- 1TaiMei Baofa Cancer Hospital, Jinan, Shandong, China
- 2Jinan Baofa Cancer Hospital, Jinan, Shandong, China
- 3Beijing Baofa Cancer Hospital, Beijing, China
- 4Immune Oncology Systems, Inc, San Diego, CA, United States
Objective: Nanotechnology offers many advantages in various fields of cancer therapy. This study describes a new method of self-forming nanoparticles (self-nano) using ferric chloride (FeCl₃) and hydrogen peroxide (H2O2) to form the self-nanoparticles in an in vivo tumor. The treatment effect is evaluated.
Method: A solution of 3% FeCl3 (0.5 mL) and 1.8% H2O2 (1.0 mL) was injected into the tumor. At various time points post-injection, tumors were collected, and sections were prepared for electron microscopy to evaluate the size of the self-nano particles. Single-cell RNA sequencing (scRNA-seq) was used to analyze the immune changes and their effect on tumor growth.
Result: The formation of self-nano in vitro was observed and confirmed, with particles averaging 421 nm in size for the FeCl3 + H2O2 solution. Over time points ranging from 1 to 14 days, the formed self-nano remained stable at a regular size of 421 ± 8 nm. The self-nano, primarily consisting of iron, induced ferroptosis under the influence of an external magnetic field, leading to tumor growth control through iron-induced cell death and immune reactions. These self-nanoparticles also showed stronger enrichment of pathways related to CD8+ T effect cells (Teff), T cell activation, and regulation of T cell proliferation.
Conclusion: The FeCl3 + H2O2 solution can form Fe₂O₃-based self-nanoparticles within tumors through H2O2-incubated oxidation of FeCl3. The self-nano remains effective for over 14 days, inducing ferroptosis and upregulating immune cells under magnetic field treatment. This method offers a novel approach for cancer treatment that can be combined with other modalities.
Introduction
Nanotechnology offers numerous advantages across scientific fields. Nanoparticles, which are essential building blocks of nanotechnology, hold great potential in medical applications (Yetisgin et al., 2020; Bae et al., 2020; Chen et al., 2020). In recent years, the advancements in nanoparticles have extended to a wide range of clinical applications, including tumor therapy. Overcoming the limitations of traditional therapies and crossing systemic, micro-environmental, and cellular biological barriers are challenges across different patient populations and solid tumors (Mitchell et al., 2021; Gavas et al., 2021). Precision therapy, which uses personalized interventions to enhance treatment outcomes, has helped address patient heterogeneity. However, the development of nanoparticles remains largely focused on optimizing the delivery platform for a universal solution (Alghamdi et al., 2022; Svensson et al., 2024). As lipid-based, polymer-based, and inorganic nanoparticles are designed with increasing specificity, they have the potential to optimize drug delivery in more personalized ways, marking the entrance into the era of precision medicine (Grzelczak et al., 2010; Mehta et al., 2023). Some nanoparticles designed for tumor treatment must be both safe and effective, often used in combination with therapeutic agents to overcome the limitations of traditional therapies or in combination with other therapeutic devices (Chehelgerdi et al., 2023; Sun et al., 2023a; Chen et al., 2019). In addition to the methods described, techniques such as vibrating sample magnetometry (VSM) and single-molecule tools are essential for sensing and monitoring the magnetic performance of ferrite nanoparticles (Thakur et al., 2020; Díez et al., 2022). VSM provides precise measurements of the magnetic properties of nanoparticles, making it a valuable tool for assessing their behavior under various conditions (Mishra and Yadav, 2024; AboGabal et al., 2023). Single-molecule tools allow for the observation and analysis of individual nanoparticles, providing insights into their magnetic responses at the molecular level (Woythe et al., 2022; Chen et al., 2021). Magnetic force microscopy (MFM) offers excellent lateral resolution and the possibility of conducting single-molecule studies like other single-probe microscopy (SPM) techniques (Vilmos et al., 2022). These techniques are particularly relevant for understanding the efficacy and safety of magnetic nanoparticles in cancer therapy (Al-Thani et al., 2024; Jungcharoen et al., 2024), which is related with hyperthermia, where the magnetic nanoparticles in the tumor cells are subjected to alternating magnetic fields to generate local heating (Robert et al., 2023). Recent studies employing these methods have demonstrated the utility of nanoparticles in enhancing our understanding of their behavior in biomedical applications. However, these nanoparticles are typically fabricated in vitro, which leads to short residence times in tumors, uneven distribution, and limited efficacy. While self-assembly of nanoparticles has been explored, in vivo assembly within tumors remains a challenge (Grover and Mackman, 2020). Therefore, this study describes a method of self-forming nanoparticles (self-nano) using ferric chloride (FeCl3) and hydrogen peroxide (H2O2) to form self-nanoparticles in vivo within tumors in mice and evaluates the effect of this treatment on tumor therapy.
The FeCl3 models of arterial thrombosis are valuable tools for investigating the cellular and molecular mechanisms that contribute to arterial thrombosis (Jungcharoen et al., 2024). Several studies have described various applications of FeCl3, such as its effects when combined with Fe-EDTA for treating psoriasis (Shi et al., 2011) and the physiochemical artifacts observed in FeCl3 thrombosis models (Neeves, 2015). When FeCl3 reacts with O2 or H2O2, it transforms into Fe2O3, enabling the facile synthesis of Fe2O3 nanoparticles (Fe2O3 NPs) via a simple sol–gel route using FeCl3 with different molarities (Shi et al., 2011). The chemical equations FeCl3 + O2 = Fe2O3 + Cl2 and FeCl3 + H2O2 = HCl + Fe2O3 + O2 describe these reactions, which readily occur, leading to the formation of Fe2O3 nanoparticles.
Cancer cells exhibit an increased iron demand compared with normal, non-cancerous cells, which supports their growth and proliferation. This iron dependency can make cancer cells more vulnerable to iron-catalyzed necrosis or iron overload, a process known as ferroptosis. Ferroptosis is a newly discovered type of cell death with unique properties and recognition functions, which is relevant to various diseases, including cancer. Recent progress in research on ferroptosis involving iron nanoparticles has highlighted their potential in cancer therapy, as they can also induce an immune response. These iron nanoparticles are, therefore, referred to as immune iron nanoparticles (imu-self-nano) (Ngadiman et al., 2015).
