al., 2022).
3.2.2.2 HDR-mediated KI
HDR is based on homologous recombination with donor DNA templates composed of two homology arms, allowing for a precise DNA repair (Filippo et al., 2008). However, since HDR is limited to the mitotic S and G2 phases of the cell cycle, it was thought to be inapplicable to non-dividing neurons. Mikuni et al. overcome this limitation by inducing CRISPR/Cas9-mediated HDR in neuronal progenitors which retain the ability of cell proliferations at the embryonic stage by in utero electroporation (IUE) (Figure 1E) (Mikuni et al., 2016). This method, termed SLENDR, allows for single-cell mEGFP KI for CaMKIIα and CaMKIIβ genes in the mouse neocortex. Likewise, other groups reported IUE-based HDR for tagging β-Actin, βIII-Tubulin, and Laminin B1 genes with mEGFP or mCherry in cortical progenitors (Tsunekawa et al., 2016; Uemura et al., 2016; Kurihara et al., 2020; Meyerink et al., 2022). Interestingly, single homology arm donors can also efficiently induce error-free, HDR-like mEGFP KI for βIII-Tubulin gene, which is termed intercellular linearized Single homology Arm donor mediated intron-Targeting Integration (SATI) (Suzuki et al., 2019). The HDR-mediated KI efficiency for GEFTs was reported to range from 1–50%, which is highly variable depending on target genes, donor DNA templates, and the timing of IUE. This KI efficiency can be improved by manipulating the DNA repair machinery (Jayathilaka et al., 2008; Kurihara et al., 2020). Unlike NHEJ-mediated methods, CRISPR/Cas9-mediated HDR enables simultaneous targeting of multiple different genes, thereby increasing the generalizability of HDR-mediated endogenous protein labeling.
IUE-based introduction of CRISPR/Cas9-mediated HDR limits its accessibility to specific brain regions and embryonic stages. To address this limitation, Nishiyama et al. expanded CRISPR/Cas9-mediated HDR to post-mitotic neurons in various ages, cell types, and brain regions. This method is based on a high titer AAV-mediated transduction with gRNA and homology donor templates, which is called vSLENDR (Nishiyama et al., 2017). HDR-mediated genome editing can potentially lead to a leaky expression of a tag sequence from the donor template, possibly due to promoter activity in the 5′ homology arm. A polyadenylation sequence, which terminates the transcription, can suppress this leaky tag expression with its insertion upstream of the 5′ homology arm in a donor DNA template (Tsunekawa et al., 2016).
4 Discussion
Recent technological advances in genetic tools and genome editing methodologies have led to the development of various techniques for single-cell endogenous protein labeling in the mammalian brain. More than 70 protein species, including key postsynaptic scaffolds or receptors crucial for synaptic plasticity, have been validated in the mouse neuronal culture or brain tissue (Table 1). These resources not only facilitate mapping of the subcellular distribution of endogenous proteins in individual neurons, but also enable interrogation of the single-cell synaptome in the mouse brain.
Single-cell synaptome mapping is valuable for examining the synaptic and cellular mechanisms that underlie circuit remodeling during the critical period. This method can detect unique subpopulations of synapses, such as silent synapses, which are thought to serve as cellular substrates for circuit remodeling during the critical period (Huang et al., 2015). The spatiotemporal architecture of the single-cell synaptome may correlate with the activity or function of the same neuron, which can be captured by activity-dependent labeling with artificial promoters (ESARE, RAM) (Kawashima et al., 2013; Sorensen et al., 2016), immediate early genes (c-fos, egr-1, Arc) (Minatohara et al., 2016), or calcium indicators (CaMPARI2) (Moeyaert et al., 2018). This correlation would help us understand activity-dependent changes in synaptic diversity at the single-neuron level, which underlie circuit remodeling during the critical period. Furthermore, advanced GEFT technology enables single-cell mapping of not only protein localization but also protein dynamics in the brain. For instance, chemical tags that irreversibly bind to fluorescently-labeled ligands (Hoelzel and Zhang, 2020) allow for mapping of protein half-lives in the brain through monitoring the decrease in the pulse-label signals for pre-existing proteins fused to chemical tags (Bulovaite et al., 2022). Importantly, single-cell synaptome mapping is applicable to broad areas of neuroscience, such as neurodegenerative diseases and traumatic brain injuries, whose pathophysiological mechanisms involve the remodeling of individual synapses (Benarroch, 2018; Jamjoom et al., 2021).
