
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nat. Prod., 12 February 2024
Sec. Natural Product Biosynthesis
Volume 3 - 2024 | https://doi.org/10.3389/fntpr.2024.1353362
Peptide natural products have a wide range of useful applications as pesticides, veterinary agents, pharmaceuticals, and bioproducts. To discover new natural products, manipulate them for analog generation, and to harness the potential of these bioactive compounds for synthetic biology, it is necessary to develop robust methods for the expression of biosynthetic genes. Cell-free synthetic biology is emerging as an important complementary approach because it is highly desirable to express protein on a more rapid timescale and does not rely upon the genetic tractability of a strain thus improving the throughput of design-build-test-learn cycles. Additionally, generating metabolites outside the cell can overcome issues such as cellular toxicity which can hamper applications like antibiotic development. In this review, we focus on the cell-free production of peptide natural products generated by non-ribosomal peptide synthetase. Nonribsomal peptides are biosynthesized by non-ribosomal peptide synthetases which are large “mega” enzymes that provide specific challenges to heterologous expression. First, we summarize NRPSs and their corresponding peptide metabolites that are expressed in cell-free systems. With that, we discuss the requirements and challenges to express such large proteins in cell-free protein synthesis as well as host machineries that have been developed for cell-free protein synthesis that could be particularly relevant to generating non-ribosomal peptide metabolites in the future. The development of cell-free systems can then be used for prototyping to accelerate efforts towards engineered biosynthesis of these complex pathways.
Peptide natural products form the basis of many important molecules including agrochemicals (Cane et al., 1998; Pereira-Dias et al., 2023), bioproducts (Tao et al., 2023), and therapeutics (Corpuz et al., 2022; Girija et al., 2022). Of these applications, the most historically significant one has been as an important source for drug leads (Atanasov et al., 2015; Harvey et al., 2015). Many peptide natural products have complex chemical scaffolds (e.g., penicillin, daptomycin, and vancomycin antibiotics) (Felnagle et al., 2008; Süssmuth and Mainz, 2017). These peptide cores are biosynthesized by two major routes: non-ribosomal peptide synthetases (NRPS) and ribosomal synthesized and post-translationally modified peptide (RiPP) biosynthetic pathways. RiPPs produce peptide precursors by utilizing the ribosome and proteinogenic amino acids. A gene cluster in RiPPs biosynthetic pathways involves a precursor peptide that is then posttranslationally modified and ultimately cleaved to form a small molecule. While RiPPs are an exciting class of natural products that have been the subject of recent synthetic biology efforts [reviewed elsewhere (Arnison et al., 2013; Zhang et al., 2018; Montalbán-López et al., 2021; Ongpipattanakul et al., 2022; Zhong et al., 2023)], this class of peptide natural product biosynthesis is beyond the scope of this review. In contrast to the ribosomal machinery, NRPSs are synthesized by a large multienzyme complex, which consists of many individual biosynthetic modules in an assembly-line process (Figure 1).
Briefly, NRPSs are comprised of modules, wherein each module consists of a set of discrete enzymatic domains that are responsible for the one cycle of the chain extension in a colinear fashion (Bozhüyük et al., 2019; Beck et al., 2020; Lu et al., 2023; Xu et al., 2023). A typical NRPS module contains a minimum of three core domains: the adenylation (A) domain, the condensation (C) domain, and the peptidyl carrier protein (PCP) otherwise known as a thiolation (T) domain. An A domain recognizes and activates an amino acid monomer as an adenylate followed by acyl transfer to a PCP (or a T) domain along with optional domains such as an epimerase (E) domain, cyclization (Cy) domain, or oxidase (Ox) domain. The activated amino acid is loaded by the PCP domain on a 4′-phosphopantetheine (4′-Ppant) arm which is then condensed via the C-domain. Chain release typically occurs by hydrolysis or macrocyclization via a thioesterase (TE) (Figure 1) (Felnagle et al., 2008; Miller and Gulick, 2016; Wenski et al., 2022) with other termination mechanisms also sometimes occurring such as reduction to the primary alcohol via a reductase. (Barajas et al., 2015; Heinemann et al., 2023). NRPSs must be activated by posttranslationally modifying the PCP domains with a phosphopantetheinyl transferase which provides a tether for the nascent peptide. Due to this colinear relationship between domain organization and metabolite structure, there has been extensive research focused on engineering pipeline of chimeric NRPS pathways including altering domain selectivity motifs (Koryakina et al., 2017; Drufva et al., 2020) or domain swapping (Khosla and Zawada, 1996; Kries et al., 2015; Sun et al., 2015; Zhang et al., 2015). For instance, existing antibiotic chemical scaffolds can be modified via engineered biosynthesis to generate analogs that have more targeted or improved activities and/or combat antibiotic-resistance.
In addition to bioengineering applications, strategies for heterologous expression are necessary to discover new peptide natural product scaffolds and to access adequate amounts for testing of bioactivity. The discovery of novel peptide natural products is hampered by low productivity or no productivity in a laboratory context (often called “cryptic” or “silent” biosynthetic gene clusters (Zarins-Tutt et al., 2016). Because not all of these bioactivities arise from organisms that are tractable to grow in the lab (Stewart, 2012) or produce natural products in high abundance, and because the structural complexity of peptide natural products means chemical synthesis is often non-trivial, the heterologous expression remains a valuable option for characterization and scaleup (Dias et al., 2012). For these reasons, dedicated campaigns have been pursued to generate complex natural products in more tractable heterologous hosts (Li et al., 2021). Heterologous hosts must be genetically manipulatable and generally contain both sufficient fluxes of metabolic precursors as well as an appropriate environment for protein folding and expression (Lawson et al., 2019). Challenges particularly associated with non-ribosomal peptide yields in cells include being compromised by the metabolic burden inhibiting host cell growth, the cytotoxicity of their products, misfolding, and the unavailability of necessary precursors in heterologous hosts (Li et al., 2018; Ji et al., 2022). As NRPSs are large, complex multi-domain proteins that are held together via flexible linkers (Farag et al., 2019), they are particularly prone to expression and solubility challenges and/or premature truncation, which is of particular importance when selecting appropriate transcriptional and translational machinery. These complexities result in an unwieldy number of parameters that can be varied when selecting an appropriate expression and screening strategy.
Cell-free protein synthesis (CFPS) system has emerged as a strategy to prototype expression as well as undergo biomanufacturing strategies that have limitations in cellular contexts for reasons like toxicity (Carlson et al., 2012; Hodgman and Jewett, 2012). CFPS systems are often divided into two main approaches, which are crude cell lysates/extracts supplemented with appropriate precursors and cofactor recycling systems and the in vitro TX-TL PURE system. The crude cell lysates are generated from cell growth and then are supplemented with cofactors and other necessary biological components for in vitro transcription and translation (TX-TL). Whereas TX-TL PURE system or PURExpress (Shimizu et al., 2001; Tuckey et al., 2014; Lavickova and Maerkl, 2019; Cui et al., 2022) is a system that has taken the minimal recombinant elements required for cell-free expression and is thus less complex than lysate-based systems. The lysate-based system has the advantage over the PURExpress system in that it allows for accessory genes (e.g., ones relevant to posttranslational modification) as well as chaperonins. However, it has the disadvantage that it is more complex and has the potential for more sources of background that can interfere with signal output and/or cofactor recycling (Wick et al., 2019; Wick and Carr, 2022). The lack of cellular survival objectives bypasses challenges with in vivo expression, and thus the protein expression process is shortened significantly (from 1 to 2 weeks in living cells to 1–2 days) (Figure 2) (Finking et al., 2002; Bogart et al., 2021; Ji et al., 2022).
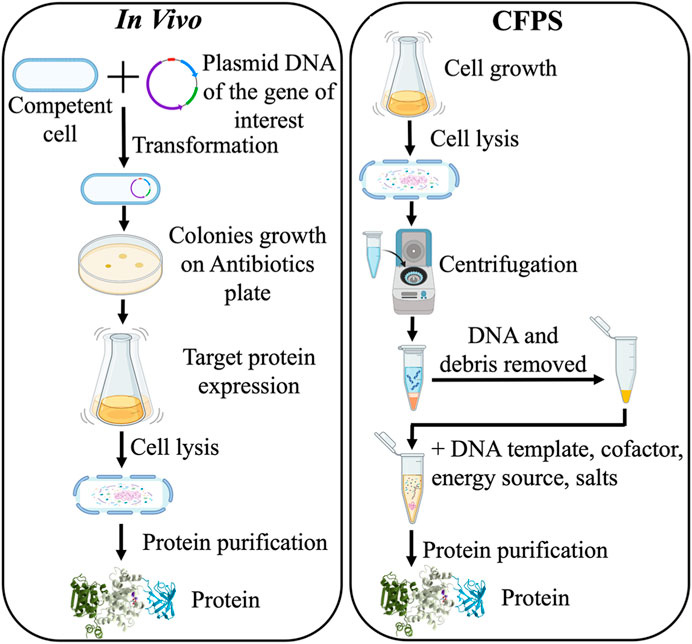
FIGURE 2. Comparison of in vivo recombinant DNA protein expression and cell-free protein synthesis (CFPS) methods.
Developing high-throughput profiling of NRPS genes in vitro platforms such as cell-free expression (CFE) systems (Li et al., 2018; Bogart et al., 2021) that include both lysate-based systems and the PURExpress system can accelerate synthetic biology research. In addition to the advantages in terms of production timeframes, cell-free affords the ability to decouple limitations that exist within cell-based systems including rapidly increasing DNA concentration as well as the introduction of non-natural components like RNA polymerases, and noncanonical amino acids, all without requiring extensive cell engineering (Finking et al., 2002; Gregorio et al., 2019; Banerjee et al., 2020). Thus, this is additionally appealing for NRPS characterization and engineering because it can accelerate design-build-test-learn (DBTL) cycles. To these ends, NRPSs have recently been expressed in both lysate-based (Goering et al., 2017; Zhuang et al., 2020; Moore et al., 2021; Dinglasan et al., 2023) and PURExpress (Siebels et al., 2020a) cell-free systems. This review highlights examples of NRPS natural products that are expressed in CFE systems in synergy with different transcriptional and translational machineries. These foundational studies can be built upon to develop more sophisticated pipelines for NRPS prototyping.
While several classes of proteins have been generated using CFPS, CFPS systems are typically optimized and developed via applying tractable and detectable reporters, predominantly GFP and other fluorescent proteins like mScarlett (Martin et al., 2018; Gregorio et al., 2019; Garenne et al., 2021). NRPSs pose more demands to a cell-free system than simple fluorescent protein reporters due to their large size, complex multidomain architecture, posttranslational modification, and cofactor requirements (e.g., ATP) and thus the efforts to express NRPS enzymes and produce non-ribosomal peptide metabolites in cell-free are somewhat limited. To date, all detection of metabolite formation from an NRPS (as opposed to the expression of the NRPS protein only) in cell-free has been performed with Escherichia coli transcriptional and translational machinery, both in lysate-based systems as well as the PURExpress system (Table 1).
Jewett and co-workers initially demonstrated the application of the lysate-based cell-free system to synthesize a cyclic dipeptide d-Phe-l-Pro diketopiperazine (DKP) (Prasad, 1995). DKP is a naturally occurring shunt product of early modules of the antibiotic gramicidin S biosynthetic pathway (Kurotsu et al., 1991). DKP is used as a model reporter because it previously has been used for a variety of domain and module experiment purposes including domain swapping studies (Kries et al., 2015), characterization of NRPS domain activity (Stanišić et al., 2021), and enzymatic redesign (Lilien et al., 2005). The antibiotic gramicidin S is produced by Brevibacillus brevis via the biosynthetic pathway of two NRPS enzymes GrsA (126 kDa, one module) and GrsB (510 kDa, four modules) (43,44). Previous reports have shown that d-Phe-l-Pro diketopiperazine can be produced via GrsA and GrsB1 in isolation (Gruenewald et al., 2004). The substrates of the first two (of five) NRPS modules of the gramicidin S pathway, GrsA, and GrsB1 (the first module of GrsB, 121 kDa), go through the spontaneous intramolecular cyclization process to form DKP (Figure 3A) (Kurotsu et al., 1991; Stachelhaus et al., 1998). GrsA converts L-phenylalanine to D-phenylalanine whereas GrsB1 creates a peptide bond between D-phenylalanine and l-proline.