In this study, we used a solution of FeCl3 and H2O2 for intratumoral injection in vivo to form the Fe2O3 nanoparticles within the tumor.
Experimental materials and methods
Preparation of animal models
Mouse ascites H22 cells that had grown well in vivo for more than two generations were prepared into a tumor cell suspension with a concentration of approximately 2×107 cells/L in normal saline and inoculated at a rate of 0.2 mL/L into the subcutaneous area of the left forelimb.
Here is a flow chart to visually represent the study design and process.

Preparation of tumors in mice, tissue disassociation, and cell collection
Mice with tumor diameters between 7 and 10 mm were selected and weighed, and their tumor size was measured. The tumor volume was calculated using the formula volume (mm3) = 1/2 (length × width2) (Ma et al., 2004). The mice were randomly divided into three groups: group 1 (Yetisgin et al., 2020), group 2 (Bae et al., 2020), and three control groups, with 13 mice in each group.
Self-formed nanoparticles
One group of mice was used to test self-nanoparticles in vitro. Preparation of all research included 3% FeCl3 solution: 30% FeCl3 stock solution, 1 mL plus 9 mL normal saline (NS) dilution obtained. Group 1 (Yetisgin et al., 2020) received an intratumoral injection of 3% FeCl3 (0.5 mL + 1.0 mL NS) as a control. Group 2 (Bae et al., 2020) received an intratumoral injection of 3% FeCl3 (0.5 mL) +1.8% H2O2 (1.0 mL) as the test group. At different time points after injection, the tumors were collected, and sections were prepared for electron microscopy to evaluate the size of the self-nano particles.
Effectiveness and survival test by scRNA-seq
Following intratumoral injection of FeCl3 (control) and FeCl3 + H2O2 (test group), the survival time of the first ten mice in each group was observed. The therapeutic method involved an intratumoral injection of 0.2 mL per tumor, followed by a second dose of 0.1 mL per tumor. Seven days after the first injection, the mice were immobilized and placed in the center of a large coil for magnetic field treatment (frequency: 0–300 kHz) with magnetic field strength under 0–15 kA/m, power: 2,000 W, and voltage: 0–300 v, once a day for 20 min each session over 10 consecutive days. The tumor volume was measured periodically, and the death of mice in each group was recorded until the natural death of the control group mice.
ScRNA-seq and data processing and analysis
Each group of four mice was used exclusively for scRNA-seq detection. The tumors were dissected 10 days after treatment, and each tumor was used for scRNA-seq analysis.
ScRNA-seq data processing and quality control
Fresh tumor samples were stored in sCelLiVE® tissue preservation solution in GEXSCOPE® (Singleron) until molecular testing (Kechin et al., 2017). Original gene expression matrix data were generated using CeleScopeR (https://github.com/singleron-RD/CeleScope) software. CeleScopeR, a single-cell data processing software developed by Singleron, was used for quality control and data filtering (Mou et al., 2019). Reads were compared with the reference genome GRCh38 using ensembl version 93, and gene annotation was carried out using STAR (version 2.6.1b) (Dobin et al., 2013).
Differentially expressed gene (DEG) analysis (Scanpy)
Differentially expressed genes (DEGs) were identified using the scanpy.tl.rank_genes_groups function based on the Wilcoxon rank-sum test with default parameters. Genes expressed in more than 10% of the cells with an average log(fold change) value greater than 1 were selected as DEGs (Manescu Paltanea et al., 2021).
Cell type annotation
Cell type identity in each cluster was determined by the expression of canonical markers found in the DEGs using the SynEcSys database (Singleron Biotechnologies, Koln, Germany) (Dobin et al., 2013).
Subtyping of major cell types and CNV detection based on scRNA-seq and pathway enrichment analysis
To investigate the potential functions of DEGs between clusters, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using the “clusterProfiler” R package 3.16.1. [9] Gene set variation analysis (GSVA) was conducted for pathway enrichment analysis (Dobin et al., 2013).
UCell gene set scoring
Gene set scoring was performed using the R package UCell v 1.1.0 (Dobin et al., 2013). UCell scores were based on the Mann–Whitney U statistic by ranking query genes in the order of their expression levels in individual cells.
Trajectory analysis
Trajectory analysis was carried out using the R package monocle (version 2.18.0) (Dobin et al., 2013) employing the DDRTree dimensionality reduction method (Dobin et al., 2013).
Transcription factor regulatory network analysis (pySCENIC)
The transcription factor network was constructed using pySCENIC (v0.11.0) with the scRNA expression matrix and transcription factors from AnimalTFDB. AUCell was also used (Dobin et al., 2013).
Cell–cell interaction analysis
Cell–cell interactions (CCIs) between different cell types were predicted based on known ligand–receptor pairs using Cellphone DB v2.1.0. Predicted interaction pairs with a p-value <0.05 and average log expression >0.1 were considered significant. Differentially activated ligand–receptor pairs between groups were visualized by dot plots in ggplot2 (Kechin et al., 2017; Dobin et al., 2013).
Results
Self-nanoparticle in vitro confirmation
Silica section was. Self-nanoparticles were formed with a regular size averaging 421 nm (Figures 1A–F) in the FeCl3 + H2O2 solution (test), while the FeCl3 solution (control) did not result in regular-sized nanoparticles (Figure 1F). At different time points ranging from 1 to 14 days, the self-nanoparticles in the FeCl₃ + H₂O₂ solution remained stable with a consistent size of 421 ± 8 nm (Figure 1).
