- 1Cluster of Excellence Physics of Life, TU Dresden, Dresden, Germany
- 2Institute of Resource Ecology, Helmholtz Zentrum Dresden Rossendorf, Dresden, Germany
- 3Faculty of Chemistry, University of Belgrade, Belgrade, Serbia
- 4Biotechnology Center (BIOTEC), Center for Molecular and Cellular Bioengineering (CMCB), TU Dresden, Dresden, Germany
Biomolecular condensation is an important mechanism of cellular compartmentalization without membranes. Formation of liquid-like condensates of biomolecules involves protein-protein interactions working in tandem with protein-water interactions. The balance of these interactions in condensate-forming proteins is impacted by multiple factors inside of a living organism. This work investigates the effects of post-translational modifications (PTMs) and salt concentration as two such perturbing factors on the protein Fused in Sarcoma (FUS), an RNA binding protein. The protein was obtained from two expression systems differing by their capability to add PTMs to the protein, bacterial and insect cell. Attenuated total reflection Terahertz spectroscopy is used to probe the solvation behavior in condensates formed from FUS protein with and without PTMs at 100 mM and 2.5 M KCl. The results show that while PTMs impact the phase-separating propensity, they do not alter protein solvation in the condensate. On the other hand, salt concentration was found to alter the stiffness of the water hydrogen bond network. These findings have implications for biomolecular condensates chemistry, showing that condensate molecular organization is perturbed by fluctuations in solvent properties.
1 Introduction
In living organisms one of the key principles of organizing matter is compartmentalization. One mechanism of sequestration that has gained importance in the life sciences is biomolecular condensation (Brangwynne et al., 2009; Banani et al., 2017). In this process, biomolecules assemble to form membrane-less organelles. Such assemblies, while separate from their surroundings, remain liquid and dynamic. Condensation influences biological processes on multiple scales, from regulating rates of reactions to buffering biomolecule concentrations in times of cellular stress (Klosin et al., 2020; Lyon et al., 2020; Alberti and Hyman, 2021). Condensates are commonly formed by proteins containing intrinsically disordered regions, where polypeptide chains do not fold into stable secondary or tertiary structures. Biomolecular condensation can be described through the framework of coupled associative and segregative transitions (Pappu et al., 2023). In this context, associative interactions describe electrostatics-based interactions between polypeptide residues, i.e., protein-protein interactions, whereas segregative interactions refer to unfavorable interactions with the solvent, i.e., protein-water interactions.
Protein-protein interactions have been shown to be essential for condensation, where multivalent, transient contacts between amino acid residues drive phase separation (Wang et al., 2018; Martin et al., 2020), resulting in the formation of a percolation network spanning the whole droplet (Farag et al., 2022). For instance, condensates of the RNA-binding protein Fused in Sarcorma (FUS) are held together by interactions between the aromatic tyrosine residues in the N-terminal domain and positively charged arginine residues in the C-terminal domain (Wang et al., 2018). Small changes in the protein sequence, such as those from post-translational modifications (PTMs), in which methylation or phosphorylation occur, impact the efficiency of phase separation (Hofweber and Dormann, 2019). In the case of FUS, arginine methylation (Qamar et al., 2018) increases the saturation concentration of phase separation. How protein sequence governs biomolecular condensation has been modeled theoretically with the stickers and spacers model. In this model, polymer chains contain patches or sequences that strongly interact, known as stickers, interspersed with regions that don’t interact called spacers (Villegas et al., 2022). Theoretical work has predicted that the effective solvation volume of the spacers impacts the effective valence and strength of the stickers in a manner analogous to changing their patterning in the protein sequence, since poor solvent behavior of the spacers brings the stickers closer to each other (Semenov and Michael, 1998; Harmon et al., 2017; Choi et al., 2020; Martin et al., 2020). The intermolecular interactions driving the formation of condensates are thus sensitive to both changes in the protein as well as its solvation environment. Such studies highlight that protein-water interactions play an equally important role as protein-protein interactions in biomolecular condensation.
Knowledge of protein hydration in condensates and how it relates to specific amino acid interactions is still lacking (Ribeiro et al., 2019). In the last years, several studies have emerged demonstrating that the liberation of hydration water from the protein solvation shell provides the entropic contribution to the free energy of condensation (Ahlers et al., 2021; Pezzotti et al., 2023; Watson et al., 2023), while the electrostatic interactions between polypeptide chains provide the enthalpic contribution (Mukherjee and Schäfer, 2023). The water retained in condensates after their formation shows significantly reduced mobility. This change in water dynamics is caused by hydrogen binding to the protein, resulting in confined water molecules in the percolation network (Lorenz-Ochoa and Baiz, 2023). However, water dynamics are not unrelated to electrostatic interactions inside condensates. The accumulation of charges from biomolecules and counterions results in the formation of unique electrochemical environments (Dai et al., 2024). Charged residues of biomolecules perturb the coordination of water molecules, creating heterogeneous water populations that depend on the biomolecule morphology (Laage et al., 2017). Recent studies have suggested that electrostatic protein interactions increase tetrahedral order of solvation water in condensates, while decreased water ordering stems from hydrophobic protein interactions (Joshi et al., 2024). For more complex, multilayered condensates pH gradients between the layers of a condensate can form, caused by differences in the amount of charged residues (King et al., 2024). Such studies indicate that solvent properties, including ion distribution, are linked to the protein biochemical makeup.