FIGURE 3. Biosynthesis of non-ribosomal peptide metabolites expressed in CFE systems. (A) Gramicidin NRPS. (B) Valinomycin NRPS. (C) Blue Pigment Synthetase A (BpsA). (D) Rhabdopeptides NRPS. (E) Tyrocidine A NRPS. (F) Pyreudione NRPS. (G) Thxatomin A NRPS.
As an initial step to establish an in vitro platform for d-Phe-l-Pro DKP biosynthesis, GrsA, and GrsB1 were expressed separately in separate lysate CFE reactions (Goering et al., 2017). E. coli BL21 Star (DE3) lysate was applied towards 20-h batch reactions resulting in full-length, soluble GrsA and GrsB1 in yields of ∼106 μg/mL and ∼77 μg/mL, respectively. To ensure adequate phophopantetheinylation, the commonly used premiscuous PPTase, Sfp from Bacillus subtilis was used and Bodipy-CoA was applied as a fluorescent labeling reagent for high throughput screening of posttranslational modification. Subsequently, instead of expressing GrsA and GrsB1 separately, these two enzymes were co-expressed in a single-pot mixture to biosynthesize d-Phe-l-Pro DKP. This process allows in vitro reconstitution of the partial NRPS assembly line for in situ product formation. By using the synthetic DKP as a reference, the target product d-Phe-l-Pro DKP with the correct d-l stereochemistry was verified by the LC-MS/MS analysis. Previously, the yield of DKP from cell-based recombinant protein expression in E. coli was reported at 9 mg/L (Gruenewald et al., 2004), which is lower compared to the cell-free production system even without optimization (up to 12 mg/L). This work was the first proof of concept that an NRPS could be expressed in a cell-free system.
Subsequent to the success of expressing the dipeptide DKP in vitro (Goering et al., 2017), the Jewett laboratory sought to investigate a total biosynthesis of a natural product instead of a small model shunt product (Zhuang et al., 2020). Their selected target, valinomycin, is a 36-membered cyclododecadepsipeptide has a wide range of bioactivities including insecticidal (Heisey et al., 1988), antifungal (Park et al., 2008), antiviral (Wu et al., 2004), anticancer efficacy (Ryoo et al., 2006), and antimicrobial (Tempelaars et al., 2011) and is naturally synthesized by various Streptomyces strains (Cheng, 2006; Magarvey et al., 2006; Matter et al., 2009). The biosynthetic pathway of valinomycin consists of Vlm1 (two modules, 370 kDa) and Vlm2 (two modules, 284 kDa). In the first module, the A domain activates α-ketoisovalerate (Kiv), which is subsequently reduced to d-2-hydroxyisovalerate (d-Hiv) by a ketoreductase (KR) domain. In the second module, l-Val is activated and converted to d-Val by the A and the epimerase (E) domain. Similarly, in module 3, another KR domain reduces activated Pyruvate (Pyr) to l-Lactate; and l-Val is activated in module 4. Three tetradepsipeptide basic units of d-α-Hiv-d-Val-l-Lac-l-Val from the four modules of valinomycin synthetase oligomerized and macrolactonized by a TE domain to form valinomycin (Figure 3B) (Huguenin-Dezot et al., 2019). Valinomycin previously was expressed heterologously in E. coli in vivo by reconstitution of the NRPS genes (vlm1 and vlm2) with the yield of 1 mg/L after a systematic bioprocess optimization (Li et al., 2015a; 2014; Jaitzig et al., 2014). As an initial step, the E. coli lysate-based CFPS system was used to co-express two enzymes Vlm1 (370 kDa) and Vlm2 (284 kDa) for valinomycin synthesis by adding two individual plasmid DNA templates harboring the genes vlm1 and vlm2 in CFPS reactions full-length and soluble in The expression of both proteins Vlm1 and Vlm2 was successfully achieved separately in a single-pot cell-free reaction by using the cell extracts prepared from E. coli BL21 Star (DE3). Sfp was once again utilized for the phosphopantetheinylation to activate to the active-holo form. To avoid the necessity for purification, the sfp gene was directly added with Vlm1 and Vlm2 to co-express in a one-pot reaction. The successful co-expression had a low yield of ∼9.8 μg/L. To further improve the biosynthetic strategy, a type II thioesterase (TE-II) was employed to regenerate the activity of Vlm1 and Vlm2 which presumably edits via hydrolyzing stalled intermediates (Li et al., 2015b). As expected, TE-II improved valinomycin biosynthesis in a one-pot CFPS reaction of all four proteins (Vlm1, Vlm2, TE-II, and Sfp) with the final yield reaching ∼37 μg/L. Taken together, this work is the first example of using an E. coli-based CFPS system to express a whole natural product gene cluster of valinomycin (>19 kb), which consists of two NRPSs (Vlm1 and Vlm2), a heterologous modification protein (Sfp), and an associated editing enzyme (TE-II). Even though this work was a significant advance in the field, the yields of valinomycin in the previous work of in vivo production in E. coli remained about three orders of magnitude higher than in CFPS reactions (Li et al., 2015b).
The co-expression of four enzymes including two large NRPSs (Vlm1 and Vlm2) together with the extended reaction duration was posited to limit resources such as energy, which was potentially responsible for the low yield of valinomycin. To overcome this constraint, cell-free metabolic engineering (CFME) was applied (Kay and Jewett, 2015; Dudley et al., 2016; Karim and Jewett, 2016), which is a platform wherein proteins are maximally expressed in cells followed by subsequent enrichment of the lysate with these proteins. Vlm1 and Vlm2 were heterologously expressed in E. coli BL21 (DE3), and E. coli BAP1, which has the sfp gene integrated into its genome (Pfeifer et al., 2001). The two lysates that enriched Vlm1 and Vlm2 proteins were then mixed with other cell-free components to initiate in vitro valinomycin biosynthesis. However, the yield of valinomycin obtained was low (∼5.5 μg/L), which led to attempts to further optimize the CFME system. Hence, the effect of supplemental cofactors such as CoA, NAD, and ATP and the mass ratio of two lysates was investigated. Valinomycin formation in CFME reactions was found best at ∼76.9 μg/L without cofactor supplementation and the mass ratio of 3:1 (cell lysate-Vlm1: cell lysate-Vlm2). Once again, the TE-II was applied for yield improvement of valinomycin by using a two-phase biosynthesis CFPS-CFME (CFPS-ME) system. In the first reaction phase, TEII was expressed by CFPS, which could be used to initiate the activity of Vlm1 and Vlm2 during the second CFME phase. Compared to the initial CFME yield (∼5.59 μg/L), this approach improved valinomycin biosynthesis significantly with a yield of ∼29.3 mg/L, which is more than 5,200 times higher. This result suggests that 1) the upstream CFPS-expressed enzyme (TEII) can efficiently activate the downstream CFME reactions (Vlm1 and Vlm2) and 2) cheap, simple precursor glucose can be converted to valinomycin substrates (Pyruvate, ketoisovalerate, and L-Val) through the glycolysis in CFPS to robustly synthesize complex natural product. Taken together, this work demonstrates the feasibility of CFPS to generate larger, more complex proteins using more complex metabolic networks within cell lysate.
The ability to transcribe and translate genomic DNA directly in cell-free systems was explored by Gringiner and coworkers via applying the PURExpress system directly from a commercially available kit. Two examples of NRPS products were generated to demonstrate feasibility. The first was Blue Pigment Synthetase A (BpsA) from Streptomyces lavendulae, which generates the pigment indigoidine (Siebels et al., 2020a). BpsA is a monomodular NRPS, consisting of an A domain that incorporates l-glutamine and undergoes spontaneous cyclization. The product is released by the oxidation (Ox) domain and thioesterase (TE) domain. BpsA activity can be directly monitored as indigoidine absorbs strongly at 590 nm (Figure 3B). The promiscuous PPTase Sfp from B. subtilis was added to the PURE system to synthesize the full-length activated BpsA. When the protein was phosphopantetheinylated prior to BpsA synthesis in a sequential reaction, the yield was at ∼28.7 μg/mL. However, when Sfp was added while the protein was synthesized in a parallel reaction, the yield decreased to ∼15.5 μg/mL. The efficiency of phosphopantetheinylation of in vitro synthesized BpsA was determined by labeling it with the fluorescent CoA analog (CoA-647). Subsequently, phosphopantetheinylated BpsA was used to in vitro biosynthesize indigoidine with the final concentration of ATP, and L-glutamine was added directly into the reaction solution at 2.65 mM and 0.95 mM respectively. The catalytic differences between BpsA expressed in cell-free versus in vivo in the IVPS system suggest that the folding environment may not be comparable. However, because the exact formulation of the IVPS kit is proprietary, it is difficult to generate and test hypotheses that explain this disparity. To further probe the IVPS system, the RXP-synthesizing NRPS KJ12ABC from the bacterium Xhenorhabdus KJ12.1 was pursued as an example of a more complex target. KJ12ABC consists of three modules, both elongation modules KJ12A (encoding a C-A-T module, 137.8 kDa) and KJ12B (encoding a C-A/MT-T module, 181.5 kDa) which prefers valine as a substrate. The KJ12C domain (62.3 kDa) encodes an unusual C-terminus standalone domain that uses phenylethylamine (PEA) for peptide chain release (Figure 3D). These three modules were expressed recombinantly in E. coli prior to being assayed for product formation. With the molar ratio of 10:1.5:1 (KJ12A:B:C) for the modules, three tetrapeptides synthesized were mV-V-mV-mV-PEA, V-V-V-mV-PEA, and V-mV-V-mV-PEA (N-methylated valine (mV), valine (V), and phenylethylamine (PEA). In addition to the examples where metabolites were generated, other proteins were expressed in the PURExpress system to test the capacity to generate large proteins from genomic DNA and were successfully labeled with the fluorescent Coa-647 analog. This includes IndC (a BpsA homolog from Photorhabdus lumincens), GrsA, and TysB1 from tyrocidine biosynthesis as several polyketide synthases which also use phosphopantetheinylated T domains. This work set the stage for future endeavors that use the PURExpress system for genome mining applications.
Despite the successful proof of concept that NRPS can be successfully expressed from the Jewett and Gringiner labs, our research group hypothesized that further optimization of the CFE conditions were likely necessary to further unlock the ability to generate NRPS in cell free systems (Jewett and Swartz, 2004; Vilkhovoy et al., 2018). Moreover, CFPS tends to use GFP as the initial protein for optimization (Jewett and Swartz, 2004); however, GFP differs substantially from biosynthetically relevant proteins such as NRPSs. For instance, the A domain uses ATP to generate an aminoacyl-AMP intermediate, which could potentially compete with CFPS components (e.g., amino acids, phosphoenolpyruvate (PEP), and magnesium ions). Additionally, with a large protein beyond the size of native E. coli proteins, premature truncation can be a concern (Goering et al., 2017; Hurst et al., 2017; Zhuang et al., 2020). Hence, our group examined different reaction conditions to probe the limitations of NRPS CFE (Dinglasan et al., 2023). Reporters were utilized, as many of the methods to evaluate full-length protein translation are low throughput (e.g., SDS-PAGE gels or proteomic evaluation via mass spectrometry) (Goering et al., 2017; Zhuang et al., 2020) This work applied a tetracysteine (TC) peptide tag expressed at the C-terminus of the expressed protein to investigate factors that limit full-length NRPS translation, and consequentially, metabolite production in an E. coli lysate-based cell-free system (Dinglasan et al., 2023) via an exogenously added organoarsenic dye that binds to the TC tag to form a fluorescent complex, which was previously established in the PURExpress cell-free system (Wick et al., 2019).