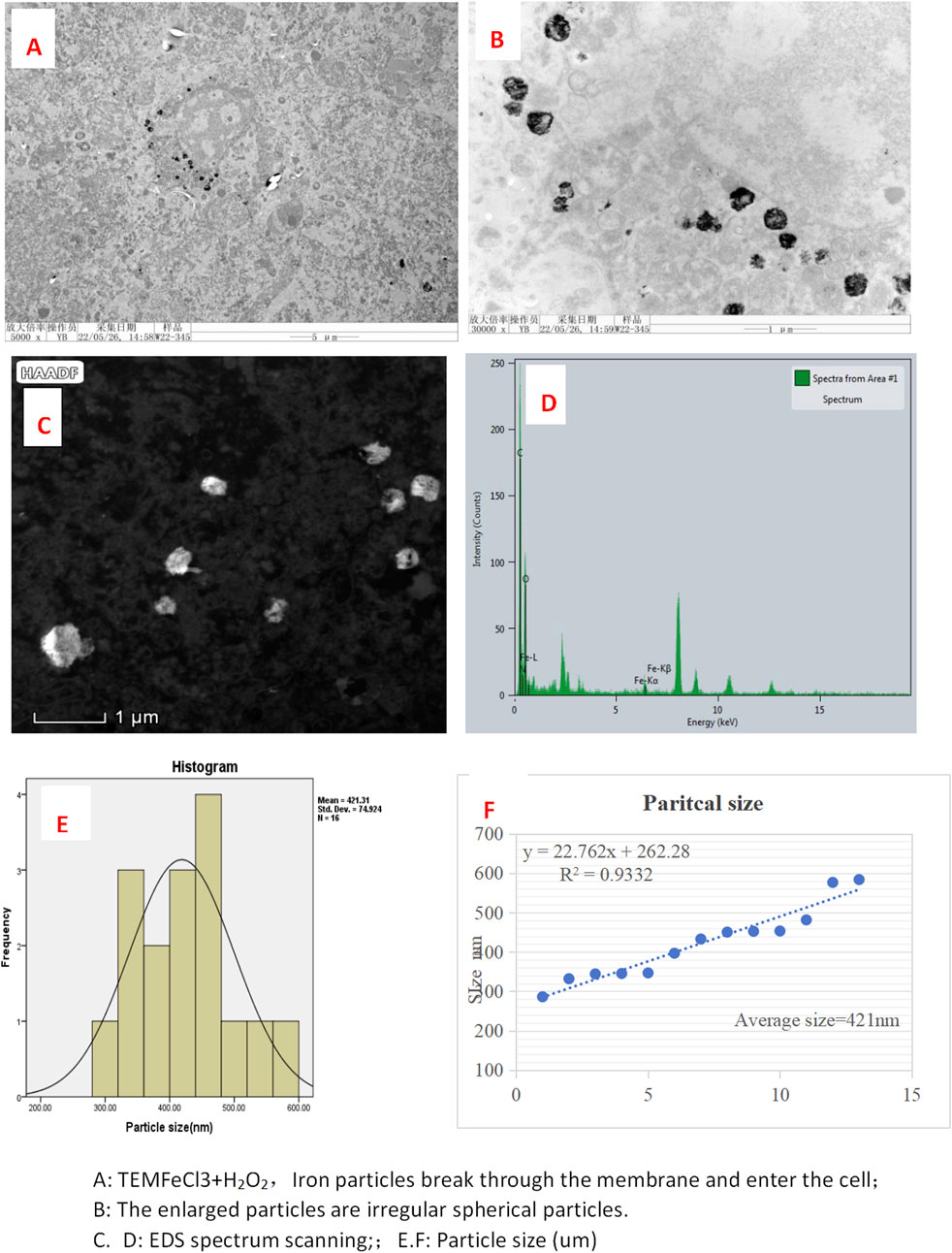
Figure 1. Electron microscopy images of FeCl3+ H2O2 in tumor in mice. (A) TEM FeC13 + H2O2, iron particles break through the membrane and enter the cell; (B) enlarged particles that are irregular spherical particles. (C, D) EDS spectrum scanning; (E, F): particle size (um).
Survival period of mice
These results indicated that Fe2Cl3 + H2O2 significantly prolonged the survival time of tumor-bearing mice. The difference between the test group and the model group was significantly greater than that in the control group (p < 0.05) (Table 1).
ScRNA-seq results
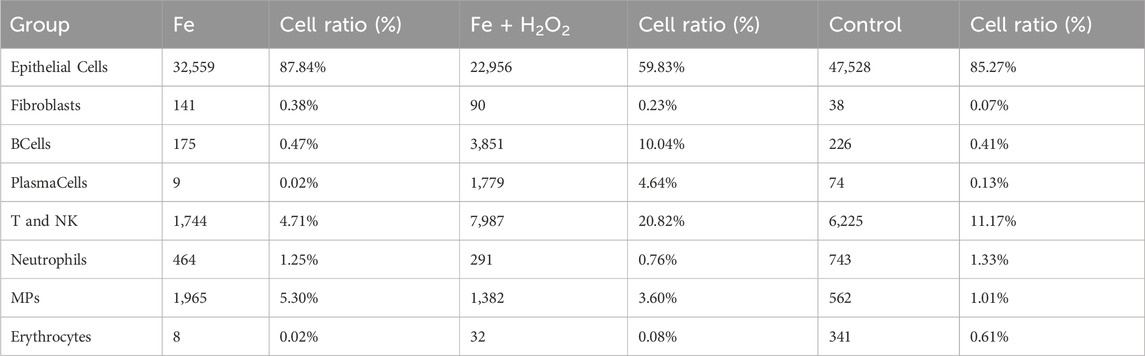
The cell atlas of nine samples was constructed, encompassing a total of 124,745 cells across eight cell types: epithelial cells (EpithelialCells), fibroblasts, B cells (BCells), plasma cells (PlasmaCells), T and NK cells (T and NK), neutrophils, mononuclear phagocytes (MPs), and erythrocytes (Table 2). Some cell types were represented in small numbers; therefore, the focus of the initial immunoassay is recommended to be on T cells, neutrophils, and mononuclear phagocytes (Figure 2).