In summary, single-cell endogenous protein labeling technologies, such as intrabodies, conditional KI, and genome-editing-mediated KI, provide a technical basis for molecular profiling of individual synapses within a single neuron in the mammalian brain tissue. This single-cell synaptome mapping would be a powerful and versatile approach for interrogating synaptic diversity within a single neuron in the brain, allowing for comprehensive and integrative understanding of the synaptic and cellular mechanisms that underpins circuit remodeling involved in neurodevelopmental and pathophysiological plasticity.
Author contributions
MU: Writing – original draft, Writing – review & editing. TM: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Grants-in-Aid for Scientific Research (20H05914, 20K21461, 20H03349, 20H05918, 23K18160, and 24K02130 to MU and 19H05206, 19K22467, 20H05055, 20H03337, 21H00188, 21F21113, 22K19355, 22H05491, 22K21353, 23H04672, 23H02574, 23K27265, 24H01229, and 24K22000 to TM), Japan Agency for Medical Research and Development (AMED) (JP19dm0207080, JP21wm0525014, and JP24wm0625117 to TM), Japan Science and Technology Agency (JST) (JPMJPR16F9 to TM and JPMJFR231M to MU), Human Frontier Science Program (CDA00043/2019-C to TM), Kowa Life Science Foundation (MU), Narishige Neuroscience Research Foundation (MU), Uehara Memorial Foundation (TM), TORAY Science Foundation (TM), Takeda Science Foundation (TM) and Brain Science Foundation (TM).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, M., Nakatsukasa, E., Natsume, R., Hamada, S., Sakimura, K., Watabe, A. M., et al. (2023). A novel technique for large-fragment knock-in animal production without ex vivo handling of zygotes. Sci. Rep. 13:2245. doi: 10.1038/s41598-023-29468-1
Bavelier, D., Levi, D. M., Li, R. W., Dan, Y., and Hensch, T. K. (2010). Removing brakes on adult brain plasticity: from molecular to behavioral interventions. J. Neurosci. 30, 14964–14971. doi: 10.1523/JNEUROSCI.4812-10.2010
Benarroch, E. E. (2018). Glutamatergic synaptic plasticity and dysfunction in Alzheimer disease. Neurology 91, 125–132. doi: 10.1212/WNL.0000000000005807
Bird, R. E., Hardman, K. D., Jacobson, J. W., Johnson, S., Kaufman, B. M., Lee, S. M., et al. (1988). Single-chain antigen-binding proteins. Science 242, 423–426. doi: 10.1126/science.3140379
Bjorklund, D. F. (2022). Children's evolved learning abilities and their implications for education. Educ. Psychol. Rev. 34, 2243–2273. doi: 10.1007/s10648-022-09688-z
Bulovaite, E., Qiu, Z., Kratschke, M., Zgraj, A., Fricker, D. G., Tuck, E. J., et al. (2022). A brain atlas of synapse protein lifetime across the mouse lifespan. Neuron 110:e4058, 4057–4073.e8. doi: 10.1016/j.neuron.2022.09.009
Cizeron, M., Qiu, Z., Koniaris, B., Gokhale, R., Komiyama, N. H., Fransen, E., et al. (2020). A brainwide atlas of synapses across the mouse life span. Science 369, 270–275. doi: 10.1126/science.aba3163
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Danner, E., Lebedin, M., de la Rosa, K., and Kühn, R. (2021). A homology independent sequence replacement strategy in human cells using a CRISPR nuclease. Open Biol. 11:200283. doi: 10.1098/rsob.200283