A common way to study macroscopic solvent effects on condensates is through the mapping of phase diagrams. Experimental parameters such as temperature, pH, protein concentration, as well as solute concentration can be easily changed in vitro, allowing for determination of what solvent conditions disrupt the delicate balance of protein-protein and protein-solvent interactions. For example, FUS is known to undergo phase separation in biologically relevant salt concentration regime, 50–150 mM. Outside this range, phase separation is suppressed due to electrostatic screening. However, the low complexity (LC) N-terminal domain of FUS, containing only the tyrosine-rich region, undergoes phase separation that is enhanced by salt (Murthy et al., 2019). For LC-FUS, hydrophobic interactions and salting-out effects contribute to driving phase separation. Interestingly, salting-out effects also have an effect on full-length FUS, which has been shown to undergo a concentration-dependent reentrant phase transition (Krainer et al., 2021). In addition to forming condensates at near-physiological salt concentrations, FUS also forms them at high salt concentrations (>2 M). Studies on model peptides have shown that hydrophobicity of arginine drives reentrant phase separation (Hong et al., 2022). Such behavior shows that even a single protein can switch between electrostatic to hydrophobic protein-protein interactions depending on its solvent environment (Hazra and Levy, 2023). Yet, how or whether protein-solvent interactions in condensates are altered by solvent conditions remains poorly understood.
The objective of this study is to determine how protein properties and solvent properties influence protein-water interactions in biomolecular condensates. To this end, FUS was utilized to prepare in vitro condensates, as it has relevance to neurodegenerative diseases and its phase diagrams have been well established. Here, protein properties of FUS were altered through PTMs and were achieved by expression of FUS in prokaryotic and eukaryotic systems (Hofweber et al., 2018; Qamar et al., 2018). Solvent properties were altered through increased salt concentration. To probe the protein-water in condensates, Terahertz (THz) spectroscopy was utilized. This method is sensitive to perturbations in the water hydrogen-bonding network and reports on structure and dynamics of solvation water (Ahlers et al., 2021). Results show that even though PTMs suppress phase separation of FUS, protein-water interactions in condensates are not perturbed by molecular level changes of the protein. Solvent conditions, however, impact the stiffness of the solvation network, where increased salt concentration softens the hydrogen-bonding network. Taken together, these results indicate that PTMs mostly influence phase separation through modification of protein-protein interactions, while solvent conditions influence protein-water interactions.
2 Materials and methods
2.1 Protein expression and purification
Bacterial expression of maltose binding protein-FUS-enhanced green fluorescent protein (MBP-FUS-eGFP) was adapted from literature (Ahlers et al., 2021). Bacterial cultures of BL-21 E. coli with a volume of 1–2 L were grown in lysogeny broth (LB) medium until reaching an optical density (600 nm) of 0.6. 0.1 mM isopropyl β-D-1-thiogalactopyranoside was added and the cultures were incubated for 22 h at 12°C. Afterwards cells were pelleted by centrifugation, resuspended in lysis buffer (50 mM Tris:HCl, pH 8, 1 M KCl, 20 mM imidazole, 1 mM dithiothreitol, 5% glycerol) with an EDTA-free proteased inhibitor cocktail, spun down in 50 mL tubes, snap-frozen, and stored at −80°C for later use. Eukaryotic expression of maltose binding protein-FUS-monomeric enhanced green fluorescent protein (MBP-FUS-mEGFP) was adapted from literature (Patel et al., 2015). For eukaryotic expression, Sf9 insect cells (Expression Systems, Cat#94-001F) with a baculovirus expression system were used. Cells were harvested 72 h after inoculation, pelleted by centrifugation and resuspended in lysis buffer of the same composition and volume ratio as bacterial cells. The sequence of the FUS protein was identical for both expression systems used to create recombinant constructs.
Before cell lysis, bacterial cells were thawed at room temperature and resuspended in lysis buffer at a ratio of 1 mL of buffer to 10 mL of the original culture. Sf9 cells were lysed immediately after harvesting. For both expression systems cell lysis was performed using a LM10 Microfluidizer (Microfluidics, Germany), and the resulting lysate was cleared from debris by centrifugation at 40,000 g for 45 min at room temperature for bacterial lysate, and 18,000 g for 60 min for insect cell lysate. The resulting supernatant was applied to a tandem of 2 Protino® Ni-NTA 5 mL columns (Macherey-Nagel, Germany) for protein purification. The protein was washed at the column with wash buffer (50 mM Tris:HCl, pH 8, 1 M KCl, 25 mM imidazole, 1 mM DTT, 5% glycerol), followed with elution buffer (50 mM Tris:HCl, pH 8.5, 1 M KCl, 250 mM imidazole, 1 mM DTT, 5% glycerol). The eluted protein fraction was collected and applied to a HiLoad™ 16/600 Superdex™ 200 pg size exclusion chromatography column (Cytiva). For the insect cell expressed protein, cleavage of the protein with 3C protease (in-house) was performed before application to the column. The protein was eluted with size exclusion buffer (50 mM Tris:HCl, pH 7.4, 500 mM KCl, 1 mM DTT, 5% glycerol), and pooled protein fractions were concentrated by centrifugation on a Vivaspin® 20 column (Sartorius, 30 kDa cutoff filter). Protein concentration was determined by absorbance at 488 nm assuming an extinction coefficient of the EGFP tag to be 55,900 M−1 cm-1, and 56,000 M−1 cm−1 for mEGFP as taken from FPBase (Lambert, 2019). The protein was then split into aliquots, snap-frozen using liquid nitrogen and stored at −80°C.
2.2 Sample preparation
KCl buffers were prepared by disolving appropriate amounts of Tris-HCl and KCl in water. Solutions were adjusted to pH 7.4 with HCl and were filtered with a 0.2 µm filter. FUS samples for experiments at low salt conditions (100 mM KCl) were prepared by diluting the FUS aliquot with 50 mM Tris buffer at pH 7.4. For 2.5 M KCl solvent an appropriate amount of 3 M KCl buffer was used for dilution. The protein samples were prepared at desired salt concentration and in the case of protein from bacterial expression, phase separation was induced by adding the TEV protease to cleave off the MBP fusion tag.