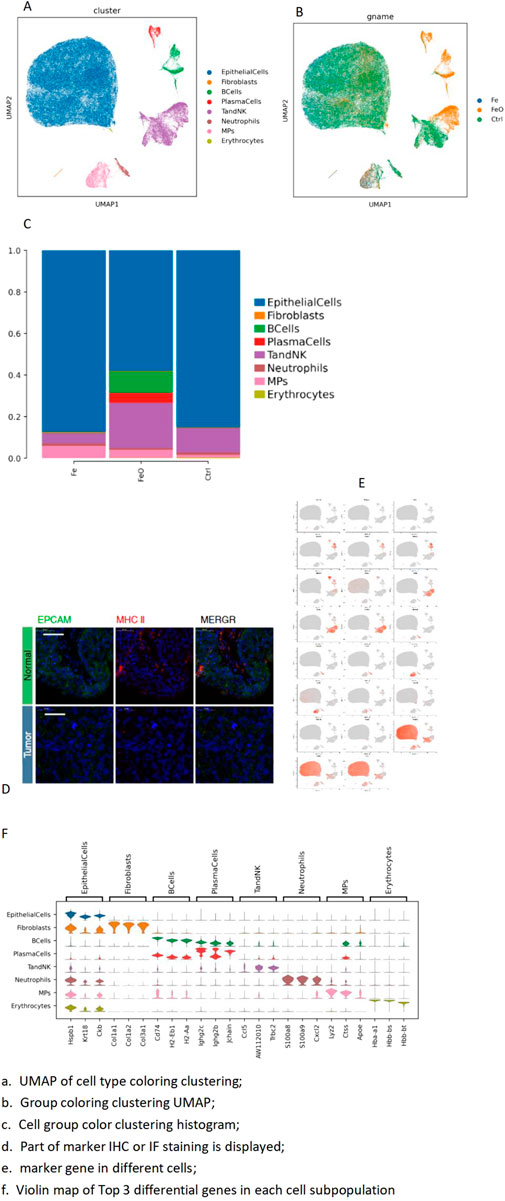
Figure 2. Integrated annotation results: top-3 marker genes. (A) UMAP of cell type coloring clustering. (B) Group coloring clustering UMAP. (C) Histogram of cell group color clustering. (D) Immunofluorescence staining markers. (E) Marker gene in different cells. (F) Violin plot map of top-3 differential genes in each cell subpopulation.
T-cell subdivision and dimension reduction
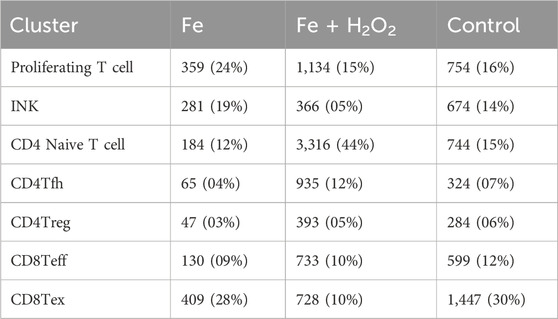
The total number of T and NK cells was 13,906, categorized into seven different subtypes. The cell differences observed between the groups were primarily driven by the S2_2 sample, which exhibited a higher number of immune cells when compared to the other samples. Compared to the control group, the test group showed decreased CD8 exhaustion and increased CD4 naive and T follicular helper (Tfh) cells (Figure 2).
T-cell differential gene analysis for inter-groups
The Ms4a4b gene expression was significantly upregulated in the control group, while the test group showed higher levels of immunoglobulin genes. The inter-group difference analyses for each subpopulation subdivision are detailed in the folders on the right, which include differential genes, differential gene enrichment, and GSEA (Figures 3A, B). Compared with the control group, the test group exhibited stronger enrichment of pathways related to T-cell activation and differentiation, along with lower negative regulation of the immune system process (Figure 3C).
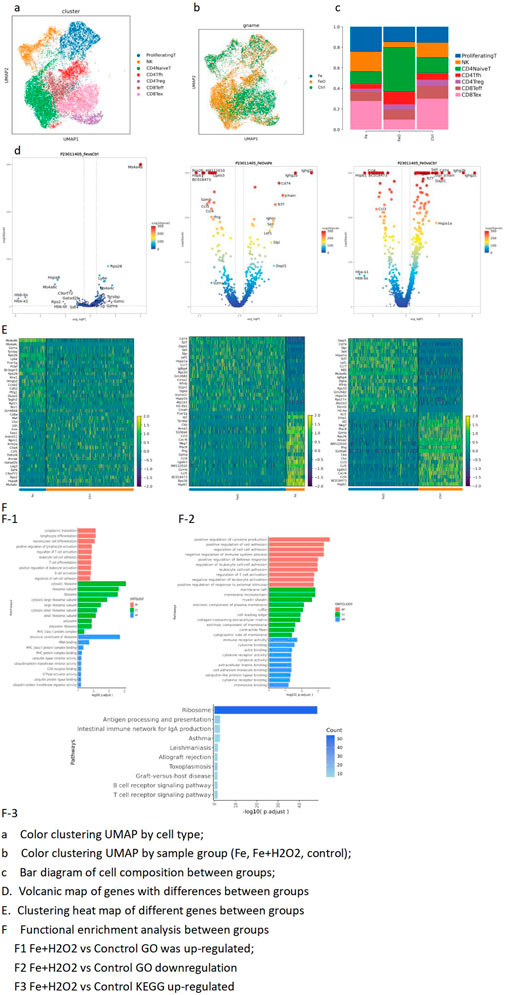
Figure 3. T-cell subdivision-dimensionality reduction and top-3 marker genes for T-cell subpopulations. (A) UMAP color clustering by cell type. (B) UMAP color clustering by sample group (Fe, Fe + H2O2, control). (C) Bar diagram of cell composition across different groups. (D) Volcano plots of differentially expressed genes between groups. (E) Heatmaps of differentially expressed genes between groups. (F) Functional enrichment analysis between groups. F1. Upregulated GO terms: Fe + H2O2 vs. control; F2. downregulated GO terms: Fe + H2O2 vs. control; F3. upregulated KEGG terms: Fe + H2O2 vs. control.
T-cell GSEA
The ribosome function was higher in the Fe + H2O2 group than in the Fe group, while the proteasome function was lower. Apoptosis and cytokine interaction decreased, and ribosome function increased in the test group (Figures 3D, E). CD4 naive GSEA showed that oxidative phosphorylation and cytokine interaction levels were lower in the test group than in the control group.
CD4 Tfh GSEA
The interaction between proteasomes and cytokines was lower in the test group than in the control group. Carbon metabolism, cytokine interaction, and endoplasmic reticulum protein processes were also decreased (Figure 3F). CD4 Treg GSEA showed decreased glycolysis in the test group compared to the control group. The interaction of apoptosis, carbon metabolism, chemokines, and cytokines decreased (Figures 3F1). CD8 Teff GSEA showed decreased amino acid synthesis and apoptosis in the test group than in the control group. Cytokine interaction and apoptosis decreased (Figures 3F2). Further CD8 Teff GSEA indicated decreased amino acid synthesis in the test group compared to the control group. The test group showed decreased cytokine interaction and apoptosis compared to the control group (Figures 3F3).