DeFelipe, J. (2010). From the connectome to the Synaptome: an epic love story. Science 330, 1198–1201. doi: 10.1126/science.1193378
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi: 10.1038/nbt.3437
Dong, J.-X., Lee, Y., Kirmiz, M., Palacio, S., Dumitras, C., Moreno, C. M., et al. (2019). A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. eLife 8:e48750. doi: 10.7554/eLife.48750
Doudna, J. A., and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Droogers, W. J., Willems, J., MacGillavry, H. D., and Jong, A. P. H. D. (2022). Duplex labeling and manipulation of neuronal proteins using sequential CRISPR/Cas9 gene editing. eNeuro 9, ENEURO.0056–ENEU22.2022. doi: 10.1523/ENEURO.0056-22.2022
Espinosa, J. S., and Stryker, M. P. (2012). Development and plasticity of the primary visual cortex. Neuron 75, 230–249. doi: 10.1016/j.neuron.2012.06.009
Fang, H., Bygrave, A. M., Roth, R. H., Johnson, R. C., and Huganir, R. L. (2021). An optimized CRISPR/Cas9 approach for precise genome editing in neurons. eLife 10. doi: 10.7554/eLife.65202
Filippo, J. S., Sung, P., and Klein, H. (2008). Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257. doi: 10.1146/annurev.biochem.77.061306.125255
Fortin, D. A., Tillo, S. E., Yang, G., Rah, J.-C., Melander, J. B., Bai, S., et al. (2014). Live imaging of endogenous PSD-95 using ENABLED: A conditional strategy to fluorescently label endogenous proteins. J. Neurosci. 34, 16698–16712. doi: 10.1523/JNEUROSCI.3888-14.2014
Fukata, Y., Dimitrov, A., Boncompain, G., Vielemeyer, O., Perez, F., and Fukata, M. (2013). Local palmitoylation cycles define activity-regulated postsynaptic subdomains. J. Cell Biol. 202, 145–161. doi: 10.1083/jcb.201302071
Gao, Y., Hisey, E., Bradshaw, T. W. A., Erata, E., Brown, W. E., Courtland, J. L., et al. (2019). Plug-and-play protein modification using homology-independent universal genome engineering. Neuron 103, 583–597.e8. doi: 10.1016/j.neuron.2019.05.047
Gross, G. G., Junge, J. A., Mora, R. J., Kwon, H.-B., Olson, C. A., Takahashi, T. T., et al. (2013). Recombinant probes for visualizing endogenous synaptic proteins in living neurons. Neuron 78, 971–985. doi: 10.1016/j.neuron.2013.04.017
Gurumurthy, C. B., Takahashi, G., Wada, K., Miura, H., Sato, M., and Ohtsuka, M. (2016). GONAD: A novel CRISPR/Cas9 genome editing method that does not require ex vivo handling of embryos. Curr. Protoc. Hum. Genet. 88, 15.8.1–15.8.12. doi: 10.1002/0471142905.hg1508s88
Hamers-Casterman, C., Atarhouch, T., Muyldermans, S., Robinson, G., Hamers, C., Songa, E. B., et al. (1993). Naturally occurring antibodies devoid of light chains. Nature 363, 446–448. doi: 10.1038/363446a0
Hayano, Y., Ishino, Y., Hyun, J. H., Orozco, C. G., Steinecke, A., Potts, E., et al. (2021). IgSF11 homophilic adhesion proteins promote layer-specific synaptic assembly of the cortical interneuron subtype. Sci. Adv. 7:eabf1600. doi: 10.1126/sciadv.abf1600
Hensch, T. K. (2004). Critical period regulation. Annu. Rev. Neurosci. 27, 549–579. doi: 10.1146/annurev.neuro.27.070203.144327
Hensch, T. K. (2005). Critical period mechanisms in developing visual cortex. Curr. Top. Dev. Biol. 69, 215–237. doi: 10.1016/S0070-2153(05)69008-4
