- NMR Laboratory, Department of Chemistry, Federal University of Paraná - UFPR, Curitiba, Brazil
Introduction: Some papers describe the presence of creatine in plants, based on a singlet signal at 3.02–3.05 ppm in the 1H NMR spectra. Although is there creatine in plants? Therefore, to answer this question, a comprehensive NMR investigation has been performed aiming the unambiguous assignment of the compound responsible for that signal.
Objective: Determine whether the compound behind the signal at 3.05 ppm is truly creatine or if it was just a misassignment, instead.
Methods: Samples of leaves and cherries from Eugenia uniflora in their natural swollen state were submitted to HR-MAS NMR analysis.
Results: It was found that the signal at 3.05 ppm was misassigned to creatine. The exhaustive NMR investigation revealed that the signal is related to the amino acid 4-hydroxy-N-methyl proline, instead.
Conclusion: The comprehensive NMR investigation revealed that there is no creatine in plants, it was just a misassignment.
Introduction
High-Resolution Magic-Angle-Spinning (HR-MAS) is a versatile technique that allow the acquisition of Nuclear Magnetic Resonance (NMR) data directly from semisolid samples, such as vegetable (e.g., leaves, flowers, fruits, etc.) and animal tissues, in their natural swollen state, without the need of any sample pre-treatment, being a valuable tool for the rapid investigation of plant chemical composition (Broberg and Kenne, 2000; Kelleher et al., 2006; Bharti et al., 2011; Alam and Jenkins, 2012; Choze et al., 2013; Farooq et al., 2013; André et al., 2014; Santos et al., 2015; Flores et al., 2018; Santos et al., 2018; Ali et al., 2020).
In a previous work, while investigating the chemical composition of Berberis laurina (Berberidaceae) by means of HR-MAS NMR spectroscopy, it was faced up a straight singlet signal at 3.02 ppm (Ali et al., 2020). This signal has been described only in NMR studies of Eugenia uniflora (Myrtaceae) (Iqbal et al., 2013; Flores et al., 2018), which is a native plant in tropical South America’s Atlantic coast countries, including Suriname, Guyana, French Guyana, Brazil, Uruguay, Paraguay, and Northern Argentina, known as Surinam cherry or Brazilian cherry. While it is currently cultivated and naturalized worldwide, mainly in tropical and subtropical regions.
Its edible fruits are botanical cherries that look like small orange-red pumpkins (Supplementary Figure S1). Known for their exotic flavor, which ranges from sweet to sour, they are primarily consumed fresh, as well as in juice, frozen pulp, jams, jellies, and other forms. Eugenia uniflora also has several significant pharmacological properties being widely used in folk medicine. Several diseases and disorders such as bronchitis, cold, cough, gout, sore throat, hypertension, headaches, influenza, hepatic diseases, painful urination, rheumatism, diarrhea, fever, stomach diseases, and other gastro-intestinal disorders are treated based on its extracts. Likewise, it is used in the treatment of obesity, and diabetes as well as to stimulate menstrual flow. Also, it was described to have diuretic, insect repelling, digestive, eupeptic, and carminative properties. Its tea is used to facilitate the process of childbirth, while its hot water extract of the fresh leaves and unripe fruits are used to treat malaria and fever (Schapoval et al., 1994; Consolini et al., 1999; Begossi et al., 2002; Bagetti et al., 2011; Celli et al., 2011; Moura et al., 2018). Besides its innumerable therapeutic properties, E. uniflora is used by the cosmetic industries (Melo et al., 2007) as well as an important colonizing species in restoration of disturbed forest areas, and to provide food for wildlife (Rodrigues and Nave, 2000; Botrel et al., 2002; Margis et al., 2002; Bianchini et al., 2003; Pinto et al., 2005).