As an initial step, BpsA-TC was expressed in E. coli BAP1 (a BL21 (DE3) derivative with a chromosomally integrated copy of sfp (Pfeifer et al., 2001)) lysates to validate that the TC tag does not abolish the catalytic function of NRPSs expressed in cell-free systems. The presence of the blue pigment indigoidine in BAP1 lysate reactions confirmed indicates full-length expression and catalytically functional BpsA-TC protein. Variant concentrations of the precursor l-glutamine, CoA, and plasmid DNA were tested, and to a maximum yield of ∼900 μM indigoidine, which is roughly 3.6 fold higher than the yield reported in the PURE cell-free system (250 μM) (Siebels et al., 2020b). Next, a protocol for TC tag detection in lysate-based CFE reactions was established by using the organoarsenic FlAsH reagent. Detecting BpsA-TC in lysates using FlAsH-based opens possibilities to investigate and optimize parameters that could constrain TX/TL processes of NRPS proteins including catalytically active A domains that consume stoichiometric ATP, precursor availability (e.g., amino acids), and require cofactors like Mg2+. To disentangle size from catalytic activity, a point mutant with an inactivated A domain was generated. Next, to probe the demands on the ATP pool, various concentrations of PEP and ATP were tested for BpsA-TC, BpsA-TC with an inactive A domain (BpsA E315A), and superfolder GFP (sfGFP). The results verified that the catalytically active A domain contributes to the additional catabolism of ATP, which causes higher energy substrate requirements for functional NRPS TX/TL compared to proteins of the same size.
In addition, magnesium ion concentrations were investigated because of their important role in almost all stages of TX/TL such as regulating the ionic strength of the lysate reaction environment and stabilizing nucleic acids and ribosomes. Once again, a difference in the expression trend was observed between the wild-type enzyme, BpsA E315A, and sfGFP implying that the active A domain impacts NRPS CFE Mg2+ requirements. Another important parameter for NRPS protein synthesis concentration of amino acids which are both the monomers of translated proteins as well as the substrate of NRPSs. A range of different amino acids concentrations were tested using the optimal concentrations of PEP (77 mM) and Mg2+ (18 mM) reaction mix for BpsA, BpsA E315A, and sfGFP. Interestingly, there are no significant differences in expression levels across testing conditions except a substantial decline at 8 mM for the wild type, which indicates the role of the A domain in disrupting this cofactor flux. To further investigate, 2 mM amino acid with increasing concentration of l-Gln was tested against BpsA-TC and BpsA-TC_E315A. The result showed that additional l-Gln in the reaction impairs the expression of both proteins. Altogether, optimal BpsA expression conditions were different compared to standard CFE conditions which were developed for small proteins like sfGFP.
After optimizing full-length NRPS expression by testing different CFE conditions with a C-terminus TC tag and the FlAsH dye, BpsA-TC, and indigoidine concentrations were identified to compare between reactions made with standard GFP optimized (containing 33 mM PEP and 12 mM Mg2+) and BpsA optimized (containing 77 mM PEP and 18 mM Mg2+) reaction mixes. Both BpsA-TC and indigoidine concentrations show improvement using the optimized mix condition. In addition, other monomodular NRPSs with similar architectures to BpsA also benefit from the total set of reaction conditions optimized. Two NRPSs were selected for testing proof of concept: pyreudione synthetase, Pys (142 kDa), which adenylates l-proline in the synthesis of amoebicidal pyreudiones (Figure 3E) and TycA (124 kDa), which incorporates and isomerizes an L-phenylalanine residue into the cyclic antibiotic decapeptide tyrocidine (Figure 3F). The improvement of expression of these two enzymes were even more dramatic than observed improvement in BpsA expression. Together, this work reveals the power of applying reporters for high throughput assaying of full-length NRPS expression and demonstrates the necessity of optimizing on systems beyond GFP.
CFPS systems have predominantly focused on E. coli, however, there are challenges with large and complex proteins like NRPSs that result in E. coli not always being the most appropriate host machinery such as poor solubility, codon usage bias mismatch, and lack of appropriate posttranslational modifications. Therefore, there has been extensive research investigating alternative CFPS host organisms transcriptional/translational machineries, many of which may be more suitable for specific NRPSs. Expressing NRPS enzymes in the cell-free system has centered around E. coli-based systems (Goering et al., 2017; Siebels et al., 2020b; Zhuang et al., 2020; Dinglasan et al., 2023). To date, the only alternate cell-free system for NRPS expression has been Streptomyces venezuelae (Moore et al., 2021). Other transcriptional/translational machineries have the potential to greatly contribute to expanding the toolkit to generate NRPSs from a variety of metabolically gifted organisms which are discussed below.
Streptomyces spp. and other closely related Actinobacteria are known for their gifted secondary metabolism and are the source of numerous clinically relevant NRPS-derived natural products including daptomycin (Debono et al., 1988; Miao et al., 2005; Gu et al., 2007), bleomycin (Shen et al., 2002), and vancomycin (van Wageningen et al., 1998; van Wageningen et al., 1998; Choroba et al., 2000; Choroba et al., 2000; Recktenwald et al., 2002). Streptomyces sp. harbor genomes known for their high guanine and cystosine (G + C) content (ranging between 67% and 72%) (Sharma, 1999; Kieser et al., 2000; Drufva et al., 2022) and utilize a myriad of complex genetic regulatory mechanisms (Zhou et al., 2020) which can cause translational issues in tractable hosts such as E. coli (Hwang et al., 2021; Sword et al., 2023). This combined with relatively cumbersome genetics, typically requiring intergenic conjugation (Mazodier et al., 1989; Flett et al., 1997) or protoplast preparation (Xu et al., 2020; Kieser et al.), has motivated the development of Streptomyces-based CFPS for rapid prototyping (Moore et al., 2023). Currently, the Streptomyces-based CFPS system is the most prevalent non-E. coli CFPS platform developed with bacterial natural product biosynthetic machinery in mind (Moore et al., 2021; 2017; Li et al., 2017; Toh et al., 2021; Xu et al., 2022b; 2022a; Moore et al., 2023). In the 1980s, initial efforts were made to develop a Streptomyces CFPS platform as a tool for biological investigation (Thompson et al., 1984). With the significant rise in fundamental understanding of Streptomyces biology over the past half century, Jewett and others sought to develop a high-yielding Streptomyces CFPS protocol in 2017 (Li et al., 2017). An optimized Streptomyces lividans system using the reporter-enhanced green fluorescent protein (eGFP) was generated (Li et al., 2017), with an initial yield of ∼45 μg/mL in batch reactions using the cell-free optimized vector pJL1-eGFP (Li et al., 2017). Additionally, other commonly used Streptomyces strains were tested to this CFPS system, which includes S. lividans B-122, S. lividans 66, S. coelicolor ISP-5233, and S. coelicolor M1152 to produce eGFP. Results showed that the S. lividans strains produced more eGFP than the S. coelicolor strains, and S. lividans B-1227 has a faster protein synthesis rate compared to the S. lividans 66. Importantly, they validated the effectiveness of using a Streptomyces-based CFPS system by expressing high G-C content genes originating from Streptomyces; these included tbrP (72% GC), tbrQ (78% GC), and tbrN (75% GC) which are tailoring genes in the biosynthesis of the non-ribosomal petide tambromycin (Goering et al., 2016) and the type II thioesterase (TEII, 64% GC) gene that is from the valinomycin BGC of S. tsusimaensis. (Li et al., 2015b). As expected, the results showed the solubility of these proteins using the Streptomyces-based CFPS system is higher compared to E. coli-based platforms despite having a lower titer (Li et al., 2015b). This implied the benefit of using a Streptomyces-based CFPS for NRPS cell-free expression but also the necessity for further optimization. By optimizing translation-related factors and creating a toolkit of promoter and ribosomal binding site libraries achieving the highest to-date reading of eGFP value of 515.7 ± 25.3 μg/mL (Xu et al., 2020; 2022b).
Another second prominent effort developed by Freemont, Moore, and others Streptomyces-based CFPS platform utilizes a S. venezuelae transcription-translation (TX-TL) system (Moore et al., 2017), achieving a yield of up to ∼1.3 µM sfGFP by optimizing the cell-extract preparation process (Moore et al., 2017; Xu et al., 2020). Subsequently, other parameters of this system were screened for the improvement of protein synthesis, which includes a promoter, minimal energy solution, ATP source, and RNase inhibitor (Moore et al., 2021). Notably, the strongest promoter observed was SP44, with 2.2 times more active than kasOp*, the initial promoter (Moore et al., 2021). This system was then used to test for the expression of high G-C content genes, which includes the oxytetracycline enzymes (OxyA, -B, -C, -D, -J, -K, -N, and -T) from Streptomyces rimosus, the thaxtomin A biosynthesis from Streptomyces scabiei (TxtA and TxtB), and an uncharacterized NRPS (NH08_RS0107360) from S. rimosus. Most of these enzymes were successfully expressed, which proves the robustness of expressing high G-C genes using this platform. Streptomyces-based CFPS systems were developed with the hope of overcoming the challenges that were faced when expressing native Streptomyces genes in E. coli CFPS, which included solubility issues, lack of post-translational modifications, codon bias, and complexity (Moore et al., 2021; Ji et al., 2022). Evidently, Streptomyces-based CFPS systems have proven to be more suitable as CFPS hosts for NRPSs than E. coli, (Li et al., 2017), especially considering many NRPSs with high GC-content genes, are derived from Streptomyces (Ji et al., 2022). Even though there is limited research expressing NRPS proteins using Streptomyces-based CFPS systems, further development, and optimization will undoubtedly accelerate natural products research.
Bacillus subtilis is a robust and efficient heterologous host that is generally used industrially for heterologous expression (Yang et al., 2021; Ejaz et al., 2022). A number of canonical NRPS pathways come from B. subtilis and other closely related Firmicutes including lipopeptides like surfactin (Sen, 2010), polymyxins (Komura and Kurahashi, 1985; Choi et al., 2009; Kim et al., 2015), bacillaene (Butcher et al., 2007), and plipastatin (Gao et al., 2018). Early reports of efforts to develop B. subtilis CFPS faced challenges due to the requirement for exogenous mRNA, protease inhibitors, DNAse treatments, or less efficient energy systems that discouraged the development of this platform (Kelwick et al., 2016). The more recent development of the B. subtilis cell-free transcription-translation platform was overcoming these shortfalls by adapting optimization techniques from the E. coli cell-free system (Kelwick et al., 2016). Initial efforts focused on B. subtilis 168, a strain that is highly characterized and heavily applied as a laboratory host which achieved a low yield of 0.011 µM GFPmut3b (a variant of GFP that is more fluorescent than wild type (Cormack et al., 1996) but not under patent protection like superfolder GFP) (Kelwick et al., 2016). It was hypothesized that B. subtilis 168 was not quite suitable for a cell-free transcription-translation reaction due to the presence of endogenously expressed proteases and a resultant degradation of translated proteins (Kelwick et al., 2016). Next, efforts were applied to a protease-deficient strain B. subtilis, WB800N which was found to result in ∼72-fold improvement in protein yield (up to 0.8 µM GFPmut3b (Kelwick et al., 2016). Despite improved expression, overall yields remain lower than those observed in E. coli cell-free transcription-translation systems (Kelwick et al., 2016). A similar B. subtilis 168 cell-free system was successfully used to diagnose the bottleneck of the N-acetylneuraminic acid (NeuAc) biosynthesis pathway in B. subtilis in vivo by finding the limiting factors through in vitro analysis (Tian et al., 2020). Subsequently, an attempt was made to compose a standardized approach to develop a cell-free platform for poorly characterized non-model organisms termed native cell-free (NCF) transcription–translation with Bacillus spp. (Moore et al., 2018). The method was applied to B. megaterium DSM319, a far less characterized Bacillus spp. (Moore et al., 2018). The Bacillus megaterium NCF system, however, is further proof of concept that NCF can be applied to non-model organisms (Moore et al., 2018).
Fast-growing organisms are of special interest as one of the main benefits of CFPS is acceleration of the timeframe of the DBTL cycle. Vibrio natriegens, a Gram-negative marine bacterium, is known for having the fastest growth rate of any non-pathogenic organism with a reported doubling time of less than 10 min (Lee et al., 2016; Weinstock et al., 2016; Fernández-Llamosas et al., 2017). Genetic engineering tools for Vibrio natriegens have been widely developed and manipulated for heterologous recombinant protein expression. (Weinstock et al., 2016; Hoffart et al., 2017). Vibrio natrigens can grow rapidly under a range of media conditions (including minimal media supplemented with various carbon sources) and under both aerobic and anaerobic conditions meaning it is an extremely versatile host (Weinstock et al., 2016; Hoffart et al., 2017). The extraordinary growth rate of V. natriegens is typically attributed to its high number of ribosomes per cell. (Aiyar et al., 2002). Furthermore, V. natriegens also contains more rRNA operons compared to E. coli (Des Soye et al., 2018).