2.3 THz spectroscopy
FTIR-THz measurements of FUS condensates were performed with a FTIR spectrometer (Bruker Vertex 80v, Germany) equipped with a mercury arc lamp source and a liquid helium cooled Si bolometer detector. The device was equipped with a single bounce diamond ATR unit (Platinum, Bruker, Germany). Spectra were measured from 30 to 680 cm−1 with a resolution of 1 cm−1 and are an average of 64 scans. Phase separation of FUS samples was prepared following literature and 10 min after induction of phase separation, 50 µL of solution was placed in an ATR flow cell (Bruker, Germany). The inlet and outlet of the flow cell were sealed with Teflon caps and parafilm to prevent evaporation. A time series of THz spectra was collected, with individual spectra recorded every 3.5 min for 90 min. Between measurements the diamond prism was cleaned with ultrapure water, 0.5 M NaOH, and isopropanol. All spectra were recorded at room temperature (22°C).
The absorption coefficient (α(ν)) of the phase-separated samples was determined from the Beer-Lambert law, as shown in Equation 1:
where I(v) and I0(v) are the intensities of the sample and reference, respectively, and dp is the penetration depth. Here, the bare diamond surface served as the reference. The wavelength (λ) dependent penetration depth was determined from the incident angle of the THz radiation (θ = 45°) and the refractive indices of the diamond (ndiamond = 2.38) and sample (nsample), as shown in Equation 2. Here it was assumed that nsample = 1.5 (Ahlers et al., 2021). The THz penetration depth in this wavelength regime is on the order of several μm (see Supplementary Figure S1).
Difference spectra, Δα, were obtained by subtracting the first spectrum in the time series (t = 0 min) from the rest of the spectra. The Δα spectra corresponding to the difference between the final and initial timepoint were used for further analysis. Spectral deconvolution was done with fitting of damped harmonic oscillators, as shown in Equation 3,
where an, w0,n, and νd,n are respectively, the amplitude, width, and center frequency of the nth peak (Schienbein et al., 2017).
2.4 Microscopy
Fluorescence images were recorded on a CSU-X spinning disk confocal microscope (Nikon) equipped with an iXon Ultra CCD camera (Andor). Samples were prepared in a 384 well PhenoPlate (Revvity) Image acquisition was done using a ×100 oil immersion objective with a numerical aperture of 1.49 (Nikon). The laser power of the argon laser source was set to 0.5% at 488 nm. 70.38 × 70.38 µm images were acquired for each condition. Image processing and droplet size quantification was done in ImageJ.
3 Results and discussion
The FUS protein was utilized as a model protein for in vitro phase separation. Previous studies have mapped the phase diagram of FUS, and it has been shown to undergo LLPS when expressed in both E. coli bacterial cells and Sf9 insect cells (Wang et al., 2018; Hofweber et al., 2018). Bacterial cells do not possess the cellular machinery needed to add PTMs to FUS (Hofweber et al., 2018), while eukaryotic cells, such as Sf9 insect cells, produce FUS containing PTMs (Qamar et al., 2018). For FUS, modifications that most commonly occur are phosphorylation and methylation (Rhoads et al., 2018a). Phosphate groups are typically added to hydrophilic serine and threonine residues, increasing the charge of the sequence (Monahan et al., 2017; Rhoads et al., 2018b; Rhoads et al., 2018a). Methylation, on the other hand, introduces hydrophobic groups primarily on positively charged arginine residues (Rhoads et al., 2018a; Dormann et al., 2012). This introduces steric hindrance to the cation-π interactions of arginine and tyrosine underlying condensate formation (Dormann et al., 2012; Qamar et al., 2018; Hofweber et al., 2018). Other common PTMs, such as glycosilation, do not readily occur for FUS (Kamemura, 2017; Kakuo et al., 2023). These modifications alter both the charge and hydrophobicity of the protein, therefore likely altering intermolecular hydrogen-bonding interactions. To determine how these modifications impact condensate morphology, fluorescence microscopy images of 7 µM GFP-labeled FUS condensates from E. coli bacterial cells and Sf9 insect cells at low salt (100 mM KCl) and high salt (2.5 M KCl) conditions were collected and are shown in Figure 1A. For simplicity, FUS without PTMs will be referred to as bacterial FUS and that with PTMs will be referred to as insect FUS. Low salt conditions are consistent with previous literature (Hofweber and Dormann, 2019; Monahan et al., 2017; Hofweber et al., 2018), where spherical-like droplets form. Droplets from insect FUS are smaller in size relative to bacterial FUS. Quantification of the total droplet area (Figure 1B) reveals that insect FUS at low salt only occupies roughly 10% of the total area while bacterial FUS occupies ∼55%. At 2.5 M KCl, where reentrant phase separation of FUS occurs, the droplet morphology of both bacterial and insect FUS is altered. Droplets are no longer spherical-like and instead coalesce into large amorphous shapes. This shape change likely stems from increased hydrophobic protein-protein interactions (Krainer et al., 2021), resulting in droplets becoming more solid-like in character. More solid-like behavior has been shown in the reentrant phase transition of another arginine-rich protein, protamine (Hong et al., 2022), suggesting that mesoscale properties are linked to molecular interactions in condensates. Quantification of the total droplet areas also shows that at high salt insect FUS covers a smaller percentage area than bacterial, ∼40% vs 65%. Interestingly, when comparing the droplet percentage areas, it is notable that the presence of PTMs causes a decrease in the condensate area in both the low and high salt regimes, indicating PTMs suppress condensation. However, PTMs suppress condensation to a greater extent in the low salt regime (55% for bacteria vs 10% for insect FUS). This suggests that protein condensation in high salt is less affected by PTMs. Previous work found that at high salt concentration, the excess concentration of ions changes the types of interactions between protein molecules from electrostatic to hydrophobic. Therefore, it seems that PTMs which introduce charge changes have a greater effect on LLPS of FUS than hydrophobic ones. A possible explanation for such an effect could be due to increased protein-water interactions by charged hydrophilic groups, such as phosphates.