To the best of our knowledge, in many papers regarding plant tissues investigation through HR-MAS NMR technique, any singlet signal has been described around 3.02–3.05 ppm, except for E. uniflora (Myrtaceae) (Iqbal et al., 2013; Flores et al., 2018) and B. laurina (Berberidaceae) (Ali et al., 2020). This signal has been assigned to the N-CH3 group of the amino acid creatine (Iqbal et al., 2013; Ali et al., 2020). However, this compound is synthesized in the liver, pancreas, and kidney from the transamination of the amino acids arginine, glycine, and methionine. Therefore, it is not produced by the kingdom plantae. There are only a few reports in which creatine was described in kingdom plantae such as in Phaseolus mungo (Fabaceae), Lens culinaris (Fabaceae), E. uniflora (Myrtaceae) (Azmat et al., 2009; Iqbal et al., 2013), and Tussilago farfara (Asteraceae) (Zhi et al., 2012).
Considering the biosynthesis of creatine, this compound might not have been found in the kingdom plantae, thus there may have a misassignment for the signal around 3.02–3.05 ppm. Therefore, in this work, NMR spectroscopy was exhaustively used to identify, in an unequivocal way, the true compound which gives rise to the signal at 3.02–3.05 ppm found in E. uniflora.
Materials and methods
Plant material
Leaves and cherries from E. uniflora L. (Myrtaceae) were collected in November 2021 from a healthy tree specimen in the backyard of the Department of Chemistry at Polytechnic Centre campus of the Federal University of Paraná, Curitiba, PR, Brazil [coordinates: 25°27′01.1″ S, 49°14′05.5″ W].
Sample preparation for NMR analysis
Fresh leaves and pitless cherries of E. uniflora were frozen in liquid nitrogen in a mortar and powdered with aid of pestle. The resulting powder (∼10.0 ± 0.5 mg) of each sample was then inserted into a 50 µL volume 4-mm zirconium oxide HR-MAS rotor, followed by the addition of 40 µL of deuterium oxide (D2O, 99.9% D, containing 0.01% w/v TMSP-d4). The sample inside the HR-MAS rotor was then homogenized with aid of a syringe needle, which also made it possible to remove air bubbles. The rotors were sealed with a small upper Kel-F spacer/Kel-F sealing screw cap to prevent the leakage of fluids due to high-spinning speed during NMR experiments, followed by a Kel-F spinning cap (Alam and Jenkins, 2012; Flores et al., 2018), and then submitted to 1H HR-MAS NMR analysis. In order to support the NMR chemical shift assignments in the HR-MAS NMR analysis, solution NMR experiments from deuterated solvent extracts of E. uniflora leaves were also performed and compared to the HR-MAS results. Therefore, each powdered plant material (i.e., 100.0 ± 1.0 mg) was submitted to extraction directly with deuterated solvent (i.e., 700 μL of D2O) in a 1.5 mL microcentrifuge tube, assisted by sonication for 10 min at 298 K (De Aquino et al., 2019). After that, samples were centrifuged at 12,000 rpm for 10 min and 600 μL of supernatant were directly transferred into a 5 mm NMR tube and submitted to solution NMR analysis.
NMR analysis
1H HR-MAS NMR spectra were acquired at 298K on a Bruker AVANCE 400 NMR spectrometer operating at 9.4 T, observing 1H and 13C nuclei at 400.13 and 100.62 MHz, respectively. The spectrometer was equipped with a 4-mm four-channel (1H/2H/13C/15N) HR-MAS probe with an actively shielded gradient field along the magic angle direction. The acquisition was performed with 5 kHz spinning speed and the spectra were acquired with 64k time-domain data points distributed over a spectral width of 20 ppm, resulting in a digital resolution of 0.24 Hz by applying single 90° excitation pulses with aid of zgpr, a water suppression pulse sequence, 1.0 s recycle delay and averaging 256 scans. The spectra were processed by applying an exponential Lorentzian multiplication with a line broadening factor of 1.0 Hz to the FID, followed by Fourier transform using a zero-filling factor of 2. Phase and baseline corrections were manually performed, and the NMR chemical shifts were referenced to TMSP-d4 signal at 0.00 ppm. Prior to the NMR analysis, the magic angle was adjusted using the 79Br signal from powdered KBr as reference. Samples were locked on the deuterium signal from D2O, and the magnetic field homogeneity was optimized for each sample. The total experiment time was 35 min, including the time required for rotor packing. Direct and long-range 1H-13C as well as 1H–1H NMR correlations experiments were acquired directly from deuterated solvent extracts on the same NMR instrument as above, although it was equipped with a 5-mm broadband solution direct detection four-channel (1H/2H/13C/31P) probe with actively shielded gradient field along z-direction, instead. The experiments were performed using standard pulse sequences from the Bruker library that include COSY, HSQC, and HMBC, and the correlation maps were processed as usual.