MP cell subdivision
A total of 3,416 macrophage (MP) cells were identified and categorized into six different subtypes. Most macrophages were divided into two groups. Moreover, significant heterogeneity was observed between the treatment group and the control group, particularly between the test and the control group. The proportion of non-classical monocytes was relatively low in both groups (Figure 4).
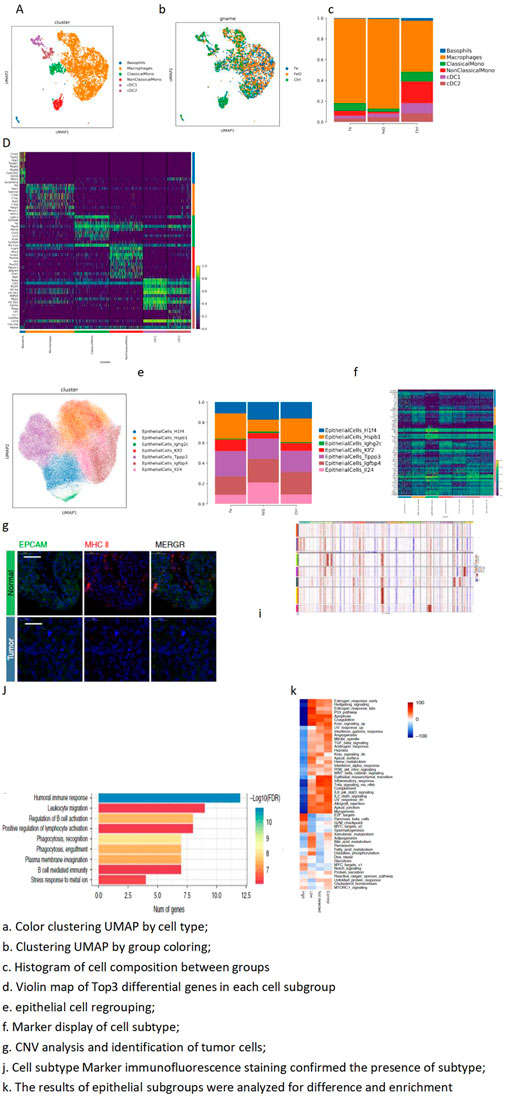
Figure 4. MP cell subdivision-dimensionality reduction and MP cell subdivision top-3 marker gene. (A) Color clustering UMAP by cell type. (B) Clustering UMAP by group coloring. (C) Histogram of cell composition between groups. (D) Violin map of top-3 differential genes in each cell subgroup. (E) Epithelial cell regrouping. (F) Marker display of cell subtype. (G) CNV analysis and identification of tumor cells. (I) Cell subtype marker immunofluorescence staining confirmed the presence of subtypes. (J) Results of epithelial subgroups were analyzed for difference and enrichment. A total of 3,416 cells were obtained from MPs, and six different isoforms were further subdivided: basophils, macrophages, classical-monocytes (ClassicalMono), non-classical monocytes (NonClassicalMono), cDC1, and cDC2. The proportion of macrophages in the Fe and FeO treatment groups was higher than that in the control group.
Tumor heterogeneity
The epithelial cells were further subdivided, and tumor cells were identified through copy number variation (CNV) analysis. The expression patterns of different tumor cells were displayed through various analyses, including differential analysis and enrichment analysis. Marker genes of tumor cells were verified through staining experiments to confirm the accuracy of tumor cell identification (Figures 4E–J).
Discussion
Nanotechnology is an integral part of medical research and holds great potential in medical applications (Yetisgin et al., 2020; Malik et al., 2023). The development of nanoparticles has overcome the limitations of conventional therapies, enabling advancements in systemic and micro-environmental treatments, thereby ushering in the era of precision medicine (4, 11, 333). This study introduces a method of self-forming nanoparticles (self-nano) using FeCl3 and H2O2 and discusses the effect of this treatment when applied in vivo for tumor therapy.
FeCl3 models of arterial thrombosis are valuable tools (Grzelczak et al., 2010). Numerous studies have explored various applications of FeCl3, such as its combination with Fe-EDTA in the treatment of psoriasis (Jungcharoen et al., 2024) and the observation of physiochemical artifacts in FeCl3 thrombosis models (Robert et al., 2023). When FeCl3 reacts with O2 or H2O2, it converts into Fe2O3, enabling the facile synthesis of Fe2O3 nanoparticles (Fe2O3 NPs) via a simple sol–gel route using FeCl3 with different molarities (Grover and Mackman, 2020). The chemical reactions FeCl3 + O2 = Fe2O3 + Cl2 and FeCl3 + H2O2 = HCl + Fe2O3 + O2 demonstrate how Fe2O3 nanoparticles are formed with ease.
In this study, we observed and confirmed the formation of self-nano of Fe2O3 with a regular size averaging 421 nm when using a solution of FeCl3 + H2O2. In contrast, a solution of FeCl3 alone did not produce nanoparticles of regular size. Over time points ranging from 1 to 14 days, the formed self-nano remained stable at a regular size of 421 ± 8 nm, exhibiting a long-acting behavior due to H2O2-induced oxidation of the tumor extracellular matrix, which led to matrix denaturation and reduced liquidity. The effectiveness and survival study showed that the test group mice exhibited longer survival compared to the control group mice under the same magnetic field treatment.
Cancer cells are more vulnerable to iron-catalyzed necrosis, a process known as ferroptosis. This rapid development of ferroptosis, particularly involving iron nanoparticles, has enhanced its application prospects in cancer treatment (Ngadiman et al., 2015; Zhang et al., 2022; Sun et al., 2023b). Magnetic nanoparticles are increasingly being researched for cancer treatment and have become one of the most significant challenges in medical research (Manescu Paltanea et al., 2021; Alromi et al., 2021). Imm-self-nano, composed mainly of iron, can play a role under the influence of an external magnetic field, leading to ferroptosis and immune reaction. Low-frequency alternating magnetic fields (MFs) have a minimal impact on immune cells. In contrast, medium-intensity and high-gradient MFs exert a greater effect, especially on larger immune cells like macrophages, which are more sensitive to high-gradient MFs (HGMFs). These MFs can skew cell polarization toward the anti-inflammatory M2 phenotype. Under varying gradient forces, elongated M2 macrophages also reshape the cytoskeleton through actin polymerization, which alters membrane receptors and ion channel gating (Lei et al., 2020; Strizova et al., 2023).