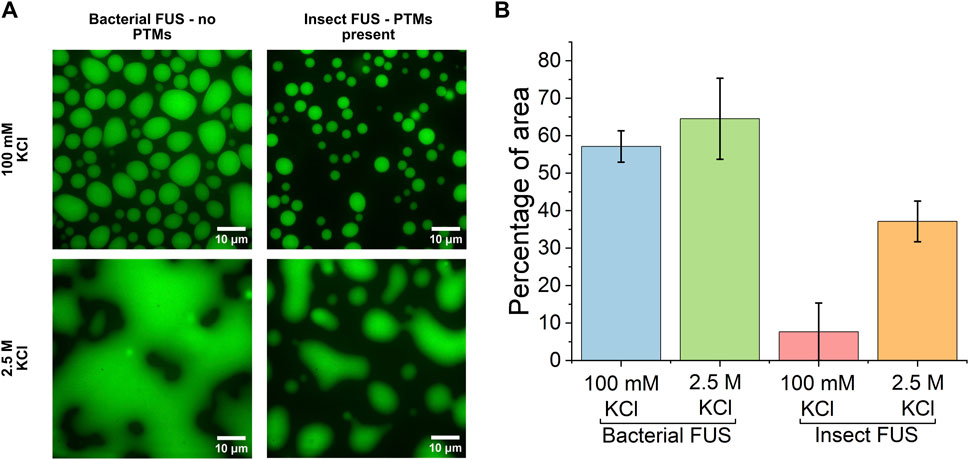
Figure 1. PTMs decrease condensate area (A). Confocal microscopy images of condensates formed by 7 µM FUS-GFP from a bacterial and insect cell expression system in 100 mM KCl and 2.5 M KCl (B). Percentage of image area covered by the condensed phase.
To understand how PTMs impact protein-water interactions, the collective behavior of water in condensates was probed by ATR-FTIR absorption spectroscopy in the THz region. Figure 2 presents the ATR-FTIR spectra of 7 µM FUS condensates from bacterial and insect expression at 100 mM KCl and 2.5 M KCl. Here, spectra are presented as the difference in the absorption coefficient of the sample at t = 0 min and t = 90 min, denoted Δα. The change in absorption with time comes from accumulation of condensates at the surface of the ATR crystal through sedimentation. The scaling of the spectrum intensity with the amount of condensates has been shown in previously published work (Ahlers et al., 2021). Only the initial (t = 0 min) and final (t = 90 min) timepoints were used for further analysis, as these represent the difference between the dilute and condensed phase. For bacterial FUS at 100 mM KCl (Figure 2A), the Δα spectrum has two prominent features with peak center frequencies at ∼130 cm−1 and ∼370 cm−1. These features are consistent with previously published literature (Ahlers et al., 2021), which looked at FUS condensates in 150 mM NaCl and phosphate buffer, and were assigned to desolvation of hydrophobic moieties on the protein (130 cm−1) and the stiff network of bound water inside the condensates (370 cm−1). For simplicity, the feature at 130 cm-1 will be referred as the desolvation peak, while the feature as 370 cm−1 will be referred to as the stiffness peak throughout the remainder of the text. This spectrum serves as a baseline case for biomolecular condensate hydration. The spectra of insect FUS (Figure 2B) at 100 mM KCl show a smaller change in the Δα, which is approximately half the intensity of bacterial FUS. This change in intensity can be ascribed to the decrease in condensate droplet area caused by the presence of PTMs, as discussed earlier. Overall, the shape of the spectrum is similar between the two expression systems.
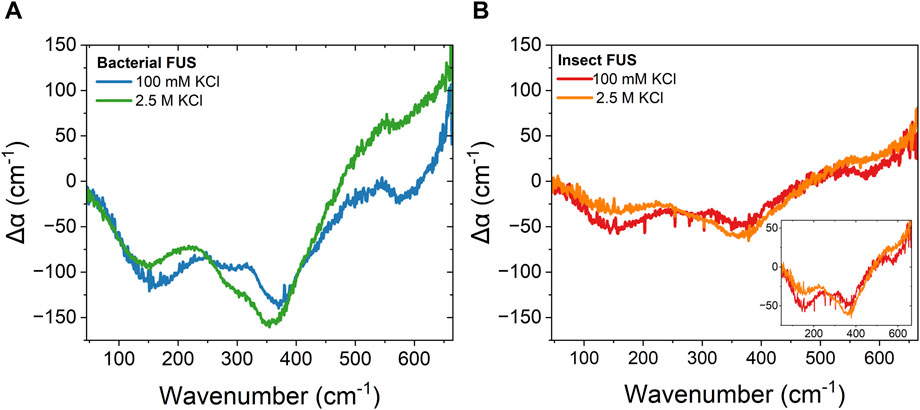
Figure 2. Reentrant phase separation gives rise to vastly different condensate hydration independent of PTMs. ATR-FTIR Δα spectra of 7 µM FUS-GFP condensates formed in 100 mM KCl and 2.5 M KCl for (A). Bacterial FUS (B). Insect FUS. Inset: insect FUS with narrowed y-axis range. The average error of all measurements is 16 cm−1.