Results and discussion
The 1H HR-MAS NMR spectra, acquired directly from both leaves and cherries of E. uniflora (Supplementary Figure S2) in their natural swollen state, showed a remarkable singlet signal at 3.05 ppm, mainly in its leaves (Figure 1). This signal has been previously assigned to creatine while investigating some plant species (Iqbal et al., 2013; Flores et al., 2018; Ali et al., 2020). However, creatine is a common amino acid found in the kingdom Animalia, but not in kingdom Plantae. NMR singlet signals around 3.00 ppm are typical of N-CH3 groups, which might have been the reason it was assigned to creatine.
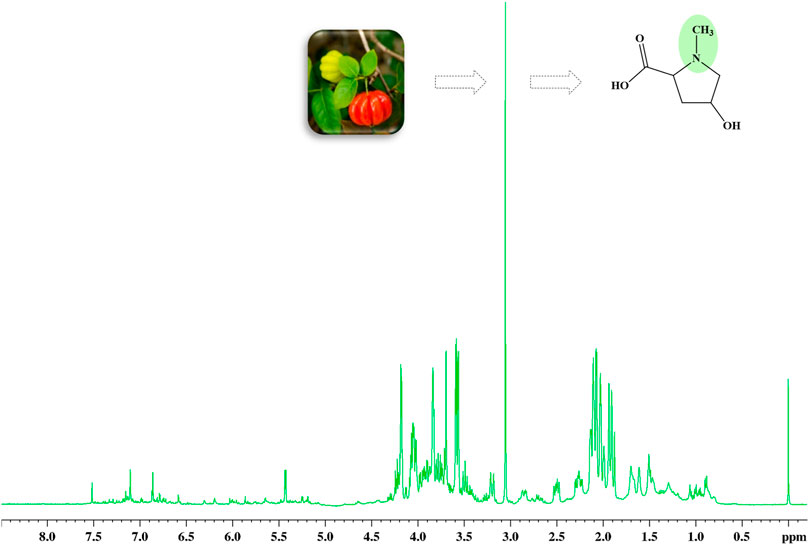
Figure 1. 1H HR-MAS NMR spectra (Natural swollen state, 400 MHz) acquired directly from the leaves of Eugenia uniflora, evidencing the singlet signal at 3.05 regarding the methyl group of 4-hydroxy-N-methyl proline (Figure 2).