This study identified a total of 124,745 cells across eight cell types, including epithelial cells (EpithelialCells), fibroblasts, B cells (BCells), plasma cells (PlasmaCells), T and NK cells (T and NK), neutrophils, mononuclear phagocytes (MPs), and erythrocytes.
The observed differences between groups were primarily due to the S2_2 sample, which contained a significantly higher number of immune cells than other samples. Compared to the control group, the test group showed decreased CD8 T cell exhaustion and increased CD4 naive and Tfh cells. The Ms4a4b gene expression was significantly upregulated in the control group, while the test group had higher levels of immunoglobulin genes. MS4a4B may, therefore, serve as a modulator in the negative-feedback regulatory loop of activated T cells (Xu et al., 2010; Permanyer et al., 2021). The test group exhibited stronger enrichment of pathways related to T-cell activation and differentiation, while MS4a4B negatively regulates T-cell proliferation.
Tumor heterogeneity was analyzed in conjunction with the results of differential enrichment between groups and subgroups (including GSVA and GSEA), inferCNV results of subgroups, and cNMF. In this study, the MKI67 gene was highly expressed in the H1f4 and Klf2 subgroups, particularly in the Klf2 subgroup. These two subgroups are also at the end of differentiation. MKI67 is associated with adaptive immunity, cell adhesion molecules (CAMs), and chemokine/immune response signal transduction pathways (Wu et al., 2021; Ren et al., 2024). The proportion of these cells was highest in the control group and decreased in the test group. These findings suggest that the synergistic effect between the two groups may result in a stronger anti-tumor response.
Additionally, the potential accumulation of magnetic nanoparticles and the possibility of hepatotoxicity were considered. While our study did not directly observe significant accumulation leading to toxic effects, it is crucial to recognize that magnetic nanoparticles can accumulate in organs like the liver, potentially leading to hepatotoxicity over time. Previous studies have reported such effects, emphasizing the importance of monitoring nanoparticle distribution and long-term safety in future investigations. Future studies should also focus on the detailed pharmacokinetics and biodistribution of these nanoparticles to ensure their safe application in clinical settings (Mitchell et al., 2021; Malik et al., 2023).
Future research should focus on further exploration of magnetic nanoparticles in cancer therapy, particularly in understanding their long-term effects and safety profiles. Studies could investigate the combination of these nanoparticles with other treatment modalities, such as chemotherapy, radiotherapy, and immunotherapy, to enhance the therapeutic outcomes (Haripriyaa and Suthindhiran, 2023; Elumalai et al., 2024). Additionally, research into the optimization of magnetic field parameters to maximize the therapeutic effects while minimizing potential side effects is crucial. Exploring the use of different magnetic nanoparticles with varied sizes, shapes, and compositions may also offer insights into improving their efficacy and biocompatibility in clinical applications (Rarokar et al., 2024; Koksharov et al., 2022).
In summary, self-nano particles can form in tumors in vitro at the site of treatment through the local injection of a solution of FeCl3 + H2O2. The H2O2 oxidation of FeCl3 produces Fe2O3, which forms self-nano particles that remain effective for over 14 days. This process induces ferroptosis and upregulates immune cells under magnetic field treatment, offering a novel approach to cancer treatment that can be combined with other modalities.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The animal studies were approved by the Shandong Baofa Cancer Research Inc. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
BY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing. YH: Methodology, Project administration, Writing–original draft, Writing–review and editing. JZ: Formal Analysis, Investigation, Methodology, Writing–original draft, Writing–review and editing. DC: Formal Analysis, Methodology, Resources, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank Dr. Wenxue Ma at the University of California, San Diego, for his critical review and editing.
Conflict of interest
Author BY was employed by Immune Oncology Systems, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CAMs, cell adhesion molecules; CNV, copy number variation; cNMF, collaborative non-negative matrix factorization; DDRTree, a dimensionality reduction method used in trajectory analysis; DEGs, differentially expressed genes; FeCl₃, ferric chloride; Fe₂O₃, iron (III) oxide; GSVA, gene set variation analysis; GO, Gene Ontology; H₂O₂, hydrogen peroxide; HGMFs, high-gradient magnetic fields; imu-self-nano, immune self-nanoparticles; KEGG, Kyoto Encyclopedia of Genes and Genomes; MFs, magnetic fields; MKI67, a gene associated with cell proliferation (also known as Ki-67); MS4A4B, a gene that modulates the cell cycle of T cells; pySCENIC, a bioinformatics tool for transcription factor regulatory network analysis; UCell, a method used for gene set scoring in single-cell RNA sequencing.