In recent years several groups have focused on optimizing the V. natriegens as a CFPS platform (Des Soye et al., 2018; Failmezger et al., 2018; Wiegand et al., 2018; Zhu et al., 2020). Of these studies, the reporter protein GFP has been applied and the highest yield achieved was 1.6 ± 0.05 g/L using the Vnat cell-free platform, which is competitive with existing E. coli CFPS platforms (Des Soye et al., 2018). Noticeably, the optimal incubation reaction temperature for maximum protein yield was found at 26°C and 30°C with extracts incubated at 30 °C produced 3 folds more protein compared to those at 37°C. The result supports the theory that the rapid growth of V. natriegens at higher temperatures is because of the larger number of ribosomes and not more efficient ribosomes (Wiegand et al., 2018). Additionally, the kinetic analysis showed that V. natriegens extracts sustained elevated protein expression rates over 60 min as compared to E. coli extracts sustained for 20 min (Wiegand et al., 2018). Vibrio natriegens CFPS demonstrated competitive yields with E. coli CFPS and observed higher sfGFP yields (∼9.3 μM) than Bacillus subtillis (0.8 μM) and S. venezuelae (1.3 μM) (Wiegand et al., 2018). Noticeably, the V. natriegens CFPS was viable with linear templates, as opposed to plasmids which also accelerates prototyping (Zhu et al., 2020). Although V. natriegens CFPS platforms have not been applied to NRPS production, the robustness of V. natriegens’ extracts in the CFPS platform means that it may be a viable alternative to E. coli, especially for NRPS from Proteobacteria.
Pseudomonas putida is a Gram-negative bacterium, that often serves as a laboratory workhorse organism for its well-characterized genome, robust culture conditions, and catabolism of a broad range of carbon sources (Loeschcke and Thies, 2015). With a complete genome sequenced, numerous genetic tools have been developed to engineer P. putida (Wang et al., 2018). There are also multiple biosynthetic gene clusters including NRPSs harbored within Pseudomonas spp. (Ackerley et al., 2003; Gao et al., 2014; González et al., 2017) and P. putida has been shown to be a robust host for heterologous expression of biosynthetic gene clusters (Domröse et al., 2017; Cook et al., 2021). An initial effort was made for the Pseudomonas organism for CFPS using the lysate crude extract from Pseudomonas fluorescens in 2004 (Nakashima and Tamura, 2004). However, it faced challenges with the complicated cell extract preparation process, which led to low protein yield. Recently, an optimized CFPS method has been developed by the Jewett laboratory using the lysate crude extract of P. putida (Wang et al., 2018). Adopting the protocol used for the E. coli CFPS system (Jewett and Swartz, 2004; Li et al., 2016), cell extract from P. putida was harvested, giving a low yield for sfGFP. Thus, this system was then optimized by manipulating reaction conditions such as lysate concentration, plasmid concentration, reaction temperature, and Mg2+ concentration. The yield achieved was ∼200 μg/mL in 4h batch reactions, which is more than ten times the un-optimized yields. Even though the current yields are significantly lower than that of the E. coli-based CFPS system (>1,000 μg/mL), they are ten times higher than the S. cerevisiae-based CFPS system (<20 μg/mL) (Schoborg et al., 2016; Wang et al., 2018). While Streptomyces-based CFPS platforms have been established to express high GC genes, P. putida also has high GC (∼60%) content. The process of harvesting crude cell extract from P. putida is less complex than for Streptomyces organisms as it does not have a clumpy mycelial physiology. (Loeschcke and Thies, 2015), which makes it a great potential for an alternative CFPS strategy with Streptomyces-based CFPS platforms to express GC-rich genes (Loeschcke and Thies, 2015). Developing P. putida-based CFPS platforms contributes to a future application for CFPS screening platforms and expands the protein synthesis toolkit for synthetic biology.
Cyanobacteria are Gram-negative oxygenic photosynthetic prokaryotes (Laxmi et al., 2023) that are capable of using solar energy to synthesize organic carbons from atmospheric carbon dioxide, which makes them an ideal target for biofuel and biochemical research (Choi et al., 2020; Choi et al., 2021). Their genomes have been known for containing a plethora of NRPS, and other natural product genes, hence, they have been a popular target for genome mining (Ehrenreich et al., 2005; Barrios-Llerena et al., 2007; Choi et al., 2021; Laxmi et al., 2023). However, as heterologous hosts, they present challenges since they can be polyploid and are slow to appropriately segregate heterologous genes (Choi et al., 2021). For these reasons, developing a cell-free system using Cyanobacteria crude extract could accelerate DBTL cycles in a fashion analogously to other hosts that have slow genetic manipulation timelines like Streptomyces sp. Recently, a CRISPR/Cas12a-based assay coupled with a cell-free system was developed as a tool for prototyping promoters using model cyanobacteria Synechocystis sp. PCC 6803, which has the potential to be applicable to other host organisms (Choi et al., 2021). The study created a promoter library and was able to use the assay to confirm a positive correlation between in vivo and in vivo performance in cyanobacteria Synechocystis (Choi et al., 2021). This method can be useful for NRPS and other natural product gene expression in cyanobacteria by significantly shortening the workflow time.
The most prominent eukaryotic expression system origin is Saccharomyces cerivisae (Yin et al., 2007). Importantly, there are many fungal NRPS pathways (Domröse et al., 2017; Zhang et al., 2023; 2022) that could be accelerated using S. cerevisiae cell-free systems. While cell-free systems for plant (Madin et al., 2000; Murota et al., 2011; Buntru et al., 2022; 2014; Buntru et al., 2021) and animal (Ezure et al., 2010; Makrydaki et al., 2021) based expression systems exist, these are out of the scope of this review, which focuses on peptide natural products from microorganisms. Notably, Saccharomyces cerevisiae is a great candidate to harness the benefit of both microbial-based and eukaryotic systems due to its fast growth characteristics (Salari and Salari, 2017). Furthermore, its capability of growing aerobically and anaerobically has contributed to its industrial success in food, brewing, biotechnology, and drug discovery (Parapouli et al., 2020). Saccharomyces cerevisiae has been extensively engineered as a heterologous host for the expression of numerous NRPSs in vivo such as β-lactam benzylpenicillin, beauvericin, bassianolide, indigoidine synthetase (BpsA), tyrocidine synthetase (TycA), and surfactin synthetase (SrfAC) (Tippelt and Nett, 2021). The ability of S. cerevisiae to heterologously express NRPSs in vivo suggests that it might be a great candidate for expressing NRPSs using S. cerevisiae CFPS systems. The most recent S. cerevisiae CFPS-based TX/TL studies achieved yields of 4.33 ± 0.37 μg/mL luciferase GFP (Hodgman and Jewett, 2014) and 17.0 ± 3.8 μg/mL sfGFP in a 10 h semi-continuous reaction (Schoborg et al., 2014). Although S. cerevisiae CFPS-based system has been successful when expressing reporters, attempts to express NRPS enzymes have not been performed yet. Optimizing conditions to express multidomain enzymes using S. cerevisiae CFPS-based platforms will open more doors to get access to “difficult-to-synthesize” proteins. Very recently, other fungal systems have been explored including Neurospora crassa and Aspergillus niger which was able to produce peroxygenases with activities reaching up to 105 U L-1 (Schramm et al., 2022). Due to the success of Aspergillus sp. as heterologous hosts in recent years (Chiang et al., 2013; Anyaogu and Mortensen, 2015; Lin et al., 2023), these systems could have broad utility for NRPS expression.
Another prominent CFPS system derived from eukaryotes developed by Alexandrov and coworkers harnesses the non-pathogenic single-celled flagellate Leishmania tarentolae (Mureev et al., 2009; Kovtun et al., 2011; Johnston and Alexandrov, 2014; Gagoski et al., 2015), which is inexpensive in bioreactors and has been successful in expressing a range of complex eukaryotic proteins (Mureev et al., 2009; Kovtun et al., 2010; Gagoski et al., 2017; Wu et al., 2022; Moradi et al., 2023). Leishmania tarentolae can be fermented on a large scale and its genetic modification process is not complicated (Kovtun et al., 2010). Additionally, many proto- and metazoan proteins were expressed by using L. tarentolae lysate (Kovtun et al., 2010). In particular, this system expressed proteins in L. tarentolae extract, and utilized Species-Independent Translation Initiation Sequence (SITS) allowing for a divergent range of applications (Kovtun et al., 2011). While the L. tarentolae system has not been applied to proteins from microbial secondary metabolism, NRPSs have been identified in some protozoan species, especially dinoflagellates such as Symbodinium tricnidorium, Cladocopium sp. (Beedessee et al., 2019) and Ostreopsis ovata (Yamazaki, 2016). The Leishmania system has been shown to be remarkably tolerant to complexity and has been shown to produce up to 300 ug/mL of recombinant protein in 2 h (Mureev et al., 2009). Furthermore, this system is suitable for high throughput screening as a result of more than 50 pure proteins obtained from the coding sequence manipulation process without cloning (Kovtun et al., 2011). This is suggestive its promise as a general host for NRPS discovery, but particularly for NRPS from understudied origins, as dinoflagellate biosynthetic gene clusters are less well characterized than bacterial or fungal ones.
With current technologies in genome sequencing and genome mining, a vast number of novel natural products have been discovered. Cell-free synthetic biology is a powerful platform that offers rapid, high-throughput, cost-effective, and robust approaches to access and harness these valuable natural products as well as generate analogues and apply the biosynthetic steps for bioproduct generation. CFPS accelerates the DBTL cycle the metabolic engineering. In this review, we focus on non-ribosomal peptide natural products that are expressed using cell-free strategies, different lysate host organisms used in CFPS (Table 1), and the potential of using CFPS to accelerate engineering non-ribosomal peptides and their analogs. The knowledge gap in using a universal platform with high yield, cost-effectiveness, inability, and scalable CFPS systems to synthesize any complex proteins such as NRPS is still a bottleneck.
With the increasing tools available to control, modify, and optimize conditions for CFPS systems with various lysate hosts, the promise of CFPS as a fruitful platform for bioengineering efforts remains high. For instance, the unique characteristics of the flexible linkers holding NRPS domains and modules together draw significant interest in studying these NRPS parts such as module and domain swapping, module and domain deletions, and insertions (Kries et al., 2015; Winn et al., 2016; Beck et al., 2020). Different approaches have been developed to engineer the A domain (Kries et al., 2015); however, expressing mutants often leads to low production or no production at all (Winn et al., 2016). Binding domains or modules are substantially longer than subdomains, hence, subdomain swapping could greatly contribute to NRPS engineering based on gene synthesis and bioinformatics research. The subdomain swapping strategy was successfully done on the A domain of the GrsA biosynthesis of gramicidin S synthetase (Kries et al., 2015). Following, the production of the novel pyoverdine derivatives proved to be transferable to different NRPS systems at high yields by exchanging the A domain (Calcott et al., 2020; Messenger et al., 2023). Another noticeable example of domain swapping is the exchanging of the T domain of the NRPS IndC with synthetic and natural T domains shows improvement in indigoidine production (Beer et al., 2014). These are a few examples of domain engineering strategies, illuminating a promising approach to generating novel molecules as well as increasing natural product production.
Taking advantage of the flexibility of cell-free systems, exogenous elements can be rapidly added to the reaction directly and manipulated, allowing the synthesis of potential peptide natural products. In recent years, many efforts have investigated non-ribosomal peptide precursors derived from amino acids (Kurusu et al., 1987; Kajimura and Kaneda, 1996; Milne et al., 2006; Han et al., 2012; Thirlway et al., 2012). CFPS affords the ability to alter host machinery, regulatory elements and rapidly engineer chimeric NRPS pathways as well as express new NRPS machineries that come from intractable organisms. In a synergistic fashion, it also affords the incorporation of unnatural building blocks to increase molecular diversity as well as overcome cellular toxicity issues that arise from heterologous expression of bioactive metabolites. While CFPS remains an emergent technology for the production of secondary metabolites, advances in lysate systems with transcriptional and translational machineries that are more appropriate to NRPS expression and cofactor balance specific to this class of natural products will afford CFPS to be applied to NRPS enzymes of increasing complexity.