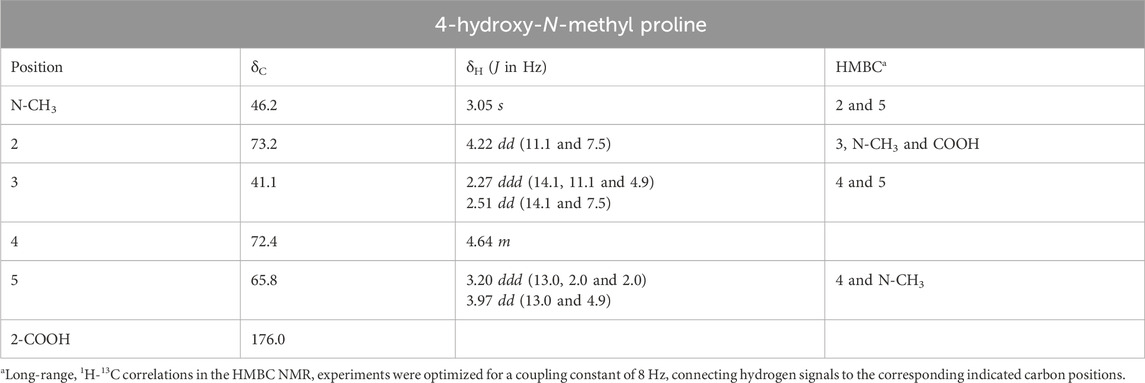
In order to confirm the identity of the compound in an unambiguous way, two-dimensional NMR experiments such as one-bond and long-range 1H-13C and 1H–1H correlation maps from HSQC, HMBC, and COSY NMR experiments were acquired from D2O extracts from both leaves and cherries of E. uniflora (Supplementary Figures S3–S5). In turn, the HSQC correlation map revealed that the hydrogen nuclei at 3.05 ppm is directly linked to a carbon at 46.2 ppm, that is typical from carbons connected to nitrogen nuclei. Moreover, the long-range 1H-13C correlation map from HMBC NMR experiment showed, for the hydrogen nuclei at 3.05 ppm, only two hydrogen-carbon correlations at 65.8 and 73.2 ppm, instead of three as usual, supporting the presence of a N-CH3 group rather than a C-CH3 group (Table 1; Supplementary Figures S3, S4).
The presence of hydrogen nuclei at 3.05 ppm connected to a carbon at 46.2 ppm is in accordance with the structure of the amino acid creatine or creatinine (Supplementary Figure S6), although the two adjacent carbons at 65.8 and 73.2 ppm do not support this hypothesis. The one-bond 1H-13C correlation map showed that there are two hydrogen nuclei linked to the carbon at 65.8 ppm at 3.20 and 3.97 ppm (i.e., a CH2 group), while for the carbon at 73.2 ppm there is only one hydrogen nucleus connect to it at 4.22 ppm (i.e., CH group). On the other hand, the hydrogen nuclei at 3.20 ppm showed long-range 1H-13C correlation with the carbons at 46.2 and 72.4 ppm, and the hydrogen nuclei at 4.22 ppm presented correlation with the carbons at 41.1, 46.2 and 176.0 ppm, revealing to be a more complex structure, which includes a carbonyl group. In addition, the one-bond 1H-13C correlation map showed that there are two hydrogen nuclei at 2.27 and 2.51 ppm connected to the carbon at 41.1 ppm, and just one hydrogen nucleus at 4.64 ppm connected to the carbon 72.4 ppm, revealing additional CH2 and CH groups in the structure (Table 1).
The comprehensive exploration of the 1H–1H correlation map from COSY NMR experiment, also allowed to establish and support a spin system consisting of the hydrogen nuclei at 4.22 (2), 2.27 and 2.51 (3), 4.64 (4), 3.20 and 3.97 ppm (5) (Figure 2; Supplementary Figure S5).
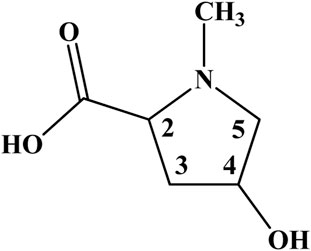
Figure 2. Chemical structure of 4-hydroxy-N-methyl proline from the leaves and cherries of Eugenia uniflora (Brazilian cherry or Surinan Cherry).
Therefore, one can conclude in an unequivocally way that, instead of creatine, the amino acid 4-hydroxy-N-methyl proline is the true compound behind the singlet signal at 3.05 in the 1H NMR spectra of E. uniflora (Figures 1, 2; Supplementary Figure S6). The misassignment may have occurred due to the hydrogen nuclei, except those for the N-CH3 group, present NMR signals in crowed regions of the spectra. In addition, they are less intense, as well as are present as multiplets, being hard to be observed.