References
AboGabal, R., Shokeir, D., and Oraby, A. H. (2023). Design and synthesis of biologically inspired biocompatible various polymeric magnetic nanoparticles for imaging and biomedical applications. Nano-Structures and Nano-Objects 36, 101048. doi:10.1016/j.nanoso.2023.101048
Alghamdi, M. A., Fallica, A. N., Virzi, N., Kesharwani, P., Pittala, V., and Greish, K. (2022). The promise of nanotechnology in personalized medicine. J. Pers. Med. 12 (5), 673. doi:10.3390/jpm12050673
Alromi, D. A., Madani, S. Y., and Seifalian, A. (2021). Emerging application of magnetic nanoparticles for diagnosis and treatment of cancer. Polym. (Basel) 13 (23), 4146. Epub 20211127. doi:10.3390/polym13234146
Al-Thani, A. N., Jan, A. G., Abbas, M., Geetha, M., and Sadasivuni, K. K. (2024). Nanoparticles in cancer theragnostic and drug delivery: a comprehensive review. Life Sci. 352, 122899. Epub 20240709. doi:10.1016/j.lfs.2024.122899
Bae, J., Parayath, N., Ma, W., Amiji, M., Munshi, N., and Anderson, K. C. (2020). Bcma peptide-engineered nanoparticles enhance induction and function of antigen-specific Cd8(+) cytotoxic T lymphocytes against multiple myeloma: clinical applications. Leukemia 34 (1), 210–223. Epub 20190819. doi:10.1038/s41375-019-0540-7
Chehelgerdi, M., Chehelgerdi, M., Allela, O. Q. B., Pecho, R. D. C., Jayasankar, N., Rao, D. P., et al. (2023). Progressing nanotechnology to improve targeted cancer treatment: overcoming hurdles in its clinical implementation. Mol. Cancer 22 (1), 169. Epub 20231009. doi:10.1186/s12943-023-01865-0
Chen, Q., Bao, Y., Burner, D., Kaushal, S., Zhang, Y., Mendoza, T., et al. (2019). Tumor growth inhibition by msteap peptide nanovaccine inducing augmented Cd8(+) T cell immune responses. Drug Deliv. Transl. Res. 9 (6), 1095–1105. doi:10.1007/s13346-019-00652-z
Chen, Q., Jia, G., Zhao, X., Bao, Y., Zhang, Y., Ozkan, C., et al. (2020). Novel survivin peptides screened with computer algorithm induce cytotoxic T lymphocytes with higher cytotoxic efficiency to cancer cells. Front. Mol. Biosci. 7, 570003. Epub 20200902. doi:10.3389/fmolb.2020.570003
Chen, Y., Wang, F., Feng, J., and Fan, C. (2021). Empowering single-molecule analysis with self-assembled DNA nanostructures. Matter 4 (10), 3121–3145. doi:10.1016/j.matt.2021.08.003
Díez, A. G., Rincón-Iglesias, M., Lanceros-Méndez, S., Reguera, J., and Lizundia, E. (2022). Multicomponent magnetic nanoparticle engineering: the role of structure-property relationship in advanced applications. Mater. Today Chem. 26, 101220. doi:10.1016/j.mtchem.2022.101220
Dobin, A., Davis, C. A., Schlesinger, F., Drenkow, J., Zaleski, C., Jha, S., et al. (2013). Star: ultrafast universal rna-seq aligner. Bioinformatics 29 (1), 15–21. Epub 20121025. doi:10.1093/bioinformatics/bts635
Elumalai, K., Srinivasan, S., and Shanmugam, A. (2024). Review of the efficacy of nanoparticle-based drug delivery systems for cancer treatment. Biomed. Technol. 5, 109–122. doi:10.1016/j.bmt.2023.09.001
Gavas, S., Quazi, S., and Karpinski, T. M. (2021). Nanoparticles for cancer therapy: current progress and challenges. Nanoscale Res. Lett. (2021) 16 (1), 173. Epub 20211205. doi:10.1186/s11671-021-03628-6
Grover, S. P., and Mackman, N. (2020). How useful are ferric chloride models of arterial thrombosis? Platelets 31 (4), 432–438. Epub 20191013. doi:10.1080/09537104.2019.1678119
Grzelczak, M., Vermant, J., Furst, E. M., and Liz-Marzan, L. M. (2010). Directed self-assembly of nanoparticles. ACS Nano 4 (7), 3591–3605. doi:10.1021/nn100869j
Haripriyaa, M., and Suthindhiran, K. (2023). Pharmacokinetics of nanoparticles: current knowledge, future directions and its implications in drug delivery. Future J. Pharm. Sci. 9 (1), 113. doi:10.1186/s43094-023-00569-y
Jungcharoen, P., Thivakorakot, K., Thientanukij, N., Kosachunhanun, N., Vichapattana, C., Panaampon, J., et al. (2024). Magnetite nanoparticles: an emerging adjunctive tool for the improvement of cancer immunotherapy. Explor Target Antitumor Ther. 5 (2), 316–331. Epub 20240423. doi:10.37349/etat.2024.00220
Kechin, A., Boyarskikh, U., Kel, A., and Filipenko, M. (2017). Cutprimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J. Comput. Biol. 24 (11), 1138–1143. Epub 20170717. doi:10.1089/cmb.2017.0096
Koksharov, Y. A., Gubin, S. P., Taranov, I. V., Khomutov, G. B., and Gulyaev, Y. V. (2022). Magnetic nanoparticles in medicine: progress, problems, and advances. J. Commun. Technol. Electron. 67 (2), 101–116. doi:10.1134/S1064226922020073
Lei, H., Pan, Y., Wu, R., and Lv, Y. (2020). Innate immune regulation under magnetic fields with possible mechanisms and therapeutic applications. Front. Immunol. 11, 582772. Epub 20201022. doi:10.3389/fimmu.2020.582772
Ma, W., Yu, H., Wang, Q., Jin, H., Solheim, J., and Labhasetwar, V. (2004). A novel approach for cancer immunotherapy: tumor cells with anchored superantigen sea generate effective antitumor immunity. J. Clin. Immunol. 24 (3), 294–301. doi:10.1023/B:JOCI.0000025451.41948.94
Malik, S., Muhammad, K., and Waheed, Y. (2023). Emerging applications of nanotechnology in healthcare and medicine. Molecules 28 (18), 6624. Epub 20230914. doi:10.3390/molecules28186624
Manescu Paltanea, V., Paltanea, G., Antoniac, I., and Vasilescu, M. (2021). Magnetic nanoparticles used in oncology. Mater. (Basel) 14 (20), 5948. Epub 20211010. doi:10.3390/ma14205948
Mehta, M., Bui, T. A., Yang, X., Aksoy, Y., Goldys, E. M., and Deng, W. (2023). Lipid-based nanoparticles for drug/gene delivery: an overview of the production techniques and difficulties encountered in their industrial development. ACS Mater Au 3 (6), 600–619. Epub 20230821. doi:10.1021/acsmaterialsau.3c00032