TS: Conceptualization, Writing–original draft, Writing–review and editing. GA: Writing–original draft, Writing–review and editing. CB: Conceptualization, Funding acquisition, Resources, Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. CB was supported by the University of Sydney School of Chemistry, Faculty of Science and is a member of the University of Sydney Drug Discovery Initiative, the University of Sydney Infectious Diseases Institute, and the University of Sydney Nanoscience Institute. TS was supported by the University of Tennessee-Knoxville, the University of Tennessee-Knoxville Oak Ridge Innovation Institute (UT-ORII) Science Alliance Advancement, Training, and Education (GATE) fellowship, and the National Institutes of Health (R15GM145182).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ackerley, D. F., Caradoc-Davies, T. T., and Lamont, I. L. (2003). Substrate specificity of the nonribosomal peptide synthetase PvdD from Pseudomonas aeruginosa. J. Bacteriol. 185, 2848–2855. doi:10.1128/JB.185.9.2848-2855.2003
Aiyar, S. E., Gaal, T., and Gourse, R. L. (2002). rRNA promoter activity in the fast-growing bacterium Vibrio natriegens. J. Bacteriol. 184, 1349–1358. doi:10.1128/JB.184.5.1349-1358.2002
Anyaogu, D. C., and Mortensen, U. H. (2015). Heterologous production of fungal secondary metabolites in Aspergilli. Front. Microbiol. 6, 77. doi:10.3389/fmicb.2015.00077
Arnison, P. G., Bibb, M. J., Bierbaum, G., Bowers, A. A., Bugni, T. S., Bulaj, G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. doi:10.1039/c2np20085f
Atanasov, A. G., Waltenberger, B., Pferschy-Wenzig, E.-M., Linder, T., Wawrosch, C., Uhrin, P., et al. (2015). Discovery and resupply of pharmacologically active plant-derived natural products: a review. Biotechnol. Adv. 33, 1582–1614. doi:10.1016/j.biotechadv.2015.08.001
Banerjee, D., Eng, T., Lau, A. K., Sasaki, Y., Wang, B., Chen, Y., et al. (2020). Genome-scale metabolic rewiring improves titers rates and yields of the non-native product indigoidine at scale. Nat. Commun. 11, 5385. doi:10.1038/s41467-020-19171-4
Barajas, J. F., Phelan, R. M., Schaub, A. J., Kliewer, J. T., Kelly, P. J., Jackson, D. R., et al. (2015). Comprehensive structural and biochemical analysis of the terminal myxalamid reductase domain for the engineered production of primary alcohols. Chem. Biol. 22, 1018–1029. doi:10.1016/j.chembiol.2015.06.022
Barrios-Llerena, M. E., Burja, A. M., and Wright, P. C. (2007). Genetic analysis of polyketide synthase and peptide synthetase genes in cyanobacteria as a mining tool for secondary metabolites. J. Ind. Microbiol. Biotechnol. 34, 443–456. doi:10.1007/s10295-007-0216-6
Beck, C., Garzón, J. F. G., and Weber, T. (2020). Recent advances in Re-engineering modular PKS and NRPS assembly lines. Biotechnol. Bioprocess Eng. 25, 886–894. doi:10.1007/s12257-020-0265-5
Beedessee, G., Hisata, K., Roy, M. C., Van Dolah, F. M., Satoh, N., and Shoguchi, E. (2019). Diversified secondary metabolite biosynthesis gene repertoire revealed in symbiotic dinoflagellates. Sci. Rep. 9, 1204. doi:10.1038/s41598-018-37792-0
Beer, R., Herbst, K., Ignatiadis, N., Kats, I., Adlung, L., Meyer, H., et al. (2014). Creating functional engineered variants of the single-module non-ribosomal peptide synthetase IndC by T domain exchange. Mol. Biosyst. 10, 1709–1718. doi:10.1039/c3mb70594c
Bogart, J. W., Cabezas, M. D., Vögeli, B., Wong, D. A., Karim, A. S., and Jewett, M. C. (2021). Cell-free exploration of the natural product chemical space. Chembiochem 22, 84–91. doi:10.1002/cbic.202000452
Bozhüyük, K. A. J., Linck, A., Tietze, A., Kranz, J., Wesche, F., Nowak, S., et al. (2019). Modification and de novo design of non-ribosomal peptide synthetases using specific assembly points within condensation domains. Nat. Chem. 11, 653–661. doi:10.1038/s41557-019-0276-z
Buntru, M., Hahnengress, N., Croon, A., and Schillberg, S. (2021). Plant-derived cell-free biofactories for the production of secondary metabolites. Front. Plant Sci. 12, 794999. doi:10.3389/fpls.2021.794999
Buntru, M., Vogel, S., Finnern, R., and Schillberg, S. (2022). Plant-based cell-free transcription and translation of recombinant proteins. Methods Mol. Biol. 2480, 113–124. doi:10.1007/978-1-0716-2241-4_8
Buntru, M., Vogel, S., Spiegel, H., and Schillberg, S. (2014). Tobacco BY-2 cell-free lysate: an alternative and highly-productive plant-based in vitro translation system. BMC Biotechnol. 14, 37. doi:10.1186/1472-6750-14-37
Butcher, R. A., Schroeder, F. C., Fischbach, M. A., Straight, P. D., Kolter, R., Walsh, C. T., et al. (2007). The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 104, 1506–1509. doi:10.1073/pnas.0610503104
Calcott, M. J., Owen, J. G., and Ackerley, D. F. (2020). Efficient rational modification of non-ribosomal peptides by adenylation domain substitution. Nat. Commun. 11, 4554. doi:10.1038/s41467-020-18365-0
Cane, D. E., Walsh, C. T., and Khosla, C. (1998). Harnessing the biosynthetic code: combinations, permutations, and mutations. Science 282, 63–68. doi:10.1126/science.282.5386.63
Carlson, E. D., Gan, R., Hodgman, C. E., and Jewett, M. C. (2012). Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 30, 1185–1194. doi:10.1016/j.biotechadv.2011.09.016
Cheng, Y.-Q. (2006). Deciphering the biosynthetic codes for the potent anti-SARS-CoV cyclodepsipeptide valinomycin in Streptomyces tsusimaensis ATCC 15141. Chembiochem 7, 471–477. doi:10.1002/cbic.200500425
Chiang, Y.-M., Oakley, C. E., Ahuja, M., Entwistle, R., Schultz, A., Chang, S.-L., et al. (2013). An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 135, 7720–7731. doi:10.1021/ja401945a
Choi, S.-K., Park, S.-Y., Kim, R., Kim, S.-B., Lee, C.-H., Kim, J. F., et al. (2009). Identification of a polymyxin synthetase gene cluster of Paenibacillus polymyxa and heterologous expression of the gene in Bacillus subtilis. J. Bacteriol. 191, 3350–3358. doi:10.1128/JB.01728-08
Choi, Y.-N., Lee, J. W., Kim, J. W., and Park, J. M. (2020). Acetyl-CoA-derived biofuel and biochemical production in cyanobacteria: a mini review. J. Appl. Phycol. 32, 1643–1653. doi:10.1007/s10811-020-02128-x
Choi, Y.-N., Shin, Y. R., Park, J. M., and Lee, J. W. (2021). Cell-free transcription-coupled CRISPR/Cas12a assay for prototyping cyanobacterial promoters. ACS Synth. Biol. 10, 1300–1307. doi:10.1021/acssynbio.1c00148
Choroba, O. W., Williams, D. H., and Spencer, J. B. (2000). Biosynthesis of the vancomycin group of antibiotics: involvement of an unusual dioxygenase in the pathway to (S)-4-Hydroxyphenylglycine. J. Am. Chem. Soc. 122, 5389–5390. doi:10.1021/ja000076v
Cook, T. B., Jacobson, T. B., Venkataraman, M. V., Hofstetter, H., Amador-Noguez, D., Thomas, M. G., et al. (2021). Stepwise genetic engineering of Pseudomonas putida enables robust heterologous production of prodigiosin and glidobactin A. Metab. Eng. 67, 112–124. doi:10.1016/j.ymben.2021.06.004
Cormack, B. P., Valdivia, R. H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. doi:10.1016/0378-1119(95)00685-0
Corpuz, J. C., Sanlley, J. O., and Burkart, M. D. (2022). Protein-protein interface analysis of the non-ribosomal peptide synthetase peptidyl carrier protein and enzymatic domains. Synthetic Syst. Biotechnol. 7, 677–688. doi:10.1016/j.synbio.2022.02.006
Cui, Y., Chen, X., Wang, Z., and Lu, Y. (2022). Cell-free PURE system: evolution and achievements. BioDesign Res. 2022, 9847014. doi:10.34133/2022/9847014
Debono, M., Abbott, B. J., Molloy, R. M., Fukuda, D. S., Hunt, A. H., Daupert, V. M., et al. (1988). Enzymatic and chemical modifications of lipopeptide antibiotic A21978C: the synthesis and evaluation of daptomycin (LY146032). J. Antibiot. 41, 1093–1105. doi:10.7164/antibiotics.41.1093
Des Soye, B. J., Davidson, S. R., Weinstock, M. T., Gibson, D. G., and Jewett, M. C. (2018). Establishing a high-yielding cell-free protein synthesis platform derived from Vibrio natriegens. ACS Synth. Biol. 7, 2245–2255. doi:10.1021/acssynbio.8b00252
Dias, D. A., Urban, S., and Roessner, U. (2012). A historical overview of natural products in drug discovery. Metabolites 2, 303–336. doi:10.3390/metabo2020303
Dinglasan, J. L. N., Sword, T. T., Barker, J. W., Doktycz, M. J., and Bailey, C. B. (2023). Investigating and optimizing the lysate-based expression of nonribosomal peptide synthetases using a reporter system. ACS Synth. Biol. 12, 1447–1460. doi:10.1021/acssynbio.2c00658
Domröse, A., Weihmann, R., Thies, S., Jaeger, K.-E., Drepper, T., and Loeschcke, A. (2017). Rapid generation of recombinant Pseudomonas putida secondary metabolite producers using yTREX. Synthetic Syst. Biotechnol. 2, 310–319. doi:10.1016/j.synbio.2017.11.001
Drufva, E. E., Hix, E. G., and Bailey, C. B. (2020). Site directed mutagenesis as a precision tool to enable synthetic biology with engineered modular polyketide synthases. Synthetic Syst. Biotechnol. 5, 62–80. doi:10.1016/j.synbio.2020.04.001
Drufva, E. E., Sword, T. T., and Bailey, C. B. (2022). “Metabolic engineering of actinomycetes for natural product discovery,” in Natural products from actinomycetes: diversity, ecology and drug discovery. Editors R. V. Rai, and J. A. Bai (Singapore: Springer Singapore), 267–307. doi:10.1007/978-981-16-6132-7_11
Dudley, Q. M., Anderson, K. C., and Jewett, M. C. (2016). Cell-free mixing of Escherichia coli crude extracts to prototype and rationally engineer high-titer mevalonate synthesis. ACS Synth. Biol. 5, 1578–1588. doi:10.1021/acssynbio.6b00154
Ehrenreich, I. M., Waterbury, J. B., and Webb, E. A. (2005). Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 71, 7401–7413. doi:10.1128/AEM.71.11.7401-7413.2005
Ejaz, S., Khan, H., Sarwar, N., Aqeel, S. M., Al-Adeeb, A., and Liu, S. (2022). A review on recent advancement in expression strategies used in Bacillus subtilis. Protein Pept. Lett. 29, 733–743. doi:10.2174/0929866529666220803163335
Ezure, T., Suzuki, T., Shikata, M., Ito, M., and Ando, E. (2010). A cell-free protein synthesis system from insect cells. Methods Mol. Biol. 607, 31–42. doi:10.1007/978-1-60327-331-2_4
Failmezger, J., Scholz, S., Blombach, B., and Siemann-Herzberg, M. (2018). Cell-free protein synthesis from fast-growing Vibrio natriegens. Front. Microbiol. 9, 1146. doi:10.3389/fmicb.2018.01146