This is similarly observed when comparing bacterial and insect FUS at 2.5 M KCl. Both spectra have similar shape, and the intensity of the insect FUS is smaller than that of the bacterial FUS. Again, this is due to the decreased droplet area in the case of insect FUS at 2.5 M KCl (see Figure 1B). However, the spectra at 2.5 M KCl are distinct from that at 100 mM. The hydrophobic desolvation feature at 130 cm-1 is less prominent relative to the high frequency peak at 370 cm-1. This shows that the population of hydrophobic hydration water that contributes to condensate formation is suppressed in the high salt regime. This likely arises from accumulation of ions at the protein interface, resulting in electrostatic screening of charged arginine groups, which then leaves less hydrophobic wrap water to be released as the normal cation-π interaction that drives FUS condensation is inhibited. The hydrophobic desolvation peak was also less prominent in liquid-solid phase separation (Ramos et al., 2023), indicating that release of solvation water is of less prominent for hydrophobically driven condensation. Considering both the low and high salt results for bacterial and insect FUS, indicates that PTMs do not significantly alter the protein-water interactions responsible for driving phase separation.
Comparison of the low and high salt spectra show spectral shifts in the peak frequencies. To quantify these shifts, spectral fitting with damped harmonics oscillators (Equation 3) was done (see Supporting Material for full fit results). Results of the spectral fitting are shown in Figure 3. At high salt conditions, both the desolvation and hydrogen-bonding stiffness peaks red shift in frequency compared to low salt conditions. For the desolvation peak (Figure 3A), this shift is 10–15 cm-1, indicating that hydrophobically driven condensation is distinguishable from electrostatically driven condensation. Such results show that the frequency of the desolvation peak can be used as a spectroscopic marker for condensation. For the water stiffness peak (Figure 3B), a red shift of ∼15 cm−1 is also observed. This high frequency peak in the condensate spectrum results from blue and red shifts of the librational peak of water, which reports on the relative stiffness of the water network in the condensate. A blue shift corresponds to a stiffer water network, while a red shift corresponds to a softer water network. Therefore, the red shift observed for reentrant phase-separated condensates indicates that the bound water inside the condensate has a softer hydrogen-bonding network compared to the low salt regime.
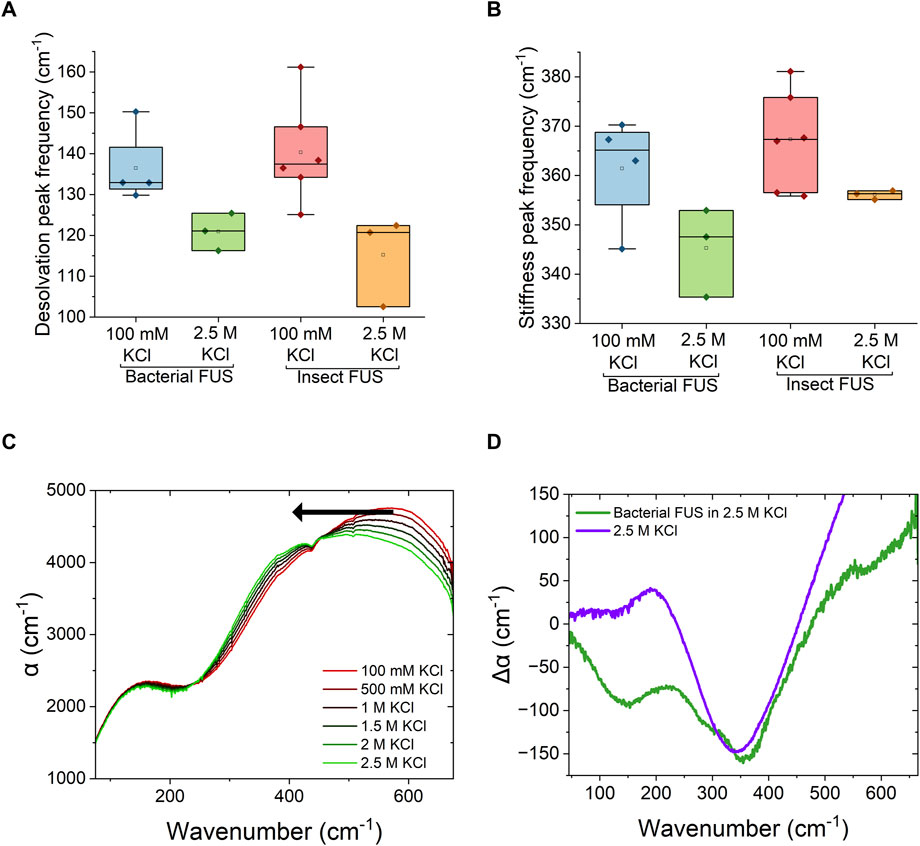
Figure 3. FUS condensation counteracts hydration changes caused by KCl (A). Fitted peak center frequencies of the low frequency feature, corresponding to protein desolvation, in the Δα spectra of FUS-GFP condensates formed in 100 mM KCl and 2.5 M KCl (B). Fitted peak center frequencies of the high frequency feature, correspond to stiffness of the hydrogen-bond network, in the Δα spectra of FUS-GFP condensates formed in 100 mM KCl and 2.5 M KCl (C). THz absorption spectra of KCl concentration series in 50 mM Tris:HCl at pH 7.4 (D). Comparison of the Δα spectra of FUS-GFP condensates formed in 2.5 M KCl with the negative of the molar absorption coefficient of 2.5 M KCl buffer obtained from data shown in panel (C).