Hoelzel, C. A., and Zhang, X. (2020). Visualizing and manipulating biological processes by using HaloTag and SNAP-tag technologies. Chembiochem 21, 1935–1946. doi: 10.1002/cbic.202000037
Huang, X., Stodieck, S. K., Goetze, B., Cui, L., Wong, M. H., Wenzel, C., et al. (2015). Progressive maturation of silent synapses governs the duration of a critical period. Proc. Natl. Acad. Sci. USA 112, E3131–E3140. doi: 10.1073/pnas.1506488112
Hubel, D. H., and Wiesel, T. N. (1970). The period of susceptibility to the physiological effects of unilateral eye closure in kittens. J. Physiol. 206, 419–436. doi: 10.1113/jphysiol.1970.sp009022
Hübener, M., and Bonhoeffer, T. (2014). Neuronal plasticity: beyond the critical period. Cell 159, 727–737. doi: 10.1016/j.cell.2014.10.035
Huston, J. S., Levinson, D., Mudgett-Hunter, M., Tai, M. S., Novotný, J., Margolies, M. N., et al. (1988). Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. 85, 5879–5883. doi: 10.1073/pnas.85.16.5879
Iascone, D. M., Li, Y., Sümbül, U., Doron, M., Chen, H., Andreu, V., et al. (2020). Whole-neuron synaptic mapping reveals spatially precise excitatory/inhibitory balance limiting dendritic and somatic spiking. Neuron 106, 566–578.e8. doi: 10.1016/j.neuron.2020.02.015
Jamjoom, A. A. B., Rhodes, J., Andrews, P. J. D., and Grant, S. G. N. (2021). The synapse in traumatic brain injury. Brain 144, 18–31. doi: 10.1093/brain/awaa321
Jayathilaka, K., Sheridan, S. D., Bold, T. D., Bochenska, K., Logan, H. L., Weichselbaum, R. R., et al. (2008). A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. 105, 15848–15853. doi: 10.1073/pnas.0808046105
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kabayama, H., Takeuchi, M., Tokushige, N., Muramatsu, S.-i., Kabayama, M., Fukuda, M., et al. (2020). An ultra-stable cytoplasmic antibody engineered for in vivo applications. Nat. Commun. 11:336. doi: 10.1038/s41467-019-13654-9
Karatan, E., Merguerian, M., Han, Z., Scholle, M. D., Koide, S., and Kay, B. K. (2004). Molecular recognition properties of FN3 Monobodies that bind the Src SH3 domain. Chem. Biol. 11, 835–844. doi: 10.1016/j.chembiol.2004.04.009
Kawashima, T., Kitamura, K., Suzuki, K., Nonaka, M., Kamijo, S., Takemoto-Kimura, S., et al. (2013). Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods 10, 889–895. doi: 10.1038/nmeth.2559
Kilisch, M., Gere-Becker, M., Wüstefeld, L., Bonnas, C., Crauel, A., Mechmershausen, M., et al. (2023). Simple and highly efficient detection of PSD95 using a Nanobody and its recombinant heavy-chain antibody derivatives. Int. J. Mol. Sci. 24:7294. doi: 10.3390/ijms24087294
Kim, J., McFee, M., Fang, Q., Abdin, O., and Kim, P. M. (2023). Computational and artificial intelligence-based methods for antibody development. Trends Pharmacol. Sci. 44, 175–189. doi: 10.1016/j.tips.2022.12.005
Koide, A., Bailey, C. W., Huang, X., and Koide, S. (1998). The fibronectin type III domain as a scaffold for novel binding proteins. J. Mol. Biol. 284, 1141–1151. doi: 10.1006/jmbi.1998.2238
Konno, K., Yamasaki, M., Miyazaki, T., and Watanabe, M. (2023). Glyoxal fixation: an approach to solve immunohistochemical problem in neuroscience research. Sci. Adv. 9:eadf7084. doi: 10.1126/sciadv.adf7084