Farag, S., Bleich, R. M., Shank, E. A., Isayev, O., Bowers, A. A., and Tropsha, A. (2019). Inter-Modular Linkers play a crucial role in governing the biosynthesis of non-ribosomal peptides. Bioinformatics 35, 3584–3591. doi:10.1093/bioinformatics/btz127
Felnagle, E. A., Jackson, E. E., Chan, Y. A., Podevels, A. M., Berti, A. D., McMahon, M. D., et al. (2008). Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5, 191–211. doi:10.1021/mp700137g
Fernández-Llamosas, H., Castro, L., Blázquez, M. L., Díaz, E., and Carmona, M. (2017). Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 7, 16046. doi:10.1038/s41598-017-16252-1
Finking, R., Solsbacher, J., Konz, D., Schobert, M., Schafer, A., Jahn, D., et al. (2002). Characterization of a new type of phosphopantetheinyl transferase for fatty acid and siderophore synthesis in Pseudomonas aeruginosa. J. Biol. Chem. 277, 50293–50302. doi:10.1074/jbc.M205042200
Flett, F., Mersinias, V., and Smith, C. P. (1997). High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes. FEMS Microbiol. Lett. 155, 223–229. doi:10.1016/s0378-1097(97)00392-3
Gagoski, D., Mureev, S., Giles, N., Johnston, W., Dahmer-Heath, M., Škalamera, D., et al. (2015). Gateway-compatible vectors for high-throughput protein expression in pro- and eukaryotic cell-free systems. J. Biotechnol. 195, 1–7. doi:10.1016/j.jbiotec.2014.12.006
Gagoski, D., Shi, Z., Nielsen, L. K., Vickers, C. E., Mahler, S., Speight, R., et al. (2017). Cell-free pipeline for discovery of thermotolerant xylanases and endo -1,4-β-glucanases. J. Biotechnol. 259, 191–198. doi:10.1016/j.jbiotec.2017.07.014
Gao, L., Guo, J., Fan, Y., Ma, Z., Lu, Z., Zhang, C., et al. (2018). Module and individual domain deletions of NRPS to produce plipastatin derivatives in Bacillus subtilis. Microb. Cell. Fact. 17, 84. doi:10.1186/s12934-018-0929-4
Gao, S.-S., Hothersall, J., Wu, J., Murphy, A. C., Song, Z., Stephens, E. R., et al. (2014). Biosynthesis of mupirocin by Pseudomonas fluorescens NCIMB 10586 involves parallel pathways. J. Am. Chem. Soc. 136, 5501–5507. doi:10.1021/ja501731p
Garenne, D., Haines, M. C., Romantseva, E. F., Freemont, P., Strychalski, E. A., and Noireaux, V. (2021). Cell-free gene expression. Nat. Rev. Methods Prim. 1, 49. doi:10.1038/s43586-021-00046-x
Girija, A., Vijayanathan, M., Sreekumar, S., Basheer, J., Menon, T. G., Krishnankutty, R. E., et al. (2022). Harnessing the natural pool of polyketide and non-ribosomal peptide family: a route map towards novel drug development. Curr. Mol. Pharmacol. 15, 265–291. doi:10.2174/1874467214666210319145816
Goering, A. W., Li, J., McClure, R. A., Thomson, R. J., Jewett, M. C., and Kelleher, N. L. (2017). In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth. Biol. 6, 39–44. doi:10.1021/acssynbio.6b00160
Goering, A. W., McClure, R. A., Doroghazi, J. R., Albright, J. C., Haverland, N. A., Zhang, Y., et al. (2016). Metabologenomics: correlation of microbial gene clusters with metabolites drives discovery of a nonribosomal peptide with an unusual amino acid monomer. ACS Cent. Sci. 2, 99–108. doi:10.1021/acscentsci.5b00331
González, O., Ortíz-Castro, R., Díaz-Pérez, C., Díaz-Pérez, A. L., Magaña-Dueñas, V., López-Bucio, J., et al. (2017). Non-ribosomal peptide synthases from Pseudomonas aeruginosa play a role in cyclodipeptide biosynthesis, quorum-Sensing regulation, and root development in a plant host. Microb. Ecol. 73, 616–629. doi:10.1007/s00248-016-0896-4
Gregorio, N. E., Levine, M. Z., and Oza, J. P. (2019). A user’s guide to cell-free protein synthesis. Methods Protoc. 2, 24. doi:10.3390/mps2010024
Gruenewald, S., Mootz, H. D., Stehmeier, P., and Stachelhaus, T. (2004). In vivo production of artificial nonribosomal peptide products in the heterologous host Escherichia coli. Appl. Environ. Microbiol. 70, 3282–3291. doi:10.1128/AEM.70.6.3282-3291.2004
Gu, J.-Q., Nguyen, K. T., Gandhi, C., Rajgarhia, V., Baltz, R. H., Brian, P., et al. (2007). Structural characterization of daptomycin analogues A21978C1-3(d-Asn11) produced by a recombinant Streptomyces roseosporus strain. J. Nat. Prod. 70, 233–240. doi:10.1021/np0605135
Han, J. W., Kim, E. Y., Lee, J. M., Kim, Y. S., Bang, E., and Kim, B. S. (2012). Site-directed modification of the adenylation domain of the fusaricidin nonribosomal peptide synthetase for enhanced production of fusaricidin analogs. Biotechnol. Lett. 34, 1327–1334. doi:10.1007/s10529-012-0913-8
Harvey, A. L., Edrada-Ebel, R., and Quinn, R. J. (2015). The re-emergence of natural products for drug discovery in the genomics era. Nat. Rev. Drug Discov. 14, 111–129. doi:10.1038/nrd4510
Heinemann, H., Zhang, H., and Cox, R. J. (2023). Reductive release from a hybrid PKS-NRPS during the biosynthesis of pyrichalasin H. H. Chem. Eur. J. 30, e202302590. doi:10.1002/chem.202302590
Heisey, R. M., Huang, J., Mishra, S. K., Keller, J. E., Miller, J. R., Putnam, A. R., et al. (1988). Production of valinomycin, an insecticidal antibiotic, by Streptomyces griseus var. flexipertum var. nov. J. Agric. Food Chem. 36, 1283–1286. doi:10.1021/jf00084a039
Hodgman, C. E., and Jewett, M. C. (2012). Cell-free synthetic biology: thinking outside the cell. Metab. Eng. 14, 261–269. doi:10.1016/j.ymben.2011.09.002
Hodgman, C. E., and Jewett, M. C. (2014). Characterizing IGR IRES-mediated translation initiation for use in yeast cell-free protein synthesis. N. Biotechnol. 31, 499–505. doi:10.1016/j.nbt.2014.07.001
Hoffart, E., Grenz, S., Lange, J., Nitschel, R., Müller, F., Schwentner, A., et al. (2017). High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl. Environ. Microbiol. 83, e01614–e01617. doi:10.1128/AEM.01614-17
Huguenin-Dezot, N., Alonzo, D. A., Heberlig, G. W., Mahesh, M., Nguyen, D. P., Dornan, M. H., et al. (2019). Trapping biosynthetic acyl-enzyme intermediates with encoded 2,3-diaminopropionic acid. Nature 565, 112–117. doi:10.1038/s41586-018-0781-z
Hurst, G. B., Asano, K. G., Doktycz, C. J., Consoli, E. J., Doktycz, W. L., Foster, C. M., et al. (2017). Proteomics-based tools for evaluation of cell-free protein synthesis. Anal. Chem. 89, 11443–11451. doi:10.1021/acs.analchem.7b02555
Hwang, S., Lee, Y., Kim, J. H., Kim, G., Kim, H., Kim, W., et al. (2021). Streptomyces as microbial chassis for heterologous protein expression. Front. Bioeng. Biotechnol. 9, 804295. doi:10.3389/fbioe.2021.804295
Jaitzig, J., Li, J., Süssmuth, R. D., and Neubauer, P. (2014). Reconstituted biosynthesis of the nonribosomal macrolactone antibiotic valinomycin in Escherichia coli. ACS Synth. Biol. 3, 432–438. doi:10.1021/sb400082j
Jewett, M. C., and Swartz, J. R. (2004). Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 86, 19–26. doi:10.1002/bit.20026
Ji, X., Liu, W.-Q., and Li, J. (2022). Recent advances in applying cell-free systems for high-value and complex natural product biosynthesis. Curr. Opin. Microbiol. 67, 102142. doi:10.1016/j.mib.2022.102142
Johnston, W. A., and Alexandrov, K. (2014). Production of eukaryotic cell-free lysate from Leishmania tarentolae. Methods Mol. Biol. 1118, 1–15. doi:10.1007/978-1-62703-782-2_1
Kajimura, Y., and Kaneda, M. (1996). Fusaricidin A, a new depsipeptide antibiotic produced by Bacillus polymyxa KT-8. Taxonomy, fermentation, isolation, structure elucidation and biological activity. J. Antibiot. 49, 129–135. doi:10.7164/antibiotics.49.129
Karim, A. S., and Jewett, M. C. (2016). A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 36, 116–126. doi:10.1016/j.ymben.2016.03.002
Kay, J. E., and Jewett, M. C. (2015). Lysate of engineered Escherichia coli supports high-level conversion of glucose to 2,3-butanediol. Metab. Eng. 32, 133–142. doi:10.1016/j.ymben.2015.09.015
Kelwick, R., Webb, A. J., MacDonald, J. T., and Freemont, P. S. (2016). Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng. 38, 370–381. doi:10.1016/j.ymben.2016.09.008
Khosla, C., and Zawada, R. J. (1996). Generation of polyketide libraries via combinatorial biosynthesis. Trends Biotechnol. 14, 335–341. doi:10.1016/0167-7799(96)10046-9
Kieser, T., Bibb, M., Buttner, M., Chater, K., and Hopwood, D. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Kim, S.-Y., Park, S.-Y., Choi, S.-K., and Park, S.-H. (2015). Biosynthesis of polymyxins B, E, and P using genetically engineered polymyxin synthetases in the surrogate host Bacillus subtilis. J. Microbiol. Biotechnol. 25, 1015–1025. doi:10.4014/jmb.1505.05036
Komura, S., and Kurahashi, K. (1985). Biosynthesis of polymyxin E by a cell-free enzyme system. J. Biochem. 97, 1409–1417. doi:10.1093/oxfordjournals.jbchem.a135195
Koryakina, I., Kasey, C., McArthur, J. B., Lowell, A. N., Chemler, J. A., Li, S., et al. (2017). Inversion of extender unit selectivity in the erythromycin polyketide synthase by acyltransferase domain engineering. ACS Chem. Biol. 12, 114–123. doi:10.1021/acschembio.6b00732
Kovtun, O., Mureev, S., Johnston, W., and Alexandrov, K. (2010). Towards the construction of expressed proteomes using a Leishmania tarentolae based cell-free expression system. PLoS ONE 5, e14388. doi:10.1371/journal.pone.0014388
Kovtun, O., Mureev, S., Jung, W., Kubala, M. H., Johnston, W., and Alexandrov, K. (2011). Leishmania cell-free protein expression system. Methods 55, 58–64. doi:10.1016/j.ymeth.2011.06.006
Kries, H., Niquille, D. L., and Hilvert, D. (2015). A subdomain swap strategy for reengineering nonribosomal peptides. Chem. Biol. 22, 640–648. doi:10.1016/j.chembiol.2015.04.015
Kurotsu, T., Hori, K., Kanda, M., and Saito, Y. (1991). Characterization and location of the L-proline activating fragment from the multifunctional gramicidin S synthetase 2. J. Biochem. 109, 763–769. doi:10.1093/oxfordjournals.jbchem.a123454
Kurusu, K., Ohba, K., Arai, T., and Fukushima, K. (1987). New peptide antibiotics LI-F03, F04, F05, F07, and F08, produced by Bacillus polymyxa. I. Isolation and characterization. J. Antibiot. 40, 1506–1514. doi:10.7164/antibiotics.40.1506
Lavickova, B., and Maerkl, S. J. (2019). A simple, robust, and low-cost method to produce the PURE cell-free system. ACS Synth. Biol. 8, 455–462. doi:10.1021/acssynbio.8b00427
Lawson, C. E., Harcombe, W. R., Hatzenpichler, R., Lindemann, S. R., Löffler, F. E., O’Malley, M. A., et al. (2019). Common principles and best practices for engineering microbiomes. Nat. Rev. Microbiol. 17, 725–741. doi:10.1038/s41579-019-0255-9