The 4-hydroxy-N-methyl proline, one of the hydroxylated proline-derivatives, has been reported in plant species of several families including Euphorbiaceae, Fabaceae, Mytaceae, Meliaceae, Leguminosae, Asteraceae, Rutaceae, Loranthaceae, Annonaceae, Berberidaceae, Lauraceae, Sapindaceae, Rhamnaceae, Tamaricaceae, Sapotaceae (Blunden et al., 2004; Blunden et al., 2006; Jones et al., 2006; Giacometti et al., 2018), as well as in marine red algae (De Aquino et al., 2017). However, to the best of our knowledge, it has not been described in E. uniflora (Myrtaceae). Indeed, it was misassigned as creatine (Supplementary Figure S7).
This amino acid is a very important compound from a biological point of view, presenting pharmacological activities, mainly anti-inflammatory properties. In fact, the anti-inflammatory properties of several plant species are attributed to 4-hydroxy-N-methyl proline (Sciuto et al., 1983; Jones et al., 2006; Ribeiro et al., 2018). Therefore, its presence in E. uniflora as well as in B. laurina (Ali et al., 2020) supports a large number of its use in folk medicine. Furthermore, HR-MAS NMR analysis unveiled substantial amounts of 4-hydroxy-N-methyl proline in the leaves of E. uniflora. In contrast, the fruits exhibited comparatively very lower amounts of the compound (Supplementary Figure S2). This information holds meaningful value for its application in traditional medicine and the cosmetic industries.
HR-MAS NMR was also used to explore some changes in the chemical composition of the cherries during ripening process. The analysis unveiled an increase in the levels of α- and β-glucose as the fruits underwent the ripening process. Conversely, stable amounts of 4-hydroxy-N-methyl proline and malic acid were consistently observed in the cherries throughout the ripening stages (Supplementary Figure S8).
Conclusion
In conclusion, the investigation of E. uniflora leaves and cherries by means of HR-MAS NMR has brought some significant insights. The initial misassignment of the singlet signal at 3.02–3.05 ppm, assigned to creatine, has been corrected through comprehensive NMR analysis. Conversely to the initial hypothesis of creatine, the compound was unequivocally identified as 4-hydroxy-N-methyl proline. This amino acid, previously unrecognized in E. uniflora, holds biological significance, known for its pharmacological activities, particularly its anti-inflammatory properties. Its presence in significant amounts in the leaves of E. uniflora, as revealed by HR-MAS NMR, underscores its potential applications in traditional medicine and cosmetic industries. Furthermore, the investigation into the fruit’s chemical composition indicated a consistency in 4-hydroxy-N-methyl proline content during ripening process. These findings not only enhance our understanding of the chemical composition of E. uniflora but also shed light on its potential applications in medicinal and cosmetic contexts. The use of HR-MAS NMR proves to be a valuable tool in unraveling such intricate chemical composition, offering important insights into the plant’s pharmacological properties.
Data availability statement
All NMR raw data are available free of charge in the UFPR public repository under the DOI number 10.5380/bdc/93, available at 10.5380/bdc/93.
Author contributions
LR: Data curation, Formal Analysis, Investigation, Writing–original draft, Writing–review and editing. SA: Data curation, Formal Analysis, Investigation, Writing–original draft. CD’O: Data curation, Investigation, Writing–review and editing, Validation. KS: Conceptualization, Data curation, Validation, Writing–original draft, Writing–review and editing. AB: Conceptualization, Methodology, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing, Data curation.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work received funds as well as scholarships from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grants 301281/2018-1 and 407499/2016-4), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, grants 23038.006541/2019-38 and 88882.344255/2019-01), Fundação Araucária and Financiadora de Estudos e Projetos (Finep).