Koopmans, F., van Nierop, P., Andres-Alonso, M., Byrnes, A., Cijsouw, T., Coba, M. P., et al. (2019). SynGO: an evidence-based, expert-curated Knowledge Base for the synapse. Neuron 103, 217–234.e4. doi: 10.1016/j.neuron.2019.05.002
Koukaroudi, D., Qiu, Z., Fransen, E., Gokhale, R., Bulovaite, E., Komiyama, N. H., et al. (2024). Sleep maintains excitatory synapse diversity in the cortex and hippocampus. Curr. Biol. 34:e3835, 3836–3843.e5. doi: 10.1016/j.cub.2024.07.032
Kurihara, T., Kouyama-Suzuki, E., Satoga, M., Li, X., Badawi, M., Thiha,, et al. (2020). DNA repair protein RAD51 enhances the CRISPR/Cas9-mediated knock-in efficiency in brain neurons. Biochem. Biophys. Res. Commun. 524, 621–628. doi: 10.1016/j.bbrc.2020.01.132
Lieber, M. R. (2010). The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211. doi: 10.1146/annurev.biochem.052308.093131
Majewska, A., and Sur, M. (2003). Motility of dendritic spines in visual cortex in vivo: changes during the critical period and effects of visual deprivation. Proc. Natl. Acad. Sci. 100, 16024–16029. doi: 10.1073/pnas.2636949100
Marchionni, I., Kasap, Z., Mozrzymas, J. W., Sieghart, W., Cherubini, E., and Zacchi, P. (2009). New insights on the role of gephyrin in regulating both phasic and tonic GABAergic inhibition in rat hippocampal neurons in culture. Neuroscience 164, 552–562. doi: 10.1016/j.neuroscience.2009.07.063
Meyerink, B. L., Kc, P., Tiwari, N. K., Kittock, C. M., Klein, A., Evans, C. M., et al. (2022). Breasi-CRISPR: an efficient genome-editing method to interrogate protein localization and protein-protein interactions in the embryonic mouse cortex. Development 149. doi: 10.1242/dev.200616
Mikuni, T., Nishiyama, J., Sun, Y., Kamasawa, N., and Yasuda, R. (2016). High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell 165, 1803–1817. doi: 10.1016/j.cell.2016.04.044
Minatohara, K., Akiyoshi, M., and Okuno, H. (2016). Role of immediate-early genes in synaptic plasticity and neuronal ensembles underlying the memory trace. Front. Mol. Neurosci. 8:78. doi: 10.3389/fnmol.2015.00078
Moeyaert, B., Holt, G., Madangopal, R., Perez-Alvarez, A., Fearey, B. C., Trojanowski, N. F., et al. (2018). Improved methods for marking active neuron populations. Nat. Commun. 9:4440. doi: 10.1038/s41467-018-06935-2
Mora, R. J., Roberts, R. W., and Arnold, D. B. (2013). Recombinant probes reveal dynamic localization of CaMKIIα within Somata of cortical neurons. J. Neurosci. 33, 14579–14590. doi: 10.1523/JNEUROSCI.2108-13.2013
Mrsic-Flogel, T. D., Hofer, S. B., Ohki, K., Reid, R. C., Bonhoeffer, T., and Hubener, M. (2007). Homeostatic regulation of eye-specific responses in visual cortex during ocular dominance plasticity. Neuron 54, 961–972. doi: 10.1016/j.neuron.2007.05.028
Nishiyama, J., Mikuni, T., and Yasuda, R. (2017). Virus-mediated genome editing via homology-directed repair in mitotic and Postmitotic cells in mammalian brain. Neuron 96, 755–768.e5. doi: 10.1016/j.neuron.2017.10.004
Qin, L., Liu, Z., Guo, S., Han, Y., Wang, X., Ren, W., et al. (2024). Astrocytic Neuroligin-3 influences gene expression and social behavior, but is dispensable for synapse number. Mol. Psychiatry 1-13. doi: 10.1038/s41380-024-02659-6