In order to determine the source of the softening of the water network in high salt condensates, ATR-FTIR spectra of KCl solutions at various concentrations were measured and are shown in Figure 3C. As the concentration of KCl increases, the librational band of water red shifts to lower frequencies. This corresponds to a decrease in tetrahedral order of water molecules (Morawietz et al., 2018). Therefore the changes in solvent stiffness in condensates are related to the stiffness of the solvent itself. Figure 3D shows the comparison of Δα spectra of bacterial FUS protein at high salt KCl with that of the Δα spectra of 2.5 M KCl buffer relative to 0 M KCl buffer. The blue shift in the FUS condensate spectrum partially reverses the induced softness of the water network caused by the solvent itself. This could stem from decreased degrees of freedom of bound water molecules in the condensate, where hydrophobic interactions between protein molecules can confine the water to a larger degree. A change in the libration region can also come from spatial confinement of the water, as THz spectra of water trapped between graphene layers have previously shown a similar blue shift in the libration region of the spectrum with increasing confinement (Ruiz-Barragan et al., 2022). However, the increased stiffness could also point to selective partitioning of ions into the dense phase. Recent results have shown that while KCl is excluded from condensates at low salt concentrations, enrichment of KCl ions in condensed phase occurs in the reentrant phase separation regime (Ausserwöger et al., 2023). Interactions of highly concentrated ions with the condensed protein might cause the ions to lose their hydration shell through forming solvent shared or contact ion pairs. Overall, such results show that independent of the solvent stiffness itself, the stiffness inside of the dense condensates phase is greater than that of the dilute phase.
As discussed earlier, a change in the desolvation and water stiffness bands were observed for both bacterial and insect FUS in the high salt regime. In order to more quantitatively assess these changes, the ratios of integrated areas of the water stiffness peak to the librational peak obtained from the damped harmonic oscillator fits were determined and are shown in Figure 4 (see Supplementary Figure S2 for example, of spectral fits). At 100 mM KCl, both bacterial and insect FUS have a peak ratio value around 1, while this value is between 1.5 and 3.6 for the high salt regime. The prominence of the high frequency peak indicates that there is a greater population of bound water in the high salt concentration regime. This is consistent with studies of hyaluronic acid and protamine coacervates, which found reduced mobility of water in high salt condensates (Hong et al., 2022). Taken all together, the results obtained here show that modification of solvent properties have a greater impact on the protein-water interactions in FUS condensates than protein level modifications.
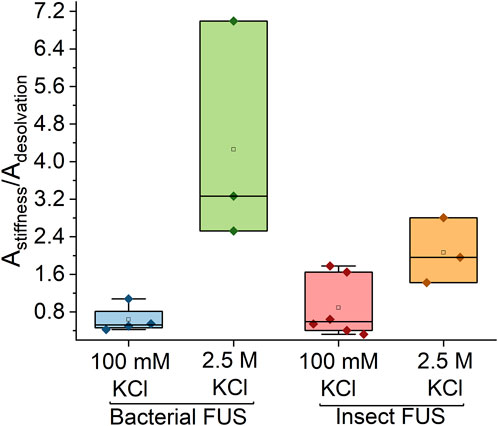
Figure 4. High salt concentration causes major shifts in the stiffness of the water network. Peak area ratios of high frequency to low frequency features. For high salt concentration ratio has a magnitude much greater than 1, while for low salt the value is close to 1.
4 Conclusion
Studies of phase-separating proteins have indicated that chemical modification of the protein through PTMs impacts the ability to form condensates. Spectroscopic measurements of FUS condensates show that these protein modifications don’t influence the solvation network inside the condensate. These PTMs likely have a larger impact on protein-water interactions in the dilute phase, i.e., before condensation occurs, and hence perturbs the saturation concentration of the phase transitions. Probing solvation properties of proteins in the low concentration limit (µM or smaller) remains a challenge, as this is below the detection limit of most methods. However, results obtained here unveil that the water hydrogen-bonding network inside of condensates is strongly impacted by the solvent environment. The water network is stiff for low salt concentrations and becomes softer at high salt concentration. Compared to the dilute phase, water inside in the condensates is always stiffer, reflective of water with suppressed degrees of freedom similar to confined or interfacial water. Such results have implications for biological processes, where fluctuations in ion concentration or pH can impact the solvation environment inside biomolecular condensates.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
AC: Data curation, Formal Analysis, Investigation, Methodology, Visualization, Writing–original draft, Writing–review and editing. AU: Investigation, Writing–review and editing. MR: Investigation, Methodology, Writing–review and editing. LP: Investigation, Methodology, Writing–review and editing. MT: Investigation, Writing–review and editing. MJ: Conceptualization, Supervision, Writing–review and editing. EA: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was support by Germany’s Excellence Strategy-EXC 2068—Projektnummer 390729961 from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) via the Cluster of Excellence Physics of Life, the Helmholtz Zentrum Dresden Rossendorf, and the DRESDEN-Concept Science and Innovation Campus.