Acknowledgments
The authors are grateful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação Araucária and Financiadora de Estudos e Projetos (Finep) for the financial support and scholarships.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fntpr.2024.1360175/full#supplementary-material
References
Alam, T. M., and Jenkins, J. E. (2012). HR-MAS NMR spectroscopy in material science. London: InTechOpen, 28. doi:10.5772/48340
Ali, S., Badshah, G., D’Oca, C. R. M., Campos, F. R., Nagata, N., Khan, A., et al. (2020). High-Resolution Magic Angle Spinning (HR-MAS) NMR-based fingerprints determination in the medicinal plant Berberis laurina. Molecules 25, 1–21. doi:10.3390/molecules25163647
André, M., Dumez, J.-N., Rezig, L., Shintu, L., Piotto, M., and Caldarelli, S. (2014). Complete protocol for slow-spinning High-Resolution Magic-Angle Spinning NMR analysis of fragile tissues. Anal. Chem. 86, 10749–10754. doi:10.1021/ac502792u
Azmat, R., Haider, S., Nasreen, H., Aziz, F., and Riaz, M. (2009). A viable alternative mechanism in adapting the plants to heavy metal environment. Pak J. Bot. 41, 2729–2738.
Bagetti, M., Facco, E. M. P., Piccolo, J., Hirsch, G. E., Rodriguez-Amaya, D., Kobori, C. N., et al. (2011). Physicochemical characterization and antioxidant capacity of pitanga fruits (Eugenia uniflora L.). Cienc. Tecnol. Alim 31, 147–154. doi:10.1590/S0101-20612011000100021
Begossi, A., Hanazaki, N., and Tamashiro, J. (2002). Medicinal plants in the Atlantic forest (Brazil): knowledge, use, and conservation. Hum. Ecol. 30, 281–299. doi:10.1023/A:1016564217719
Bharti, S. K., Bhatia, A., Tewari, S. K., Sidhub, O. P., and Roy, R. (2011). Application of HR-MAS NMR spectroscopy for studying chemotype variations of Withania somnifera (L.) Dunal. Magn. Reson Chem. 49, 659–667. doi:10.1002/mrc.2817
Bianchini, E., Popolo, R. S., Dias, M. C., and Pimenta, J. A. (2003). Diversidade e estrutura de espécies arbóreas em área alagável do município de Londrina, sul do Brasil. Acta Bot. Bras. 17, 405–419. doi:10.1590/S0102-33062003000300008
Blunden, G., Patel, A. V., Adrian-Romero, M., and Meléndez, P. (2004). The accumulation of trans-4-hydroxy-N-methylproline and N-methylproline by some plant species. Biochem. Syst Ecol 32, 1153–1158. doi:10.1016/j.bse.2004.04.008
Blunden, G., Patel, A. V., Armstrong, N., and Romero, M. A. (2006). Distribution and chemotaxonomic significance of N-Methylprolines in selected plant families. NPC 1, 1934578X0600100–130. doi:10.1177/1934578X0600100208
Botrel, R. T., Oliveira-Filho, A. T., Rodrigues, L. A., and Curi, N. (2002). Influência do solo e topografia sobre as variações da composição florística e estrutura da comunidade arbóreo-arbustiva de uma floresta estacional semidecidual em Ingaí, MG. Rev. Bras. Bot. 25, 195–213. doi:10.1590/S0100-84042002000200008