Laxmi, S., Singh, A., and Asthana, R. K. (2023). “Molecular biology of non-ribosomal peptide (NRP) and polyketide (PK) biosynthesis in cyanobacteria,”. Cyanobacterial biotechnology in the 21st century. Editors B. Neilan, M. R. Z. Passarini, P. K. Singh, and A. Kumar (Singapore: Springer Nature Singapore), 133–147. doi:10.1007/978-981-99-0181-4_8
Lee, H. H., Ostrov, N., Wong, B. G., Gold, M. A., Khalil, A., and Church, G. M. (2016). Vibrio natriegens, a new genomic powerhouse. BioRxiv. doi:10.1101/058487
Li, J., Jaitzig, J., Hillig, F., Süssmuth, R., and Neubauer, P. (2014). Enhanced production of the nonribosomal peptide antibiotic valinomycin in Escherichia coli through small-scale high cell density fed-batch cultivation. Appl. Microbiol. Biotechnol. 98, 591–601. doi:10.1007/s00253-013-5309-8
Li, J., Jaitzig, J., Lu, P., Süssmuth, R. D., and Neubauer, P. (2015a). Scale-up bioprocess development for production of the antibiotic valinomycin in Escherichia coli based on consistent fed-batch cultivations. Microb. Cell. Fact. 14, 83. doi:10.1186/s12934-015-0272-y
Li, J., Jaitzig, J., Theuer, L., Legala, O. E., Süssmuth, R. D., and Neubauer, P. (2015b). Type II thioesterase improves heterologous biosynthesis of valinomycin in Escherichia coli. J. Biotechnol. 193, 16–22. doi:10.1016/j.jbiotec.2014.10.037
Li, J., Lawton, T. J., Kostecki, J. S., Nisthal, A., Fang, J., Mayo, S. L., et al. (2016). Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J. 11, 212–218. doi:10.1002/biot.201500030
Li, J., Wang, H., Kwon, Y.-C., and Jewett, M. C. (2017). Establishing a high yielding streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng. 114, 1343–1353. doi:10.1002/bit.26253
Li, J., Zhang, L., and Liu, W. (2018). Cell-free synthetic biology for in vitro biosynthesis of pharmaceutical natural products. Synthetic Syst. Biotechnol. 3, 83–89. doi:10.1016/j.synbio.2018.02.002
Li, L., Maclntyre, L. W., and Brady, S. F. (2021). Refactoring biosynthetic gene clusters for heterologous production of microbial natural products. Curr. Opin. Biotechnol. 69, 145–152. doi:10.1016/j.copbio.2020.12.011
Lilien, R. H., Stevens, B. W., Anderson, A. C., and Donald, B. R. (2005). A novel ensemble-based scoring and search algorithm for protein redesign and its application to modify the substrate specificity of the gramicidin synthetase a phenylalanine adenylation enzyme. J. Comput. Biol. 12, 740–761. doi:10.1089/cmb.2005.12.740
Lin, S.-Y., Oakley, C. E., Jenkinson, C. B., Chiang, Y.-M., Lee, C.-K., Jones, C. G., et al. (2023). A heterologous expression platform in Aspergillus nidulans for the elucidation of cryptic secondary metabolism biosynthetic gene clusters: discovery of the Aspergillus fumigatus sartorypyrone biosynthetic pathway. Chem. Sci. 14, 11022–11032. doi:10.1039/d3sc02226a
Loeschcke, A., and Thies, S. (2015). Pseudomonas putida-a versatile host for the production of natural products. Appl. Microbiol. Biotechnol. 99, 6197–6214. doi:10.1007/s00253-015-6745-4
Lu, Z., Liu, X.-H., Yuan, X., Liu, F., and Wang, T. (2023). Engineered biosynthesis through the adenylation domains from nonribosomal peptide synthetases. Curr. Top. Med. Chem. 23, 1973–1984. doi:10.2174/1568026623666230601142757
Madin, K., Sawasaki, T., Ogasawara, T., and Endo, Y. (2000). A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. U. S. A. 97, 559–564. doi:10.1073/pnas.97.2.559
Magarvey, N. A., Ehling-Schulz, M., and Walsh, C. T. (2006). Characterization of the cereulide NRPS α-hydroxy acid specifying Modules: activation of α-keto acids and chiral reduction on the assembly line. J. Am. Chem. Soc. 128, 10698–10699. doi:10.1021/ja0640187
Makrydaki, E., Marshall, O., Heide, C., Buldum, G., Kontoravdi, C., and Polizzi, K. M. (2021). Cell-free protein synthesis using Chinese hamster ovary cells. Meth. Enzymol. 659, 411–435. doi:10.1016/bs.mie.2021.08.004
Martin, R. W., Des Soye, B. J., Kwon, Y.-C., Kay, J., Davis, R. G., Thomas, P. M., et al. (2018). Cell-free protein synthesis from genomically recoded bacteria enables multisite incorporation of noncanonical amino acids. Nat. Commun. 9, 1203. doi:10.1038/s41467-018-03469-5
Matter, A. M., Hoot, S. B., Anderson, P. D., Neves, S. S., and Cheng, Y.-Q. (2009). Valinomycin biosynthetic gene cluster in Streptomyces: conservation, ecology and evolution. PLoS ONE 4, e7194. doi:10.1371/journal.pone.0007194
Mazodier, P., Petter, R., and Thompson, C. (1989). Intergeneric conjugation between Escherichia coli and Streptomyces species. J. Bacteriol. 171, 3583–3585. doi:10.1128/jb.171.6.3583-3585.1989
Messenger, S. R., McGuinniety, E. M. R., Stevenson, L. J., Owen, J. G., Challis, G. L., Ackerley, D. F., et al. (2023). Metagenomic domain substitution for the high-throughput modification of nonribosomal peptides. Nat. Chem. Biol. doi:10.1038/s41589-023-01485-1
Miao, V., Coëffet-LeGal, M.-F., Brian, P., Brost, R., Penn, J., Whiting, A., et al. (2005). Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry. Microbiol. Read. Engl. 151, 1507–1523. doi:10.1099/mic.0.27757-0
Miller, B. R., and Gulick, A. M. (2016). Structural biology of nonribosomal peptide synthetases. Methods Mol. Biol. 1401, 3–29. doi:10.1007/978-1-4939-3375-4_1
Milne, C., Powell, A., Jim, J., Al Nakeeb, M., Smith, C. P., and Micklefield, J. (2006). Biosynthesis of the (2S,3R)-3-methyl glutamate residue of nonribosomal lipopeptides. J. Am. Chem. Soc. 128, 11250–11259. doi:10.1021/ja062960c
Montalbán-López, M., Scott, T. A., Ramesh, S., Rahman, I. R., van Heel, A. J., Viel, J. H., et al. (2021). New developments in RiPP discovery, enzymology and engineering. Nat. Prod. Rep. 38, 130–239. doi:10.1039/d0np00027b
Moore, S. J., Lai, H.-E., Chee, S.-M., Toh, M., Coode, S., Chengan, K., et al. (2021). A streptomyces venezuelae cell-free toolkit for synthetic biology. ACS Synth. Biol. 10, 402–411. doi:10.1021/acssynbio.0c00581
Moore, S. J., Lai, H.-E., Li, J., and Freemont, P. S. (2023). Streptomyces cell-free systems for natural product discovery and engineering. Nat. Prod. Rep. 40, 228–236. doi:10.1039/d2np00057a
Moore, S. J., Lai, H.-E., Needham, H., Polizzi, K. M., and Freemont, P. S. (2017). Streptomyces venezuelae TX-TL - a next generation cell-free synthetic biology tool. Biotechnol. J. 12. doi:10.1002/biot.201600678
Moore, S. J., MacDonald, J. T., Wienecke, S., Ishwarbhai, A., Tsipa, A., Aw, R., et al. (2018). Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria. Proc. Natl. Acad. Sci. U. S. A. 115, E4340–E4349. doi:10.1073/pnas.1715806115
Moradi, S. V., Wu, Y., Walden, P., Cui, Z., Johnston, W. A., Petrov, D., et al. (2023). In vitro reconstitution and analysis of SARS-CoV-2/host protein-protein interactions. ACS Omega 8, 25009–25019. doi:10.1021/acsomega.3c01625
Mureev, S., Kovtun, O., Nguyen, U. T. T., and Alexandrov, K. (2009). Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 27, 747–752. doi:10.1038/nbt.1556
Murota, K., Hagiwara-Komoda, Y., Komoda, K., Onouchi, H., Ishikawa, M., and Naito, S. (2011). Arabidopsis cell-free extract, ACE, a new in vitro translation system derived from Arabidopsis callus cultures. Plant Cell. Physiol. 52, 1443–1453. doi:10.1093/pcp/pcr080
Nakashima, N., and Tamura, T. (2004). Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter. Biochem. Biophys. Res. Commun. 319, 671–676. doi:10.1016/j.bbrc.2004.05.034
Ongpipattanakul, C., Desormeaux, E. K., DiCaprio, A., van der Donk, W. A., Mitchell, D. A., and Nair, S. K. (2022). Mechanism of action of ribosomal synthesized and post-translationally modified peptides. Chem. Rev. 122, 14722–14814. doi:10.1021/acs.chemrev.2c00210
Parapouli, M., Vasileiadis, A., Afendra, A.-S., and Hatziloukas, E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–32. doi:10.3934/microbiol.2020001
Park, C. N., Lee, J. M., Lee, D., and Kim, B. S. (2008). Antifungal activity of valinomycin, a peptide antibiotic produced by Streptomyces sp. Strain M10 antagonistic to Botrytis cinerea. J. Microbiol. Biotechnol. 18, 880–884. https://koreascience.kr/article/JAKO200822179194503.page
Pereira-Dias, L., Oliveira-Pinto, P. R., Fernandes, J. O., Regalado, L., Mendes, R., Teixeira, C., et al. (2023). Peptaibiotics: harnessing the potential of microbial secondary metabolites for mitigation of plant pathogens. Biotechnol. Adv. 68, 108223. doi:10.1016/j.biotechadv.2023.108223
Pfeifer, B. A., Admiraal, S. J., Gramajo, H., Cane, D. E., and Khosla, C. (2001). Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792. doi:10.1126/science.1058092
Prasad, C. (1995). Bioactive cyclic dipeptides. Peptides 16, 151–164. doi:10.1016/0196-9781(94)00017-Z
Recktenwald, J., Shawky, R., Puk, O., Pfennig, F., Keller, U., Wohlleben, W., et al. (2002). Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules the GenBank/EMBL/DDBJ accession number for the balhimycin biosynthetic gene sequence reported in this paper is Y16952. Microbiol. Read. Engl. 148, 1105–1118. doi:10.1099/00221287-148-4-1105
Ryoo, I.-J., Park, H.-R., Choo, S.-J., Hwang, J.-H., Park, Y.-M., Bae, K.-H., et al. (2006). Selective cytotoxic activity of valinomycin against HT-29 Human colon carcinoma cells via down-regulation of GRP78. Biol. Pharm. Bull. 29, 817–820. doi:10.1248/bpb.29.817
Salari, R., and Salari, R. (2017). Investigation of the best Saccharomyces cerevisiae growth condition. Electron Physician 9, 3592–3597. doi:10.19082/3592
Schoborg, J. A., Clark, L. G., Choudhury, A., Hodgman, C. E., and Jewett, M. C. (2016). Yeast knockout library allows for efficient testing of genomic mutations for cell-free protein synthesis. Synthetic Syst. Biotechnol. 1, 2–6. doi:10.1016/j.synbio.2016.02.004
Schoborg, J. A., Hodgman, C. E., Anderson, M. J., and Jewett, M. C. (2014). Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J. 9, 630–640. doi:10.1002/biot.201300383
Schramm, M., Friedrich, S., Schmidtke, K.-U., Kiebist, J., Panzer, P., Kellner, H., et al. (2022). Cell-free protein synthesis with fungal lysates for the rapid production of unspecific peroxygenases. Antioxidants (Basel), 11. doi:10.3390/antiox11020284
Sen, R. (2010). Surfactin: biosynthesis, genetics and potential applications. Adv. Exp. Med. Biol. 672, 316–323. doi:10.1007/978-1-4419-5979-9_24
Sharma, A. (1999). “Streptomyces,” in Encyclopedia of food microbiology (Elsevier), 2134–2138. doi:10.1006/rwfm.1999.1545
Shen, B., Du, L., Sanchez, C., Edwards, D. J., Chen, M., and Murrell, J. M. (2002). Cloning and characterization of the bleomycin biosynthetic gene cluster from Streptomyces verticillus ATCC15003. J. Nat. Prod. 65, 422–431. doi:10.1021/np010550q