Acknowledgments
We thank Dorothee Dormann for providing the FUS plasmids for bacterial expression. The plasmids for insect cell expression were made by Andrey Pozniakovsky (Hyman lab). We thank Nagaraja Chappidi (Alberti lab) for kindly providing us with FUS from insect cell expression. We also thank Quang Minh Thai for experimental support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnano.2025.1556384/full#supplementary-material
References
Ahlers, J., Adams, E. M., Bader, V., Pezzotti, S., Winklhofer, K. F., Tatzelt, J., et al. (2021). The key role of solvent in condensation: mapping water in liquid-liquid phase-separated FUS. Biophysical J. 120 (7), 1266–1275. doi:10.1016/j.bpj.2021.01.019
Alberti, S., and Hyman, A. A. (2021). Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell. Biol. 22 (3), 196–213. doi:10.1038/s41580-020-00326-6
Ausserwöger, H., Qian, D., Krainer, G., de Csilléry, E., Welsh, T. J., Sneideris, T., et al. (2023). Quantifying collective interactions in biomolecular phase separation. BioRxiv 2023, 543137. doi:10.1101/2023.05.31.543137
Banani, S. F., Lee, H. O., Hyman, A. A., and Rosen, M. K. (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell. Biol. 18 (5), 285–298. doi:10.1038/nrm.2017.7
Brangwynne, C. P., Eckmann, C. R., Courson, D. S., Rybarska, A., Hoege, C., Gharakhani, J., et al. (2009). Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324, 1729–1732. doi:10.1126/SCIENCE.1172046
Choi, J.Mo, Holehouse, A. S., and Pappu, R. V. (2020). Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133. doi:10.1146/ANNUREV-BIOPHYS-121219-081629
Dai, Y., Wang, Z.-G., and Zare, R. N. (2024). Unlocking the electrochemical functions of biomolecular condensates. Nat. Chem. Biol. 20 (11), 1420–1433. doi:10.1038/s41589-024-01717-y
Dormann, D., Madl, T., Valori, C. F., Bentmann, E., Tahirovic, S., Abou-Ajram, C., et al. (2012). Arginine methylation next to the PY-nls modulates transportin binding and nuclear import of FUS. EMBO J. 31 (22), 4258–4275. doi:10.1038/emboj.2012.261
Farag, M., Cohen, S. R., Borcherds, W. M., Bremer, A., Mittag, T., and Pappu, R. V. (2022). Condensates formed by prion-like low-complexity domains have small-world network structures and interfaces defined by expanded conformations. Nat. Commun. 13 (1), 7722–7815. doi:10.1038/s41467-022-35370-7
Harmon, T. S., Holehouse, A. S., Rosen, M. K., and Pappu, R. V. (2017). Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. ELife 6 (November), e30294. doi:10.7554/ELIFE.30294
Hazra, M. K., and Levy, Y. (2023). Cross-talk of Cation−π interactions with electrostatic and aromatic interactions: a salt-dependent trade-off in biomolecular condensates. J. Phys. Chem. Lett. 14 (38), 8460–8469. doi:10.1021/ACS.JPCLETT.3C01642
Hofweber, M., and Dormann, D. (2019). Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294 (18), 7137–7150. doi:10.1074/JBC.TM118.001189
Hofweber, M., Hutten, S., Bourgeois, B., Spreitzer, E., Niedner-Boblenz, A., Schifferer, M., et al. (2018). Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 173 (3), 706–719.e13. doi:10.1016/J.CELL.2018.03.004
Hong, Y., Najafi, S., Casey, T., Shea, J. E., Han, S. I., and Hwang, D. S. (2022). Hydrophobicity of arginine leads to reentrant liquid-liquid phase separation behaviors of arginine-rich proteins. Nat. Commun. 13 (1), 7326–7415. doi:10.1038/s41467-022-35001-1
Joshi, A., Avni, A., Walimbe, A., Rai, S. K., Sarkar, S., and Mukhopadhyay, S. (2024). Hydrogen-bonded network of water in phase-separated biomolecular condensates. J. Phys. Chem. Lett. 15 (30), 7724–7734. doi:10.1021/ACS.JPCLETT.4C01153
Kakuo, M., Horii, T., Tonomura, N., Sato, R., Ogawa, M., Okajima, T., et al. (2023). Evidence that only EWS among the FET proteins acquires a low partitioning property for the hyperosmotic stress response by O-GlcNAc glycosylation on its low-complexity domain. Exp. Cell. Res. 424 (1), 113504. doi:10.1016/J.YEXCR.2023.113504
Kamemura, K. (2017). O-GlcNAc glycosylation stoichiometry of the FET protein family: only EWS is glycosylated with a high stoichiometry. Biosci. Biotechnol. Biochem. 81 (3), 541–546. doi:10.1080/09168451.2016.1263148
King, M. R., Ruff, K. M., Lin, A. Z., Pant, A., Farag, M., Lalmansingh, J. M., et al. (2024). Macromolecular condensation organizes nucleolar sub-phases to set up a PH gradient. Cell. 187 (8), 1889–1906.e24. doi:10.1016/J.CELL.2024.02.029
Klosin, A., Oltsch, F., Harmon, T., Honigmann, A., Jülicher, F., Hyman, A. A., et al. (2020). Phase separation provides a mechanism to reduce noise in cells. Science 367 (6476), 464–468. doi:10.1126/SCIENCE.AAV6691
Krainer, G., Welsh, T. J., Joseph, J. A., Espinosa, J. R., Wittmann, S., de Csilléry, E., et al. (2021). Reentrant liquid condensate phase of proteins is stabilized by hydrophobic and non-ionic interactions. Nat. Commun. 12, 1085–1114. doi:10.1038/s41467-021-21181-9
Laage, D., Elsaesser, T., and Hynes, J. T. (2017). Water dynamics in the hydration shells of biomolecules. Chem. Rev., 117 (16), 10694–10725. doi:10.1021/ACS.CHEMREV.6B00765
Lambert, T. J. (2019). FPbase: a community-editable fluorescent protein database. Nat. Methods 16, 277–278. doi:10.1038/s41592-019-0352-8