Broberg, A., and Kenne, L. (2000). Use of high-resolution magic angle spinning nuclear magnetic resonance spectroscopy for in situ studies of low-molecular-mass compounds in red algae. Anal. Biochem. 284, 367–374. doi:10.1006/abio.2000.4722
Celli, G. B., Pereira-Netto, A. B., and Beta, T. (2011). Comparative analysis of total phenolic content, antioxidant activity, and flavonoids profile of fruits from two varieties of Brazilian cherry (Eugenia uniflora L.) throughout the fruit developmental stages. Food Res. Int. 44, 2442–2451. doi:10.1016/j.foodres.2010.12.036
Choze, R., Alcantara, G. B., Alves Filho, E. G., Silva, L. M. A., Faria, J. C., and Lião, L. M. (2013). Distinction between a transgenic and a conventional common bean genotype by 1H HR-MAS NMR. Food Chem. 141, 2841–2847. doi:10.1016/j.foodchem.2013.05.123
Consolini, A. E., Baldini, O. A. N., and Amat, A. Ì. G. (1999). Pharmacological basis for the empirical use of Eugenia uniflora L. (Myrtaceae) as antihypertensive. J. Ethnopharmacol. 66, 33–39. doi:10.1016/s0378-8741(98)00194-9
De Aquino, P. E. A., Magalhães, T. R., Nicolau, L. A. D., Leal, L. K. A. M., De Aquino, N. C., Santos, S. M., et al. (2017). The anti-inflammatory effects of N-methyl-(2S,4R)-trans-4-hydroxyl-proline from Syderoxylon obtusifolium are related to its inhibition of TNF-alpha and inflammatory enzymes. Phytomedicine 24, 14–23. doi:10.1016/j.phymed.2016.11.010
De Aquino, P. E. A., Souza, T. F. G., Santos, F. A., Viana, A. F. S. C., Louchard, B. O., Leal, L. K. A. M., et al. (2019). The wound healing property of N-Methyl-(2S,4R)-trans-4-Hydroxy-L-Proline from Sideroxylon obtusifolium is related to its anti-inflammatory and antioxidant actions. JEBIM 24, 1–11. doi:10.1177/2515690X19865166
Farooq, H., Courtier-Murias, D., Soong, R., Bermel, W., Kingery, W. M., and Simpson, A. J. (2013). HR-MAS NMR spectroscopy: a practical guide for natural samples. Curr. Org. Chem. 17, 3013–3031. doi:10.2174/13852728113179990126
Flores, I. S., Martinelli, B. C. B., Pinto, V. S., Queiroz, L. H. K., and Lião, L. M. (2018). Important issues in plant tissues analyses by HR-MAS NMR. Phytochem. Anal. 30, 5–13. doi:10.1002/pca.2785
Giacometti, J., Žauhar, G., and Žuvic, M. (2018). Optimization of ultrasonic-assisted extraction of major phenolic compounds from olive leaves (Olea europaea L.) using response surface methodology. Foods 7, 149–214. doi:10.3390/foods7090149
Iqbal, M., Mushtaq, M. Y., Ali, K., Korthout, H. A. A. J., Verpoorte, R., Cardozo, M. L., et al. (2013). NMR spectroscopy coupled with multivariate data analysis to assess antiinflammatory activities of Eugenia uniflora fruits in different developmental stages. Leiden: Faculty of Science - Leiden University, 19.