Mishra, S., and Yadav, M. D. (2024). Magnetic nanoparticles: a comprehensive review from synthesis to biomedical frontiers. Langmuir 40, 17239–17269. Epub 20240812. doi:10.1021/acs.langmuir.4c01532
Mitchell, M. J., Billingsley, M. M., Haley, R. M., Wechsler, M. E., Peppas, N. A., and Langer, R. (2021). Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20 (2), 101–124. Epub 20201204. doi:10.1038/s41573-020-0090-8
Mou, Y., Wang, J., Wu, J., He, D., Zhang, C., Duan, C., et al. (2019). Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J. Hematol. Oncol. 12 (1), 34. Epub 20190329. doi:10.1186/s13045-019-0720-y
Neeves, K. B. (2015). Physiochemical artifacts in Fecl3 thrombosis models. Blood 126 (6), 700–701. doi:10.1182/blood-2015-05-644708
Ngadiman, N. H., Idris, A., Irfan, M., Kurniawan, D., Yusof, N. M., and Nasiri, R. (2015). γ-Fe2O3 nanoparticles filled polyvinyl alcohol as potential biomaterial for tissue engineering scaffold. J. Mech. Behav. Biomed. Mater 49, 90–104. Epub 20150509. doi:10.1016/j.jmbbm.2015.04.029
Permanyer, M., Bosnjak, B., Glage, S., Friedrichsen, M., Floess, S., Huehn, J., et al. (2021). Efficient il-2r signaling differentially affects the stability, function, and composition of the regulatory T-cell pool. Cell Mol. Immunol. 18 (2), 398–414. Epub 20210106. doi:10.1038/s41423-020-00599-z
Rarokar, N., Yadav, S., Saoji, S., Bramhe, P., Agade, R., Gurav, S., et al. (2024). Magnetic nanosystem a tool for targeted delivery and diagnostic application: current challenges and recent advancement. Int. J. Pharm. 7, 100231. doi:10.1016/j.ijpx.2024.100231
Ren, Y., Liang, H., Huang, Y., Miao, Y., Li, R., Qiang, J., et al. (2024). Key candidate genes and pathways in T lymphoblastic leukemia/lymphoma identified by bioinformatics and serological analyses. Front. Immunol. 15, 1341255. Epub 20240223. doi:10.3389/fimmu.2024.1341255
Robert, W., Miguel, C., Margaret, A., Harald, P., and Marcuello, C. (2023). A review of the current state of magnetic force microscopy to unravel the magnetic properties of nanomaterials applied in biological systems and future directions for quantum technologies. Nanomaterials 13 (18), 2585. doi:10.3390/nano13182585
Shi, Q., Yang, X. Q., and Cui, X. (2011). The effects of fecl(3) and Fe-edta on the development of psoriasis. Biol. Trace Elem. Res. 140 (1), 73–81. Epub 20100330. doi:10.1007/s12011-010-8671-8
Strizova, Z., Benesova, I., Bartolini, R., Novysedlak, R., Cecrdlova, E., Foley, L. K., et al. (2023). M1/M2 macrophages and their overlaps - myth or reality? Clin. Sci. (Lond) 137 (15), 1067–1093. doi:10.1042/CS20220531
Sun, L., Liu, H., Ye, Y., Lei, Y., Islam, R., Tan, S., et al. (2023a). Smart nanoparticles for cancer therapy. Signal Transduct. Target Ther. 8 (1), 418. Epub 20231103. doi:10.1038/s41392-023-01642-x
Sun, S., Shen, J., Jiang, J., Wang, F., and Min, J. (2023b). Targeting ferroptosis opens new avenues for the development of novel therapeutics. Signal Transduct. Target Ther. 8 (1), 372. Epub 20230921. doi:10.1038/s41392-023-01606-1
Svensson, E., von Mentzer, U., and Stubelius, A. (2024). Achieving precision healthcare through nanomedicine and enhanced model systems. ACS Mater Au 4 (2), 162–173. doi:10.1021/acsmaterialsau.3c00073
Thakur, P., Chahar, D., Taneja, S., Bhalla, N., and Thakur, A. (2020). A review on mnzn ferrites: synthesis, characterization and applications. Ceram. Int. 46 (10), 15740–15763. Epub 20200407. doi:10.1016/j.ceramint.2020.03.287
Vilmos, V. A., Csaba, M. C., Petra, P. B., István, A., and Szabó, A. (2022). Structural and magnetic characterisation of a biocompatible magnetic nanoparticle assembly. J. Magnetism Magnetic Mater. 545 (1 March 2022), 168772. doi:10.1016/j.jmmm.2021.168772
Woythe, L., Madhikar, P., Feiner-Gracia, N., Storm, C., and Albertazzi, L. (2022). A single-molecule view at nanoparticle targeting selectivity: correlating ligand functionality and cell receptor density. ACS Nano 16 (3), 3785–3796. Epub 20220311. doi:10.1021/acsnano.1c08277
Wu, S. Y., Liao, P., Yan, L. Y., Zhao, Q. Y., Xie, Z. Y., Dong, J., et al. (2021). Correlation of Mki67 with prognosis, immune infiltration, and T cell exhaustion in hepatocellular carcinoma. BMC Gastroenterol. 21 (1), 416. Epub 20211101. doi:10.1186/s12876-021-01984-2
Xu, H., Yan, Y., Williams, M. S., Carey, G. B., Yang, J., Li, H., et al. (2010). Ms4a4b, a Cd20 homologue in T cells, inhibits T cell propagation by modulation of cell cycle. PLoS One 5 (11), e13780. Epub 20101101. doi:10.1371/journal.pone.0013780
Yetisgin, A. A., Cetinel, S., Zuvin, M., Kosar, A., and Kutlu, O. (2020). Therapeutic nanoparticles and their targeted delivery applications. Molecules 25 (9), 2193. Epub 20200508. doi:10.3390/molecules25092193
Keywords: nanoparticles, self-nano, hydrogen peroxide, ferroptosis, ferric chloride, iron(III) oxide, tumor therapy, magnetic field treatment
Citation: Yu B, Han Y, Zhang J and Chen D (2024) Formation of self-nanoparticles and the immune effect on tumors after injection of ferric chloride with H2O2 under magnetic field therapy. Front. Nanotechnol. 6:1465888. doi: 10.3389/fnano.2024.1465888
Received: 17 July 2024; Accepted: 26 August 2024;
Published: 12 September 2024.
Edited by:
Mohammad Tavakkoli Yaraki, Macquarie University, AustraliaReviewed by:
Marzieh Ramezani Farani, Inha University, Republic of KoreaCarlos Marcuello, Instituto de Nanociencia y Materiales de Aragón (INMA), Spain
Copyright © 2024 Yu, Han, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Baofa Yu, YmZ5dWNoaW5hQDEyNi5jb20=
 Baofa Yu
Baofa Yu Yan Han2
Yan Han2