Lorenz-Ochoa, K. A., and Baiz, C. R. (2023). Ultrafast spectroscopy reveals slow water dynamics in biocondensates. J. Am. Chem. Soc. 145 (50), 27800–27809. doi:10.1021/JACS.3C10862
Lyon, A. S., Peeples, W. B., and Rosen, M. K. (2020). A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell. Biol. 22 (3), 215–235. doi:10.1038/s41580-020-00303-z
Martin, E. W., Holehouse, A. S., Peran, I., Farag, M., Jeremias Incicco, J., Bremer, A., et al. (2020). Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 367 (6478), 694–699. doi:10.1126/science.aaw8653
Monahan, Z., Ryan, V. H., Janke, A. M., Burke, K. A., Rhoads, S. N., Zerze, G. H., et al. (2017). Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 36 (20), 2951–2967. doi:10.15252/EMBJ.201696394
Morawietz, T., Marsalek, O., Pattenaude, S. R., Streacker, L. M., Ben-Amotz, D., and Markland, T. E. (2018). The interplay of structure and dynamics in the Raman spectrum of liquid water over the full frequency and temperature range. J. Phys. Chem. Lett. 9 (4), 851–857. doi:10.1021/ACS.JPCLETT.8B00133
Mukherjee, S., and Schäfer, L. V. (2023). Thermodynamic forces from protein and water govern condensate formation of an intrinsically disordered protein domain. Nat. Commun. 14 (1), 5892–5913. doi:10.1038/s41467-023-41586-y
Murthy, A. C., Dignon, G. L., Kan, Y., Zerze, G. H., Parekh, S. H., Mittal, J., et al. (2019). Molecular interactions underlying Liquid−liquid phase separation of the FUS low-complexity domain. Nat. Struct. and Mol. Biol. 26 (7), 637–648. doi:10.1038/s41594-019-0250-x
Pappu, R. V., Cohen, S. R., Dar, F., Farag, M., and Kar, M. (2023). Phase transitions of associative biomacromolecules. Chem. Rev. 123 (14), 8945–8987. doi:10.1021/ACS.CHEMREV.2C00814
Patel, A., Lee, H. O., Maharana, S., Jahnel, M., Hein, M. Y., Stoynov, S., et al. (2015). A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 162 (5), 1066–1077. doi:10.1016/j.cell.2015.07.047
Pezzotti, S., König, B., Ramos, S., Schwaab, G., and Havenith, M. (2023). Liquid-liquid phase separation? Ask the water. J. Phys. Chem. Lett. 14 (6), 1556–1563. doi:10.1021/ACS.JPCLETT.2C02697
Qamar, S., Wang, G. Z., Randle, S. J., Simone Ruggeri, F., Varela, J. A., Lin, J. Q., et al. (2018). FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 173 (3), 720–734.e15. doi:10.1016/J.CELL.2018.03.056
Ramos, S., Kamps, J., Pezzotti, S., Winklhofer, K. F., Tatzelt, J., and Havenith, M. (2023). Hydration makes a difference! How to tune protein complexes between liquid–liquid and liquid–solid phase separation. Phys. Chem. Chem. Phys. 25 (41), 28063–28069. doi:10.1039/D3CP03299J
Rhoads, S. N., Monahan, Z. T., Yee, D. S., Leung, A. Y., Newcombe, C. G., O’Meally, R. N., et al. (2018a). The prionlike domain of FUS is multiphosphorylated following DNA damage without altering nuclear localization. Mol. Biol. Cell. 29 (15), 1786–1797. doi:10.1091/mbc.e17-12-0735
Rhoads, S. N., Monahan, Z. T., Yee, D. S., and Shewmaker, F. P. (2018b). The role of post-translational modifications on prion-like aggregation and liquid-phase separation of FUS. Int. J. Mol. Sci. 2018 (3), 886. doi:10.3390/IJMS19030886
Ribeiro, S. S., Samanta, N., Simon, E., and Marcos, J. C. (2019). The synergic effect of water and biomolecules in intracellular phase separation. Nat. Rev. Chem. 3 (9), 552–561. doi:10.1038/s41570-019-0120-4
Ruiz-Barragan, S., Sebastiani, F., Schienbein, P., Abraham, J., Schwaab, G., Nair, R. R., et al. (2022). Nanoconfinement effects on water in narrow graphene-based slit pores as revealed by THz spectroscopy. Phys. Chem. Chem. Phys. 24 (40), 24734–24747. doi:10.1039/D2CP02564G
Schienbein, P., Schwaab, G., Forbert, H., Havenith, M., and Marx, D. (2017). Correlations in the solute-solvent dynamics reach beyond the first hydration shell of ions. J. Phys. Chem. Lett. 8 (11), 2373–2380. doi:10.1021/ACS.JPCLETT.7B00713
Semenov, A. N., and Michael, R. (1998). Thermoreversible gelation in solutions of associative polymers. 1. Statics. Macromolecules 31 (4), 1373–1385. doi:10.1021/MA970616H
Villegas, J. A., Meta, H., and Levy, E. D. (2022). Molecular and environmental determinants of biomolecular condensate formation. Nat. Chem. Biol. 18 (12), 1319–1329. doi:10.1038/s41589-022-01175-4
Wang, J., Jeong, Mo C., Holehouse, A. S., Lee, H. O., Zhang, X., Jahnel, M., et al. (2018). A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 174 (3), 688–699.e16. doi:10.1016/J.CELL.2018.06.006
Keywords: biomolecular condensates, protein solvation, THz spectroscopy, reentrant phase separation, post-translational modifications
Citation: Czajkowski A, Udayabanu A, Raj M, Pulibandla LCP, Tursunović M, Jahnel M and Adams EM (2025) Protein modifications and ionic strength show the difference between protein-mediated and solvent-mediated regulation of biomolecular condensation. Front. Nanotechnol. 7:1556384. doi: 10.3389/fnano.2025.1556384
Received: 06 January 2025; Accepted: 05 February 2025;
Published: 05 March 2025.
Edited by:
Giancarlo Franzese, University of Barcelona, SpainCopyright © 2025 Czajkowski, Udayabanu, Raj, Pulibandla, Tursunović, Jahnel and Adams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ellen M. Adams, ZWxsZW5fbWFyaWUuYWRhbXNAdHUtZHJlc2Rlbi5kZQ==
 Artur Czajkowski
Artur Czajkowski