Queiroz Zetune Villa Real, K., Mougios, N., Rehm, R., Sograte-Idrissi, S., Albert, L., Rahimi, A. M., et al. (2023). A versatile Synaptotagmin-1 Nanobody provides perturbation-free live synaptic imaging and low linkage-error in super-resolution microscopy. Small Methods 7:218. doi: 10.1002/smtd.202300218
Ran, F. A., Hsu, P. D., Wright, J., Agarwala, V., Scott, D. A., and Zhang, F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308. doi: 10.1038/nprot.2013.143
Ranson, A., Cheetham, C. E., Fox, K., and Sengpiel, F. (2012). Homeostatic plasticity mechanisms are required for juvenile, but not adult, ocular dominance plasticity. Proc. Natl. Acad. Sci. USA 109, 1311–1316. doi: 10.1073/pnas.1112204109
Rimbault, C., Breillat, C., Compans, B., Toulmé, E., Vicente, F. N., Fernandez-Monreal, M., et al. (2024). Engineering paralog-specific PSD-95 recombinant binders as minimally interfering multimodal probes for advanced imaging techniques. eLife 13:e69620. doi: 10.7554/eLife.69620
Slaymaker, I. M., Gao, L., Zetsche, B., Scott, D. A., Yan, W. X., and Zhang, F. (2016). Rationally engineered Cas9 nucleases with improved specificity. Science 351, 84–88. doi: 10.1126/science.aad5227
Son, J.-H., Keefe, M. D., Stevenson, T. J., Barrios, J. P., Anjewierden, S., Newton, J. B., et al. (2016). Transgenic FingRs for live mapping of synaptic dynamics in genetically-defined neurons. Sci. Rep. 6:18734. doi: 10.1038/srep18734
Sorensen, A. T., Cooper, Y. A., Baratta, M. V., Weng, F. J., Zhang, Y., Ramamoorthi, K., et al. (2016). A robust activity marking system for exploring active neuronal ensembles. eLife 5. doi: 10.7554/eLife.13918
Spence, E. F., Dube, S., Uezu, A., Locke, M., Soderblom, E. J., and Soderling, S. H. (2019). In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton-mediated synaptic maturation. Nat. Commun. 10:386. doi: 10.1038/s41467-019-08288-w
Sun, Y. J., Espinosa, J. S., Hoseini, M. S., and Stryker, M. P. (2019). Experience-dependent structural plasticity at pre- and postsynaptic sites of layer 2/3 cells in developing visual cortex. Proc. Natl. Acad. Sci. USA 116, 21812–21820. doi: 10.1073/pnas.1914661116
Suzuki, K., Tsunekawa, Y., Hernandez-Benitez, R., Wu, J., Zhu, J., Kim, E. J., et al. (2016). In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144–149. doi: 10.1038/nature20565
Suzuki, K., Yamamoto, M., Hernandez-Benitez, R., Li, Z., Wei, C., Soligalla, R. D., et al. (2019). Precise in vivo genome editing via single homology arm donor mediated intron-targeting gene integration for genetic disease correction. Cell Res. 29, 804–819. doi: 10.1038/s41422-019-0213-0
Tomas-Roca, L., Qiu, Z., Fransén, E., Gokhale, R., Bulovaite, E., Price, D. J., et al. (2022). Developmental disruption and restoration of brain synaptome architecture in the murine Pax6 neurodevelopmental disease model. Nat. Commun. 13:6836. doi: 10.1038/s41467-022-34131-w
Trimmer, J. S. (2022). Genetically encoded INTRABODIES as high-precision tools to visualize and manipulate neuronal function. Semin. Cell Dev. Biol. 126, 117–124. doi: 10.1016/j.semcdb.2021.11.004