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755. doi:10.1038/90802
Siebels, I., Nowak, S., Heil, C. S., Tufar, P., Cortina, N. S., Bode, H. B., et al. (2020a). Cell-free synthesis of natural compounds from genomic DNA of biosynthetic gene clusters. ACS Synth. Biol. 9, 2418–2426. doi:10.1021/acssynbio.0c00186
Siebels, I., Nowak, S., Heil, C. S., Tufar, P., Cortina, N. S., Bode, H. B., et al. (2020b). Cell-free synthesis of natural compounds from genomic DNA of biosynthetic gene clusters. BioRxiv 9, 2418–2426. doi:10.1021/acssynbio.0c00186
Stachelhaus, T., Mootz, H. D., Bergendahl, V., and Marahiel, M. A. (1998). Peptide bond formation in nonribosomal peptide biosynthesis. J. Biol. Chem. 273, 22773–22781. doi:10.1074/jbc.273.35.22773
Stanišić, A., Hüsken, A., Stephan, P., Niquille, D. L., Reinstein, J., and Kries, H. (2021). Engineered nonribosomal peptide synthetase shows opposite amino acid loading and condensation specificity. ACS Catal. 11, 8692–8700. doi:10.1021/acscatal.1c01270
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi:10.1128/JB.00345-12
Sun, H., Liu, Z., Zhao, H., and Ang, E. L. (2015). Recent advances in combinatorial biosynthesis for drug discovery. Drug Des. devel. Ther. 9, 823–833. doi:10.2147/DDDT.S63023
Süssmuth, R. D., and Mainz, A. (2017). Nonribosomal peptide synthesis-principles and prospects. Angew. Chem. Int. Ed. 56, 3770–3821. doi:10.1002/anie.201609079
Sword, T. T., Barker, J. W., Spradley, M., Chen, Y., Petzold, C. J., and Bailey, C. B. (2023). Expression of blue pigment synthetase a from Streptomyces lavenduale reveals insights on the effects of refactoring biosynthetic megasynthases for heterologous expression in Escherichia coli. Purif 210, 106317. doi:10.1016/j.pep.2023.106317
Tao, X. B., LaFrance, S., Xing, Y., Nava, A. A., Martin, H. G., Keasling, J. D., et al. (2023). ClusterCAD 2.0: an updated computational platform for chimeric type I polyketide synthase and nonribosomal peptide synthetase design. Nucleic Acids Res. 51, D532–D538. doi:10.1093/nar/gkac1075
Tempelaars, M. H., Rodrigues, S., and Abee, T. (2011). Comparative analysis of antimicrobial activities of valinomycin and cereulide, the Bacillus cereus emetic toxin. Appl. Environ. Microbiol. 77, 2755–2762. doi:10.1128/AEM.02671-10
Thirlway, J., Lewis, R., Nunns, L., Al Nakeeb, M., Styles, M., Struck, A.-W., et al. (2012). Introduction of a non-natural amino acid into a nonribosomal peptide antibiotic by modification of adenylation domain specificity. Angew. Chem. Int. Ed. 51, 7181–7184. doi:10.1002/anie.201202043
Thompson, J., Rae, S., and Cundliffe, E. (1984). Coupled transcription--translation in extracts of Streptomyces lividans. Mol. Gen. Genet. 195, 39–43. doi:10.1007/BF00332721
Tian, R., Wang, M., Shi, J., Qin, X., Guo, H., Jia, X., et al. (2020). Cell-free synthesis system-assisted pathway bottleneck diagnosis and engineering in Bacillus subtilis. Synthetic Syst. Biotechnol. 5, 131–136. doi:10.1016/j.synbio.2020.06.006
Tippelt, A., and Nett, M. (2021). Saccharomyces cerevisiae as host for the recombinant production of polyketides and nonribosomal peptides. Microb. Cell. Fact. 20, 161. doi:10.1186/s12934-021-01650-y
Toh, M., Chengan, K., Hanson, T., Freemont, P. S., and Moore, S. J. (2021). A high-yield streptomyces transcription-translation toolkit for synthetic biology and natural product applications. J. Vis. Exp. doi:10.3791/63012
Tuckey, C., Asahara, H., Zhou, Y., and Chong, S. (2014). Protein synthesis using a reconstituted cell-free system. Curr. Protoc. Mol. Biol. 108, 1–22. doi:10.1002/0471142727.mb1631s108
van Wageningen, A. M., Kirkpatrick, P. N., Williams, D. H., Harris, B. R., Kershaw, J. K., Lennard, N. J., et al. (1998). Sequencing and analysis of genes involved in the biosynthesis of a vancomycin group antibiotic. Chem. Biol. 5, 155–162. doi:10.1016/s1074-5521(98)90060-6
Vilkhovoy, M., Horvath, N., Shih, C.-H., Wayman, J. A., Calhoun, K., Swartz, J., et al. (2018). Sequence specific modeling of E. coli cell-free protein synthesis. ACS Synth. Biol. 7, 1844–1857. doi:10.1021/acssynbio.7b00465
Wang, H., Li, J., and Jewett, M. C. (2018). Development of a Pseudomonas putida cell-free protein synthesis platform for rapid screening of gene regulatory elements. Synth. Biol. (Oxf) 3, ysy003. doi:10.1093/synbio/ysy003
Weinstock, M. T., Hesek, E. D., Wilson, C. M., and Gibson, D. G. (2016). Vibrio natriegens as a fast-growing host for molecular biology. Nat. Methods 13, 849–851. doi:10.1038/nmeth.3970
Wenski, S. L., Thiengmag, S., and Helfrich, E. J. N. (2022). Complex peptide natural products: biosynthetic principles, challenges and opportunities for pathway engineering. Synthetic Syst. Biotechnol. 7, 631–647. doi:10.1016/j.synbio.2022.01.007
Wick, S., and Carr, P. A. (2022). Measurement of transcription, translation, and other enzymatic processes during cell-free expression using PERSIA. Methods Mol. Biol. 2433, 169–181. doi:10.1007/978-1-0716-1998-8_10
Wick, S., Walsh, D. I., Bobrow, J., Hamad-Schifferli, K., Kong, D. S., Thorsen, T., et al. (2019). PERSIA for direct fluorescence measurements of transcription, translation, and enzyme activity in cell-free systems. ACS Synth. Biol. 8, 1010–1025. doi:10.1021/acssynbio.8b00450
Wiegand, D. J., Lee, H. H., Ostrov, N., and Church, G. M. (2018). Establishing a cell-free Vibrio natriegens expression system. ACS Synth. Biol. 7, 2475–2479. doi:10.1021/acssynbio.8b00222
Winn, M., Fyans, J. K., Zhuo, Y., and Micklefield, J. (2016). Recent advances in engineering nonribosomal peptide assembly lines. Nat. Prod. Rep. 33, 317–347. doi:10.1039/c5np00099h
Wu, C.-Y., Jan, J.-T., Ma, S.-H., Kuo, C.-J., Juan, H.-F., Cheng, Y.-S. E., et al. (2004). Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U. S. A. 101, 10012–10017. doi:10.1073/pnas.0403596101
Wu, Y., Cui, Z., Huang, Y.-H., de Veer, S. J., Aralov, A. V., Guo, Z., et al. (2022). Towards a generic prototyping approach for therapeutically-relevant peptides and proteins in a cell-free translation system. Nat. Commun. 13, 260. doi:10.1038/s41467-021-27854-9
Xu, D., Zhang, Z., Yao, L., Wu, L., Zhu, Y., Zhao, M., et al. (2023). Advances in the adenylation domain: discovery of diverse non-ribosomal peptides. Appl. Microbiol. Biotechnol. 107, 4187–4197. doi:10.1007/s00253-023-12585-2
Xu, H., Liu, W.-Q., and Li, J. (2020). Translation related factors improve the productivity of a streptomyces-based cell-free protein synthesis system. ACS Synth. Biol. 9, 1221–1224. doi:10.1021/acssynbio.0c00140
Xu, H., Liu, W.-Q., and Li, J. (2022a). A streptomyces-based cell-free protein synthesis system for high-level protein expression. Methods Mol. Biol. 2433, 89–103. doi:10.1007/978-1-0716-1998-8_5
Xu, H., Yang, C., Tian, X., Chen, Y., Liu, W.-Q., and Li, J. (2022b). Regulatory Part Engineering for high-yield protein synthesis in an all-streptomyces-based cell-free expression system. ACS Synth. Biol. 11, 570–578. doi:10.1021/acssynbio.1c00587
Yamazaki, T. (2016). A monofunctional nonribosomal peptide synthase might be involved in the biosynthesis of marine toxins of the palytoxin family. FASEB J. 30. doi:10.1096/fasebj.30.1_supplement.lb118
Yang, H., Qu, J., Zou, W., Shen, W., and Chen, X. (2021). An overview and future prospects of recombinant protein production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 105, 6607–6626. doi:10.1007/s00253-021-11533-2
Yin, J., Li, G., Ren, X., and Herrler, G. (2007). Select what you need: a comparative evaluation of the advantages and limitations of frequently used expression systems for foreign genes. J. Biotechnol. 127, 335–347. doi:10.1016/j.jbiotec.2006.07.012
Zarins-Tutt, J. S., Barberi, T. T., Gao, H., Mearns-Spragg, A., Zhang, L., Newman, D. J., et al. (2016). Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Nat. Prod. Rep. 33, 54–72. doi:10.1039/c5np00111k
Zhang, H., Li, Z., Zhou, S., Li, S.-M., Ran, H., Song, Z., et al. (2022). A fungal NRPS-PKS enzyme catalyses the formation of the flavonoid naringenin. Nat. Commun. 13, 6361. doi:10.1038/s41467-022-34150-7
Zhang, J., Yan, Y.-J., An, J., Huang, S.-X., Wang, X.-J., and Xiang, W.-S. (2015). Designed biosynthesis of 25-methyl and 25-ethyl ivermectin with enhanced insecticidal activity by domain swap of avermectin polyketide synthase. Microb. Cell. Fact. 14, 152. doi:10.1186/s12934-015-0337-y
Zhang, L., Wang, C., Chen, K., Zhong, W., Xu, Y., and Molnár, I. (2023). Engineering the biosynthesis of fungal nonribosomal peptides. Nat. Prod. Rep. 40, 62–88. doi:10.1039/d2np00036a
Zhang, Y., Chen, M., Bruner, S. D., and Ding, Y. (2018). Heterologous production of microbial ribosomal synthesized and post-translationally modified peptides. Front. Microbiol. 9, 1801. doi:10.3389/fmicb.2018.01801
Zhong, G., Wang, Z.-J., Yan, F., Zhang, Y., and Huo, L. (2023). Recent advances in discovery, bioengineering, and bioactivity-evaluation of ribosomal synthesized and post-translationally modified peptides. ACS Bio Med. Chem. Au 3, 1–31. doi:10.1021/acsbiomedchemau.2c00062
Zhou, Q., Ning, S., and Luo, Y. (2020). Coordinated regulation for nature products discovery and overproduction in Streptomyces. Synthetic Syst. Biotechnol. 5, 49–58. doi:10.1016/j.synbio.2020.04.002
Zhu, B., Gan, R., Cabezas, M. D., Kojima, T., Nicol, R., Jewett, M. C., et al. (2020). Increasing cell-free gene expression yields from linear templates in Escherichia coli and Vibrio natriegens extracts by using DNA-binding proteins. Biotechnol. Bioeng. 117, 3849–3857. doi:10.1002/bit.27538
Keywords: non-ribosomal peptide synthetase, modular megaenzyme, peptide natural products, synthetic biology, cell-free protein synthesis (CFPS)
Citation: Sword TT, Abbas GSK and Bailey CB (2024) Cell-free protein synthesis for nonribosomal peptide synthetic biology. Front. Nat. Produc. 3:1353362. doi: 10.3389/fntpr.2024.1353362
Received: 10 December 2023; Accepted: 18 January 2024;
Published: 12 February 2024.
Edited by:
Jian Li, ShanghaiTech University, ChinaReviewed by:
Hajo Kries, Leibniz Institute for Natural Product Research and Infection Biology, GermanyCopyright © 2024 Sword, Abbas and Bailey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Constance B. Bailey, Q29uc3RhbmNlLkJhaWxleUBTeWRuZXkuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.