
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
EDITORIAL article
Front. Netw. Physiol. , 13 December 2022
Sec. Systems Interactions and Organ Networks
Volume 2 - 2022 | https://doi.org/10.3389/fnetp.2022.1076702
This article is part of the Research Topic Coupling in Biological Systems: Definitions, Mechanisms, and Implications View all 6 articles
Editorial on the Research Topic
Coupling in biological systems: Definitions, mechanisms, and implications
Biological systems exhibit an enormous complexity. Their temporal evolution ubiquitously depends on non-linear interactions between non-identical, heterogeneous entities such as molecules, cells, tissues, organs, or organisms. As a result, the observed dynamics at the ensemble level show emergent phenomena that can only occur at the macroscopic scale and cannot be understood solely from the intrinsic behavior of its individual constituents, limiting the success of traditional reductionist approaches (Wolkenhauer and Green, 2013). The entities involved can exist in different positions of space and time and may span across various scales.
The process of interaction or information exchange between two or more entities in a given physical, biological or chemical system is often referred to as “coupling”. Using mathematical parlance, the dynamical evolution of a given part in a coupled system depends on the present or former state of other parts in the overall system.
In multicellular systems of neurons, the emergent dynamics is the neural network computation (Figure 1 upper right), governed by schemes of synaptic coupling (i.e., connectome). Likewise, the macroscopic output of physiology is a result of multi-organ coupling throughout the body (Figure 1 middle). At the organismic level, coupling between the environment and the body enables adaptation (Figure 1 left). On the lowest end of the biological hierarchy, coupling among molecular components creates feedback networks within a single cell (Figure 1 bottom right).
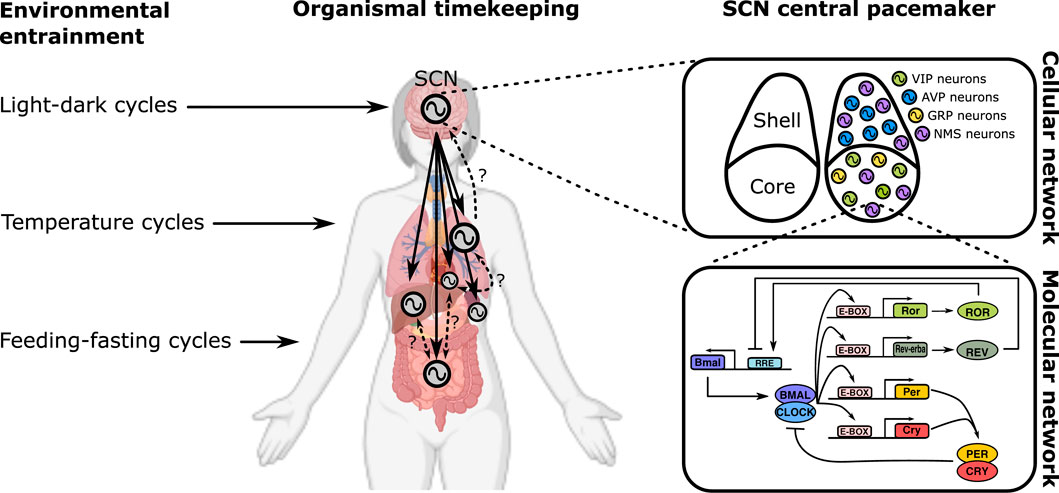
FIGURE 1. Multiscale organization of circadian clock networks. Circadian clocks exist in all scales of biology, and so does the coupling among them. The scale spans from the whole organism (far left) to molecules within a single cell (bottom right).
In this Research Topic, we are particularly interested in coupled biological oscillators, i.e., systems composed of constituents that show rhythmically recurring patterns. Circadian oscillation is a ubiquitous biological phenomenon well suited for studying coupling across spatiotemporal scales. These self-sustained rhythms, with a period of approximately 1 day, are maintained in neuronal and non-neuronal systems at both cellular and organ levels.