Tsunekawa, Y., Terhune, R. K., Fujita, I., Shitamukai, A., Suetsugu, T., and Matsuzaki, F. (2016). Developing ade novotargeted knock-in method based onin uteroelectroporation into the mammalian brain. Development 143, 3216–3222. doi: 10.1242/dev.136325
Uemura, T., Mori, T., Kurihara, T., Kawase, S., Koike, R., Satoga, M., et al. (2016). Fluorescent protein tagging of endogenous protein in brain neurons using CRISPR/Cas9-mediated knock-in and in utero electroporation techniques. Sci. Rep. 6:35861. doi: 10.1038/srep35861
Vakulskas, C. A., Dever, D. P., Rettig, G. R., Turk, R., Jacobi, A. M., Collingwood, M. A., et al. (2018). A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 24, 1216–1224. doi: 10.1038/s41591-018-0137-0
van Oostrum, M., Blok, T. M., Giandomenico, S. L., Tom Dieck, S., Tushev, G., Furst, N., et al. (2023). The proteomic landscape of synaptic diversity across brain regions and cell types. Cell 186:e5423, 5411–5427.e23. doi: 10.1016/j.cell.2023.09.028
Watanabe, M., Fukaya, M., Sakimura, K., Manabe, T., Mishina, M., and Inoue, Y. (1998). Selective scarcity of NMDA receptor channel subunits in the stratum lucidum (mossy fibre-recipient layer) of the mouse hippocampal CA3 subfield. Eur. J. Neurosci. 10, 478–487. doi: 10.1046/j.1460-9568.1998.00063.x
Willems, J., de Jong, A. P. H., Scheefhals, N., Mertens, E., Catsburg, L. A. E., Poorthuis, R. B., et al. (2020). ORANGE: A CRISPR/Cas9-based genome editing toolbox for epitope tagging of endogenous proteins in neurons. PLoS Biol. 18:e3000665. doi: 10.1371/journal.pbio.3000665
Xiang, X., Corsi, G. I., Anthon, C., Qu, K., Pan, X., Liang, X., et al. (2021). Enhancing CRISPR-Cas9 gRNA efficiency prediction by data integration and deep learning. Nat. Commun. 12:3238. doi: 10.1038/s41467-021-23576-0
Yau, C. N., Lai, H. M., Lee, K., Kwok, A. J., Huang, J., and Ko, H. (2023). Principles of deep immunohistochemistry for 3D histology. Cell Rep Methods 3:100458. doi: 10.1016/j.crmeth.2023.100458
Yoon, B.-J., Smith, G. B., Heynen, A. J., Neve, R. L., and Bear, M. F. (2009). Essential role for a long-term depression mechanism in ocular dominance plasticity. Proc. Natl. Acad. Sci. 106, 9860–9865. doi: 10.1073/pnas.0901305106
Zhong, H., Ceballos, C. C., Massengill, C. I., Muniak, M. A., Ma, L., Qin, M., et al. (2021). High-fidelity, efficient, and reversible labeling of endogenous proteins using CRISPR-based designer exon insertion. eLife 10:e64911. doi: 10.7554/eLife.64911
Keywords: endogenous proteins, synapse, synaptome, single cell, critical period plasticity, intrabody, CRISPR/Cas9, genome editing
Citation: Uchigashima M and Mikuni T (2024) Single-cell synaptome mapping: its technical basis and applications in critical period plasticity research. Front. Neural Circuits. 18:1523614. doi: 10.3389/fncir.2024.1523614
Edited by:
Takuya Takahashi, Yokohama City University, JapanReviewed by:
Minjie Shen, Fudan University, ChinaCopyright © 2024 Uchigashima and Mikuni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Motokazu Uchigashima, dWNoaWdhc2hpbWFAYnJpLm5paWdhdGEtdS5hYy5qcA==; Takayasu Mikuni, dG1pa3VuaUBicmkubmlpZ2F0YS11LmFjLmpw
 Motokazu Uchigashima
Motokazu Uchigashima Takayasu Mikuni
Takayasu Mikuni