Jones, G. P., Naidu, B. P., Waisel, Y., Solomon, A., and Paleg, L. G. (2006). Occurrence and stress response of N-methylproline compounds in Tamarix species. Phytochemistry 67, 156–160. doi:10.1016/j.phytochem.2005.10.027
Kelleher, B. P., Simpson, M. J., and Simpson, A. J. (2006). Assessing the fate and transformation of plant residues in theterrestrial environment using HR-MAS NMR spectroscopy. Geochim. Cosmochim. Acta 70, 4080–4094. doi:10.1016/j.gca.2006.06.012
Margis, R., Felix, D., Caldas, F. J., Salgueiro, F., De Araujo, D. S. D., Breyne, P., et al. (2002). Genetic differentiation among three neighboring Brazil Cherry (Eugenia uniflora L.) populations within the Brazilian Atlantic rain forest. Biodivers. Conserv. 11, 149–163. doi:10.1023/A:1014028026273
Melo, R. M., Corrêa, V. F. S., Amorim, A. C. L., Miranda, A. L. P., and Rezende, C. M. (2007). Identification of impact aroma compounds in Eugenia uniflora L. (Brazilian Pitanga) leaf essential oil. J. Braz Chem. Soc. 18, 179–183. doi:10.1590/S0103-50532007000100020
Moura, G. S., Oliveira, I. J., Bonome, L. T., and Franzener, G. (2018). Eugenia uniflora L.: potential uses as a bioactive plant. Arq. Inst. Biol. 85, 1–9. doi:10.1590/1808-1657000752017
Pinto, J. R. R., Oliveira-Filho, A. T., and Hay, J. D. V. (2005). Influence of soil and topography on the composition of a tree community in a central Brazilian valley Forest. Edinb J. Bot. 62, 69–90. doi:10.1017/S0960428606000035
Ribeiro, V. P., Arruda, C., El-Salam, M. A., and Bastos, J. K. (2018). Brazilian medicinal plants with corroborated antiinflammatory activities: a review. Pharm. Biol. 56, 253–268. doi:10.1080/13880209.2018.1454480
Rodrigues, R. R., and Nave, A. G. (2000). Heterogeneidade florística das matas ciliares. São Paulo, 45–71. EDUSP.
Santos, A. D. C., Fonseca, F. A., Dutra, L. M., Santos, M. F. C., Menezes, L. R. A., Campos, F. R., et al. (2018). 1H HR-MAS NMR-based metabolomics study of different persimmon cultivars (Diospyros kaki) during fruit development. Food Chem. 239, 511–519. doi:10.1016/j.foodchem.2017.06.133
Santos, A. D. C., Fonseca, F. A., Lião, L. M., Alcantara, G. B., and Barison, A. (2015). High-resolution magic angle spinning nuclear magnetic resonance in foodstuff analysis. Trends Anal. Chem. 73, 10–18. doi:10.1016/j.trac.2015.05.003
Schapoval, E. E. S., Silveira, S. M., Miranda, M. L., Alice, C. B., and Henriques, A. T. (1994). Evaluation of some pharmacological activities of Eugenia uniflora L. J. Ethnopharmacol. 44, 137–142. doi:10.1016/0378-8741(94)01178-8
Sciuto, S., Chillemi, R., Piattelli, M., and Impellizzeri, G. (1983). The identification of 4-hydroxy-N-methylproline in the red alga, Chondria coerulescens-spectral information. Phytochemistry 22, 2311–2312. doi:10.1016/S0031-9422(00)80168-5
Keywords: NMR, HR-MAS NMR, Eugenia uniflora, 4-hydroxy-N-methyl proline, creatine, Brazilian cherry
Citation: Novais LMRd, Ali S, D’Oca CDRM, Salome KS and Barison A (2024) Is there creatine in plants? The true compound behind the 1H NMR signal at 3.05 ppm in plant extracts. Front. Nat. Produc. 3:1360175. doi: 10.3389/fntpr.2024.1360175
Received: 22 December 2023; Accepted: 22 July 2024;
Published: 16 August 2024.
Edited by:
Renata Araujo, Federal University of Rio Grande do Norte, BrazilReviewed by:
Takeshi Kodama, University of Toyama, JapanLuzineide Wanderley Tinoco, Federal University of Rio de Janeiro, Brazil
Alvicler Magalhaes, Federal University of Rio de Janeiro, Brazil
Copyright © 2024 Novais, Ali, D’Oca, Salome and Barison. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andersson Barison, YW5kZXJubXJAdWZwci5icg==
 Leice Milla Ribeiro de Novais
Leice Milla Ribeiro de Novais Sher Ali
Sher Ali Caroline Da Ros Montes D’Oca
Caroline Da Ros Montes D’Oca Kahlil Schwanka Salome
Kahlil Schwanka Salome Andersson Barison
Andersson Barison