An organism’s circadian rhythm must synchronize with the daily cycle most strongly defined by ambient light (called Zeitgeber, or “time-giver”). This uni-directional coupling between a Zeitgeber and a forced pacemaker is the simplest form of coupling, yet it can yield an astonishing variety of dynamical phenomena (Heltberg et al., 2021), in particular in response to seasonal variations of day length and luminosity (Schmal et al., 2020; Burt et al., 2021). Healy et al. extend the concept of Zeitgeber to include non-photic cues such as sleep, feeding-fasting cycles, or physical activity. A host of molecular pathways are suggested, which can independently and/or synergistically entrain the clock through different molecular components such as PER1/2, CRY1/2, BMAL1/CLOCK, and DBP. Grabe et al. mathematically explore this in a two-oscillator system simultaneously entrained by photic and non-photic Zeitgebers. The molecular circadian clock network is composed of two main loops of transcriptional-translational feedback: One centers on transcriptional regulation of E-box (controls, notably, Per/Cry) and the other on RRE (drives transcription of Bmal1) elements. Mammals have a central circadian clock that resides in the hypothalamic suprachiasmatic nucleus (SCN), which receives light signals directly from the retina. Using an evolutionary game theoretic framework, Spencer et al. seek for coupling topologies of the SCN that sustain circadian synchronization at minimal “cost”. The evolutionary mechanism can drive the SCN network to adopt sparse coupling against the metabolically expensive all-to-all coupling. Gu et al. consider the heterogeneous structure of the SCN network that can be divided into a dorsomedial (DM; shell) and a ventrolateral (VL; core) subregion. The authors suggest the possibility that the sparse network of the SCN can attain synchronization through small-world/scale-free coupling. The brain contains several circadian clock loci outside the SCN. Chrobok et al. discuss coupling in one stream of these clocks along the subcortical visual system (SVS). The SVS exhibits various timescales of neural firing, and circadian coupling is thought to enable multiplexing different frequency bands.
The circadian system provides an ideal context to study coupling in biological systems, which maintain oscillations in various scales of space and time. Although coupling is often a conceptualization of indirect interactions that simplifies a series of underlying processes, its consequence can be directly explored through mathematical modeling to produce experimentally testable predictions. Mathematical modeling can also provide a platform to classify the strength, directionality, and polarity of coupling in a wide range of spatiotemporal dynamics of biology while being neutral to exact details.
CS and JM drafted the manuscript. CS created the figure and JM revised it. All authors reviewed and agreed on the manuscript.
CS acknowledges support from the Deutsche Forschungsgemeinschaft (DFG) through grant SCHM3362/2-1—Project-ID 414704559. JM is financially supported by the Higher Education Sprout Project by the Ministry of Education (MOE) and in part by National Science and Technology Council (NSTC) in Taiwan under grants 110-2311-B-038-003, 110-2314-B-038-162, and 111-2314-B-038-008.
We thank Anna-Marie Finger for the initial arrangements of the Research Topic.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Burt, P., Grabe, S., Madeti, C., Upadhyay, A., Merrow, M., Roenneberg, T., et al. (2021). Principles underlying the complex dynamics of temperature entrainment by a circadian clock. iScience 24 (11), 103370. doi:10.1016/j.isci.2021.103370
Heltberg, M. L., Krishna, S., Kadanoff, L. P., and Jensen, M. H. (2021). A tale of two rhythms: locked clocks and chaos in biology. Cell Syst. 12 (4), 291–303. doi:10.1016/j.cels.2021.03.003
Schmal, C., Herzel, H., and Myung, J. (2020). Clocks in the wild: entrainment to natural light. Front. Physiol. 11, 272. doi:10.3389/fphys.2020.00272
Keywords: circadian clock, coupling, biological network, entrainment, zeitgeber, suprachiasmatic nucleus (SCN)
Citation: Schmal C, Hong S, Tokuda IT and Myung J (2022) Editorial: Coupling in biological systems: Definitions, mechanisms, and implications. Front. Netw. Physiol. 2:1076702. doi: 10.3389/fnetp.2022.1076702
Received: 21 October 2022; Accepted: 30 November 2022;
Published: 13 December 2022.
Edited and reviewed by:
Plamen Ch. Ivanov, Boston University, United StatesCopyright © 2022 Schmal, Hong, Tokuda and Myung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christoph Schmal, Y2hyaXN0b3BoLnNjaG1hbEBodS1iZXJsaW4uZGU=; Jihwan Myung, amlod2FuQHRtdS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.